Abstract
The speed at which a molecular motor operates is critically important for the survival of a virus or an organism but very little is known about the underlying mechanisms. Tailed bacteriophage T4 employs one of the fastest and most powerful packaging motors, a pentamer of gp17 that translocates DNA at a rate of up to ∼2000-bp/s. We hypothesize, guided by structural and genetic analyses, that a unique hydrophobic environment in the catalytic space of gp17-adenosine triphosphatase (ATPase) determines the rate at which the ‘lytic water’ molecule is activated and OH− nucleophile is generated, in turn determining the speed of the motor. We tested this hypothesis by identifying two hydrophobic amino acids, M195 and F259, in the catalytic space of gp17-ATPase that are in a position to modulate motor speed. Combinatorial mutagenesis demonstrated that hydrophobic substitutions were tolerated but polar or charged substitutions resulted in null or cold-sensitive/small-plaque phenotypes. Quantitative biochemical and single-molecule analyses showed that the mutant motors exhibited 1.8- to 2.5-fold lower rate of ATP hydrolysis, 2.5- to 4.5-fold lower DNA packaging velocity, and required an activator protein, gp16 for rapid firing of ATPases. These studies uncover a speed control mechanism that might allow selection of motors with optimal performance for organisms’ survival.
INTRODUCTION
Tailed bacteriophages are considered as the most abundant forms of life on the planet (1). They employ powerful molecular motors to condense the viral genomes inside a capsid shell (2–5). These motors belong to ASCE superfamily of multimeric ring-type NTPases that perform numerous and diverse tasks in cellular organisms such as transport of cargo between compartments, unwinding of DNA to allow replication, unfolding of proteins for degradation and segregation of chromosomes during cell division (6,7). One of the critical aspects of motor function is the speed at which the motor performs a task. Motors such as chromatin modeling Adenosine triphosphatases (ATPases) (8) operate at a slow speed compared to DNA helicases (9) which must unwind DNA at a much higher speed in order to keep up with the rate of DNA synthesis.
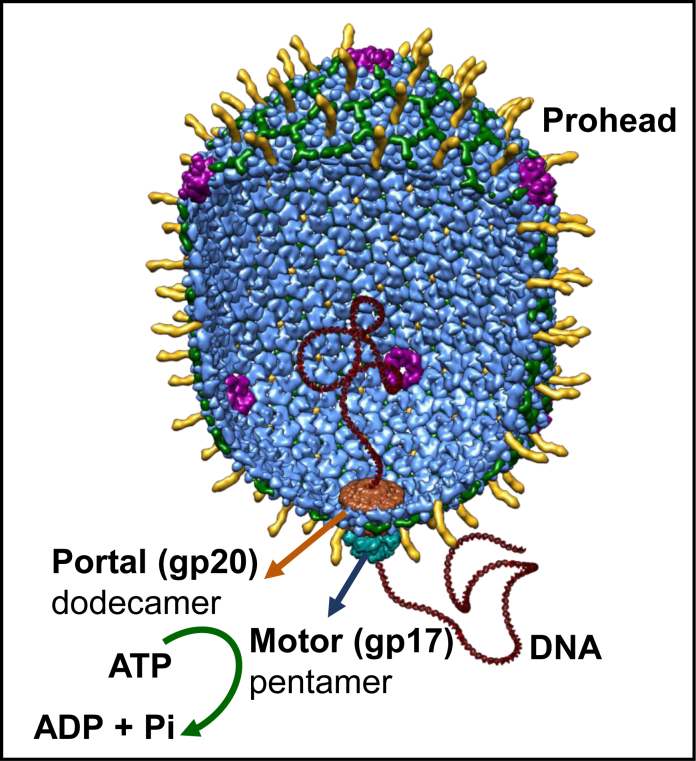
The tailed bacteriophage T4 belongs to the family of Myoviridae. It is a relatively large phage and an important model in molecular biology to elucidate basic mechanisms. During assembly, its 120 × 86 nm icosahedral head (capsid) is packaged with ∼171-kb, 56 μm-long DNA to near crystalline density (10). An oligomeric motor containing five subunits of gp17 ‘large terminase’ (TerL) docks on the special dodecameric portal (gp20) vertex of the capsid (Figure 1) (11,12). gp17 consists of an N-terminal ATPase domain that provides energy for packaging and a C-terminal nuclease/translocase domain that generates an end and translocates the genome (13). The ATPase domain has two subdomains; subdomain I (Nsub I) that contains all the canonical signatures including Walker A, Walker B and catalytic carboxylate, and a smaller subdomain II (Nsub II) containing sites that regulate ATP hydrolysis. The ATPase and translocase domains are linked by a flexible hinge and several charge pairs at the interface. The motor subunits are proposed to alternate between two conformational states, Extended (or Relaxed) and Compact (or Tensed), generating electrostatic force that powers translocation DNA, 2 bp at a time (11,14).
Figure 1.
A structural model of the phage T4 DNA packaging machine. It consists of the pentameric motor (gp17) assembled at the dodecameric portal vertex (gp20) of the prohead (11). The model is based on the cryo-EM structure of the prohead–motor complex and functional studies (11).
In addition to motor and portal, the phage packaging machine consists of a regulator, the 11- or 12-meric gp16 ‘small terminase’ (TerS) that interacts with gp17 to form a holo-terminase complex and regulates gp17 functions (15). TerS is essential for recognition and cutting of concatemeric viral genome in vivo, although the TerL motor alone is sufficient to package an already-cut DNA in vitro starting from an end (13). In addition, gp16 stimulates gp17-ATPase which is thought to be important for rapid firing of motor subunits when the newly created end is inserted into the motor channel for packaging initiation (16,17).
The phage T4 DNA packaging motor is the fastest and most powerful reported to date (18). It can package up to 2000 bp/s generating a power density of 5000 kW/m3, twice that of an automobile engine. A fast motor may have evolved to enable packaging of phage T4’s 171-kb genome in the same amount of time in one infection cycle as other phages that package shorter genomes. Single-molecule analyses determined that the packaging velocity of phage motors is roughly proportional to the size of the genome the motor packages; 2,000-bp/sec for T4 that packages 171-kb genome, 800-bp/s for λ that packages 48.5 kb genome, and 200-bp/s for phi29 that packages 20-kb genome (18–20). We proposed that the speed of the motor may be evolutionarily scaled to the size of the genome it packages, in order to optimize the motor’s performance for phage survival (18).
How is the motor speed controlled? Very little information is available on the basic mechanism. Evidence suggests that residues in vicinity of the Walker B motif and the γ-phosphate of bound ATP in the ATPase pocket are important. The Walker B motif is highly conserved in ATPase motors (21), consisting of a β-strand of 4–6 hydrophobic amino acids (aa) tucked into the Rossmann fold followed by a loop containing a strictly conserved aspartate that coordinates with the ATP–Mg complex, and a catalytic glutamate that acts as a base by accepting a proton from a ‘lytic’ water molecule to generate the OH− nucleophile. In chromatin remodeling ATPase mkTIP49, a D346N mutation residing at ∼8Å distance from the Walker B aspartate and ATP γ-phosphate resulted in 2-fold increase in the rate of ATP hydrolysis (8). In the phage λ packaging motor, mutation of residues located in the predicted helix connecting Walker B and ATPase coupling C-motifs (T194M, G212S) resulted in 3- to 10-fold reduction in packaging velocity (22). In SpoIII, an Escherichia coli DNA translocase that is involved in chromosome segregation, mutation of D584 located at a similar position resulted in 2.5-fold reduction in chromosome transport rate (23). Mutational analysis of phage T4 motor revealed that the presence of a hydrophobic aa next to Walker B catalytic glutamate (E256) is essential for function (24). This hydrophobic aa requirement is conserved in ∼93% of large terminase sequences and ∼99% of helicase sequences in the data base.
We hypothesize that the hydrophobic microenvironment of the catalytic space surrounding the catalytic glutamate and the γ-phosphate of bound ATP is a critical determinant of the rate of ATP hydrolysis and motor speed. Since the catalytic glutamate must accept, but not hydrogen-bond with, a proton in order to generate an OH− nucleophile from the polarized water molecule (8), hydrophobicity of this microenvironment as provided by the aa sidechains oriented into this space would be critical. These would help exclude non-specific water molecules from this space and create an appropriate microenvironment for polarization of the lytic water molecule (9,25–26). The OH− nucleophile then attacks the γ-phosphate, resulting in cleavage of ATP to ADP and Pi. These steps determine the rate of ATP hydrolysis and speed of the motor.
We tested the above hypothesis by changing the hydrophobicity of the catalytic space by mutation. Substitution of M195 and F259 residues located at 5–6Å distance from the catalytic glutamate with less hydrophobic, polar or charged aa resulted in null phenotype or partially defective cold sensitive/small plaque (cs/sp) phenotype. Remarkably, a slight increase in the hydrophobicity of the sidechain by a single carbon restored partial function. The cs/sp mutants (M195T, E and V) showed 3- to 66-fold reduction in infectious virus particles, 70–90% loss of ATPase activity and in vitro DNA packaging activity. Furthermore, single-molecule optical tweezers data showed that the M195E/V motors were unable to fire at the rate necessary to initiate DNA packaging unless assisted by the ATPase activator protein gp16. Once initiated, the mutant motors translocated DNA at 2.5- to 4.5-fold lower rate than the wild-type (WT) motors.
Thus, our studies for the first time addressed a critical aspect of the biological energy transductions by formulating a defined hypothesis. The results uncovered a speed control mechanism for the phage T4 DNA packaging motor, which may be broadly extended to other packaging and molecular motors that are critical for life.
MATERIALS AND METHODS
Bacteria, phage and plasmids
Escherichia coli P301 (sup−) was a non-suppressor strain used for recombinational genetic rescue. Escherichia coli B40 (sup+), a serine-suppressor, was used to make S194am phage stock and E. coli Y-suppresssor (sup+) (27), a tyrosine-suppressor, was used to make Y253am phage stock (21). Escherichia coli strains, DH5α and BL21 (DE3) (Novagen) were used as a host strains for pET-28b plasmids (Novagen) carrying gene 17 (g17).
Structure analyses
Analysis of amino acids residues in vicinity of E256 and γ-phosphate and distance measurements were performed using Pymol (The PyMOL Molecular Graphics System, version 1.3, Schrödinger LLC) and Chimera (the Resource for Biocomputing, Visualization and Informatics, version 1.10.2, University of California, San Fransisco, CA, USA).
Primers
Amplification primers
All the nucleotides listed below correspond to the coding sequence of g17. The g17 end primers containing NheI ends and BamHI ends that were used to amplify g17 DNA are: 5′-forward primer containing NheI restriction site (shown in bold), 5′-AAAGCTAGC ATGGAACAACCGATTAATGTATTAAATG-3′ (nt 1–28) and 3′-reverse primer containing BamHI restriction site (shown in bold), 5′- AAAGGATCC TTATACCA TTGACATACCATGAGATAC-3′ (nt 1797–1833). The italicized nucleotides represent the tag sequence added to the 5′-end for efficient cutting at the adjacent NheI or BamHI sequence.
Sequencing Primers
The primers used to amplify the DNA flanking at the targeted mutation site for sequencing are as follow: for M195 mutants, 5′-forward: 5′-CGTGACTATCAGCGTGATATGC-3′ (nt 418–439); for F259 mutants, 5′-forward: 5′- GCAAGCAATTGAACTGCTTCC-3′ (nt 612–632).
Mutagenesis primers
The primers used for combinatorial mutagenesis libraries are as follow: M195 mutants: forward mutant primer: 5′- GCACACAAAGGCTCANNNTCTGCGGAAGTTTTA-3′ (nt 568–600), reverse mutant primer: 5′- TAAAACTTCCGCAGANNNTGAGCCTTTGTGTGC-3′; F259 mutants: forward mutant primer: 5′-ATTGACGAATGTGCGNNNATTCCAAA CTTCCAT-3′ (nt 760–792), reverse mutant primer: 5′-ATGGAAGTTTGGAATNNNCGCACA TTCGTCAAT-3′. The mutation sites are shown in bold; NNN represents randomization of nucleotides.
Combinatorial mutagenesis
Met195 and Phe259 mutants were constructed by combinatorial mutagenesis. Combinatorial mutagenesis was performed by the splicing by overlap extension polymerase chain reaction (SOE-PCR) strategy (27,28). SOE-PCR substitutes a codon at the mutation site to other codons so that every possible amino acid substitution is generated. Combinatorial mutagenesis allowed construction of a pool of full-length gp17 mutants with all possible mutations at the targeted amino acid position. This was accomplished by two successive PCRs with four oligonucleotides. In the first PCR, two reactions were set up at the same time. The first reactions contained g17 forward primer and mutant reverse primer. The second reaction contained g17 reverse primer and mutant forward primer. The second round of PCR is the gene fusion step in which two PCR products from the first round of PCR were stitched together to reconstitute the full length of DNA by overlap extension. For example, to construct M195 mutant library, the following primers were used: primer 1 (g17 5′-forward): 5′-AAAGCTAGCATGGAACAACCGATTAATGTATTAAATG-3′; primer 2 (g17 mutant reverse): 5′-GTCTAAAACTTCCGCAGANNNTGAGC CTTTGTGTGCAAG-3′; primer 3 (g17 mutant forward): 5′-CTTGCACACAAAGGCTCANNNTCTGCGGAAGTTTTAGAC-3′; primer 4 (g17 3′-reverse): 5′-AAAGGATCCTTATACCATTGACATACCATGAGATAC-3′. Primer 1 and 4 are 5′ and 3′ ends of g17, and primers 2 and 3 are complementary primers corresponding to the regions that flank the M195 residue. To have a library of all possible mutations, primers 2 and 3 were synthesized by randomization of nucleotides at the mutant site.
The PCR-amplified DNA was digested with the restriction enzymes NheI and BamHI (Thermo Fisher) for directional cloning. The pET28-b vector was also linearized with the same restriction enzymes, purified from the agarose gel and ligated with the mutant DNAs. Ligation inserts the mutant g17 DNA in-frame with a 25 amino acid tag containing hexa-histidine sequence at the N-terminus of g17 coding sequence. The ligation mixture was transformed into the high efficiency DH5α E. coli competent cells (NEB) on LB-kanamycin plates. Individual transformants were grown on LB-kanamycin liquid medium and plasmid DNAs were extracted using the miniprep DNA purification kit (Qiagen). The plasmid DNAs were digested with NheI and BamHI to confirm the presence of g17 inserts.
Genetic recombination and marker rescue
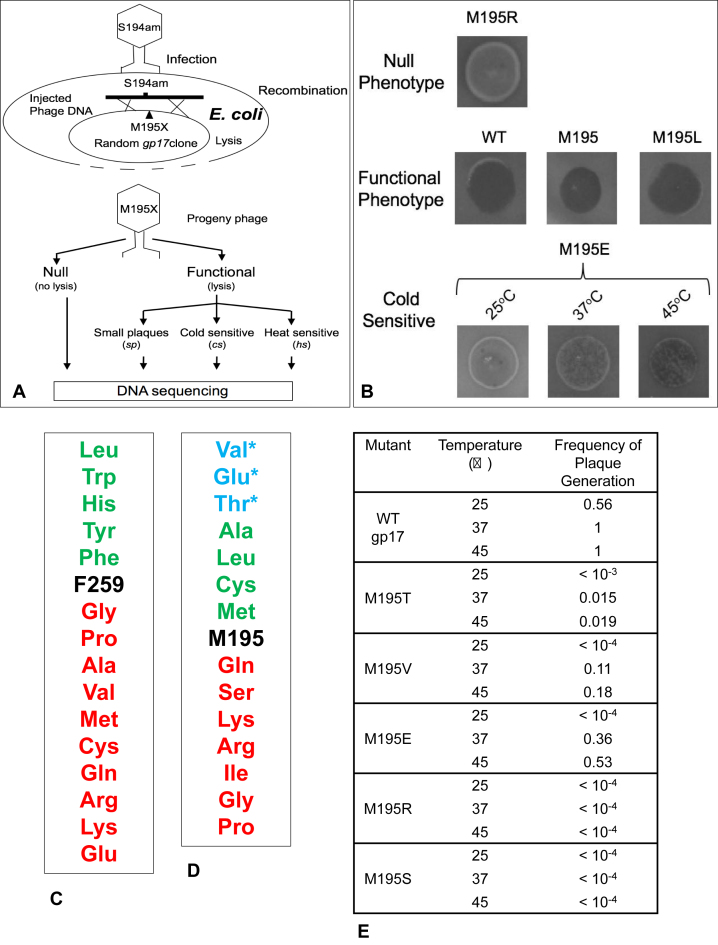
After confirming the presence of DNA with correct size, plasmid DNAs were transformed into E. coli BL21 (DE3) and spread on LB-kanamycin plates. Plates were incubated at 37°C overnight. Individual colonies were inoculated into 1 ml of LB-kanamycin medium and grown at 37°C overnight. To test the phenotype of each mutant, recombinational genetic rescue was performed using amber phages. An amber phage is a phage that contains amber stop codon at a particular amino acid position. A recombinational genetic rescue can exchange the genome between the mutant and the amber phage, so that the mutant sequence will now be part of the phage genome. We used two amber mutant phages, S194am and Y253am, which are very close to the mutational sites M195 and F259 respectively, so that genetic recombination between the amber phage and the mutant would almost always exchange the mutant into phage genome. The phenotype of the mutant phage is tested by its ability to lyse E. coli and produce a plaque. Top agar containing E. coli P301 (sup-) was plated on LB plate and 3 μl of each mutant E. coli clone was spotted. Then, 3 μl of amber phage [2.5 × 104 plaque forming units (pfu)/μl] was spotted on the top of the mutant spot and the plates were incubated overnight in 37°C, 25°C and 45°C. Four controls were also spotted on the same plates, WT gp17 clone, amber phage only, BL21 cells and a non-specific (gp37) clone. The presence of gp17 insert was confirmed by using another g17 amber phage, H436am, in which the amber mutation is far away from the M195 and F259 sites. All the g17 inserts regardless of the mutation at M195 or F259 would give positive marker rescue with this amber phage. See Results and Figure 3 for additional details.
Figure 3.
M195 and F259 residues are critical for phage T4 DNA packaging. (A) Schematic of combinatorial mutagenesis to determine the functional consequence of substitution with each amino acid. (B) Examples of phenotypes resulting from various substitutions followed by genetic rescue. (C) Phenotypes of amino acid substitutions at F259 and M195 residues. Substitutions below the native sequence resulted in null phenotype and those above resulted in functional phenotype. The asterisk ‘*’ represents sp/cs phenotype. (D) Frequency of plaque generation at different temperatures following genetic rescue of various amino acid substitutions at M195 residue. See Supplementary Materials and Methods for details.
Purification of gp17 proteins
The gp17 proteins were over-expressed and purified according to the protocols described previously (16). Briefly, the pET-28b plasmid constructs containing the inserts for gp17 (WT, E195 R195, S195, T195 and V195) were transformed into E. coli BL21 (DE3) pLys-S cells. Recombinant cells were grown to log phase at 37°C. isopropyl β-D-1-thiogalactopyranoside (IPTG) (1 mM) was added to induce over-expression of the recombinant protein and the culture was transferred to 29°C for 3 h. The cells were pelleted and resuspended in 40 ml of binding buffer A [20 mM Tris–HCl pH 8, 1 mM ATP, 5 mM Mg2+, 100 mM NaCl, 10 mM imidazole and protease inhibitor cocktail (Roche)]. The cells were lysed by French Press and the supernatant was applied to a Histrap column (AKTA-Prime, GE Healthcare). The hexa-his tagged gp17 was eluted using a linear gradient of 50–500 mM imidazole in buffer A and the peak fractions were applied to a 16 × 100 cm Superdex-200 size-exclusion column pre-equilibrated with buffer B (20 mM Tris–HCl pH 8, 30 mM NaCl). The peak fractions were concentrated and stored at −80°C.
In vivo nuclease assay
Recombinant E. coli BL21 (DE3) pLys-S cells containing cloned gp17 constructs were grown and gp17 expression was induced with 1 mM IPTG as above (29). Two ml aliquots were taken at 0 and 180 min after induction. Plasmid DNA was isolated by the alkaline lysis procedure (Qiagen) and resuspended in 50 μl water. A total of 10 μl of the DNA were electrophoresed on a 1% (w/v) agarose gel at 100 V for 2 h, stained with ethidium bromide and analyzed by Bio-Rad Chemi-DOC imaging system (30). Presence of a diffused smear of cleaved DNA throughout the lane is a hallmark of in vivo nuclease activity.
ATPase assay
The purified gp17 proteins (0.2–0.6 μM for WT, 1.6–2.0 μM for mutants) either alone or with equimolar amounts of gp16 oligomer were incubated in a reaction mixture (20 μl) containing (0.005–8 mM) cold ATP and 1 μCi of [γ-32P] ATP (Sp. Act. 3000 Ci mmol-1; Perkin Elmer) at 37°C in ATPase buffer (50 mM Tris–HCl pH 7.5, 0.1 M NaCl and 5 mM MgCl2) for 10 min. The reaction was terminated by adding ethylenediaminetetraacetic acid (EDTA) to 50 mM and the products were separated by thin layer chromatography on PEI plates (Sorbent Technologies). The 32Pi was quantified by Phosphor-imaging (Storm 820, Molecular dynamics) (16). Catalytic rates were extracted by fitting the data traces of 32Pi versus time to the expression A(t) = A∞(1 – exp(-kt)), and determining the initial slope (t = 0). This was done to mitigate effects due to limited substrate on the estimates of the catalytic rates.
In vitro DNA packaging
The purified gp17 proteins (0.5–2 μM) and empty heads (31) were incubated in a reaction mixture (20 μl) containing 400 ng XbaI-linearized pAAV-GFP DNA (5.4 kb), 30 mM Tris–HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2 and 1 mM ATP for 30 min at 37°C (13). The reaction was terminated by adding DNase I (Sigma) to a final concentration of 10 μg/ml and incubated for 30 min at 37°C. Proteinase K (Thermo Scientific), 50 mM EDTA and 0.2% sodium dodecyl sulphate (SDS) were added and the reaction mixture was incubated for 30 min at 60°C. The samples were then electrophoresed on 1% (w/v) agarose gel for 2 h at 100 V and stained with ethidium bromide. The amount of packaged DNA was quantified by Bio-Rad Chemi-DOC imaging system (13).
Single molecule optical tweezers assays
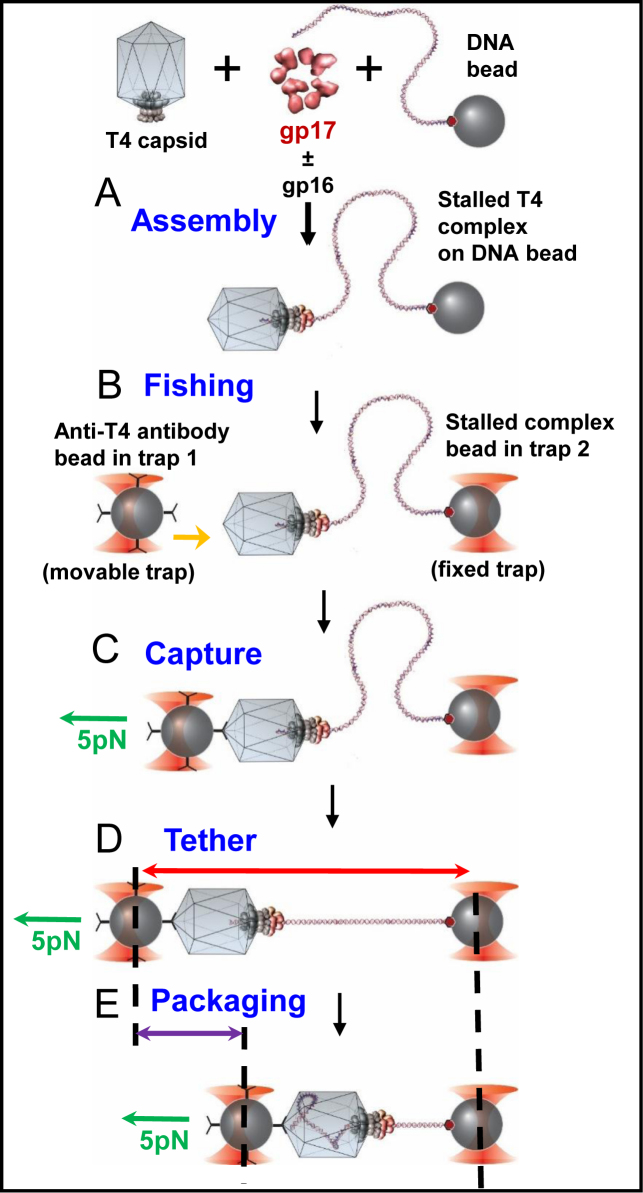
Single molecule assays were conducted on high resolution dual trap optical tweezers that were set up and calibrated as described previously (5) (see Figure 6 and ‘Results’ section for more details). DNA beads were prepared by incubating a 3.4-kb ds DNA that had a biotin tag on one end, along with 1 μm streptavidin beads (Spherotech), for 30 min at RT. Stalled complexes were assembled by incubating gp17 (2 μM) (WT, E195 or V195) with DNA beads and ∼1010 empty head particles in the presence of 0.25 mM ATP for 10 s, followed by 2 mM ATP-γ-S at room temperature for 30 min. In experiments where gp16 was added, the stalled complexes were assembled in the presence of an equimolar amount of gp16 N-C fusion (2 μM). No gp16 (or gp17 was present in any of the subsequent steps. Tethers were formed by ‘fishing’ (32) for the stalled capsid using Protein G beads (Spherotech) coated with anti-T4 antibodies in the presence of packaging buffer (30 mM Tris–HCl pH 7.6, 5 mM MgCl2, 80 mM NaCl), as well as an oxygen scavenging system (100 mg/ml glucose oxidase, 20 mg/ml catalase and 4 mg/ml glucose to prevent damage by the reactive singlet oxygen species) with 0.2 mM ATP-γ-S, and packaging was restarted by moving the entire tether into packaging buffer containing 1 mM ATP (33).
Figure 6.
Schematic of single-molecule DNA packaging using optical tweezers. DNA beads were prepared by incubating Streptavidin-coated polystyrene beads with 3.4-kb dsDNA that had biotin on one end. Stalled complexes were assembled by mixing DNA beads with T4 capsids and gp17 in the presence of 1 mM ATP for 10 s, followed by addition of 2 mM ATP-γS. Where needed, gp16 N-C fusion was added. Tethers were formed by moving the antibody bead close to the DNA bead for 1 s and then pulling apart until a force of 5 pN was felt, which indicated formation of a tether. The tether was held at 5 pN force and moved to the ATP channel where packaging (indicated by the shortening of the tether) was recorded against 5 pN of applied force.
RESULTS
Architecture of the catalytic center in ATPase motors
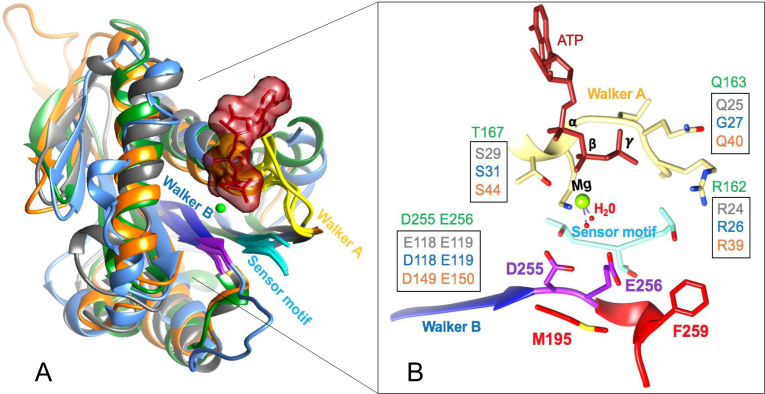
Active sites of enzymes provide unique microenvironments for interaction with substrate(s) and the chemistry that follows during catalysis. Structural analyses of gp17-ATPase, and other ATPase motors such as PcrA helicase and F1-ATPase (β-subunit) show that the ATP binding pocket consists predominantly of a hydrophobic environment, in particular the catalytic space surrounding the β and γ-phosphates of bound ATP. In the gp17 ATPase structure (11,34), ∼84% of the aa sidechains at the distance of 8Å or closer from the β and γ-phosphates of the bound ATP molecule are either hydrophobic or neutral. For PcrA helicase and F1-ATPase, the percentage is 78 and 92%, respectively (35,36).
Oriented in this hydrophobic environment are a cluster of charged and polar residues which interact with bound ATP, specifically the β- and γ-phosphates (Figure 2A). Among these are the highly conserved signature residues: lysine and serine/threonine of Walker A motif, aspartate of Walker B motif, catalytic glutamate and arginine finger (2,7–9,11,34) (Figure 2B) ((37). These residues create a network of interactions by coordinating with the Mg2+ ion and generally two to four water molecules. All other water molecules from bulk solvent are excluded from this space. One of the bound water molecules is referred to as ‘lytic’ water as it is polarized and split into H+ and OH− during catalysis. The catalytic glutamate acts as a base and accepts the proton whereas the OH− nucleophile ‘attacks’ the γ-phosphate resulting in ATP hydrolysis.
Figure 2.
The ATP binding pocket of T4 and other phage packaging ATPases. (A) Structural alignment of the packaging ATPases from phages T4 (green) (2o0H) (11), phi29 (cyan) (5HD9) (41), Sf6 (gray) (4IFE) (40) and P74–26 (orange) (4ZNL) (39). (B) Signature residues of Walker A, Walker B and sensor (Motif III) (37) motifs that are oriented into the ATPase catalytic center are shown. Note that the WT aa D255 and E256 (shown in purple) are modeled because the crystal structure was that of a mutant in which these two residues were switched. The residues M195 and F259 shown in red are predicted to contribute to the hydrophobicity of the microenvironment in the catalytic space.
This basic architecture is conserved in the ATPase catalytic centers, including the ATPase (large terminase) subunits of all phage packaging machines (Figure 2A and B) (11,38–41). Hydrophobicity in this catalytic space, especially that surrounding the catalytic glutamate and γ-phosphate is thought to affect the activation energy for activation and polarization of lytic water and in turn, the rate of ATP hydrolysis and speed of the motor (8). Therefore, altering the hydrophobicity, such as introduction of a charged or polar sidechain into this space, could alter the hydrogen bond network either by allowing a competing water molecule to enter this space, or by creating competition for catalytic glutamate by hydrogen bonding with the lytic water molecule. These would lead to an increase in activation energy for splitting the lytic water, decrease in the rate of ATP hydrolysis, and decrease in the speed of the motor.
Residues M195 and F259 located in the catalytic space are critical for motor function
In the crystal structure of gp17 ATPase–ATP complex, we found two hydrophobic amino acids, M195 and F259, with their sidechains oriented toward the catalytic carboxylate E256 at the distance of 5.3 and 6.4 Å, respectively (Figure 2B and Supplementary Figure S1A). These residues likely contribute to the optimal hydrophobicity needed to activate the lytic water molecule. If so, substitution with a less hydrophobic, polar or charged residue should lower the speed of the phage T4 DNA packaging motor. Consistent with this prediction, (i) both the M195 and F259 residues are strictly conserved in the T4 family large terminases (Supplementary Figure S2), and (ii) in the 8–10 times slower packaging motor from phage phi29 that packages ∼20-kb DNA, the corresponding residues at spatially equivalent positions are polar and charged (Supplementary Figure S1B) (19,41); a similar pattern was also observed in the packaging ATPase motor of phage Sf6 that packages ∼39-kb DNA (Supplementary Figure S3) (40), although the packaging velocity of this motor has not been determined.
The above hypothesis was tested by combinatorial mutagenesis (Figure 3A) (27). Two gp17 ATPase libraries were constructed by introducing every possible codon at each of these positions (see Supplementary Materials and Methods). The mutant plasmids were transformed into sup- E. coli and infected with either S194am (for M195 library) or Y253am (for F259 library) phages to determine which substitutions can rescue the amber mutation by genetic recombination. Rescue was performed at three temperatures (25°C, 37°C and 45°C) and the phenotypes were scored as null (no lysis of E. coli), functional (complete lysis), small plaque (sp) or cold sensitive (cs) (Figure 3B).
Of the 136 F259 mutants screened, 28% were found to be functional and 72% were null. Sequencing of about 60 mutant clones showed that, in addition to Phe, other aromatic and hydrophobic aa substitutions (Tyr, Tryp, His or Leu) were tolerated (Figure 3C). Examination of the plaque size showed no significant differences between the functional mutants and the WT (Phe substitution) at any of the temperatures tested.
On the other hand, analysis of the Met195 library revealed multiple phenotypes. Of the 138 mutants screened, 57% were functional and 43% were null. From about 80 mutants sequenced, we recovered a series of functional (Ala, Leu, Cys) and null (Gln, Ser, Lys, Arg, Ile, Gly or Pro) mutants. Interestingly, however, three substitutions, Val, Glu or Thr, resulted in partially functional cs/sp phenotype (Figure 3D).
Since partially functional phenotypes are good candidates to evaluate motor speed that could affect head filling, the efficiency of plaque generation by these mutants was quantified. Escherichia coli cells carrying the mutant plasmids were infected with S194am phage and the frequency at which the mutant gp17 protein (rescued by recombination) could generate an infectious virus particle was compared with that of the WT gp17. Consistent with the phenotype, the cs/sp mutant proteins showed greatly reduced number of plaques at 37°C when compared to the WT and the plaque number was further reduced, or abolished, at 25°C (Figure 3E). For instance, Thr substitution resulted in nearly two orders of magnitude fewer plaques at 37°C and more than three orders of magnitude fewer plaques at 25°C. The Glu and Val substitutions reduced plaque formation by 3- and 10-fold respectively at 37°C, and more than four orders of magnitude at 25°C. The null phenotypes, as expected, produced no plaques at any temperature. The above data demonstrate that the cs/sp M195 mutant motors produce fewer infectious virus particles than the WT gp17, which is further worsened at a lower temperature.
M195 mutants are defective in ATPase and DNA packaging
gp17 exhibits three activities in vitro that are related to motor function; ATPase activity that provides energy for DNA translocation, DNA packaging activity that includes packaging initiation and DNA translocation, and DNA cleavage activity that generates cuts in DNA at the time of packaging initiation and termination (headful nuclease) (10,16). To determine which of these functions are affected by mutation, we purified five mutant proteins (Figure 4A) and analyzed their biochemical activities. These include two null mutants, R195 and S195, and three cs/sp mutants, T195, V195 and E195.
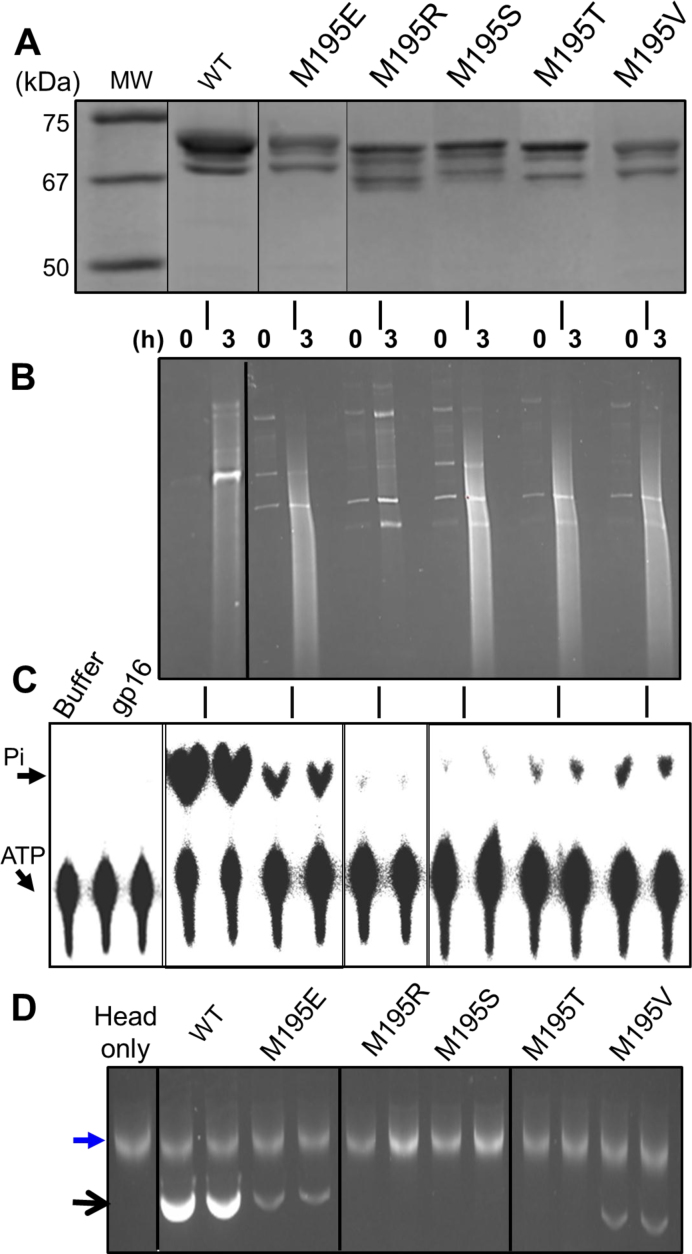
Figure 4.
Residues Met195 and Phe259 are critical for function. (A) SDS-polyacrylamide gel showing the purity of gp17 proteins. Note that the full-length protein was non-specifically cleaved at the C-terminus to produce 2–3 shorter bands. All the species retained DNA packaging activity. (B) in vivo nuclease activity assays. The numbers ‘0’ and ‘3’ at the top of the lanes represent hours after induction with IPTG. (C) ATPase assays. (D) in vitro DNA packaging assays. Upper arrow represents a small amount of DNA associated with the heads. Lower arrow represents the packaged DNA. The vertical lines between the panels A and D indicate the gp17 protein used in each assay, as shown at the top of panels A and D. The lanes corresponding to the ATPase and DNA packaging activities of WT or mutant gp17 lanes in panels C and D represent duplicate assays. The black lines within a panel indicate the boundaries of the sliced and pasted lanes. This was done to maintain the same order going from panel A to panel D. See Supplementary Materials and Methods for details.
Our data show that all the mutants retained similar levels of in vivo nuclease activity, as evident from the appearance of a cleaved DNA smear throughout the lane (30), when compared to the WT (Figure 4B). This result and the fact that several of these mutant motors can package full-length phage genome and produce infectious virus particles in vivo indicated that the mutations did not cause gross perturbation of gp17 structure. The mutant proteins, however, showed major differences in the ATPase and DNA packaging activities. The null mutants, the charged aa substitution R195 and the polar aa substitution S195, lost ∼90% of the ATPase activity (Figure 4C) and nearly all the DNA packaging activity (Figure 4D). The V195 and Glu195 mutants, consistent with their cs/sp phenotype, were partially functional, retaining ∼20–30% of the ATPase and DNA packaging activities. The T195 mutant, however, exhibited a more severe defect, losing ∼90% ATPase activity and nearly all the DNA packaging activity. This is consistent with the minute plaques produced by this mutant (Figure 3E).
E195 and V195 mutants show reduced steady state velocity of ATP hydrolysis
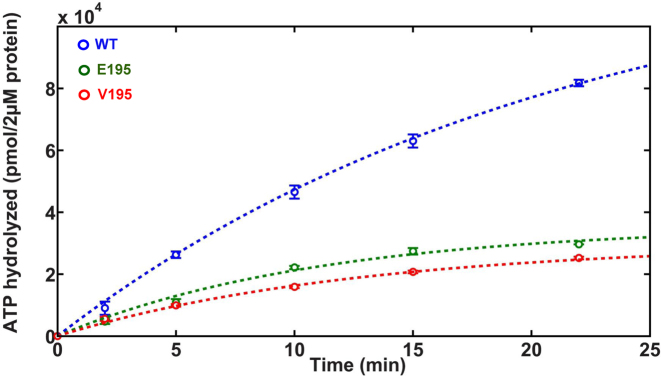
The steady state velocity of ATP hydrolysis was quantified for the E195 and V195 mutant proteins relative to WT gp17. The activity of T195 mutant was too low to perform detailed characterizations. The proteins were incubated with 1 mM ATP and the amount of Pi formed with time was quantified (Figure 5). The data clearly showed that the hydrolysis of ATP by the mutants is slower when compared to the WT gp17. Catalytic rates were extracted by fitting these data to the expression A(t) = A∞(1 – exp(-kt) and determining the initial slope (at t = 0). This was done to mitigate any effects such as the changing substrate concentration with time on the estimates of the catalytic rates. From the slopes, we determined that the E195 and V195 mutants showed 1.8- to 2.5-fold lower rate of ATP hydrolysis when compared to WT gp17.
Figure 5.
E195 and V195 mutants show reduced rate of ATP hydrolysis. The amount of Pi formed with time was quantified for WT (blue), E195 (green) and V195 (red) gp17s using the standard ATPase assay. Data were fit to the expression A(t) = A∞(1 – exp(-kt)) (dashed lines) and the rates determined from the slope at t = 0. See Supplementary Materials and Methods for details.
The DNA packaging velocity of E195 and V195 mutant motors is slower than the WT motor
Single-molecule experiments using high resolution optical tweezers (32) were performed to determine motor speed (Figure 6) (32). Unlike the bulk assays that give an average velocity of an ensemble of gp17 molecules, the single-molecule assay determines the velocity of the individual motors in real time.
DNA-coated beads were prepared by incubating 1-μm streptavidin-coated polystyrene beads with 3.4-kb dsDNA with biotin at one end. Packaging motor complexes were assembled by incubating the DNA beads with purified empty heads and gp17 proteins in the presence of 25 μM ATP for 10 s to initiate packaging. Packaging was then stalled by adding excess ATP-γS (2 mM) (Figure 6, (A)). A single bead containing DNA attached to one end was used to capture the phage head of the stalled complex at the other end by using a second polystyrene bead coated with T4 antibody (B). As a result, a single stalled packaging motor complex was captured between the two beads (C). The DNA tethered between the beads was then stretched by applying a force of 5 pN (D) and moved into an area of the microfluidic chamber containing ATP (1 mM) to re-start packaging (E). Motor speed was determined by measuring the decrease in tether length with time at a constant force of 5 pN (‘force-clamp’ mode) (20,32).
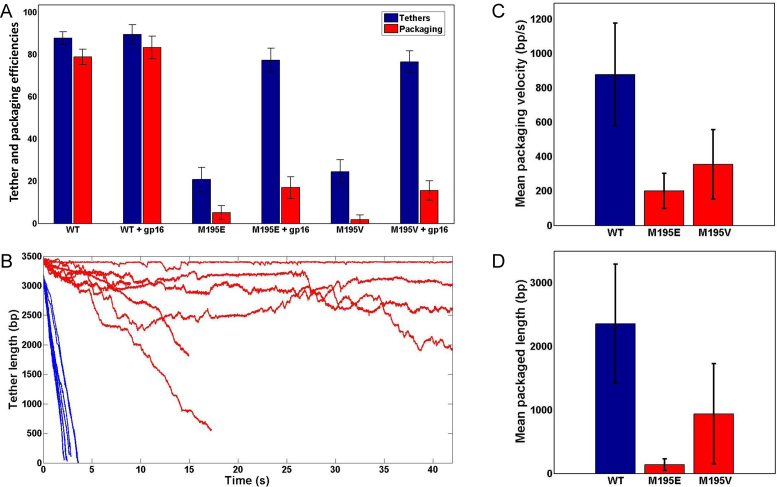
The data show that the mutant motors, unlike the WT motor, formed tethers poorly (Figure 7A). The tethering efficiency of WT motor was ∼85% whereas that of V195 and E195 motors was ∼20% (Figure 7A). We suspected that this was because the mutant motors could not efficiently initiate DNA packaging and form the stalled complex (see Figure 6A). Furthermore, <1% of the mutant motor tethers packaged DNA whereas >80% of WT motor tethers ended up packaging DNA (Figure 7A). Clearly, the mutant motors were defective in initiating DNA packaging. Our previous data suggested that packaging initiation is energetically costly because the motor ATPases must fire in rapid succession (17) in order to translocate the initial ∼30-bp of DNA deep into the 100-nm long motor/portal channel such that it will not come back out. Thus, the E195 and Val195 motors were unable to fire ATPases at a high rate.
Figure 7.
The speed of E195 and V195 mutant motors is slower than the WT motor. (A) Frequency of tether formation (blue) and packaging (red) of WT, E195 and V195 motors. (B) Representative traces showing packaging of WT (blue), E195 or V195 (red) motors (n = 17). Each line represents packaging by one individual packaging motor. (C) Mean packaging velocity of WT (blue) or mutant (red) motors. The velocity was calculated from the areas when the motor is engaged in packaging and by disregarding the pauses. Error bars are standard deviations. (D) Mean packaged length in bp of WT (blue) or mutant (red) motors. Error bars are standard deviations.
We found that the tethering efficiency of mutant motors could be enhanced by adding the small terminase protein gp16, the gp17-ATPase activator protein. It has been well established that gp16 stimulates the rate of ATP hydrolysis by gp17 ATPase up to ∼50-fold (16). This stimulation appears to be essential for efficient ackaging initiation in vivo (16), because initiation must follow immediately after cutting the concatemeric DNA by the holo-terminase (TerL–TerS) complex followed by assembly of the motor on the prohead portal (2). Otherwise, the newly generated end could slip out of the motor complex. However, since cutting is not required for in vitro DNA packaging, gp17 alone can attach to an already cut end and initiate packaging without the assistance of the ATPase activator protein gp16. Since the E195 and V195 mutants exhibited lower rates of ATP hydrolysis and were unable to form tethers efficiently, we hypothesized that this could be compensated by stimulating the ATPase activity of the mutant proteins with gp16.
By screening various gp16 mutants, we identified a mutant, gp16 N-C fusion, in which the central oligomerization domain was deleted and the N- and C-domains were fused, being the most effective in enhancing tether formation. In previous studies (15), the same mutant, like the WT gp16, also stimulated the gp17-ATPase activity. But unlike the WT gp16 which inhibited DNA packaging in bulk assays (42), the N-C fusion stimulated DNA packaging (15). Therefore, the gp16 N-C fusion mutant was used to stimulate packaging initiation so that the speed of the mutant motors could be measured while the DNA is being translocated. The above experiments were repeated by assembling the stalled complexes in the presence of both gp17 and gp16 N-C fusion (Figure 6A). The stalled complex was then captured by the DNA-bead and suspended in a buffer containing no gp16 and the DNA between the capsid-bead and DNA-bead was stretched to determine tethering efficiency (Figure 6B–D). Under these conditions, the tethering efficiency of the mutant motors was greatly increased, from ∼20 to ∼80%, whereas the already high ∼85% tethering efficiency of WT motor increased only marginally (Figure 7A). In fact, this is the first direct demonstration that gp16 (N-C fusion) increases the efficiency of packaging initiation.
The packaging speed, defined as the number of bp translocated per second, was determined after packaging was initiated as above. Significantly, only ∼25% of the mutant motor tethers re-initiated packaging whereas ∼90% of the WT motor tethers could re-initiate (Figure 7A). Of note is that gp16 is no longer available in the packaging buffer and it is unlikely that gp16 added earlier was still associated with the motor complex because the unbound material was washed off in the chamber. Furthermore, previous studies showed that the interaction between gp16 and gp17 is very weak and a gp16–gp17 complex could not be isolated (43). Of the motors that did initiate packaging (Figure 7B), the V195 and E195 mutant motors showed 2.5–4.5 times slower speed than the WT motor. In addition, while the WT motors packaged at a fairly uniform rate, the mutant motors exhibited frequent pausing where the motor temporarily stopped packaging. The E195 motor exhibited even more frequent pausing than the V195 motor. During pausing, the mutant motors allowed unpackaging, a previously observed behavior where the packaged DNA was slowly released under 5 pN force, probably due to the motor's weakened grip on the DNA (Figure 7B) (32). Comparing motor speeds only when the motor was engaged in packaging (by disregarding pauses), the WT motors packaged at an average rate of about 900 bp/s, whereas the E195 and V195 motors packaged at 200 bp/s and 375 bp/s, respectively (Figure 7C). The mean packaged length between two successive pauses was also reduced, from 2.3-kb for WT to 125-bp for the E195 motor and 900 bp for the V195 motor (Figure 7D).
DISCUSSION
The speed at which a motor performs its task is critical for optimal functioning of a metabolic process and ultimately for the survival of an organism. Our previous studies (18) indicated that the speed of different phage packaging motors may be evolutionarily optimized to accommodate the size of the genome the virus packages. Otherwise, a phage such as T4 that packages a large genome may not be able to effectively compete with another phage packaging much smaller genome if both phages infected the same cell. In addition, packaging sequesters the newly replicated genome inside the capsid, protecting it from degradation by nonspecific nucleases. Controlling motor speed would be even more complicated in cellular organisms where, often, multiple motors must work together for a desired outcome. However, there have been no attempts to address this critical point. Here, we addressed this question by formulating a defined hypothesis and directly testing it using a combination of structural, biochemical, genetic and single-molecule biophysical analyses. These led to a speed control mechanism that could be broadly applicable to numerous molecular motors that play critical roles in living organisms.
Our genetic and structural analyses revealed that the hydrophobic microenvironment in the catalytic space encompassing Walker B catalytic glutamate and γ-phosphate of ATP might be a critical determinant of motor speed. Bulk solvent is excluded from this space, except for, generally, two to four water molecules of which one is the lytic water (8,38). Polarization of lytic water molecule and acceptance of a proton by the glutamate base creates an OH− nucleophile that then attacks the γ-phosphate (9,25). This architecture provides a large window to modulate hydrophobicity by introducing combinations of sidechains into this space, thereby controlling the rate at which the nucleophile is generated. The rate of ATP hydrolysis and the speed of the motor could be manipulated in this way, which allows for selection of an optimally performing motor.
Our structural analyses identified two hydrophobic residues, M195 and F259, oriented at the distance of 5.3 and 6.4 Å respectively from catalytic glutamate, which we predicted would contribute to the hydrophobicity. These residues are strictly conserved in the T4 related large terminases and notably, in the slow-packaging phage phi29 motor, the equivalently positioned residues are polar and/or charged (19). Combinatorial mutagenesis of these residues showed that, at both the positions, hydrophobic substitutions were tolerated whereas polar or charged substitutions resulted in null or partially functional phenotypes.
The phenotypic pattern of M195 substitutions was particularly instructive. Indeed, the difference between Ser and Thr mutants was striking, considering that these amino acids are generally interchangeable without resulting in any phenotypic difference (2,27). The serine mutant with a polar hydroxyl group in the sidechain was null whereas the Thr mutant containing, in addition, a hydrophobic carbon was partially functional (cs/sp). This means that while introduction of a polar group into the microenvironment was detrimental, it could be compensated by slightly increasing the hydrophobicity. Similarly, a Glu substitution with two hydrophobic carbons in the sidechain was partially functional even though it has a terminal negative charge while an aspartic acid containing one less hydrophobic carbon was null. Our data further suggest that the hydrophobic sidechain must align or fit into the hydrophobic pocket appropriately. Leucine, having a linear sidechain, was functional whereas Val substitution with a branched sidechain was cs/sp and an Ile with the same branch but with a bulkier sidechain was null.
Biochemical and single-molecule analyses demonstrated that the primary functional defect in the mutants was reduction in the rate of ATP hydrolysis. In bulk assays, the E195 and V195 mutants showed 1.8- to 2.5-fold lower rate of ATP hydrolysis when compared to WT gp17. These mutants, in single molecule experiments, showed a remarkable drop in the frequency of DNA tethering to the packaging motor. As reported previously (20,32), tethering is directly related to packaging initiation and energetically costly. Rapid firing of the motor ATPases is essential to translocate the DNA end sufficiently into the capsid such that it would remain stably inserted in the capsid. The slow ATPase mutants apparently could not attain this high turnover rate necessary to achieve successful initiation. The 5 pN force applied to stretch the DNA probably imposed even more energy burden on the mutants to initiate packaging. These data are in agreement with the poor packaging efficiency of the mutants observed in bulk assays which also measures packaging initiation efficiency of an ensemble of molecules.
In experiments where the speed of the individual motors was directly measured, after the packaging was initiated, the mutant motors exhibited 2.5- to 4.5-fold lower DNA packaging velocity than the WT motors. Here, the packaging velocity was determined by excluding the pauses such that the measurements reflect the speed of the motor while it is actively translocating the DNA. This could mean that other functions of the motor such as the hinge movement, force generation and DNA movement that are coupled to ATP hydrolysis in the proposed electrostatic force mechanism (11,14) may adjust such that the DNA is translocated at a slower rate. Our data showed that the slow-packaging mutant motors frequently pause, often accompanied by unpackaging. This behavior is reminiscent of the pause-unpackaging state of the WT motor when ATP is limiting (32). Hence, the high frequency of pausing and unpackaging in the speed mutants might be because the mutant motor is more frequently unoccupied with the nucleotide when compared to the WT motor even when the ATP concentration was not limiting. As was observed in our previous studies (32), an unoccupied motor subunit pauses and also has a weaker grip on the DNA, allowing slow unpackaging under the influence of 5 pN force. Thus, it is possible that the speed mutations also had lowered the binding affinity for ATP. Alternatively, since the rate of OH− nucleophile generation is presumably slowed in the mutants, the off-rate of ATP increased leading to transient unoccupancy, resulting in pausing. Further analyses on the packaging dynamics of the motor need to be performed in order to resolve these questions.
In conclusion, our studies identified modulation of hydrophobicity in the predominantly hydrophobic nucleotide binding pocket as one potential mechanism by which the speed of a molecular motor can be controlled. The nucleotide binding pocket is one of the most conserved and ancient motifs (44) in living organisms. While the conserved residues provide structural and catalytic framework, and are rarely replaceable (2,27), the role of many non-conserved residues that are also part of the ATPase pocket remained unknown. Our results show that some of these non-conserved residues, specifically the hydrophobic residues located in the catalytic space, might fulfill important regulatory functions, such as optimization of motor speed by controlling the number of water molecules allowed into this space and adjusting their network of interactions. Selection of optimal speed variants that confer fitness would then drive the evolution of a virus or an organism. The phenotypic difference observed between T195 and S195 mutants is a good example of how subtle changes can affect survival. We propose that in icosahedral phages such a mechanism co-evolved with genome size allowing the selection of a fast DNA packaging motor for the large genome containing phage T4.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB-1411989 to V.B.R.]; National Institutes of Health [AI 081726 to V.B.R., GM118817 to Y.R.C., in part]; American Society for Microbiology Undergraduate Summer Research Fellowship (2015) (to S.L.). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hendrix R.W., Smith M.C., Burns R.N., Ford M.E., Hatfull G.F.. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rao V.B., Feiss M.. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2015; 2:351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rao V.B., Feiss M.. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008; 42:647–681. [DOI] [PubMed] [Google Scholar]
- 4. Casjens S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011; 9:647–657. [DOI] [PubMed] [Google Scholar]
- 5. Chemla Y.R., Smith D.E.. Single-molecule studies of viral DNA packaging. Adv. Exp. Med. Biol. 2012; 726:549–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer L.M., Leipe D.D., Koonin E.V., Aravind L.. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004; 146:11–31. [DOI] [PubMed] [Google Scholar]
- 7. Erzberger J.P., Berger J.M.. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006; 35:93–114. [DOI] [PubMed] [Google Scholar]
- 8. Afanasyeva A., Hirtreiter A., Schreiber A., Grohmann D., Pobegalov G., McKay A.R., Tsaneva I., Petukhov M., Kas E., Grigoriev M. et al. Lytic water dynamics reveal evolutionarily conserved mechanisms of ATP hydrolysis by TIP49 AAA+ ATPases. Structure. 2014; 22:549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 10. Black L.W., Rao V.B.. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 2012; 82:119–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun S., Kondabagil K., Draper B., Alam T.I., Bowman V.D., Zhang Z., Hegde S., Fokine A., Rossmann M.G., Rao V.B.. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008; 135:1251–1262. [DOI] [PubMed] [Google Scholar]
- 12. Sun L., Zhang X., Gao S., Rao P.A., Padilla-Sanchez V., Chen Z., Sun S., Xiang Y., Subramaniam S., Rao V.B. et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat. Commun. 2015; 6:7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kondabagil K.R., Zhang Z., Rao V.B.. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006; 363:786–799. [DOI] [PubMed] [Google Scholar]
- 14. Migliori A.D., Keller N., Alam T.I., Mahalingam M., Rao V.B., Arya G., Smith D.E.. Evidence for an electrostatic mechanism of force generation by the bacteriophage T4 DNA packaging motor. Nat. Commun. 2014; 5:4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun S., Gao S., Kondabagil K., Xiang Y., Rossmann M.G., Rao V.B.. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leffers G., Rao V.B.. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 2000; 275:37127–37136. [DOI] [PubMed] [Google Scholar]
- 17. Vafabakhsh R., Kondabagil K., Earnest T., Lee K.S., Zhang Z., Dai L., Dahmen K.A., Rao V.B., Ha T.. Single-molecule packaging initiation in real time by a viral DNA packaging machine from bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15096–15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuller D.N., Raymer D.M., Kottadiel V.I., Rao V.B., Smith D.E.. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16868–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith D.E., Tans S.J., Smith S.B., Grimes S., Anderson D.L., Bustamante C.. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001; 413:748–752. [DOI] [PubMed] [Google Scholar]
- 20. Fuller D.N., Raymer D.M., Rickgauer J.P., Robertson R.M., Catalano C.E., Anderson D.L., Grimes S., Smith D.E.. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007; 373:1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitchell M.S., Rao V.B.. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J. Biol. Chem. 2006; 281:518–527. [DOI] [PubMed] [Google Scholar]
- 22. Tsay J.M., Sippy J., DelToro D., Andrews B.T., Draper B., Rao V., Catalano C.E., Feiss M., Smith D.E.. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010; 285:24282–24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burton B.M., Marquis K.A., Sullivan N.L., Rapoport T.A., Rudner D.Z.. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007; 131:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kondabagil K., Dai L., Vafabakhsh R., Ha T., Draper B., Rao V.B.. Designing a nine cysteine-less DNA packaging motor from bacteriophage T4 reveals new insights into ATPase structure and function. Virology. 2014; 468-470:660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Story R.M., Steitz T.A.. Structure of the recA protein-ADP complex. Nature. 1992; 355:374–376. [DOI] [PubMed] [Google Scholar]
- 26. Yoshida M., Amano T.. A common topology of proteins catalyzing ATP-triggered reactions. FEBS Lett. 1995; 359:1–5. [DOI] [PubMed] [Google Scholar]
- 27. Rao V.B., Mitchell M.S.. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J. Mol. Biol. 2001; 314:401–411. [DOI] [PubMed] [Google Scholar]
- 28. Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R.. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989; 77:61–68. [DOI] [PubMed] [Google Scholar]
- 29. Studier F.W., Rosenberg A.H., Dunn J.J., Dubendorff J.W.. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990; 185:60–89. [DOI] [PubMed] [Google Scholar]
- 30. Bhattacharyya S.P., Rao V.B.. A novel terminase activity associated with the DNA packaging protein gp17 of bacteriophage T4. Virology. 1993; 196:34–44. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Z., Kottadiel V.I., Vafabakhsh R., Dai L., Chemla Y.R., Ha T., Rao V.B.. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011; 9:e1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kottadiel V.I., Rao V.B., Chemla Y.R.. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:20000–20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landry M.P., McCall P.M., Qi Z., Chemla Y.R.. Characterization of photoactivated singlet oxygen damage in single-molecule optical trap experiments. Biophys. J. 2009; 97:2128–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun S., Kondabagil K., Gentz P.M., Rossmann M.G., Rao V.B.. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007; 25:943–949. [DOI] [PubMed] [Google Scholar]
- 35. Soultanas P., Dillingham M.S., Velankar S.S., Wigley D.B.. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 1999; 290:137–148. [DOI] [PubMed] [Google Scholar]
- 36. Bowler M.W., Montgomery M.G., Leslie A.G., Walker J.E.. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 A resolution. J. Biol. Chem. 2007; 282:14238–14242. [DOI] [PubMed] [Google Scholar]
- 37. Mitchell M.S., Matsuzaki S., Imai S., Rao V.B.. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002; 30:4009–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parke C.L., Wojcik E.J., Kim S., Worthylake D.K.. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 2010; 285:5859–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hilbert B.J., Hayes J.A., Stone N.P., Duffy C.M., Sankaran B., Kelch B.A.. Structure and mechanism of the ATPase that powers viral genome packaging. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3792–E3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao H., Christensen T.E., Kamau Y.N., Tang L.. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8075–8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mao H., Saha M., Reyes-Aldrete E., Sherman M.B., Woodson M., Atz R., Grimes S., Jardine P.J., Morais M.C.. Structural and molecular basis for coordination in a viral DNA packaging motor. Cell Rep. 2016; 14:2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Zahrani A.S., Kondabagil K., Gao S., Kelly N., Ghosh-Kumar M., Rao V.B.. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J. Biol. Chem. 2009; 284:24490–24500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao S., Rao V.B.. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J. Biol. Chem. 2011; 286:3944–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walker J.E., Saraste M., Runswick M.J., Gay N.J.. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982; 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.