Abstract
To date, little is known about the evolution of plastid genomes (plastomes) in Lauraceae. As one of the top five largest families in tropical forests, the Lauraceae contain many species that are important ecologically and economically. Lauraceous species also provide wonderful materials to study the evolutionary trajectory in response to parasitism because they contain both nonparasitic and parasitic species. This study compared the plastomes of nine Lauraceous species, including the sole hemiparasitic and herbaceous genus Cassytha (laurel dodder; here represented by Cassytha filiformis). We found differential contractions of the canonical inverted repeat (IR), resulting in two IR types present in Lauraceae. These two IR types reinforce Cryptocaryeae and Neocinnamomum—Perseeae–Laureae as two separate clades. Our data reveal several traits unique to Cas. filiformis, including loss of IRs, loss or pseudogenization of 11 ndh and rpl23 genes, richness of repeats, and accelerated rates of nucleotide substitutions in protein-coding genes. Although Cas. filiformis is low in chlorophyll content, our analysis based on dN/dS ratios suggests that both its plastid house-keeping and photosynthetic genes are under strong selective constraints. Hence, we propose that short generation time and herbaceous lifestyle rather than reduced photosynthetic ability drive the accelerated rates of nucleotide substitutions in Cas. filiformis.
Keywords: laurel, Lauraceae, hemiparasitic plant, plastid genome, substitution rate, genome rearrangement
Introduction
The primary function of plastids is photosynthesis, which produces essential carbon resources and allows for autotrophy. Plastid genomes (plastomes) of seed plants are highly conserved in size, organization, and gene content (Jansen and Ruhlman 2012). They typically have a quadripartite structure with a pair of canonical inverted repeats (IRA, IRB) separated by large single copy (LSC) and small single copy (SSC) regions. However, when plants encounter lost or decreased photosynthetic (PS) ability, their plastomes are subjected to relaxation of selective constraints on the organization and gene content. As a result, reductive and rearranged plastomes are often a hallmark of parasitic plants (Krause 2011; Wicke etal. 2011).
Parasitic plants provide excellent materials to study plastomic evolution in response to altered lifestyles. Previous studies have shown gene loss and plastome reduction in relation to the degree of parasitism in Cuscuta (Braukmann etal. 2013) and Orobanchaceae (Wicke etal. 2016). Parasitic plants have developed a differential organ, the haustorium, to penetrate hosts for exploiting water and nutrients. In contrast, mycoheterotrophic plants do not penetrate another plant and obtain living resources with help from fungi. Despite their different ways of survival, these two groups share a similar pattern related to loss of plastid genes (Barrett etal. 2014; Wicke etal. 2016), which indicates the convergence of plastomic evolution.
There are ∼4,500 parasitic plants in 20 flowering plant families. It was proposed that flowering parasites have evolved at least 11 times independently (Barkman etal. 2007). Despite low efficiency, most flowering parasites are still able to perform photosynthesis (Heide-Jørgesen 2008). Thus, PS parasites are all hemiparasitic. However, only a tiny fraction, circa 390 species, of flowering parasites are holoparasitic. These holoparasites lack PS ability and require hosts to complete their life cycle. Retention of PS ability in hemiparasites was thought to have advantages in providing energy before parasitizing hosts or to support development of flowers and fruit (Těšitel 2016).
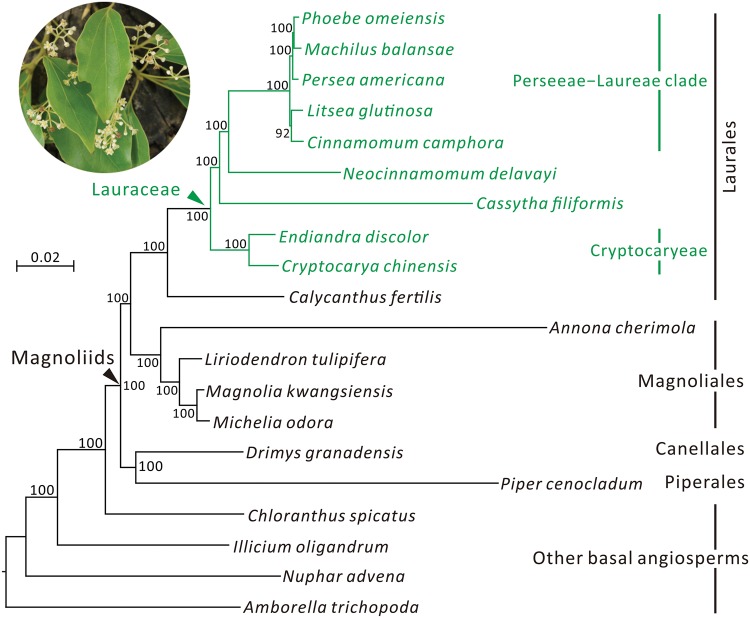
Lauraceae (laurel family) is the largest family of Laurales, grouped in magnoliids of the “basal angiosperms” clade (Jansen etal. 2007). The laurel family includes ∼3,000 species (Christenhusz and Byng 2016). They are woody trees or shrubs, except for the herbaceous vine genus Cassytha, the laurel dodder, which contains a dozen hemiparasitic species. Although many Lauraceous species are the major components of tropical and subtropical forests, the availability of their plastomes is limited to nine species that represent only six of the 45 genera (http://www.ncbi.nlm.nih.gov/genbank/, last accessed on February 2017). Previously, molecular phylogenetics of Lauraceae was mostly based on a single or a few loci (Rohwer 2000; Chanderbali etal. 2001; Rohwer and Rudolph 2005; Wang etal. 2010). The plastid phylogenomic approach, which constructs trees from most of the genes encoded in plastomes, was rarely used for estimating intergeneric relationships of Lauraceae. In addition, unlike the other parasitic family of magnoliids, Hydnoraceae, in which all species are parasitic, the sole herbaceous and parasitic genus, Cassytha, allows for within-family comparisons of plastomes between parasitic and nonparasitic species and between herbaceous and woody plants.
To this end, we have sequenced the plastomes of Cassytha filiformis and its three nonparasitic relatives. With these and other available plastomes, we aimed to 1) use plastid phylogenomics to infer the phylogeny of Lauraceous genera, 2) uncover the dynamics of plastomic organizations and nucleotide substitution rates in the laurel family, and 3) compare the plastomes between parasitic and nonparasitic species within Lauraceae to understand the evolution of traits relevant to altered lifestyles in the parasitic Cas. filiformis.
Materials and Methods
Collection of Plant Materials
Fresh stems of Cas. filiformis (voucher Chaw 1498) were collected from an individual growing at the seashore in Tainan City. For Cinnamomum camphora (voucher Chaw 1510), Cryptocarya chinensis (voucher Chaw 1500), and Neocinnamomum delavayi (voucher Chaw 1499), fresh leaves were collected separately, with the former two growing in Taipei Botanical Garden and the latter in Kunming Botanical Garden.
DNA Extraction and Plastome Sequencing and Assembly
For each species, 2 g ground tissue was used for DNA extraction with the CTAB method (Stewart and Via 1993). The yielded DNA was sheared into 450 bp fragments for library construction. For Cas. filiformis, Cin. camphora, and Cry. chinensis, sequencing was conducted on an Illumina Hiseq 2500 platform at Yourgene Bioscience (New Taipei City), and ∼2 Gb of 126 bp paired-end reads was obtained for each species. For Neo. delavayi, ∼2 Gb of 150 bp paired-end reads were sequenced by using the Illumina NextSeq 500 platform at Genomics (New Taipei City).
We used CLC Genomics Workbench 4.9 (CLC Bio, Arhus, Denmark) for quality trimming and de novo assembling. The thresholds for trimming were error probability < 0.01, ambiguous nucleotides = 1, and removal of <15 bp reads, and parameters for assembling were word size = 31, bubble size = 50, minimum contig length = 1 kb, and mapping reads to contigs. Contigs with ≥ 50× sequencing depths were used in BLAST searches against the plastome sequences of Machilus balansae (NC028074), and those with <10−10E-values were considered plastomic contigs. Gaps between the plastomic contigs were closed with sequences of PCR amplicons obtained by using specific primers (supplementary table S1, Supplementary Material online). The complete plastome sequences were imported into CLC Genomics Workbench to estimate average sequencing depths with the parameters of mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, and similarity fraction = 0.8. Plastome maps were visualized by using Circos 0.67 (Krzywinski etal. 2009).
Plastome Annotation
Gene prediction was performed in DOGMA (Wyman etal. 2004) and tRNA genes were predicted by using tRNAscan-SE 1.21 (Schattner etal. 2005). The boundaries of predicted genes/exons were confirmed by aligning them with their orthologous genes/exons of other magnoliid species. Repeats were identified with BlastN (NCBI) by searching each plastome against itself. We discarded the repeats with sequence identity < 90%.
Sequence Alignment and Phylogenetic Tree Construction
Sequences of the 67 shared protein-coding genes were extracted from the plastomes of the 20 examined basal angiosperms (supplementary table S1, Supplementary Material online). For each genes, MUSCLE (Edgar 2004) implemented in MEGA 6.06 (Tamura etal. 2013) was used for sequence alignment with the option of aligning codons. All gene alignments were concatenated into a supermatrix. We used MEGA to remove ambiguous sites and gaps with the option “Exclude sites with missing/ambiguous data and gap”. The best-fit substitution model for the concatenated genes was assessed by use of Jmodeltest 2.1.10 (Darriba etal. 2012) with the AIC method to compare the likelihood scores. We used RAxML v8.2.4 (Stamatakis 2014) to analyze a maximum likelihood (ML) tree based on the concatenated genes under a GTRGAMMAI model. Amborella trichopoda was the outgroup. Bootstrap support values for the tree nodes were assessed with 1,000 nonparametric replicates. A maximum parsimony (MP) tree was built using MEGA with the Tree Bisection-Reconnection method and 1,000 bootstrap replicates.
Identification of Syntenic Nongenic Loci
To examine whether shrinkage of nongenic loci (including intergenic and intronic loci) contributed to plastome reduction, we compared the lengths of syntenic nongenic loci among Cas. filformis, Cin. camphora, Cry. chinensis, and Neo. delavayi. A nongenic region was identified as a syntenic locus when its flanking genes were the same among the examined laurels.
Estimation of Nucleotide Substitution Rates
According to the criterion described in Wicke etal. (2016), the 67 shared genes were divided into house-keeping (HK) and PS genes. In addition, accD, clpP, ycf1, and ycf2 genes for a group of “other pathway” in Wicke etal. (2016) were classified as HK genes in our study. Both nonsynonymous (dN) and synonymous (dS) substitution rates for the concatenated HK or PS genes, respectively, were estimated by using the codeml program of PAML 4.8 (Yang 2007). The parameters were runmode = 0, seqtype = 1, CodonFreq = 2, estFreq = 0, and model = 1, and the constrained tree was the ML tree shown in figure 2.
Fig. 2.
—ML tree inferred from concatenation of the 67 plastid protein-coding genes with Amborella trichopoda as the outgroup. The Lauraceae are highlighted with green. Bootstrap support values estimated from 1,000 replicates are shown near nodes. The scale bar for branches denotes 0.02 substitutions per site.
Analyses of Divergence Times and Absolute Substitution Rates
The hypothesis of equal evolutionary rates was rejected by the Test Molecular Clock (ML) program implemented in MEGA, so we used r8s 1.81 (Sanderson 2003) to estimate the divergence times with the ML tree (fig. 2) and the parameters of method = PL (penalized likelihood), algorithm = TN (truncated Newton optimization), and smoothing = 3. Constrained ages were based on the database of TimeTree (Hedges etal. 2015). To estimate the 95% confidence interval of the divergence times, we generated 100 bootstrapping data sets from concatenation of the 67 genes by using the seqboot program in Phylip 3.695 (Felsenstein 1989). Each of the data sets was used to build a ML tree for estimating the divergence times in r8s 1.81 with the same parameters above.
To estimate absolute nucleotide substitution rates, we generated dN and dS trees for each of the 67 genes by using PAML 4.8. For the nine examined Lauraceous species, absolute dN and dS substitution rates were obtained by dividing dN and dS branch lengths by their estimated divergence times, respectively.
Measurement of Chlorophyll Content
Cassytha filiformis is leafless; therefore, 0.25 g fresh stems were collected for extraction and measurement of its chlorophyll (chl) content with the method described in Yang etal. (1998). This method was also used to measure chlorophyll content in Cin. camphora and Cry. chinensis with 0.25 g fresh leaves for each. The chlorophyll content was determined by the values of Chl a + b with the equation: 17.76A646.6 + 7.34A663.6, where A646.6 and A663.6 are absorption peaks.
Results
Plastomic Reduction in Cassytha
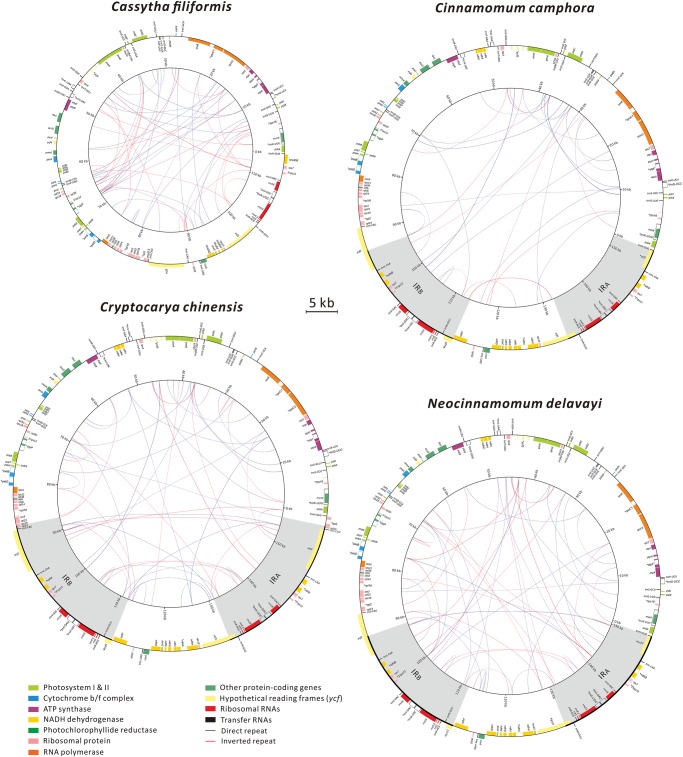
The plastomes of Cas. filiformis, Cin. camphora, Cry. chinensis, and Neo. delavayi were assembled as circular molecules and deposited in GenBank under accession numbers LC210517, LC228240, LC212965, and LC213014, respectively. Their assemblies are supported by high sequencing coverages that are all >350× (table 1). The plastome of Cas. filiformis lacks a pair of the canonical IRs, whereas the remaining three species feature a typical seed plant plastomic structure, with IRA and IRB separated by LSC and SSC regions (fig. 1). Although these four plastomes vary in length from 114,622 to 157,718 bp, their gene densities are similar, estimated to be ∼0.846 genes per kb on average (table 1). Cassytha filiformis has relatively decreased GC composition in the nongenic loci, so its GC content is the lowest among the examined species (table 1). However, Cas. filiformis has the highest repeat density, estimated to be ∼0.942 repeats per kb, and Cin. camphora has the lowest at 0.455 repeats per kb.
Table 1.
Plastome Assemblies and Characteristics of the Four Newly Sequenced Laurels
| Species | Cassytha filiformis | Cinnamomum camphora | Cryptocarya chinensis | Neocinnamomum delavayi |
|---|---|---|---|---|
| ×-Coverage | 996.7 | 953.2 | 660.0 | 394.6 |
| Plastome length (bp) | 114,622 | 152,739 | 157,718 | 150,850 |
| IR length (bp) | –a | 20,115 | 24,615 | 20,257 |
| GC content (%)b | 36.93 | 39.12 | 39.07 | 38.84 |
| Genic region (%) | 39.92 | 41.28 | 40.97 | 41.01 |
| Nongenic region (%)c | 33.31 | 36.58 | 36.56 | 36.21 |
| Gene density (genes/kb) | 0.881 | 0.838 | 0.824 | 0.842 |
| Repeat density (repeats/kb) | 0.942 | 0.445 | 0.723 | 0.716 |
IR loss.
Whole plastome.
Intron and intergenic space.
Fig. 1.
—Plastome maps of the four newly sequenced laurels. The maps were drawn to scale. Color boxes inside and outside the outermost circle are genes with clockwise and counterclockwise transcriptional direction, respectively. Copies of directed and inverted repeats are linked with blue and red lines in the innermost circle, respectively. IRA and IRB are highlighted in grey.
Our dot-plot analysis indicates that Cin. camphora and Neo. delavayi have an identical plastomic organization (supplementary fig. S1a, Supplementary Material online). As compared with Cin. camphora, Cry. chinensis has a relatively long IR that encompasses also the fragment 5′ycf2–rpl23 (supplementary fig. S1b, Supplementary Material online). In contrast, Cin. camphora contains two fragments (i.e., 3′ndhB–Ψycf2 in IRA and ndhB–Ψycf1 in IRB) that are absent from Cas. filiformis (supplementary fig. S1c, Supplementary Material online). Our PCR results verified that both of these fragments are also absent from another laurel dodder, Cas. pubescens, restricted to Australia (supplementary fig. S2, Supplementary Material online). These data suggest that 1) contractions of IRA and IRB together contributed to loss of IRs from Cas. filiformis and that 2) IR loss might be common to laurel dodders, but more taxa are required to test this speculation.
No functional NADH dehydrogenase (ndh) gene was detected in the plastome of Cas. filiformis, in which ndhA, C, F–K are completely lost and ndhB, D–E are pseudogenized (supplementary fig. S3, Supplementary Material online). In Cas. filiformis, rpl23 is also pseudogenized because of two premature stop codons. Other plastid HK genes, PS genes, and structural RNA genes are all retained in Cas. filiformis, resembling those in nonparasitic laurels. Moreover, syntenic nongenic loci are not significantly shrunk in the Cas. filiformis plastome (supplementary fig. S4, Supplementary Material online). As a result, losses of IRs and many genes rather than shrinkage of nongenic loci have led to the plastome of Cas. filiformis being extremely reduced, ∼0.73–0.76 times shorter than those of its three nonparasitic relatives (table 1).
Plastid Phylogenomics and IR Evolution in Lauraceae
With concatenation of the 67 shared protein-coding genes, our phylogenetic analysis yielded a robust ML tree with > 90% bootstrap support for all nodes (fig. 2). This ML tree supports that within magnoliids, Laurales and Magnoliales form a monophyletic group, which is sister to the Canellales–Piperales clade. Within Lauraceae, two separate clades are supported; one contains Cry. chinensis and Endiandra discolor (Cryptocaryeae) and the other Cas. filiformis, Neo. delavayi, and the Perseeae–Laureae clade (Chanderbali etal. 2001). Using this phylogenetic tree, we evaluated the IR evolution across Lauraceae.
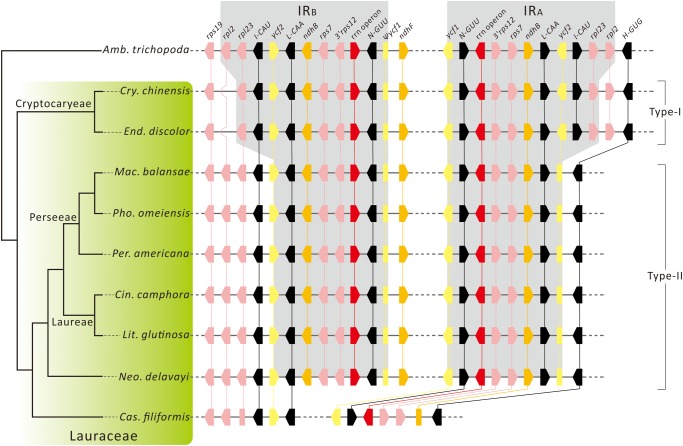
Figure 3 compares IR regions between the reference species (Amb. trichopoda) and the examined laurels, except for Ca. filiformis, whose IR has been lost. The IRs of the examined laurels is reduced and they can readily be divided into two types: Type-I and Type-II IRs. The former is specific to the two Cryptocaryeae species, with one copy of rpl2 lost due to contraction of the IRB boundary. In contrast, Type-II IR resulted from loss of not only rpl2 but also rpl23, trnI-CAU, and 5′ycf2 from the IRA of Neo. delavayi and the Perseeae–Laureae clade (fig. 3). Therefore, at least two independent events of IR reduction occurred in the plastome evolution of Lauraceae.
Fig. 3.
—IR evolution in Lauraceae. The tree in the left panel was modified from the ML tree in figure 2. Genes located in or adjacent to IRs are shown in the right panel, and those of Amborella trichopoda are references for comparisons. Genes with their transcriptional orientations are color-coded according to their functions. Orthologous genes are linked with vertical lines. The rrn operon includes trnV-GAC, rrn16, trnI-GAU, trnA-UGC, rrn23, rrn4.5, rrn5, and trnR-ACG. Genes and their relative positions were not drawn to scale.
The IRs were lost/reduced after Cassytha diverged from Neo. delavay and the Perseeae–Laureae clade. We propose two possible scenarios that are equally parsimonious. One is that the common ancestor of Cassytha, Neo. delavayi, and the Perseeae–Laureae clade lost a copy of the fragment (i.e., rpl2–5′ycf2) due to contraction of the IRA, and subsequent contractions of the IRA and IRB resulted in IR loss from Cassytha (supplementary fig. S5a, Supplementary Material online). Alternatively, Cassytha independently lost the IR due to contractions of the IRA and IRB, whereas the common ancestor of Neo. delavayi and the Perseeae–Laureae clade experienced contraction of the IRA to loss a copy of the fragment rpl2–5′ycf2 (supplementary fig. 5b, Supplementary Material online).
Evolution of Nucleotide Substitution Rates in Lauraceae
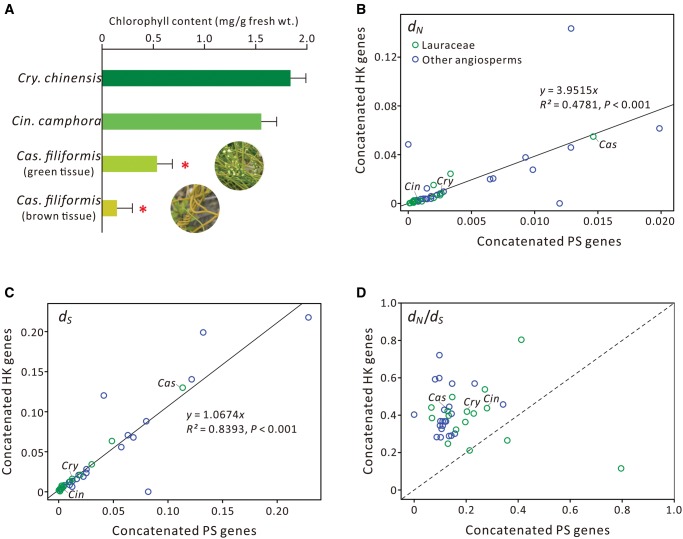
Chlorophyll content in the leaf tissues of Cry. chinensis and Cin. camphora was estimated at 1.84 ± 0.68 (mg/g fresh wt.) and 1.55 ± 0.21, respectively. In contrast, chlorophyll content was only 0.54 ± 0.15 and 0.15 ± 0.06 for the green and brown tissues of Cas. filiformis, respectively (fig. 4a). In terms of chlorophyll content, Cas. filiformis was always statistically lower than Cry. chinensis and Cin. camphora, regardless of using green or brown tissue for the comparison (all P < 0.001, two-sided Student’s t-test). These results reinforce that the PS ability of Cas. filiformis is reduced but not absent. We examined whether this significantly reduced chlorophyll content affected only the plastid PS genes by comparing nucleotide substitution rates between HK and PS genes across the 20 species shown in the ML tree (fig. 2).
Fig. 4.
—Chlorophyll content of three laurels and comparison of nucleotide substitution rates between plastid house-keeping (HK) and photosynthetic (PS) genes across the 20 basal angiosperms. (a) Estimated chlorophyll content in fresh leaves of Cryptocarya chinensis and Cinnamomum camphora and in green and brown tissues of Cassytha filiformis. Red asterisks indicate significantly lower content in Cas. filiformis than in Cry. chinensis and Cin. camphora (P < 0.001, Student’s t-test). Regression analysis of HK versus PS genes for dN (b) and dS (c), respectively. (d) Comparison of the dN/dS values between HK and PS genes. The dashed line indicates that HK and PS genes have the same dN/dS values. Cas, Cas. filiformis; Cry, Cry. chinensis; Cin, Cin. camphora.
Because the reduced photosynthesis should only affect PS genes, we would expect Cas. filiformis to differ from nonparasitic laurels. The dN and dS trees consistently revealed Cas. filiformis with the longest branch among the examined laurels, regardless of the concatenated HK or PS genes used in the analysis (supplementary fig. S6, Supplementary Material online). The linear regression of HK to PS genes for all branches in the trees yielded a significant correlation in dN (R2 = 0.4781, P < 0.001) and dS (R2 = 0.8393, P < 0.001; fig. 4b and c). For both dN and dS, Cas. filiformis, resembling other laurels, adhered to the regression line. Therefore, Cas. filiformis and its nonparasitic relatives, such as Cry. chinensis and Cin. camphora, have retained the same relation of HK to PS genes for dN as well as dS.
In most sampled taxa (including Cas. filiformis), dN/dS values are lower for PS than HK genes (fig. 4d), which indicates stronger negative selection for the former than the latter in general. The dN/dS values for PS genes were lower for Cas. filiformis than Cry. chinensis and Cin. camphora (fig. 4d). However, these three laurels are similar in dN/dS values for HK genes. Although Cas. filiformis has significantly decreased chlorophyll content, its PS genes are under stronger functional constraints than its HK genes.
Accelerated Nucleotide Substitution Rates in Ca. filiformis
To investigate whether accelerated substitution rates caused the long branch of Cas. filiformis (supplementary fig. S6, Supplementary Material online), we estimated the absolute nonsynonymous (RN) and synonymous (RS) substitution rates for each of the 67 shared genes. This allowed us to compare substitution rates between Cas. filiformis and other laurels at the same timescale. Our dating results suggest that Cas. filiformis diverged from its nonparasitic relatives after 98.85 Ma (supplementary fig. S7, Supplementary Material online). Neocinnamomum has diverged from its sister group, the Perseeae–Laureae clade, for ∼94 Myr, in agreement with the previous estimate (Nie etal. 2007). Overall, divergence times among the examined laurels were estimated to vary between 28.3 and 98.85 Ma.
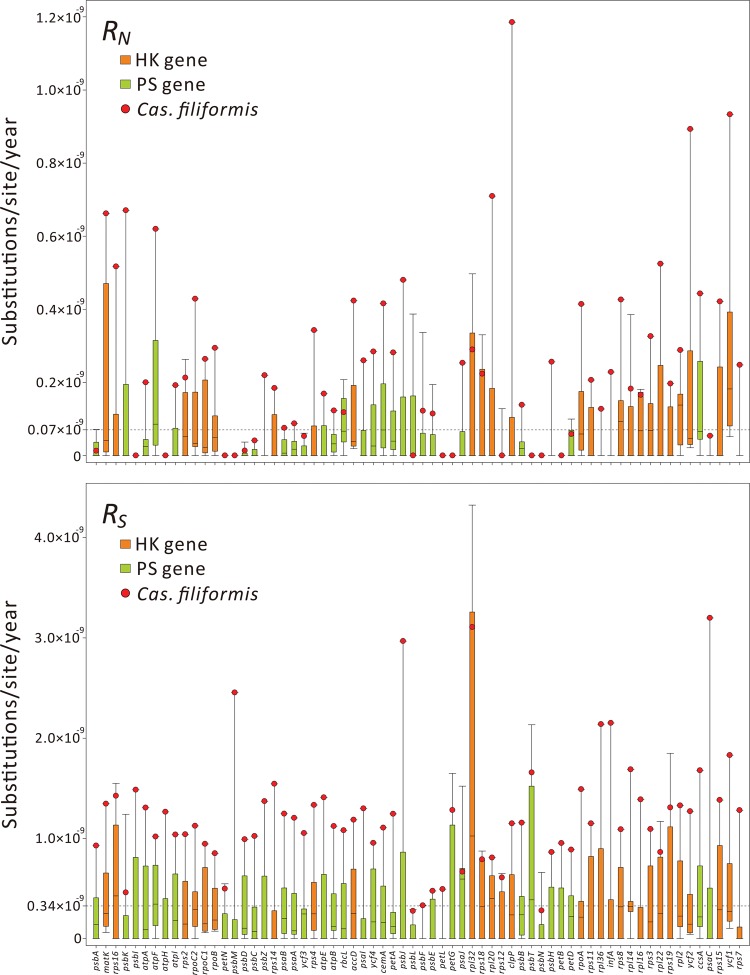
Figure 5 shows variation in RN and RS among the 67 genes for all examined laurels. The mean RN of laurels was estimated at 0.07 × 10−9 (substitutions/site/year), whereas the mean RS was 0.34 × 10−9, ∼5 times higher than the RN. Cassytha filiformis is an outlier in most of the examined genes, particularly in RS (fig. 5). The mean RN and RS for Cas. filiformis are 0.26 × 10−9 and 1.23 × 10−9 (substitutions/site/year), respectively. These rates are ∼6.5- and 5.3-fold faster than those for other laurels (RN = 0.04 × 10−9; RS = 0.23 × 10−9). Moreover, in comparison to other laurels, both RN and RS of Cas. filiformis are significantly elevated in HK (both P < 0.001, two-sided Wilcoxon rank-sum tests), PS (both P < 0.001), or all 67 genes (both P < 0.001). Collectively, these data strongly suggest that Ca. filiformis has an accelerated rate of nucleotide substitutions in plastid protein-coding genes.
Fig. 5.
—Box-plot comparisons of absolute nonsynonymous (RN) and synonymous (RS) rates between Cassytha filiformis and other laurels for each of the 67 plastid genes. Dashed horizontal lines indicate the average rates of the 67 genes across all examined laurels.
Discussion
Previously, using the MP method and the partial sequence of matK, Rohwer (2000) placed Cassytha sister to the rest of Lauraceous genera except for Hypodaphnis. On the basis of five plastid loci, Chanderbali etal. (2001) constructed a MP tree and revealed that Cassytha was closer to Neocinnamomum than other laurels, although this phylogeny was likely an artifact of long-branch attraction (LBA). A sister-relationship between Cassytha and Neocinnamomum was also recovered in the MP tree inferred from the 67 shared plastid genes (supplementary fig. S8, Supplementary Material online). In contrast, our ML tree strongly supports a sister relationship between Cassytha and the clade consisting of Neocinnamomum and the Perseeae–Laureae clade (fig. 2), which agrees with the Bayesian inference (BI) trees inferred from trnK (Rohwer and Rudolph 2005) and combined two plastid loci and nuclear ITS sequences (Wang etal. 2010). MP trees are highly susceptible to LBA when examined taxa contain extremely heterogeneous rates of sequence substitutions (Philippe etal. 2005; Wu etal. 2011, 2013). We show that the nucleotide substitution rate is significantly accelerated in Cas. filiformis (figs. 4 and 5). Therefore, the distinctively different rates between Cassytha and other laurels might have led to the misleading MP trees here and previously reported.
Our comparative analyses revealed that the plastomic organizations of laurels are generally conserved except for the IR-LSC boundary (supplementary fig. S1, Supplementary Material online). In Lauraceae, IR evolution involves the complete loss from Cassytha and boundary shifts in other genera (fig. 3; supplementary figs. S1 and S2, Supplementary Material online). We discovered two IR types in Lauraceae (fig. 3) and they are synapomorphic trait that reinforces our phylogenomic inferences (see the left panel of fig. 3). For instance, the common possession of Type-I IR supports the monophyly of Cryptocaryeae taxa. In contrast, the Neocinnamomum—Perseeae–Laureae clade is supported by sharing Type-II IR. There are 45 genera in Lauraceae (Christenhusz and Byng 2016), but we only sampled 10, which by no means is sufficient to illustrate a comprehensive picture of IR evolution in Lauraceae. Including more taxa, particularly the earliest divergent genus Hypodaphnis (Rohwer 2000; Chanderbali etal. 2001; Rohwer and Rudolph 2005), would greatly improve our understanding of IR evolution in Lauraceae.
To study how the plastome has evolved in response to parasitism, comparing the parasite with its nonparasitic relatives is required. Here, we identified a number of plastomic features that are specific to Cassytha. First, we document IR loss in this hemiparasitic genus. The IR loss is not common in parasites. For example, IRs are retained in many parasitic plants in diverse families, including Orobanchaceae (Wolfe etal. 1992; Wicke etal. 2013; Cusimano and Wicke 2016; Samigullin etal. 2016), Convolvulaceae (Funk etal. 2007; McNeal etal. 2007), Hydnoraceae (Naumann etal. 2016), Santalaceae and Viscaceae (Petersen etal. 2015), and Schoepfiaceae (Su and Hu 2016). It was hypothesized that the presence of IRs might have benefits in plastome stability (Palmer and Thompson 1982), and when repeats are rare, intramolecular recombination might be confined to IRs, thus decreasing plastomic rearrangements (Blazier etal. 2016; Ruhlman etal. 2017). Indeed, some IR-lacking plastomes are characterized by extensive inversions in legumes (Palmer and Thompson 1982; Perry and Wolfe 2002; Schwarz etal. 2015) and conifers (Wu and Chaw 2014, 2016; Hsu etal. 2016; Qu etal. 2017). However, no plastomic inversion was detected in Cas. filiformis when Cin. camphora was used as the reference (supplementary fig. S1, Supplementary Material online). Of note, the former lacks IRs and has diverged from the latter since the Cretaceous (supplementary fig. S7, Supplementary Material online).
Cassytha filiformis has higher repeat density than its nonparasitic confamilial taxa (fig. 1, table 1). A positive correlation between repeat density and degrees of parasitism in Orobanchaceae was reported (Wicke etal. 2013), and repeat density is much higher in obligate Viscum than facultative Osyri, although both are members of Santalales (Petersen etal. 2015). Repeats are mutagenic elements capable of triggering illegitimate recombination, ultimately resulting in genome rearrangements (Maréchal and Brisson 2010). Moreover, repeat content and plastomic rearrangements are positively correlated (Weng etal. 2014; Blazier etal. 2016). However, why the IR-lacking plastome of Cas. filiformis includes abundant repeats but lacks any inversion remains an enigma.
Rpl23 is the only ribosomal protein gene that is pseudogenized in the plastome of Cas. filiformis (supplementary fig. S3, Supplementary Material online). Because other plastid ribosomal protein genes are retained in Cas. filiformis, a nuclear RPL23 must have been imported to facilitate assembly of the plastid ribosome. In contrast, 11 plastid ndh genes encoding the NDH complex in regulating PS electron flow under photo-oxidative stresses (Yamori and Shikanai 2016) are either wanting or pseudogenized in Cas. filiformis (supplementary fig. S3, Supplementary Material online). Although plastid ndh genes were proposed to be essential for plants in adapting to terrestrial environments (Martín and Sabater 2010), they have been lost multiple times during seed plant evolution (Ruhlman etal. 2015). Activity of the NDH complex is dispensable under nonstressed conditions (Peltier and Cournac 2002; Ruhlman etal. 2015), and loss of plastid ndh genes is an early syndrome in the shift of autotrophy to heterotrophy (Barrett etal. 2014; Wicke etal. 2016). In a broad sampling of taxa, including some heterotrophic plants, the lost plastid ndh genes were nottransferred to the nucleus (Ruhlman et al 2015; Lin etal. 2017). After parasitizing hosts, Cassytha stops developing stomata (Heide-Jørgesen 2008) and possibly also reduces its chlorophyll content and photosynthesis (fig. 4a). Therefore, in Cas. filiformis, the loss of stress-responsive ndh genes likely is associated with parasitism.
In plastomes of parasites, accelerated nucleotide substitution rates have often been linked with relaxation of selective constraints (Wicke etal. 2013, 2016; Petersen etal. 2015). We found Cas. filiformis with elevated substitution rates at both dN and dS sites (fig. 5). However, our results also indicate that both HK and PS genes of Cas. filiformis have experienced strong selective constraints, as evidenced by their dN/dS values not deviating from those of nonparasitic laurels (fig. 4d). Of note, Cas. filiformis fits within the regression lines well (fig. 4b and c), so the dN is approximately four times faster in HK than PS genes (fig. 4b: slope = 3.9515). However, in Cas. filiformis, these two functional groups of genes have a nearly identical dS (fig. 4c: slope = 1.0674). Hence, Cas. filiformis has lower dN/dS values in PS than HK genes (fig. 4d), despite its significantly low chlorophyll content (fig. 4a). Previously, strong selection for retention of photosynthesis was documented in dodder, Cuscuta, a stem-parasitic genus (McNeal etal. 2007). The species of Cassytha are also stem-parasitic and they must perform photosynthesis before parasitizing their hosts (Heide-Jørgesen 2008). Accordingly, strong selection for retention of PS genes is required for Cas. filiformis.
Lineage effects, such as generation time (Smith and Donoghue 2008; Gaut etal. 2011; De La Torre etal. 2017) and body size (Lanfear etal. 2013), contribute to nucleotide substitution rates. Cassytha, the sole herbaceous genus of Lauraceae, is considered perennial (Heide-Jørgesen 2008). Nonetheless, we observed that in Taiwan, Cas. filiformis produced seeds annually and its populations disappeared in winter. The nuclei, mitochondria, and plastids of diverse parasitic plants, including Cassytha, have co-accelerated nucleotide substitution rates (Bromham etal. 2013). This highlights the lineage effects overwhelmingly shaping nucleotide evolution of Cassytha. In conclusion, we propose that in Cas. filiformis, the accelerated nucleotide substitution rates are likely associated with two distinctive features, short generation time and herbaceous lifestyle, rather than decreased PS capability.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Drs Ting-Shuang Yi and Xiao-Jian Qu and Mr Jing-Yi Lu for providing plant materials of Neo. delavayi and Cas. pubescens, respectively. We are indebted to Dr Robert K. Jansen for his critical comments and valuable suggestions. This work was supported by research grants from the Ministry of Science and Technology, Taiwan (MOST 103-2621-B-001-007-MY3), and from the Biodiversity Research Center of Academia Sinica to S.M.C.
Literature Cited
- Barkman TJ, et al. 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 7:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Blazier JC, et al. 2016. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 117(7):1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T, Kuzmina M, Stefanović S.. 2013. Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J Exp Bot. 64(4):977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R.. 2013. Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evol Biol. 13:126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanderbali AS, van der Werff H, Renner SS.. 2001. Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Missouri Bot Gard. 88(1):104–134. [Google Scholar]
- Christenhusz MJM, Byng JW.. 2016. The number of known plants species in the world and its annual increase. Phytotaxa 261:201–217. [Google Scholar]
- Cusimano N, Wicke S.. 2016. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 210(2):680–693. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Li Z, Van de Peer Y, Ingvarsson PK.. 2017. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol Biol Evol. 34(6):1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP: phylogeny inference package. Cladistics 5:164–166. [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K.. 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B, Yang L, Takuno S, Eguiarte LE.. 2011. The patterns and causes of variation in plant nucleotide substitution rates. Annu Rev Ecol Evol Syst. 42(1):245–266. [Google Scholar]
- Heide-Jørgesen H. 2008. Parasitic flowering plants. Leiden (Netherlands: ): Koninklijke Brill NV. [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Wu CS, Chaw SM.. 2016. Birth of four chimeric plastid gene clusters in Japanese umbrella pine. Genome Biol Evol. 8(6):1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104(49):19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA.. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Netherlands: Springer; p. 103–126. [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K. 2011. Piecing together the puzzle of parasitic plant plastome evolution. Planta 234(4):647–656. [DOI] [PubMed] [Google Scholar]
- Lin CS, et al. 2017. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90(5):994–1006. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N.. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186(2):299–317. [DOI] [PubMed] [Google Scholar]
- Martín M, Sabater B.. 2010. Plastid ndh genes in plant evolution. Plant Physiol Biochem. 48(8):636–645. [DOI] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW.. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie ZL, Wen J, Sun H.. 2007. Phylogeny and biogeography of Sassafras (Lauraceae) disjunct between eastern Asia and eastern North America. Plant Syst Evol. 267(1–4):191–203. [Google Scholar]
- Naumann J, et al. 2016. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol Evol. 8(2):345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29(2):537–550. [DOI] [PubMed] [Google Scholar]
- Peltier G, Cournac L.. 2002. Chlororespiration. Annu Rev Plant Biol. 53:523–550. [DOI] [PubMed] [Google Scholar]
- Perry AS, Wolfe KH.. 2002. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 55(5):501–508. [DOI] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Seberg O.. 2015. Plastome evolution in hemiparasitic mistletoes. Genome Biol Evol. 7(9):2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F.. 2005. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 5:50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Wu CS, Chaw SM, Yi TS.. 2017. Insights into the existence of isomeric plastomes in Cupressoideae (Cupressaceae). Genome Biol Evol. 9(4):1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer JG. 2000. Toward a phylogenetic classification of the Lauraceae: evidence from matK sequences. Syst Bot. 25(1):60–71. [Google Scholar]
- Rohwer JG, Rudolph B.. 2005. Jumping genera: the phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Ann Missouri Bot Gard. 92:153–178. [Google Scholar]
- Ruhlman TA, et al. 2015. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15:100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman TA, Zhang J, Blazier JC, Sabir JSM, Jansen RK.. 2017. Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am J Bot. 104(4):559–572. [DOI] [PubMed] [Google Scholar]
- Samigullin TH, Logacheva MD, Penin AA, Vallejo-Roman CM.. 2016. Complete plastid genome of the recent holoparasite Lathraea squamaria reveals earliest stages of plastome reduction in Orobanchaceae. PLoS ONE. 11(3):e0150718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EN, et al. 2015. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J Syst Evol. 53(5):458–468. [Google Scholar]
- Smith SA, Donoghue MJ.. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322(5898):86–89. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CN, Via LE.. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14(5):748–750. [PubMed] [Google Scholar]
- Su HJ, Hu JM.. 2016. The complete chloroplast genome of hemiparasitic flowering plant Schoepfia jasminodora. Mitochondrial DNA. 1(1):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Těšitel J. 2016. Functional biology of parasitic plants: a review. Plant Ecol Evol. 149:5–20. [Google Scholar]
- Wang ZH, Li J, Conran JG, Li HW.. 2010. Phylogeny of the Southeast Asian endemic genus Neocinnamomum H. Liu (Lauraceae). Plant Syst Evol. 290(1–4):173–184. [Google Scholar]
- Weng ML, Blazier JC, Govindu M, Jansen RK.. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31(3):645–659. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D.. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 25(10):3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113(32):9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD.. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 89(22):10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM.. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM, Huang YY.. 2013. Chloroplast phylogenomics indicates that Ginkgo biloba is sister to cycads. Genome Biol Evol. 5(1):243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM.. 2014. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol J. 12(3):344–353. [DOI] [PubMed] [Google Scholar]
- Wu CS, Chaw SM.. 2016. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (cupressophytes). Genome Biol Evol. 8:3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang CM, Chang KW, Yin MH, Huang HM.. 1998. Methods for the determination of the chlorophylls and their derivatives. Taiwania 43:116–122. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yamori W, Shikanai T.. 2016. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol. 67(1):81–106. [DOI] [PubMed] [Google Scholar]
- Zanne AE, Allen AP.. 2013. Taller plants have lower rates of molecular evolution. Nat Commun. 4:1879.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.