Abstract
All eukaryotes have three essential nuclear multisubunit RNA polymerases, abbreviated as Pol I, Pol II and Pol III. Plants are remarkable in having two additional multisubunit RNA polymerases, Pol IV and Pol V, which synthesize noncoding RNAs that coordinate RNA-directed DNA methylation for silencing of transposons and a subset of genes. Based on their subunit compositions, Pols IV and V clearly evolved as specialized forms of Pol II, but their catalytic properties remain undefined. Here, we show that Pols IV and V differ from one another, and Pol II, in nucleotide incorporation rate, transcriptional accuracy and the ability to discriminate between ribonucleotides and deoxyribonucleotides. Pol IV transcription is considerably more error-prone than Pols II or V, which may be tolerable in its synthesis of short RNAs that serve as precursors for siRNAs targeting non-identical members of transposon families. By contrast, Pol V exhibits high fidelity transcription, similar to Pol II, suggesting a need for Pol V transcripts to faithfully reflect the DNA sequence of target loci to which siRNA–Argonaute silencing complexes are recruited.
INTRODUCTION
In all eukaryotes, three nuclear multisubunit RNA polymerases are essential for viability: RNA Polymerase I (Pol I), which synthesizes precursors for the three largest ribosomal RNAs, Pol II, which transcribes thousands of mRNAs and noncoding RNAs, and Pol III, which is primarily required for 5S ribosomal RNA and tRNA biogenesis. Remarkably, plants have two additional nuclear RNA polymerases, Pol IV and Pol V that are each composed of 12 subunits (1), at least seven of which are shared with Pol II (2–5). In the siRNA-directed DNA methylation (RdDM) pathway, which primarily silences transposons, viruses and transgenes, Pols IV and V have non-redundant functions. Pol IV partners with RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) to generate short double-stranded RNAs that are then diced into 24 nt siRNAs (6–8). These siRNAs are incorporated into an Argonaute family protein, primarily AGO4, and guide cytosine methylation and repressive chromatin modifications at loci transcribed by Pol V (9–11). There is also evidence that RNA-directed DNA methylation at sites of Pol V transcription can be guided by 21 nucleotide siRNAs derived from degraded transposon mRNAs or by undiced Pol IV and RDR2-dependent RNAs (12–15).
Pols IV and V apparently have fewer constraints on their evolution than other polymerases, allowing their subunit compositions to vary (5) and their catalytic subunits to experience amino acid substitution rates that are ten to twenty times greater than for Pol II (3). More than 140 amino acid positions that are invariant in the catalytic subunits of Pols I, II, and III have diverged in Pols IV and V (Supplementary Figure S1A and B and Table S1) (16). These include substitutions and deletions within elements that are thought to be critically important for polymerase function, including the trigger loop and bridge helix (Supplementary Figure S1A) (17). In Pol II and other polymerases, conformational changes in the trigger loop and bridge helix result in the transition from an open state, which allows nucleotide triphosphate (NTP) entry into the catalytic center, to a closed state in which the NTP has been positioned adjacent to the 3′ end of the nascent RNA chain to enable phosphodiester bond formation (18,19). Ratchet-like transitions between the open and closed conformations are thought to be linked to RNA translocation, affecting elongation rate as well as the accuracy (fidelity) of NTP incorporation (20,21).
Despite deletions and substitutions within the trigger loop, bridge helix, and other conserved domains, Pols IV and V are functional and have been shown to synthesize RNA in vitro (22). However, their accuracy and speed, relative to Pol II, has not been studied. Here, we report initial measurements of several parameters of Pol II, Pol IV and Pol V catalytic activity. We show that Pols IV and V catalyze RNA synthesis more slowly than Pol II and we show that Pol IV transcription is notably error-prone. Surprisingly, Pol V is less error-prone than Pol II, at least in terms of misincorporating ribonucleoside triphosphates (rNTPs) mismatched to the DNA template. However, both Pol IV and Pol V exhibit reduced ability, relative to Pol II, to discriminate between ribonucleosides and deoxyribonucleosides. The implications of Pol IV and Pol V’s enzymatic properties are discussed with respect to what is known about their functions.
MATERIALS AND METHODS
Amino acid sequences for the largest and second-largest subunits of RNA polymerases I, II, III, IV and V, from multiple species, were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). Sequences were aligned using Clustal Omega (23,24).
Pols II, IV, and V were immunoprecipitated from leaf tissue of 3 week old transgenic lines expressing FLAG epitope-tagged RNA polymerase subunits, NRPB2-FLAG (Pol II), NRPD1-FLAG (Pol IV), or NRPE1-FLAG (Pol V), as previously described (22). Leaf tissue was ground to a powder in liquid nitrogen using a mortar and pestle and then resuspended in 3.5 ml extraction buffer (20 mM Tris–HCl, pH 7.6, 300 mM sodium sulfate, 5 mM magnesium sulfate, 5 mM DTT, 1 mM PMSF, 1% plant protease inhibitor (Sigma) per gram of tissue. The homogenate was subjected to centrifugation at 16 000 × g for 15 min, 4°C. The supernatant was collected and subjected to a second round of centrifugation using the same conditions. Fifty microliter anti-FLAG agarose resin (Sigma) was added to each lysate and incubated at 4°C for 3 h on a rotating mixer. The resin was washed twice with 10 ml of wash buffer (20 mM Tris–HCl, pH 7.6, 300 mM sodium sulfate, 5 mM magnesium sulfate, 5 mM DTT, 1 mM PMSF, 0.5% IGEPAL CA-630 detergent) and once with 10 ml CB100 (25 mM HEPES–KOH, pH 7.9, 20% glycerol, 100 mM KCl, 1 mM DTT, 1 mM PMSF). Immunoprecipitated polymerases were used immediately for in vitro transcription reactions. Pol II immunoprecipitated from 1 g of leaf tissue expressing NRPB2-FLAG was sufficient for 20 in vitro transcription reactions, whereas 4 g of NRPD1-FLAG or NRPE1-FLAG leaf tissue was needed for a single reaction. Pols II, IV and V purified by FLAG tag-enabled affinity capture are free of cross-contamination by one another, or other polymerases, as determined by mass spectrometry (22).
in vitro transcription reactions were conducted using a 17 nt RNA primer hybridized to various 32 nt ssDNA oligos, as previously described (22), with minor modifications. RNA primers (2 μM) were end-labelled using T4 polynucleotide kinase and γ-32P-ATP, and excess γ-32P-ATP was removed using Performa spin columns according to the manufacturer's protocol (Edge Bio). RNA–DNA hybrid templates were generated by combining equimolar amounts of end-labelled RNA primer and unlabeled DNA template in 1× annealing buffer (100 mM potassium acetate, 30 mM HEPES–KOH, pH 7.5), placed in a boiling water bath, and allowed to cool to room temperature. Template sequences are provided in Supplementary Table S2.
50 μl of resuspended, washed polymerase, still bound to affinity resin, was used for each transcription reaction. Washed NRPB2-FLAG resin (Pol II) was resuspended in 1 ml CB100 buffer, enough for 20 reactions. Washed Pol IV and Pol V resins were resuspended in a final volume of 50 μl per transcription reaction. Fifty microliters of 2× transcription reaction mix containing 21.7 μl of the 250 nM, annealed RNA-DNA template solution, 100 μM (unless otherwise noted) high purity rNTPs (GE Healthcare), 120 mM ammonium sulfate, 40 mM HEPES–KOH pH 7.9, 24 mM magnesium sulfate, 20 μM zinc sulfate, 20 mM DTT, 20% glycerol and 1.6 U/μl Ribolock (Thermo Fisher Scientific) was then added. In rNTP:dNTP discrimination experiments, the total NTP concentration was 100 μM. The high purity NTPs (GE Healthcare) used in the reactions are free of other NTPs or RNAse activity; use of high purity NTPs that are not cross-contaminated by other NTPs is important for misincorporation assays (25). Reactions were incubated at room temperature for 1 h on a rotating mixer. Reactions were desalted using Performa spin columns according to the manufacturer's protocol (Edge Bio), then precipitated with 1/10 volume 3 M sodium acetate, 20 μg GlycoBlue (Thermo Fisher Scientific) and an equal volume of isopropanol. Pellets were resuspended in 5 μl RNA gel loading dye (47.5% formamide, 0.01% SDS, 0.01% bromophenol blue, 0.005% xylene cyanol, 0.5 mM EDTA), heated at 70°C for 3 min, and subjected to electrophoresis on a 15% polyacrylamide, 7 M urea sequencing gel.
Km* and rate assays were performed similar to in vitro transcription assays described above, with minor modifications. Transcription was initiated by adding 50 μl 2× transcription reaction mix containing 0.1 μM of the first nucleotide complementary to the template (rGTP), and incubation for 20 min. The elongation complexes were then washed with 800 μl 1X transcription buffer lacking NTPs (60 mM ammonium sulfate, 20 mM HEPES–KOH pH 7.9, 12 mM magnesium sulfate, 10 μM zinc sulfate, 10 mM DTT, 10% glycerol). 0.8 U/μl of RNase inhibitor was added to washed elongation complex resin. For Km* reactions, 100 μl washed resin was distributed to 1.5 ml tubes containing appropriate volumes of the next complementary nucleotide, or a non-complementary nucleotide, to achieve the substrate concentration being tested for that nucleotide. Reactions were incubated at room temperature, on a rotating mixer, for 30 min. For rate assays, 500 nM of the complementary nucleotide (UTP) was added to the washed elongation complexes and the reactions were incubated at room temperature for the range of times indicated in the figure. Reactions were cleaned and analyzed as described for in vitro transcription assays above.
To determine error rates for Pol II or Pol IV transcripts generated from M13mp18 single-stranded DNA (Bayou Biolabs) as the template, transcription reactions were conducted as previously described, but in a transcription reaction mix consisting of 7.5 nM M13mp18 (Bayou Biolabs), 1 mM ATP, 1 mM GTP, 1 mM CTP, 1 mM UTP, 60 mM ammonium sulfate, 20 mM HEPES–KOH pH 7.9, 12 mM magnesium sulfate, 10 μM zinc sulfate, 10 mM DTT, 10% glycerol, and 0.8 U/μl Ribolock (Thermo Fisher Scientific).
Reaction products were purified using Performa spin columns according to the manufacturer's protocol (Edge Bio), then precipitated with 1/10 volume 3 M sodium acetate, 20 μg GlycoBlue (Thermo Fisher Scientific) and an equal volume of isopropanol. Pellets were resuspended in 5 μl nuclease-free water and DNase treated using a Turbo DNA-free kit (Thermo Fisher Scientific) according to the manufacturer's protocol. RNAs were then treated with RNA 5′ pyrophosphohydrolase (RppH from New England Biolabs) to convert 5′-end triphosphates to 5′ monophosphates. Reactions were cleaned with Oligo Clean & Concentrator columns (Zymo Research) according to the manufacturer's protocol. Without any fragmentation, RNAs were circularized with RNA ligase 1 (NEB, M0204S) according to the manufacture's guidelines. Circularized RNA templates were then reverse transcribed in a rolling-circle reaction according to the protocol described by Acevedo et al., with the exception that the incubation time at 42°C was extended from 2 to 20 min (26,27). Second strand synthesis and the remaining steps for the library preparation were then performed using a NEBNext Ultra RNA Library Pre Kit for Illumina (E7530L) and the NEBNext Multiplex Oligos for Illumina (E7335S, E7500S), according to the manufacturer's protocols. A size selection for amplified products longer than 300 nt was performed before sequencing and 300 nt single-end reads were then generated using an Illumina HiSeq instrument. Following the autocorrelation-based method and Bayesian approach described by Lou and Hussman et al., the structure of repeats within a read was identified and the consensus sequence of a repeat was constructed (28). Because of the random-priming approach used for rolling-circle reverse transcription, the 5′ end of the consensus sequence can be any nucleotide of the circularized RNA template. To reorganize the consensus sequence and make the ends correspond to the 5′ and 3′ of the original RNA transcript, we first constructed a tandem duplicate of the consensus sequence and mapped it back to the M13mp18 reference by BWA (29). Therefore, the longest continuous mapping region of the duplicated consensus sequence corresponds to the original RNA transcript. Because mapping can be ambiguous at the first and last few nucleotides, we excluded the 4 nucleotides at each end of the reorganized consensus sequence prior to subsequent transcript analyses to minimize potential false positives. The reconstructed consensus sequence was then mapped to the M13mp18 reference sequence, with transcription errors called for mismatches present in tandem copies of the RNA, and a frequency of mismatch no larger than 1%. The data were deposited in NCBI with the BioProject Number PRJNA393568.
RESULTS
Accuracy of Pol II, IV and V transcription
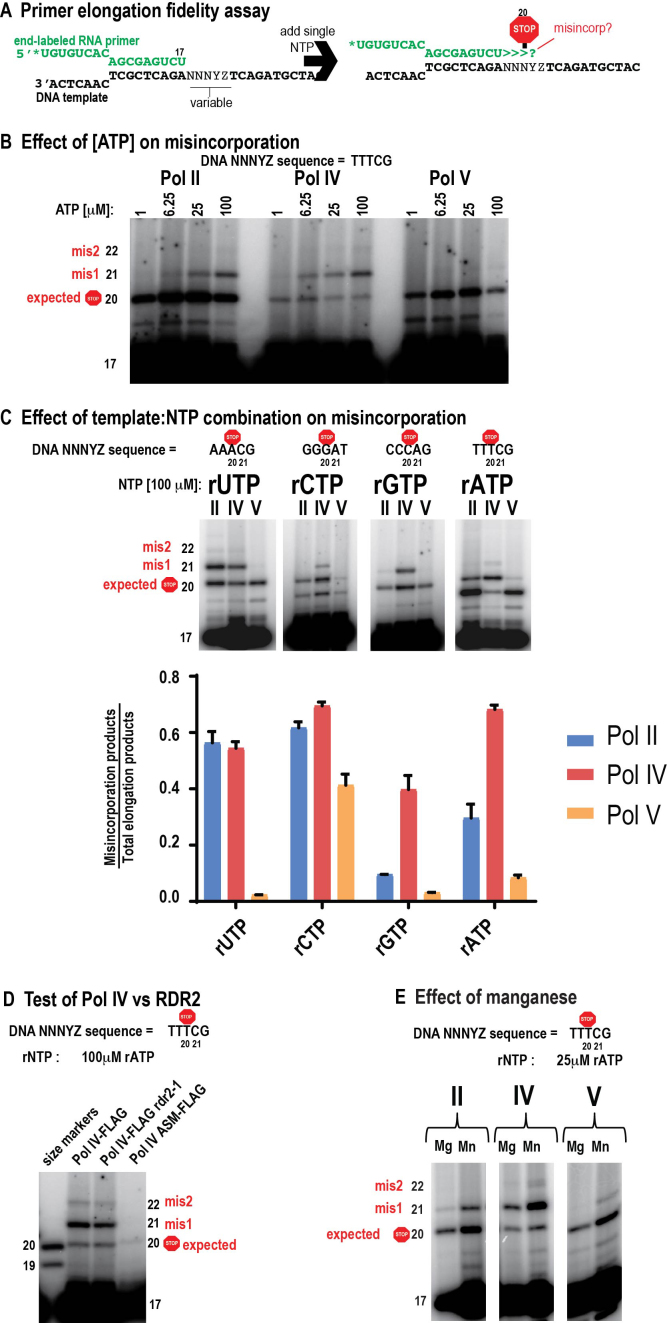
Affinity-purified Pols II, IV or V were tested in assays in which a 32 nt DNA template is annealed to a 17 nt RNA whose 3′ end is complementary to the DNA template, yielding a 9 bp DNA-RNA hybrid (Figure 1A). The RNA serves as a primer that can be elongated in a templated fashion by all three polymerases (22). The single-stranded DNA templates have three identical nucleotides located immediately adjacent to the 3′ end of the primer such that addition of a single rNTP allows elongation of the RNA by 3 nt, to a length of 20 nt. Generation of 21 nt, or longer, RNAs indicates misincorporation of the NTP across from non-complementary nucleotides of the template. Importantly, the assay depends on the use of high purity (HPLC-purified) NTPs to avoid false-positive signals resulting from contamination by other NTPs (25). Use of a single-stranded DNA template circumvents complications due to differences in the abilities of Pols II, IV and V to displace the non-template strand of duplex DNA (22).
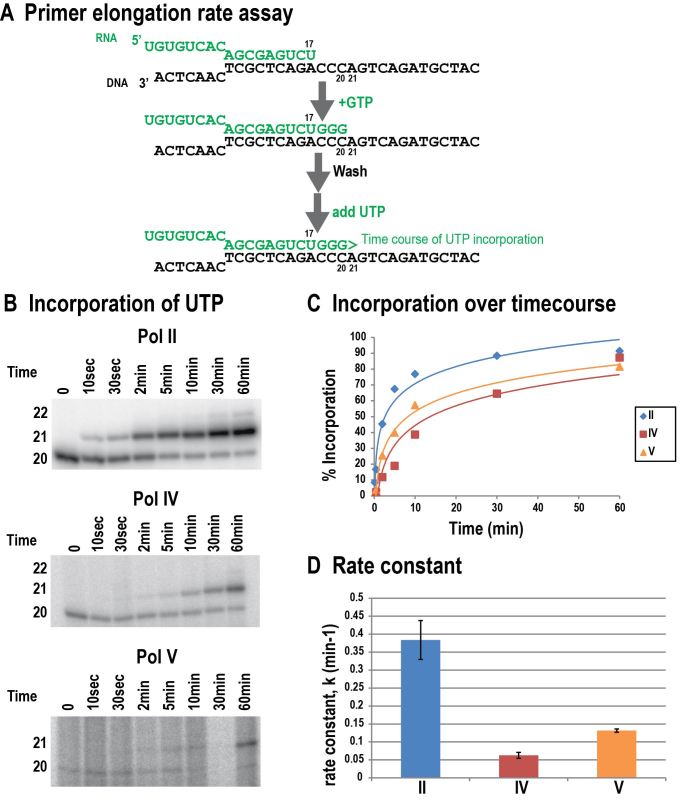
Figure 1.
Pol IV and Pol V have altered fidelities relative to Pol II. (A) Design of the assay. (B) Pol II, IV, and V primer elongation products visualized by phosphorimaging following electrophoresis on a 15% PAGE gel. (C) Effect of NTP: DNA template combination on Pol II, IV, or V misincorporation. (D) Primer elongation fidelity assay comparing Pol IV immunoprecipitated from a FLAG-tagged NRPD1 transgenic line in which RDR2 co-immunoprecipitates with Pol IV (Pol IV-FLAG), Pol IV immunoprecipitated from an rdr2 null mutant line (Pol IV-FLAG, rdr2–1), or Pol IV immunoprecipitated from a transgenic line expressing an active site mutant form of the NRPD1 subunit (ASM-FLAG). (E) Effect of manganese in the transcription assay, in place of magnesium.
In primer elongation reactions involving adenosine incorporation, templated by Ts in the DNA, Pols II, IV, and V primarily synthesize the expected 20 nt RNA products when the ATP concentration is low (1 μM) (Figure 1B). However, as the rATP concentration is increased (6.25, 25 or 100 μM), RNAs of 21 and 22 nt are synthesized as a result of misincorporating adenosine across from the ensuing cytosine (misincorporation event 1; mis1) or guanosine (misincorporation event 2) bases of the DNA template. Comparing the ratio of properly arrested (20 nt) to misincorporation products (>20 nt), reveals that Pol IV generates the most misincorporation products, and Pol V the fewest (Figure 1B). Subsequent tests comparing misincorporation frequency in the presence of 100 μM UTP, rCTP, rGTP or rATP confirmed that Pol IV is considerably more error-prone than Pols II or V, and that Pol V is the least error-prone of the three enzymes; this was true for all template and NTP combinations tested (Figure 1C). The results also reveal that misincorporation varies considerably depending on the template-NTP combination, particularly for Pol II.
The RNA-dependent RNA polymerase, RDR2, physically associates with Pol IV (22,30) and might plausibly contribute to NTP misincorporation. To test this possibility, we compared Pol IV isolated from wild-type RDR2 plants, Pol IV isolated from a rdr2–1 null mutant, and Pol IV that is inactivated as a result of clustered point mutations in the Metal A site of the catalytic center yet expressed in a wild-type RDR2 background (16). Pol IV isolated from the rdr2–1 mutant misincorporated rNTPs to the same extent as Pol IV isolated from wild type plants, whereas the Pol IV active site mutant lacked significant activity (Figure 1D). Based on these controls, we conclude that Pol IV, and not RDR2, is responsible for the RNA transcripts generated in the assays.
Magnesium ions are important for NTP positioning at the active site of RNA and DNA polymerases such that substitution by bulkier manganese ions typically makes RNA and DNA polymerases error-prone (31,32). We tested whether Pol IV and Pol V active sites are similarly sensitive to manganese. Indeed, substituting manganese for magnesium in the reaction buffer substantially increased nucleotide misincorporation by Pols IV and V, as for Pol II (Figure 1E). These results suggest that the catalytic centers of Pols IV and V are similar to Pol II in their sensitivity to manganese ions.
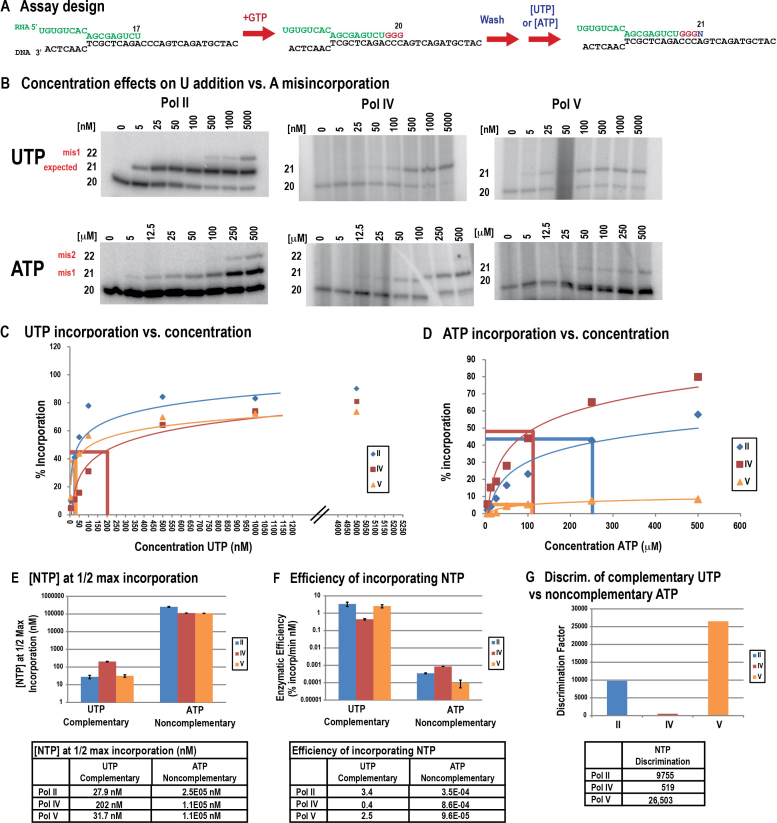
To assess the contribution of nucleotide selectivity to Pol II, IV and V transcriptional fidelity, we compared the relative affinities of the enzymes for complementary versus non-complementary nucleotides during RNA elongation (18). For these assays, the 17 nt RNA primer was annealed to a template having CCCAG as the variable sequence downstream of the primer, such that addition of 0.1 μM rGTP resulted in polymerase-engaged elongation complexes that contain 20 nt RNAs (Figure 2A). Importantly, no misincorporation into 21 nt or longer products is detected using this low rGTP concentration (compare to the 0 μM UTP reactions in Figure 2B; see also Supplementary Figure S2). The elongation complexes were then washed to remove templates not engaged by the resin-immobilized RNA polymerases, as well as free rGTP, and were then incubated with UTP, the nucleotide complementary to the next template position, or with rATP, which is non-complementary. Production of 21 nt or longer extension products was then monitored over a range of rNTP concentrations, from 0 to 5 μM (5000 nM) for the complementary nucleotide (UTP) or 0 to 500 μM for the non-complementary nucleotide (rATP) (Figure 2B). The band intensity for elongation products that are 21 nt or longer was then divided by the value of the total intensity for all products of 20 nt or longer and this ratio (expressed as %) was plotted versus UTP or rATP concentration (Figure 2C and D). From these plots, we can obtain an estimated pseudo Km (Km*; considered a pseudo Km because we are not measuring velocity in these fixed-time reactions) as well as maximum incorporation (Imax) values, using linear regression to fit the data to the equation y = (Imax*x)/(Km*+x), as in the study of Wang et al. (18). Figure 2E shows the estimated Km*s for Pols II, IV and V, for both complementary and non-complementary rNTPs. All three polymerases have much higher (∼1000-fold) estimated Km*s for the non-complementary (rATP) versus complementary ribonucleotide (UTP), indicating that the polymerases have much higher affinities for the correct versus incorrect rNTP. Pol II and Pol V have similar Km*s for the complementary rNTP (28 nM and 32 nM, respectively), but Pol IV has a much higher Km* (202 nM), suggesting decreased affinity for the correct rNTP compared to Pols II or V (Figure 2E). For the non-complementary nucleotide, Pol IV and Pol V have similar estimated Km*s (∼112 μM) that are ∼2 fold lower than for Pol II (252 μM) (Figure 2E).
Figure 2.
Differences in complementary versus non-complementary nucleotide incorporation contributes to differences in Pol IV and Pol V fidelities, relative to Pol II. (A) Overview of the primer elongation assay. (B) Representative gels showing the incorporation of the complementary nucleotide (UTP) by Pols II, IV and V over a range of UTP concentrations. (C) Percent incorporation, calculated as the intensity of products of 21 and 22 nt divided by the total intensity of products 20 nt or longer) is plotted versus nucleotide concentration, with linear regression used to fit the curves to the equation y = (Imax*x)/(Km*+x). (D) Percent incorporation (intensity of 21 and 22 nt products divided by the total intensity of products 20 nt and longer) plotted versus nucleotide concentration, with linear regression used to fit the curves to the equation y = (Imax*x)/(Km*+x). (E) Km* values (nucleotide concentration at 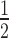 maximum incorporation (Imax)) of Pols II, IV and V for complementary and non-complementary NTPs. Data is representative of two replicate experiments (see Supplementary Figure S2). (F) Efficiency of nucleotide incorporation (Imax/Km*). (G) Discrimination factor, calculated as complementary NTP efficiency/non-complementary NTP efficiency.
maximum incorporation (Imax)) of Pols II, IV and V for complementary and non-complementary NTPs. Data is representative of two replicate experiments (see Supplementary Figure S2). (F) Efficiency of nucleotide incorporation (Imax/Km*). (G) Discrimination factor, calculated as complementary NTP efficiency/non-complementary NTP efficiency.
Pol IV’s decreased affinity for the correct rNTP and increased affinity for a non-complementary NTP is consistent with Pol IV’s propensity for misincorporation, as shown in Figure 1. Pol V, on the other hand, has a Km* for the complementary ribonucleotide that is similar to Pol II, but has a lower Km* for the non-complementary ribonucleotide, suggesting that Pol V has a higher affinity for the wrong nucleotide compared to Pol II. This was unexpected given that Pol V produces fewer misincorporation products than Pol II in the fidelity assays of Figure 1. The explanation for this apparent paradox comes from considering enzymatic efficiency, which in conventional Michaelis–Menten analyses is estimated by dividing Vmax (the concentration of substrate at which the enzymes active site is saturated) by the Km, substituted in our case by Imax and Km* (Figure 2F). Pol II and Pol V are similarly efficient at incorporating the complementary nucleotide, whereas Pol IV is relatively inefficient. By contrast, Pol IV is the most efficient at incorporating the non-complementary ribonucleotide, whereas Pol V is least efficient. Collectively, these experiments indicate that although Pol V has a higher affinity for the non-complementary rNTP compared to Pol II (Figure 2E), it has a lower Imax (Figure 2D) such that the overall efficiency of incorporating a wrong nucleotide is lower than for Pol II (Figure 2F). Pol V’s 3-fold increased propensity to incorporate correct versus incorrect nucleotides, compared to Pol II, and Pol IV’s 19-fold decreased ability compared to Pol II (Figure 2G), are consistent with the misincorporation results of Figure 1.
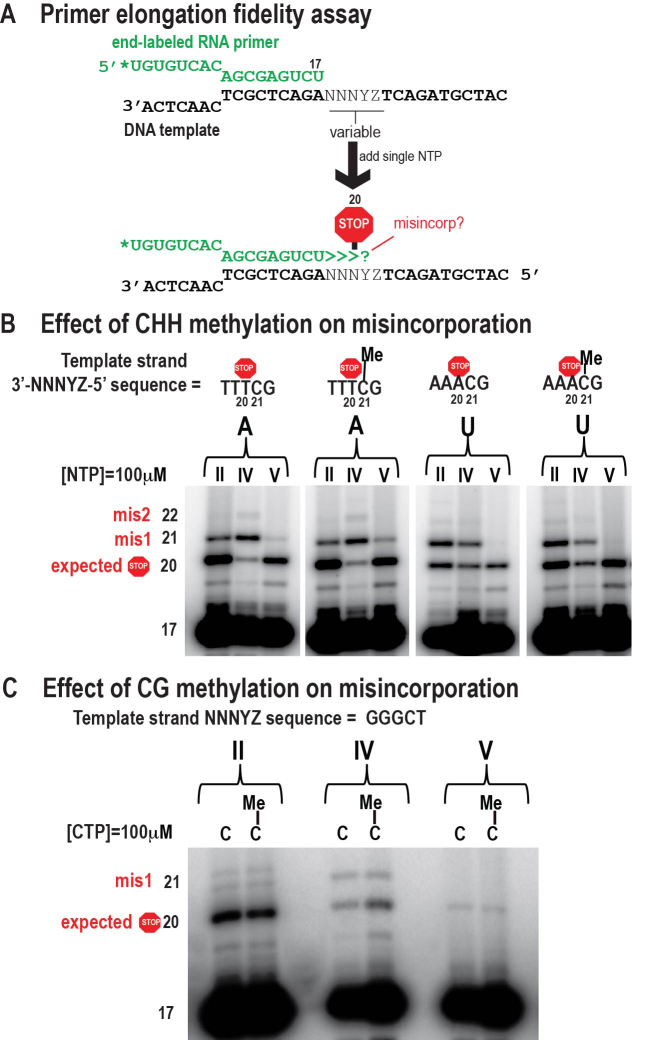
We tested whether ribonucleotide misincorporation is enhanced by cytosine methylation. Comparing methylated and unmethylated DNA template oligonucleotides in primer elongation experiments (Figure 3A), we observed no increase in misincorporation of rATP, UTP or rCTP caused by methylation of template cytosines, in either 5′ meCHH or 5′ meCG sequence contexts, compared to unmethylated template cytosines (Figure 3B and C; note that template sequences are written 3′ to 5′). These results suggest that misincorporation of rNTPs at methylcytosine positions of the template is not an intrinsic property of Pols II, IV or V.
Figure 3.
Effect of DNA template cytosine methylation on Pol II, IV or V transcriptional fidelity. (A) Overview of primer elongation assay. (B) Tests for effects of cytosine methylation in the CHH context (5′-CTT-3′ or 5′-CAA-3′) on ATP or UTP misincorporation. (C) Tests of cytosine methylation in the CG context (5′ to 3′) on CTP misincorporation.
Discrimination between rNTPs and dNTPs
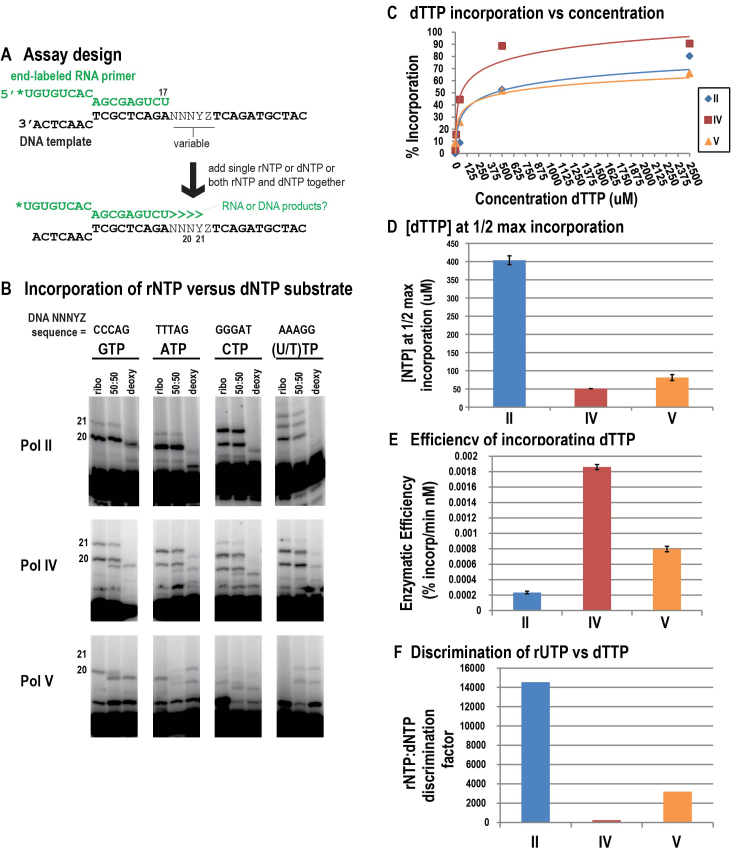
To test the abilities of Pol IV and Pol V to discriminate between ribo- and deoxyribo-nucleoside triphosphates, primer elongation experiments were conducted in the presence of 100% rNTP, 100% dNTP or a 50:50% mix of rNTP and dNTP (Figure 4A). Primers that are elongated by incorporating dNTPs migrate faster than rNTP-elongated primers when subjected to electrophoresis on 15% denaturing polyacrylamide sequencing gels (Figure 4B). When provided a 50:50 mix of rNTP and dNTP, Pol II makes products whose mobility is the same as when only rNTP is provided, demonstrating a strong preference for rNTPs over dNTPs (Figure 4B). Pol IV prefers rNTPs, but dNTP elongation products are also detected in reactions containing equal amounts of both types of NTP (Figure 4B). Pol V, surprisingly, displays similar incorporation of rNTPs or dNTPs when either is provided alone, and preferentially incorporates the dNTPs when provided with a 50:50 mix (Figure 4B). These results show that Pols IV and V have a reduced ability, compared to Pol II, to discriminate between rNTPs and dNTPs. In order to better understand the basis for this loss of discrimination, we determined the affinities of Pols II, IV and V for a dNTP in the same way that rNTP affinity was assessed in Figure 2 (Supplementary Figure S3A and B). We found that Pols IV and V have increased affinities, relative to Pol II, for dNTPs (Figure 4C–F).
Figure 4.
Pols IV and V misincorporate dNTPs to a greater extent than Pol II. (A) Assay design. (B) Primer elongation assays in the presence of only rNTP, only dNTP, or a 50:50 rNTP:dNTP mix. (C) Percent incorporation (calculated by dividing the intensity of products resulting from incorporation of dTTP (21 nt) by the total intensity of products 20 nt and longer) is plotted versus nucleotide concentration, with linear regression used to fit the curves to the equation y = (Imax*x)/(Km*+x). (D) Pseudo Km's (Km*, nucleotide concentration at 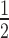 maximum incorporation (Imax)) of Pols II, IV and V for dTTP. Note that Pols IV and V have higher affinity for dTTP than Pol II. (E) Efficiency at incorporating dTTP is calculated as Imax/Km*. Note that Pols IV and V are more efficient at incorporating dTTP than Pol II, with Pol IV being the most efficient. (F) rNTP:dNTP discrimination factor, calculated as UTP efficiency (see Figure 2)/dTTP efficiency. Pol II is over 60 times better than Pol IV and 4.5 times better than Pol V at discriminating between UTP and dTTP. Note that the increased incorporation of dNTPs by Pols IV and V relative to Pol II appears to result from their increased affinity for the dNTP.
maximum incorporation (Imax)) of Pols II, IV and V for dTTP. Note that Pols IV and V have higher affinity for dTTP than Pol II. (E) Efficiency at incorporating dTTP is calculated as Imax/Km*. Note that Pols IV and V are more efficient at incorporating dTTP than Pol II, with Pol IV being the most efficient. (F) rNTP:dNTP discrimination factor, calculated as UTP efficiency (see Figure 2)/dTTP efficiency. Pol II is over 60 times better than Pol IV and 4.5 times better than Pol V at discriminating between UTP and dTTP. Note that the increased incorporation of dNTPs by Pols IV and V relative to Pol II appears to result from their increased affinity for the dNTP.
Rates of nucleotide incorporation
Relative rates of nucleotide incorporation for Pols II, IV and V were assessed by elongating a 17 nt primer to 20 nt RNA using a low concentration of GTP as in Figure 2 (Figure 5A), then conducting a time-course of 21 nt RNA production upon addition of 500 nM UTP (Figure 5B). The percentage of 21 nt product, relative to total 20 + 21 nt product, was plotted against time (Figure 5C) and the data were fitted to the equation c(t) = A × (1 – exp[-k – t]) to calculate the rate constant, k, as described in Sydow et al. (25). Pol IV and Pol V both have decreased rates of nucleotide incorporation relative to Pol II (Figure 5D). Pol IV is the slowest of the three enzymes, with a rate that is ∼6 times slower than Pol II (k values of 0.06 versus 0.38, respectively), and Pol V is ∼3 times slower than Pol II (k values of 0.13 versus 0.38, respectively) (Figure 5D).
Figure 5.
Relative nucleotide incorporation rates of Pols II, IV and V. (A) Assay design. (B) Representative gels showing the incorporation of UTP by Pols II, IV and V versus time (UTP concentration = 500 nM). (C) Percent incorporation (intensity of elongation products divided by total products) versus time, with linear regression used to fit the curves to the equation c(t) = A × (1 – exp[–k × t]). (D) Elongation rate constants for Pols II, IV and V. Data is representative of two replicate experiments (see Supplementary Figure S4).
Transcription error rates in de novo synthesized RNAs
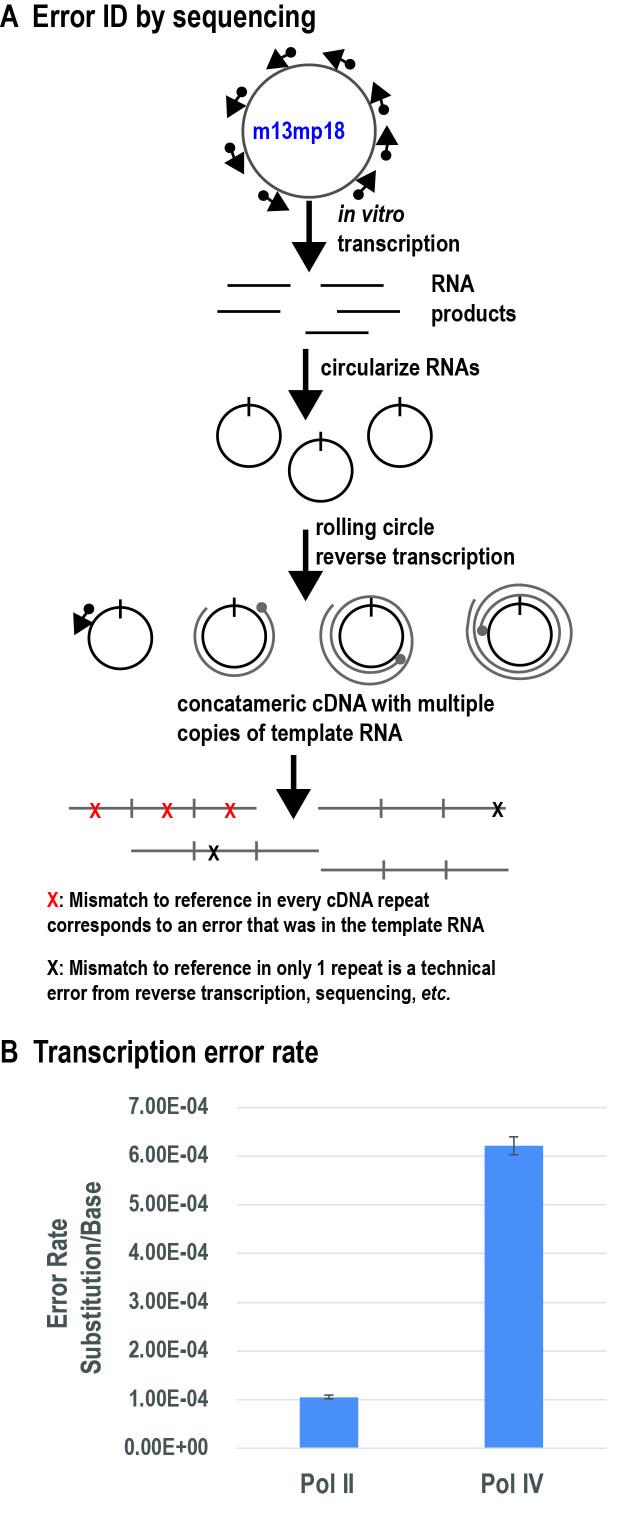
To test whether differences in fidelity observed using defined template oligonucleotides and primer extension with single NTPs are also observed for transcripts initiated de novo in a primer-independent manner and in the presence of all four NTPs, we sequenced RNAs generated by Pol II or Pol IV using single-stranded, circular bacteriophage M13mp18 DNA as the template. Pol V did not generate sufficient numbers of transcripts from this template to be included in these analyses. Pols II and IV initiate at more than two thousand distinct start sites within the ∼7.2 kb M13 genome sequence, providing a diverse set of transcripts (6). The resulting Pol II and Pol IV transcripts were subjected to ‘circle sequencing’ (Figure 6A) (28). In this method, the 5′ and 3′ ends of transcripts are ligated to form circles prior to cDNA synthesis using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). This enzyme has strand displacement activity, allowing it to reverse-transcribe the RNA circle multiple times, producing a cDNA concatamer composed of multiple DNA copies of the original RNA template. True transcription errors present in the RNA are present in each repeat of the concatamer whereas sporadic errors introduced by reverse transcriptase, or the DNA polymerases used for PCR amplification or sequencing polymerase will not be common to each repeat (see diagram in Figure 6A). Variations of this method have been used to identify genetic subtypes in RNA viral populations (26,27) and to measure the transcriptional fidelities of bacterial RNA polymerases (33).
Figure 6.
Pol IV exhibits increased error rate relative to Pol II in RNAs initiated de novo (primer-independent), using single-stranded M13 DNA as the template. (A) Assay design for circle sequencing of cDNA concatamers. (B) Overall error rates of Pol II and Pol IV, expressed as substitution frequency per base sequenced.
Using the circular sequencing approach, transcription error rates for RNAs generated by Pol II or Pol IV using the M13 template were calculated as the number of errors divided by the total number of nucleotides sequenced. Pol IV’s in vitro error rate was found to be roughly six times greater than the transcription error rate for Pol II (6.4 × 10−4 and 1.1 × 10−4, respectively) (Figure 6B). These results support our primer elongation results using single nucleotides, which also showed that Pol IV has an increased propensity for misincorporation relative to Pol II. The observed Pol II in vitro error rate is also consistent with a previously estimated in vitro error rate for Pol II isolated from wheat germ, which ranged from 10−4 to 10−6 depending on the combination of NTP and DNA template (34).
DISCUSSION
Collectively, our initial investigation of Pol IV and Pol V catalytic properties provides several insights concerning the biochemical characteristics of the enzymes. Pol IV and Pol V differ from Pol II, and from one another, in a number of enzymatic properties, including accuracy and catalytic rate. Pol IV is the slowest of the three enzymes, and is also the most error-prone. Pol IV/RDR2-dependent precursor RNAs are only ∼30–40 nt in length, just long enough to encode single 24 nt siRNAs (6,7), such that Pol IV may not need to be fast. Based on bioinformatic analyses of Pol IV and RDR2-dependent RNAs isolated from plant lysates, Pol IV has been hypothesized to misincorporate NTPs at positions of methylated cytosines, potentially inducing transcriptional termination (7). In direct tests of this hypothesis in vitro, DNA methylation has no measurable effect on Pol IV fidelity (Figure 3). This indicates that misincorporation at methylcytosines is not an intrinsic property of Pol IV, but does not preclude the possible involvement of other proteins or activities.
Because 24 nt siRNAs primarily guide the silencing of transposons whose family members are not necessarily identical, error-prone transcription by Pol IV may be tolerated, and possibly even beneficial. The accuracy of Pol IV transcription would, however, affect whether an siRNA anneals to a target with perfect or imperfect complementarity, potentially influencing whether AGO4 might slice, or just bind, a Pol V-transcribed target, similar to miRNAs (35). There is evidence that slicing activity is important at some, but not all, RdDM loci (36).
Pol V makes longer transcripts than Pol IV. Although their precise size remains undefined, RT-PCR analyses indicate that they can be 200 nt or more (37–39). Our results indicate that Pol V transcription is highly accurate, suggesting that accuracy is important for Pol V transcript function. Pol V makes RNAs at loci to be silenced by RNA-directed DNA methylation, and its transcripts are thought to provide scaffolds for the binding of siRNA-AGO complexes that then recruit additional chromatin modifying activities (11,37), consistent with studies in fission yeast and other organisms (40). A need for precise basepairing between siRNAs and Pol V transcripts might result in selective pressure to maintain Pol V fidelity. A recent study has suggested that siRNA-AGO4 complexes may also bind directly to DNA at Pol V-transcribed loci (41). If the act of transcription is all that is needed for Pol V to function, it is not clear why Pol V would need to generate RNAs that faithfully match the sequence of transcribed loci. One possibility might be that siRNAs first bind Pol V transcripts prior to binding the corresponding DNA sequence. Another possibility might be that Pol V transcripts are used to generate R-loops at transcribed loci, thereby enabling siRNA-AGO interactions with the displaced DNA strand (42), and that precise basepairing of the RNA and DNA might somehow be important.
Some substitutions of highly conserved amino acids in Pols IV and V occur at positions known to affect RNA polymerase fidelity. For example, mutation of N479 in the S. cerevisiae RNA Pol II largest subunit, Rpb1 results in reduced discrimination between NTPs and dNTPs (18). Both Pol IV and Pol V have substitutions at the position corresponding to yeast N479, consistent with their incorporation of dNTPs to a greater extent than Pol II. However, the details of altered rNTP:dNTP discrimination differs in the yeast N479 Pol II mutant versus Pols IV and V. The N479S Pol II mutant has decreased affinity for rNTPs, rather than increased affinity for dNTPs (18). In contrast, Pol IV and Pol V have increased dNTP affinity (Supplementary Figure S3). Additional diverged amino acids of Pols IV and V presumably contribute to these differences.
The rNTP concentrations used in this study are within the range of reported physiological concentrations in cells, but the concentration of dNTPs we used in Figure 4B are ∼3–20 times greater than published cellular dNTP levels (43). The nucleotide concentration at which Pol II dNTP incorporation is half-maximum is 12 times greater than reported cellular dTTP concentrations; whereas, the Pol IV and Pol V dNTP misincorporation is half-maximal at dTTP concentrations only 2- or 3-fold greater than reported in vivo, respectively. Therefore, it is possible that Pol IV and Pol V may be incorporating dNTPs into their products in vivo, although it is not clear what the biological consequences of incorporating dNTPs into Pol IV or Pol V transcription products would be. Misincorporation of dNTPs into RNA by a T7 RNA polymerase mutant has been found to block translation of the dNTP-carrying transcripts (44). Incorporating dNTPs into Pol IV or Pol V transcripts might similarly inhibit their translation, but this seems unnecessary for Pol IV and Pol V RNAs, which are thought to act in the nucleus. Incorporation of dNTPs might potentially affect binding or processing of Pol IV or Pol V transcripts. Pol IV and Pol V have also been implicated in DNA double-strand break repair, such that synthesizing transcripts that include dNTPs might somehow be important (45). Similar to misincorporation of a non-complementary base, misincorporation of a dNTP reduces the likelihood of adding a subsequent nucleotide, potentially inducing pausing or termination (18).
The trigger loop within the largest subunit of yeast RNA Pol II is thought to play an important role in transcriptional fidelity. Amino acids within the trigger loop (Leu1081, Gln1078, His1083, and Asn 1082) contact the base, phosphate, and ribose of an incoming NTP to facilitate precise positioning and catalysis (18). Pols IV and V have amino acid substitutions at three of these four positions (Supplementary Figure S1A). In addition, A. thaliana Pol IV, which has reduced accuracy relative to Pol II, has diverged at a trigger loop position whose mutation in the yeast Pol II largest subunit, Rpb1 (position E1103) results in increased NTP misincorporation (20). The E1103G mutation is thought to destabilize the active site open conformation, causing increased misincorporation due to greater sequestration of non-complementary nucleotides within the closed conformation (20). It has been suggested that other conditions that reduce Pol II fidelity, such as deletion of the ninth subunit, or the presence of manganese, similarly promote a closed trigger loop conformation (18,20,46,47). Opening and closing of the trigger loop is also important for elongation, such that mutations alter the catalytic rate (48,49). Given their altered fidelities (Figures 1, 2 and 4), rates of nucleotide incorporation (Figure 5), and extensive sequence divergence (or deletion) in the trigger loop region (Supplementary Figure S1), we speculate that the Pol IV active center may naturally adopt a more closed structure, whereas the Pol V active center may resemble the Pol II open conformation. These speculations underscore a need for high-resolution structural models for Pol IV and Pol V. Such studies would help assess the impact of changes in highly conserved amino acids as well as the potential role of compensatory changes at other amino acid positions, potentially helping preserve structural features important for catalytic functions.
Supplementary Material
ACKNOWLEDGEMENTS
C.S.P. is an Investigator of the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute (HHMI) [Investigator funds to C.S.P.]; Gordon and Betty Moore Foundation [Investigator funds to C.S.P.]; National Institutes of Health [GM077590 to C.S.P.; GM036827 to M.L., training grant T32GM007757 support of M.M]. Funding for open access charge: HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolic L., Pikaard C.S.. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell. 2009; 33:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang Y., Kendall T., Forsythe E.S., Dorantes-Acosta A., Li S., Caballero-Perez J., Chen X., Arteaga-Vazquez M., Beilstein M.A., Mosher R.A.. Ancient origin and recent innovations of RNA polymerase IV and V. Mol. Biol. Evol. 2015; 32:1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo J., Hall B.D.. A multistep process gave rise to RNA polymerase IV of land plants. J. Mol. Evol. 2007; 64:101–112. [DOI] [PubMed] [Google Scholar]
- 4. Tucker S.L., Reece J., Ream T.S., Pikaard C.S.. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb. Symp. Quant. Biol. 2010; 75:285–297. [DOI] [PubMed] [Google Scholar]
- 5. Haag J.R., Brower-Toland B., Krieger E.K., Sidorenko L., Nicora C.D., Norbeck A.D., Irsigler A., LaRue H., Brzeski J., McGinnis K. et al. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep. 2014; 9:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blevins T., Podicheti R., Mishra V., Marasco M., Tang H., Pikaard C.S.. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife. 2015; 4:e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhai J., Bischof S., Wang H., Feng S., Lee T.F., Teng C., Chen X., Park S.Y., Liu L., Gallego-Bartolome J. et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell. 2015; 163:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X.. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015; 25:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matzke M.A., Mosher R.A.. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014; 15:394–408. [DOI] [PubMed] [Google Scholar]
- 10. Wendte J.M., Pikaard C.S.. The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta. 2016; 1860:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S.. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009; 41:630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang D.L., Zhang G., Tang K., Li J., Yang L., Huang H., Zhang H., Zhu J.K.. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 2016; 26:66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye R., Chen Z., Lian B., Rowley M.J., Xia N., Chai J., Li Y., He X.J., Wierzbicki A.T., Qi Y.. A Dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell. 2016; 61:222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K.. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013; 162:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K.. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015; 34:20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haag J.R., Pontes O., Pikaard C.S.. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS One. 2009; 4:e4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landick R. Functional divergence in the growing family of RNA polymerases. Structure. 2009; 17:323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D.. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006; 127:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vassylyev D.G., Vassylyeva M.N., Zhang J., Palangat M., Artsimovitch I., Landick R.. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007; 448:163–168. [DOI] [PubMed] [Google Scholar]
- 20. Kireeva M.L., Nedialkov Y.A., Cremona G.H., Purtov Y.A., Lubkowska L., Malagon F., Burton Z.F., Strathern J.N., Kashlev M.. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell. 2008; 30:557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feig M., Burton Z.F.. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys. J. 2010; 99:2577–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haag J.R., Ream T.S., Marasco M., Nicora C.D., Norbeck A.D., Pasa-Tolic L., Pikaard C.S.. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell. 2012; 48:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R.. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010; 38:W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sydow J.F., Brueckner F., Cheung A.C., Damsma G.E., Dengl S., Lehmann E., Vassylyev D., Cramer P.. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell. 2009; 34:710–721. [DOI] [PubMed] [Google Scholar]
- 26. Acevedo A., Andino R.. Library preparation for highly accurate population sequencing of RNA viruses. Nat. Protoc. 2014; 9:1760–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Acevedo A., Brodsky L., Andino R.. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature. 2014; 505:686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lou D.I., Hussmann J.A., McBee R.M., Acevedo A., Andino R., Press W.H., Sawyer S.L.. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19872–19877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Law J.A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E.. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goodman M.F., Keener S., Guidotti S., Branscomb E.W.. On the enzymatic basis for mutagenesis by manganese. J. Biol. Chem. 1983; 258:3469–3475. [PubMed] [Google Scholar]
- 32. El-Deiry W.S., Downey K.M., So A.G.. Molecular mechanisms of manganese mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:7378–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Traverse C.C., Ochman H.. Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:3311–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Mercoyrol L., Corda Y., Job C., Job D.. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. Eur. J. Biochem. 1992; 206:49–58. [DOI] [PubMed] [Google Scholar]
- 35. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J.. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006; 443:1008–1012. [DOI] [PubMed] [Google Scholar]
- 37. Wierzbicki A.T., Haag J.R., Pikaard C.S.. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008; 135:635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wierzbicki A.T., Cocklin R., Mayampurath A., Lister R., Rowley M.J., Gregory B.D., Ecker J.R., Tang H., Pikaard C.S.. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012; 26:1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bohmdorfer G., Sethuraman S., Rowley M.J., Krzyszton M., Rothi M.H., Bouzit L., Wierzbicki A.T.. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife. 2016; 5:e19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wendte J.M., Pikaard C.S.. Targeting Argonaute to chromatin. Genes Dev. 2016; 30:2649–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lahmy S., Pontier D., Bies-Etheve N., Laudie M., Feng S., Jobet E., Hale C.J., Cooke R., Hakimi M.A., Angelov D. et al. Evidence for ARGONAUTE4-DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 2016; 30:2565–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pikaard C.S., Haag J.R., Pontes O.M., Blevins T., Cocklin R.. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb. Symp. Quant. Biol. 2013; 77:205–212. [DOI] [PubMed] [Google Scholar]
- 43. Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994; 140:1–22. [DOI] [PubMed] [Google Scholar]
- 44. Sousa R., Padilla R.. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995; 14:4609–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y.. A role for small RNAs in DNA double-strand break repair. Cell. 2012; 149:101–112. [DOI] [PubMed] [Google Scholar]
- 46. Walmacq C., Kireeva M.L., Irvin J., Nedialkov Y., Lubkowska L., Malagon F., Strathern J.N., Kashlev M.. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J. Biol. Chem. 2009; 284:19601–19612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaplan C.D., Larsson K.M., Kornberg R.D.. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol. Cell. 2008; 30:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E.. A ratchet mechanism of transcription elongation and its control. Cell. 2005; 120:183–193. [DOI] [PubMed] [Google Scholar]
- 49. Zakharova N., Bass I., Arsenieva E., Nikiforov V., Severinov K.. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta' subunit inhibit transcript cleavage and transcript elongation. J. Biol. Chem. 1998; 273:24912–24920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.