Abstract
Background and Aims Despite the recent wealth of studies targeted at contact zones of cytotypes in various species, some aspects of polyploid evolution are still poorly understood. This is especially the case for the frequency and success rate of spontaneous neopolyploidization or the temporal dynamics of ploidy coexistence, requiring massive ploidy screening and repeated observations, respectively. To fill this gap, an extensive study of spatio-temporal patterns of ploidy coexistence was initiated in the widespread annual weed Tripleurospermum inodorum (Asteraceae).
Methods DNA flow cytometry along with confirmatory chromosome counts was employed to assess ploidy levels of 11 018 adult individuals and 1263 ex situ germinated seedlings from 1209 Central European populations. The ploidy screening was conducted across three spatial scales and supplemented with observations of temporal development of 37 mixed-ploidy populations.
Key Results The contact zone between the diploid and tetraploid cytotypes has a diffuse, mosaic-like structure enabling common cytotype coexistence from the within-population to the landscape level. A marked difference in monoploid genome size between the two cytotypes enabled the easy distinction of neotetraploid mutants from long-established tetraploids. Neotetraploids were extremely rare (0·03 %) and occurred solitarily. Altogether five ploidy levels (2x–6x) and several aneuploids were discovered; the diversity in nuclear DNA content was highest in early ontogenetic stages (seedlings) and among individuals from mixed-ploidy populations. In spite of profound temporal oscillations in cytotype frequencies in mixed-ploidy populations, both diploids and tetraploids usually persisted up to the last census.
Conclusions Diploids and tetraploids commonly coexist at all spatial scales and exhibit considerable temporal stability in local ploidy mixtures. Mixed-ploidy populations containing fertile triploid hybrids probaby act as effective generators of cytogenetic novelty and may facilitate inter-ploidy gene flow. Neopolyploid mutants were incapable of local establishment.
Keywords: aneuploidy, annual plant, cytotype coexistence, flow cytometry, Matricaria perforata, mixed-ploidy population, neopolyploid, ploidy screening, temporal dynamics, Tripleurospermum inodorum, triploid
INTRODUCTION
Polyploidy (whole-genome duplication) is widely considered one of the most important evolutionary forces driving the diversification of flowering plants (Soltis et al., 2009; Weiss-Schneeweiss et al., 2013). It triggers both genotypic and phenotypic novelty (Levin, 1983; Flagel and Wendel, 2009; Hegarty et al., 2013) and serves as an ‘instant’ mechanism of sympatric speciation (Coyne and Orr, 2004; Wood et al., 2009). Many plant species retain individuals of two or more different ploidy levels in certain parts of their distributional range (e.g. Kliber and Eckert, 2005; Castro et al., 2012; Godsoe et al., 2013; Kolář et al., 2013) or even within their populations (e.g. Kao, 2007; Duchoslav et al., 2010; Sonnleitner et al., 2010; Trávníček et al., 2011). Local coexistence of different cytotypes may arise either from an in-situ polyploidization event, resulting in the sympatric occurrence of genetically close di- and polyploids (i.e. primary contact) or through cytotype immigration into a population of another ploidy (i.e. secondary contact; Petit et al., 1999). Mixed-ploidy populations serve as a convenient microcosm for the study of factors affecting the origin and establishment of polyploid derivatives (Husband, 2004; Hanzl et al., 2014). In addition, they offer an opportunity to investigate the evolution of already established cytotypes that may be further subjected to inter-ploidy gene flow and transfer of adaptations (Chapman and Abbott, 2010; Arnold et al., 2015; Hülber et al., 2015).
According to theoretical predictions, the coexistence of two or more cytotypes within a population is unstable, representing only a transitory state before one of the cytotypes is locally fixed (Levin, 1975; Fowler and Levin, 1984). This stems from the fact that inter-ploidy mating results in progeny with reduced vitality, fertility or both (Ramsey and Schemske, 1998), making cytotype fitness dependent on frequency. The less abundant cytotype suffers more from the shortage of appropriate mates and is ultimately eliminated from the population (i.e. minority cytotype exclusion principle; Levin, 1975). Such a frequency-dependent mating disadvantage has been repeatedly observed both in experimental and in natural mixed-ploidy populations (Hagberg and Ellerström, 1959; Husband, 2000; Baack, 2005a; Mráz et al., 2012). However, there are several mechanisms that may compensate for the minority cytotype disadvantage (Rodríguez, 1996; Husband, 2004; Li et al., 2004; Rausch and Morgan, 2005; Oswald and Nuismer, 2011). Firstly, it is only the outcome of sexual reproduction that is affected, while many species also rely on other modes of reproduction, such as vegetative propagation (Hroudová and Zákravský, 1993; Castro et al., 2007) and apomixis (Kao, 2007; Mráz et al., 2008). Secondly, the likelihood of a minority cytotype receiving compatible pollen increases through assortative (non-random) mating (Husband and Sabara, 2003). This can be achieved by shifts in flowering phenology of the cytotypes (Lumaret et al., 1987; Petit et al., 1997), different pollinator preferences (Segraves and Thompson, 1999; Kennedy et al., 2006) or an increased rate of autogamy (Barringer, 2007). Assortative mating may also be promoted by spatial clustering of cytotypes (Baack, 2005b) as a result of ploidy-specific (micro-)habitat preferences (Lumaret et al., 1987; Ramsey, 2011), human-mediated introduction of one cytotype into populations of another (Meirmans et al., 1999; Mráz et al., 2012; Otisková et al., 2014) or a limited dispersal capacity (Baack, 2005b; Hanzl et al., 2014). The persistence of a minority cytotype in mixed-ploidy populations can also be enhanced by its recurrent origins (Peckert and Chrtek, 2006; Ramsey, 2007), repeated immigration (Levin, 1975), superior competitive ability (Keeler and Davis, 1999) or better resistance to herbivore and parasite attacks (Nuismer and Thompson, 2001; Arvanitis et al., 2008). Temporal oscillations in population size (resulting, for instance, from periodic disturbances) may also disrupt frequency-dependent selection and prevent the local fixation of one of the cytotypes (Rausch and Morgan, 2005; Halverson et al., 2008).
Recent years have seen a considerable number of studies describing patterns in contact zones between different cytotypes of various species, examining processes allowing cytotype coexistence and improving our knowledge of the mechanisms of polyploid establishment (Husband and Sabara, 2003; Weiss-Schneeweiss et al., 2013). However, we still know very little about the temporal aspect of ploidy coexistence, for which repeated observations are required, such as the fate of mixed-ploidy populations and the spatio-temporal dynamics of contact zones. The main challenge stems from the fact that the vast majority of heteroploid species subjected to detailed investigations are (long-lived) perennials (e.g. Lumaret et al., 1987; Petit et al., 1997; Keeler and Davis, 1999; Baack, 2005a; Kao, 2007; but see Buggs and Pannell, 2007; Manzaneda et al., 2012), which precludes the assessment of their evolutionary dynamics in a reasonable time frame. Further understudied topics of polyploid evolution are rare evolutionary events such as the frequency of neopolyploid origins and the rate of successful polyploid establishment demanding massive ploidy screening both within and among populations (Halverson et al., 2008). The relative contribution of recurrent polyploid origins to other processes stabilizing cytotype coexistence in situ is yet to be adequately addressed.
To fill these gaps, we focused on the spatio-temporal dynamics of ploidy coexistence in the common annual weed Triplerospermum inodorum (Asteraceae). The species includes two morphologically indistinguishable cytotypes (Kay, 1969). Whereas diploids predominate in the western part of Europe, tetraploids are more common in its eastern parts, with a diffuse contact zone extending from France to Poland (Rottgardt, 1956; Kay, 1969; Lankosz-Mróz, 1976). Our pilot surveys revealed frequent coexistence of the two cytotypes in the Czech Republic, which provides a unique opportunity to study the diversity and distribution of cytotypes across several spatial scales – from within-population to landscape level. Using massive flow-cytometric screening across multiple life stages, spatial scales and years, we addressed the following questions: (1) What are the cytotype distribution patterns at various spatial scales and how common is ploidy coexistence? (2) Is there any consistent trend in the temporal development of local ploidy mixtures? (3) Is cytotype diversity at the seedling stage comparable with that in adult plants? (4) Given the differences in monoploid genome size between di- and tetraploids (see Results), what is the frequency of spontaneous polyploid mutants in natural populations?
MATERIALS AND METHODS
Study species
Scentless mayweed [Tripleurospermum inodorum (L.) Sch. Bip., syn. Matricaria perforata Mérat, Asteraceae] is a common and widely distributed weed of arable land and other human-disturbed sites. Its native range covers most of Europe and Western Asia and it has been introduced to North America and some other temperate regions (Kay, 1976, 1994; Woo et al., 1991).
Tripleurospermum inodorum is an annual herb with finely dissected tripinnate leaves and 0·2- to 0·7-m-tall flowering stems terminated by one to a few hundred capitula (Kubát, 2004). The species is insect-pollinated and usually self-incompatible (Kay, 1965; Woo et al., 1991); however, self-pollination in some populations has been reported (Kay, 1969). A typical plant produces thousands of achenes that lack any obvious adaptations for long-distance dispersal. Sexual reproduction is the only means of propagation, as the scentless mayweed is incapable of vegetative reproduction (Kay, 1994). Its achenes can survive for at least 10 years buried in soil (Kay, 1994), which suggests that it develops a permanent soil seed bank (Bowes et al., 1995). Germination and the formation of leaf rosettes takes place both in autumn and in spring, resulting in a winter annual, spring annual or, less frequently, short-lived perennial life cycle (Woo et al., 1991; Kubát, 2004).
Two morphologically indistinguishable cytotypes, diploid (2n = 18) and tetraploid (2n = 36), have been reported from both the native and the introduced ranges (Kay, 1969; Woo et al., 1991). Diploids predominate in western (i.e. oceanic) parts of Europe whereas tetraploids prevail in more continental, eastern parts of the continent (Kay, 1969). A diffuse contact zone of the two cytotypes extends from north-eastern France through Germany to Poland, but no mixed-ploidy populations have been reported (Rottgardt, 1956; Kay, 1969; Lankosz-Mróz, 1976). Interploidy breeding barriers seem to be weak, as artificial crosses of di- and tetraploids resulted in triploid seeds (Kay, 1965). An autopolyploid origin has been suggested for the tetraploid cytotype (Arora and Madhusoodanan, 1981).
Landscape, regional and local ploidy screening
Ploidy screening at the landscape level was conducted in Central Europe between 2011 and 2015, mainly focusing on the Czech Republic, where pilot data confirmed the occurrence of both diploids and tetraploids. The sampling was designed to cover regularly the study area and include various habitats. For each population, we recorded its position using a handheld GPS unit, briefly described the habitat and collected fresh leaves from at least ten randomly chosen individuals (whenever possible). Collected leaves were placed into plastic zip-lock bags and kept refrigerated until flow-cytometric analysis. Due to close resemblance between T. inodorum and Matricaria spp. at the stage of leaf rosettes, we attempted to sample unequivocally identifiable flowering individuals. If leaf rosettes were collected (<3 % of populations), only samples that could be reliably identified based on their genome size were included.
To capture the structure of contact zones at a regional level, a detailed survey was carried out in three areas in the Czech Republic with common co-occurrence of di- and tetraploids: (1) in the surroundings of the town of Mariánské Lázně (MAR; 30 × 30 km; coordinates of the centroid 49°58′28″N, 12°45′29″E), (2) in the surroundings of the town of Rakovník (RAK; 28 × 31 km; 50°09′59″N, 13°43′15″E) and (3) east of the town of Soběslav (SOB; 21 × 23 km; 49°16′59″N, 14°52′13″E). The sampling strategy generally followed that of the landscape ploidy screening, but the sampling was much denser, and considerable effort was made to representatively cover all major habitat types.
The fine-scale distribution of di-, tetraploids and minority cytotypes was recorded in five mixed-ploidy populations (for more details, see Table 1). The size of each plot was adjusted to include >50 plants. The positions of all T. inodorum individuals were recorded, and one leaf per plant was collected for ploidy estimation.
Temporal dynamics of ploidy mixtures
We used two different approaches to describe temporal changes in ploidy composition of mixed-ploidy sites. At one of the fine-scale study plots (population no. 22), established in 2011, the distribution of cytotypes was recorded during the four consecutive years. Unfortunately, this approach for various reasons (low-abundance populations, sites on arable land unsuitable for the establishment of permanent plots, etc.) could not be applied to other mixed-ploidy sites.
As an alternative, we selected 36 mixed-ploidy populations discovered during the regional screening in 2011–2014 and resampled them in 2015. We only focused on populations where it was possible to repeat the sampling at exactly the same site and within approximately the same spatial bounds. Whenever possible, >20 plants per population were cytotyped in 2015.
Cytotype diversity of offspring
The ploidy screening of adult plants was complemented by analysis of ex-situ germinated seedlings. This approach enabled the capture of cytotype diversity at early ontogenetic stages of T. inodorum and thus to search for minority cytotypes with lower viability. Achenes were collected in one diploid, one tetraploid and five mixed-ploidy populations during autumn 2013. In each population, we pooled open-pollinated flower heads from >50 randomly chosen individuals, except for the two least abundant ones. The flower heads were stored in paper bags at room temperature until April 2014, when a few thousand achenes of each source population were sown in seedling trays filled with common garden soil. After 3 weeks, the ploidy level of at least 60 randomly selected seedlings per population was analysed, and only ploidy-variable populations were subjected to further sampling.
Due to extensive variability in DNA content of the offspring analysed, resulting in more-or-less continuous variation, we set arbitrary boundaries between the particular euploid cytotypes and aneuploids for a better overview of the results (Supplementary Data Fig. S1). Sixteen plants with known chromosome numbers were used as reference points. Aneuploid offspring was divided into three groups according to the nearest euploid ploidy levels: 2x–3x, 3x–4x and 4x–5x.
Flow cytometry
Relative genome size of T. inodorum individuals was inferred from fluorescence intensities of DAPI-stained nuclei using flow cytometry (FCM). Sample preparation generally followed the two-step procedure using Otto buffers (Doležel et al., 2007). Briefly, approx. 0·5 cm2 of fresh sample leaf tissue and an appropriate amount of internal reference standard Bellis perennis L. (2C = 3·38 pg DNA; Schönswetter et al., 2007) were chopped together in 0·5 mL of ice-cold extraction buffer (0·1 m citric acid, 0·5 % Tween 20). The resulting suspension was filtered through a 42-μm nylon mesh, incubated for approx. 5 min. and then mixed with 1 mL of a staining solution (0·4 m Na2HPO4.12H2O, 4 μg DAPI, 2 μL β-mercaptoethanol). The fluorescence intensity of 3000 particles was analysed using a Partec PA II flow cytometer (Partec GmbH, Münster, Germany) equipped with a UV LED chip. Up to ten individuals were pooled together during the ploidy screening, as our pilot analyses proved that minority cytotypes can be reliably detected even at low frequencies (<10 %). Each plant was analysed separately when mixed-ploidy samples were encountered or if the quality of the resulting histograms was insufficient (i.e. coefficient of variation of the G0/G1 peak of the sample >4 %). The interpretation of FCM results was based on chromosome counts obtained from reference plants in cultivation.
Chromosome counts
A subset of seedlings from mixed-ploidy populations representing the main categories of nuclear DNA content were transplanted into pots and cultivated in a greenhouse. Chromosomes were counted in somatic mitoses in root tips. Root tips were sampled in the morning, pretreated either with a saturated water solution of α-bromonaphtalene or with 0·002 m 8-hydroxyquinoline for 3·5–4 h at room temperature, rinsed with water, fixed overnight in a cold mixture of ethanol and acetic acid (3 : 1) and stored in 70 % ethanol at 4 °C until required. The root tips were then hydrolysed in 1 m HCl at 60 °C for 7 min, rinsed with water, and their meristematic tissue was squashed in a drop of lacto-propionic orceine (Dyer, 1963). Chromosomes were counted in at least five metaphases per plant using an Olympus BX-51 microscope (total magnification ×1000).
Data analysis
Differences in monoploid genome size (i.e. 1Cx values; Suda et al., 2006) between diploids and tetraploids were compared using an analysis of variance (ANOVA) of 50 randomly selected flow-cytometric estimates for each of the cytotypes. Only single-individual analyses where the coefficient of variation of the T. inodorum sample was <3 % were included.
Applied to the dataset of 36 mixed-ploidy populations revisited after 1–4 years, a chi-square test was used to compare the numbers of populations turning uniformly diploid and uniformly tetraploid. The Kruskal–Wallis rank sum test was used to test whether the time between the two sequential visits had a significant influence on a population becoming ploidy-uniform or remaining mixed. An effect of habitat type on the fate of ploidy mixtures was tested using Fisher’s exact test. Fisher’s exact tests were also used for comparisons of cytotype composition (abundance of 2x, 3x and 4x individuals) at each site between the two visits, and by analogy, to evaluate between-year changes in cytotype composition of the mixed-ploidy population no. 22.
Spatial segregation of diploids and tetraploids within mixed-ploidy populations was assessed using the Mantel test (Mantel, 1967) implemented in the R package ‘ade4’ (Dray and Dufour, 2007). A pairwise distance matrix derived from the position of individuals in each study plot was compared with a pairwise binary matrix coding cytotype identity. Triploids were omitted from spatial analyses due to their low abundance. Significance levels were estimated using a permutation test with 9999 replicates.
RESULTS
Cytotype diversity and the origin of new tetraploid mutants
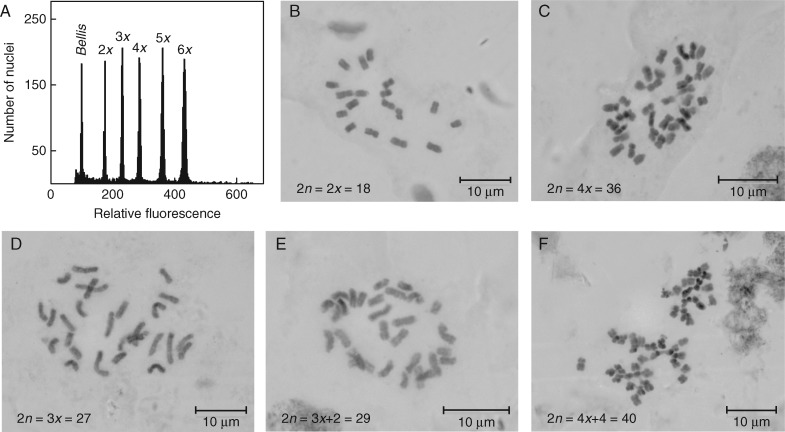
Flow-cytometric analyses of 11 018 individuals from 1209 populations of T. inodorum yielded five distinct euploid cytotypes, ranging from the diploid to the hexaploid level (Fig. 1A). All ploidy levels with the exception of pentaploids were corroborated by chromosome counts: diploids (2n = 2x = 18; Fig. 1B), triploids (2n = 3x = 27; Fig. 1D), tetraploids (2n = 4x = 36; Fig. 1C) and hexaploids (2n = 6x = 54). Apart from diploids and tetraploids, triploids were the most common cytotype, occurring at a mean rate of 7·7 % (s.e. = 0·95 %) in mixed-ploidy populations. The other two minority cytotypes were extremely rare in natural populations, as only one pentaploid and one hexaploid were found (each constituting 0·01 % of sampled individuals). The pentaploid grew in mixed-ploidy population no. 510 along with diploids and tetraploids, while the hexaploid occurred in the otherwise tetraploid population no. 1057 (Supplementary Data Table S1).
Fig. 1.
Flow-cytometric (FCM) estimates of relative genome size and confirmatory karyological analyses of Tripleurospermum inodorum. (A) Simultaneous FCM analysis of five individuals representing the five euploid cytotypes (2x–6x) distinguished during ploidy screening, with Bellis perennis as the internal standard. (B–D) Photomicrographs of metaphase chromosomes of individuals belonging to the three most common ploidy levels: diploid (2n = 18; B), triploid (2n = 27; D) and tetraploid (2n = 36; C). (E,F) Metaphase chromosomes of two individuals identified as putative aneuploids in FCM analyses, and later confirmed as being hypertriploid (2n = 29; E) and hypertetraploid (2n = 40; F), respectively.
Interestingly, the relative genome size of tetraploids (2·90 ± 0·004 s.e., mean 1Cx = 0·726) did not equal double the diploid value (1·72 ± 0·008 s.e., mean 1Cx = 0·862; F1,98 = 2,242, P ≪ 0·001). The monoploid genome size of tetraploids was on average approx. 19 % lower than that of diploids; the consistency of these differences was verified on selected individuals using propidium iodide FCM (data not shown). This lower monoploid genome size of established tetraploids enabled the easy detection of new polyploid mutants (i.e. neotetraploids) in routine flow-cytometric analyses (Fig. 2). Only three individuals with nuclear DNA content corresponding to neotetraploids (relative genome sizes of 3·26, 3·42 and 3·58) were identified in natural populations, constituting approx. 0·03 % of the samples analysed. These individuals were found singly in mixed-ploidy populations nos. 1052 and 1182, while the collection at site no. 187 consisted of one presumably neotetraploid plant from an unpaved field road, without any other T. inodorum individual in its proximity (Table S1).
Fig. 2.
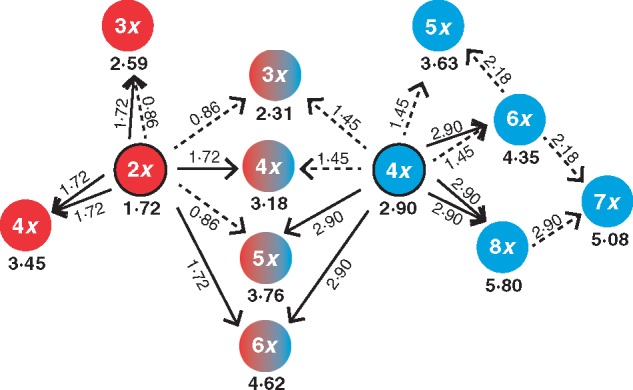
A theoretical overview of crosses involving reduced (dashed arrows) and unreduced (solid arrows) gametes of diploid (red with black stroke) and tetraploid (blue with black stroke) parents and cytotypes of the resulting offspring. Numbers refer to relative genome sizes of particular gametes (regular font) and cytotypes (bold font). Notice that some cytotypes have more than one way of origin and these can be easily distinguished based on relative genome size in Tripleurospermum inodorum. For simplicity, other possible crosses involving derived cytotypes are not shown.
The relative genome size of triploids (2·28 ± 0·003 s.e.), intermediate between diploids and established tetraploids, is in accordance with their hybrid origin. Theoretically, the expected genome size of such hybrids is 2·31, contrary to the value of 2·59 expected for triploid mutants originating from a fusion of reduced and unreduced gametes of diploids (Fig. 2). On the other hand, the relative genome size of the single pentaploid individual (3·75) suggests its origin from a fusion of an unreduced gamete of an established tetraploid with a reduced gamete of a diploid (expected genome size = 3·76) rather than from hybridization between established tetraploid and hexaploid parents (expected genome size = 3·63). Aside from euploid cytotypes, five presumably aneuploid individuals were detected during our flow-cytometric ploidy screening, occurring singly in mixed-ploidy populations (nos. 189, 780 and 1094) or in otherwise uniformly tetraploid populations (nos. 373 and 381; Table S1). The nuclear DNA content of three of these putative aneuploids exceeded the tetraploid level whereas two other individuals were intermediary between diploids and triploids, and between triploids and tetraploids, respectively.
Spatial patterns of diploid–tetraploid coexistence
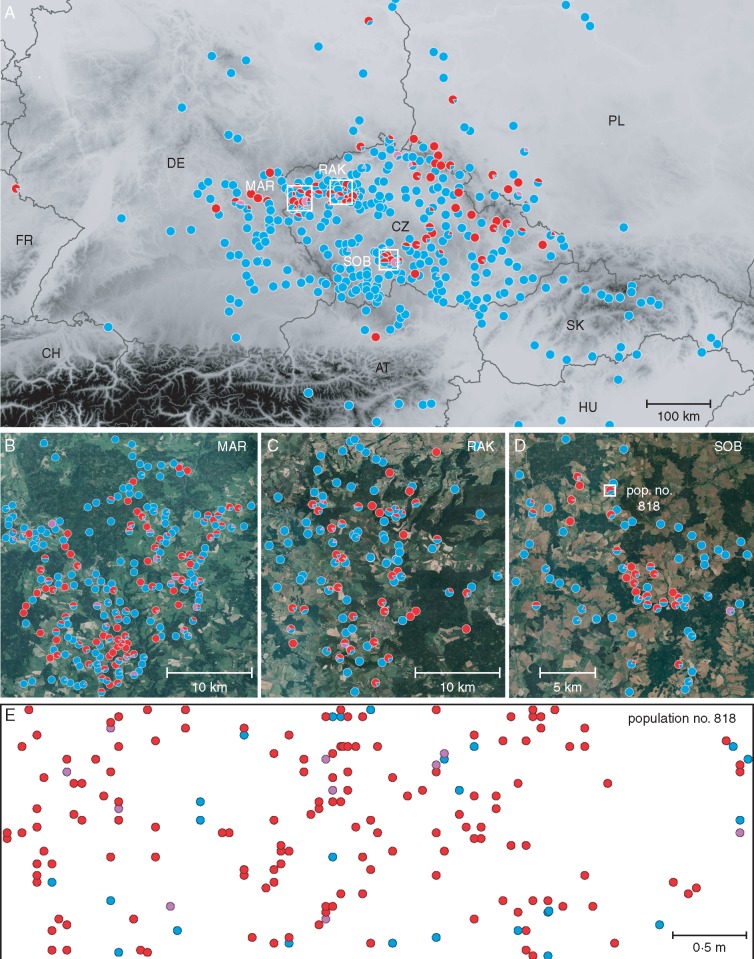
A total of 1209 populations were screened for cytotype composition; most of them were purely tetraploid (823; 68 %), mixed-ploidy populations were less frequent (271; 22 %) and pure diploid populations were the rarest (115; 10 %). While the predominant tetraploids occur in most of the sampled regions, diploids are much more scattered and presumably absent from some areas (e.g. the south-east part of Central Europe and several regions within the Czech Republic; Fig. 3A). The contact zone of the two cytotypes has a diffuse, mosaic-like character.
Fig. 3.
Structure of the contact zone between cytotypes of Tripleurospermum inodorum illustrating the coexistence of diploids (red), tetraploids (blue) and their triploid hybrids (violet) across three different spatial scales. Mixed-ploidy populations in A–D are presented as pie charts. (A) Cytotype distribution patterns in Central European populations (11 018 analysed individuals). (B–D) Three regions subjected to more detailed ploidy screening. (E) Small-scale distribution of di-, tri- and tetraploid individuals in one of the mixed-ploidy populations studied.
More intensive ploidy screening was targeted at three regions within the Czech Republic (Fig. 3B–D). In western Bohemia (MAR; Fig. 3B), 358 populations were investigated, of which 184 (51·4 %) were tetraploid, 120 (33·5 %) mixed-ploidy and 54 (15·1 %) diploid. A further 157 populations were screened in north-west Bohemia (RAK; Fig. 3C), with 85 (54·1 %) tetraploid, 52 (33·1 %) mixed-ploidy and 20 (12·7 %) diploid populations. Finally, 95 populations were visited in southern Bohemia (SOB; Fig. 3D), consisting of 46 (48·4 %) tetraploid, 41 (43·2 %) mixed-ploidy and only eight (8·4 %) diploid populations. The cytotype distribution patterns are similar among all three regions: tetraploid populations were the most common, and diploids were more frequently (2–5 times) found in mixed-ploidy populations than in pure diploid ones.
At the most detailed (intra-population) scale, the relative abundance of diploids and tetraploids varied across the sampled populations. Three were diploid-dominated whereas two exhibited similar frequencies of the two major cytotypes (Table 1). Triploid hybrids were discovered in four of these populations, in proportions ranging from 2·3 to 14·6 %. Diploid and tetraploid individuals were more or less randomly scattered within the study plots (Figs 3E and 4; Supplementary Data Fig. S2). Results of the Mantel test, however, suggested a slightly positive segregation of cytotypes in three of the five mixed-ploidy populations (Table 1).
Table 1.
Abundance and relative proportions of cytotypes and spatial segregation of diploids and tetraploids in five mixed-ploidy populations studied in detail
| Population* | Cytotype composition of population [count (ratio)] |
Spatial segregation† |
|||
|---|---|---|---|---|---|
| 2x | 3x | 4x | rM | P | |
| 818 | 144 (80·4 %) | 10 (5·6 %) | 25 (14·0 %) | 0·0568 | 0·060 |
| 22 | 60 (46·2 %) | 3 (2·3 %) | 67 (51·5 %) | 0·0907 | <0·001 |
| 150 | 58 (75·3 %) | 3 (3·9 %) | 16 (20·8 %) | −0·0293 | 0·803 |
| 916 | 23 (56·1 %) | 6 (14·6 %) | 12 (29·3 %) | 0·1289 | 0·028 |
| 889 | 40 (44·9 %) | 0 (0·0 %) | 49 (55·1 %) | 0·0479 | 0·014 |
Population codes correspond to sequential numbers in the complete list of populations under study (Table S1).
Mantel correlation coefficient (rM) and its significance level (P) showing spatial association of georeferenced diploid and tetraploid individuals (low-abundance triploids were omitted).
Temporal changes in ploidy composition
Of the 36 mixed-ploidy populations in which temporal development of the cytotype composition was assessed after 1–4 years, 26 populations remained mixed-ploidy, four turned into purely diploid and six became uniformly tetraploid. The two cytotypes did not differ in the probability of them prevailing in the initially mixed-ploidy population (χ2 = 0·4, d.f. = 1, P = 0·527), and the fate of ploidy mixtures did not depend on the time between the two sequential visits (Kruskal–Wallis test, χ2 = 0·209, d.f. = 1, P = 0·648) or the type of habitat (Fisher’s exact test, P = 0·331). However, significant temporal changes in ploidy composition were recorded only in ten of the 36 populations, four of them turned to be purely tetraploid and six remained mixed-ploidy (Table 2).
Table 2.
Temporal shifts in ploidy composition at 36 initially mixed-ploidy sites sampled during our 2011–2014 field campaign and re-collected in 2015
| Population* | Initial ploidy screening |
Ploidy screening in 2015 |
Fisher’s test |
Habitat type | |||||
|---|---|---|---|---|---|---|---|---|---|
| Season | 2x | 3x | 4x | 2x | 3x | 4x | P | ||
| 34 | 2012 | 6 | – | 1 | 18 | – | – | 0·280 | Field |
| 547 | 2014 | 4 | – | 2 | 17 | – | – | 0·059 | Field |
| 725 | 2014 | 10 | – | 3 | 20 | – | – | 0·052 | Field |
| 665 | 2014 | 18 | – | 1 | 20 | – | – | 0·487 | Field |
| 46 | 2012 | 5 | 1 | 4 | – | – | 7 | 0·035 | Ruderal site |
| 323 | 2013 | 3 | 4 | 19 | – | – | 44 | <0·001 | Pasture |
| 313 | 2013 | – | 1 | 16 | – | – | 12 | 1 | Railway |
| 900 | 2014 | 3 | – | 7 | – | – | 12 | 0·078 | Ruderal site |
| 884 | 2014 | 10 | – | 15 | – | – | 20 | 0·001 | Field |
| 932 | 2014 | 8 | 3 | 8 | – | – | 20 | <0·001 | Field |
| 83 | 2011 | 3 | – | 14 | 4 | 1 | 26 | 0·801 | Ruderal site |
| 602 | 2012 | 8 | – | 2 | 22 | 1 | – | 0·085 | Ruderal site |
| 181 | 2013 | 3 | 3 | 1 | 25 | 2 | 16 | 0·021 | Field |
| 228 | 2013 | 17 | 2 | – | 17 | – | 2 | 0·237 | Field |
| 226 | 2013 | 2 | 1 | 1 | 7 | 3 | 12 | 0·452 | Roadside |
| 337 | 2013 | 11 | – | 10 | 4 | 2 | 27 | 0·004 | Field |
| 215 | 2013 | 11 | 1 | 25 | 17 | – | 10 | 0·016 | Roadside |
| 309 | 2013 | – | 1 | 8 | 1 | – | 15 | 0·600 | Field |
| 293 | 2013 | 8 | – | 1 | 35 | – | 1 | 0·364 | Field |
| 544 | 2014 | 4 | – | 7 | 14 | 1 | 17 | 0·800 | Forest drive |
| 710 | 2014 | 3 | 1 | 10 | 11 | 1 | 18 | 0·537 | Fallow |
| 556 | 2014 | 8 | – | 5 | 4 | 1 | 2 | 0·562 | Field |
| 736 | 2014 | 6 | 1 | 5 | 4 | 2 | 34 | 0·004 | Field |
| 802 | 2014 | 5 | 1 | 5 | 1 | – | 14 | 0·011 | Forest drive |
| 471 | 2014 | 9 | 1 | 3 | 9 | 3 | 18 | 0·037 | Fallow |
| 431 | 2014 | 7 | 2 | 1 | 14 | 3 | 13 | 0·106 | Field drive |
| 671 | 2014 | 12 | 1 | 1 | 7 | 1 | 3 | 0·506 | Field |
| 775 | 2014 | 2 | 3 | 12 | 1 | 1 | 18 | 0·383 | Field drive |
| 778 | 2014 | 1 | 3 | 1 | 7 | 4 | 1 | 0·335 | Field drive |
| 920 | 2014 | 12 | – | 2 | 19 | – | 1 | 0·555 | Field |
| 1,095 | 2014 | 5 | 3 | 5 | 11 | 1 | 4 | 0·258 | Field |
| 891 | 2014 | 18 | 1 | 1 | 26 | 2 | 2 | 1 | Field |
| 889 | 2014 | 13 | 1 | 3 | 13 | 2 | 4 | 1 | Fallow |
| 830 | 2014 | 4 | – | 16 | 4 | 1 | 17 | 1 | Roadside |
| 819 | 2014 | 10 | – | 5 | 7 | – | 4 | 1 | Ruderal site |
| 855 | 2014 | 9 | – | 6 | 4 | – | 4 | 0·685 | Field |
Significant changes (α = 0·05) in cytotype abundance between the two visits are highlighted in bold.
Population codes correspond to sequential numbers in the complete list of populations under study (Table S1).
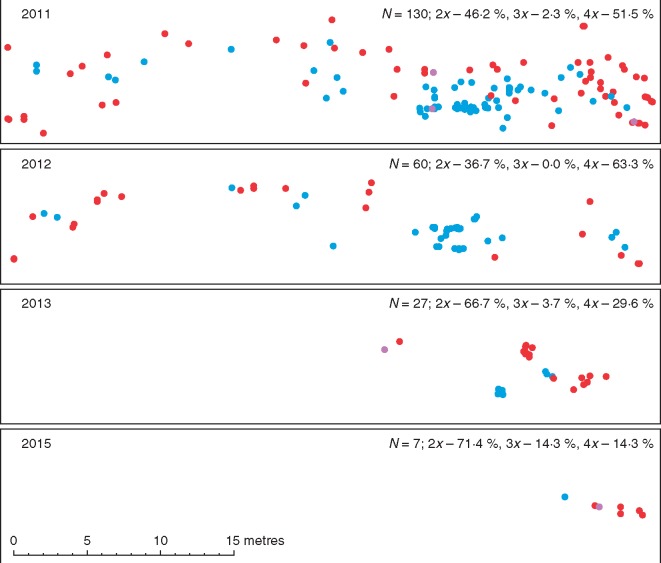
In one mixed-ploidy plot (population no. 22) that was re-collected yearly over five consecutive seasons, both diploids and tetraploids persisted until the last census in 2015. Interestingly, this was possible even despite a striking decrease in the overall abundance of the species and the fact that no plants were found at the locality in 2014 (Fig. 4). Between-year shifts in cytotype composition were significant only between 2012 and 2013 (Fisher’s exact test, P = 0·007).
Fig. 4.
Spatio-temporal changes in the occurrence of diploids (red), triploids (violet) and tetraploids (blue) within a study plot set in the mixed-ploidy population no. 22. The plot was visited once a year from 2011 to 2015, and the position and ploidy level was assessed for all Tripleurospermum inodorum individuals within its bounds. No plants were found in 2014.
Cytotype composition of offspring in mixed-ploidy populations
Flow-cytometric analyses of 1263 seedlings yielded all five euploid cytotypes and a substantial number of putative aneuploids (Table 3). Although no minority cytotypes or aneuploids were identified among the progeny of one uniformly diploid population studied, a single hexaploid (1·6 %) was detected in a uniformly tetraploid population. The greatest diversity in nuclear DNA content was found among offspring from mixed-ploidy populations. All these populations contained diploid, triploid and tetraploid offspring in various proportions, and three of them also included aneuploids (Table 3). Interestingly, the nuclear DNA content of the aneuploid offspring formed an almost perfect continuum spanning the 3x–4x and 4x–5x levels (Fig. S1); note that a few of these putative aneuploids might be in fact triploid or tetraploid mutants originating from crosses involving unreduced gametes. Five randomly selected aneuploid individuals were cultivated and subjected to chromosome counting, which revealed a hyper-triploid (2n = 3x+2 = 29; Fig. 1E) and four hyper-tetraploids (2n = 4x+1–5 = 37–41; Fig. 1F, Supplementary Data Table S2). The overall incidence of putatively aneuploid offspring ranged from 0 to 34 % across the populations under study. Additionally, one pentaploid and three hexaploid individuals were identified in two of the mixed-ploidy populations (Table 3).
Table 3.
Diversity in nuclear DNA content among 1263 seedlings germinated from pooled samples of open-pollinated achenes collected in five mixed-ploidy populations, one reference diploid population (no. 858) and one reference tetraploid population (no. 299); the offspring was assigned either to one of five euploid cytotypes (2x, 3x, 4x, 5x, 6x), or to one of three arbitrarily set classes of putative aneuploids (delimited by the closest lower and higher euploid cytotype: 2x–3x, 3x–4x, 4x–5x)
| Population* | Ploidy composition | Cytotypes at the level of seedlings [count (ratio)] |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2x | 2x–3x | 3x | 3x–4x | 4x | 4x–5x | 5x | 6x | |||
| 858 | 2x | 76 (100·0 %) | – | – | – | – | – | – | – | 76 |
| 299 | 4x | – | – | – | – | 62 (98·4 %) | – | – | 1 (1·6 %) | 63 |
| 66 | 2x, 3x, 4x | 57 (32·4 %) | – | 9 (5·1 %) | – | 110 (62·5 %) | – | – | – | 176 |
| 46 | 2x, 3x, 4x | 25 (9·6 %) | – | 37 (14·2 %) | 1 (0·4 %) | 197 (75·8 %) | – | – | – | 260 |
| 22 | 2x, 3x, 4x | 98 (77·8 %) | – | 4 (3·2 %) | – | 24 (19·0 %) | – | – | – | 126 |
| 220 | 2x, 3x, 4x | 7 (8·5 %) | 1 (1·2 %) | 34 (41·5 %) | 26 (31·7 %) | 12 (14·6 %) | 1 (1·2 %) | 1 (1·2 %) | – | 82 |
| 323 | 2x, 3x, 4x | 18 (3·8 %) | – | 7 (1·5 %) | 4 (0·8 %) | 436 (90·8 %) | 12 (2·5 %) | – | 3 (0·6 %) | 480 |
Population codes correspond to sequential numbers in the complete list of populations under study (Table S1).
DISCUSSION
Cytotype coexistence at various spatial scales
Our extensive flow-cytometric ploidy screening documented a diffuse, mosaic-like contact zone of the diploid and tetraploid cytotypes of T. inodorum in Central Europe. It revealed that the two cytotypes commonly coexist at all spatial scales under study (Fig. 3). Although there are reports of diploids and tetraploids coexisting within the same regions in parts of Germany and Poland (Rottgardt, 1956; Kay, 1969; Lankosz-Mróz, 1976), mixed-ploidy populations have never been reported, and only the tetraploid cytotype had until now been known from the Czech Republic (Kubát, 2004). This discrepancy probably stems from the much denser sampling in our study. Within three regions of the Czech Republic (medium-scale ploidy screening), mixed-ploidy populations were very common, reaching 33–43 %. These values are greater than those documented in most other mixed-ploidy plant systems (e.g. McArthur and Sanderson, 1999; Baack, 2004; Stuessy et al., 2004; Kolář et al., 2009; Treier et al., 2009; Duchoslav et al., 2010; Castro et al., 2012; Koutecký et al., 2012; Manzaneda et al., 2012). However, comparable or even higher frequencies of mixed-ploidy populations have been reported for some species such as Andropogon gerardii (McAllister et al., 2015), Arnica cordifolia (Kao, 2007), Galax urceolata (Burton and Husband, 1999), Gymnadenia conopsea (Trávníček et al., 2011) and Senecio carniolicus (Sonnleitner et al., 2010).
At both the large and the medium scale, tetraploid populations of T. inodorum were clearly the most common in Central Europe. Very interesting is the striking shortage of uniformly diploid populations, which in all cases were 2–5 times less frequent than mixed-ploidy populations (composed of both diploids and tetraploids). The observed pattern of cytotype distribution could partly result from an interplay between migration and forces stabilizing cytotype coexistence in ploidy mixtures. Seed transfer is probably very common in this annual weed and may occasionally bring the two cytotypes together to form a mixed-ploidy population. Although the achenes of T. inodorum lack any obvious adaptations for long-distance dispersal, they can become effectively dispersed together with mud or soil adhering to the wheels of farm vehicles or the hooves of farm animals (Kay, 1994). Our study area is located at the eastern border of the Europe-wide contact zone, in tetraploid-dominated regions adjoining uniformly tetraploid parts of the T. inodorum range (Fig. 3A). Most immigrants are therefore likely to be tetraploid and many new populations will thus be founded by tetraploids. Tetraploid immigrants may also lead to conversion of diploid populations into ploidy mixtures and to the gradual fixation of the tetraploid cytotype in mixed-ploidy populations. Potentially, this scenario might lead to a gradual shift in cytotype composition at the landscape level in favour of tetraploids and thus result in the movement of the contact zone (sensuBuggs, 2007). The only well-documented case of a moving contact zone of cytotypes has been reported from Spanish populations of Mercurialis annua, where diploids were displacing hexaploids because of ploidy-specific differences in their sexual systems (Buggs and Pannell, 2006). In T. inodorum, persistent soil seed banks may slow the local turnover of the cytotypes driven by immigration, and theoretically, migration of diploids from the opposite direction would counteract the spread of the tetraploid. However, a long-term study of population dynamics at numerous sites (both ploidy-uniform and mixed) is necessary to address the potential instability of the contact zone in this species.
Nonetheless, there are also other factors that may influence the spatial structure of the contact zone. The two cytotypes may differ in their habitat requirements. If tetraploids have a wider ecological amplitude than diploids and the spectrum of habitats suitable for diploids mostly overlaps with that of tetraploids, then the number of uniformly diploid populations should indeed be much lower than the number of mixed or purely tetraploid populations. However, apart from a relationship with the macroclimate (Kay, 1969, 1994; Woo et al., 1991), which is unlikely to shape the distribution of cytotypes at the medium-scale level (30 × 30 km in our case), we did not observe any differences between the cytotypes in a detailed vegetation survey (M. Čertner et al., unpubl. res.). Finally, recurrent origins of tetraploids from diploids could have turned most diploid populations into mixtures of diploids and tetraploids. However, genome size data disprove the frequent formation of tetraploid mutants, as discussed below.
Rates of spontaneous polyploidization in natural populations
While the frequency of gametic non-reduction, as the main driver of polyploidization, is reasonably well understood (Bretagnolle and Thompson, 1995; Ramsey and Schemske, 1998; Levin, 2002; Sora et al., 2016), the actual rate with which new polyploid mutants emerge and persist in natural populations is still mostly unknown (but see Ramsey, 2007). This rate can be approximated from the occurrence of higher-ploidy minority cytotypes in populations of their lower-ploid progenitors. However, certain cytotypes may originate both from crosses involving unreduced gametes in uniform-ploidy populations of progenitors and through inter-ploidy hybridization of established cytotypes (see Fig. 2), only the former pathway of cytotype formation being a reliable proxy of spontaneous polyploidization in natural populations.
In this respect, the profound 19 % difference in monoploid genome sizes of diploids and tetraploids makes T. inodorum a unique study system. Given that the relative genome size of progeny is the sum of parental genome sizes (considering scenarios involving both reduced and unreduced gametes; Fig 2), we were able to distinguish between the alternative scenarios of polyploid formation and to confidently assess the rate of spontaneous polyploidization in populations of T. inodorum. In spite of our thorough sampling, polyploid mutants were extremely rare, as only one hexaploid and three neotetraploids were found among 11 018 adult individuals from 1209 populations. The hexaploid originated in a tetraploid population by fusion of a reduced and an unreduced gamete. For the tetraploid mutants, two possible modes of origin were assumed (Fig. 2). Two individuals with relative genome sizes of 3·58 (population no. 187) and 3·42 (population no. 1092) could have resulted from a fusion of two unreduced gametes of diploids (expected relative genome size = 3·45). A single plant with a relative genome size of 3·26 (population no. 1227) might have originated from a fusion of an unreduced gamete of a diploid with a reduced gamete of a tetraploid (expected relative genome size = 3·18). Moreover, the overall rate of neotetraploid occurrence in natural populations may be even lower because the genome size of neotetraploids overlaps with that of certain aneuploids and we were unable to verify their status using chromosome counts.
The roughly similar incidence of neotetraploids and hexaploids suggests that the rates of spontaneous polyploidization in diploid and tetraploid parental populations are comparable. Neopolyploids always occurred solitarily, without any signs of their further spread, and were probably restricted to first generation. The mostly annual life cycle of T. inodorum along with the virtual lack of clonal propagation suggests that such neopolyploids are facing a strong minority disadvantage (Levin, 1975; Husband, 2000) and that their only chance of producing a new generation would be through autogamy. However, the self-compatibility of neopolyploids remains to be tested.
To our knowledge, the present study is the first to use pronounced differences in monoploid genome size of cytotypes to systematically screen for new polyploid mutants in diploid–polyploid contact zones. Generally, differences in monoploid genome sizes are indicators of independent evolution of particular lineages (Greilhuber, 2005), suggesting that tetraploids of T. inodorum either underwent substantial diversification following their origin or that they originated from a diploid lineage other than the one with which they are currently sympatric. In both scenarios, the diploid–tetraploid contact zone of scentless mayweed represents an example of secondary cytotype contact (sensuPetit et al., 1999).
Mixed-ploidy populations as generators of cytogenetic novelty
Our intensive screening of 1263 seedlings from seven T. inodorum populations revealed not only all the five ploidy levels but also several aneuploids (spanning the 2x–3x, 3x–4x and 4x–5x ploidy levels), thus capturing more cytotype diversity than was found among 11 018 adult individuals from across Central Europe. Specifically, aneuploids were more frequent among seedlings than among adult plants (4·0 % vs. 0·3 %, respectively; note that only individuals from mixed-ploidy populations are considered here), and more diverse in their relative genome size. Similarly, the incidence of hexaploid mutants was higher among seedlings (0·5 % vs. 0·01 %; relative to the overall number of tetraploids). Unfortunately, the more or less continuous variation in the genome size of analysed aneuploid seedlings (Fig. S1) hindered such comparisons for triploid and tetraploid mutants.
Although the common use of pooled samples during our large- and medium-scale ploidy screening could have masked aneuploids differing by one or just a few chromosomes from the closest euploid level, the shortage of both aneuploids with more divergent genome sizes and neopolyploid mutants among adult plants is still evident. One possible explanation is that aneuploids and neopolyploids suffer from lower vitality and become outcompeted during later stages of the plant life cycle. However, a subset of aneuploids and hexaploids grown under greenhouse conditions exhibited vigorous growth to maturity. We therefore attribute the deficiency of these cytotypes solely to the lower frequency of their emergence (Table 3). Given the massive seed production in T. inodorum (Kay, 1994) along with strong over-compensating density dependence (Buckley et al., 2001), chance events are likely to prevent aneuploids and polyploid mutants from reaching maturity in many natural populations. The minority cytotype exclusion principle might further hinder their survival to subsequent generations (Levin, 1975) although it has no effect on cytotype diversity forming recurrently within each generation.
The degree of cytotype diversity at the seedling stage was tightly linked with the cytotype composition of populations. While only uniform-ploidy progeny was obtained by germinating seeds from both a uniformly diploid and a uniformly tetraploid population (with the exception of a single hexaploid individual in the tetraploid population), a minimum of three cytotypes were always present among progeny originating from mixed-ploidy populations and sometimes also aneuploids (Table 3). Aneuploid progeny most probably results from crosses in which either one or both parents are triploids. Triploids and other odd-level polyploids are known to produce gametes with a varying, unbalanced number of chromosomes (Ramsey and Schemske, 1998), giving rise to aneuploid progeny, as has been documented by experimental crosses (Krahulcová and Jarolímová, 1993; Norrmann et al., 1997) and in natural mixed-ploidy populations (Keeler, 2004). Partial fertility of T. inodorum triploids was previously reported by Kay (1965), who observed triploids attaining a certain, albeit low, seed set when used as the egg parent in manipulated crosses with diploids or tetraploids. We also noticed triploid plants setting seed in natural mixed-ploidy populations during our field sampling.
Reproductive interactions between triploid hybrids and their parental cytotypes make mixed-ploidy populations of T. inodorum important generators of cytogenetic novelty. In a longer time frame, these processes might facilitate inter-ploidy gene flow between diploids and tetraploids (Comai, 2005), especially if also aneuploids are at least partly fertile. This would in turn strongly impact further evolution of the established cytotypes meeting in a secondary contact zone. Given the high frequency of mixed-ploidy populations and the considerable numbers of triploids within them (mean rate approx. 8 %), the common coexistence of T. inodorum cytotypes might have larger evolutionary significance, beyond a mere cytogeographical fact, and confer an adaptive advantage to the species. Manipulated pollinations and molecular-genetic analyses are currently in progress to study reproductive interactions among various euploid and aneuploid cytotypes of T. inodorum and their potential for mediating inter-ploidy gene flow.
Temporal stability of ploidy mixtures
The main constraints of local cytotype coexistence are inter-cytotype reproductive interactions (Levin, 1975; Husband, 2000; Li et al., 2004; Rausch and Morgan, 2005) and resource competition among cytotypes (Maceira et al., 1993; Collins et al., 2011). The strength of these mechanisms is generally influenced by specific life-traits of the given plant species. In T. inodorum, a lack of means of clonal reproduction (Kay, 1994) along with self-incompatibility reported from some populations (Kay, 1969) decrease reproductive assurance of a minority cytotype and thus the probability of cytotype coexistence. On the other hand, the spatial aggregation of plants of the same cytotype that we observed in three out of the five mixed-ploidy populations under study could increase non-random pollen transfer and restrict inter-cytotype competition (Baack, 2005b; Kennedy et al., 2006). Given the lack of clonal growth, limited seed dispersal may be responsible for the spatial clumping of T. inodorum cytotypes. In addition, we have not noticed any striking ploidy-specific differences in plant size, vigour or reproductive effort (our personal observations) that could favour the persistence of one of the cytotypes. However, due to probable interactions between factors facilitating cytotype coexistence on the one hand and factors leading to the fixation of one of the cytotypes on the other, the most reliable way of assessing the fate of ploidy mixtures are temporal observations of their development.
Despite our initial expectations of rapid fixation of one of the cytotypes in mixed-ploidy populations of this annual weed, our temporal observations suggest surprisingly high stability of ploidy coexistence. In one mixed-ploidy population screened annually over a 5-year period, both diploids and tetraploids persisted at the site until the last season, in spite of a severe gradual reduction of population size and even an absence of adults and seedlings in one year (Fig. 4). Similarly, most of the 36 mixed-ploidy sites re-analysed for ploidy composition (72 %) contained both diploids and tetraploids after 1–4 years (Table 2). It is likely that permanent soil seed banks were established at these sites in the first year of our study or even earlier. Subsequent germination of variably aged seeds made the cytotype composition of populations in each given year more or less independent of the year before. The effect of permanent soil seed banks is often neglected in polyploid studies (but see Hahn et al., 2012), but it may play an important role in stabilizing mixed-ploidy populations, especially of annual and short-lived plants. Moreover, repeated immigration of seeds of particular cytotypes from surrounding populations may have a similar effect. Finally, the fixation of one of the cytotypes in mixed-ploidy populations might be slowed down or even hampered by disturbances that prevent the dominant cytotype from attaining the highest population densities (Halverson et al., 2008).
To our knowledge, this study is the first to document temporal changes in cytotype composition in situ, at the within-population scale. We are aware of the relatively short observation periods in our study, which precludes reaching more general conclusions. Also, the fact that our tests showed no effect of habitat and/or time between the two sequential visits on the fate of ploidy mixtures could be partially attributed to their low statistical power. More comprehensive assessments of temporal changes in cytotype composition of mixed-ploidy sites are currently being undertaken, along with a targeted study of potentially stabilizing mechanisms.
CONCLUSIONS
Extensive flow-cytometric screening of over 11 000 adult individuals from 1209 populations supplemented with more than 1200 ex situ germinated seedlings was used to survey a diploid–tetraploid contact zone of T. inodorum in Central Europe. The contact zone has a diffuse, mosaic-like structure enabling the common coexistence of the two cytotypes at all spatial scales studied. Mixed-ploidy populations, containing both diploids and tetraploids, then probably act as effective generators of cytogenetic novelty. This was especially apparent from the striking variation in DNA content of offspring from mixed-ploidy sites, which could be attributed to ongoing reproductive interactions between relatively numerous triploid hybrids and their parental cytotypes. In a longer time frame, such reproductive interactions might even facilitate inter-ploidy gene flow from diploids to tetraploids and vice versa. The local coexistence of diploids and long-established tetraploids meeting in certain parts of their overlapping distributional range might thus leave significant traces in the cytotypes’ evolutionary history or even confer an adaptive advantage to the species (e.g. through the transfer of adaptations).
Our study species has two unique aspects when compared with other well-documented mixed-ploidy systems. Firstly, the profound 19 % difference in monoploid genome size between diploids and tetraploids enables the easy distinction of neotetraploid mutants from long-established tetraploids during routine flow-cytometric ploidy screening. Neotetraploid mutants were extremely rare among adult individuals of T. inodorum (approx. 0·03 %); they never formed uniform populations but occurred solitarily, without any sign of their local establishment and further spread. Secondly, aside from capturing the spatial distribution of ploidy diversity, the predominantly annual life cycle of the species also allows the monitoring of trends in the temporal development of this diversity. In spite of considerable oscillations in cytotype frequencies and the overall abundance of mixed-ploidy populations, our observations suggest that the local coexistence of diploids and tetraploids is surprisingly stable. The persistence of mixed-ploidy populations is probably driven by a combined effect of permanent soil seed banks, repeated immigration of seeds of particular cytotypes from surrounding populations and recurring local disturbances.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: the complete list of populations sampled in our study. Table S2: relative genome size and chromosome counts of ex-situ germinated seedlings. Figure S1: distribution of relative genome size among seedlings and arbitrary delimitation of euploid and aneuploid cytotypes. Figure S2: small-scale distribution of cytotypes in three mixed-ploidy populations.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to T. Urfus, J. Prančl, M. Ducháček, L. Ekrt, Z. Kaplan, A. Knotek, A. Nedomová, D. Čertnerová, V. Otisková, M. Pospíšilová, M. Štech, P. Schönswetter and B. Frajman for their help with collecting samples in the field. This work was partly funded by the Czech Science Foundation (project 14-18870S) and partly by the Charles University (project GAUK 913213). Additional support was provided by the Czech Academy of Sciences (long-term research development project no. RVO 67985939) and in the form of institutional resources provided by the Ministry of Education, Youth and Sports of the Czech Republic for the support of science and research.
LITERATURE CITED
- Arnold B, Kim ST, Bomblies K.. 2015. Single geographic origin of a widespread autotetraploid Arabidopsis arenosa lineage followed by interploidy admixture. Molecular Biology and Evolution 32: 1382–1395. [DOI] [PubMed] [Google Scholar]
- Arora OP, Madhusoodanan KJ.. 1981. Nature of tetraploidy in Matricaria inodora L. Cytologia 46: 773–779. [Google Scholar]
- Arvanitis L, Wiklund C, Ehrlén J.. 2008. Plant ploidy level influences selection by butterfly seed predators. Oikos 117: 1020–1025. [Google Scholar]
- Baack EJ. 2004. Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus: Ranunculaceae). American Journal of Botany 91: 1783–1788. [DOI] [PubMed] [Google Scholar]
- Baack EJ. 2005a. Ecological factors influencing tetraploid establishment in snow buttercups (Ranunculus adoneus, Ranunculaceae): minority cytotype exclusion and barriers to triploid formation. American Journal of Botany 92: 1827–1835. [DOI] [PubMed] [Google Scholar]
- Baack EJ. 2005b. To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity 94: 538–546. [DOI] [PubMed] [Google Scholar]
- Barringer BC. 2007. Polyploidy and self-fertilization in flowering plants. American Journal of Botany 94: 1527–1533. [DOI] [PubMed] [Google Scholar]
- Bowes GG, Thomas AG, Lefkovitch LP.. 1995. Changes with time in the germination of buried scentless chamomile (Matricaria perforata Mérat) seeds. Canadian Journal of Plant Science 75: 277–281. [Google Scholar]
- Bretagnolle F, Thompson JD.. 1995. Tansley review no. 78. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolypoid plants. New Phytologist 129: 1–22. [DOI] [PubMed] [Google Scholar]
- Buckley YM, Hinz HL, Matthies D, Rees M.. 2001. Interactions between density-dependent processes, population dynamics and control of an invasive plant species, Tripleurospermum perforatum (scentless chamomile). Ecology Letters 4: 551–558. [Google Scholar]
- Buggs RJA. 2007. Empirical study of hybrid zone movement. Heredity 99: 301–312. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Pannell JR.. 2006. Rapid displacement of a monoecious plant lineage is due to pollen swamping by a dioecious relative. Current Biology 16: 996–1000. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Pannell JR.. 2007. Ecological differentiation and diploid superiority across a moving ploidy contact zone. Evolution 61: 125–140. [DOI] [PubMed] [Google Scholar]
- Burton TL, Husband BC.. 1999. Population cytotype structure in the polyploid Galax urceolata (Diapensiaceae). Heredity 82: 381–390. [DOI] [PubMed] [Google Scholar]
- Castro S, Loureiro J, Santos C, Ater M, Ayensa G, Navarro L.. 2007. Distribution of flower morphs, ploidy level and sexual reproduction of the invasive weed Oxalis pes-caprae in the western area of the Mediterranean region. Annals of Botany 99: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro S, Loureiro J, Procházka T, Münzbergová Z.. 2012. Cytotype distribution at a diploid-hexaploid contact zone in Aster amellus (Asteraceae). Annals of Botany 110: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Abbott RJ.. 2010. Introgression of fitness genes across a ploidy barrier. New Phytologist 186: 63–71. [DOI] [PubMed] [Google Scholar]
- Collins AR, Naderi R, Mueller-Schaerer H.. 2011. Competition between cytotypes changes across a longitudinal gradient in Centaurea stoebe (Asteraceae). American Journal of Botany 98: 1935–1942. [DOI] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews. Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour AB.. 2007. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22: 1–20. [Google Scholar]
- Duchoslav M, Šafářová L, Krahulec F.. 2010. Complex distribution patterns, ecology and coexistence of ploidy levels of Allium oleraceum (Alliaceae) in the Czech Republic. Annals of Botany 105: 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer AF. 1963. The use of lacto-propionic orcein in rapid squash methods for chromosome preparations. Stain Technology 38: 85–90. [Google Scholar]
- Flagel LE, Wendel JF.. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Fowler NL, Levin DA.. 1984. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. The American Naturalist 124: 703–711. [Google Scholar]
- Godsoe W, Larson MA, Glennon KL, Segraves KA.. 2013. Polyploidization in Heuchera cylindrica (Saxifragaceae) did not result in a shift in climatic requirements. American Journal of Botany 100: 496–508. [DOI] [PubMed] [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg A, Ellerström S.. 1959. The competition between diploid, tetraploid and aneuploid rye: theoretical and practical aspects. Hereditas 45: 369–416. [Google Scholar]
- Hahn MA, Buckley YM, Müller-Schärer H.. 2012. Increased population growth rate in invasive polyploid Centaurea stoebe in a common garden. Ecology Letters 15: 947–954. [DOI] [PubMed] [Google Scholar]
- Halverson K, Heard SB, Nason JD, Stireman JO.. 2008. Origins, distribution, and local co-occurrence of polyploid cytotypes in Solidago altissima (Asteraceae). American Journal of Botany 95: 50–58. [DOI] [PubMed] [Google Scholar]
- Hanzl M, Kolář F, Nováková D, Suda J.. 2014. Nonadaptive processes governing early stages of polyploid evolution: insights from a primary contact zone of relict serpentine Knautia arvensis (Caprifoliaceae). American Journal of Botany 101: 935–945. [DOI] [PubMed] [Google Scholar]
- Hegarty M, Coate J, Sherman-Broyles S, Abbott R, Hiscock S, Doyle J.. 2013. Lessons from natural and artificial polyploids in higher plants. Cytogenetic and Genome Research 140: 204–225. [DOI] [PubMed] [Google Scholar]
- Hroudová Z, Zákravský P.. 1993. Ecology of two cytotypes of Butomus umbellatus II: reproduction, growth and biomass production. Folia Geobotanica & Phytotaxonomica 28: 413–424. [Google Scholar]
- Hülber K, Sonnleitner M, Suda J, et al. 2015. Ecological differentiation, lack of hybrids involving diploids, and asymmetric gene flow between polyploids in narrow contact zones of Senecio carniolicus (syn. Jacobaea carniolica, Asteraceae). Ecology and Evolution 5: 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC. 2000. Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proceedings of the Royal Society of London, Series B – Biological Sciences 267: 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC. 2004. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society 82: 537–546. [Google Scholar]
- Husband BC, Sabara HA.. 2003. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytologist 161: 703–713. [DOI] [PubMed] [Google Scholar]
- Kao RH. 2007. Asexuality and the coexistence of cytotypes. New Phytologist 175: 764–772. [DOI] [PubMed] [Google Scholar]
- Kay QON. 1965. Experimental and comparative ecological studies of selected weeds. PhD Thesis, University of Oxford, UK.
- Kay QON. 1969. The origin and distribution of diploid and tetraploid Tripleurospermum inodorum (L.) Schultz Bip. Watsonia 7: 130–141. [Google Scholar]
- Kay QON. 1976. 60. Matricaria L In: Tutin TG, Heywood VH, Burges NA, et al. eds. Flora Europaea, Vol. 4. Cambridge: Cambridge University Press, 165–167. [Google Scholar]
- Kay QON. 1994. Tripleurospermum inodorum (L.) Schultz Bip. Journal of Ecology 82: 681–697. [Google Scholar]
- Keeler KH. 2004. Impact of intraspecific polyploidy in Andropogon gerardii (Poaceae) populations. The American Midland Naturalist 152: 63–74. [Google Scholar]
- Keeler KH, Davis GA.. 1999. Comparison of common cytotypes of Andropogon gerardii (Andropogoneae, Poaceae). American Journal of Botany 86: 974–979. [PubMed] [Google Scholar]
- Kennedy BF, Sabara HA, Haydon D, Husband BC.. 2006. Pollinator-mediated assortative mating in mixed ploidy populations of Chamerion angustifolium (Onagraceae). Oecologia 150: 398–408. [DOI] [PubMed] [Google Scholar]
- Kliber A, Eckert CG.. 2005. Interaction between founder effect and selection during biological invasion in an aquatic plant. Evolution 59: 1900–1913. [PubMed] [Google Scholar]
- Kolář F, Štech M, Trávníček P, et al. 2009. Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Annals of Botany 103: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolář F, Lučanová M, Vít P, et al. 2013. Diversity and endemism in deglaciated areas: ploidy, relative genome size and niche differentiation in the Galium pusillum complex (Rubiaceae) in Northern and Central Europe. Annals of Botany 111: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutecký P, Štěpánek J, Baďurová T.. 2012. Differentiation between diploid and tetraploid Centaurea phrygia: mating barriers, morphology and geographic distribution. Preslia 84: 1–32. [Google Scholar]
- Krahulcová A, Jarolímová V.. 1993. Ecology of two cytotypes of Butomus umbellatus I. Karyology and breeding behaviour. Folia Geobotanica & Phytotaxonomica 28: 385–411. [Google Scholar]
- Kubát K. 2004. 33. Tripleurospermum Schultz Bip. - heřmánkovec In: Slavík B, Štěpánková J, eds. Květena České republiky, Vol. 7. Praha: Academia, 248–250. [Google Scholar]
- Lankosz-Mróz M. 1976. Karyological investigations on Tripleurospermum maritimum (L.) Koch ssp. inodorum (L.) ex Vaarama from Poland. Acta Biologica Cracoviensia (Series Botanica) 19: 93–105. [Google Scholar]
- Levin DA. 1975. Minority cytotype exclusion in local plant populations. Taxon 24: 35–43. [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. The American Naturalist 122: 1–25. [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. Oxford: Oxford University Press. [Google Scholar]
- Li BH, Xu XM, Ridout MS.. 2004. Modelling the establishment and spread of autotetraploid plants in a spatially heterogeneous environment. Journal of Evolutionary Biology 17: 562–573. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Guillerm JL, Delay J, Ait Lhaj Loutfi A, Izco J, Jay M.. 1987. Polyploidy and habitat differentiation in Dactylis glomerata L. from Galicia (Spain). Oecologia 73: 436–446. [DOI] [PubMed] [Google Scholar]
- Maceira NO, Jacquard P, Lumaret R.. 1993. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytologist 124: 321–328. [DOI] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a general regression approach. Cancer Research 27: 209–220. [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T.. 2012. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). The New Phytologist 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C, Blaine R, Kron P, et al. 2015. Environmental correlates of cytotype distribution in Andropogon gerardii (Poaceae). American Journal of Botany 102: 92–102. [DOI] [PubMed] [Google Scholar]
- McArthur ED, Sanderson SC.. 1999. Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae). American Journal of Botany 86: 1754–1775. [PubMed] [Google Scholar]
- Meirmans PG, Calame FG, Bretagnolle F, Felber F, den Nijs JCM.. 1999. Anthropogenic disturbance and habitat differentiation between sexual diploid and apomictic triploid Taraxacum sect. Ruderalia. Folia Geobotanica 34: 451–469. [Google Scholar]
- Mráz P, Šingliarová B, Urfus T, Krahulec F.. 2008. Cytogeography of Pilosella officinarum (Compositae): altitudinal and longitudinal differences in ploidy level distribution in the Czech Republic and Slovakia and the general pattern in Europe. Annals of Botany 101: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mráz P, Španiel S, Keller A, et al. 2012. Anthropogenic disturbance as a driver of microspatial and microhabitat segregation of cytotypes of Centaurea stoebe and cytotype interactions in secondary contact zones. Annals of Botany 110: 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrmann GA, Quarín CL, Keeler KH.. 1997. Evolutionary implications of meiotic chromosome behavior, reproductive biology, and hybridization in 6x and 9x cytotypes of Andropogon gerardii (Poaceae). American Journal of Botany 84: 201–207. [PubMed] [Google Scholar]
- Nuismer SL, Thompson JN.. 2001. Plant polyploidy and non-uniform effects on insect herbivores. Proceedings of the Royal Society of London, Series B – Biological Sciences 268: 1937–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald BP, Nuismer SL.. 2011. Neopolyploidy and diversification in Heuchera grossulariifolia. Evolution 65: 1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otisková V, Koutecký T, Kolář F, Koutecký P.. 2014. Occurrence and habitat preferences of diploid and tetraploid cytotypes of Centaurea stoebe in the Czech Republic. Preslia 86: 67–80. [Google Scholar]
- Peckert T, Chrtek J.. 2006. Mating interactions between coexisting diploid, triploid and tetraploid cytotypes of Hieracium echioides (Asteraceae). Folia Geobotanica 41: 323–334. [Google Scholar]
- Petit C, Lesbros P, Ge X, Thompson JD.. 1997. Variation in flowering phenology and selfing rate across a contact zone between diploid and tetraploid Arrhenatherum elatius (Poaceae). Heredity 79: 31–40. [Google Scholar]
- Petit C, Bretagnolle F, Felber F.. 1999. Evolutionary consequences of diploid-polyploid hybrid zones in wild species. Trends in Ecology and Evolution 14: 306–311. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2007. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae). Heredity 98: 143–150. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- Rausch JH, Morgan MT.. 2005. The effect of self-fertilization, inbreeding depression, and population size on autopolyploid establishment. Evolution 59: 1867–1875. [PubMed] [Google Scholar]
- Rodríguez DJ. 1996. A model for the establishment of polyploidy in plants. The American Naturalist 147: 33–46. [Google Scholar]
- Rottgardt K. 1956. Morphologische, cytologische und physiologische Untersuchungen von Ökotypen in Schleswig-Holstein. Beiträge zur Biologie der Pflanzen 32: 225–278. [Google Scholar]
- Segraves KA, Thompson JN.. 1999. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution 53: 1114–1127. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann C.. 2007. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Molecular Phylogenetics and Evolution 42: 92–103. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. 2009. Polyploidy and angiosperm diversification. American Journal of Botany 96: 336–348. [DOI] [PubMed] [Google Scholar]
- Sonnleitner M, Flatscher R, Escobar García P, et al. 2010. Distribution and habitat segregation on different spatial scales among diploid, tetraploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps. Annals of Botany 106: 967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sora D, Kron P, Husband BC.. 2016. Genetic and environmental determinants of unreduced gamete production in Brassica napus, Sinapis arvensis and their hybrids. Heredity 117: 440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuessy TF, Weiss-Schneeweiss H, Keil DJ.. 2004. Diploid and polyploid cytotype distribution in Melampodium cinereum and M. leucanthum (Asteraceae, Heliantheae). American Journal of Botany 91: 889–898. [DOI] [PubMed] [Google Scholar]
- Suda J, Krahulcová A, Trávníček P, Krahulec F.. 2006. Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon 55: 447–450. [Google Scholar]
- Trávníček P, Kubátová B, Čurn V, et al. 2011. Remarkable coexistence of multiple cytotypes of the Gymnadenia conopsea aggregate (the fragrant orchid): Evidence from flow cytometry. Annals of Botany 107: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier UA, Broennimann O, Normand S, et al. 2009. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 90: 1366–1377. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM.. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SL, Thomas AG, Peschken DP, et al. 1991. The biology of Canadian weeds. 99. Matricaria perforata Mérat (Asteraceae). Canadian Journal of Plant Science 71: 1101–1119. [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH.. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.