Abstract
Hemipteran insects are well-known in their ability to establish symbiotic relationships with bacteria. Among them, heteropteran insects present an array of symbiotic systems, ranging from the most common gut crypt symbiosis to the more restricted bacteriome-associated endosymbiosis, which have only been detected in members of the superfamily Lygaeoidea and the family Cimicidae so far. Genomic data of heteropteran endosymbionts are scarce and have merely been analyzed from the Wolbachia endosymbiont in bed bug and a few gut crypt-associated symbionts in pentatomoid bugs. In this study, we present the first detailed genomic analysis of a bacteriome-associated endosymbiont of a phytophagous heteropteran, present in the seed bug Henestaris halophilus (Hemiptera: Heteroptera: Lygaeoidea). Using phylogenomics and genomics approaches, we have assigned the newly characterized endosymbiont to the Sodalis genus, named as Candidatus Sodalis baculum sp. nov. strain kilmister. In addition, our findings support the reunification of the Sodalis genus, currently divided into six different genera. We have also conducted comparative analyses between 15 Sodalis species that present different genome sizes and symbiotic relationships. These analyses suggest that Ca. Sodalis baculum is a mutualistic endosymbiont capable of supplying the amino acids tyrosine, lysine, and some cofactors to its host. It has a small genome with pseudogenes but no mobile elements, which indicates middle-stage reductive evolution. Most of the genes in Ca. Sodalis baculum are likely to be evolving under purifying selection with several signals pointing to the retention of the lysine/tyrosine biosynthetic pathways compared with other Sodalis.
Keywords: Lygaeoidea, endosymbiosis, taxonomy, comparative genomics, metabolism, molecular evolution
Introduction
Most insects have established specific associations with bacterial symbionts. These associations show a broad range of symbiotic interactions, ranging from parasitism to mutualism. Bacterial symbionts can be found on the surface of the insects but also inside their bodies (e.g. the gut system). Often, mutualistic symbionts and insects establish a more intimate relationship, where the symbionts are maintained inside specialized host cells, called bacteriocytes, that can form an organ-like termed bacteriome (Buchner 1965). These intracellular symbionts (hereafter endosymbionts) are usually defined as primary, or obligate, if the insect requires the symbiotic relationship for survival, and secondary, or facultative, if the relationship is not essential for its survival. However, in some cases, a secondary endosymbiont can act as a coprimary one, if its presence is also essential for the insect or the primary endosymbiont (Sudakaran et al. 2017). Although different bacterial lineages are capable of establishing a stable endosymbiotic relationship with insects, representatives of the Bacteroidetes as well as Alpha-, Beta-, and Gammaproteobacteria, especially Enterobacteriaceae, are the most prominent groups (Moya et al. 2008; Moran et al. 2008; Sabree et al. 2009; Husník et al. 2011; Sudakaran et al. 2017).
Among others, species of the Sodalis group (Gammaproteobacteria: Enterobacterales: Pectobacteriaceae) offer a spectrum of various types of endosymbiosis. The eponymous strain was originally described as a secondary endosymbiont of the tsetse fly Glossina morsitans (Dale and Maudlin 1999). Because then, numerous different Sodalis or Sodalis-allied species were found in several insect groups, such as weevils (Heddi et al. 1999; Oakeson et al. 2014), hippoboscid louse fly (Nováková and Hypša 2007; Chrudimský et al. 2012), chewing lice (Fukatsu et al. 2007; Smith et al. 2013) and seal lice (Boyd et al. 2016). In addition, hemipteran insects such as aphids (Burke et al. 2009), psyllids (Sloan and Moran 2012; Arp et al. 2014), scale insects (Gruwell et al. 2010; Husník and McCutcheon 2016), spittlebugs (Koga and Moran 2014), and stinkbugs (Kaiwa et al. 2010, 2011; Matsuura et al. 2014; Hosokawa et al. 2015) frequently harbor Sodalis endosymbionts. Recently, a Sodalis-allied bacterial strain was also isolated from a human wound infection (Clayton et al. 2012), possibly representing a free-living ancestral state of Sodalis. This Sodalis, named Sodalis praecaptivus, and the one from G. morsitans are the only species cultivable so far.
Based on their pattern of occurrence in different ecological niches and insects, the “characteristics” of each Sodalis species and their specific effects on their hosts are quite diverse. For example, Sodalis species are often described as facultative endosymbionts, but have also been found in insect bacteriocytes, showing strict mutualistic obligatory relationship with their weevil hosts (Oakeson et al. 2014), or as copartners, complementing missing metabolic functions of an obligatory endosymbiont in the Carsonella-psyllid system (Sloan and Moran 2012). This illustrates that representatives of the genus Sodalis, or allied bacteria, cover a broad spectrum, ranging from free-living species, over facultative commensals to obligate mutualists of insects. The phylogeny and taxonomy of Sodalis-allied symbionts, mainly derived from analyses of their 16S rRNA and few other gene sequences, present several inconsistencies produced by events of horizontal transmission and new hosts acquisition (Dale et al. 2001; Snyder et al. 2011; Smith et al. 2013).
Numerous primary and secondary endosymbiotic bacteria, and hosts’ structures that harbor them, were described in stinkbugs or true bugs (Heteroptera) (Buchner 1965). Sodalis-allied endosymbionts were also detected in some members, for the first time in the superfamily Pentatomoidea (Heteroptera: Pentatomomorpha), more specifically in the families Acanthosomatidae, Pentatomidae, Scutelleridae, and Urostylididae (Kaiwa et al. 2010, 2011, 2014; Matsuura et al. 2014; Hosokawa et al. 2015). It is generally argued that Sodalis endosymbionts do not play an essential role in the biology of most of their heteropteran hosts, although such functions could not be completely excluded in urostylidid stinkbugs, due to the high infection rates in these species (Kaiwa et al. 2014; Hosokawa et al. 2015). Until present, no Sodalis symbiont has been found in the superfamily Lygaeoidea (reviewed in Sudakaran et al. 2017). The reason for this is not clear, because most lygaeoid bugs also harbor a broad range of endosymbiotic bacteria accommodated in specific structures, ranging from midgut crypts to bacteriomes, depending on the (sub)families (Kuechler et al. 2010, 2011, 2012; Kikuchi et al. 2011; Matsuura et al. 2012).
One of these bacteriome-associated endosymbiosis was also described in Henestaris halophilus, a member of the lygaeoid subfamily Henestarinae (Heteroptera: Lygaeoidea: Geocoridae), but has not been analyzed in detail so far (Kuechler et al. 2012). The subfamily Henestarinae is mainly distributed in southern Palearctic and African regions and contains about 19 species placed in 3 genera (Schuh and Slater 1995). All species, mainly characterized by their stalked eyes, live in saline-affected habitats both inshore and inland. The genus Henestaris is phytophagous and H. halophilus mainly feeds on seeds and infructescence of halophytes, like Plantago maritima, Artemisia maritima, Aster trifolium or Atriplex spp., (Wachmann et al. 2007), but occasionally also on grasses, especially Puccinella distans (Hiebsch 1961).
In the present work, we provide the first detailed description of the bacteriome-associated endosymbiont of H. halophilus, identified as a member of the Sodalis group, including molecular characterization, ultrastructural morphology and localization and transmission route. We also present the endosymbiont’s complete genomic sequence which is characterized by a reduced genome size and a very low coding density. Our metabolic reconstruction analysis suggests that the main contribution of the endosymbiont to its insect host involves processes related to cuticle hardening and the production of vitamins. Finally, several Sodalis-allied species were compared at both the metabolic and sequence levels, and the taxonomic status of the whole Sodalis group was revisited.
Materials and Methods
Insect Material
Adults and larval stages of Henestaris halophilus were collected from their natural habitat in Talamone (Italy) and Sülldorf (Saxony-Anhalt, Germany). Live individuals were brought to the laboratory and maintained at 25 °C under long day conditions (16:8 h) on sunflower seeds and distilled water enriched with 0.05% ascorbic acid. Laid eggs were carefully collected and allowed to develop at 25 °C. Developing eggs were extracted for fixation (eggs were dissected in 90% [vol/vol] ethanol to remove chorion and vitelline membrane) followed by whole mount fluorescence in situ hybridization (wFISH) analysis. Insect bacteriomes and ovaries were dissected in Ringer's solution (8.0 g NaCl, 0.4 g KCl, 0.4 g CaCl2, and 1.0 g Hepes per liter, pH 7.2).
Microscopy Analysis
For wFISH, freshly dissected bacteriomes, ovaries, and embryos were incubated overnight at room temperature in Carnoy’s solution (ethanol:chloroform:acetate, 6:3:1) and then washed in an ascending ethanol series (70%, 90%, and 2× 100%). After washing, the fixed samples were stored at −20 °C until use. Afterwards, all samples were washed with PBSTw [PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) containing 0.3% Tween 20] three times for 10 min. After thorough washing, the samples were equilibrated with hybridization buffer [30% (vol/vol) formamide, 0.02 M Tris–HCl (pH 8.0), 0.9 M NaCl, 0.01% SDS] three times for 10 min, followed by overnight incubation at 28 °C in hybridization buffer containing 1% of 10 nmol/µl symbiont specific probe Hen500 (5'-Cy3-CCATTGTCTTCTTCTCCGCC-3') and helper probe Hen500_H1 (5'-GAAAGTGCTTTACAACCCTAAGG-3'). Next, the samples were incubated for 20 min at 42 °C in hybridization buffer without probe. The samples were washed again with PBSTw three times for 15 min, and then incubated with 1% (vol/vol) SYBR Green I (1:10,000). The staining was stopped by washing in PBSTw. At the final step, the samples were mounted onto glass slides using antifade solution (citifluor) and glycerol (1:1) containing medium. The samples were examined under an SP5 confocal laser-scanning microscope (Leica). Electron microscopy was performed as described by Kuechler et al. (2012).
DNA Extraction, Sequencing, and Genome Annotation
A pool of bacteriomes dissected from 25 females was utilized for total genomic DNA extraction using the PureLink Genomic DNA Mini Kit (Invitrogen). Six independent whole-genome amplification reactions (GenomiPhi v2, GE Healthcare) were performed following manufacturer instructions. Because chimera formation seems to be a random process, samples were mixed to maintain possible chimeras at a low ratio relative to the amplified nonchimeric DNA. Amplified DNA was used for sequencing by the Illumina HiSeq2000 (350-bp paired-end library and 2× 100 bp) platform at Macrogen, Inc. Genome assembly and annotation procedures are presented in Supplementary Material online.
Genome and Metabolism Comparisons
Several Sodalis genomes and allied-species genomes were downloaded from NCBI and other sources (see table 1). Pantoea ananatis LMG 5342 (NC_016816) was used as an outgroup to allow topology comparisons (Husník and McCutcheon 2016). The proteomes of the above species were used as input for OrthoMCL v2.0.9 (1.5 inflation value) using USEARCH v9.1 (ublast -id 0.5 -maxhits 10,000 -acceptall -evalue 1e−5 -accel 1 -weak_evalue 0.1) (Li et al. 2003; Edgar 2010). The orthologous clusters of proteins (hereafter OCPs) output from OrthoMCL (supplementary files: phylogenomics) was used to calculate the number of clusters composing the core genome, pangenome, pairwise shared clusters and strain specific clusters in Python (supplementary table S1, Supplementary Material online). Cluster of Orthologous (COG) and KEGG groups were assigned to each species using DIAMOND v0.8 (e-value 1e−5, Buchfink et al. 2015) and MEGAN6 Community Edition (Huson et al. 2016) using the RefSeq database (accessed: July 5, 2016) clustered at 98% identity with CD-HIT v4.6 (Fu et al. 2012). Pathway tools v19 (Karp et al. 2002) was used to reconstruct, and compare, the metabolism of each Sodalis endosymbiont (supplementary files: pathway-tools-databases).
Table 1.
Genome Features of Several Sodalis Symbionts Ordered by Genome Size
| Organism | Host | Short Name | Accession | Contigs | Genome (Mb) | GC (%) | CDS|ψ | CDS (%) | rRNAs|tRNAs|ncRNAs |
|---|---|---|---|---|---|---|---|---|---|
| Ca. Mikella endobia | Mealybug | MiEn | LN999831 | 1 | 0.35 | 30.6 | 273|7 | 75, 5 | 3|41|6 |
| Ca. Moranella endobia PCVAL | Mealybug | MoEn | NC_021057 | 1 | 0.54 | 43.5 | 411|15 | 76, 2 | 5|41|1 |
| Ca. Moranella endobia PCIT | Mealybug | MoPC | CP002243 | 1 | 0.54 | 43.5 | 406|29 | 77 | 5|41|0 |
| Ca. Hoaglandella endobia | Mealybug | HoEn | LN999835 | 1+2 | 0.64c | 42.8 | 517|16 | 80, 4 | 3|41|10 |
| Ca. Doolittlea endobia | Mealybug | DoEn | LN999833 | 1+1 | 0.85c | 44.2 | 568|99 | 59, 8 | 3|41|11 |
| Ca. Gullanella endobia | Mealybug | GuEn | LN999832 | 1 | 0.94 | 28.9 | 461|29 | 48, 1 | 3|39|7 |
| S-endosymbiont of Heteropsylla cubana | Psyllid | SoHc | NC_018420 | 1 | 1.12 | 28.9 | 532|19 | 47, 3 | 3|38|2 |
| Sodalis-like symbiont of Philaneus spumarius PSPU | Froghopper | SoPSb | BASS01000000 | 562 | 1.38 | 54.1 | 1400|NA | NA | 4|39|44 |
| S-endosymbiont of Ctenarytaina eucalypti | Psyllid | SoCe | NC_018419 | 1 | 1.44 | 43.3 | 758|21 | 47, 9 | 3|40|2 |
| P-endosymbiont of Henestaris halophilus | True bug | SoBaa | PRJEB12882 | 1 | 1.62 | 44.5 | 713|166 | 37, 3 | 3|42|10 |
| Sodalis-like endosymbiont of Proechinophthirus fluctus str. SPI-1 | Seal louse | SoPf | LECR01000000 | 92 | 2.18 | 50 | 695|683 | NA | 16|40|2 |
| Sodalis glossinidius str. “morsitans” | Tsetse fly | SoGl | NC_007712-15 | 1+3 | 4.29c | 54.7 | 3177|1280 | 52, 9 | 22|72|1 |
| Ca. Sodalis pierantonius str. SOPE | Weevil | SOPE | CP006568 | 1 | 4.51 | 56 | 2309|1771 | 46, 2 | 9|55|3 |
| Ca. Sodalis melophagi | Hippoboscid louse fly | SoMeb | http://users.prf.jcu.cz/novake01/d | 236 | 4.57 | 50.8 | 4545|NA | NA | NA |
| Sodalis praecaptivus | Human wound | SoHS | NZ_CP006569-70 | 1 | 4.16c | 57.5 | 4429|25 | 81 | 23|76|1 |
The acronym refers to the proposed name Ca. Sodalis baculum sp. nov. strain kilmister see below. It is introduced here to have a consistent abbreviation in each part.
No annotation available, annotation was done using prokkav1.12 with default parameters plus gram negative and metagenome options (Seemann et al. 2014).
Including plasmids.
Last accessed September 29, 2017.
TyrA Protein Analysis
Tridimensional structure plays a crucial role in protein activity. To predict if TyrA protein of Henestaris endosymbiont is likely to be still functional, its tridimensional (tertiary) structure was modeled with the I-TASSER server (Yang et al. 2015). Putative dimerization (quaternary structure) of TyrA was modeled with the COTH server (Mukherjee and Zhang 2011). Pdb files were viewed, aligned, and compared with UCSF Chimera v1.11.2 (Pettersen et al. 2004) (supplementary files: tyrA_analysis).
Phylogenomic Analysis
A core set of 153 single copy proteins were codon-aligned with a Perl wrapper using MAFFT v7.215 (Katoh et al. 2002), Transeq (EMBOSS: 6.6.0.0, Rice et al. 2000), PAL2NAL v14 (Suyama et al. 2006), and Gblocks v0.91b (codon data with no gaps allowed) (Castresana 2000). Alignments with more than 70% of the columns present in all the species were selected and screened for the saturation of the phylogenetic signal with a custom R script (R Core Team 2016). Briefly, saturation was measured using the correlation coefficient between raw genetic distances and the corrected distances (K80) (supplementary files: phylogenomics). Only protein alignments that showed a coefficient greater than 0.7 at the codon-level were selected for further analysis (77 proteins).
Maximum-Likelihood (ML) phylogenetic tree reconstruction was performed on IQ-TREE v1.5.5 (Nguyen et al. 2015) using ModelTest (Kalyaanamoorthy et al. 2017) with seven partition schemes: 1) a single partition (concatenated alignment), 2) fully partitioned (each protein as a partition) with each partition having its own evolutionary model, 3) as 2) with different branch lengths, 4) as 2) but allowing partition mixing, 5) as 3) but allowing partition mixing, 6) a single partition with JTT + CAT20 (profile mixture models), and 7) partitions obtained in 2) but with CAT20. In addition, a Bayesian posterior consensus tree was inferred with MrBayes v3.2.2 (4 chains, 2,000,000 generations, 1,000 sampling frequency, 1,000 burn-in) (Ronquist et al. 2012). The standard deviation of split frequencies was below 0.01 in the four chains and their convergence was checked with Tracer v1.6. The approximately unbiased (AU) test (Shimodaira 2002) implemented in IQ-TREE was used to select the best possible tree under three partitions model: a single partition, full partitioned and partitioned with a mixing strategy. The selected tree was plotted with Figtree v1.4.3 and modified with InkScape v0.92.
Averaged Nucleotide Identity/Averaged Amino Acid Identity Values Calculation
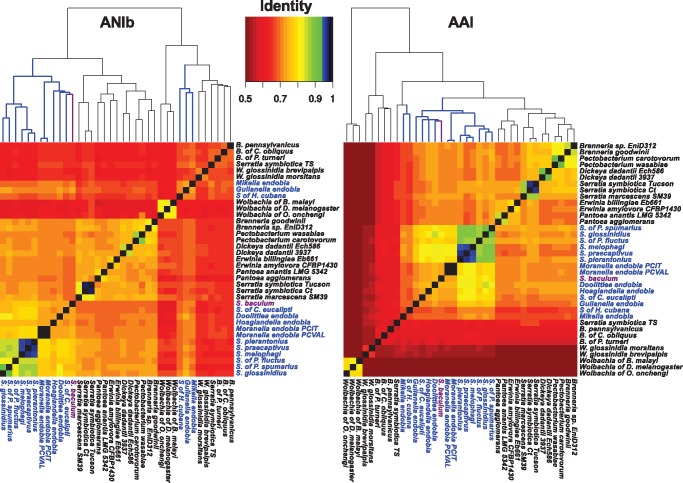
The aforementioned genomes (table 1) plus some phylogenetically related genomes, including some endosymbionts, were downloaded (Dickeya, Pantoea, Serratia, Brenneria, Pectobacterium, Erwinia, Wigglesworthia, and Blochmannia; see supplementary table S2, Supplementary Material online) were used for calculating the averaged nucleotide identity (ANI) and averaged amino acid identity (AAI). Some Wolbachia strains were used as representatives of a non Gammaproteobacteria endosymbiont genus. ANI values were calculated with JSpecies v1.2.1 (Richter and Rosselló-Móra 2009). AAI values were obtained with the enveomics toolbox using USEARCH v9.1 (ublast -id 0.1 -maxhits 1,000 -acceptall -evalue 1e−5 -accel 1) as alignment algorithm (Edgar 2010; Rodriguez-R and Konstantinidis 2016). Heatmaps and hierarchical clustering (Euclidean distances and complete clustering) were produced with the gplots package from R (R Core Team 2016).
Molecular Evolution within the Sodalis Genus
Codeml from PAML v4.7 package (Yang 2007) was used to calculate dS, dN, and their omega ratio (ω) values in the different OCPs. Divergence times between different Sodalis species were standardized using a triplet approach, which utilized the species of interest, one reference Sodalis (S. glossinidius or S. praecaptivus) from the opposite branch of the species selected (see fig. 4 for more details) and Pantoea ananatis as an outgroup. This set-up allowed us to fix the time, in the common branch, from P. ananatis to the Sodalis last common ancestor, making the time because divergence of the Sodalis species equal (e.g. S. melophagi—S. glossinidius—P. ananatis or Sodalis of Heteropsylla cubana—S. praecaptivus—P. ananatis).
Fig. 4.
—Hierarchical clustering of pairwise Average Nucleotide Identity (ANI, left) and Average Amino Acid Identity (AAI, right). Clusters containing Sodalis-allied species are highlighted in blue. SoBa is highlighted in purple. Values greater than 95% start at blue in the color scale.
For each orthologous group in each triplet, three branch models were computed: m0 (one ω), m1 (free ω ratios in each branch) and m2 (two ω setting the species of interest the foreground branch). Each model was computed three times and the iteration with the greater likelihood was stored. The best model was selected using the likelihood ratio test (LTR) and comparing first the m1 against the m2, and the winner against the m0. P-values of LTR tests were corrected using a Bonferroni method (two tests). Python and related scripts are presented in the supplementary files: dNdS_analysis. COG groups were assigned using the output from MEGAN6.
All statistical tests were performed in R. In general, statistical tests were performed on OCPs with ω values below 1, as most of the genes were evolving at this ratio. Only few genes had ω values greater than 1. Some of these values should be taken cautiously, as they can represent alignment artifacts (e.g. open reading frames from fragmented genes in draft genomes). Briefly, raw and log transformed data were checked for normality (Shapiro’s test and QQ-plots) and heteroscestaticity (Levene’s test). Parametric tests were used on normal (or close to normal) and homoscedastic data while nonparametric tests were used in case of heteroscedasticity data. Ordinary Linear Modeling (OLM) was used to detect significant correlation in single Sodalis symbionts. Phylogenetic generalized least squares (PGLS) was used to detect significant correlations across species as it accounts for phylogenetic autocorrelations. All the statistical analyses are presented as an Rmd file (supplementary files: dNdS_analysis).
Results
Bacteriome Characterization
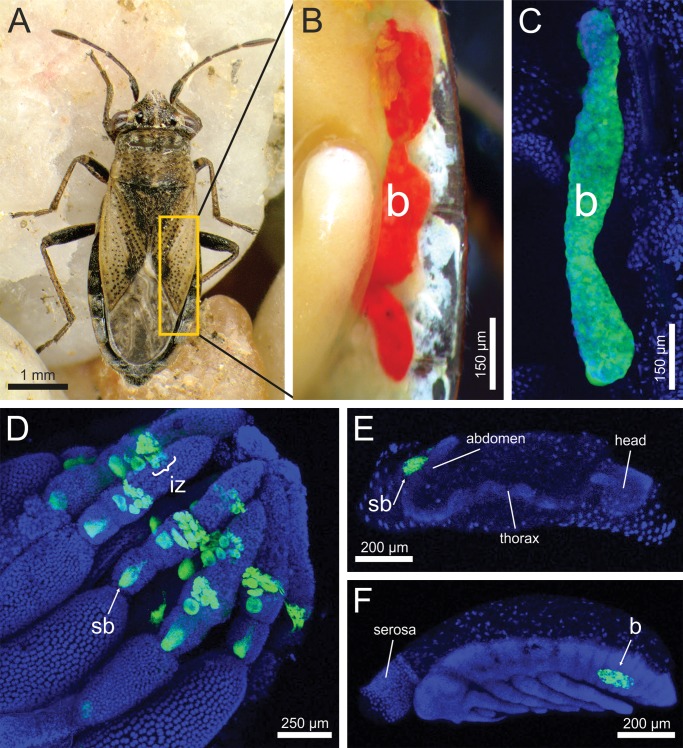
All dissected individuals of Henestaris halophilus (fig. 1A) possessed a pair of elongated, tubular-shaped, red-colored bacteriomes, located on either side of the abdomen adjacent to the gonads (fig. 1B). The bacteriomes extended in adults from the second to the fourth abdominal segment and were subdivided into three sections, not completely separated from each other. Male individuals often presented slender bacteriomes.
Fig. 1.
The endosymbiotic system of Henestaris halophilus (A) Adult female. (B) Dissected bacteriome (b) of tubular shape on the right side of the abdomen. The paired bacteriomes are slightly separated into three parts by contractions. (C) Fluorescence in situ hybridization (FISH) of the Sodalis endosymbiont inside the bacteriome, stained with the specific probe Hen500 (Cy3; green) and SYBR Green I (blue). (D) Extensive signals were also detected in the ovaries. The symbionts are located in ovarial bacteriocytes forming an “infection zone” (iz), where from symbionts are transferred into the developing oocyte by an emerging “symbiont ball” (sb). (E) During early embryogenesis (∼36 h after egg deposition), the symbiont ball is attached to abdomen, followed by infection of the embryo. (F) After katatrepsis, an embryonic back flip within the egg, the symbionts are already located inside the bacteriome.
Fluorescence in situ hybridization (FISH) was used for localization of the H. halophilus endosymbionts. A specific endosymbiont signal was detected in the tubular-shaped bacteriomes (fig. 1C). In addition, fluorescent activity was detected in the ovaries (fig. 1D), where several bacteriocytes formed an infection zone, and in the developing embryos. At the beginning of the embryonic development (∼36 h), a symbiont mass, in general described as a “symbiont ball,” was observed on the anterior pole side of the egg (fig. 1E). After embryonic katatrepsis, the developing bacteriomes were recorded at the same position in the abdomen as described for adults (fig. 1F). Initially, bacteriomes were of spherical shape, but were extended to their final tubular shape during the postkatatrepsis embryonic development (data not shown). These observations strongly indicate that the described endosymbiont is transferred to offspring via vertical maternal transmission.
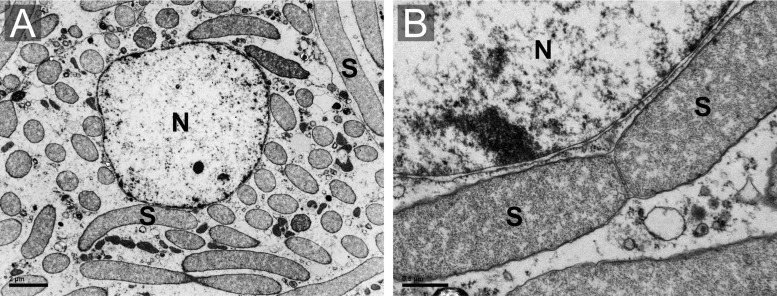
Ultrastructural examinations by electron microscopy (TEM) revealed that the bacteriocytes present a single nucleus and are densely filled with rod-shaped bacteria, presenting the typical gammaproteobacterial structure and three membranes (the bacteria cell wall and a host-derived one) (fig. 2A and B).
Fig. 2.
—Transmission electron microscopy (TEM) micrographs of the Sodalis endosymbiont of Henestaris halophilus. (A) Overwiew of a bacteriocyte completely filled by rod-shaped endosymbiont (S). (B) Enlarged image of dividing endosymbionts showing the characteristic gammaproteobacterial cell structure. Nucleus (N).
Endosymbiont Identification
A 1.5 kb 16S rRNA bacterial gene fragment was amplified by PCR from DNA samples of H. halophilus bacteriomes, derived from geographically distant localities. Cloning and sequencing indicated that all nucleotide sequences are nearly identical (99.6–100%). Comparison with GenBank databases indicated that the bacteriome-associated endosymbiont of H. halophilus is related to the gammaproteobacterial Sodalis cluster (supplementary fig. S1, Supplementary Material online). The 16S rRNA sequence showed the highest similarity (94–95%) to sequences of Sodalis-allied endosymbionts of scale insects from the Coelostomidiidae family and Sodalis-allied endosymbionts of stinkbugs and weevils. The complete 16S rRNA sequence of the H. halophilus bacteriome-associated endosymbiont was obtained by genome sequencing (see below). Sequences of two additional bacteria, Ca. Lariskella arthropodarum and Rickettsia sp. were also detected in the Illumina genomic reads, but with very low coverage. However, no FISH signals of Lariskella and Rickettsia were detected in the analyzed bacteriomes and ovaries (data not shown), suggesting that these endosymbionts might have sporadic appearance or that they are present in H. halophilus in very low amounts.
Comparative Genomics of H. Halophilus Endosymbiont and Related Species
The genome of the bacteriome-associated endosymbiont of H. halophilus was assembled as a single closed circular chromosome with a coverage of 527×. The genome was found to be of intermediate size (1.62 Mb), showed no AT enrichment (45% GC content) and displayed low coding density (37.3%) (table 1). In addition, it presented a reduced number of coding genes (713), pseudogenes (166), no active mobile elements, a single rRNA operon, and a reduced set of tRNA genes (42). Among the pseudogenes, several transcription factors (11), cell wall and transporter genes (29), genes encoding enzymes involved in amino acid and cofactors metabolism (19) or genes related to the replication, transcription and translation machinery (50) were identified (supplementary fig. S2 and table S3, Supplementary Material online). Comparisons against 14 related Sodalis and Sodalis-allied endosymbionts genomes suggested an intermediate to advanced stage of reductive evolution (supplementary table S1, Supplementary Material online).
Three duplicated segments, remnants of two duplication events, of 12, 10, and 2 kb, including the groS–groL operon among other genes, were found. For most of these duplicated genes, one of the duplicated copies is pseudogenized while the other (or the two others, in the case of groS and groL) retains the functionality.
Orthologous clusters of proteins (OCPs) were computed for the Sodalis endosymbiont of H. halophilus, the 14 Sodalis-allied species and P. ananatis (supplementary table S1, Supplementary Material online). The Sodalis core genome, mainly driven from the most reduced Sodalis, harbors 166 OCPs, 75% of them belonging to the J, K, L, and O COG categories (translation, transcription, replication, and post-translational machinery, respectively). Among the other categories, three OCPs were classified as E (amino acid metabolism). From them, two were related to the Fe–S sulfur cluster protein biosynthesis (IscS, SufS) and one to the chorismate pathways (AroK). Three OCPs were classified as H (coenzyme metabolism), including LipA and LipB that compose the complete salvage lipoate pathway, and RibE/H, which is an intermediate reaction in the riboflavin pathway. The rest of OCPs were found to belong to other COG categories (21) or remained without an ascription to a specific COG (12).
The Sodalis endosymbiont of H. halophilus presented 146 strain-specific OCPs, but only three of them were annotated as nonhypothetical proteins: HBA_0606 (DeaD division protein), HBA_0622 (a duplicated GroS), and HBA_0766 (secretion monitor precursor SecM). Most of the hypothetical proteins were short proteins (60 amino acids in average) without hits in the databases used for their annotation (see Supplementary Material online). Also, these proteins were not classified to a COG category. One possibility is that these proteins are open reading frames (ORFs) derived from unrecognizable pseudogenes or small proteins with an unidentified function.
Taxonomy of the Sodalis Clades
A phylogenomic tree, based on 77 single copy core proteins belonging to all 15 analyzed Sodalis species, was obtained. The tree clearly indicated the presence of two main clades, with the two cultivable species of Sodalis, S. glossinidius, and S. praecaptivus, being associated with clade A and clade B, respectively. The Sodalis endosymbiont of H. halophilus was placed in clade B (fig. 3). In addition, no clear association was observed between the phylogeny of the Sodalis species and the taxonomy of their insect hosts (fig. 3).
Fig. 3.
—Phylogenomic tree of several Sodalis and Sodalis-allied species. The two clades used in subsequent analysis are denoted by the letters A (blue) and B (green). Only the best topology found by the AU-test is displayed: ML tree with a single partition schema under JTT+CAT20 model. Node legends denote ML boostrap and Bayesian posterior probabilities; * in Bayesian posterior probabilities denotes alternatives topologies found in MrBayes partitioned reconstruction (S-endosymbiont of H. cubana together with Mikella endobia and Sodalis-like of P. fluctus as a basal clade of S-endosymbiont of C. eucalypti).
The presence of the two cultivable species of Sodalis in different clades made us question the taxonomic status of the other 13 Sodalis-allied species, by utilizing a genome comparison approach (using ANI and AAI methods). As a reference, free-living and endosymbiotic bacterial species, belonging to eight additional genera of Gammaproteobacteria (which are phylogenetically related to Sodalis) and one Alphaproteobacteria (outgroup) were considered. Multiple comparisons indicated that a restrictive threshold of ∼80% (75-81%) AAI, discriminates well between the eight genera used as reference.
When clustering analysis was applied to the ANI/AAI matrices, one large cluster including almost all the Sodalis-allied species was recovered for both, with the exception of the three fast-evolving lineages (Mikella, Gullanella, and S. of H. cubana) (fig. 4). AAI values among the five Sodalis species with the largest genomes: S. glossinidius, Sodalis of P. spumarius (clade A), S. praecaptivus, S. pierantonius, and S. melophagi (clade B), ranged between 85 and 96%. Moreover, the averaged AAI values between the aforementioned five Sodalis species and the remaining Sodalis-allied species (except Mikella and the symbiont of H. cubana), were higher or close to 80%, clearly suggesting that all analyzed species belong to one Sodalis genus. In comparison, AAI values between the Sodalis group and the reference genera showed a range of values lower than 70% AAI.
Applying the 95% ANI threshold (Konstantinidis and Tiedje 2005), the Sodalis endosymbiont of H. halophilus was identified as a new Sodalis species. It also confirmed the strain status of the two M. endobia species and indicated that although S. praecaptivus, S. pierantonius, and S. melophagi are likely to be undergoing a speciation process within their different hosts, they can still be considered strains of the same species.
Based on our phylogenomics and the ANI/AAI results, we propose the name Candidatus Sodalis baculum sp. nov. strain kilmister for the newly described endosymbiont of H. halophilus. The species epitheton refers to the structure of the bacteriome. The slender, tubular-shaped appearance is similar to a baculum (penis bone) that can be found in many placental mammals. The strain name is proposed in honor of the musician Ian “Lemmy” Fraser Kilmister (1945–2015).
Metabolic Capabilities of Candidatus Sodalis Baculum
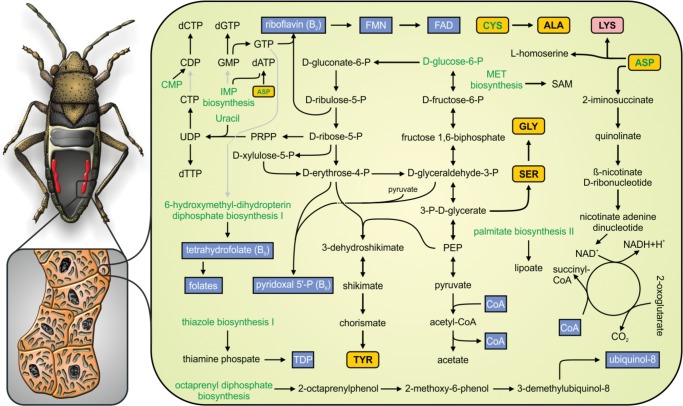
The full metabolism of Ca. Sodalis baculum (hereafter abbreviated as SoBa) was reconstructed (fig. 5). Despite the low coding density of its genome, SoBa still harbors a complete glycolytic pathway and a functional pentose phosphate pathway that produces several intermediate metabolites and reducing agents (NADPH). Furthermore, SoBa is capable of producing its own cell wall, reflected by its rod shaped cell appearance (fig. 2), which is comparable to free-living related species.
Fig. 5.
—Metabolic reconstruction of Ca. Sodalis baculum. Intact pathways are shown in solid black lines, while incomplete ones are shown in gray. Essential, nonessential amino acids and cofactors are shown in rose, yellow, and blue boxes, respectively. Green lettering was applied to biosynthetic steps and precursors that are not executed or formed in the endosymbiont.
In contrast, the SoBa genome does not contain all the required pathways for the synthesis of nucleotides. The genes encoding for enzymes synthesizing inosine monophosphate from 5-phosphoribosyl 1-pyrophosphate (PRPP) have been lost or pseudogenized, while the genes involved in the synthesis of uridine monophosphate from uracil using PRPP have been retained. Consequently, the capability of synthesizing pyrimidines importing/using only uracil is still present, while purines have to be imported from the insect host.
Furthermore, SoBa has lost most of the genes encoding enzymes required for amino acid biosynthesis, limiting its capabilities to the production of five amino acids (alanine, glycine, lysine, serine, and tyrosine). Alanine may be produced in a single step, probably as a byproduct of the transfer of sulfur to tRNAs, from imported cysteine (ABC transporter CydD). The presence of the enzyme glycine/serine hydroxymethyltransferase (encoded by glyA) might reflect an ability to produce glycine from serine or vice versa, but also to produce tetrahydrofolate, which serves as a one-carbon carrier of the biosynthesis of purines and other compounds.
The tyrosine and lysine biosynthetic pathways are present in SoBa (fig. 5). The tyrosine pathway is partially shared by the phenylalanine and tryptophan pathways, but the loss of one phenylalanine and several tryptophan biosynthetic genes significantly reduces the possibility that SoBa is capable of synthesizing these amino acids. The essential amino acid lysine is synthesized using aspartate, which is likely imported from the hosts’ cytosol by the glutamate/aspartate transporter GltP. Although the argD gene encoding succinyldiaminopimelate transaminase is missing, the specific catalytic reaction might be performed by phosphoserine aminotransferase (SerC) as reported in Escherichia coli (Lal et al. 2014). The synthesis of l-homoserine is theoretically possible, but the conservation of this pathway is more likely to be associated with the fact that the thrA and asd genes encoded enzymes are required also in the lysine biosynthetic pathway.
In addition to the biosynthesis of intermediate metabolites and amino acids, SoBa preserves the complete pathways for the biosynthesis of several cofactors and vitamins, such as acetyl-CoA, lipoate, NAD, riboflavin and its derivatives, pyridoxal 5-phosphate (vitamin B6), thiamin diphosphate (TDP), ubiquinol-8, S-adenosyl-l-methionine (SAM), and tetrahydrofolate (vitamin B9). Finally, the SoBa genome also contains the whole Fe–S biosynthesis pathway cluster and is capable of producing glutathione.
Metabolic Pathways Comparisons among Sodalis
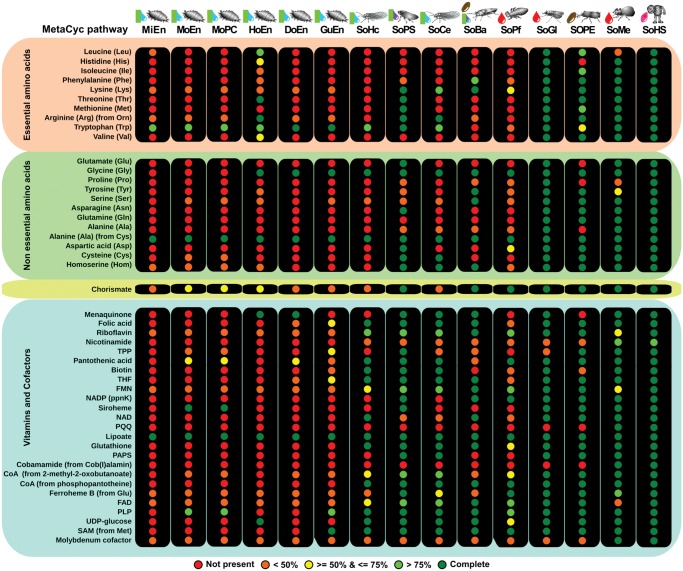
Amino acid and cofactors biosynthetic potential of each Sodalis-allied species was explored at the pathway level (fig. 6). With respect to the ability to synthesize essential amino acids, we found that tryptophan can be produced by all of the Sodalis-allied species of hosts that feed exclusively on plant sap. In hosts feeding on other diets, tryptophan can probably be obtained in other ways, as indicated by the loss of the pathway in SoBa, Sodalis endosymbiont of Proechinophthirus fluctus and S. pierantonius. The lysine biosynthetic pathway, which is present in SoBa, was lost in all Sodalis-allied species present in mealybugs, the psyllid H. cubana and the louse P. fluctus. The Sodalis from psyllids, mealybugs and the froghopper retain some genes that complement the essential amino acid production of their hosts’s primary endosymbionts. Sodalis of P. fluctus cannot produce any essential amino acid, but is still able to produce several vitamins. Only the recently acquired Sodalis maintain the ability to produce most (8 or more) of the 10 essential amino acids, including tryptophan and lysine.
Fig. 6.
—Amino acid and cofactors metabolism of several Sodalis and Sodalis-allied species. Circles represent complete MetaCyc pathways colored according to their completeness. Sodalis species are ordered by genome size (increasing order). See table 1 for organism acronyms.
The analysis of the synthesis of nonessential amino acids in Sodalis indicated that Sodalis-allied species with reduced genomes only rarely synthesize these amino acids. Moreover, the nonessential amino acids that are produced are probably byproducts of essential pathways for the symbiotic relationship. This phenomenon could be explained by the settlement of these endosymbionts in the host environment, acquiring most of the amino acids from their hosts’ cytosol. Tyrosine biosynthesis was found to be conserved only in SoBa, while all the other Sodalis-allied species with reduced genomes have lost this ability. In addition, a functional chorismate biosynthetic pathway was only detected in Sodalis-allied species that are capable of producing tryptophan, tyrosine, or phenylalanine (fig. 6).
Cofactors and vitamins biosynthesis pathways are mainly lost in the Sodalis of mealybugs with the exception of lipoate. The rest of the Sodalis species show the conservation of a larger cofactor/vitamins biosynthetic potential, with Sodalis of H. cubana, Sodalis of Ctenarytaina eucalypti, and SoBa being an exception. Comparisons to other Sodalis outside the mealybug group showed that SoBa has lost the ability to synthesize some important cofactors such as panthotenate, biotin, and siroheme. On the other hand, the riboflavin pathway is maintained in SoBa, while other Sodalis species, with a broad range of genome sizes and diets, are likely to have lost it (fig. 6).
Patterns of Molecular Evolution in the Sodalis Genus
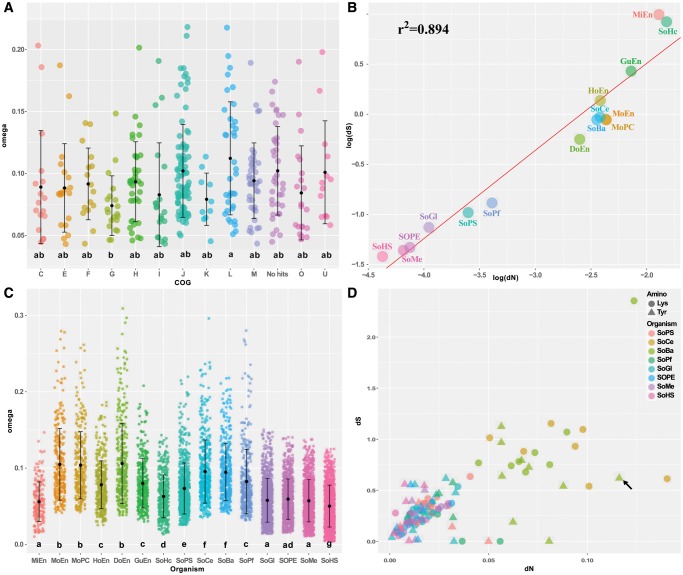
The evolutionary trends of the different COG categories in SoBa were analyzed. According to their ω values, the fastest evolving COG categories were L (Replication) and J (Translation), while the slowest evolving category was G (Carbohydrates metabolism) (fig. 7A).
Fig. 7.
—Molecular evolution in different Sodalis species. (A) SoBa omega single gene values across different COG groups. (B) dN/dS correlation across different Sodalis lineages. Each dot represents the median dN/dS of all analyzed genes in each Sodalis lineage. (C) Omega single gene values across the different Sodalis species. (D) Scatter plot showing the dN and dS values for the OCPs belonging to the Tyr and Lys pathways in several Sodalis. The tyrA gene of SoBa is denoted by a black arrow. Only OCPs with omega values below 1 were used on (A) and (C). Lowercase letters in (A) and (C) represent the statistical significant groups obtained. Regression line on (B) was calculated using the PGLS method under a Brownian model of evolution.
When all the Sodalis species were compared, on average, the values of both dN and dS were very different among lineages although the evolutionary time of all branches was forced to be identical (see Materials and Methods for more details). Relatively to the free-living S. praecaptivus, four major groups were identified (fig. 7B): Sodalis lineages that evolve at nearly similar rate as S. praecaptivus (S. glossinidius, S. melophagi, S. pierantonius, Sodalis of P. fluctus, and Sodalis of P. spumarius), those evolving at medium accelerated rate (Doolittlea, Moranella PCVAL, Moranella PCIT, SoBa, Sodalis of C. eucalypti), and two groups showing high (Gullanella) and very high substitution rates (Mikella and Sodalis of H. cubana). A strong positive linear relationship, on both linear (not shown, see supplementary files: dNdS_analysis) and log–log scales (PGLS P-value < 0.05, r2 = 0.89, fig. 7B), exists between the average genomic dS and dN values. However, most of the linear relationships between dN and dS values of individual genes in lineages with highly reduced genomes were nonsignificant. A slightly different picture was observed when dN and dS values of individual genes were obtained from free-living (S. praecaptivus) and Sodalis with genomes larger than 1 Mb. Although the linear relationships were positive and significant in most of these Sodalis (OLM P-value <0.05), the variance explained by the linear models was higher in free-living and recently acquired endosymbionts (e.g. S. praecaptivus r2 = 0.34, S. glossinidius, r2=0.24; see supplementary files: dNdS_analysis), than the variance explained in endosymbionts with a longer relationships with their hosts (e.g. SoBa r2=0.05, Sodalis of C. eucalypti r2=0.04), possibly suggesting the presence of a general mechanism affecting both substitution rates simultaneously and some slight effect of natural selection on synonymous codon usage in the free-living and less reduced genomes (see discussion part below).
Although the averaged ω were significantly different between the various Sodalis (fig. 7C), most of the genes showed ω values below 1 (purifying selection), while only 232 genes had ω values greater than 1 (positive or relaxed selection). Most of the genes with an ω greater than 1 were present in the Sodalis with larger genomes (see supplementary files: dNdS_analysis). It should be noted that some of the ω > 1 values need to be interpreted carefully. These genes presented low dS values (203 genes with dS values below 0.01) which produced the high ω values reported (greater than 10), which can reflect calculation/alignment artifacts. For example, only five of the 17 genes with ω > 1 in SoBa presented dS values greater than 0.01 and ω values lower than 10: slyA, pdxB, manX, nadE, and mreD. Details of the conducted analyses are presented in the supplementary files: dnds_analysis, Supplementary Material online.
Evolution of the Tyrosine and Lysine Pathways in Sodalis
Genes from the tyrosine and lysine biosynthetic pathways showed different evolutionary patterns (fig. 7D and supplementary files: dNdS_analysis, Supplementary Material online). Genes from the lysine pathway presented a positive and significant linear relationship between dN and dS values across Sodalis species (PGLS P-value <0.05, r2 = 0.95), but also within most of the analyzed species (e.g. OLS SoBa P-value <0.05, r2=0.77), suggesting the action of some mechanisms (maybe synergistic to natural selection) acting on both nonsynonymous and synonymous changes for pathway conservation. This is supported by the data from Sodalis of P. fluctus and C. eucalypti, which have an incomplete lysine biosynthetic pathway and present a negative, although no significant, linear relationship between dN and dS (higher accumulation of dN than dS).
In contrast, genes from the tyrosine pathway presented a positive and significant linear relationships between dN and dS values across species (PGLS P-value <0.05, r2 = 0.96) but only few positive significant correlations were found within species (S. melophagi and S glossinidius). SoBa presented a nonsignificant negative correlation between dN and dS for the tyrosine pathway. From all the genes in this pathway, the tyrA gene of SoBa was identified as an outlier, showing a larger dN value than all other tyrosine biosynthetic genes (fig. 7D, black arrow). Under the possibility that this pathway is being lost in SoBa, we tested if the predicted accumulation of nonsynonymous substitutions in tyrA of SoBa could affect the protein functionality. For that, comparisons of TyrA 3 D structures of SoBa, S. pierantonius and S. praecaptivus were performed (supplementary fig. S3 and files tyrA_analysis, Supplementary Material online). The active pocket, composed by a set of β-sheets, was found to be maintained in all compared TyrA proteins. The N-terminal region was found to be highly polymorphic between the three species. Specific differences were detected at the end of the PDH domain and at the C-terminal region of the TyrA enzyme of SoBa, when compared with both the S. pierantonius and S. praecaptivus enzymes. Moreover, comparisons of the C-terminal region of the TyrA enzyme of SoBa to that of E. coli, S. pierantonius, and S. praecaptivus, indicated that only in SoBa, this region contains more changes relatively to the rest of the protein (28% versus 12% on average, window size of 50 amino acids). Some mutations previously described in E. coli were detected in TyrA of SoBa. Among these, we identified two mutations that are expected to reduce the inhibition of the enzyme by Tyr (A354T and F357C) and one mutation that is expected to interfere with the binding of the inhibitor Tyr (Y303C). Despite these differences, our 3D predictions suggested that TyrA of SoBa is still capable of forming an active quaternary (homodimeric) structure (supplementary fig. S3 and files: tyrA_analysis, Supplementary Material online).
Discussion
The Bacteriome-Associated Endosymbiosis of H. halophilus
This work presents the first molecular characterization of a bacteriome-associated symbiotic system harbored by the lygaeoid bug Henestaris halophilus. Our phylogenetic and genomic characterization revealed that the endosymbiont belongs to the genus Sodalis (Gammaproteobacteria). Sodalis-allied species were already found to be associated with different heteropteran taxa, especially in species of the superfamily Pentatomoidea (Hosokawa et al. 2015). Moreover, it is generally assumed that these symbiotic associations are common in stinkbugs and are of facultative nature, as infection rates are usually found to be low (less than 15% of individuals harboring the Sodalis symbiont, with the exception of the family Urostylididae which shows 95% infection rate) (Hosokawa et al. 2015). It is important to note that until now, no Sodalis-allied symbionts were detected in the superfamily Lygaeoidea (Kikuchi et al. 2011; Hosokawa et al. 2015), but this could be related to the low number of species screened so far. In addition, it is not clear if the bacteriome-associated symbiosis we found in H. halophilus is a singularity within the Henestarinae subfamily (∼20 species). No bacteriome or any Sodalis-allied endosymbionts could be detected in the sister species H. laticeps, although this species is morphologically similar to H. halophilus and can be jointly found in the same habitats. One possibility of course is that the bacteriome was lost in H. laticeps and that other uncharacterized Henestarinae species harbor bacteriome-associated symbiosis systems as well. Further screening of additional Henestarinae species is likely to provide more insights on this currently remaining “open issue.”
Genome Reduction in the Sodalis Endosymbiont of H. halophilus
The genome of SoBa displays several typical features of endosymbionts that are in an intermediate genome reduction stage: genome size below 2 Mb, no AT bias, and low coding density. Some other characteristics of the SoBa genome were found to be closer to genomes of endosymbionts that are in an advanced reduction stage: reduced set of protein coding and tRNA genes, one rRNA operon, only two annotations of potentially transposase pseudogenes and one phage integrase (Toft and Andersson 2010). Analyses of four newly established endosymbiont species (S. praecaptivus, S. melophagi, S. pierantonius, and S. glossinidius) suggested that the putative free-living ancestral Sodalis genome (i.e. before the switch to an intracellular life-stage), should have been larger than 4 Mb and had a GC content >50%.
In contrast to the facultative species S. glossinidius (Toh et al. 2006; Belda et al. 2010) or the recent-obligatory species S. pierantonius (Oakeson et al. 2014), the number of pseudogenes in the SoBa genome is estimated to be very low. However, due to the low coding density in the genome, it could be possible that the intergenic regions of SoBa contain DNA from some pseudogenes that have lost their nucleotide identity to other orthologous genes during the long evolutionary period. Once these regions will be lost, the size of the SoBa genome will probably drop below 700 kb with a coding density higher than 70%, as observed in other advanced endosymbiont systems (table 1) (Moran et al. 2008; Moya et al. 2008).
A clear indication that the reductive evolution process is in an intermediate stage in SoBa comes from the presence of two duplicated functional copies of the groS and groL genes and one groL pseudogene. Endosymbionts that are in an advanced stage of genome reduction, such as Buchnera, contain only one copy of each gene (Shigenobu et al. 2000). In contrast, endosymbionts in an ongoing genome reduction process could contain more than one copy, such as in S. pierantonius (Oakeson et al. 2014). The presence of pseudogenes also supports our argument on the ongoing genome reduction process in SoBa. As found before in other endosymbionts that went through a genome reduction process, parts of DNA replication and repair machinery (topoisomerase IV, uvrABC, recA, and rarA), transcription factors, energy production (cyoABE), specific transporters, and components of cell wall are lost (supplementary table S3, Supplementary Material online). For example, we found that a key gene in the synthesis of Kdo-lipid A and several genes in the lipid A-core synthesis were lost or pseudogenized, leading to a less virulent capacity, an important feature of mutualistic endosymbiotic life (Toft and Andersson 2010).
Finally, the OCPs comparisons indicates that SoBa is only losing genetic material instead of gaining, as 565 from the 711 protein coding genes in the SoBa genome are shared with other Sodalis species. Moreover, 98% of the strain specific clusters were found to be hypothetical proteins. Taken together, these findings reinforce the hypothesis that the SoBa gene content is just a subset of its free-living ancestor (Silva et al. 2001).
New Hints on the Sodalis-Allied Species Relationships
Our phylogenomics analysis suggested that the Sodalis-allied species can be divided into two major clusters, without clear signals of cospeciation events. This is in agreement with the recently reported phylogenomic analysis of Husník and McCutcheon (2016), which showed that Sodalis-allied strains of mealybugs do not form a monophyletic group but are found interspersed in different clades harboring also Sodalis present in psyllids and spittlebugs, clearly suggesting multiple Sodalis acquisitions in different insect lineages.
In addition, we followed Richter and Rosselló-Móra (2009) and used ANI/AAI values, complemented with phylogenomics, for endosymbionts taxonomic classification. ANI/AAI values are computational methods that show a strong correlation with the DNA–DNA hybridization technique used so far to define bacterial species (Konstantinidis and Tiedje 2005; Goris et al. 2007). Using Enterobacteriaceae genomes, comparisons of free-living and symbiotic species from the same genus placed the threshold (for within genus similarity) to ≥ ≈80% AAI (fig. 4), with the exception of the closely related genera of Brenneria and Pectobacterium. Moreover, Wolbachia or Serratia, two genera that contain endosymbionts, showed values similar to those observed here for Sodalis (fig. 4). Our results strongly suggest that all the Sodalis-allied species analyzed in this work belong to the same genus and therefore, should be renamed accordingly (e.g. Ca. Sodalis mikella) (Dale and Maudlin 1999). Alternatively, designation of the genus followed by the name of its insect host (e.g. Sodalis endosymbiont of Paracoccus marginatus) could also be considered (Ramírez-Puebla et al. 2015; Lindsey et al. 2016).
Ca. Sodalis Baculum as a Mutualistic Endosymbiont
The metabolic capacities of SoBa suggest an important role in complementing its host diet. Among the amino acids synthesized by SoBa, two large pathways have been preserved for the production of the essential amino acid lysine and the nonessential amino acid tyrosine. The most plausible reason why natural selection has preserved the lysine and tyrosine pathways, in spite of the strong reductive evolution, is that large amounts of these amino acids are required for the insect host, at least in some period of its life cycle. While the high requirements for lysine cannot be compensated by the insect metabolism, tyrosine may be directly synthesized by the insect phenylalanine hydroxylase if the substrate phenylalanine is available in sufficient amounts (PAH, E.C. 1.14.16.1). In insects, the metabolism of tyrosine is involved in, at least, three types of physiological processes: neurotransmission, melanin formation and sclerotization (cuticle hardening). For the latter, large amounts of several dopamine derivatives are required. These compounds act as cross-linking agents of cuticular proteins through their covalent binding to amino acid residues of these proteins (Andersen 2010; Suderman et al. 2010). The requirement for high amounts of lysine may also be related to the hardening of the cuticle, as lysine, potentially present in H. halophilus cuticular proteins, is known to be involved in creating adducts between cuticular proteins and dopamine derivatives (Suderman et al. 2010).
Higher tyrosine quantities are likely to be needed for sclerotization, as was demonstrated in the pea aphid Acyrthosiphon pisum, where the endosymbiont Buchnera delivers precursors such as phenylpyruvate and phenylanalanine, which are converted by the insect metabolism to tyrosine (Rabatel et al. 2013). The RNAi-mediated disruption of the insect phenylalanine hydroxylase activity produces, among other effects, an impairment in embryonic development which may not be compensated by Buchnera as it does not have the ability to synthesize tyrosine (Simonet et al. 2016). The endosymbiont S. pierantonius also provides its weevil host with phenylalanine and tyrosine, needed for the production of catecholamines involved in cuticle synthesis (Wicker and Nardon 1982; Oakeson et al. 2014; Vigneron et al. 2014).
Following the argument of a high tyrosine demand by H. halophilus, our results suggest a similar strategy to the one reported for Buchnera in A. pisum. In Buchnera, the prephenate dehydratase PheA (a related TyrA enzyme) shows a feedback inhibition insensitiveness to phenylalanine (Jiménez et al. 2000). The prephenate dehydrogenase TyrA have the same regulatory mechanism, being its function inhibited by high tyrosine concentrations. The allosteric inhibition region in TyrA was reported to be in the C-terminus using E. coli mutation analysis (Chen et al. 2003; Lütke-Eversloh and Stephanopoulos 2005; Raman et al. 2014). Interestingly, the prephenate dehydrogenase gene (tyrA) from SoBa presents higher nonsynonymous substitution rates, mainly at its C-terminus, compared with other genes from the tyrosine biosynthetic pathway. Changes in this region could cause the SoBa prephenate dehydrogenase to be continuously active at high tyrosine concentrations, due to the loss of the allosteric inhibition, producing high amounts of this amino acid.
Amino Acids and Cofactors Production in the Sodalis Genus
In general, the ability of the different Sodalis species to synthesize amino acids and cofactors is correlated with their genome sizes and their symbiotic status (primary, coprimary, or secondary). Pathways in which more than 75% of the reactions still appear to be functional in the endosymbiont are likely to be complemented by the host cells, as observed in several symbiotic systems (e.g. Wilson et al. 2010), or by a symbiotic partner (if present). However, when complementation takes place between two bacterial symbionts, many different combinations of a shared pathway can evolve (Sloan and Moran 2012; Husník et al. 2013; Koga and Moran 2014; Husník and McCutcheon 2016). As expected, recent acquired Sodalis present the most complete set of metabolic pathways, and this is independent of their symbiotic status (primary or secondary) (Toh et al. 2006; Oakeson et al. 2014; Nováková et al. 2015). SoBa has a metabolic potential close to the coprimary Sodalis from psyllids and the seal louse P. fluctus (Sloan and Moran 2012; Boyd et al. 2016) with some specific signatures: the ability to produce lysine, tyrosine, and riboflavin. SoBa is the only endosymbiont with reduced genome that is able to produce tyrosine and, with the exception of Sodalis from P. spumarius, also the amino acid lysine. As indicated earlier, these two amino acids are likely to play an important role in H. halophilus–endosymbiont interaction.
It has been demonstrated that the provisioning of riboflavin by endosymbiotic bacteria is essential to aphid’s growth (Nakabachi and Ishikawa 1999). Moreover, the ability to provision riboflavin has likely played a major role in the establishment of Ca. Serratia symbiotica as a coprimary endosymbiont in some aphid lineages (Manzano-Marín et al. 2016). In this context, it is interesting to note the presence of a complete riboflavin biosynthetic pathway (including yigB) in SoBa. This is in contrast to Sodalis of mealybugs, psyllids and cicadas where the pathway is almost lost or incomplete (fig. 6). The possibility of complementation by the insect host (by horizontally acquired genes) or by an endosymbiotic partner in mealybgus, psyllids, and cicadas cannot be ignored, although so far, no yigB or ybjI orthologous genes have been reported yet in these groups (Husník et al. 2013; Sloan et al. 2014; Husník and McCutcheon 2016). In any case, the ability to produce riboflavin might have played an important role, in addition to the ability to produce lysine and tyrosine, in the establishment of the H. halophilus–SoBa relationship.
We notice that lipoate, an essential cofactor in many oxidative reactions, including pyruvate decarboxylation, but also an important antioxidant (Spalding and Prigge 2010; Cronan 2016), is present in all the Sodalis analyzed. Lipoate can be acquired by de novo biosynthesis or by scavenging (Spalding and Prigge 2010). Maintenance of both pathways has been proposed as a signature of pathogenic (if a lipoamidase is present) or gut-associated bacteria, which scavenges lipoate only when it is available from the environment (Spalding and Prigge 2010). As many other endosymbionts, most Sodalis produce de novo lipoate from acetyl-CoA (fatty acids biosynthesis pathway), or other intermediate metabolites (Mikella and Hoaglandella use acetoacetyl). Recently acquired Sodalis present both the biosynthetic and the scavenging pathways, while only the biosynthetic one is maintained in Sodalis endosymbionts with reduced genomes. It therefore seems that the loss of the scavenging pathway together with the lpd gene, reflects in the Sodalis genus, a change from a putative pathogenic or gut-associated bacteria to a mutualistic endosymbiont. This way, the competition with the host/mitochondria for lipoate is avoided both by maintaining the ability to de novo synthetize lipoate and by losing the ability to exploit the host lipoate by scavenging (Spalding and Prigge 2010).
Molecular Evolutionary Trends in the Sodalis Genus
The overall ω values in the different Sodalis species (0.05-0.11) indicated a strong effect of natural (purifying) selection for preserving the amino acid sequences of the retained genes. Our analysis indicated large (and significant) differences among lineages in dS and dN values. The averaged values of these parameters were highly correlated, although this correlation was not extended within each lineage to individual genes, except for recently acquired Sodalis species. Correlation in individual genes suggests a selection of synonymous codon usage in highly expressed genes, which have been almost completely lost in Sodalis species with longer times of coevolution with their hosts. The large differences among nucleotide divergence rates in different Sodalis species and the correlation between averaged dN and dS values may be explained by among-lineage differences in: 1) the efficiency of the replication and repair machineries: the diversity, concentration, error rate, and activity of DNA replication and repair enzymes, 2) the endosymbiont generation time: species with shorter generation times are expected to have larger rates of mutations per year because the larger numbers of DNA replications per unit of time generate larger numbers of mutations, and 3) the control of the endosymbiont by its host cell: mutations in genes coding for enzymes involved in replication and repair may be compensated by the import of host-encoded enzymes (Silva and Santos-Garcia 2015).
Conclusions
Based on the structure of the H. halophilus bacteriome and the phylogenetic placement of its endosymbiont Sodalis baculum, the symbiosis of H. halophilus can be typified as a rare event within the Lygaeoidea. Based on the low coding density and several other evolutionary characteristics of the S. baculum genome, it can be concluded that it is still on an ongoing genome reduction process. Sodalis baculum is not only the first Sodalis to be described in lygaeoid bugs, but is also the first Sodalis, within heteropteran insects, that may hold a mutualistic relationship with its host, mainly supplying tyrosine, lysine, and some cofactors. Finally, our results allow us to propose the reunification of all the Sodalis-allied species known to date into a single genus.
Supplementary Material
Supplementary Material includes supplementary Material and Methods, supplementary figures S1 to S3 and supplementary tables S1 to S3 and is are available at Genome Biology and Evolution online. Supplementary Files Supplementary Files can be found at http://dx.doi.org/10.17632/n38gkjry35.1
Authors’ Contributions
S.M.K. conceived the study, collected the insect material, designed, and performed the microscopic experiments and characterized the endosymbiont. F.J.S. designed the molecular evolutionary analysis. D.S-G. performed the bioinformatic analysis. D.S-G. and S.M. designed the statistical analysis. S.M.K., F.J.S., and D.S.-G. analyzed the data and wrote the manuscript with inputs from S.M. K.D. advised in the experimental design and contributed with material and reagents. All authors participated in the revision of the manuscript.
Supplementary Material
Acknowledgments
We thank Peter Göricke and Peer Schnitter for their assistance in insect collection, Stefan Geimer and Rita Grotjahn for their help in electron microscopy, Nico Jacob and Laura Schneider for supporting rearing experiments and imaging analysis and two anonymous reviewers for their suggestions on early drafts of the manuscript. D.S.-G. is recipient of a PostDoctoral Fellowship from The Hebrew University of Jerusalem. This publication was funded by the German Research Foundation (DFG) and the University of Bayreuth in the funding progamme Open Access Publishing.
Literature Cited
- Andersen SO. 2010. Insect cuticular sclerotization: a review. Insect Biochem Mol Biol. 40(3): 166–178. [DOI] [PubMed] [Google Scholar]
- Arp A, Munyaneza JE, Crosslin JM, Trumble J, Bextine B.. 2014. A global comparison of Bactericera cockerelli (Hemiptera: Triozidae) microbial communities. Environ Entomol. 43(2): 344–352. [DOI] [PubMed] [Google Scholar]
- Belda E, Moya A, Bentley S, Silva FJ.. 2010. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11: 449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, et al. 2016. Two bacterial genera, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus fluctus (Phthiraptera: Anoplura). Appl Environ Microbiol. 82(11): 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1): 59–60. [DOI] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of Animals With Plant Microorganisms. New York (NY): Interscience Publishers. [Google Scholar]
- Burke GR, Normark BB, Favret C, Moran NA.. 2009. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 75(16): 5328–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4): 540–552. [DOI] [PubMed] [Google Scholar]
- Chen S, Vincent S, Wilson DB, Ganem B.. 2003. Mapping of chorismate mutase and prephenate dehydrogenase domains in the Escherichia coli T-protein. Eur J Biochem. 270(4): 757–763. [DOI] [PubMed] [Google Scholar]
- Chrudimský T, Husník F, Nováková E, Hypša V.. 2012. Candidatus Sodalis melophagi sp. nov.: phylogenetically independent comparative model to the tsetse fly symbiont Sodalis glossinidius. PLoS One 7(7): e40354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, et al. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8(11): e1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. 2016. Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol Mol Biol Rev. 80(2): 429–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Maudlin I.. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol. 49(1): 267–275. [DOI] [PubMed] [Google Scholar]
- Dale C, Young SA, Haydon DT, Welburn SC.. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A. 98(4): 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19): 2460–2461. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23): 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, et al. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl Environ Microbiol. 73(20): 6660–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, et al. 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 57(Pt 1): 81–91. [DOI] [PubMed] [Google Scholar]
- Gruwell ME, Hardy NB, Gullan PJ, Dittmar K.. 2010. Evolutionary relationships among primary endosymbionts of the mealybug subfamily phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl Environ Microbiol. 76(22): 7521–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P.. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci U S A. 96(12): 6814–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebsch H. 1961. Faunistisch-ökologische Untersuchungen an den Salzstellen bei Hecklingen und westlich der Numburg mit Angaben über die Biologie von Henestaris halophilus (Burm.) [Dissertation]. Universität Halle.
- Hosokawa T, Kaiwa N, Matsuura Y, Kikuchi Y, Fukatsu T.. 2015. Infection prevalence of Sodalis symbionts among stinkbugs. Zoological Lett. 1: 5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V.. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9: 87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153(7): 1567–1578. [DOI] [PubMed] [Google Scholar]
- Husník F, McCutcheon JP.. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A. 113(37): E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, et al. 2016. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 12(6): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez N, González-Candelas F, Silva FJ.. 2000. Prephenate dehydratase from the aphid endosymbiont (Buchnera) displays changes in the regulatory domain that suggest its desensitization to inhibition by phenylalanine. J Bacteriol. 182(10): 2967–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa N, et al. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol. 76(11): 3486–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa N, et al. 2011. Bacterial symbionts of the giant jewel stinkbug Eucorysses grandis (Hemiptera: Scutelleridae). Zool Sci. 28(3): 169–174. [DOI] [PubMed] [Google Scholar]
- Kaiwa N, et al. 2014. Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr Biol. 24(20): 2465–2470. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14(6): 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley S, Romero P.. 2002. The pathway tools software. Bioinformatics 18(Suppl 1): S225–S232. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14): 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T.. 2011. An ancient but promiscuous host–symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5(3): 446–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA.. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8(6): 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM.. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 102(7): 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler S, Dettner K, Kehl S.. 2010. Molecular characterization and localization of the obligate endosymbiotic bacterium in the birch catkin bug Kleidocerys resedae (Heteroptera: Lygaeidae, Ischnorhynchinae). FEMS Microbiol Ecol. 73: 408–418. [DOI] [PubMed] [Google Scholar]
- Kuechler S, Dettner K, Kehl S.. 2011. Characterization of an obligate intracellular bacterium in midgut epithelium of bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae). Appl Environ Microbiol. 77(9): 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler SM, Renz P, Dettner K, Kehl S.. 2012. Diversity of symbiotic organs and bacterial endosymbionts of lygaeoid bugs of the families Blissidae and Lygaeidae (Hemiptera: Heteroptera: Lygaeoidea). Appl Environ Microbiol. 78(8): 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal PB, Schneider BL, Vu K, Reitzer L.. 2014. The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol Microbiol. 94(4): 843–856. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9): 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey ARI, Bordenstein SR, Newton ILG, Rasgon JL.. 2016. Wolbachia pipientis should not be split into multiple species: a response to Ramírez-Puebla et al., “Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola’, ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups”. Syst Appl Microbiol. 39(3): 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Stephanopoulos G.. 2005. Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: generation and characterization of tyrosine-insensitive mutants. Appl Environ Microbiol. 71(11): 7224–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Simon J-C, Latorre A.. 2016. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera – Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol. 8(5): 1440–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, et al. 2014. Bacterial symbionts of a devastating coffee plant pest, the stinkbug Antestiopsis thunbergii (Hemiptera: Pentatomidae). Appl Environ Microbiol. 80(12): 3769–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, et al. 2012. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. 6(2): 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42: 165–190. [DOI] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A.. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 9(3): 218–229. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Zhang Y.. 2011. Protein–protein complex structure prediction by multimeric threading and template recombination. Structure 19(7): 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H.. 1999. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 45(1): 1–6. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E, Hypša V.. 2007. A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett. 269(1): 131–135. [DOI] [PubMed] [Google Scholar]
- Nováková E, Husník F, Šochová E, Hypša V.. 2015. Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl Environ Microbiol. 81(18): 6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 6(1): 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 25(13): 1605–1612. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2016. R: A Languaje and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Autria.
- Rabatel A, et al. 2013. Tyrosine pathway regulation is host-mediated in the pea aphid symbiosis during late embryonic and early larval development. BMC Genomics 14: 235.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Rogers JK, Taylor ND, Church GM.. 2014. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci U S A. 111(50): 17803–17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Puebla ST, et al. 2015. Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola’, ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups. Syst Appl Microbiol. 38(6): 390–399. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6): 276–277. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 106(45): 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT.. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. Peer J Prepr. 4: e1900v1. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3): 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree ZL, Kambhampati S, Moran NA.. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A. 106(46): 19521–19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh RT, Slater JA.. 1995. True Bugs of the World (Hemiptera: Heteroptera): Classification and Natural History. Ithaca (NY: ): Cornell University Press. [Google Scholar]
- Seemann T.. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14): 2068–2069. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H.. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407(6800): 81–86. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3): 492–508. [DOI] [PubMed] [Google Scholar]
- Silva FJ, Latorre A, Moya A.. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17(11): 615–618. [DOI] [PubMed] [Google Scholar]
- Silva FJ, Santos-Garcia D.. 2015. Slow and fast evolving endosymbiont lineages: positive correlation between the rates of synonymous and non-synonymous substitution. Front Microbiol. 6: 1279.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet P, et al. 2016. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci Rep. 6: 34321.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA.. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 29(12): 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31(4): 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WA, et al. 2013. Phylogenetic analysis of symbionts in feather-feeding lice of the genus Columbicola: evidence for repeated symbiont replacements. BMC Evol Biol. 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AK, McMillen CM, Wallenhorst P, Rio RV.. 2011. The phylogeny of Sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol Lett. 317(2): 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Prigge ST.. 2010. Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev. 74(2): 200–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Kost C, Kaltenpoth M.. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25(5): 375–390. [DOI] [PubMed] [Google Scholar]
- Suderman RJ, Dittmer NT, Kramer KJ, Kanost MR.. 2010. Model reactions for insect cuticle sclerotization: participation of amino groups in the cross-linking of Manduca sexta cuticle protein MsCP36. Insect Biochem Mol Biol. 40(3): 252–258. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue): W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft C, Andersson SG.. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 11(7): 465–475. [DOI] [PubMed] [Google Scholar]
- Toh H, et al. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16(2): 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, et al. 2014. Insects recycle endosymbionts when the benefit is over. Curr Biol. 24(19): 2267–2273. [DOI] [PubMed] [Google Scholar]
- Wachmann E, Melber A, Deckert J.. 2007. Wanzen 3 – Tierwelt Deutschlands. Keltern: Goecke and Evers. [Google Scholar]
- Wicker C, Nardon P.. 1982. Development responses of symbiotic and aposymbiotic weevils Sitophilus oryzae L. (Coleoptera, Curculionidae) to a diet supplemented with aromatic amino acids. J Insect Physiol. 28(12): 1021–1024. [Google Scholar]
- Wilson AC, et al. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 19: 249–258. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8): 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2015. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12(1): 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.