Abstract
The discovery of structured non-coding RNAs (ncRNAs) in bacteria can reveal new facets of biology and biochemistry. Comparative genomics analyses executed by powerful computer algorithms have successfully been used to uncover many novel bacterial ncRNA classes in recent years. However, this general search strategy favors the discovery of more common ncRNA classes, whereas progressively rarer classes are correspondingly more difficult to identify. In the current study, we confront this problem by devising several methods to select subsets of intergenic regions that can concentrate these rare RNA classes, thereby increasing the probability that comparative sequence analysis approaches will reveal their existence. By implementing these methods, we discovered 224 novel ncRNA classes, which include ROOL RNA, an RNA class averaging 581 nt and present in multiple phyla, several highly conserved and widespread ncRNA classes with properties that suggest sophisticated biochemical functions and a multitude of putative cis-regulatory RNA classes involved in a variety of biological processes. We expect that further research on these newly found RNA classes will reveal additional aspects of novel biology, and allow for greater insights into the biochemistry performed by ncRNAs.
INTRODUCTION
The last 15 years has seen a tremendous increase in our understanding of the biological significance and functional diversity of bacterial non-coding RNAs (ncRNAs) (1–5). For example, recent bacterial ncRNA discoveries have revealed unexpected resistance systems for fluoride and guanidine (6,7) and a greatly expanded number of self-cleaving ribozymes (8,9). Thus, the detection of novel ncRNAs has the potential to expand our biochemical understanding of RNA, as well as shed light on biological processes that are associated with these RNAs.
A highly successful strategy to find bacterial ncRNAs has been a comparative genomics approach based on identifying conserved nucleotide sequences and establishing the existence of conserved structures via analyzing nucleotide covariation. Structured RNAs exhibit covariation, in which a mutation that disrupts the RNA secondary structure is followed by a compensatory mutation that restores the affected base-pair. With many examples of such structure-preserving mutations in a multiple-sequence alignment, strong evidence emerges that the aligned sequences form RNA structures that are necessary for their biological function, and therefore function as ncRNAs.
This covariation analysis strategy has a long history of supporting and generating high-quality predictions of RNA secondary structure (10–12), and has been applied to discover novel RNA structures (13–19). This idea has been particularly successful in finding riboswitches and ribozymes, as most known metabolite- or ion-binding riboswitch classes have been found using this basic strategy (20), along with 5 of the 14 currently validated natural ribozyme classes (8,9,21).
In previous work (18), we grouped non-coding intergenic regions (IGRs) based on BLAST comparisons and inferred a conserved secondary structure within each group of putatively related sequences using the CMfinder program (22). We ran this analysis on several lineages of related bacteria and archaea, as well as metagenomic sequences from various environments, analyzing the IGRs in each lineage or metagenome separately. This effort produced 104 predicted structured RNAs, including the fluoride (6), ZMP/ZTP (23), c-di-GMP-II (24), THF (25), glutamine (26), azaaromatic (27) guanidine-III (28) and SAM/SAH (18) riboswitch classes. However, any remaining undiscovered RNA classes are likely to be rare (i.e. have few representatives in available sequences), and thus not as easy to find using this method. So, an ongoing challenge is to develop strategies to find increasingly uncommon RNA classes.
One possibility would be to analyze all IGRs at once, thus ensuring that all representatives of a rare RNA are available in the dataset analyzed. However, the complete set of available genomic and metagenomic IGRs totals 37 billion base pairs, which presents two difficulties. First, the computational time and memory required for analysis expands with many IGRs. More crucially, the false positive rate when IGRs are compared increases with their total number, making it difficult to extract rare RNAs. Our previous strategy (Supplementary Figure S1) can be viewed as a way of analyzing subsets of these IGRs that concentrate related RNAs. Specifically, in this earlier work (18), we reasoned that related RNAs are likely to be enriched in related bacteria that would be found in a similar lineage or metagenomic environment.
We have speculated that other methods of selecting IGR subsets might also help to enrich previously undetected RNAs (Supplementary Figure S1). For example, we recently observed that self-cleaving ribozymes in bacteria are often located nearby to specific gene classes (8). We exploited this fact to focus our searches on IGRs near these genes, as these IGRs are enriched for self-cleaving ribozymes (9). The key insight is that the comparative genomics method can take any subset of IGRs as input, and find RNAs enriched within those IGRs that were previously too rare to detect. Using this approach to concentrate self-cleaving ribozymes is only one possible application. In the current study, we found several additional tactics to select promising sets of input IGRs that enrich RNAs in other ways (see ‘Materials and Methods’ section). We demonstrate that these other measures also led to the discovery of new RNA classes.
During the course of this work, the functions of some novel structured RNAs identified via this approach were solved and published separately. These were the NiCo (29) and preQ1-III riboswitches (30) and the twister (8), twister sister, pistol and hatchet ribozymes (9), as well as variant hammerhead ribozymes with a short stem III (31). We also detected a permuted form of group II introns (Roth,A., Weinberg,Z., Vanderschuren,K., Murdock,M.H., Poiata,E., Breaker,R.R., unpublished data) that we will publish separately.
MATERIALS AND METHODS
Databases and general methods
A previously analyzed set of bacterial and archaeal genomes and metagenomes (32) (and see Supplementary Methods) was also analyzed in the present study. In total, we analyzed 237 Gb (237 × 109 bp) of sequences, with 54 Gb genomic and 183 Gb metagenomic sequences. These sequences contained 37 Gb of IGRs. Genes and RNAs were annotated as before, with protein domains predicted with the Conserved Domain Database version 2.25 (33), and RNAs by the Rfam database version 12.1 (34). The analyzed sequences chiefly come from RefSeq (35), IMG/M (36), MG-RAST (37) and GenBank (38). Some predicted conserved RNA structures were analyzed using earlier versions of databases. We searched for eukaryotic or viral homologs of new RNA structures (Supplementary Methods).
To identify conserved RNA structures from within a set of IGRs, our approach (Supplementary Figure S1) is based on a previously established method (18). The essential part of this earlier method is to cluster similar subsequences within the IGRs based on BLAST scores (17), and then infer RNA structural alignments from the sequences in each cluster using CMfinder (22). In the current study, we used a reimplementation of the clustering algorithm (Weinberg,Z., unpublished open-source software, available at http://weinberg-overcluster2.sourceforge.io). This software implements single-linkage clustering on subsequences of IGRs, instead of the full IGR sequence (as would happen with, e.g. blastclust). Thus, subsequences in a given IGR can participate in separate clusters. We also used an updated CMfinder, version 0.4.1, that extends the previous software version primarily to handle fragmentary sequence contigs in which RNA sequences are sometimes truncated (Weinberg,Z., unpublished open-source software, available at http://weinberg-cmfinder.sourceforge.io). Rho-independent transcription terminators were predicted using RNIE (39), and RNA structures were drawn using R2R (40) and Adobe Illustrator.
Candidate structured RNAs were scored (see Supplementary Methods for details) using a combination of an updated RNAphylo (41), and a new method called hmmpair. Both methods are implemented in the updated CMfinder package (above). The main change to RNAphylo was that it also now handles truncated RNAs. The basic idea of hmmpair is to evaluate an alignment by analyzing whether the alignment of covarying base pairs is supported by adjacent sequence conservation, and this analysis is conducted using a profile HMM model of the alignment.
All 224 predicted RNA classes were compared to Rfam version 12.2 (34) to ensure that they are not known (see Supplementary Methods). We used RNAcode (42) to find potential coding regions within all predicted RNAs. The best P-value was ∼0.001. With 224 motifs, this corresponds to an E-value of ∼0.22, which is not convincing.
Methods to select subsets of IGRs
Technical details of these methods and numbers of IGR subsets analyzed are in Supplementary Methods.
Lineage and environment strategies
Previously used strategies (18) are to select all IGRs within a lineage of bacteria (e.g. Enterobacteria), or IGRs within samples from a particular environment (e.g. hot springs). These methods were also applied here.
Large IGRs
Relatively large RNAs are of particular interest in bacteria, because they often catalyze chemical reactions, or have other unusual functions (12). These RNAs are necessarily in larger IGRs, and so in this strategy, only IGRs of at least 600 bp were considered (Supplementary Methods). The resulting RNA structures were generally larger, but for example CyVA-1 RNAs average only 78 nt (Supplementary Table S1).
Cluster by downstream gene
A given class of riboswitches (or other cis-regulatory RNA) is often located upstream of a characteristic gene class or small number of gene classes. For example, while purine riboswitches are known to reside upstream of 143 different gene classes in the sequences we analyzed, 22% are upstream of genes encoding a COG2252 domain (a permease) and 18% regulate genes encoding a TIGR01744 domain (xanthine phosphoribosyltransferase). Thus, the IGRs upstream of COG2252 or TIGR01744 genes will be enriched for purine riboswitches. So, we cluster all IGRs by domains in their immediately downstream gene and then further cluster each domain-specific set of IGRs as usual using BLAST and overcluster2. Conserved protein domains are taken from the Conserved Domain Database (see above), and all available domains are used (Supplementary Methods).
Nearby to genetic elements
Certain RNA classes are often located nearby to specific genetic elements. For example, bacterial self-cleaving ribozymes are often located within 6 Kb of certain gene classes (9). We also searched nearby to various other gene sets (Supplementary Methods).
No recognized genes
To explore unusual sequences, we selected metagenomic contigs that were at least 3 Kb, but did not have any gene that was longer than 800 bp or that matched a known conserved domain with an E-value better than 10−4. All IGRs within such contigs were used.
Transcription terminators
Cis-regulatory RNAs such as riboswitches often use Rho-independent transcription terminators as part of their regulatory mechanism (43,44). Therefore, the presence of a terminator in an IGR could indicate an upstream riboswitch or other RNA. However, terminators are also commonly used to end transcription of individual mRNAs and operons. We used the distances from the terminator to the upstream and downstream flanking genes to try to distinguish such cases. In addition to these distances, we exploited the fact that structured RNAs often have higher G+C content than other sequences (45), and applied Support Vector Machines (SVMs) to make predictions (46) (Supplementary Methods).
BLAST RefSeq against metagenomes
Some lineages are not well represented in sequenced organisms in RefSeq, but are common in certain environments. For example, the phylum Fibrobacteres has only two sequenced organisms in RefSeq version 63, but is well represented in cow rumen. Therefore we used all IGRs in known Fibrobacteria from RefSeq, and all IGRs in metagenome samples in which at least 0.01% of contigs were predicted as coming from this phylum, according to MetaPhlAn (47). In our other strategies, all IGRs are compared against all other IGRs using BLAST. However, in this strategy, RefSeq IGRs were compared against all other RefSeq IGRs and metagenomic IGRs, but metagenomic IGRs were not compared to other metagenomic IGRs. This reduces the number of comparisons, and thus the false positive rate. Our IGR clustering strategy easily accommodates these partial BLAST comparisons.
Experimental methods
In some cases, candidate conserved RNAs were tested for ligand binding using in-line probing assays with a previously established protocol (48). Self-cleavage assays were conducted as previously described (9). RNA molecules tested and concentrations of potential ligands are listed in Supplementary Text.
RESULTS
Detection of motifs
The strategies we implemented (see ‘Materials and Methods’ section) resulted in many computational predictions of structured RNAs. These predictions were evaluated manually, and we analyzed the most promising candidates in a process that centered on additional homology searches and attempts to improve structural predictions with help from CMfinder, as previously described (18). Promising candidates were those exhibiting strong evidence for a conserved RNA secondary structure in the form of covariation. Evaluating covariation is not simply a matter of looking for covarying base-pairs. Unfortunately, non-biological covariation can result, for example, from aligning sequences that are not homologous, or by aligning non-homologous regions of homologous sequences. Thus, when evaluating evidence of covariation, it is important to consider alignment mistakes that might have been introduced in the original computational alignment, or in subsequent editing of the alignment. Indeed, most candidate ncRNAs initially predicted lacked convincing covariation and were not further pursued. Successful candidate ncRNAs, along with their alignments and conserved secondary structures, are termed ‘motifs’.
We compared our predictions to previously known RNAs in the Rfam database (34), and did not report motifs with significant matches (Supplementary Methods). Two motifs are related to previously predicted RNAs. The c4–2 motif is likely a distant homolog of a previously described RNA (Supplementary Text) and one representative of a DUF1646 RNA was predicted, though the homologs and structure were not previously elucidated (see below). Otherwise, none of the 224 motifs have any apparent homology to previously predicted RNAs.
Extensive supplementary data is provided on all motifs. All 224 novel motifs detected in this work are summarized in Supplementary Table S1 and drawn in Supplementary File 1. Supplementary File 2 provides alignments of all motifs, with the locations, taxonomy and nearby genes of their representatives in printable form, while Supplementary Files 3 and 4 contain motif alignments in a machine-readable format. Due to space limitations, not all motifs are described in the main text, and so most motifs are described only in Supplementary Text. Additionally, this Supplementary Text contains further details on motifs that are more briefly described in the main text, and also describes assays on some motifs for riboswitch or ribozyme function.
Genetic features that surround a candidate motif often yield clues to the function of the RNA. In bacteria, cis-regulatory elements typically reside in the 5′ untranslated regions (UTRs) of protein-coding mRNAs, which they regulate. However, gene annotations are imperfect and therefore the exact locations of 5′ UTRs are not reliably known. So, we refer to a ‘potential 5′ UTR’ that is the non-coding region upstream of a gene, which likely corresponds to the 5′ UTR of the transcript (18). Thus, a motif that usually occurs in the potential 5′ UTRs of genes likely functions as a riboswitch or another type of cis-regulatory RNA.
Cis-regulatory RNAs use a variety of mechanisms to regulate genes in cis. In bacteria, the most common mechanisms (5) include formation of Rho-independent transcription terminator hairpins (43,44) and sequestration of the Shine-Dalgarno sequence (SD). Therefore, we pay particular attention to possible transcription terminators and SDs when analyzing likely motif functions.
In the following text, we use available data, e.g. genetic contexts and sequence features, to suggest likely hypotheses for each motif. This analysis forms a possible basis for future experimental work to elucidate the motifs’ functions.
Biochemically sophisticated RNAs
Many RNAs perform important biological tasks using modes of action that are relatively easy for RNA to implement, such as binding proteins or binding other RNAs via Watson-Crick base pairing. However, RNA also has biochemical capabilities approaching those of proteins. In particular, riboswitch RNAs specifically bind small molecules or ions to regulate gene expression (1), and ribozyme RNAs catalyze chemical reactions (2).
Some of the 224 motifs possess characteristics that suggest they perform biochemically challenging tasks, such as those performed by riboswitches and ribozymes. For such challenging functions, only a restricted set of sequences work. Thus, nucleotides will be highly conserved in critical regions, such as the ligand-binding core of a riboswitch or the active site of a ribozyme. By contrast, less challenging biochemical functions permit more sequence and structural variability. For example, nucleotides in antisense RNAs can change through mutation when their RNA targets change.
Extreme levels of nucleotide conservation are most obvious when RNAs are highly diverged, and yet some nucleotide positions are highly conserved. Many RNAs, on the other hand, are restricted to a narrow phylogenetic range, and may appear to exhibit conservation simply because a shorter amount of time has elapsed in which to mutate. We consider RNAs present in more than one bacterial phylum to be significantly diverged, and multiple nucleotides that are highly (at least 97%) conserved in such diverged RNAs suggest a demanding function.
The implementation of such challenging functions typically requires precise placement of nucleotides, which often leads to a more complicated secondary structure. In the absence of atomic-resolution structures, we assume that the presence of a pseudoknot (49) or multistem junction indicates a complex structure, as previously proposed (12). A pseudoknot occurs when two stems overlap such that only one side of each stem is within the other stem. A multistem junction is a loop that includes three or more base pairs. Thus, RNAs present in multiple phyla, with at least one pseudoknot or multistem junction and multiple 97%-conserved nucleotides will typically perform sophisticated biochemical tasks.
Several riboswitches and ribozymes are only known in a narrow phylogenetic range, e.g. preQ1-III riboswitches (30) and hatchet ribozymes (9), and a few have simple secondary structures, e.g. SAM/SAH riboswitches (18) and guanidine-II riboswitches (50), and hatchet ribozymes (9). Thus, some motifs that fail to satisfy these criteria could nonetheless implement unusual biochemistry. On the other hand, some motifs that satisfy all three criteria could perform straightforward biochemistry, such as binding another RNA via Watson-Crick base pairing. However, only three such cases arise (Supplementary Table S2) in the ‘seed’ alignments of the Rfam Database (34) version 12.1, out of 637 entries present in Bacteria. Thus, it is very likely that motifs meeting the above criteria indeed carry out complex functions.
Thirteen motifs detected in this work match the above characteristics typical of known ncRNAs with complex functions, and therefore are likely to perform sophisticated functions (Supplementary Table S3). Curiously, all 13 such motifs do not exhibit unequivocal evidence of ribozyme or riboswitch function based on nearby genetic features. This result raises the possibility that new types of sophisticated RNA functions exist. In the rest of this manuscript, we highlight potentially interesting motifs, with a focus on motifs that might perform interesting biochemistry. Additional structured ncRNA motifs predicted by our bioinformatics pipeline are presented in Supplementary Text, Table S1 and Files 1 and 2.
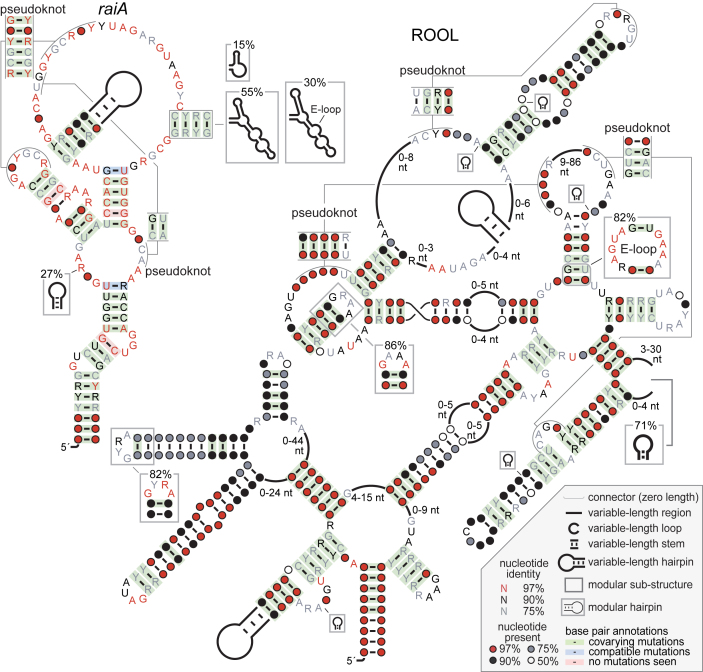
ROOL motif
Large and complex-folded ncRNAs in bacteria, while rare, are especially likely to have exotic biochemical functions, such as catalyzing chemical reactions (12). The Rumen-Originating, Ornate, Large (ROOL) motif (Figure 1) was originally found in cow rumen metagenome data. ROOL RNAs average 581 nt (Supplementary Table S1), and conserve a complex structure consisting of six multistem junctions and three pseudoknots (Figure 1). In bacteria, only ribosomal RNAs, GOLLD RNA and group II introns are longer and more structurally complicated (12) than ROOL. However, in comparison to some other large bacterial RNAs, the ROOL motif has fewer nucleotides that are highly (≥97%) conserved. Nonetheless, there are 10 highly conserved nucleotides and the motif is widespread, with at least one member present in three phyla. Thus, ROOL RNAs have the potential to perform unusual sophisticated biochemistry.
Figure 1.
Consensus features of ROOL and raiA RNAs. The legend (lower, right) applies to all consensus diagrams in this work. An expanded drawing of the raiA motif is available (Supplementary File 1). A comprehensive set of consensus diagrams for all 224 motifs is in Supplementary File 1.
ROOL RNAs have similar properties to those of the previously reported GOLLD RNA motif (12), and these similarities could suggest a related function. The GOLLD motif is a large bacterial RNA that is often located nearby to tRNAs and in prophages, but also in regions that do not appear to be prophages. ROOL RNAs share all of these properties. However, although the common genomic contexts of these two motifs suggest a similar biological function, the commonalities could arise for other reasons. The GOLLD and ROOL motifs might both be used sometimes by phages, and their proximity to tRNAs could arise if these ncRNAs are often co-transcribed.
raiA motif
The raiA motif (Figure 1) is present in two phyla: Firmicutes and Actinobacteria (Supplementary File 2). Motif representatives are often found in the potential 5′ UTRs of protein-coding genes, which is consistent with a cis-regulatory function. However, these genes are often quite far (e.g. >600 bp) from the raiA RNAs and raiA RNAs are, in fact, often closer to the upstream gene. This arrangement, which is not typical of other cis-regulatory motifs, casts some doubt on this possible function for raiA motif RNAs.
Genes located downstream of raiA RNAs frequently encode specific protein domains in both Firmicutes and Actinobacteria (Supplementary File 2). Since these associations are strongly evident in two phyla, we presume that these domains are related to the function of raiA RNAs. These domains include RaiA and periplasmic binding protein (PBP). RaiA binds to ribosomes and inhibits translation under stress conditions, while PBP domains function in transporters whose specificity is not predicted by available models.
Genes with ComFC domains (COG1040) often occur nearby and upstream of raiA RNAs, also in both Actinobacteria and Firmicutes. These comFC genes typically are oriented in the same direction as the RNA, but sometimes are in opposite directions. However, the gene located immediately upstream of each raiA RNA is usually encoded on the same strand as the RNA. Many cis-regulatory RNAs in bacteria regulate the downstream genes, but we are not aware of cases where they are also associated with an upstream gene. This observation suggests that raiA RNAs are not cis regulators of the downstream gene. Moreover, it is unusual for bacterial cis-regulatory RNAs to reside in a 3′ UTR. Therefore it also seems unlikely that raiA motif RNAs regulate expression of the upstream ORF.
ComFC is implicated in genetic competence (33). This prediction is supported by other genes often located adjacent to comFC genes and upstream of raiA RNAs. These genes include comFA genes, which encode a helicase required for competence. We also notice many upstream genes associated with sporulation and flagella, which could be related. The raiA motif has the characteristics expected for RNAs with sophisticated functions (Supplementary Table S3), but its apparent association with upstream genes makes it doubtful that it functions in a manner similar to most riboswitches. In vitro experiments did not provide evidence of the RNA binding small molecules (Supplementary Text).
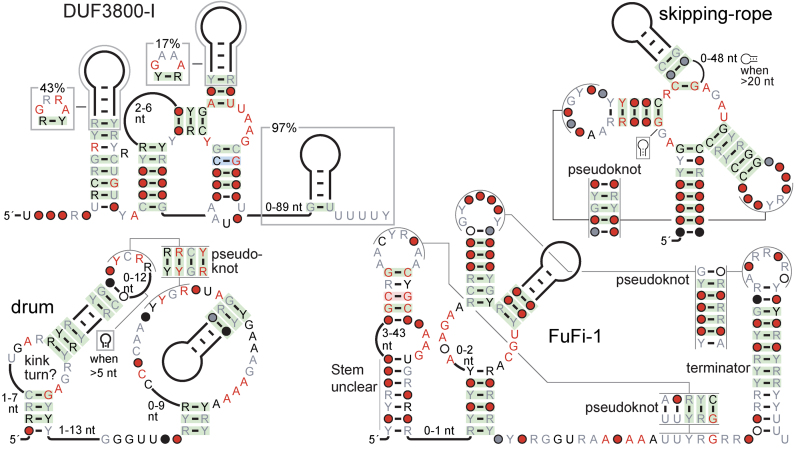
Highly conserved motifs with 3′ terminators
We found four motifs with characteristics suggestive of sophisticated biochemical function (Supplementary Table S3) that have predicted Rho-independent transcription terminators on their 3′ ends (Supplementary File 2). These motifs are called DUF3800-I, drum, FuFi-1 (Fusobacteria/Firmicutes-1) and skipping-rope (Figure 2).
Figure 2.
Consensus features of motifs with properties that suggest a sophisticated biochemical function that are followed by Rho-independent terminators: the DUF3800-I, drum, FuFi-1 and skipping-rope motifs. Annotations are as for Figure 1.
Although the motifs have structural properties typical of riboswitches and self-cleaving ribozymes, they are improbable riboswitches since they are only rarely found in potential 5′ UTRs. Similarly, their downstream terminators are at odds with a self-cleaving function. Indeed, experiments on DUF3800-I, FuFi-1 and skipping-rope RNAs did not reveal self-cleavage activity (Supplementary Text). Additionally DUF3800-I, skipping-rope and drum RNAs have a compelling association with proteins that are encoded nearby, in either orientation and on either side of the RNA (Supplementary Text, see ‘Additional comments on drum, DUF3800-I and skipping-rope motifs’). Association with one specific protein domain, especially when the domain is not encoded on the same strand, would be unusual for self-cleaving ribozymes and also for self-splicing ribozymes. Thus, these motifs could perform a biochemically challenging task that is distinct from that of known RNAs.
Other DUF3800-associated motifs
After finding that the DUF3800-I motif associates with genes encoding the DUF3800 domain, we noticed a possible relationship between these genes and genes associated with drum and skipping-rope RNAs (Supplementary Text). We then searched for additional RNA motifs around DUF3800-encoding genes in bacteria and archaea, and found ten additional motifs, named DUF3800-II to DUF3800-XI (Supplementary File 1). The 11 DUF3800 RNA motifs do not resemble each other in any obvious way, although the trinucleotide sequence UAA seems subjectively to be somewhat common (Supplementary File 1). It is remarkable that 11 different structures presumably achieve the same biological function in conjunction with DUF3800 genes. Many of the DUF3800-associated motifs are present in multiple phyla and some display complex structural features (Supplementary Tables S1 and 3). Several are followed by Rho-independent terminators (Supplementary File 2). Unfortunately, no specific hypothesis for the function of these curious motifs is obvious to us.
RT (reverse transcriptase) motifs
The RT-1 through RT-19 motifs (Supplementary File 1) are found nearby to reverse transcriptase (RT) genes. RT genes in bacteria have been observed in group II introns, retrons and diversity-generating retroelements (DGRs) (51), and the RT RNA motifs might function as part of these elements.
The RT motifs do not seem to match the typical properties of group II introns. Group II introns are large (at least 300 nt) and form RNA structures both 5′ and 3′ to an ORF that encodes an RT domain (51). However, we do not find cases where RT motif RNAs are present on both sides of one RT gene. Moreover, the RT RNA motifs are smaller than known self-splicing RNAs. Therefore, these motifs do not have characteristics expected of self-splicing introns, although additional conserved structure might perhaps remain undetected.
Some of the RT motifs could function as retrons. Retrons consist of an RT gene adjacent to an RNA structure that acts as a primer for the RT reaction (51). The biological function of retrons is unknown, but they produce a covalently bound RNA/DNA hybrid (51). We speculate that many of the RT RNA motifs participate in retrons.
At least one of the new motifs, the RT-10 motif, is likely to function as part of a DGR. The best-studied DGR is present in Bordetella phage BPP-1, and creates a high rate of mutation in a phage tail protein that can thus rapidly evolve to bind different proteins on the host's cell surface (51). This DGR contains two approximate repeats, termed VR and TR, that flank a short gene. The TR is located nearby to an RT gene. The RT-10 motif is present in the region between the TR and the RT gene in phage BPP-1, where no RNA structure was previously proposed. Since RNA structures in retrons act as primers, RT-10 RNAs might perform this role for this DGR. We speculate that additional RT RNA motifs function in other DGRs.
DUF1874 motif and integrons
The DUF1874 motif (Supplementary Figure S2) is a multistem structure with an internal loop that has many highly conserved nucleotides. It is usually present in the potential 5′ UTRs of protein-coding genes. However, the motif is frequently present in consecutive IGRs, i.e. a DUF1874 RNA is followed by an ORF, which is followed by another DUF1874 RNA, then another ORF, etc.
This genetic arrangement is typical of attC sites, which are components of integrons (52). attC sites function at the level of DNA as recombination sites, allowing the shuffling and incorporation of genes as cassettes. Characterized attC sites exhibit little sequence conservation and adopt hairpin structures formed of single-stranded DNA (ssDNA) (52). If the DUF1874 motif is an unusual attC site, it would be a rare example of an ssDNA with a more complex structure than a simple hairpin.
Our strategy for finding structured RNAs would apply equally well to finding ssDNA structures. Indeed, given the similarities between ssDNA and ssRNA, it would be difficult to reliably distinguish such motifs bioinformatically. So, we reanalyzed our previously reported motifs, and found that the Transposase-resistance motif (18) matches the already-known attC sites of class 1 integrons (53). Thus, our methods are capable of finding attC sites.
Integrons commonly have integrase genes at their 5′ ends, and the enzymes encoded by these genes recombine the genetic cassettes within the integron (52). We did not observe an integrase gene at the likely 5′ end of a run of DUF1874 representatives. However, some integrons lack integrases (54). Nonetheless, it is possible that the DUF1874 motif's striking pattern of occurrence in consecutive IGRs results from a biological role other than an attC site.
Plasmid-associated motifs
We found three motifs that frequently occur in plasmids (Supplementary Table S4), and could play roles in plasmid replication. All three are moderately long, with average lengths of 223–349 nt (Supplementary Table S1). If the motifs participate in plasmid replication, they could function as ssDNAs. As just noted, our method can also discover ssDNA motifs. The plasmid motifs also might play regulatory roles, or perform functions often associated with plasmids, as do toxin/anti-toxin systems (55).
The Area Required for Replication in a Plasmid Of Fusobacterium (ARRPOF) motif (Supplementary Figure S3) occurs in Fusobacterium nucleatum plasmid pKH9. Although this plasmid has a predicted rep (replication) gene, which presumably can replicate the plasmid, the plasmid can also replicate when the rep gene is deleted (56). A 1 Kb region that lacks predicted genes (Supplementary Text) was found to be sufficient for replication (56). This region was cut into two, and each sub-region individually failed to allow replication. One of these sub-regions contains an ARRPOF RNA, so the motif likely plays an important role in plasmid replication.
One instance of the GC-Enriched, Between Replication Origins (GEBRO) motif (Supplementary Figure S4) is found in the Streptococcus mutans plasmid pUA140, in a region of high G+C content, between the predicted single-stranded and double-stranded origins (57). This motif might function to help or regulate the replication of plasmids like pUA140 that use rolling-circle replication. Since such replication involves ssDNA, there is an added possibility that the GEBRO motif functions as ssDNA.
Finally, the Plasmid-Associated gamma-Proteobacteria Especially Vibrionales (PAGEV) motif (Supplementary Figure S5) is found in Vibrionales bacteria and other gamma-Proteobacterial organisms. A PAGEV RNA is found in a region that is predicted as a replication origin (58) because of its similarity to an experimentally determined replicon whose mechanism is unknown (Supplementary Text).
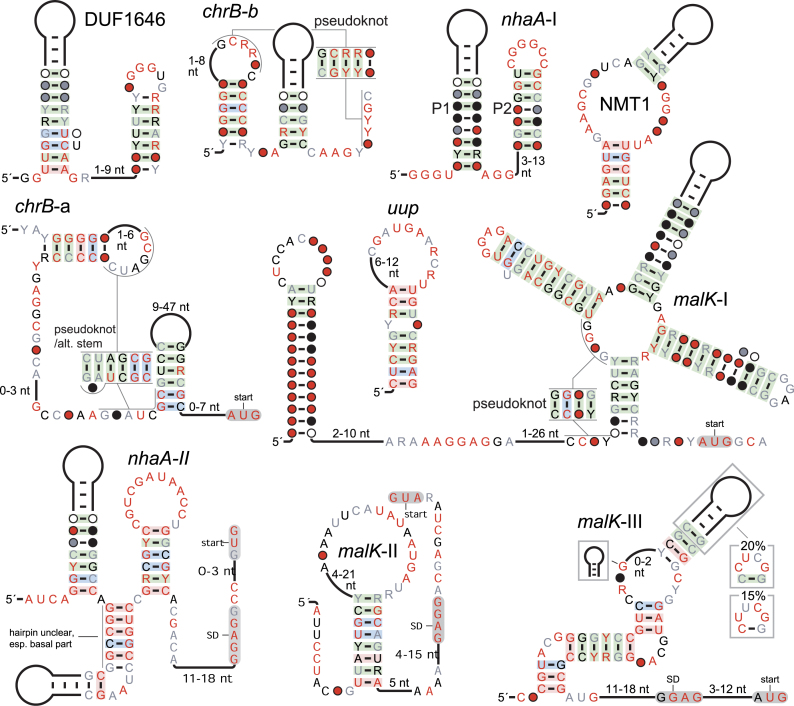
Sodium-related motifs
The remaining motifs presented in this manuscript are all strong candidates for cis-regulatory function. Although these motifs do not have all three of the properties indicative of biochemically sophisticated functions, some might function as riboswitches. The DUF1646 and nhaA-I and -II motifs (Figure 3) are implicated in regulation related to sodium. DUF1646 RNAs are positioned so as to regulate a variety of genes (Supplementary File 2) encoding proteins that are not homologous, yet generally have functions related to Na+ ion transport. The most common classes are nhaA-like Na+/H+ antiporters and DUF1646, whose function is unknown. Other regulated gene classes encode adenosine triphosphate (ATP)-dependent transporters of sodium and oxaloacetate carboxyltransferase, whose reaction is coupled to transmembrane sodium ion transport (59). The RNA also occurs rarely upstream of c-di-AMP riboswitches (60) that are in turn upstream of either dapB or kamA, two genes involved in lysine metabolism and known to be regulated in a c-di-AMP-dependent manner. While these genes have no direct relationship to sodium, c-di-AMP riboswitches commonly control genes whose expression is related to the osmotic shock response (60).
Figure 3.
Consensus diagrams of selected motifs that are likely to be cis-regulatory: the chrB-a and -b, DUF1646, malK-I, -II and -III, nhaA-I and -II, NMT1 and uup motifs. Annotations are the same as in Figure 1.
A DUF1646 RNA in Enterococcus hirae ATCC 9790 is upstream of the ntp operon, which encodes a sodium-exporting ATPase. The expression of this operon is increased with higher Na+ levels, especially under circumstances that hinder the generation of proton-motive force (61). Although the region implicated in this regulation does not overlap the DUF1646 RNA (Supplementary Text), it is still possible that DUF1646 RNAs implement sodium-based regulation, or regulate genes based on a complementary stimulus (Supplementary Text).
A DUF1646 RNA in Enterococcus faecalis was predicted as being a cis-regulatory RNA by term-seq (Supplementary Text), a procedure that uses RNA-seq to find products of Rho-independent termination (62). These complementary data, and the predicted transcription terminators we generally observe downstream of DUF1646 RNAs (Supplementary Text), support the conclusion that this motif is cis-regulatory.
The nhaA-I and -II motifs are also typically found in the presumed 5′ UTRs of genes that encode Na+/H+ antiporters. nhaA-I RNAs are also very rarely found upstream of signaling, peptidoglycan-related and DUF1646 genes, among others. DUF1646 is implicated in sodium transport by its association with the DUF1646 motif, whose likely relationship with sodium was just described. nhaA-I RNAs are occasionally found nearby to another nhaA-I RNA, an arrangement that could produce a tighter ligand dose-response curve (63,64). NhaA-II RNAs are also typically found upstream of nhaA genes, but one nhaA-II RNA is upstream of a SAM-dependent methyltransferase.
chrB-a and -b motifs
These related motifs (Figure 3) occur in a variety of alpha-Proteobacteria in the potential 5′ UTRs of multiple protein-coding gene classes with a direct relationship to chromate resistance (Supplementary Text). These data suggest that chrB RNAs function as chromate sensors, although in vitro experiments did not provide evidence of binding to chromate or structurally related chemicals (Supplementary Text).
malK-I, -II and -III motifs
Three motifs with distinct structures (Figure 3) were discovered that are predominantly found in the potential 5′ UTRs of malK genes that encode the ATPase domains of sugar transporters, e.g. for maltose or glycerol-3-phosphate. In all three motifs, the RNA structure is located nearby to the SD of the malK gene. Unfortunately, it is difficult to predict the substrate of the transporter from protein sequence alone. Therefore, if these motifs function as riboswitches, identification of the ligand might require the experimental screening of a variety of ligand candidates. We did not observe any evidence that the RNA recognized any of the few ligands that we examined (Supplementary Text).
uup motif
The uup motif (Figure 3) lies in potential 5′ UTRs, and the most commonly regulated gene classes encode ATPases that are annotated either as being a part of ABC-type transporters or as fungal elongation factor 3. Thus, the specific biochemical function of these genes is unclear. In rare cases, DUF2992 domains are encoded by the second gene in an operon that is likely regulated by an uup RNA. DUF2992 genes are associated with yjdF motif RNAs (48), which were recently found to respond to a variety of azaaromatic compounds (27). uup RNAs are found in three phyla and have several conserved nucleotides. However, the motif's secondary structure is simpler than that of most known riboswitches.
NMT1 motif
Multiple gene classes are predicted to be regulated by NMT1 RNAs (Figure 3). The most common is NMT1, which is thought to be required for thiamin synthesis and regulated by thiamin in yeast (33). Also common are genes encoding dioxygenases of various or uncertain specificities. A less-common class of genes is probably related to isoxanthopterin deaminases. A subset of amidohydrolases were found to function in vitro as isoxanthopterin deaminases, although their natural substrate is unclear (65). A group of similar proteins had ambiguous features that made it unclear whether they also could work on isoxanthopterin (65), and members of this ambiguous group are apparently regulated by NMT1 RNAs. In-line probing of NMT1 RNAs in the presence of various small molecules did not reveal a ligand for this motif (Supplementary Text).
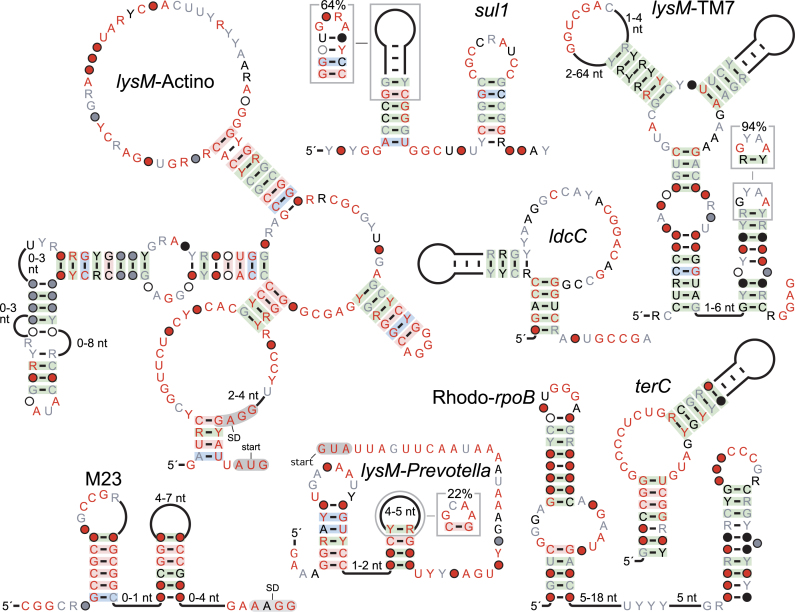
ldcC motif
This motif is widespread in Firmicutes, and present in two species of Spirochaetes, and has multiple highly conserved nucleotides (Figure 4). Its secondary structure consists of a hairpin with a bulge. However, there is a possible pseudoknot stem whose nucleotides do not vary, thus eliminating the possibility of covariation, which would more reliably indicate the presence of the pseudoknot. If the pseudoknot is conserved, this motif would satisfy the criteria to likely be a sophisticated RNA.
Figure 4.
Consensus diagrams of selected motifs that are likely to be cis-regulatory: the ldcC, lysM-Actino, -Prevotella and -TM7, M23, Rhodo-rpoB, sul1 and terC motifs. Annotations are the same as in Figure 1.
The ldcC motif is usually predicted in the 5′ UTRs of genes, most of which are predicted to encode decarboxylases of metabolites such as arginine, ornithine or SAM. Other genes include those coding for endopeptidase C39A and PotA, a spermidine/putrescine transporter. The RNA might thus play a role related to polyamine homeostasis.
c-di-AMP-related motifs
We found four motifs (Figure 4) upstream of genes that are commonly regulated by riboswitches that sense the signaling molecule c-di-AMP (60), so these motifs might also function in c-di-AMP signaling. Three motifs, named lysM-TM7, lysM-Actino and lysM-Prevotella were found in distinct phyla in the potential 5′ UTRs of lysM genes. This gene encodes the ‘lysin’ domain, which is involved in degradation of the cell wall. The fourth motif, named the M23 motif, is typically in the potential 5′ UTRs of M23 peptidase genes. In one case, it is upstream of nadE, which encodes NAD synthetase.
We did not observe binding to c-di-AMP for these motifs (Supplementary Text). Nonetheless, we did observe possible binding of an M23 RNA to an unknown compound in dialyzed yeast extract (data not shown). However, as no binding was observed for c-di-AMP, the motif was not further pursued.
sul1 motif
The sul1 motif (Figure 4) is virtually always positioned in the potential 5′ UTRs of various non-homologous genes, and is probably a cis-regulatory element. By far the most commonly associated gene class, sul1, encodes predicted sulfate transporters. Other genes encode serine acetyltransferase, cyclopropane fatty acid synthase, SAM-dependent methyltransferase or glycosyltransferase. Since these genes are implicated in sulfur metabolism or the related methionine pathway, sul1 RNAs might play a role in sulfur metabolism. We did not observe binding between sul1 RNAs and ions such as sulfate (Supplementary Text).
terC motif
terC RNAs (Figure 4) are positioned in a manner consistent with cis-regulatory elements. The most common gene class apparently regulated by terC RNAs encodes TIGR03717 and TIGR03718. These domain are homologous with TerC, a membrane-bound protein that confers resistance to tellurium (66). These domains are also the most commonly regulated by Mn2+ riboswitches (67–69), and COG3809 genes are also common to both terC RNAs and Mn2+ riboswitches. These data suggest that terC RNA’s function might relate to Mn2+ or to another metal cation, although we did not identify such a cation that induced structural modulation in the RNA (Supplementary Text).
Rhodo-rpoB motif
This motif (Figure 4) is consistently found upstream of rpoB genes, which encode the beta subunit of RNA polymerase. The motif is largely restricted to the Rhodobacterales, an order of alpha-Proteobacteria. A motif called Lacto-rpoB was previously discovered upstream of rpoB genes (18). However, the Lacto-rpoB and Rhodo-rpoB motifs occur in different phyla and have unrelated secondary structures. Thus, a wide variety of bacteria might use distinct RNA structures to regulate their RNA polymerase genes.
DISCUSSION
The discovery of 224 new structured motifs, of which the vast majority are undoubtedly formed by RNAs, suggests that many such RNA classes might still remain hidden in biology. If genomic and metagenomic DNA sequence datasets continue to grow, ever rarer RNA classes could appear in this sequence data in sufficient numbers to be discovered by using similar bioinformatics approaches. However, it might be necessary to design new algorithms or approaches to be able to detect rare RNA classes even with a constantly growing sequence data collection.
Recently, a strategy was described to detect cis-regulatory RNAs in bacteria by an RNA-seq-based technique that selects transcripts produced by Rho-independent termination (62), as terminators are often used by cis-regulatory RNAs (43,44). This ‘term-seq’ strategy analyzed three genomes and predicted potential novel regulatory RNAs. Surprisingly, only one motif out of our 224 is also predicted by term-seq (62). This intersection is a DUF1646 RNA in E. faecalis. However, a second DUF1646 RNA in the same organism is not predicted by term-seq, possibly because it is not associated with a strong transcription terminator stem (Supplementary Text). Indeed, only three of 224 motifs are present in any of the three organisms studied by term-seq and none of the 224 motifs overlap metagenomic predictions from term-seq (Supplementary Methods).
These results suggest that many methods have increased difficulty in detecting the rare RNA classes that presumably remain undetected. In our method, rare RNAs are difficult to cluster together and may not collectively provide strong evidence of covariation, due to a limited diversity of representatives. A method like term-seq does not have these problems, but finding rare RNAs likely requires screening large numbers of organisms to find those in which a given rare RNA occurs. Screening of metatranscriptomes disfavors RNAs transcribed at lower levels or in organisms that are rare in the given environment. Additionally, individual RNAs may not be detectable with the term-seq method, due to biochemical issues or inevitable false negative rates. With common RNA classes, there are many opportunities to discover an example, but with rare classes, the class might be missed.
Fortunately, the low overlap between our results and those of term-seq supports the notion that alternate methods may be largely complementary. Thus, there is the exciting potential to find many more RNA classes, which might be rare, through a combination of multiple methods, each with their own strengths and weaknesses.
The 224 motifs presented in this paper present a starting point for experimental efforts to uncover their functions. Several motifs were revealed whose properties suggest an exciting biochemical function, such as the 581-nt ROOL RNA, and widespread motifs such as raiA, drum, DUF3800-I and FuFi-1. Moreover, the variety of RNA motifs found overall suggests involvement in a wide range of biological roles.
NOTE ADDED IN PROOF
After acceptance of our manuscript, we became aware of a publication providing data on the ncRNA we named ROOL (Cousin,F.J., et al., 2017, Microbial Genomics, doi: 10.1099/mgen.0.000126).
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Bjornson and N. Carriero for assisting us in using Yale computing clusters at the Yale Center for Research Computing, Rob Knight, Folker Meyer, Mihai Pop and HMP for pre-publication data, and N. Sudarsan and other Breaker lab members for helpful discussions. We also thank Paul Gardner for discussions about the use of terminators for finding riboswitches. This project extensively used sequence data from the US Department of Energy (www.jgi.doe.gov) and its user community.
Footnotes
Present addresses:
Zasha Weinberg, Bioinformatics Group, Department of Computer Science and Interdisciplinary Centre for Bioinformatics, Universität Leipzig, Härtelstraße 16–18, D-04107 Leipzig, Germany.
Christina E. Lünse, Institute of Biochemistry, Universität Leipzig, Brüderstraße 34, D-04103 Leipzig, Germany.
Tyler D. Ames, Phosplatin Therapeutics, 1350 Avenue of the Americas, Third floor, New York, NY 10019-4703, USA.
James W. Nelson, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA 02139; Howard Hughes Medical Institute.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [RR19895, GM022778]; Howard Hughes Medical Institute (HHMI). Funding for open access charge: HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1. Meyer M.M. The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip. Rev. RNA. 2017; 8:e1370. [DOI] [PubMed] [Google Scholar]
- 2. Jimenez R.M., Polanco J.A., Lupták A.. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015; 40:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barquist L., Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015; 49:367–394. [DOI] [PubMed] [Google Scholar]
- 4. Gottesman S., Storz G.. RNA reflections: converging on Hfq. RNA. 2015; 21:511–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wachter A. Gene regulation by structured mRNA elements. Trends Genet. 2014; 30:172–181. [DOI] [PubMed] [Google Scholar]
- 6. Baker J.L., Sudarsan N., Weinberg Z., Roth A., Stockbridge R.B., Breaker R.R.. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012; 335:233–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson J.W., Atilho R.M., Sherlock M.E., Stockbridge R.B., Breaker R.R.. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell. 2017; 65:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roth A., Weinberg Z., Chen A.G., Kim P.B., Ames T.D., Breaker R.R.. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014; 10:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinberg Z., Kim P.B., Chen T.H., Li S., Harris K.A., Lünse C.E., Breaker R.R.. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015; 11:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pace N.R., Thomas B.C., Woese C.R.. Gesteland RF, Cech TR, Atkins JF. Probing RNA structure, function, and history by comparative analysis. The RNA World. 1999; 2nd edn, NY: Cold Spring Harbor Laboratory Press; 113–141. [Google Scholar]
- 11. Michel F., Westhof E.. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990; 216:585–610. [DOI] [PubMed] [Google Scholar]
- 12. Weinberg Z., Perreault J., Meyer M.M., Breaker R.R.. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009; 462:656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rivas E., Eddy S.R.. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001; 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Washietl S., Hofacker I.L., Stadler P.F.. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao Z., Barrick J., Weinberg Z., Neph S., Breaker R., Tompa M., Ruzzo W.L.. A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007; 3:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinberg Z., Barrick J.E., Yao Z., Roth A., Kim J.N., Gore J., Wang J.X., Lee E.R., Block K.F., Sudarsan N. et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007; 35:4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tseng H.H., Weinberg Z., Gore J., Breaker R.R., Ruzzo W.L.. Finding non-coding RNAs through genome-scale clustering. J. Bioinform. Comput. Biol. 2009; 7:373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weinberg Z., Wang J.X., Bogue J., Yang J., Corbino K., Moy R.H., Breaker R.R.. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010; 11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heyne S., Costa F., Rose D., Backofen R.. GraphClust: alignment-free structural clustering of local RNA secondary structures. Bioinformatics. 2012; 28:i224–i232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun E.I., Rodionov D.A.. Computational analysis of riboswitch-based regulation. Biochim. Biophys. Acta. 2014; 1839:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R.. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004; 428:281–286. [DOI] [PubMed] [Google Scholar]
- 22. Yao Z., Weinberg Z., Ruzzo W.L.. CMfinder–a covariance model based RNA motif finding algorithm. Bioinformatics. 2006; 22:445–452. [DOI] [PubMed] [Google Scholar]
- 23. Kim P.B., Nelson J.W., Breaker R.R.. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol. Cell. 2015; 57:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee E.R., Baker J.L., Weinberg Z., Sudarsan N., Breaker R.R.. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010; 329:845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ames T.D., Rodionov D.A., Weinberg Z., Breaker R.R.. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem. Biol. 2010; 17:681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ames T.D., Breaker R.R.. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011; 8:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li S., Hwang X.Y., Stav S., Breaker R.R.. The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA. 2016; 22:530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherlock M.E., Breaker R.R.. Biochemical validation of a third guanidine riboswitch class in bacteria. Biochemistry. 2017; 56:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furukawa K., Ramesh A., Zhou Z., Weinberg Z., Vallery T., Winkler W.C., Breaker R.R.. Bacterial riboswitches cooperatively bind Ni(2+) or Co(2+) ions and control expression of heavy metal transporters. Mol. Cell. 2015; 57:1088–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCown P.J., Liang J.J., Weinberg Z., Breaker R.R.. Structural, functional, and taxonomic diversity of three preQ1 riboswitch classes. Chem. Biol. 2014; 21:880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lünse C.E., Weinberg Z., Breaker R.R.. Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers. RNA Biol. 2016; doi:10.1080/15476286.2016.1251002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinberg Z., Nelson J.W., Lünse C.E., Sherlock M.E., Breaker R.R.. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2077–E2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015; 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J. et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015; 43:D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Markowitz V.M., Chen I.M., Chu K., Szeto E., Palaniappan K., Pillay M., Ratner A., Huang J., Pagani I., Tringe S. et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014; 42:D568–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A. et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008; 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2013; 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gardner P.P., Barquist L., Bateman A., Nawrocki E.P., Weinberg Z.. RNIE: genome-wide prediction of bacterial intrinsic terminators. Nucleic Acids Res. 2011; 39:5845–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weinberg Z., Breaker R.R.. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao Z. Genome Scale Search of Noncoding RNAs: Bacteria to Vertebrates. 2008; Seattle: University of Washington. [Google Scholar]
- 42. Washietl S., Findeiß S., Müller S.A., Kalkhof S., von Bergen M., Hofacker I.L., Stadler P.F., Goldman N.. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011; 17:578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gusarov I., Nudler E.. The mechanism of intrinsic transcription termination. Mol. Cell. 1999; 3:495–504. [DOI] [PubMed] [Google Scholar]
- 44. Yarnell W.S., Roberts J.W.. Mechanism of intrinsic transcription termination and antitermination. Science. 1999; 284:611–615. [DOI] [PubMed] [Google Scholar]
- 45. Rivas E., Eddy S.R.. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000; 16:583–605. [DOI] [PubMed] [Google Scholar]
- 46. Chang C.-C., Lin C.-J.. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011; 2:27. [Google Scholar]
- 47. Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C.. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012; 9:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 49. Staple D.W., Butcher S.E.. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005; 3:e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sherlock M.E., Malkowski S.N., Breaker R.R.. Biochemical validation of a second guanidine riboswitch class in bacteria. Biochemistry. 2017; 56:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zimmerly S., Wu L.. An unexplored diversity of reverse transcriptases in bacteria. Microbiol. Spectr. 2015; 3, doi:10.1128/microbiolspec.MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 52. Escudero J.A., Loot C., Nivina A., Mazel D.. The integron: adaptation on demand. Microbiol. Spectr. 2015; 3, doi:10.1128/microbiolspec.MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 53. Moura A., Soares M., Pereira C., Leitao N., Henriques I., Correia A.. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009; 25:1096–1098. [DOI] [PubMed] [Google Scholar]
- 54. Cury J., Jove T., Touchon M., Neron B., Rocha E.P.. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016; 44:4539–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003; 301:1496–1499. [DOI] [PubMed] [Google Scholar]
- 56. Bachrach G., Haake S.K., Glick A., Hazan R., Naor R., Andersen R.N., Kolenbrander P.E.. Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system. Appl. Environ. Microbiol. 2004; 70:6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zou X., Caufield P.W., Li Y., Qi F.. Complete nucleotide sequence and characterization of pUA140, a cryptic plasmid from Streptococcus mutans. Plasmid. 2001; 46:77–85. [DOI] [PubMed] [Google Scholar]
- 58. Stavrinides J., Guttman D.S.. Nucleotide sequence and evolution of the five-plasmid complement of the phytopathogen Pseudomonas syringae pv. maculicola ES4326. J. Bacteriol. 2004; 186:5101–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hase C.C., Fedorova N.D., Galperin M.Y., Dibrov P.A.. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 2001; 65:353–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nelson J.W., Sudarsan N., Furukawa K., Weinberg Z., Wang J.X., Breaker R.R.. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013; 9:834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yasumura K., Igarashi K., Kakinuma Y.. Promoter analysis of the sodium-responsive V-ATPase (ntp) operon in Enterococcus hirae. Arch. Microbiol. 2002; 178:172–179. [DOI] [PubMed] [Google Scholar]
- 62. Dar D., Shamir M., Mellin J.R., Koutero M., Stern-Ginossar N., Cossart P., Sorek R.. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016; 352:aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welz R., Breaker R.R.. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007; 13:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 65. Hall R.S., Agarwal R., Hitchcock D., Sauder J.M., Burley S.K., Swaminathan S., Raushel F.M.. Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase. Biochemistry. 2010; 49:4374–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Toptchieva A., Sisson G., Bryden L.J., Taylor D.E., Hoffman P.S.. An inducible tellurite-resistance operon in Proteus mirabilis. Microbiology. 2003; 149:1285–1295. [DOI] [PubMed] [Google Scholar]
- 67. Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dambach M., Sandoval M., Updegrove T.B., Anantharaman V., Aravind L., Waters L.S., Storz G.. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol. Cell. 2015; 57:1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Price I.R., Gaballa A., Ding F., Helmann J.D., Ke A.. Mn(2+)-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell. 2015; 57:1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.