Abstract
Integrating DNA delivery systems hold promise for many applications including treatment of diseases; however, targeted integration is needed for improved safety. The piggyBac (PB) transposon system is a highly active non-viral gene delivery system capable of integrating defined DNA segments into host chromosomes without requiring homologous recombination. We systematically compared four different engineered zinc finger proteins (ZFP), four transcription activator-like effector proteins (TALE), CRISPR associated protein 9 (SpCas9) and the catalytically inactive dSpCas9 protein fused to the amino-terminus of the transposase enzyme designed to target the hypoxanthine phosphoribosyltransferase (HPRT) gene located on human chromosome X. Chimeric transposases were evaluated for expression, transposition activity, chromatin immunoprecipitation at the target loci, and targeted knockout of the HPRT gene in human cells. One ZFP-PB and one TALE-PB chimera demonstrated notable HPRT gene targeting. In contrast, Cas9/dCas9-PB chimeras did not result in gene targeting. Instead, the HPRT locus appeared to be protected from transposon integration. Supplied separately, PB permitted highly efficient isolation of Cas9-mediated knockout of HPRT, with zero transposon integrations in HPRT by deep sequencing. In summary, these tools may allow isolation of ‘targeted-only’ cells, be utilized to protect a genomic locus from transposon integration, and enrich for Cas9-mutated cells.
INTRODUCTION
Homologous directed recombination (HDR) dependent methods of DNA integration and repair require targeted DNA cleavage or nicking for transgene integration or gene editing. Contemporary methods include the use of zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and the newer CRISPR/Cas9 system (1). All of these systems exhibit off-target effects and none of them enzymatically integrate DNA (2). The PB transposon system is being developed for potential therapeutic application in genetic modification of clinically relevant cell types (3–8). The system actively integrates DNA into chromosomes; however, the native PB transposase is not targeted in its DNA delivery which poses a potential safety concern for certain applications (8). HDR-mediated mechanisms of transgene integration or gene repair such as those initiated by targeted nucleases may not work well in adult non-dividing tissues, which are important targets for genomic therapies (2). Given that PB actively integrates DNA and is not dependent on HDR, we sought to engineer PB for targeted integration into a user-defined locus in human cells independent of HDR.
A chimeric PB transposase was first shown to be capable of biasing integration in plasmid based transposition assays (9). Owens et al. fused a Gal4 DNA binding domain to PB and observed biasing PB integration toward the 56 898 UAS-like cognate Gal4 binding sites (CGGNNNNNNNNNNNCCG) in the human genome (10). However, in order to target PB to a unique user-defined chromosomal location, the locus must be chosen and an engineered DNA binding domain must be generated to target the user-chosen locus. Chimeric ZFP-PB transposase targeting of a unique genomic locus has not yet been demonstrated despite engineering ZFPs designed to target the checkpoint kinase-2 (CHK2) gene, the ROSA26 locus, or the l-gulono-g-lactone oxidase (GULOP) pseudogene (11,12). Owens et al. subsequently used a TALE domain for targeting PB integration into the C–C motif receptor 5 (CCR5) locus in human cells demonstrating the one isolated report of chimeric-PB transposase mediated targeted integration into a unique endogenous locus (13). Subsequently, Ye et al. engineered a TALE targeting the first exon of the fibroin light-chain gene in Bombyx mori L (14). Although this TALE-PB chimera increased transposition efficiency, no targeted integration was observed (14). The CRISPR/Cas9 system has been shown capable of gene editing in vitro and in vivo (15–18). Though not yet tested in a chimeric transposase configuration, this system would be highly attractive due to the ease of RNA-guided targeting that requires a simple search and cloning step to put the target sequence into the guide RNA plasmid. Flexible targeting of integration would be extremely attractive for a number of experimental and therapeutic uses. A catalytically inactive version, dCas9, has been successfully fused to protein domains resulting in targeted gene activation or repression (19). However, dCas9 or Cas9 fusion to PB or other transposases has not yet been reported. Therefore, given the lack of success with ZFP-PB chimeras targeting an endogenous locus (11,12), conflicting reports of targeting with TALE-PB chimeras (13,14), and no reports of dCas9- or Cas9-PB chimeras, we sought to perform a side-by-side systematic comparison of ZFP-, TALE- or Cas9/dCas9-PB chimeras targeting a single genetic locus.
The hypoxanthine phosphoribosyltransferase (HPRT) gene located on chromosome X has been used as a target for single gene targeting using viral vectors (20). Targeted knockout of HRPT enables selection of cells with targeted events through the use of 6-thioguanine (6-TG) which kills cells expressing active HPRT protein. In particular, targeted manipulation of HPRT using adeno-associated viral vectors (AAV) has led to mechanistic understanding of gene-targeting using AAV and its improvement (21–23). The HPRT locus has long been considered in refining gene transfer methodologies and remains a site of clinical interest as one can select out gene-targeted cells (24). We undertook the current study to perform a side-by-side comparison of ZFP-, TALE- and dCas9-PB chimeras for single gene targeting of the HPRT locus in human cells.
MATERIALS AND METHODS
Plasmid constructs
The PB-SB-SA-βGeo plasmid, carrying the PB terminal repeat sequences flanking a splice acceptor followed by the beta-galactosidase-neomycin resistance fusion protein gene (βGeo), was provided by Dr Allan Bradley (25). Four separate HPRT-targeted ZFPs were designed in silico using http://www.scripps.edu/barbas/zfdesign/zfdesignhome.php (26). Engineered HPRT-gene targeted ZFP-PB cDNAs were synthesized by the GenScript (Piscataway, NJ, USA). HPRT-targeted TALENs were provided by Dr. Dan Voytas (27). The SpCas9-humanized (and dCas9 variant), gRNA-humanized, and pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmids were obtained from Addgene (44 246, 44 248 and 42 230, respectively) (28,29). Standard molecular biology techniques were used to subclone the various ZFPs, TALEs, Cas9 and dCas9 upstream of the PB transposase cDNA in pCMV-ZFP-piggyBac to create chimeric transposase fusion proteins (11). All chimeras contained a hemagglutinin epitope (HA) tag and the following protein linker sequence between the added DNA binding domain and the PB transposase: GGSGGSGGSGGSGTS. The added nuclear localization sequences (NLS) to the ZFP, TAL, Cas9 and dCas9 vectors consisted of one copy of the SV40 NLS at locations indicated in the Supplementary Information. Guide RNAs (gRNA) for use with dCas9 targeting HPRT were designed using http://crispr.mit.edu/ (30). All plasmids were confirmed by DNA sequencing.
Colony count assays
HT-1080 cells were seeded into 100 mm dishes at one million cells per dish one day before transfection and were maintained in minimum essential medium alpha (containing 10% fetal bovine serum and penicillin-streptomycin without ribonucleosides and deoxyriboneucleosides) (ThermoFisher, Walthman, MA, USA). Cells were transfected with 2 μg PB-SB-SA-βGeo and 1 μg transposase plasmid using FuGENE-6 (Promega, Madison, WI, USA) according to manufacturer's instructions. For gRNA delivery, if not supplied on the transposase vector, cells were also transfected with 1 μg of hSpCas9 sgRNA plasmid. Two days after transfection, cells were trypsinized and split at a 1:100 dilution into media containing 500 μg/ml of geneticin for selection. Ten days to two weeks post-transfection, colonies were fixed and stained with 1% methylene blue in phosphate buffered saline (PBS) as described previously (31). To evaluate for the formation of 6-TG resistant colonies, one million of neomycin-resistant HT-1080 cells were seeded into 100 mm dishes with MEM alpha containing 30 μM 6-TG (Sigma-Aldrich, St. Louis, MO, USA). Ten days to two weeks after selection, 6-TG-resistant colonies were fixed and counted as described above.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) of transfected HT-1080 cells was performed using a kit available from Sigma Aldrich (CHP 1; St Louis, MO) according to the manufacturer's directions. Immunoprecipitation was performed using an anti-HA-tag antibody (HA.11, Biolegend, San Deigo, CA, USA) according to the manufacturer's directions and as described previously (11). Final DNA was resuspended and used for PCR using ChIP primers listed in the Supplementary Information.
PCR recovery transposon integration sites in HPRT and Illumina sequencing
HT-1080 cells were transfected and selected as described above. After selection with 6-TG for 14 days, HT-1080 cells were harvested and genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). Nested PCR was carried out with forward HPRT gene specific primers and reverse primers of specific to 5′TR and 3′TR of PB (Supplementary Information). After the first run PCR, 1 μl of the PCR product was used as a template for amplification with the second run PCR primers and the second PCR products were separated on a 1.0% agarose gel. The amplified PCR fragments were excised, purified with Gel Purification Kit (Qiagen) and inserted into the pCR 4-TOPO vector provided from the TOPO TA cloning Kit for sequencing (ThermoFisher, Walthman, MA). One shot MAX efffciency DH1α-T1 Escherichia coli (ThermoFisher, Walthman, MA, USA) were transformed with the ligated pCR 4-TOPO vector and inserted PCR fragments confirmed with restriction enzyme EcoR I were sequenced by GENEWIZ (South Plainfield, NJ, USA). Integration sites were mapped using UCSC Human Genome BLAT.
For next generation sequencing, DNA libraries were prepared and sequenced as described by others (12,32). DNA libraries containing unique barcodes (ATAC, CTAT, TATC, ATAG, CTAG or TATG) were obtained from three to five independent experiments, mixed and sequenced using HiSeq, and analyzed. FASTQ data were generated using Illumina's CASAVA software, version 1.8.2. The first four bases of each sequence in the FASTQ files were examined. Using the list of the six barcodes, FASTQ reads were binned according to if and how the first four bases of the read sequence matched the tetramers in the list. Thus for each sample a total of seven FASTQ files were created: six based on barcode binning and one consisting of reads whose first four bases did not match any provided barcodes. The first forty bases were trimmed off of each read in each binned FASTQ file. Sickle was used to trim 3′ bases based on quality score using a quality threshold of 20. Trimmed FASTQ data was aligned against the hg19 reference using bwa 0.6.2 with default parameters. A program was written to extract the chromosome, position, and mapping orientation for each read in the alignment. The regions of existing mappings were cached along with the coverage within the region. If a read's mapping overlapped the coordinates of existing mappings, then the existing coordinates in the cache were extended accordingly. After all the reads in a BAM were examined, the ends of each alignment region were trimmed to remove any edge position with <10% of the maximum coverage within the region. For each trimmed region, the four reference bases upstream (for forward alignments) or downstream (for reverse alignments) were extracted. If a region overlapped any of the groups in any of the BED regions described above, this was noted. The four extracted bases, the region coordinates, and the alignment direction were exported to a CSV file. The total number of reads evaluated, the total number of reads having the TTAA motif at the mapping start position, the number of TTAA motif reads within the HPRT gene, and number of reads overlapping each BED group are shown at the end of the same CSV file (see Supplementary Information). Evaluating for potential Z3 or TAL2 binding sites neighboring hotspots of integration on chromosome X was done by searching the results table for regions within that chromosome containing >1000 reads. The HPRT region coordinates were each padded by 10 000 bases on the 5′ and 3′ ends. The sequences for the padded regions were extracted and a pairwise2 method of the Biopython library was used to perform a local alignment of the Z3-PB fusion and TAL2-PB fusion DNA binding domain site sequences (Table 1). This method did not reveal any candidates having two or fewer mismatched or clipped bases or fewer than two bases worth of insertions or deletions. Quantitative PCR was used to determine the number of transposon insertions in comparison to the genomic RnaseP gene as described previously (11).
Table 1. Sequences targeted in the human HPRT gene.
| Transposases | Target HPRT sequence | X chromosome location |
|---|---|---|
| Z1-PB | AACAGGGTAATGGACTGG | 133607777–794 |
| Z2-PB | AGGGTAATGGACTGGGGC | 133607780–797 |
| Z3-PB | CACAACATTGACACTGTG | 133609934–951 |
| Z4-PB | ATTGACACTGTGGATGAA | 133609939–956 |
| TAL1-PB | GCATACCTAATCATTAT | 133607429–445 |
| TAL2-PB | AGGGTGTTTATTCCTCATG | 133607461–479 |
| TAL3-PB | CCATTCCTATGACTGTAGAT | 133609351–370 |
| TAL4-PB | AGCTATTGTGTGAGTAT | 133609386–402 |
| gRNAs for dCas9-PB | ||
| 1 | CTAGTATTGTTGGGTAATCT | 134462908–927 |
| 2 | ATCTGGGGTGAGACAAACTT | 134462939–958 |
| 3 | CAAGATTACCCAACAATACT | 134462906–925 |
| E2 | TTATGCTGAGGATTTGGAAA | 134473412–431 |
| E3 | GTAGCCCTCTGTGTGCTCAA | 134475233–252 |
The names of the chimeric transposases or gRNAs are depicted in the left column. The unique genomic sequences targeted in the human HPRT gene are shown in the middle column with the chromosomal locations on chromosome X in the right column using the February 2009 human genome assembly (GRCh37/hg19).
Surveyor assay
HT-1080 cells were transfected with 3 μg gRNA-pCMV-hSpCas9, 1 μg HA-PB and 1 μg PB-SB-SA-βGeo plasmids or 1 μg gRNA-pCMV-Cas9-PB and 1 μg PB-SB-SA-βGeo plasmids with Fugene 6. One day following transfection, cells were diluted into media for G418 selection at 30°C in 5% CO2 incubator for 2 days and then transferred to culture at 37°C for 10 days (33,34). Genomic DNA was prepared from G418 selected cells and amplified with HPRT gene specific primers (Supplementary Information). Cas9 or Cas9-PB introduced mutations were detected with IDT Surveyor Mutation detection kit (Integrated DNA Technologies, Coralville, IA, USA).
RESULTS
Engineering chimeric PB transposases for targeted knockout of HPRT
We evaluated three different potential targeting technologies via fusion with the PB transposase for targeted knockout of HPRT in human cells. In silico, we designed four different ZFPs targeting HPRT that were subsequently synthesized and fused to PB. We obtained two TALEN pairs previously engineered using Golden Gate assembly to create a double stranded break (DSB) in exon 2 or exon 3 of human HPRT in HEK293T cells (27). Using molecular biology techniques, we fused the TALE (subsequently referred to as TAL) cDNA without the nuclease domain to the N-terminus of the PB. We also fused catalytically active humanized Cas9 (Cas9) and the inactive variant (dCas9) to the N-terminus of PB. This permitted testing the targeting of dCas9/Cas9-PB to HPRT using gRNAs targeted to the HPRT locus. We compared delivering the gRNA for dCas9-PB on the same or separate vector. All engineered chimeric proteins contained an identical linker sequence between the DNA targeting domain and the PB transposase and a HA-tag for detection of expression and ChIP (11). Ultimately, we were able to compare four ZFPs, four TALs, Cas9 and dCas9 with various gRNAs for their ability to direct PB transposon integration into the HPRT locus. Schematics for all vectors used are described in (Supplementary Information).
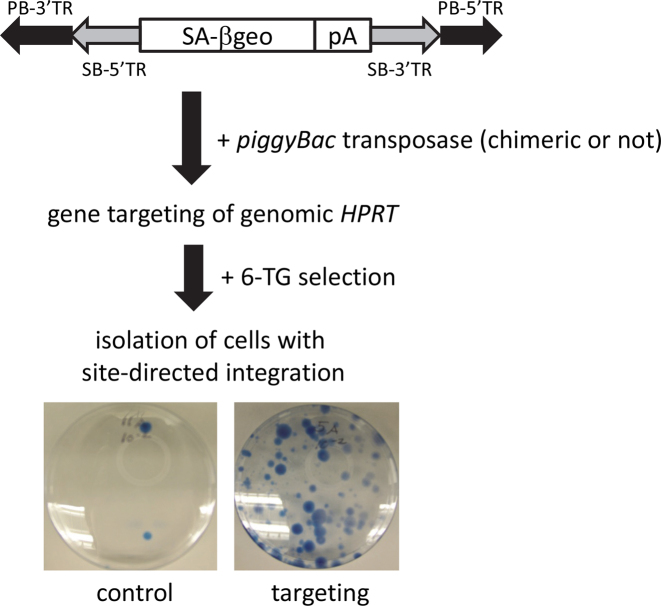
Targeted knockout of HPRT was achieved using a transposon containing a splice acceptor followed by the βGeo fusion protein gene sequence (PB-SB-SA-βGeo) (25). We used HT-1080 cells as they contain one copy of HPRT and have been utilized for gene targeting strategies using other vector systems (24,35). Integration of the PB-SB-SA-βGeo transposon into any expressed gene gives neomycin resistance via splicing resulting in production of the neomycin resistance transgene product. Active HPRT protein metabolizes 6-TG into a toxic product resulting in cell death, so targeted integration into HPRT results in 6-TG resistance (Figure 1). Therefore, we used neomycin resistance as a proxy for transposon integration in general, whereas 6-TG resistance served as a proxy for targeted knockout of HPRT. The locations of the DNA sequences targeted by the ZFP-, TAL- and Cas9/dCas9-PB chimeras are listed in Table 1.
Figure 1.
Quantitation of gene targeting efficiency by targeted knockout of HPRT. The PB-SB-SA-βGeo transposon was transfected with chimeric or native PB transposase into HT1080 cells. G418 selection was used as a proxy for transposase activity whereas 6-TG selection allowed isolation of cells with targeted integrations due to targeted knockout of HPRT.
ZFP-PB mediated gene targeting of HPRT
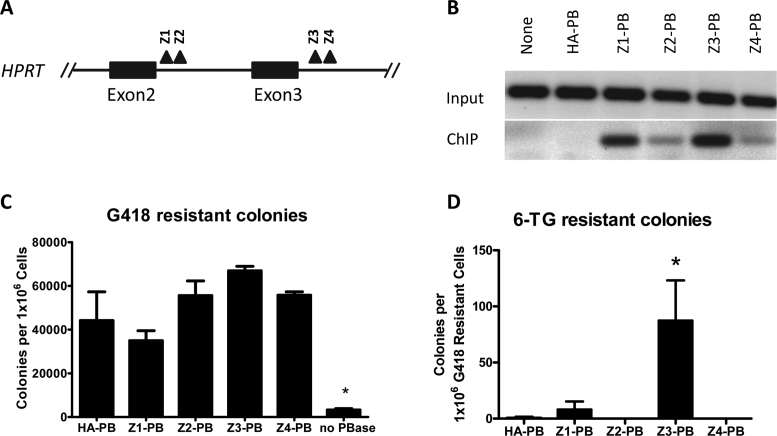
We evaluated four different ZFP-PB chimeras for targeting the HPRT locus (Figure 2A). These ZFPs were engineered to target unique 18 bp sequences within the endogenous HPRT gene sequence. Expression of each protein was confirmed in HEK293 cells by western blot analysis (Supplementary Information). We next used ChIP to determine if the ZFP-PB chimeras targeted binding to their cognate loci in the genome of cells. We observed no binding of HA-PB at the ZFP targeted loci (negative control); however, all ZFP-PB chimeras were immunoprecipitated at their target sequences, although with varying efficiency (Figure 2B). All ZFP-PB chimeras exhibited transposition efficiency similar to that of HA-PB and significantly higher than the no transposase control as observed in the G418-resistant colony counts (Figure 2C). Two of the ZFP-PB chimeras exhibited 6-TG resistant colonies. Z3-PB demonstrated significantly more 6-TG resistant colonies compared to the other ZFP-PB chimeras (Figure 2D). Although previous reports have not demonstrated the ability to engineer ZFP-PB to target integration into an endogenous locus in human cells (11,12), our results demonstrate that engineered ZFP-PB chimeras can be generated which bind to target loci in the HPRT locus of human cells, retain transposition activity, and direct integration into the HPRT locus as measured by quantitative readout of 6-TG resistant cells.
Figure 2.
ZFP-PB mediated gene targeting of HPRT. (A) four different ZFPs were engineered as described in the Materials and Methods to target HPRT. The approximate binding locations are depicted by the black triangles. (B) ChIP of the differing ZFP-PB chimeras at their cognate target sites in the human genome. HA-tagged PB, HA-PB, is depicted as a negative control. None represents the absence of transfected transposase. (C) G418 resistant colonies produced after co-transfection of the PB-SB-SA-βGeo plasmid with the varying ZFP-PB chimeras. ANOVA followed by Bonferroni post-test analysis revealed no PBase (no transposase) to be the only condition significantly different from HA-PB (the positive control) (N = 4, ±SEM). (D) Colony number after replating G418 resistant cells into and selecting with 6-TG. ANOVA with Bonferroni post-test revealed Z3-PB to be significantly different from HA-PB (N = 4, ±SEM).
TAL-PB mediated targeting of HPRT
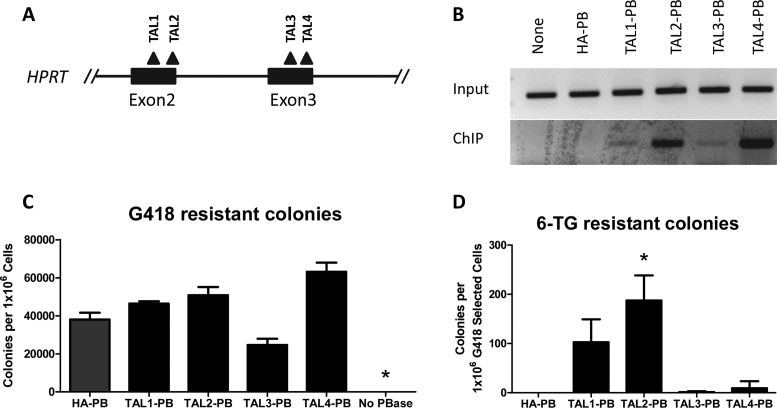
TALs represent another class of DNA binding domain proteins which can be user-engineered. We evaluated the ability of our four engineered TAL-PB chimeras, using the same process as outlined for the ZFP-PB chimeras, to target the HPRT locus (Figure 3A). These TALs were engineered to target unique 17–20 bp sequences within the HPRT gene and were previously validated as effective TALENs by others (27). We confirmed expression of the TAL-PB fusions with Western blot analysis (Supplementary Information), binding at their target loci by ChIP and transposition activity by colony assay (Figure 3B and C). Three of the TAL-PB fusions (TAL1-, TAL2- and TAL4-PB) exhibited 6-TG resistant colonies (Figure 3B). TAL2-PB produced the most 6-TG resistant colonies in the range of 2.1-fold more than Z3-PB (Figures 2D and 3D). Therefore, TAL-PB chimeras were also generated to target the HPRT locus, permitting isolation of cells with targeted integration events.
Figure 3.
TAL-PB mediated gene targeting of HPRT. (A) four different TALs were engineered with the approximate binding locations are depicted by the black triangles. (B) ChIP of the differing TAL-PB chimeras at their cognate target sites in the human genome. HA-PB, is depicted as a negative control. None represents the absence of transfected transposase. (C) G418 resistant colonies produced after co-transfection of the PB-SB-SA-βGeo plasmid with the varying TAL-PB chimeras. ANOVA followed by Bonferroni post-test analysis revealed no PBase (no transposase) to be the only condition significantly different from HA-PB (the positive control) (N = 4, ±SEM). (D) Colony number after replating G418 resistant cells into and selecting with 6-TG. ANOVA with Bonferroni post-test revealed TAL2-PB to be significantly different from HA-PB (N = 4, ±SEM).
Attempting dCas9/Cas9-PB targeting of HPRT
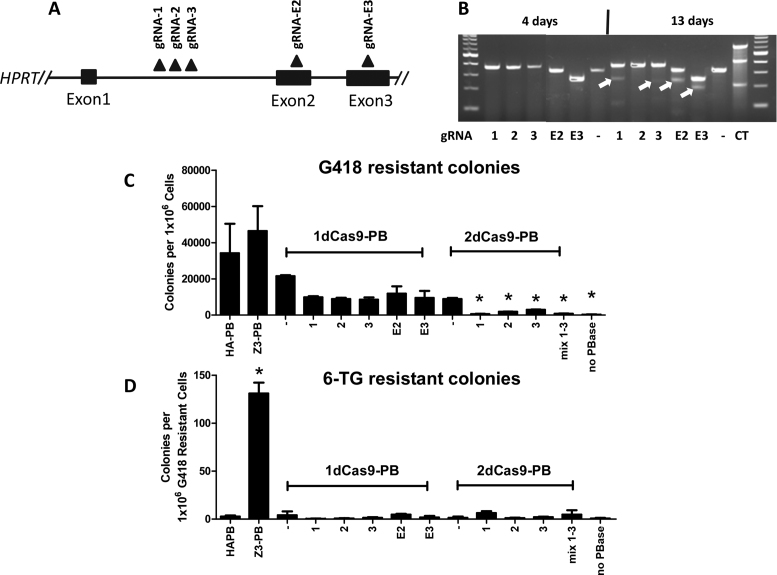
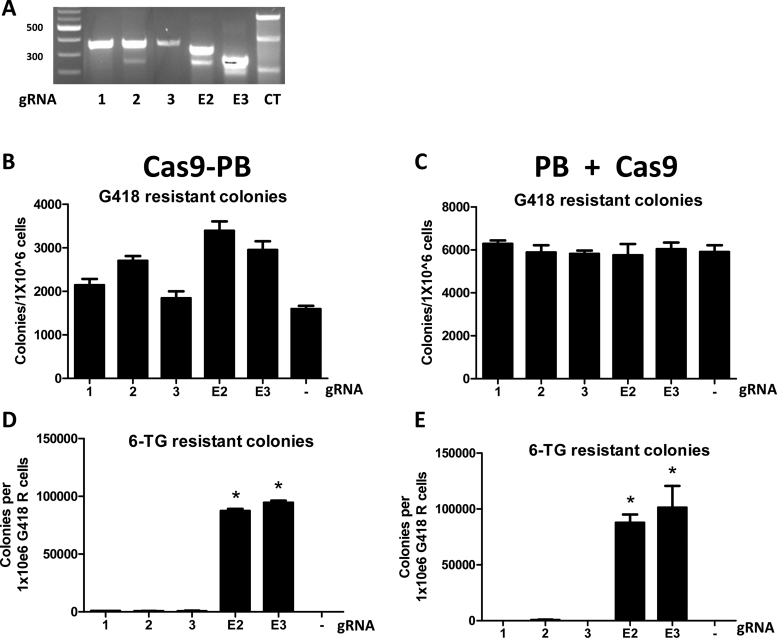
The CRISPR/Cas9 system represents a more recent technology enabling targeting for a variety of purposes. dCas9, a ‘dead’ SpCas9 mutant resultant from mutation of catalytic residues, has been fused to activator, repressor and nuclease domains leading to targeted gene activation, repression, or DNA DSBs (19,36,37). Therefore, we sought to fuse the dCas9 protein to PB to create a chimeric protein that would potentially employ gRNA-mediated targeted transposition into a user-defined chromosomal locus. gRNAs targeting HPRT were used with 1dCas9-PB by supplying them within separate DNA vectors (gRNAs 1, 2 and 3) or in the same vector (gRNAs E2 and E3) (Figure 4A). We evaluated 1dCas9-PB expression in HEK293 cells by western blot. We were unable to visualize full-length 1dCas9-PB expression in direct protein lysates; however, we could confirm expression in human cells by immunoprecipitation of 1dCas9-PB using the HA-tag (Supplementary Information). We were also unable to reliably ChIP 1dCas9-PB at target loci using the various gRNAs as we were unable to differentiate 1dCas9-PB without a gRNA from with the various gRNAs (unpublished data). Therefore, to determine if our gRNAs were functional for targeting of Cas9 to the various HPRT sequences, a surveyor assay was used to confirm targeted DNA cleavage (34,38). Catalytically active Cas9 (hSpCas9) was co-transfected with PB transposase and transposon to allow for selection of stably transfected cells and enrich for recovery of cells with targeted DNA cleavage. We observed targeted DNA cleavage mediated by four out of the five gRNAs tested by surveyor assay at 13 days post-transfection (Figure 4B). Therefore, four out of the five gRNAs were capable of directing Cas9 to the target loci in the HPRT gene. The 1dCas9-PB chimera with and without gRNAs exhibited transposition activity as measured by G418 colony count; however, there was a trend of less G418-resistant colonies whether or not gRNAs were included (Figure 4C). We were unable to reliably recover 6-TG resistant colonies using HPRT targeted gRNAs with 1dCas9-PB, whereas HA-PB (negative control) and Z3-PB (positive control) produced the expected results (Figure 4D).
Figure 4.
Attempted dCas9-PB mediated gene targeting of HPRT. (A) five different gRNAs were selected to target HPRT and were used supplying the gRNA on a separate (2,4,15) or the same (E2, E3) vector as dCas9-PB. The approximate binding locations are depicted by the black triangles. (B) Surveyor assay using the five gRNAs with catalytically active hSpCas9 and co-transfected with the PB-SB-SA-βGeo plasmid. Cells were selected in G418 for 4 days (left) and 13 days (right) and then assayed for the presence of targeted DNA cleavage using the surveyor assay. (C) G418 resistant colonies produced after co-transfection with the PB-SB-SA-βGeo plasmid with 1dCas9-PB/2dCas9-PB and the varying gRNAs. ANOVA followed by Bonferroni post-test analysis revealed no PBase (no transposase) and 2dCas9-PB to be the conditions significantly different from HA-PB (the positive control) (N = 4, ±SEM), though there was a trend to decreased G418 colony count when using 1dCas9-PB with the gRNAs. (D) Colony number after replating G418 resistant cells into and selecting with 6-TG. ANOVA with Bonferroni post-test revealed Z3-PB to be significantly different from HA-PB (N = 4, ±SEM). None of the gRNAs tested in combination with dCas9-PB resulted in a significant number of 6-TG resistant colonies.
Given our inability to target transposon integration into the HPRT locus using 1dCas9-PB, we sought to evaluate localization of 1dCas9-PB in transfected cells. We used immunofluorescence to evaluate the localization of HA-tagged native PB, Z3-PB, TAL2-PB, 1dCas9 alone and 1dCas9-PB (Supplementary Information). We observed strong nuclear localization for HA-PB, Z3-PB and TAL2-PB. However, 1dCas9-PB was mostly excluded from the nucleus. This was not due to PB fusion as dCas9 alone also exhibited the same localization pattern. Nonetheless, Cas9 must enter the nuclei of cells at some level as we observed targeted DNA cleavage (Figure 4B). Limited nuclear import may have impaired the overall transposition activity of 1dCas-PB in human cells (Figure 4C). We subsequently engineered a dCas9-PB construct containing added NLSs at both the N- and C-terminal sequence of the dCas9 protein in fusion with piggyBac (2dCas9-PB) (Supplementary Information). 2dCas9-PB demonstrated strong nuclear localization compared to 1dCas9-PB evaluated via immunofluorescence (Supplementary Information); however, the added NLS did not improve transposition activity or gene targeting of HPRT (Figure 4C and D). Some have reported combinatorial effects when using more than one gRNA simultaneously with dCas9-fusions (28). Therefore, we evaluated using gRNAs 1–3 together with 2dCas9-PB but found no improvement in transposition or gene targeting compared to 2dCas9-PB with a single gRNA (Figure 4C and D).
We next evaluated the potential for gene targeting of HPRT using catalytically active hSpCas9 fused to PB (Cas9-PB). We used the same gRNAs as were evaluated with dCas9-PB and performed the surveyor assay for targeted DNA cleavage by the fusion protein. Three of the gRNAs resulted in targeted DNA cleavage of HPRT (gRNAs 2, E2 and E3; Figure 5A); however, the results slightly differed when compared to using Cas9 alone for targeted DNA cleavage. gRNAs 1, 3, E2 and E3 demonstrated cleavage with Cas9 alone, whereas gRNAs 2, E2 and E3 manifested DNA cleavage with Cas9-PB (Figures 4B and 5A). These results imply that gRNAs that are functional with Cas9 alone may not necessarily be effective when using a Cas9-PB fusion and vice versa. However, this result did demonstrate that a Cas9-PB fusion can result in targeted DNA cleavage, so the Cas9 portion of the fusion was active. Cas9-PB was also able to function as a transposase as measured by the number of G418 resistant colonies produced; however, it transposed less efficiently when compared to the PB, ZFP-PB, and TAL-PB proteins (compare Figures 2C, 3C and 5B). Separating Cas9 from PB and transfecting them separately also resulted in fewer G418 resistant colonies, implying that Cas9 hindered PB transposition whether it was fused to PB or supplied separately (Figures 2C, 3C and 5C). We recovered a very high number of 6-TG resistant colonies when using the E2 and E3 gRNAs with Cas9-PB or Cas9 + PB, presumably due to knockout of HPRT via Cas9-mediated DNA cleavage and mutagenesis of exon 2 or 3 (Figure 5D and E).
Figure 5.
Attempted Cas9-PB mediated gene targeting of HPRT. (A) Surveyor assay using the five gRNAs as depicted in Figure 4A with catalytically active hSpCas9 fused to PB and co-transfected with the PB-SB-SA-βGeo plasmid. (B) G418 resistant colonies produced after co-transfection with the PB-SB-SA-βGeo plasmid with Cas9-PB and the varying gRNAs. ANOVA followed by Bonferroni post-test analysis revealed no difference between conditions (N = 4, ±SEM). (C) G418 resistant colonies produced after co-transfection with the PB-SB-SA-βGeo plasmid with Cas9 and PB on separate plasmids with or without the varying gRNAs. ANOVA followed by Bonferroni post-test analysis revealed no difference between conditions (N = 4, ±SEM). (D) Cas9-PB colony number after replating G418 resistant cells into and selecting with 6-TG. ANOVA with Bonferroni post-test revealed E2 and E3 to be significantly different from no gRNA (N = 4, ±SEM). (E) Cas9 + PB (supplied separately) colony number after replating G418 resistant cells into and selecting with 6-TG. ANOVA with Bonferroni post-test revealed E2 and E3 to be significantly different from no gRNA (N = 4, ±SEM).
Evaluation of targeted integrations into HPRT
In order to map integrations into HPRT, we first used PCR to recover integrations from genomic DNA isolated from 6-TG resistant cells created using Z3-PB and TAL2-PB. We used a series of HPRT specific primers that were staggered across the gene in PCR reactions with PB terminal repeat specific primers (Supplementary Information). All recovered integrations were sequenced to confirm targeting within HPRT. Although some integrations were recovered within 5kb of the target sites for the ZFP or TAL, many were recovered even at further distances (Supplementary Information).
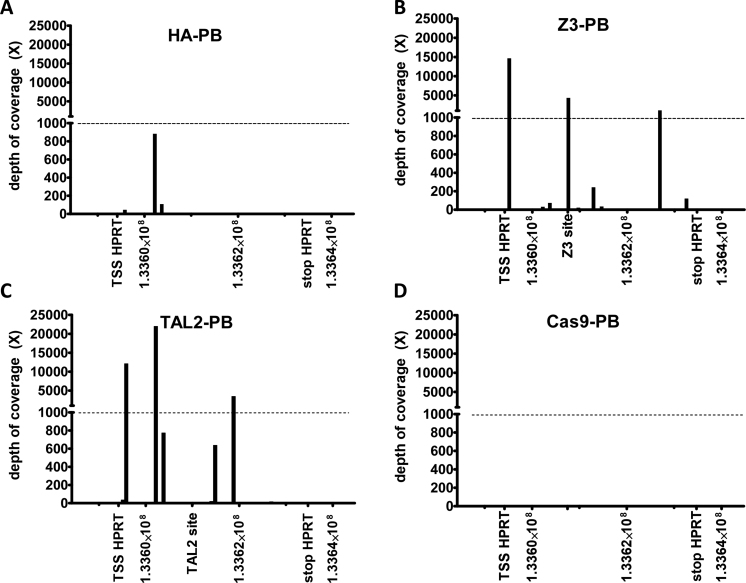
We next used Illumina HiSeq sequencing to quantitate integrations throughout the genome, particularly in HPRT (Figure 6). DNA libraries recovered from 6-TG resistant cells after transfection with HA-PB, Z3-PB and TAL2-PB were used. The 6-TG resistant cells recovered after HA-PB transfection likely represent a background integration rate of HA-PB in HT-1080 cells as we observed an enrichment of 80 to 188 fold more 6-TG resistant colonies when using Z3-PB and TAL2-PB respectively (Figures 2D and 3D). Z3-PB demonstrated a greater depth of coverage for transposon integrations into HPRT compared to HA-PB. The same was observed for TAL2-PB mediated integrations. Both Z3-PB and TAL2-PB exhibited sites of integration with >1000 reads in HPRT by deep sequencing, whereas HA-PB did not exhibit any sites with >1000 reads in HPRT (Figure 6). One of the integration hotspots for Z3-PB was within 200 bp of the Z3 binding site, whereas TAL2-PB hotspots were more dispersed throughout HPRT. Additionally, PB has been shown to have a predilection for integrating near transcription start sites of genes (31,32). Both Z3-PB and TAL2-PB showed hotspots of integration near the transcription start site of HPRT. The overall percentage of transposon integrations into the HPRT gene was 0.01%, 0.45%, and 0.97% respectively for HA-PB, Z3-PB, and TAL2-PB in 6-TG resistant cells (Table 2). Quantitative PCR of the neomycin resistance part of the transposon was used to determine the average number of total PB transposon integrations per copy of genomic RnaseP in 6-TG resistant cells. We observed an increased number of PB transposon integrations in the 6-TG resistant HT-1080 cells using Z3-PB and TAL2-PB compared to HA-PB (Table 2). Therefore, more 6-TG resistant cells were obtained due to targeting; however, we also observed an increased number of overall transposon integrations in 6-TG resistant cells when using these chimeric transposases. We evaluated the other hotspots of integration on chromosome X for Z3-PB and TAL2-PB outside of HPRT with >1000 reads. We did not find any potential Z3 or TAL2 binding sites (with two or fewer mismatches or deletions) within 10 kb either direction of these hotspots.
Figure 6.
Deep sequencing of transposon integrations into HPRT on chromosome X. The depth of coverage of recovered integrations is depicted throughout the length HPRT with an additional 10 000 bases beyond the transcription start site (TSS) and stop sequence. Both Z3-PB (B) and TAL2-PB (C) demonstrated increased integrations into HPRT compared to control HA-PB (A). Whereas, Cas9-PB demonstrated zero integrations into HPRT using gRNA E2 or E3 (D).
Table 2. Analysis of transposon targeting into HPRT.
| gRNA | Number of unique sites | Total sequencing reads | Reads in HPRT | % targeted into HPRT | Average # of copies/RnaseP | |
|---|---|---|---|---|---|---|
| HA-PB | N/A | 16683 | 7840617 | 1168 | 0.01 | 8.5±2.5 |
| Z3-PB | N/A | 19224 | 4959877 | 20624 | 0.42 | 36±12 |
| TAL2-PB | N/A | 21879 | 4010639 | 39054 | 0.97 | 49±10 |
| Cas9-PB | E2 | 51570 | 10288878 | 0 | 0 | ND |
| E3 | 59658 | 8956385 | 0 | 0 | ND | |
| PB + Cas9 | E2 | 25601 | 9036304 | 0 | 0 | ND |
| E3 | 64514 | 9007196 | 0 | 0 | ND |
HiSeq was used to recover thousands of unique sites using the various transposases from 6-TG resistant cells. The ‘% targeted into HPRT’ represents the sequencing reads of confirmed integrations with the cognate TTAA target site recovered between the 5′ and 3′ untranslated regions of the full genomic sequence of human HPRT in BLAT. The percentage represents the depth of coverage of recovered integration sites within HPRT divided by the total number of integrations recovered. The ‘Average # of copies/RnaseP’ represents the number of copies of the neomycin resistance gene (contained within the transposon) divided by the copies of RnaseP (number of copies of transposon per haploid genome) measured using qPCR.
We also evaluated for integrations in HPRT when using Cas9-PB with the gRNAs E2 and E3 (Table 2 and Figure 6D). Interestingly, we found zero integrations of PB into HPRT when using Cas9-PB or PB + Cas9 and these gRNAs (Supplementary Information and Figure 6D). Therefore, the 6-TG resistant colonies we observed were from Cas9-mediated cleavage and mutagenesis of HPRT rather than targeted integration of PB transposons into HPRT (Figure 5D). These results also imply that DNA binding or cleavage by Cas9 inhibits PB integration nearby. Therefore, PB can be used in combination with active Cas9 for efficient isolation of cells with targeted Cas9 cleavage, while the fusion protein prevents PB insertions into genomic locations as specified by the gRNA.
DISCUSSION
We sought to directly compare the available targeting technologies in combination with the PB transposon system for gene targeting of human HPRT. Our methodology allowed us to count the number of 6-TG resistant colonies as a quantitative proxy for the efficiency of each transposase chimera for knocking out HPRT. In comparing four different engineered ZFPs, four different TALs, and five different gRNAs with dCas9/Cas9-PB, we observed the highest rate of gene targeting with one ZFP-PB and one TAL-PB chimera (Z3-PB and TAL2-PB respectively). TAL2-PB performed the best based on the number of 6-TG resistant colonies recovered as well as the number of integrations within HPRT in those resistant cells. Our analysis indicates that in considering chimeric-PB for gene targeting, one should evaluate multiple different DNA binding domains simultaneously to maximize the chance of success.
In human cells, a few different DNA binding domains have been fused to the PB transposase with the goal of targeting chromosomal loci with varying success. In a former study, we fused a previously validated ZFP with high specificity to the checkpoint kinase-2 (CHK2) gene to the N-terminus of PB (11). Given a lack of TTAA nucleotide elements surrounding the target site of the ZFP in the promoter of CHK2 gene, we engineered an artificial target site with surrounding TTAAs and observed biased integration in human cells (11). Owens et al. found that fusion of a Gal4 DNA binding domain to PB was effective at biasing PB integration toward the 56 898 UAS-like cognate Gal4 binding sites (CGGNNNNNNNNNNNCCG) in the human genome (10). Others have fused PB to the Rep protein of adeno-associated virus, but they observed an apparent lack of biased integration near Rep recognition sequences in the human genome (39). Subsequently, Li et al. created ZFP-PB fusions targeting human ROSA26 or the L-gulono-g-lactone oxidase (GULOP) pseudogene (12). Although these engineered ZFPs could restore integration of an excision-active/integration-defective transposase (iPB7R327A/K375A/D450N), no targeted integration into ROSA26 or GULOP was observed (12). Of the DNA binding domain-PB fusions mentioned above, only those targeting ROSA26 or GULOP were user-designed protein sequences to target unique DNA sequences within the human genome; however, neither of those resulted in measurable targeting of transposon integrations. This is the first report of a user-designed ZFP-PB chimera successfully targeting a gene in human cells by directing integration into human HPRT (Figure 2).
TAL DNA binding domains offer a different technology than designer ZFPs for gene targeting. Owens et al. engineered a TAL domain and tested it in different configurations for targeting PB integration into the C–C motif receptor 5 (CCR5) locus in human cells (13). They observed targeting efficiency of 0.01–0.015% in the CCR5 locus of transfected cells with their best construct. However, Ye et al. observed a lack of targeting integration with a different TAL-PB chimera (14). We compared four different TALs designed to target HPRT and observed different targeting efficiencies based on the number of 6-TG resistant colonies. When plating 1 million G418 resistant cells, TAL2-PB resulted in 188 ± 51 (N = 4, ±SEM) 6-TG resistant colonies while HA-PB produced zero. This was 2.14 times more 6-TG resistant cells than those resulting from the best performing ZFP-PB chimera (Z3-PB, 87 ± 35 6-TG resistant colonies (N = 4, ±SEM)). Our results reveal targeting in 0.019% of G418 resistant cells, in the range of that observed by Owens et al. using a different TAL targeting CCR5 (13). Our analysis involved deep sequencing of recoverable PB insertions throughout the genome, whereas Owens et al. used PCR to recover only insertions near the genomic TAL binding site (13).
The CRISPR/Cas9 system represents a more recently discovered gene targeting method that has become widely utilized in a short period of time due to target flexibility made possible by computationally directed gRNA selection and simple cloning procedures. Although dCas9 has been fused to transcriptional activators and repressors and nuclease domains (19,36,37), fusion of dCas9 to PB resulted in decreased transposition activity and a lack of targeting of HPRT (Figure 4). This lack of observable targeting was not due to gRNA selection, as the tested gRNAs produced DSBs when combined with catalytically active hSpCas9 (Figure 4B). The fusion of 1dCas9 to PB changed the localization of the transposase from nuclear to predominantly cytoplasmic (Supplementary Information) (28). Nonetheless, adding another NLS to create 2dCas9-PB improved nuclear localization but did not improve transposition activity or gene targeting (Supplementary Information and Figure 4). The linker sequence between the DNA binding domain and PB was the same between the ZFPs, TALs and dCas9/Cas9. Perhaps a smaller version of Cas9, such as SaCas9, may be more amenable to fusion to PB (17). Because PB prefers to integrate into open chromatin, it may not be able to penetrate dCas9/Cas9 targeted DNA. A transposase or recombinase from another family may be better able to integrate into the targeted DNA, so fusion to other enzymes should be considered. Alternatively, different linker sequences and lengths could be attempted between dCas9 and PB, as well as other nuclear localization sequences.
Evaluating a Cas9-PB fusion demonstrated that Cas9 cleavage of a target site appears to inhibit PB nearby as we were unable to recover PB integrations into HPRT with deep sequencing analysis when using Cas9 + PB or Cas9-PB with the gRNAs E2 and E3 (Table 2). Perhaps the scanning DNA binding activity of Cas9 prevents PB availability for transposition (40). Nonetheless, PB represents a powerful platform for selection of Cas9 mediated DNA cleavage events in cells. In particular, excision-active/integration-defective transposase (iPB7R327A/K375A/D450N) could be used after selection Cas9-altered cells to generate cells with targeted DNA alterations and no other exogenous DNA (12). Additionally, targeted DNA cleavage could be used to de-target PB integration from a particular location which could be useful when using PB to select CRISPR/Cas9 modified cells as one can have some confidence that targeted disruptions result from CRISPR/Cas9 and not PB transposition.
Overall, our analysis of ZFP-, TAL- and Cas9/dCas9-PB chimeras designed to target HPRT would suggest that TAL-PB was most effective based on the number of 6-TG resistant colonies recovered as well as the % of recovered deep sequencing reads for PB insertions into HPRT (Figures 3D, 6C and Table 2). Of note, we began our comparison by using TALs already validated as TALENs for HPRT (27). Given publicly available information, we were unable to design ZFP-PB chimeras to target those overlapping TAL binding sites; however, we were able to design and synthesize ZFPs capable of targeting the HPRT locus permitting our comparison. As others have demonstrated in Drosophila melanogaster (41), it might be possible to re-engineer TAL-PB fusions designed to bind the overlapping genomic target site of our Z3-PB fusion for a direct comparison of chimeric piggyBac targeting of a very specific genomic sequence within HPRT rather than the overall HPRT gene locus.
Although we could achieve gene targeting of HPRT without relying on HDR, there remains room for improvement. Both Z3-PB and TAL2-PB exhibited gene targeting as measured by 6-TG resistant cells; however, both also exhibited a higher number of transposon integrations per cell in those 6-TG resistant cells. Ultimately, one would want to achieve specificity defined as only targeted integrations in the absence of off-target integrations. The PB transposase is highly active in human cells. Therefore, mutations could be made to decrease activity or make the transposase integration activity more dependent on the fused DNA binding domain. Inducible expression of the transposase or the use of a weak promoter may decrease the number of integrations per cell while increasing targeting efficiency. Additionally, modifications of the transposon vector have not been tested with regards to improving targeting efficiency. Gogöl-Doring et al. recently identified PB interacting proteins, including cellular bromodomain and extraterminal (BET) domain proteins such as BRD4, which could potentially be exploited to improve PB mediated targeted DNA delivery to host chromosomes (42).
Transposon vectors offer an alternative to gene editing strategies as they actively integrate DNA and are not dependent upon homologous recombination (2). Gene editing strategies using Cas9 have recently shown promise in vivo (43–46). For ornithine transcarbamylase deficiency, Cas9 delivery to newborn mice was effective while for adult mice it was toxic (45). Cas9 has also been shown to create chromosomal translocations in cultured cells via off-target DSBs (47). The PB transposon system is being utilized for genetic modification of clinically relevant cell types (3–5,7,48,49). Further refinements to improve targeting of transposon integration would improve the usefulness and safety of all these applications. Our system of using targeting of HPRT to isolate gene targeted cells should allow further refinement and improvement of targeting integration with PB.
Supplementary Material
ACKNOWLEDGEMENTS
The TALENs were provided by Dan Voytas. The anti-piggyBac transposase antibody was a gift from Malcolm Fraser.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [DK093660 to M.H.W.] (in part); Department of Veterans Affairs [BX002190 to M.H.W., BX002797 to L.E.W.]; Vanderbilt Center for Kidney Disease (to M.H.W.). Funding for open access charge: NIH [DK093660, VA BX002190].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 2. Gaj T., Gersbach C.A., Barbas C.F. 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013; 31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grabundzija I., Irgang M., Mates L., Belay E., Matrai J., Gogol-Doring A., Kawakami K., Chen W., Ruiz P., Chuah M.K. et al. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010; 18:1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Manuri P.V.R., Wilson M.H., Maiti S.N., Mi T., Singh H., Olivares S., Dawson M.J., Huls H., Lee D.A., Rao P.H. et al. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum. Gene Ther. 2010; 21:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakazawa Y., Huye L.E., Dotti G., Foster A.E., Vera J.F., Manuri P.R., June C.H., Rooney C.M., Wilson M.H.. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J. Immunother. 2009; 32:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakazawa Y., Huye L.E., Salsman V.S., Leen A.M., Ahmed N., Rollins L., Dotti G., Gottschalk S.M., Wilson M.H., Rooney C.M.. PiggyBac-mediated cancer immunotherapy using EBV-specific cytotoxic T-cells expressing HER2-specific chimeric antigen receptor. Mol. Ther. 2011; 19:2133–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.Q., Paschon D.E., Miranda E., Ordonez A., Hannan N.R., Rouhani F.J. et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011; 478:391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woodard L.E., Wilson M.H.. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015; 33:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maragathavally K.J., Kaminski J.M., Coates C.J.. Chimeric Mos1 and piggyBac transposases result in site-directed integration. FASEB J. 2006; 20:1880–1882. [DOI] [PubMed] [Google Scholar]
- 10. Owens J.B., Urschitz J., Stoytchev I., Dang N.C., Stoytcheva Z., Belcaid M., Maragathavally K.J., Coates C.J., Segal D.J., Moisyadi S.. Chimeric piggyBac transposases for genomic targeting in human cells. Nucleic Acids Res. 2012; 40:6978–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kettlun C., Galvan D.L., George A.L. Jr, Kaja A., Wilson M.H.. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. 2011; 19:1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B. Jr et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Owens J.B., Mauro D., Stoytchev I., Bhakta M.S., Kim M.S., Segal D.J., Moisyadi S.. Transcription activator like effector (TALE)-directed piggyBac transposition in human cells. Nucleic Acids Res. 2013; 41:9197–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye L., You Z., Qian Q., Zhang Y., Che J., Song J., Zhong B.. TAL effectors mediate high-efficiency transposition of the piggyBac transposon in silkworm Bombyx mori L. Sci. Rep. 2015; 5:17172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho S.W., Kim S., Kim J.M., Kim J.S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013; 31:230–232. [DOI] [PubMed] [Google Scholar]
- 16. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K.. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013; 31:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sander J.D., Joung J.K.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014; 32:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russell D.W., Hirata R.K.. Human gene targeting by viral vectors. Nat. Genet. 1998; 18:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirata R., Chamberlain J., Dong R., Russell D.W.. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002; 20:735–738. [DOI] [PubMed] [Google Scholar]
- 22. Khan I.F., Hirata R.K., Wang P.R., Li Y., Kho J., Nelson A., Huo Y., Zavaljevski M., Ware C., Russell D.W.. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010; 18:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trobridge G., Hirata R.K., Russell D.W.. Gene targeting by adeno-associated virus vectors is cell-cycle dependent. Hum. Gene Ther. 2005; 16:522–526. [DOI] [PubMed] [Google Scholar]
- 24. Caskey C.T., Kruh G.D.. The HPRT locus. Cell. 1979; 16:1–9. [DOI] [PubMed] [Google Scholar]
- 25. Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., Liu P.. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9290–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mandell J.G., Barbas C.F. 3rd. Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006; 34:W516–W523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F.. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011; 39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilson M.H., Coates C.J., George A.L. Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007; 15:139–145. [DOI] [PubMed] [Google Scholar]
- 32. Burnight E.R., Staber J.M., Korsakov P., Li X., Brett B.T., Scheetz T.E., Craig N.L., McCray P.B. Jr. A hyperactive transposase promotes persistent gene transfer of a piggyBac DNA transposon. Mol. Ther. Nucleic Acids. 2012; 1:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doyon Y., McCammon J.M., Miller J.C., Faraji F., Ngo C., Katibah G.E., Amora R., Hocking T.D., Zhang L., Rebar E.J. et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008; 26:702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moriarity B.S., Rahrmann E.P., Beckmann D.A., Conboy C.B., Watson A.L., Carlson D.F., Olson E.R., Hyland K.A., Fahrenkrug S.C., McIvor R.S. et al. Simple and efficient methods for enrichment and isolation of endonuclease modified cells. PLoS One. 2014; 9:e96114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doetschman T., Gregg R.G., Maeda N., Hooper M.L., Melton D.W., Thompson S., Smithies O.. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987; 330:576–578. [DOI] [PubMed] [Google Scholar]
- 36. Guilinger J.P., Thompson D.B., Liu D.R.. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014; 32:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K.. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014; 32:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doyon Y., Choi V.M., Xia D.F., Vo T.D., Gregory P.D., Holmes M.C.. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods. 2010; 7:459–460. [DOI] [PubMed] [Google Scholar]
- 39. Ammar I., Gogol-Doring A., Miskey C., Chen W., Cathomen T., Izsvak Z., Ivics Z.. Retargeting transposon insertions by the adeno-associated virus Rep protein. Nucleic Acids Res. 2012; 40:6693–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beumer K.J., Trautman J.K., Christian M., Dahlem T.J., Lake C.M., Hawley R.S., Grunwald D.J., Voytas D.F., Carroll D.. Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene targeting in Drosophila. G3 (Bethesda). 2013; 3:1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gogol-Doring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W., Uckert W., Schulz T.F., Izsvak Z., Ivics Z.. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T Cells. Mol. Ther. 2016; 24:592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016; 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016; 351:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016; 34:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016; 34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K.. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009; 458:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yusa K., Rad R., Takeda J., Bradley A.. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009; 6:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.