Abstract
Comparative genomics studies typically limit their focus to single nucleotide variants (SNVs) and that was the case for previous comparisons of woolly mammoth genomes. We extended the analysis to systematically identify not only SNVs but also larger structural variants (SVs) and indels and found multiple mammoth-specific deletions and duplications affecting exons or even complete genes. The most prominent SV found was an amplification of RNase L (with different copy numbers in different mammoth genomes, up to 9-fold), involved in antiviral defense and inflammasome function. This amplification was accompanied by mutations affecting several domains of the protein including the active site and produced different sets of RNase L paralogs in four mammoth genomes likely contributing to adaptations to environmental threats. In addition to immunity and defense, we found many other unique genetic changes in woolly mammoths that suggest adaptations to life in harsh Arctic conditions, including variants involving lipid metabolism, circadian rhythms, and skeletal and body features. Together, these variants paint a complex picture of evolution of the mammoth species and may be relevant in the studies of their population history and extinction.
Keywords: woolly mammoth, elephant, comparative genomics, evolution, viral defense
1. Introduction
The woolly mammoth (Mammuthus primigenius) was the last surviving species of the mammuthus genus with the last known population on Wrangel Island about 4,000 years ago.1,2 Woolly mammoth is one of the most studied extinct species, although much is still unknown, e.g. why they became extinct, how they evolved, and how they differ from elephants, their closest living relatives. Perhaps the most common theories for the cause of their extinction are a warming climate, hunting by humans, or both. Woolly mammoths lived in a cold, dry steppe-tundra where average winter temperatures ranged from -30°C to -50°C, much different from the tropical and subtropical environments of modern African and Asian elephants.3 Mammoths had many anatomical adaptations minimizing heat loss in its harsh environment, such as thick fur, small ears, and small tails (compared with modern elephants), and a thick layer of fat under the skin to reduce heat loss and possibly serve as a heat source or fat reservoir for the winter.4–6 Mitochondrial analysis has suggested there were three clades (I–III) with clade I surviving ∼30,000 years after the extinction of clades II and III despite sharing overlapping territory with clade II in Northeastern Siberia and possibly clade III in Europe.7
In 2008, the first whole genome sequencing (WGS) (<1×) of a woolly mammoth was published.8 However, only recently have high coverage WGS datasets become available from two studies that identified SNVs unique to woolly mammoths to infer the genetic basis of adaptations to the Arctic (Lynch study)9 and to analyse species diversity prior to extinction (Palkopoulou study).1 Our combined dataset included mammoths M4 and M25 from the Lynch study, and mammoths Wrangel and Oimyakon from the Palkopoulou study. We used the combined dataset, as well as four Asian elephant WGS datasets,9,10 to analyse structural variants (SVs), copy number variants (CNVs) and indels, as well as SNVs, to further investigate genetic adaptations and diversity. Combining datasets also enabled a comparison of clades, as each study had one clade I (M4, Wrangel) and one clade II (M25, Oimyakon) mammoth. All mammoths remains originated from northern Siberia (M4, ∼20,000 years ago; M25, ∼60,000 years ago; Oimyakon, ∼44,800 years ago) or Wrangel Island, off the coast of northern Siberia (Wrangel, ∼4,300 years ago), although the exact location is unknown for M4.1,9,11
In this paper, we analysed the patterns of variation across the genomes of these four mammoths and compared them to the available elephant genomes. Using algorithms from our GROM (Genome Rearrangement Omni-Mapper) suite, GROM-RD12 and GROM (in preparation), we systematically identified variants, ranging from single nucleotide changes and short indels to deletions and amplifications of regions encompassing gene fragments and complete genes. These variants reveal the signs of evolutionary adaptation of mammoths to the harsh and cold environment. We identified changes in many parts of the genome, including genes associated with metabolism, circadian rhythms, immunity and skeletal/body shape. Taken together, they describe a rich evolutionary history of the woolly mammoth species and may shed light on causes of their extinction.
2. Materials and methods
2.1. WGS data
WGS fasta files for woolly mammoths M4 and M25 and Asian Elephants Asha, Parvathy, and Uno were downloaded from the Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra (9 February 2017, date last accessed) (project accession number: PRJNA281811). WGS fasta files for the Wrangel and Oimyakon mammoths were downloaded from the European Nucleotide Archive (ENA), http://www.ebi.ac.uk/ena (9 February 2017, date last accessed) (accession number: ERP008929). WGS fasta files for the Asian elephant Emelia were downloaded from ENA (accession: ERP004241). WGS fasta files were mapped to the African reference genome loxAfr3, downloaded from UCSC (https://genome.ucsc.edu, http://hgdownload.soe.ucsc.edu/goldenPath/loxAfr3/bigZips/ (9 February 2017, date last accessed)), using BWA MEM,13 version 0.7.4, with default parameters. Duplicates were removed using SAMtools,14 version 0.1.19.
2.2. Variant detection and analysis
We limited analysis to supercontigs/scaffolds ≥1,000,000 bases. Visual inspection of the mapped files using IGV15 indicated an increase in low mapping quality reads and highly variable coverages for smaller (<1,000,000) supercontigs. For GROM-RD,12 this threshold was set to 5,000,000 bases to provide adequate sampling and increase specificity. GROM-RD identifies regions of abnormal coverage that are unlikely to occur by chance based on analysis of read depth variance with nucleotide composition and mapping quality across the genome. This reduces false positives in potentially biased, highly variable ancient DNA datasets. GROM-RD measures read-depth variance for various window sizes (100–10,000 bases). A benefit of this approach is limiting spikes in false positives with decreasing CNV length. CNVs were detected using GROM-RD with default parameters. Homozygous deletions (GROM-RD copy number estimate <0.5) and duplications (copy number estimate >3.5) found in all woolly mammoths (80% reciprocal CNV overlap) were analysed using GROM-RD and filtered if the GROM-RD copy number estimate was <1.5 (deletions) or >2.5 (duplications) in any of the Asian elephants. Indels and SNVs were detected using our variant calling framework GROM (manuscript in preparation, software will be made publicly available as stipulated by the funding grant). Additionally, GROM-RD calls were filtered if CNVnator (run with default parameters) detected the CNV in any Asian elephant (20% reciprocal overlap) or did not detect the CNV in all mammoths (50% reciprocal overlap). A simplified SNV/indel caller module in GROM was implemented to replicate, with a few differences, the SNV detection method used in a previous mammoth study.9 Indels and SNVs predicted as being heterozygous (<80%, indels, or <90%, SNVs, of overlapping reads contained the variant) or supported by <4 reads with mapping quality ≥ 20 in any of the mammoths, or with at least one variant-supporting read (no mapping quality threshold) in any of the Asian elephants were filtered. We also required each Asian elephant to have at least 4× coverage at the indel or SNV site predicted in mammoth. These requirements were reciprocated when calling elephant-specific variants. Fixed, derived CNVs, indels, and SNVs were then uploaded to VEP (Variant Effect Predictor, www.ensembl.org) to identify variants potentially affecting genes. GO term and KO phenotypes obtained from the Gene Ontology Consortium (geneontology.org), and MGI (www.informatics.jax.org). GO term, KEGG pathway, and KO phenotype enrichment for woolly mammoth fixed, derived amino acid substitutions was analysed using PANTHER (http://pantherdb.org/ (9 February 2017, date last accessed)),16 WebGestalt (http://www.webgestalt.org/ (9 February 2017, date last accessed)),17 and Vlad (http://proto.informatics.jax.org/prototypes/vlad/ (9 February 2017, date last accessed)), respectively. Using a set of SNVs (four variant-supporting reads, mapping quality ≥20, allele frequency ≥0.4) found in at least one mammoth or Asian elephant, FST estimates were calculated, as described in a prior publication,18 with VCFtools,19 version 0.1.12, using the –weir-fst-pop function with a 100,000 base window size (–fst-window-size) and 10,000 base sliding window (–fst-window-step). Protein multiple sequence alignments produced using T-Coffee20 for protein sequences shorter than the recommend limit of 2,500 amino acids or MUSCLE21 for longer protein sequences.
3. Results
3.1. CNVs
As noted in Lynch et al.,9 mammoths and Asian elephants diverged after branching from African elephants (reference genome), thus fixed CNVs in mammoths are considered derived. Using an intersection of GROM-RD12 and CNVnator22 calls, we found 56 fixed, derived mammoth CNVs, including 55 deletions and one amplification. According to VEP (ensembl.org), three deletions with putative loss of function and one amplification with potential gain of function occurred in the exons of protein-coding genes, and two deletions affected RNA genes (Table 1).
Table 1.
Fixed, derived woolly mammoth CNVs
| CNV | Location | Gene | Consequence | Biotype |
|---|---|---|---|---|
| DUP | scaffold_16:17919954-17969811 | RNASEL | Transcript amplification | Protein coding |
| DEL | scaffold_2:62406050-62418121 | 5S_rRNA | Transcript ablation | rRNA |
| DEL | scaffold_21:14388123-14390331 | CD44 | Feature truncation | Protein coding |
| DEL | scaffold_4:20346665-20394204 | ENSLAFG00000031480 | Transcript ablation | Protein coding |
| DEL | scaffold_48:12765543-12778793 | U6 | Transcript ablation | snRNA |
| DEL | scaffold_55:5626970-5637970 | ENSLAFG00000027547 | Transcript ablation | Protein coding |
Ensembl gene ID used when gene symbol not available.
Perhaps the most famous CNV in the elephant genome is the TP53 gene.23 However, analysis with GROM-RD and visual inspection with IGV15 did not reveal any copy number change relative to elephants in the corresponding genome regions of woolly mammoths.
One deletion occurred in an exon of CD44. The CD44 gene is expressed in multiple tissues including the central nervous system, lung, epidermis, liver, and pancreas.24 Its product is involved in multiple functions, including cellular adhesion, hyaluronate degradation, lymphocyte activation, lymph node homing, angiogenesis, and the release of cytokines.24 CD44 contributes to the maintenance of stem cell features, such as apoptosis resistance.25 Intriguingly, the deleted exon is the first of the ten so-called variable exons of CD44, whose splicing and histone mark deposition has been shown to be modulated by Argonaute proteins and strongly affected in Ago2-/- mouse embryonic fibroblasts,26 thus its loss may affect the functionality and tissue distribution of CD44 isoforms. Among a multitude of CD44 GO terms and knockout (KO) mammalian phenotypes listed in Supplementary Table S1, there are two phenotypes of special interest to woolly mammoths, increased diameter of the tibia and short tibia. Mammoth limb bones were much greater in diameter than the limb bones of modern day elephants.27 Additionally, the hind leg to fore leg ratio was smaller in mammoths compared with elephants,27 with their body size and stature decreasing towards the end of the last glacial advance in the Pleistocene.27 On the basis of a small sample, the tibia to femur ratio of mammoths (0.57 ± 0.04, n = 5) was less than the tibia to femur ratio of African elephants (0.60 ± 0.02, n = 5).27
One may argue that deletions detected with an alignment of reads against an evolutionary distant reference may represent false positives arising due to rapid evolution and low sequence similarity in the respective regions. Even if that was true, this argument does not change the relevance of our logic on the possible effects on the exon of CD44, since any significant sequence alteration would also modify the function of the region in woolly mammoths, compared with African and Asian elephants. Further, the large sizes of the detected deletions (Table 1) suggest extreme changes, not seen in other genes. Other deletions were observed outside of the coding regions and are thus also outside the scope of the study and do not affect our conclusions.
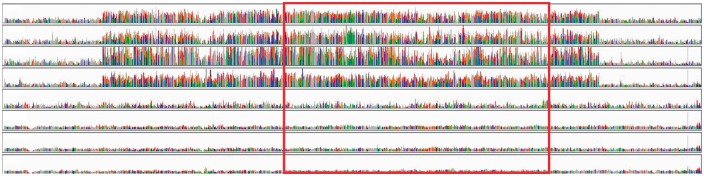
We detected a fixed, derived woolly mammoth amplification encompassing the RNase L gene (scaffold_16:17939571-17966869, forward strand), including ∼20,000 bases upstream. GROM-RD predicted five (Wrangel, Oimyakon), six (M25), or nine (M4) copies of RNase L (6.6 copies, stdev 0.8) in the mammoths. Figure 1 shows normalized coverages and the location of RNase L within the CNV. RNase L has several critical roles including antiviral response, adipocyte differentiation, tumorigenesis, cell proliferation, innate immune response, and apoptosis.28 Antiviral response involves endonucleolytic cleavage of single-stranded foreign RNAs, ribosomal RNAs, and mRNAs by activated RNase L.28–30
Figure 1.
RNase L amplification unique to woolly mammoths. Additional 10,000 bases shown upstream and downstream of duplication. Read coverage normalized (y axis maximum = 6× average genome read depth). Top four tracks show mammoths (Wrangel, Oimyakon, M4, M25). Bottom four tracks show Asian elephants (Emelia, Asha, Parvathy, Uno). Box indicates region containing RNase L exons.
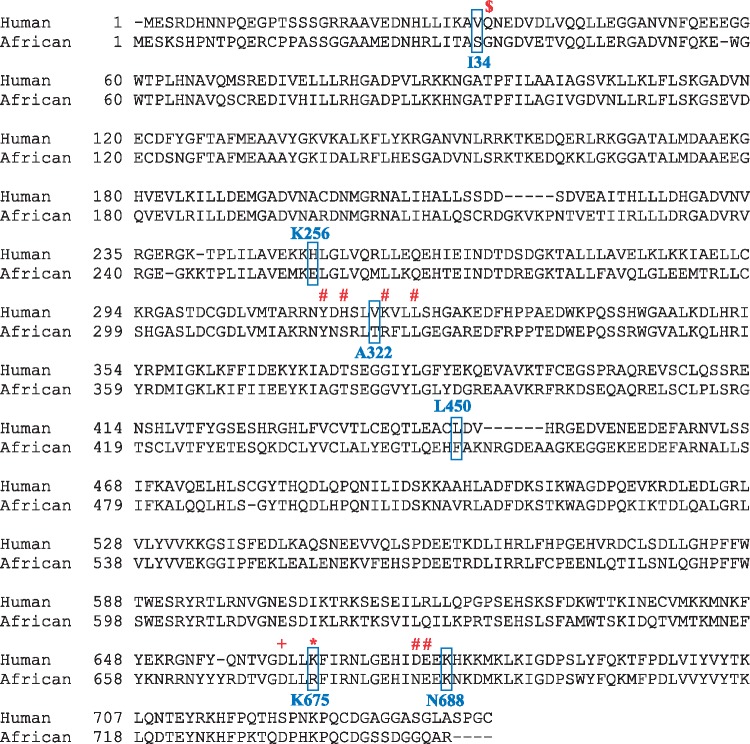
Additionally, we identified nine derived SNVs predicted to occur in 3–9 of the RNase L copies and with no evidence in any of the Asian elephant samples. Six of these were non-synonymous (Table 2) and may reflect adaptation in woolly mammoths. We used T-Coffee20 to align the RNase L protein sequence for mammoth with the protein sequence of 16 other species, including human, mouse, and cold-adapted species polar bear, alpine marmot, and walrus (Supplementary Fig. S1). A more concise alignment of human and African elephant RNase L protein sequence is shown in Fig. 2. Substitution S34I is adjacent to residue G35 (Supplementary Fig. S1), which is involved in 2–5A interaction,31 needed for dimerization of RNase L and activation of its antiviral activity. V34 was most prevalent with S34 occurring in African and Asian elephants, T34 in dolphin, and I34 in mammoth, Alpine marmot, and platypus. T322A is close to several residues involved in self-domain dimerization (Y310, S312, R316, and L319)31 and 2-5A sensing (Y308, Y310).32 V322 was predominant with I322 occurring in Anole lizard, T322 in African and Asian elephants, and A322 in mammoth, cow, sheep, armadillo, dolphin, and platypus. Of interest, woolly mammoth substitution R675K was identified as a ribonuclease active site in pigs by Huang et al.,31 who has shown that mutant pK672A (mammoth 675) results in a defective RNase L. In this position, K occurs in all of the aligned species except African and Asian elephants, which have R, and in a previous alignment by Huang, R occurs in chicken.31 Both R and K occur in the mammoth. Together with the significant amplification of the locus in the mammoth genomes, these variants likely reflect adaptation processes related to RNase L functionality and specificity.
Table 2.
Amino acid variants in mammoth RNase L
| Location | Codons (elephant/mammoth) | Amino acids (elephant/mammoth) | Protein position | Domain |
|---|---|---|---|---|
| scaffold_16:17939894 | aGt/aTt | S/I | 34 | ANK |
| scaffold_16:17940559 | Gag/Aag | E/K | 256 | ANK |
| scaffold_16:17940757 | Aca/Cca | T/A | 322 | – |
| scaffold_16:17941141 | Ttt/Ctt | F/L | 450 | Protein kinase |
| scaffold_16:17965935 | aGg/aAg | R/K | 675 | RNase |
| scaffold_16:17965975 | aaG/aaC | K/N | 688 | RNase |
Variants occur in 3–9 copies of RNase L. Mammoth predicted to have 5–9 copies of RNase L.
Figure 2.
African elephant RNase L alignment to human RNase L. Wolly mammoth residue in bold text are shown next to the corresponding elephant and human residues (boxed). Residues of interest near or coinciding with woolly mammoth amino acid substitutions marked above with the following signs: $ (2-5A interaction site), # (self-domain dimerization), or * (ribonuclease active site). RNases L cleavage site H683 (hH672) is marked with a plus sign (+).
RNase L is able to selectively target specific cellular RNA28 and is involved in antiviral activity against numerous virus families.33 RNase L activation in virus-infected cells was shown to trigger the NLRP3 inflammasome,34 the latter being implicated in the host response to many different types of RNA and DNA viruses, including herpesviridae.35 Notably, elephant endotheliotropic herpesviruses (EEHV) can cause a highly fatal hemorrhagic disease when transmitted to young Asian elephants: two available genomes of Asian elephants were obtained postmortem from the animals affected by this disease.10 The EEHV genotypes found in African elephants appear to be generally less virulent.36
Arctic conditions may be of additional significance with regard to the antiviral action of this protein. Temperature-dependent transmission of rotavirus (family Reoviridae) has been shown in humans, with a 13 percent decrease in infections per 1°C increase in temperature above 5°C.37 In river water in Japan, the peak reovirus level was found in winter during the cold weather months.38 Influenza virus was found to favor cold and dry conditions for transmission in guinea pigs.39 Macaques infected with SRV-4, family Retroviridae, had less SRV-4 antibodies in cold weather.40 Mammoths lived in cold dry steppe tundra, and likely in close matriarch-led groups similar to modern elephants, possibly increasing pressure for the species to evolve defenses against viruses adapted to their environment. Not surprisingly, RNase L KO mammalian phenotypes included increased susceptibility to viral infection, as well as abnormal thymus morphology, enlarged thymus, and thymus hyperplasia (Supplementary Table S1).
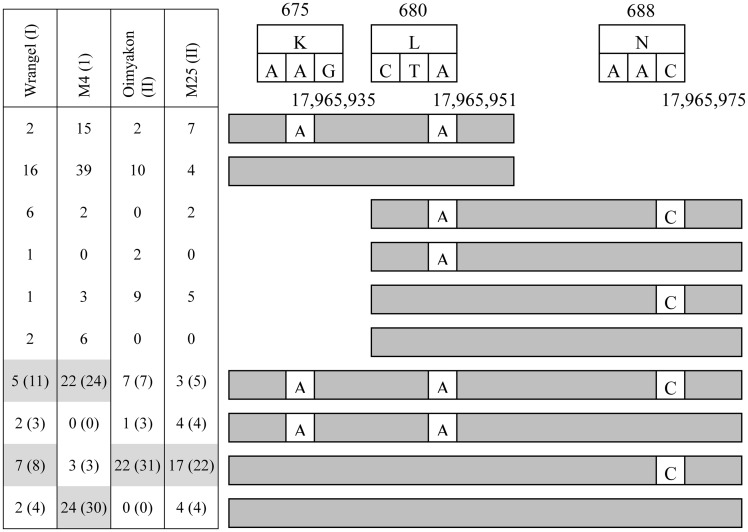
Given the close arrangement of the SNVs near the ribonuclease active site, we further analysed their co-occurrence in the individual reads (Fig. 3). For all mammoths, non-synonymous A allele (R675K, aGg/aAg, scaffold 16:17,965,935) always co-occurred with synonymous A allele (H683, scaffold_16:17,965,951) for reads containing both nucleotide locations (Fig. 3). The patterns of co-occurrence of the non-synonymous C allele (K688N, scaffold_16:17,965,975) with the A alleles at 17,965,935 and 17,965,951 appeared to be linked with the clade structure. The nine RNase L copies of M4 showed almost equal occurrence of GGG (same as the African elephant reference) or AAC (all three nucleotides substituted compared with the reference) haplotypes at these three positions. Another clade I member, Wrangel, showed prevalence of AAC and GGC in the reads containing the three SNV locations, while GGC haplotype was strongly preferred in the clade II genomes.
Figure 3.
Occurrences of woolly mammoth SNV combinations in reads for RNase L domain variants. Read counts for Wrangel, M4, Oimyakon, and M25 are left of reads. Read counts in parentheses indicate addition of inferred counts based on the observation that SNVs for residues 675 and 680 always co-occur. Most frequent haplotypes highlighted in gray. Residue number above reads. SNV combinations with no occurrences are not shown.
3.2. SNVs
We found 836,806 (606,176 intergenic; 224,483 intronic; 6,147 exonic) fixed, derived woolly mammoth SNVs, including 2,283 fixed, derived amino acid substitutions in 1,888 protein-coding genes, four protein-coding genes with a start loss and 18 protein-coding genes with pre-mature stop codons (Supplementary Material S1). We note that Lynch et al.9 has used similar criteria (see Materials and methods) to find a higher total of fixed, derived woolly mammoth SNVs (1.4 million compared with 0.8 million). This was expected considering our study had more samples (four woolly mammoths, four Asian elephants) compared with the Lynch study (two woolly mammoths, three Asian elephants), likely decreasing false positives. However, Lynch et al. had fewer fixed, derived woolly mammoth amino acid substitutions (2,020 compared with 2,283). Several factors may have contributed to the discrepancy. For instance, we did not hard-mask potential cytosine deamination sites or filter short reads, and instead used a higher mapping quality threshold (>20) compared with Lynch et al.’s threshold (>0). Ancient DNA is known to have miscodings of C to T and G to A.41 However, Briggs et al.41 has suggested that with sufficient coverage nucleotide misincorporations should not prevent a reliable Neandertal or mammoth genome sequence from being determined. We reasoned that our potentially less stringent thresholds (to increase sensitivity) would be adequate considering we analysed four mammoth samples, each with ≥10× coverage, and filtered SNVs using four Asian elephants (see Supplementary Table S2 for coverages). We tested for ancient DNA biases by comparing fixed, non-synonymous variants in woolly mammoths (2,283), Asian elephants (2,634), and the Lynch study woolly mammoths (2,020). We found that rates (as a fraction of all substitutions) for fixed, non-synonymous variants in the mammoths were similar for substitutions corresponding to the most common potential miscodings, C → T (0.216, present study; 0.211, Lynch study) and G → A (0.227, present study; 0.209, Lynch study) (Table 3), and thus our variant collection was unlikely to have been biased by these miscodings compared with the variants in the Lynch study. The rates in mammoths were only moderately higher than those in Asian elephants (Table 3).
Table 3.
Nucleotide substitution comparison
| Change | Mammoth (this study) |
Asian elephant |
Mammoth (Lynch) |
|||||
|---|---|---|---|---|---|---|---|---|
| Occurrences |
Occurrences |
Occurrences |
||||||
| Raw | Normalized | Raw | Normalized | % Chg | Raw | Normalized | % Chg | |
| A/C | 81 | 0.035 | 103 | 0.039 | −9.3 | 78 | 0.038 | −6.1 |
| A/G | 286 | 0.125 | 417 | 0.158 | −20.9 | 266 | 0.129 | −2.8 |
| A/T | 42 | 0.018 | 63 | 0.024 | −23.1 | 50 | 0.024 | −24.1 |
| C/A | 96 | 0.042 | 87 | 0.033 | 27.3 | 92 | 0.045 | −5.7 |
| C/G | 111 | 0.049 | 173 | 0.066 | −26.0 | 112 | 0.054 | −10.4 |
| C/T | 493 | 0.216 | 500 | 0.190 | 13.8 | 436 | 0.211 | 2.2 |
| G/A | 518 | 0.227 | 436 | 0.166 | 37.1 | 431 | 0.209 | 8.7 |
| G/C | 140 | 0.061 | 156 | 0.059 | 3.5 | 126 | 0.061 | 0.5 |
| G/T | 77 | 0.034 | 103 | 0.039 | −13.7 | 90 | 0.044 | −22.7 |
| T/A | 46 | 0.020 | 70 | 0.027 | −24.2 | 53 | 0.026 | −21.5 |
| T/C | 312 | 0.137 | 432 | 0.164 | −16.7 | 261 | 0.126 | 8.1 |
| T/G | 81 | 0.035 | 94 | 0.036 | −0.6 | 69 | 0.033 | 6.1 |
Compared mammoth nucleotide substitutions with Asian elephant and previously identified mammoth9 (Lynch et al., 2015) substitutions. Comparison using fixed, derived non-synonymous SNVs. Most common miscodings for ancient DNA in bold.
In agreement with an earlier analysis of KEGG pathways and KO phenotypes,9 we analysed fixed, derived woolly mammoth amino acid substitutions and found amongst 58 enriched KEGG pathways complement and coagulation cascades (P = 5.5 × 10−9, adjP = 4.2 × 10−7), fat digestion and absorption (P = 0.0038, adjP = 0.021), and circadian rhythm – mammal (P = 0.034, adjP = 0.092) enrichment, and our GO term analysis revealed 284 enrichments including lipid metabolic process (P = 9.5 × 10−4) and homeostatic process (P = 3.2 × 10−4) (Supplementary Material S1). Additionally, we were interested in identifying genes under positive selection. Given our limited sample population, we identified genes with maximum FST values (top 5%).42 We found 544 of 2,283 (24%) mammoth substitutions, affecting 446 of 1,888 (24%) protein-coding genes, were in the top 5% of FST values (Supplementary Material S1). Rather than repeat previous observations by Lynch et al. (which we also detected), we report a few interesting findings from our analysis, expanding the list of likely selected variants fixed across the mammoth genomes.
We found three fixed, derived non-synonymous woolly mammoth variants (R381K, A424V, I2640V) in APOB (0.99 FST rank). Multiple alignment, using MUSCLE,21 of the APOB protein sequence for mammoth and 15 other species indicated the V substitution at mammoth residue position 424 was shared with the cold-adapted walrus, the cold-blooded Anole lizard, and platypus (Supplementary Fig. S2). Residue I was pre-dominant at mammoth residue position 2,640 except for a V substitution in mammoth, the cold-adapted Alpine marmot, and the cold-blooded Anole lizard. APOB codes an apolipoprotein for chylomicrons and LDL particles. High levels of the APOB protein have been linked to atherosclerotic plaques.43 In a comparison of polar bears and brown bears, a recent study44 noted nine non-synonymous APOB mutations in polar bears and suggested that a carnivorous diet of pre-dominantly fatty acids induced adaptive changes in APOB. Mammoths likely had a plant-based diet similar to modern elephants.45 Lower ApoB/ApoA1 plasma levels have been noted in cold-adapted human swimmers.46 In carp, cold adaptation included up-regulation of six apolipoprotein genes including APOB.47 We also found three non-synonymous variants in TRPM8, a protein that was briefly noted in Lynch et al. and is responsible for sensitivity to noxious cold. TRPM8 transmembrane domain S2 influences menthol binding and mutation Y745H (mouse) results in a loss of sensitivity to menthol.48 In mammoth, mutations were R368H (h364), G710S (h706), and C711S (h707). On the basis of multiple protein sequence alignment, using T-Coffee, with 16 other organisms, mammoth was the only organism with a substitution of the consensus amino acid at these locations (Supplementary Fig. S3). G710S and C711S occur in S1 (residues 697–716).
Modifications to genes responsible for circadian rhythms have also been identified uniquely in the woolly mammoth. As previously reported by Lynch et al.,9 woolly mammoth had a fixed, non-synonymous variant in PER2. We also found previously unreported fixed, non-synonymous woolly mammoth variants in several important clock genes: ARNTL (BMAL1) of 0.25 FST rank, CRTC1 (1.0 FST rank), KDM5A (0.86 FST rank), and KMT2A (1.0 FST rank). The ARNTL gene is of particular importance as a core circadian oscillator in mammals. ARNTL controls not only the innate 24-h cycle of the organism, but also the transcription of several circadian dependent genes. BMAL1 KO mice have been shown to lose core circadian control, and therefore become arrhythmic.49 Although loss of control may not be beneficial, a weakening of this core oscillator could potentially aid the mammoth in adjusting its behaviour and compensating for the large differences of daylight experienced in summer compared with winter. Another important molecular component of the circadian clock, CRTC1, has a substitution identified in woolly mammoths. This protein is an important member of the signaling pathway for light response and setting the circadian clock of an organism.50 Alterations of this gene can possibly help to force the innate 24-h cycle of an organisms to adapt to the continually variable light–dark cycle of its environment.50 This may have enabled the mammoth to more easily entrain to light and elicit a varied wake response throughout the changing seasons. KDM5A acts on the transcription of the BMAL1/CLOCK complex and works to dampen circadian oscillators.51 KMT2A also plays an essential role in altering the chromatin state of circadian controlled genes.52 These adaptations are possibly due to the increased latitude that woolly mammoths inhabited, forcing a more flexible circadian clock as seasonal changes introduced an extremely varied cycle of light and darkness through the course of the year. Another highly mutated gene was NCKAP5 (0.98 FST rank). The function of NCKAP5 is unknown, but SNPs in NCKAP5 have been linked to hypersomnia.53
KEGG analysis indicated enrichment of ‘pathways in cancer’ (P = 9.6 × 10−5, adjP = 0.0021), which lead us to investigate several cancer-related genes that had multiple non-synonymous variants. BRCA1 (0.98 FST rank), BRCA2 (0.52 FST rank), and PARP14 (0.84 FST rank) had three, seven, and seven non-synonymous variants, respectively. Although BRCA1 and BRCA2 have vital functions, rapid evolution of BRCA1 and BRCA2 has been observed in mammals.54 Citing that ‘nearly all known cases of recurrent positive selection in primate genomes involve genes in one of three categories: (i) immunity, (ii) environmental perception (such as odorant and taste receptors), or (iii) sexual selection and mate choice’,54–56 Lou et al. proposed that rapid BRCA1 and BRCA2 evolution is due to adaptation to viruses. A recent study hypothesized that PARP14 is involved in host–virus defense due to the gene’s strong positive selection in primates.57 This may also explain the high number of fixed, derived woolly mammoth non-synonymous variants we found in PARP14, also involved in DNA repair. Although speculative, possible changes in the repair pathways in mammoth genomes in response to viruses might lend further support to our observation of active development of antiviral mechanisms via amplification of RNase L.
We also investigated the 18 genes with pre-mature stop codons, eight having gene symbols. Genes with pre-mature stop codons had mutations associated with ancient DNA bias (C → T, G → A) in 14 of 18 (78%) cases, considerably higher than the rate observed in Asian elephants (36%). However, one gene with a stop gain, ABCC11 (1.0 FST rank), had four non-synonymous variants, of which two occurred downstream of the stop gain, W703* (Table 4). ABCC11 has been shown to determine wet or dry ear wax58 in humans, and the ear wax single nucleotide polymorphism, rs17822931-G/A (G180R), was shown to be absent in Africans and increase in prevalence with absolute latitude.59 Ohashi et al. also indicated that rs17822931-A resulted in loss of function. ABCC11 also has roles involving bile acids, conjugated steroids, and cyclic nucleotides.60,61
Table 4.
Fixed, derived non-synonymous woolly mammoth variants in ABCC11
| Location | Variant | Protein position | Amino acids (elephant/mammoth) | Codons (elephant/mammoth) |
|---|---|---|---|---|
| scaffold_43:16997813 | Missense variant | 115 | S/G | Agt/Ggt |
| scaffold_43:16985516 | Missense variant | 359 | M/L | Atg/Ctg |
| scaffold_43:16957501 | Stop gained | 703 | W/* | tgG/tgA |
| scaffold_43:16957475 | Missense variant | 712 | G/E | gGa/gAa |
| scaffold_43:16919280 | Missense variant | 1,278 | Q/P | cAa/cCa |
Mitochondrial phylogenetic analysis has suggested two primary clades of woolly mammoth, clades I and II, that are thought to have evolved in isolation on opposite sides of the Bering Strait.7 Our dataset contained two mammoths from each major clade, clade I (Wrangel and M4) and clade II (Oimyakon and M25). Clade II disappeared ∼30,000–40,000 years prior to the extinction of clade I. We investigated the differences between the genomes of clades I and II and found four fixed, derived clade I CNVs, three deletions and one duplication, none of which occurred in exonic regions. Similarly, there were two fixed, derived clade II CNVs, both duplications, but neither occurred in exons. We found 57 fixed, derived clade I indels (11 insertions and 46 deletions), and 65 fixed, derived clade II indels (24 insertions and 41 deletions), none of which occurred in exons. For SNVs, we found 1,215 fixed, derived clade I variants, of which 279 and five occurred in introns and exons, respectively. Four were non-synonymous variants in protein-coding genes (Table 5). Clade II had 584 fixed SNVs, 125 and three occurred in introns and exons, respectively. Two non-synonymous clade II variants occurred in protein-coding genes (Table 5).
Table 5.
Fixed, derived non-synonymous clade variants
| Clade | Location | SYMBOL | Protein position | Amino acids (ele/mam) | Codons (ele/mam) |
|---|---|---|---|---|---|
| I | scaffold_25:32550639 | ZDHHC23 | 129 | K/E | Aag/Gag |
| I | scaffold_3:68117492 | ENSLAFG00000010153 | 891 | R/L | cGt/cTt |
| I | scaffold_40:10330726 | SULT6B1 | 115 | R/Q | cGa/cAa |
| I | scaffold_64:11532808 | SPTBN5 | 901 | G/R | Ggg/Agg |
| II | scaffold_125:2369691 | CCDC94 | 292 | P/L | cCg/cTg |
| II | scaffold_81:784525 | ENSLAFG00000032374 | 232 | T/M | aCg/aTg |
Non-synonymous clade variants were homozygous in the clade and had no evidence of the variant in the Asian elephants or the other clade. Ensembl gene ID used when gene symbol not available.
Clade I genes with fixed, derived amino acid substitutions included ZDHHC23, SULT6B1, and SPTBN5. Of most interest, SULT6B1 is a sulfotransferase that utilizes 3-phospho-5-adenylyl sulphate (PAPS) to catalyze the sulphate conjugation of thyroxine and is involved in the metabolism of thyroxine.62 Unlike other SULTs, SULT6B1 is uniquely specific for thyroxine, suggesting a role in regulation of thyroxine.62 Thyroxine (T4) is a thyroid hormone involved in growth, development, differentiation, and basal metabolic homeostasis, as well as the regulation of protein, fat, and carbohydrate metabolism.63–65 Also, it is involved in facultative thermogenesis (heat production in response to cold or overeating).66 Administering thyroid hormones results in a severe drop in body temperature.67
Clade II had a fixed, derived amino acid substitution in CCDC94. CCDC94 knockdown in zebrafish increased sensitivity of ionizing radiation-induced apoptosis and CCDC94 protects cells from ionizing radiation-induced apoptosis by repressing expression of p53 mRNA.68
GO terms and KO mammalian phenotypes for genes with fixed, derived non-synonymous clade I and II variants are summarized in Supplementary Tables S3 and S4, respectively. Interestingly, fixed, derived non-synonymous clade I variant ZDHHC23 and clade II variant CCDC94 had KO phenotypes ‘increased caudal vertebrae number’ and ‘decreased caudal vertebrae number’, respectively. At the time of writing, it was unknown to us if clade I and clade II differed in their number of tail bones. The caudal vertebrae number of woolly mammoths has been estimated to be 21 but has not been ‘confidently established’27 as few complete skeletons have been found. Modern elephants have 28–33 caudal bones.27 Woolly mammoth shorter tail adaptation likely reduced heat loss and frostbite.69
3.3. Indels
We found 20,576 fixed, derived woolly mammoth indels, 2,413 insertions and 18,163 deletions. Of these, 597 insertion indels and 5,174 deletion indels overlapped introns. Exons were found to harbor four insertion and 16 deletion indels (Table 6). All fixed, derived woolly mammoth indels were <3 bases in length resulting in frameshifts and were classified as high impact variants by VEP (ensembl.org). A number of protein-coding gene regions disrupted by an indel showed interesting functions, potentially relevant to woolly mammoth adaptation. Supplementary Table S5 lists GO terms and KO phenotypes for fixed, derived woolly mammoth indels affecting exonic regions. WWC1 functions as a tumor suppressor regulating Hippo signaling70,71 and has GO term ‘negative regulation of organ growth’. In agreement with SNV analysis in the present study and by Lynch et al.,9 we found an abundance of fixed, derived woolly mammoth variants in genes involved with lipid metabolism, possibly an adaptation for storing fat (e.g., ETNK1, lipid metabolic process).
Table 6.
Fixed, derived woolly mammoth indels occurring in exons
| Indel | Location | Gene | Consequence | Type |
|---|---|---|---|---|
| DEL | scaffold_1:91019166-91019166 | WWC1 | Frameshift | Protein |
| DEL | scaffold_2:12181244-12181244 | ETNK1 | Frameshift | Protein |
| DEL | scaffold_4:40567579-40567579 | ENSLAFG00000029865 | Frameshift | Protein |
| DEL | scaffold_6:80415826-80415826 | ADAMTSL1 | Frameshift | Protein |
| DEL | scaffold_6:82071600-82071600 | ENSLAFG00000027513 | Frameshift, stop lost | Protein |
| DEL | scaffold_7:76407941-76407941 | ARHGEF28 | Frameshift | Protein |
| DEL | scaffold_10:17436611-17436611 | PAX2 | Frameshift | Protein |
| DEL | scaffold_15:54351650-54351650 | ENSLAFG00000028486 | Frameshift | Protein |
| DEL | scaffold_153:798742-798742 | SBNO2 | Frameshift | Protein |
| DEL | scaffold_16:21968006-21968006 | RALGPS2 | Frameshift | Protein |
| DEL | scaffold_35:4581294-4581294 | GPR83 | Frameshift | Protein |
| DEL | scaffold_36:4137479-4137479 | ARPP21 | Frameshift | Protein |
| DEL | scaffold_63:11761905-11761905 | PGAM2 | Frameshift | Protein |
| DEL | scaffold_63:13307853-13307854 | SNORA5 | Non coding exon | snoRNA |
| DEL | scaffold_68:424956-424956 | ENSLAFG00000026930 | Frameshift | Protein |
| DEL | scaffold_91:278361-278361 | ENSLAFG00000027842 | Frameshift | Protein |
| INS | scaffold_7:56774332-56774332 | ENSLAFG00000027421 | Coding sequence | Protein |
| INS | scaffold_43:286385-286385 | KIFC3 | Coding sequence | Protein |
| INS | scaffold_96:4058660-4058660 | ENSLAFG00000032317 | Coding sequence | Protein |
| INS | scaffold_107:2600881-2600881 | NOL8 | Coding sequence | Protein |
Ensembl gene ID used when gene symbol not available.
Of particular interest, GPR83 is a member of the G protein-coupled receptor subfamily72 that includes several receptors linked to the regulation of metabolism.GPR83 is expressed in the hypothalamus and regulated by nutrient availability.73 GPR83 knock-out mice have shown a significant increase in food intake compared with wild-type mice but appeared to be protected from obesity and glucose intolerance and have normal insulin sensitivity, regardless of diet type.73 Another study found that GPR83 knockdown mice had increased levels of weight gain compared with wild-type mice, despite consuming an identical amount of food. Additionally, the knockdown of GPR38 promoted a decrease in core body temperature during the daily activity period.74 Increases in weight gain and lower core body temperatures have shown to be advantageous to mice when presented with lower ambient environmental temperatures. Mice that had developed these traits were shown to have a stable metabolic rate and more easily maintained core body temperatures in ambient cold temperatures compared with their wild-type counterparts.75 These studies suggest a possible role for modification or loss-of-function of GPR83 transcripts in the survival of mammoths considering the harsh environment of their habitat.
Another affected gene, RALGPS2, had one mammalian KO phenotype, ‘increased rib number’. In mammals, the number of cervical vertebrae is highly conserved at seven. Approximately 90 percent of humans with a cervical rib die before reaching reproductive age,76 due to a strong association with multiple major congenital abnormalities. However, a recent study77 found abnormal cervical rib numbers in Late Pleistocene mammoths (33%), a rate 10 times higher than in extant elephants (3.6%), which appears consistent with our finding. The abnormal numbers were due to large cervical ribs on the seventh vertebra and mammoth cervical ribs were relatively large compared with cervical ribs in humans.77
Several genes with fixed, derived woolly mammoth indels had mammalian KO phenotypes related to decreased body size: decreased birth body size (GPR83), decreased body length (ARHGEF28), and decreased body weight (NOL8), and as noted earlier, CD44 KO was linked to short tibias. Towards the end of their existence mammoths decreased in size.27 All samples in this study were near the end of the mammoths existence, from 60,000 years ago or more recent. The cause of the extinction of woolly mammoths is unknown but is often regarded to be due to a warming climate and hunting by humans. We note that several genes with fixed, derived woolly mammoth indels had detrimental reproductive KO phenotypes, such as NOL8 (pre-weaning lethality), SBNO2 (pre-weaning lethality), and PAX2 (postnatal lethality and perinatal lethality). ARHGEF28 had the mammalian KO phenotype ‘reproductive system’. Additionally, we found that all genes with fixed, derived woolly mammoth indels (and with a gene symbol) occurred in one or more recent screens of gene essentiality (Table 7).
Table 7.
Essentiality of genes with fixed, derived woolly mammoth indels occurring in protein-coding regions
| Gene symbol | Indel | Studies indicating gene essential |
|---|---|---|
| KIFC3 | INS | a, b |
| NOL8 | INS | a, b, c |
| WWC1 | DEL | a, b |
| ETNK1 | DEL | a, b |
| ADAMTSL1 | DEL | a |
| ARHGEF28 | DEL | a |
| PAX2 | DEL | a |
| RALGPS2 | DEL | a |
| GPR83 | DEL | a |
| ARPP21 | DEL | a |
| PGAM2 | DEL | a |
| SBNO2 | DEL | a |
4. Discussion
We performed systematic detection of variants, ranging from single nucleotide changes and short indels to deletions and amplifications of regions encompassing gene fragments and complete genes in four woolly mammoth genomes, comparing them to the elephant genomes. Extending the findings of previous SNV-only analyses, we found many unique woolly mammoth genetic variants involved in processes relevant to living in a harsh, cold environment, such as lipid metabolism and thermogenesis. We also observed changes in the core circadian oscillator genes that may have occurred as woolly mammoths adapted to the extremely varied cycle of light and darkness through the course of the year in the Arctic. Further, we noticed many immunity-related variants, most strikingly the copy number amplification of RNase L, important in antiviral response. Immunity tends to evolve rapidly and woolly mammoth genomes display signs of adaptation to the differences in environmental threats they faced compared with their tropical/subtropical elephant counterparts. The haplotype patterns in RNase L gene described above show similarities in clade II but differences in clade I, likely reflecting the history of development of multiple defense mechanisms, which appear to be absent (or reduced) in the modern elephants. Do multiple RNase L paralogs and multiple non-synonymous changes in the repair pathways indicate that mammoths faced much deadlier viral threats? Were those threats further exacerbated by the cold Arctic conditions? Was this possibly linked to the ultimate demise of mammoths?
African elephants have been shown to have at least 20 copies of the tumor suppressor TP53, possibly explaining their low cancer mortality despite their large size and long life span.23 Such TP53 locus amplification is also present in woolly mammoth genomes making RNase L their second largest CNV. It is worth noting that RNase L may also play a role in tumor suppression78 and has been linked to prostate cancer.79
Recently, other avenues of analysis of ancient DNA have been proposed, related to CpG methylation and corresponding epigenetic patterns (as summarized in a recent review80). Sampling based on methylated binding domains (MBD) has been performed in a number of organisms, including two other woolly mammoths.81 We analysed these MDB mammoth samples but found the genome coverage to be too fragmentary (as estimated by total SNP counts from all MDB- and MBD+ samples <0.3% of SNPs in a typical mammoth genome and by nearly negligible overlap with a union of SNPs from our study) to draw any meaningful conclusions.
And as for the differences between clades I and II, although only a few are reported here, some variants may represent continuing evolution of adaptations to a cold environment. Fixed non-synonymous variants in clade I (ZDHHC23) and clade II (CCDC94) have been shown to affect the number of tail bones and clade I non-synonymous variant SULT6B1 plays a role in thyroid hormone regulation. Since clade II disappeared during the last glacial period prior to rising temperatures, these variants might have contributed to the longer survival of clade I mammoths.
Taken together, these observations point to a rich population history of these iconic animals extending beyond the description based on three clades.
Supplementary data
Supplementary data are available at DNARES Online.
Supplementary Material
Acknowledgement
This work was supported by the National Science Foundation (award DBI-1458202 to A.G.).
Conflict of interest
None declared.
References
- 1. Palkopoulou E., Mallick S., Skoglund P., et al. 2015, Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol.: CB, 25, 1395–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vartanyan S.L., Arslanov K.A., Karhu J.A., Possnert G., Sulerzhitsky L.D.. 2008, Collection of radiocarbon dates on the mammoths (Mammuthus primigenius) and other genera of Wrangel Island, northeast Siberia, Russia. Quatern. Res., 70, 51–59. [Google Scholar]
- 3. MacDonald G.M., Beilman D.W., Kuzmin Y.V., et al. 2012, Pattern of extinction of the woolly mammoth in Beringia. Nat. Commun., 3, 893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fisher D.C., Tikhonov A.N., Kosintsev P.A., Rountrey A.N., Buigues B., van der Plicht J.. 2012, Anatomy, death, and preservation of a woolly mammoth (Mammuthus primigenius) calf, Yamal Peninsula, northwest Siberia. Quatern. Int, 255, 94–105. [Google Scholar]
- 5. Hill C.L. 2009, Mammoths: giants of the ice age. Revised edition. Adrian Lister and Paul Bahn, 2007, University of California Press, Berkeley, CA, 192 pp., $29.95 (hardcover). Geoarchaeology, 24, 117–119. [Google Scholar]
- 6. Repin V.E., Taranov O.S., Ryabchikova, Tikhonov A.N., Pugachev V.G.. 2004, Sebaceous glands of the woolly mammoth, Mammothus primigenius Blum: histological evidence. Doklady Biol. Sci.: Proc. Acad. Sci. USSR, Biol. Sci. Sect./Transl. Russ., 398, 382–384. [DOI] [PubMed] [Google Scholar]
- 7. Palkopoulou E., Dalen L., Lister A.M., et al. 2013, Holarctic genetic structure and range dynamics in the woolly mammoth. Proc. R. Soc. B-Biol. Sci., 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller W., Drautz D.I., Ratan A., et al. 2008, Sequencing the nuclear genome of the extinct woolly mammoth. Nature, 456, 387–390. [DOI] [PubMed] [Google Scholar]
- 9. Lynch V.J., Bedoya-Reina O.C., Ratan A., et al. 2015, Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the arctic. Cell Rep., 12, 217–228. [DOI] [PubMed] [Google Scholar]
- 10. Dastjerdi A., Robert C., Watson M.. 2014, Low coverage sequencing of two Asian elephant (Elephas maximus) genomes. Gigascience, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert M.T., Drautz D.I., Lesk A.M., et al. 2008, Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc. Natl. Acad. Sci. U. S. A., 105, 8327–8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith S.D., Kawash J.K., Grigoriev A.. 2015, GROM-RD: resolving genomic biases to improve read depth detection of copy number variants. PeerJ, 3, e836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Handsaker B., Wysoker A., et al. 2009, The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson J.T., Thorvaldsdottir H., Winckler W., et al. 2011, Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas P.D., Campbell M.J., Kejariwal A., et al. 2003, PANTHER: a library of protein families and subfamilies indexed by function. Genome Res., 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang B., Kirov S., Snoddy J.. 2005, WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res., 33, W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weir B.S., Cockerham C.C.. 1984, Estimating F-statistics for the analysis of population-structure. Evol. Int. J. Organ. Evol., 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 19. Danecek P., Auton A., Abecasis G., et al. 2011, The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Notredame C., Higgins D.G., Heringa J.. 2000, T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 21. Edgar R.C. 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res., 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abyzov A., Urban A.E., Snyder M., Gerstein M.. 2011, CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res., 21, 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abegglen L.M., Caulin A.F., Chan A., et al. 2015, Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA, 314, 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sneath R.J., Mangham D.C.. 1998, The normal structure and function of CD44 and its role in neoplasia. Mol. Pathol.: MP, 51, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zoller M. 2015, CD44, hyaluronan, the hematopoietic stem cell, and leukemia-initiating cells. Front. Immunol., 6, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ameyar-Zazoua M., Rachez C., Souidi M., et al. 2012, Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol., 19, 998–1004. [DOI] [PubMed] [Google Scholar]
- 27. Haynes G. 1993, Mammoths, Mastodons and Elephants – Biology, Behavior and the Fossil Record. Cambridge University Press: New York. [Google Scholar]
- 28. Brennan-Laun S.E., Ezelle H.J., Li X.L., Hassel B.A.. 2014, RNase-L control of cellular mRNAs: roles in biologic functions and mechanisms of substrate targeting. J. Interf. Cytok. Res., 34, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fabre O., Salehzada T., Lambert K., et al. 2012, RNase L controls terminal adipocyte differentiation, lipids storage and insulin sensitivity via CHOP10 mRNA regulation. Cell Death Differ., 19, 1470–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salehzada T., Cambier L., Thi N.V., Manchon L., Regnier L., Bisbal C.. 2009, Endoribonuclease L (RNase L) regulates the myogenic and adipogenic potential of myogenic cells. PLoS ONE, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang H., Zeqiraj E., Dong B.H., et al. 2014, Dimeric structure of pseudokinase RNase L bound to 2–5A reveals a basis for interferon-induced antiviral activity. Mol. Cell, 53, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han Y.C., Donovan J., Rath S., Whitney G., Chitrakar A., Korennykh A.. 2014, Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science, 343, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ezelle H.J., Malathi K., Hassel B.A.. 2016, The roles of RNase-L in antimicrobial immunity and the cytoskeleton-associated innate response. Int. J. Mol. Sci., 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakrabarti A., Banerjee S., Franchi L., et al. 2015, RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe, 17, 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nour A.M., Reichelt M., Ku C.C., Ho M.Y., Heineman T.C., Arvin A.M.. 2011, Varicella-Zoster virus infection triggers formation of an interleukin-1 beta (IL-1 beta)-processing inflammasome complex. J. Biol. Chem., 286, 17921–17933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richman L.K., Zong J.C., Latimer E.M., et al. 2014, elephant endotheliotropic herpesviruses EEHV1A, EEHV1B, and EEHV2 from cases of hemorrhagic disease are highly diverged from other mammalian herpesviruses and may form a new subfamily. J. Virol., 88, 13523–13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Atchison C.J., Tam C.C., Hajat S., van Pelt W., Cowden J.M., Lopman B.A.. 2010, Temperature-dependent transmission of rotavirus in Great Britain and The Netherlands. Proc. Biol. Sci./R. Soc., 277, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tani N., Dohi Y., Kurumatani N., Yonemasu K.. 1995, Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol., 39, 577–580. [DOI] [PubMed] [Google Scholar]
- 39. Lowen A.C., Mubareka S., Steel J., Palese P.. 2007, Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog., 3, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zao C.L., Ward J.A., Tomanek L., Cooke A., Berger R., Armstrong K.. 2011, Virological and serological characterization of SRV-4 infection in cynomolgus macaques. Arch. Virol., 156, 2053–2056. [DOI] [PubMed] [Google Scholar]
- 41. Briggs A.W., Stenzel U., Johnson P.L.F., et al. 2007, Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U. S. A., 104, 14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou X., Meng X., Liu Z., et al. 2016, population genomics reveals low genetic diversity and adaptation to hypoxia in Snub-nosed monkeys. Mol. Biol. Evol., 33(10), 2670–2681. [DOI] [PubMed] [Google Scholar]
- 43. McQueen M.J., Hawken S., Wang X.Y., et al. 2008, Lipids, lipoproteins, and apolipoproteins as risk markers of myocardial infarction in 52 countries (the INTERHEART study): a case–control study. Lancet, 372, 224–233. [DOI] [PubMed] [Google Scholar]
- 44. Liu S., Lorenzen E.D., Fumagalli M., et al. 2014, Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell, 157, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwartz-Narbonne R., Longstaffe F.J., Metcalfe J.Z., Zazula G.. 2015, Solving the woolly mammoth conundrum: amino acid (1)(5)N-enrichment suggests a distinct forage or habitat. Sci. Rep., 5, 9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lesna I.K., Rychlikova J., Vavrova L., Vybiral S.. 2015, Could human cold adaptation decrease the risk of cardiovascular disease? J. Therm. Biol., 52, 192–198. [DOI] [PubMed] [Google Scholar]
- 47. Gracey A.Y., Fraser E.J., Li W., et al. 2004, Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Natl. Acad. Sci. U. S. A., 101, 16970–16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montell C. 2006, A mint of mutations in TRPM8 leads to cool results. Nat. Neurosci., 9, 466–468. [DOI] [PubMed] [Google Scholar]
- 49. Lowrey P.L., Takahashi J.S.. 2011, Genetics of circadian rhythms in mammalian model organisms. Adv. Genet., 74, 175–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jagannath A., Butler R., Godinho S.I., et al. 2013, The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell, 154, 1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DiTacchio L., Le H.D., Vollmers C., et al. 2011, Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science, 333, 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katada S., Sassone-Corsi P.. 2010, The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol., 17, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khor S.S., Miyagawa T., Toyoda H., et al. 2013, Genome-wide association study of HLA-DQB1*06:02 negative essential hypersomnia. PeerJ, 1, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lou D.I., McBee R.M., Le U.Q., et al. 2014, Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evol. Biol., 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Clark A.G., Glanowski S., Nielsen R., et al. 2003, Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science, 302, 1960–1963. [DOI] [PubMed] [Google Scholar]
- 56. Vallender E.J., Lahn B.T.. 2004, Positive selection on the human genome. Hum. Mol. Genet., 13, R245–254. [DOI] [PubMed] [Google Scholar]
- 57. Daugherty M.D., Young J.M., Kerns J.A., Malik H.S.. 2014, Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host–virus conflicts. PLoS genetics, 10, e1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoshiura K., Kinoshita A., Ishida T., et al. 2006, A SNP in the ABCC11 gene is the determinant of human earwax type. Nat. Genet., 38, 324–330. [DOI] [PubMed] [Google Scholar]
- 59. Ohashi J., Naka I., Tsuchiya N.. 2011, The impact of natural selection on an ABCC11 SNP determining earwax type. Mol. Biol. Evol., 28, 849–857. [DOI] [PubMed] [Google Scholar]
- 60. Chen Z.S., Guo Y., Belinsky M.G., Kotova E., Kruh G.D.. 2005, Transport of bile acids, sulfated steroids, estradiol 17-beta-D-glucuronide, and leukotriene C4 by human multidrug resistance protein 8 (ABCC11). Mol. Pharmacol., 67, 545–557. [DOI] [PubMed] [Google Scholar]
- 61. Guo Y.P., Kotova E., Chen Z.S., et al. 2003, MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2ʹ,3ʹ-dideoxycytidine and 9ʹ-(2ʹ-phosphonylmethoxyethyl) adenine. J. Biol. Chem., 278, 29509–29514. [DOI] [PubMed] [Google Scholar]
- 62. Takahashi S., Sakakibara Y., Mishiro E., et al. 2009, Molecular cloning, expression and characterization of a novel mouse SULT6 cytosolic sulfotransferase. J. Biochem., 146, 399–405. [DOI] [PubMed] [Google Scholar]
- 63. Bates P.C., Holder A.T.. 1988, The anabolic actions of growth hormone and thyroxine on protein metabolism in Snell dwarf and normal mice. J. Endocrinol., 119, 31–41. [DOI] [PubMed] [Google Scholar]
- 64. Boelen A. 2009, Thyroid hormones and glucose metabolism: the story begins before birth. Exp. Physiol., 94, 1050–1051. [DOI] [PubMed] [Google Scholar]
- 65. Pucci E., Chiovato L., Pinchera A.. 2000, Thyroid and lipid metabolism. Int. J. Obes. Relat. Metab. Disord.: J. Int. Assoc. Study Obes., 24(Suppl. 2), S109–112. [DOI] [PubMed] [Google Scholar]
- 66. Silva J.E. 2001, The multiple contributions of thyroid hormone to heat production. J. Clin. Investig., 108, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Doyle K.P., Suchland K.L., Ciesielski T.M., et al. 2007, Novel thyroxine derivatives, thyronamine and 3-iodothyronamine, induce transient hypothermia and marked neuroprotection against stroke injury. Stroke; J. Cereb. Circ., 38, 2569–2576. [DOI] [PubMed] [Google Scholar]
- 68. Sorrells S., Carbonneau S., Harrington E., et al. 2012, Ccdc94 protects cells from ionizing radiation by inhibiting the expression of p53. PLoS Genet., 8, e1002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Campbell K.L., Roberts J.E., Watson L.N., et al. 2010, Substitutions in woolly mammoth hemoglobin confer biochemical properties adaptive for cold tolerance. Nat. Genet., 42, 536–540. [DOI] [PubMed] [Google Scholar]
- 70. Xiao L., Chen Y.H., Ji M., Dong J.X.. 2011, KIBRA regulates hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem., 286, 7788–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu J.Z., Zheng Y.G., Dong J.X., Klusza S., Deng W.M., Pan D.J.. 2010, Kibra functions as a tumor suppressor protein that regulates hippo signaling in conjunction with Merlin and expanded. Dev. Cell., 18, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oh D.Y., Kim K., Kwon H.B., Seong J.Y.. 2006, Cellular and molecular biology of orphan G protein-coupled receptors. Int. Rev. Cytol., 252, 163–218. [DOI] [PubMed] [Google Scholar]
- 73. Muller T.D., Muller A., Yi C.X., et al. 2013, The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat. Commun., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dubins J.S., Sanchez-Alavez M., Zhukov V., et al. 2012, Downregulation of GPR83 in the hypothalamic preoptic area reduces core body temperature and elevates circulating levels of adiponectin. Metab.: Clin. Exp., 61, 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kaplan M.L., Leveille G.A.. 1974, Core temperature, O2 consumption, and early detection of ob-ob genotype in mice. Am. J. Physiol., 227, 912–915. [DOI] [PubMed] [Google Scholar]
- 76. Galis F., Van Dooren T.J., Feuth J.D., et al. 2006, Extreme selection in humans against homeotic transformations of cervical vertebrae. Evol.; Int. J. Organ. Evol., 60, 2643–2654. [PubMed] [Google Scholar]
- 77. Reumer J.W., Ten Broek C.M., Galis F.. 2014, Extraordinary incidence of cervical ribs indicates vulnerable condition in Late Pleistocene mammoths. PeerJ, 2, e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chakrabarti A., Jha B.K., Silverman R.H.. 2011, New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res.: Off. J. Int. Soc. Interferon Cytokine Res., 31, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meyer M.S., Penney K.L., Stark J.R., et al. 2010, Genetic variation in RNASEL associated with prostate cancer risk and progression. Carcinogenesis, 31, 1597–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Morozova I., Flegontov P., Mikheyev A.S., et al. 2016, Toward high-resolution population genomics using archaeological samples. DNA Res., 23, 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Seguin-Orlando A., Gamba C., Der Sarkissian C., et al. 2015, Pros and cons of methylation-based enrichment methods for ancient DNA. Sci. Rep., 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang T., Birsoy K., Hughes N.W., et al. 2015, Identification and characterization of essential genes in the human genome. Science, 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hart T., Chandrashekhar M., Aregger M., et al. 2015, High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell, 163. [DOI] [PubMed] [Google Scholar]
- 84. Blomen V.A., Majek P., Jae L.T., et al. 2015, Gene essentiality and synthetic lethality in haploid human cells. Science, 350, 1092–1096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.