Abstract
While skeletal myogenesis is tightly coordinated by myogenic regulatory factors including MyoD and myogenin, chromatin modifications have emerged as vital mechanisms of myogenic regulation. We have previously established that bexarotene, a clinically approved agonist of retinoid X receptor (RXR), promotes the specification and differentiation of skeletal muscle lineage. Here, we examine the genome-wide impact of rexinoids on myogenic differentiation through integral RNA-seq and ChIP-seq analyses. We found that bexarotene promotes myoblast differentiation through the coordination of exit from the cell cycle and the activation of muscle-related genes. We uncovered a new mechanism of rexinoid action which is mediated by the nuclear receptor and largely reconciled through a direct regulation of MyoD gene expression. In addition, we determined a rexinoid-responsive residue-specific histone acetylation at a distinct chromatin state associated to MyoD and myogenin. Thus, we provide novel molecular insights into the interplay between RXR signaling and chromatin states pertinent to myogenic programs in early myoblast differentiation.
INTRODUCTION
While functional DNA sequences in the non-coding portion of the genome offer an important framework for gene regulation, the epigenome exhibits remarkable lineage-specificity and plays a critical role in differential gene expression (1–3). The findings in recent years that certain DNA elements are associated with distinct histone modifications have provided a new pathway to identify networks of regulatory loci whose activities underpin the control of gene expression. As such, lineage-specific enhancers can be identified by promoter-distal enrichment in H3K4me1 and/or the recruitment of histone acetyltransferases (HATs) (4,5). Furthermore, two classes of enhancers have been described, active enhancers marked additionally by H3K27ac and poised enhancers marked by the presence of H3K4me1 but lack of H3K27ac (6,7). These studies also reveal that poised enhancers can be activated during differentiation by gaining H3K27ac, and as a result are able to mediate the expression of proximal genes. Therefore, enhancers marked by histone acetylation upon differentiation may reflect the activation of distinct gene programs regulated by lineage-specific transcription factors.
During myogenic differentiation, the commitment and development of skeletal muscle lineage is regulated by complex signaling pathways that induce a sequential expression of the muscle regulatory factors (MRFs). Myf5 and MyoD are the first MRFs to be expressed during development and have overlapping roles in the commitment of progenitor cells into the skeletal muscle cell lineage (8–10). Myogenin functions downstream of Myf5 and MyoD, and has been identified in particular as a fundamental regulator of myoblast differentiation (11). The MRFs generally recognize a highly similar DNA motif, referred to as the E-box, to which they bind and dimerize with other basic helix-loop-helix transcription factors (10). How MRFs regulate gene expression during early myoblast differentiation is consequently affected by their DNA binding partner and through their association with the HATs (12–14).
C2C12 is a non-transformed myogenic cell line obtained by continuous passaging of primary myoblasts isolated from mouse limb muscle (15). They not only closely resemble proliferating myoblasts that express the MyoD determination factors, abide genetic manipulation in that selected stable clones retain their ability to differentiate, but also provide a more homogenous population compared to primary myoblasts (16,17). Furthermore, studies of gene expression in this well characterized and widely used model of myogenesis provide results consistent with that obtained from primary tissue cells (18,19).
Retinoid X receptors (RXRs) belong to the nuclear receptor (NR) superfamily. There are three subtypes of RXRs, namely RXRα, RXRβ and RXRγ, which are bound constitutively to DNA, regardless of ligand, but act as transcriptional activators upon agonist activation (20–22). The function of RXRα is very important for early development (23–25). Specifically, RXRα null mice die in utero and present with myocardial and ocular malformation (25,26). We have previously reported that bexarotene, a selective RXR agonist, promotes the specification and differentiation of skeletal myoblasts through the function of RXRα (27,28). As a potential dimerizing partner of many nuclear receptors, RXR is involved in the regulation of a wide array of genetic targets, but it is unclear how rexinoids affect global myogenic expression.
In this study, we examined the rexinoid responsive transcriptional program and the interplay of rexinoid signaling with myoblast-specific chromatin state. We found that bexarotene coordinates muscle-related gene expression and bexarotene-promoted myoblast differentiation is largely transmitted through MyoD expression and coupled with MyoD- and myogenin-associated residue-specific histone acetylation at a distinct subset of enhancers. Our study thus provides novel molecular insights into the regulation of myogenic expression at the early stage of myoblast differentiation.
MATERIALS AND METHODS
Cell culture and reagents
Cells of the murine myoblasts cell line C2C12 (ATCC) were maintained in growth medium (GM), Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), at 37°C with 5% CO2. To induce differentiation, the medium of 80% confluent cell cultures was changed to differentiation medium (DM), DMEM supplemented with 2% HS (29) and 50 nM of bexarotene was used for treatment conditions. Bexarotene was purchased from the LC Laboratories and UVI 3003 was from Tocris. The RXRα shRNA knockdown cells have been described previously (28).
RNA-seq and data processing
Total RNA was extracted using RNeasy kit (Qiagen) according to manufacturer's instructions and sequenced by the McGill University Genome Quebec Innovation Centre in two biological repeats. Sequencing reads were aligned to the mouse genome build mm9 using Tophat (30) and guided by transcripts from the Ensembl 67 database. Transcript assembly was performed using Cufflinks and was followed by Cuffdiff (31) to estimate normalized gene expression and perform differential expression testing. To identify genes showing differential expression between conditions for downstream analysis, genes with a fold change in expression greater than ±1.5-fold and below a false discover rate (FDR) of 5% were selected. GO (gene ontology) terms significantly associated (P-value ≤ 0.05, Bonferroni) with genes that were differentially expressed were identified using the database for annotation, visualization and integrated discovery (32). For visualization and hierarchical clustering, z-scores were calculated from raw FPKM values (Fragments Per Kilobase of transcript per Million mapped reads), which were log2-transformed and scaled to the row mean and standard deviation using the following formula: [log2 (FPKM)-mean of row]/(standard deviation of row). These z-scores were also used to calculate the Pearson correlation coefficients between conditions.
ChIP-seq and data processing
Cells were crosslinked and sonicated followed by chromatin immunoprecipitation as previously described (29). Antibodies against H3K9ac, H3K18ac, H3K27ac, H4K8ac and H3K27me3 were obtained from Abcam (ab4441, ab1191, ab4729, ab15823 and ab6002), RXRα, and p300 from Santa Cruz (sc-553x and sc-32758x). DNA was purified using the MinElute Spin Columns Kit (Qiagen) and input chromatin DNA was used as control. Purified DNA was sequenced by the McGill University Genome Quebec Innovation Centre with Illumina HiSeq 2000. Sequencing reads were mapped to the mouse genome build mm9 using Bowtie, allowing for three mismatches and reporting the single best alignment per 50 bp read. Picard was used to filter out replicated reads (http://picard.sourceforge.net/), and BAM files were converted into BED files with the BEDTools suite (33). For visualization of ChIP-seq signals in the Integrative Genomics Viewer, aligned reads were extended by 125 bp at their 3′ end and basewise signal intensity was computed. Local peaks in read enrichment were identified using MACS (34) (v1.0.0) with a P-value threshold of 1 × 10−5.
Chromatin state model
ChIP-seq datasets were obtained from the NCBI Gene Expression Omnibus (GEO) for RNA Pol II under the accession number GSM721286 and for H3K4me1 under GSM721288 (16), while for H3K4me3 under GSM918415 and for H3K36me3 under GSM918417 (35). The corresponding input for Pol II and H3K4me1 were obtained under GSM721306 (16), and for H3K4me3 and H3K36me3 under GSM918421 (35). Genome-wide binding sites for MyoD and myogenin were obtained for MyoD_GM under accession number GSM915186, for MyoD_24h under GSM915183 and for myogenin_24h under GSM915159 (35). The chromatin state model was generated using ChromHMM (36). The enrichment of transcription start sites (TSSs), Pol II binding sites and highly conserved non-coding elements (HCNCEs) was calculated as a ratio between the fraction of nucleotides overlapping between the feature and state and the joint probability of observing the feature and state. HCNCEs for the mm9 build were identified using genomic evolutionary rate profiling (37).
Analysis of histone enrichment and transcription factor binding sites
The enrichment of histone acetylation at the MRF binding sites was calculated with ngsplot (38), which calculates the coverage vectors for each query region based on specified alignment files. Following normalization and transformation on the coverage, an average profile is created as the number of reads in 20 bp bins within a 2 kb region, centered at the peaks and normalized to the total number of mapped reads (in millions) in the dataset. Two-sided Wilcoxon signed-rank test was used for statistical analysis. Homer (39) was used to perform de novo motif analysis for RXRα ChIP-seq peaks, allowing for motif identification within 100 bp region from the peak center. Overlapping peaks were calculated as being located within 100 bp of one another with the mergePeaks tool within Homer.
Reverse transcription qPCR analysis
Reverse transcription was performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time PCR was conducted using SYBR® Green PCR Master Mix and HotStarTaq DNA polymerase (Qiagen) on a CFX96 Touch Real-Time PCR Detection System (BioRad). Quantification of the targets, normalized to endogenous reference and relative to a calibrator control was calculated using the formula 2–ΔΔCT. MyoD, myogenin, Akt2 and Angptl4 gene specific primers have been described previously (28,29,40,41) and primers for Cdkn1a, Sntb1, Tnnc1 and Tmem38a are listed in Supplementary Table S2.
Quantitative ChIP analysis
Following ChIP, purified DNA was amplified in triplicate real-time PCR reactions with locus-specific primers described previously (16,28,29,42) or in Supplementary Table S2. Input DNA was used to create a standard curve in the PCR amplification for each set of primers. Quantification was analyzed as the abundance of locus-specific DNA in percentage of input DNA (enrichment as the percentage of input). MyoD antibody was from Santa Cruz (sc-584x) and myogenin from the Developmental Studies Hybridoma Bank (F5D).
Western analysis
Whole cell extracts were prepared as described previously (43). Bio-Rad ChemiDoc MP System was used to capture chemiluminescent images, and ImageJ software was used for densitometry quantification.
RESULTS
Rexinoids promote distinct transcriptional programs during myoblast differentiation
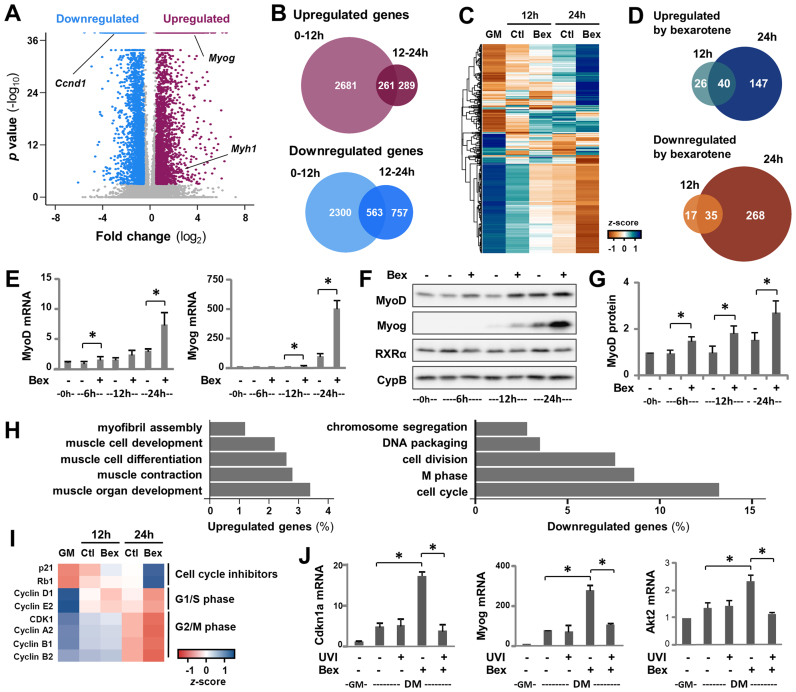
Given that little is known about the gene programs modulated by rexinoid signaling during myogenesis, we interrogated the transcriptome associated with the action of bexarotene, a selective RXR agonist. C2C12 myoblasts were differentiated in the presence of bexarotene for 12 or 24 h with untreated differentiating or proliferating myoblasts as controls, and then subjected to RNA-seq analysis in two biological repeats (GSE94560, Supplementary Table S1). Cuffdiff analysis revealed that in untreated differentiating myoblasts, several thousand genes were up- or downregulated in comparison to their expression in proliferating myoblasts (FC ≥ ±1.5-fold, FDR ≤ 5%) (Figure 1A and B).
Figure 1.
Bexarotene coordinates differentiation-related gene expression. (A) C2C12 myoblasts were differentiated for 12 or 24 h and subjected to RNA-seq analysis with proliferating myoblasts as control. Volcano plot analysis of differentially expressed genes by 24 h of differentiation among all annotated Ensembl genes. Genes with a significant fold change in expression greater than 1.5-fold are shown in magenta (upregulated) and blue (downregulated), while the remaining genes are in gray. (B) Union analysis for genes differentially expressed during the first 12 h (0–12 h) and/or between 12 and 24 h (12–24 h) of differentiation. (C) Hierarchical clustering of bexarotene responsive genes on standardized values of expression in proliferating myoblasts (GM) and myoblasts differentiated for 12 or 24 h in the absence or presence of bexarotene (Ctl or Bex 50 nM). FPKM values were log2-transformed and scaled to the mean and standard deviation of the row to give row-based z-scores. (D) Overlap between genes that were responsive to bexarotene. (E) Levels of MyoD and myogenin transcripts were examined by a time course of RT-qPCR and presented as fold change relative to proliferating myoblasts (0 h), normalized to internal control (error bars: SEM; n = 3; *, P < 0.05). (F) Western analysis of MyoD, myogenin and RXRα protein with cyclophilin-B as a loading control. (G) Quantification of MyoD blots was presented as fold change relative to proliferating myoblasts (error bars: SEM; n = 5). (H) GO terms associated with genes that were responsive to bexarotene. (I) Heat map of bexarotene-responsive cell cycle-related genes presented as standardized z-scores in proliferating myoblasts (GM), and myoblasts differentiated with or without bexarotene for 12 or 24 h. (J) C2C12 myoblasts were differentiated with bexarotene in the presence or absence of RXR antagonist UVI 3003 (UVI, 1 μM) for 24 h. The mRNA levels of myogenin, Akt2, Tmem8c and Cdkn1a were determined by RT-qPCR and presented as the fold change in relation to proliferating myoblasts (error bars: SEM; n = 3).
Consistent with previous microarray studies (44,45), many of the upregulated genes were involved in muscle development and function, including myogenin (Myog) and skeletal muscle specific myosin heavy chain (Myh1), while downregulated genes were involved largely in cell cycle regulation and progression, such as Ccnd1, reflecting simultaneous exit from the cell cycle and the activation of myogenic expression (Figure 1A and Supplementary Figure S1). Notably, much of the differential expression occurring over the 24 h of differentiation took place during the first 12-h period, indicating a rapid switch in transcriptional programs at the onset of myoblast differentiation (Figure 1B).
We also identified 533 genes that were differentially expressed in bexarotene-treated differentiating myoblasts compared to the untreated by 24 h of differentiation (Figure 1C and D). Hierarchical clustering of the bexarotene responsive genes on standardized values of expression revealed distinct groups of genes that appeared to be differentially coregulated across treated and untreated conditions (Figure 1C). Evidently, most of the bexarotene responsive genes were not affected during the first 12-h period of differentiation, suggesting that the majority of these genes may be regulated as an indirect response to bexarotene treatment (Figure 1D).
Nonetheless, MyoD, a key regulator of myogenic differentiation, was identified as an early genetic target of bexarotene through our RNA-seq analysis. Time course of RT-qPCR analysis revealed that MyoD transcripts were significantly upregulated by bexarotene starting from the 6-h time-point, while myogenin transcripts from the 12-h time-point (Figure 1E). The levels of MyoD and myogenin protein emulated their sequential upregulation during early differentiation and were further augmented following bexarotene treatment (Figure 1F and G). In comparison, levels of RXRα protein remained largely steady (Figure 1F).
Interestingly, GO analysis illustrated that bexarotene affected the expression of many genes that are associated with muscle cell differentiation and cell cycle regulation, indicative of targeted genes involved in the coordinated progression of myoblast differentiation (Figure 1H and I). For example, Cdkn1a, a gene that was upregulated at the onset of myoblast differentiation, was further augmented by bexarotene as validated by RT-qPCR analysis (Figure 1J). More importantly, co-treatment of differentiating myoblasts with RXR antagonist UVI 3003 (28) attenuated bexarotene-enhanced Cdkn1a expression (Figure 1J). Similarly, the RXR antagonist also impeded the positive effects of bexarotene on gene expression of Akt2 and myogenin which have been identified previously as rexinoid responsive genes (28). Taken together, our data suggests that bexarotene-enhanced myogenic expression is mediated through RXR selective signaling and rexinoid action occurs through the coordination of myoblast differentiation, from the exit of the cell cycle to the activation of myogenic expression.
RXR signaling regulates MyoD directly to promote myoblast differentiation
We have previously shown that bexarotene acts through the function of RXRα to enhance muscle-related gene expression (28). To identify direct genetic targets of RXR signaling in early myoblast differentiation, we performed RXRα ChIP-seq analysis with myoblasts differentiated for 24 h in the absence or presence of bexarotene (GSE94558, Supplementary Table S1). To profile histone acetylation coupled with rexinoid action, we also performed ChIP-seq for H4K8ac, H3K9ac, H3K18ac and H3K27ac in matching conditions (GSE94558, Supplementary Table S1).
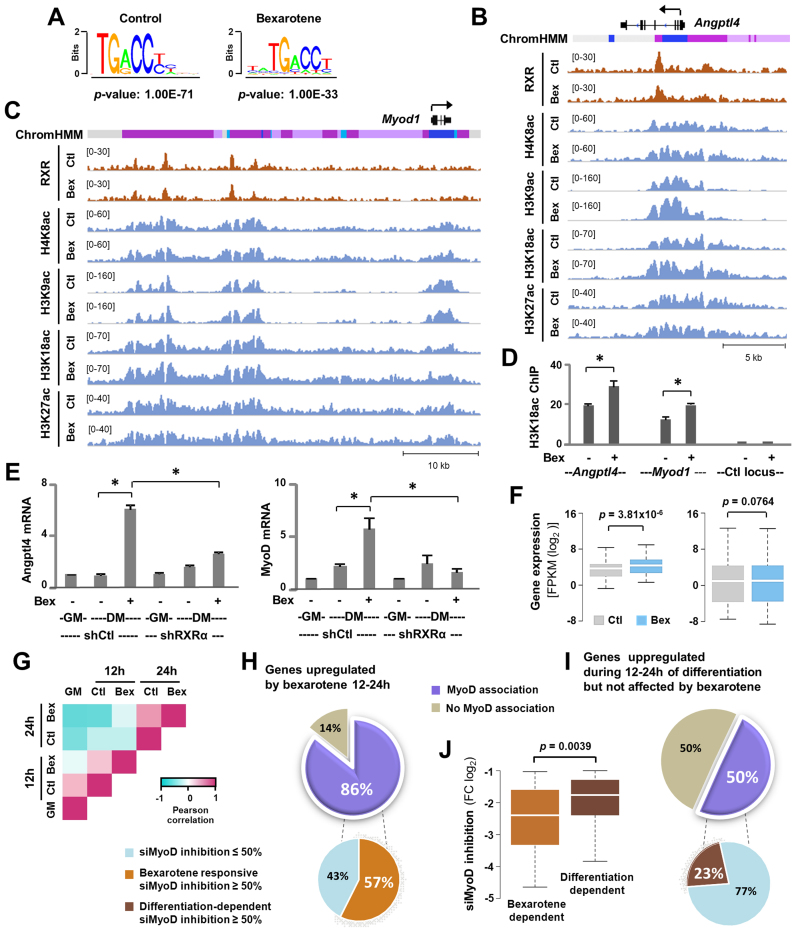
Through MACS analysis of the RXRα read enrichment signals, 627 and 1207 high confidence peaks were identified in the treated and untreated condition, respectively (Figure 2 and Supplementary Figure S2). As determined by de novo motif analysis, the nuclear receptor motif was found to be the top ranking binding motif amongst the RXRα peaks in both treated and untreated conditions (Figure 2A), in agreement with previous studies that RXRs are bound to their DNA sites constitutively, despite the presence of ligand (46,47). Particularly, RXRα occupancy was evident at a previously identified locus of the gene encoding angiopoietin-like factor 4 (Angptl4), a model target of RXR signaling (48), and the Angptl4 locus exhibited an apparent increase in H3K18ac enrichment following bexarotene treatment compared to untreated myoblasts (Figure 2B).
Figure 2.
RXR regulates MyoD expression. (A) Consensus binding sequences of nuclear receptors were discovered through de novo motif analysis of RXRα peaks in myoblasts differentiated for 24 h in the absence or presence of bexarotene (Ctl or Bex 50 nM). (B) Genome browser view of RXRα and histone acetylation signals at the Angptl4 locus. Black bars show Refseq gene position and ChromHMM track colors correspond to color designated to each chromatin state as in Figure 3A. (C) Genome browser view of the Myod1 locus. (D) H3K18ac enrichments at the Angptl4 and Myod1 locus were examined by qChIP analysis with an intergenic region as control. Enrichment was quantified as percentage of input (error bars: SEM; n = 3; *, P < 0.05). (E) RXRα-knockdown (shRXRα) myoblasts were differentiated with or without bexarotene for 24 h with proliferating myoblasts (GM) as controls. A non-silencing shRNA (shCtl) was used in parallel as control. The mRNA levels of Angptl4 and MyoD were assessed by RT-qPCR and plotted as fold change in relation to proliferating myoblasts (error bars: SEM; n = 3). (F) Expression levels of bexarotene responsive genes assigned to the RXRα peaks in differentiating myoblasts and that of all RXRα associated genes in differentiating myoblasts is presented as FPKM measured by RNA-seq. (G) The heatmap of Pearson correlation coefficients calculated between the row-based z-scores for bexarotene responsive genes. (H and I) MyoD association is categorized for the 147 genes upregulated by bexarotene between 12 and 24 h of differentiation and the 246 genes upregulated in the same period but not affected by bexarotene. A pie chart below displays the percentage of genes which exhibited a >50% of inhibition in C2C12 myoblasts differentiated for 24 h following siMyoD knockdown, compared to a non-specific silencing control. (J) Expression of MyoD-associated genes (the 86 and 50% group respectively) from the siMyoD knockdown microarrays is presented as fold change in relation to a non-specific silencing control.
Interestingly, the core enhancer region (CER) of Myod1 (49,50), ∼20 kb upstream of the TSS, as well as three additional regions (about 26, 34 and 38 kb upstream of the TSS) were also occupied by RXRα (Figure 2C). Moreover, H3K18ac at the CER was further enriched following bexarotene treatment as compared to the untreated condition (Figure 2C). ChIP-qPCR analysis validated that H3K18ac signals at the Angptl4 and Myod1 loci increased significantly following bexarotene treatment (Figure 2D), which is consistent with previous reports that H3K18ac is often associated with nuclear receptor signaling and rexinoid action (28,51).
Additionally, the mRNA levels of Angptl4 and MyoD in differentiating myoblasts were indeed augmented after 24 h of bexarotene treatment and the positive effects of bexarotene on their gene expression were impeded by RXRα shRNA knockdown, indicating that bexarotene-enhanced gene expression is mediated through the function of RXRα as a transcription factor (Figure 2E and Supplementary Figure S2B). On average, the expression levels of RXRα-associated bexarotene responsive genes increased significantly following bexarotene treatment, whereas that of ENSEMBL genes assigned to RXRα en bloc was rather comparable in treated and untreated conditions (Figure 2F). Taken together, our data suggests that a common mode of molecular regulation may mediate differential gene expression observed in rexinoid-enhanced myoblast differentiation, in that it may be reconciled largely through the regulation of MyoD gene expression.
Pearson correlation analysis revealed that the expression levels of bexarotene responsive genes at 12-h of the untreated condition not only positively correlated to the treated condition at the same time point, but also to the proliferating state (Figure 2G), suggesting that these genes may be globally coregulated at the early stage of differentiation. Likewise, the lack of positive correlation between 24-h treated condition with the 12-h untreated and proliferating state again suggests that the late response to bexarotene treatment may be an indirect outcome. We therefore analyzed in greater detail the association of MyoD with bexarotene responsive genes to explore the potential of rexinoid signaling through MyoD to enhance myogenic expression, since MyoD is a direct target of RXRα (Figure 2C).
We found that 86% of the late bexarotene upregulated genes (12–24 h) were associated with MyoD (Figure 2H). In comparison, only 50% of the genes upregulated during the 12–24 h period of differentiation but not affected by bexarotene, were associated with MyoD (Figure 2I). In addition, 57% of MyoD associated bexarotene upregulated genes were significantly inhibited (FC ≥ 0.5) by MyoD siRNA knockdown (52), whereas only 23% of the differentiation-dependent bexarotene-nonresponsive genes were impacted in the same degree (Figure 2I). Finally, the expression of bexarotene upregulated genes was inhibited more significantly by MyoD siRNA in comparison to the differentiation dependant genes (Figure 2J). Taken together, our study suggests that MyoD is a major mediating factor in rexinoid responsive gene expression at the early stage of myoblast differentiation.
Mapping of genomic loci through chromatin states in proliferating myoblasts
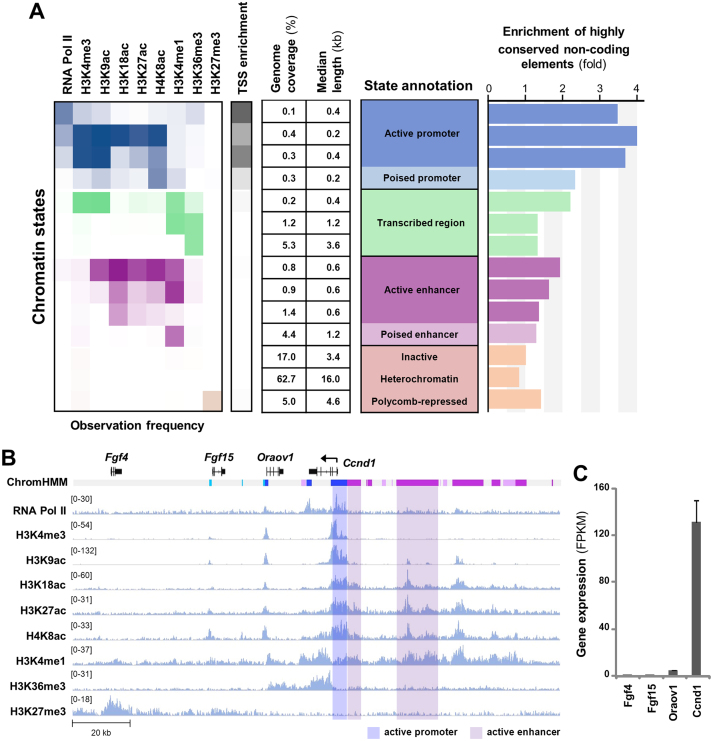
To further delineate the molecular pathways of rexinoid signaling in myogenic modulation, we generated a chromatin state model based on genome-wide co-occurrence of different epigenetic marks in committed proliferating myoblasts, using a hidden Markov model-based method. Incorporating published ChIP-seq datasets for the promoter-associated mark H3K4me3 and RNA polymerase II (RNA Pol II), enhancer-associated mark H3K4me1, transcription-associated mark H3K36me3 and the repressive mark H3K27me3 together with our own H3K9ac, H3K18ac, H3K27ac and H4K8ac ChIP-seq data in proliferating C2C12 myoblasts (GSE94558, Supplementary Table S1), we established a model with 14 chromatin states reflecting diverse activities in gene expression prior to the onset of myoblast differentiation (Figure 3).
Figure 3.
Characterization of the epigenome in proliferating myoblasts. (A) The 14-state chromatin state model was generated based on global ChIP-seq read enrichment for RNA Pol II, H3K4me3, H3K9ac, H3K18ac, H3K27ac, H4K8ac, H3K4me1, H3K36me3 and H3K27me3. TSS enrichment was calculated as the ratio between the fraction of bases in the genome overlapping the feature and state and the joint probability that a base would overlap with the feature and state. Enrichment of highly conserved non-coding elements was calculated similarly. (B) Genome browser view of the ChIP-seq signals and chromatin states at the Ccnd1 locus. Black bars show Refseq gene positions and ChromHMM track colors correspond to the colors designated for each chromatin state as in panel A. (C) Expression levels of the Refseq genes in proliferating myoblasts is presented as FPKM measured by RNA-seq analysis.
These chromatin states were categorized as promoters, transcribed regions, enhancers and regions that were either inactive or repressed (Figure 3A). Briefly, promoter states were identified by enrichment of H3K4me3, TSSs, RNA Pol II and histone acetylation (Figure 3A). Such is the case of Ccnd1 promoter (53), reflecting the fact that Ccnd1 is readily accessible to transcriptional machinery and actively transcribed in proliferating myoblasts (Figure 3B and C). On the other hand, enhancers were identified by the presence of H3K4me1 and absence of H3K4me3 and H3K36me3, and further classified as active or poised based on levels of histone acetylation (Figure 3A). For example, upstream region of the gene encoding Ccnd1 (54), a critical regulator of cellular proliferation, were marked by abundant of H3K4me1 and histone acetylation coupled to a high level of Ccnd1 transcripts, in contrast to neighbouring genes (Figure 3B and C).
As previously reported for other cell types (55), the majority of the myoblast genome (∼80%) was devoid of histone modifications analyzed (Figure 3A). Promoter and enhancer regions covered about 1 and 8% of the genome respectively, and had shorter median lengths than inactive or repressed regions, indicating that only a small portion of the genome is utilized to actively regulate gene expression within proliferating myoblasts (Figure 3A). Active promoters and enhancers were also enriched in HCNCEs (Figure 3A), as previously identified through genomic evolutionary rate profiling (37). Thus, distinct chromatin states characterized in proliferating myoblasts may be used to identify lineage-specific regulatory loci pertinent to myogenic differentiation.
MyoD and myogenin associate with distinct chromatin states
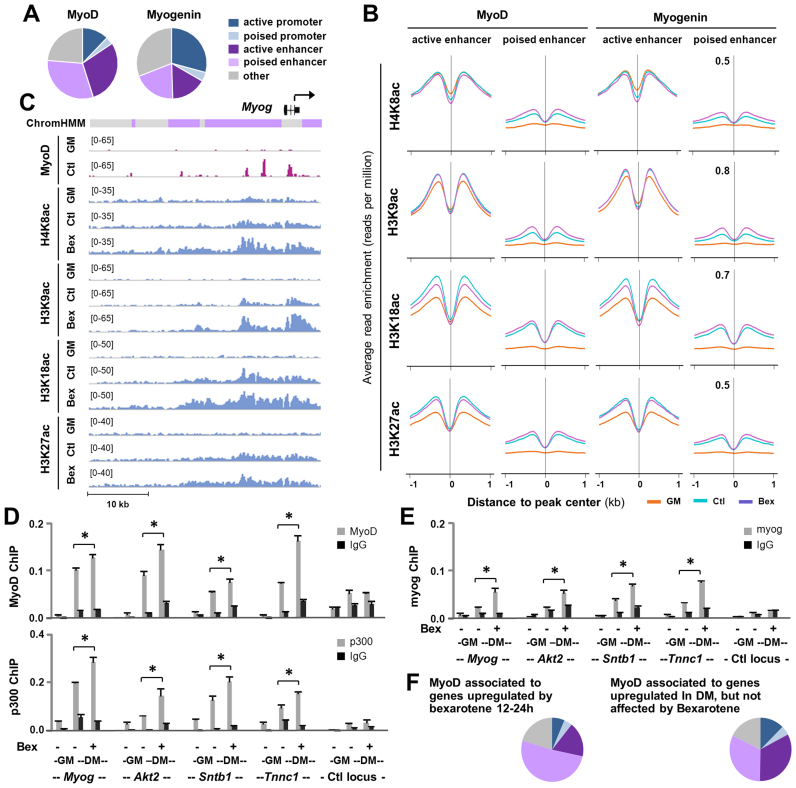
Since rexinoids appear to act through MyoD expression and MyoD and myogenin play sequential roles in the control of myogenic differentiation, we used published datasets (35) on genome-wide myogenin- and MyoD binding sites (Supplementary Table S1) in differentiating myoblasts to examine changes in residue-specific histone acetylation that may be linked to rexinoid signaling during early myogenic differentiation. As predicted by our 14 chromatin state model for proliferating myoblasts, 33 and 36% of myogenin peaks were associated to the promoter and enhancer regions respectively, whereas MyoD binding sites (∼61%) were largely found at the enhancer regions (Figure 4A). Although myogenin and MyoD sites at active enhancers displayed a differentiation-dependent increase in H3K18ac and H3K27ac signals, their binding sites at poised enhancers exhibited an apparent enrichment in each of the acetylation marks profiled including H4K8ac and H3K9ac (Figure 4B). Most interestingly, H4K8ac, H3K9ac, H3K18ac and H3K27ac were further enriched following bexarotene treatment at MyoD- and myogenin-associated poised enhancers, while no such increase was found at the active enhancers (Figure 4B). Our analyses thus suggest a possible role for MyoD and myogenin in the regulation of poised enhancers particularly in the context of rexinoid signaling.
Figure 4.
Association of MyoD and myogenin with distinct chromatin states. (A) Genome wide MyoD and myogenin binding sites in myoblasts differentiated for 24 h were associated with distinct chromatin states. (B) The average read enrichment profiles for H4K8ac, H3K9ac, H3K18ac and K3K27ac corresponding to ±1 kb of the MyoD and myogenin peaks associated to active and poised enhancers in proliferating myoblasts (GM), and myoblasts differentiated for 24 h in the absence or presence bexarotene (Ctl or Bex 50 nM). (C) The genome browser view of the ChIP-seq signals for MyoD and indicated histone acetylation at the Myogenin locus. Black bars show Refseq gene position and ChromHMM track colors correspond to the color designated for each chromatin state. (D and E) ChIP-qPCR analysis was performed for poised enhancers identified for Myog, Akt2, Sntb1 and Tnnc1 using antibodies against MyoD, p300 or myogenin. Normal IgG antiserum and a random locus (Ctl) were used as negative controls. Quantification is presented as the percentage of enrichment in relation to the input chromatin DNA (error bars: SEM; n = 3; *, P < 0.05). (F) Chromatin state distribution of MyoD peaks associated with genes upregulated by bexarotene and with genes upregulated during differentiation but not affected by bexarotene (the 86 and 50% group in Figure 2H and I, respectively).
In differentiating myoblasts, the activation of myogenin expression is coupled with the binding of MyoD to the promoter and upstream enhancer regions of the Myog locus (56) which was characterized as poised or inactive regions in proliferating myoblasts based on our 14 chromatin state model (Figure 4C). While myogenin has been identified previously (28) as a bexarotene responsive gene (Figure 1), RXR occupancy was not detected at the Myog locus (GSM2478304, GSM2478305). However, MyoD associated H4K8ac, H3K9ac and H3K18ac signals at the poised enhancers were not only enriched by 24 h of differentiation, but also further augmented following bexarotene treatment (Figure 4C). To delineate the potential mechanisms of MyoD mediated rexinoid responsive gene expression, we conducted ChIP-qPCR analysis to examine the association of MyoD and p300 to multiple poised enhancers as indicated by our chromatin state model, including a previously identified Myog enhancer (16). Normal IgG antiserum and a random locus were used as negative controls in the analysis. As shown in Figure 4D, the association of MyoD and p300 to these poised enhancers were markedly enriched by 24 h of differentiation. More importantly, the enrichment of MyoD and p300 at these poised enhancers were further significantly augmented with the addition of bexarotene (Figure 4D). Interestingly, myogenin was also significantly enriched at these poised enhancers to a similar degree following bexarotene treatment of the differentiating myoblasts (Figure 4E). RT-qPCR revealed that the expression of genes associated to these poised enhancers emulated the degree of MyoD, myogenin and p300 enrichments (Figure 1 and Supplementary Figure S1). Thus, bexarotene responsive gene expression is mediated through the activation of poised enhancers by MyoD and myogenin as transcription factors and p300 as a HAT.
By and large, 51% of the MyoD peaks associated with the late bexarotene upregulated genes (12–24 h) were found at poised enhancers (Figure 4F). In comparison, only 32% of MyoD peaks associated with genes upregulated during the 12–24 h period of differentiation but not affected by bexarotene were found at the poised enhancers, which was similar to the chromatin state distribution of all MyoD peaks (compare Figure 4F with A). Taken together, our results highlight the importance of MyoD-coupled histone acetylation for the activation of myogenic expression at the onset of myoblast differentiation and for transmitting rexinoid signaling in differentiating myoblasts through poised enhancers, as MyoD is a direct genetic target of RXRα (Figure 2C) and important for myogenin expression (8,56).
Overlapping of myogenin and MyoD in bexarotene responsive histone acetylation
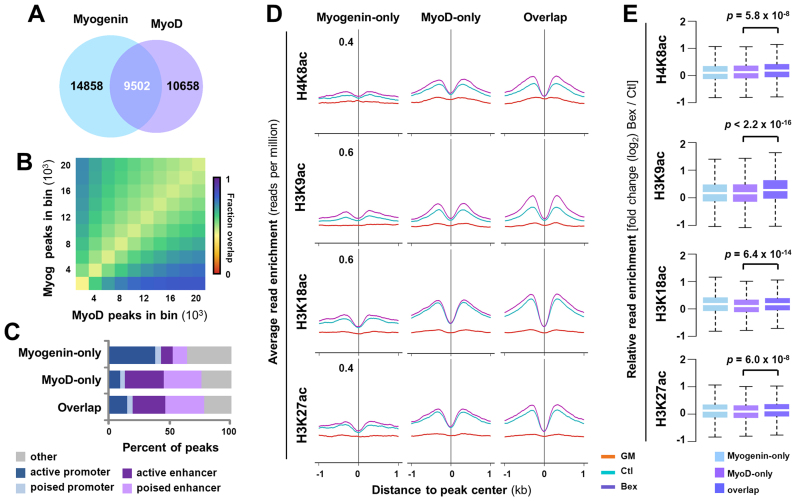
Since bexarotene responsive genes may be globally coregulated at the early stage of differentiation and MyoD and myogenin play critical roles in muscle development, we further explored their interplay with rexinoid signaling. We first grouped genome-wide myogenin binding sites based on their colocalization with MyoD and found that ∼39% of myogenin sites overlapped with MyoD (Figure 5A and B), similar to a previous observation in which about 42% of myogenin sites overlap with MyoD at promoter regions (18). Interestingly, about 38% of myogenin-only sites were found in active promoters, but only 12% were found in poised enhancers (Figure 5C). On the other hand, ∼31% of MyoD sites were associated with poised and active enhancers, regardless of their colocalization with myogenin (Figure 5C). Additionally, the pattern of chromatin state association for sites overlapped by myogenin and MyoD is similar to that bound by MyoD-only but markedly different from that bound by myogenin-only (Figure 5C). Collectively, these results suggest that while myogenin and MyoD may share some overlapping targets, they each bind to a distinct set of loci associated with different chromatin states.
Figure 5.
Overlapping of myogenin with MyoD in histone acetylation. (A) Union analysis of MyoD and myogenin ChIP-seq signals in myoblasts differentiated for 24 h. (B) The top 20,000 MyoD and myogenin peaks were ranked by read enrichment, and grouped into bins with increments of 2000. Each box represents the fraction of sites bound by MyoD and myogenin in that bin. (C) The association of unique and overlapping MyoD and myogenin peaks with distinct chromatin states. (D) The average ChIP-seq signal profiles for H4K8ac, H3K9ac, H3K18ac and H3K27ac at poised enhancers bound by MyoD- or myogenin-only, or both in proliferating myoblasts (GM), and myoblasts differentiated for 24 h in the absence or presence bexarotene (Ctl or Bex 50 nM). (E) Quantification of log2-fold change in histone acetylation.
More interestingly, poised enhancers bound by MyoD-only or overlapped by MyoD and myogenin showed a similar increase in differentiation-dependent histone acetylation (Figure 5D). However, histone acetylation in the two classes of binding sites differed in their response to rexinoid signaling. The MyoD binding sites overlapped by myogenin displayed a more pronounced increase in histone acetylation following bexarotene treatment as compared to those bound by MyoD-only, particularly in H3K9ac (Figure 5D and E). Taken together, our analyses suggest that while MyoD may lead the localization of MyoD–myogenin complexes across the enhancer regions, myogenin possibly cooperates with MyoD in rexinoid responsive histone acetylation at poised enhancers.
DISCUSSION
We have examined the molecular pathways associated with rexinoid-enhanced myogenic differentiation using integral RNA-seq and ChIP-seq analyses. Our findings show that rexinoid signaling acts through the coordination of myogenic expression, from the exit of the cell cycle to the activation of muscle-related genes. Interestingly, bexarotene-responsive gene expression is mediated through RXR as a transcription factor and reconciled largely through a direct regulation of MyoD gene expression. While histone acetylation increases with differentiation at MyoD binding sites associated with different chromatin states, bexarotene augments the enrichments of H3K9ac and H4K8ac particularly at poised enhancers. Our studies thus provide novel molecular insights into the interplay between rexinoid signaling and MRFs-associated chromatin states during early myogenic differentiation.
Intercellular signaling is vital for the intricate coordination of cell cycle regulation and muscle-specific expression at the early stages of myoblast differentiation. The transition between proliferation and differentiation is dependent on a rapid exit from the cell cycle and simultaneous expression of early MRFs. We found that Myod1 is an early rexinoid responsive gene, the active enhancer regions of Myod1 are occupied by RXRα, and knockdown of RXRα impedes MyoD gene expression (Figures 1 and 2), suggesting that MyoD is a direct genetic target of RXR in early myogenic differentiation. At the onset of differentiation, MyoD activates the expression of both muscle-specific genes and cell cycle inhibitors, allowing irreversible exit from the cell cycle and progression into differentiation (57,58). Given that the expression of MyoD is upregulated by bexarotene and RXRα is important for the positive effect of bexarotene on MyoD expression (Figures 1 and 2) and myoblast differentiation (28), a common mode of molecular regulation may mediate differential gene expression observed in bexarotene-enhanced myoblast differentiation.
Characterization of the chromatin states in proliferating C2C12 myoblasts reveals a muscle-specific usage of regulatory DNA elements, and the activity of muscle-specific enhancers is marked by local changes in residue-specific histone acetylation at the early stage of myoblast differentiation (Figures 3–5). While H3K9ac is generally considered as a global mark of promoter activity, we identify an enrichment of H3K9ac at poised enhancers coupled by MyoD at the early stage of differentiation (Figures 4 and 5). Interestingly, this enrichment of H3K9ac was further augmented at myogenin and MyoD overlapping sites following bexarotene treatment (Figure 5). Thus, H3K9ac may reflect the control of a discrete set of genes through the function of MyoD with cooperation of myogenin in early myogenic expression and MyoD plays important roles in the activation of poised enhancers particularly in milieu of rexinoid action. Besides H3K9ac, the enrichments of H4K8ac are also strongly coupled with MyoD and myogenin at the poised enhancers (Figures 4 and 5), suggesting that MyoD and myogenin may play important roles in residue-specific histone acetylation.
A recent study found that although Myf5 and MyoD bind to a largely shared set of DNA binding sites, they differ in their potential to activate gene transcription (59). We found here that myogenin is preferentially associated with promoters, while MyoD has an affinity to enhancers (Figure 4). In addition, MyoD appears to be governing the localization of myogenin and MyoD complexes across enhancers, and myogenin may cooperate with MyoD in rexinoid responsive histone acetylation at poised enhancers primed with MyoD binding (Figure 5). More interestingly, rexinoid-upregulated genes are preferentially associated with MyoD as compared to genes that are upregulated during the same period but not affected by rexinoid, while only a subset of MyoD associated genes are affected by rexinoids (Figure 2). Taken together, our findings support a role for MyoD in the activation of poised enhancers at the onset of myogenic differentiation and in the interplay with rexinoid signaling to further promote myoblast differentiation.
A variety of tissue-related diseases are characterized by muscle wasting, including cerebral palsy, inflammatory myopathies, muscular dystrophies (60–62). Particularly in Duchenne muscular dystrophy patients, the type II skeletal muscle fibers are more prone to damage (63). Interestingly, nuclear receptors, such as PPARδ which regulate transcription by heterodimerizing with RXR, play a key role in the regulation of mitochondrial respiration (64), skeletal muscle lipid oxidation as well as the determination of skeletal muscle fiber types, where an activated form of PPAR was shown to increase the proportion of type I fibers (65), suggesting that RXR agonists may be able to promote muscle generation. Thus, the application of specific nuclear receptor agonists may be explored in muscle-related diseases to promote myofiber numbers and oxidative capacity (66).
In this study, we demonstrate that specific targeting of RXR signaling promotes normal regulation of gene expression occurring during myoblast differentiation, and describe a novel molecular regulation of MyoD gene expression through the activation of RXR. A potential direction will be therefore to determine if rexinoids are able to similarly regulate myogenic transcription in a specific physiological context or disease models. Moreover, the model of rexinoid-enhanced myogenesis also offers an excellent system to identify additional genetic targets and molecular interactions for therapeutic development toward muscle-related diseases.
DATA AVAILABILITY
All RNA-seq and ChIP-seq data from this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, assigned as SuperSeries GSE94561. This SuperSeries is composed of two SubSeries assigned as GSE94560 (RNA-seq data) and GSE94558 (ChIP-seq data), respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs C. Porter and T. Perkins for assistance in transcript assembly of the RNA-seq reads and Mr H. AlSudais for RNA isolation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research Society Operating Grant [#21126 to Q.L.]; Natural Sciences and Engineering Research Council of Canada Graduate Scholarship (to S.K.); Umm Al-Qura University of Saudi Arabia (to M.H.). Funding for open access charge: Cancer Research Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Nord A.S., Blow M.J., Attanasio C., Akiyama J.A., Holt A., Hosseini R., Phouanenavong S., Plajzer-Frick I., Shoukry M., Afzal V. et al. . Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013; 155:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. . The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou V.W., Goren A., Bernstein B.E.. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011; 12:7–18. [DOI] [PubMed] [Google Scholar]
- 4. Hon G.C., Hawkins R.D., Ren B.. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009; 18:R195–R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krebs A.R., Karmodiya K., Lindahl-Allen M., Struhl K., Tora L.. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell. 2011; 44:410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. . Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R.. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993; 75:1351–1359. [DOI] [PubMed] [Google Scholar]
- 9. Davis R.L., Weintraub H., Lassar A.B.. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987; 51:987–1000. [DOI] [PubMed] [Google Scholar]
- 10. Buckingham M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001; 11:440–448. [DOI] [PubMed] [Google Scholar]
- 11. Edmondson D.G., Olson E.N.. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989; 3:628–640. [DOI] [PubMed] [Google Scholar]
- 12. Puri P.L., Sartorelli V., Yang X.J., Hamamori Y., Ogryzko V. V, Howard B.H., Kedes L., Wang J.Y., Graessmann A., Nakatani Y. et al. . Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997; 1:35–45. [DOI] [PubMed] [Google Scholar]
- 13. Sartorelli V., Huang J., Hamamori Y., Kedes L.. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 1997; 17:1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roth J.-F., Shikama N., Henzen C., Desbaillets I., Lutz W., Marino S., Wittwer J., Schorle H., Gassmann M., Eckner R.. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 2003; 22:5186–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yaffe D., Saxel O.. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977; 270:725–727. [DOI] [PubMed] [Google Scholar]
- 16. Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D.. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E149–E158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falcieri E., Burattini S., Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 2004; 48:223–234. [PubMed] [Google Scholar]
- 18. Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D.. An initial blueprint for myogenic differentiation. Genes Dev. 2005; 19:553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L. et al. . Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010; 18:662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P. et al. . The nuclear receptor superfamily: the second decade. Cell. 1995; 83:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gampe R.T., Montana V.G., Lambert M.H., Wisely G.B., Milburn M. V, Xu H.E.. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000; 14:2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol. Endocrinol. 2005; 19:1418–1428. [DOI] [PubMed] [Google Scholar]
- 23. Sapin V., Dollé P., Hindelang C., Kastner P., Chambon P.. Defects of the chorioallantoic placenta in mouse RXRalpha null fetuses. Dev. Biol. 1997; 191:29–41. [DOI] [PubMed] [Google Scholar]
- 24. Sucov H.M., Dyson E., Gumeringer C.L., Price J., Chien K.R., Evans R.M.. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994; 8:1007–1018. [DOI] [PubMed] [Google Scholar]
- 25. Kastner P., Grondona J.M., Mark M., Gansmuller A., LeMeur M., Decimo D., Vonesch J.L., Dollé P., Chambon P.. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994; 78:987–1003. [DOI] [PubMed] [Google Scholar]
- 26. Krezel W., Dupé V., Mark M., Dierich A., Kastner P., Chambon P.. RXR gamma null mice are apparently normal and compound RXR alpha +/-/RXR beta -/-/RXR gamma -/- mutant mice are viable. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9010–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le May M., Mach H., Lacroix N., Hou C., Chen J., Li Q.. Contribution of retinoid X receptor signaling to the specification of skeletal muscle lineage. J. Biol. Chem. 2011; 286:26806–26812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. AlSudais H., Aabed K., Nicola W., Dixon K., Chen J., Li Q.. Retinoid X receptor-selective signaling in the regulation of Akt/Protein Kinase B isoform-specific expression. J. Biol. Chem. 2016; 291:3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamed M., Khilji S., Chen J., Li Q.. Stepwise acetyltransferase association and histone acetylation at the Myod1 locus during myogenic differentiation. Sci. Rep. 2013; 3:2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D. et al. . A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014; 515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davydov E. V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S., Margulies E., Cooper G., Asimenos G., Thomas D. et al. . Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010; 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen L., Shao N., Liu X., Nestler E., Metzker M., Daniel C.K., Karyn M.S., David E.L., Richard K.W., Mardis E. et al. . ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaplan R., Gan X., Menke J.G., Wright S.D., Cai T.-Q.. Bacterial lipopolysaccharide induces expression of ABCA1 but not ABCG1 via an LXR-independent pathway. J. Lipid Res. 2002; 43:952–959. [PubMed] [Google Scholar]
- 41. Kersten S., Lichtenstein L., Steenbergen E., Mudde K., Hendriks H.F.J., Hesselink M.K., Schrauwen P., Muller M.. Caloric restriction and exercise increase plasma ANGPTL4 levels in humans via elevated free fatty acids. Arterioscler. Thromb. Vasc. Biol. 2009; 29:969–974. [DOI] [PubMed] [Google Scholar]
- 42. Daniel B., Nagy G., Hah N., Horvath A., Czimmerer Z., Poliska S., Gyuris T., Keirsse J., Gysemans C., Van Ginderachter J.A. et al. . The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014; 28:1562–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen J., St-germain J.R., Li Q.. B56 regulatory subunit of protein phosphatase 2A mediates valproic acid-induced p300 degradation B56 regulatory subunit of protein phosphatase 2A mediates valproic acid-induced p300 degradation. Mol. Cell. Biol. 2005; 25:525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moran J.L., Li Y., Hill A.A., Mounts W.M., Miller C.P.. Gene expression changes during mouse skeletal myoblast differentiation revealed by transcriptional profiling. Physiol. Genomics. 2002; 10:103–111. [DOI] [PubMed] [Google Scholar]
- 45. Lionikas A., Cheng R., Lim J.E., Palmer A.A., Blizard D.A., Vandenbergh D.J., Blizard D.A., Lionikas A., Cheng R., Lim J. et al. . Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol. Genomics. 2010; 42:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pazin M.J., Kadonaga J.T.. What's up and down with histone deacetylation and transcription. Cell. 1997; 89:325–328. [DOI] [PubMed] [Google Scholar]
- 47. Torchia J., Glass C., Rosenfeld M.G.. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 1998; 10:373–383. [DOI] [PubMed] [Google Scholar]
- 48. Yoon J.C., Chickering T.W., Rosen E.D., Dussault B., Qin Y., Soukas A., Friedman J.M., Holmes W.E., Spiegelman B.M.. Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Mol. Cell. Biol. 2000; 20:5343–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goldhamer D.J., Faerman A., Shani M., Emerson C.P.. Regulatory elements that control the lineage-specific expression of myoD. Science. 1992; 256:538–542. [DOI] [PubMed] [Google Scholar]
- 50. Goldhamer D.J., Brunk B.P., Faerman A., King A., Shani M., Emerson C.P.. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995; 121:637–649. [DOI] [PubMed] [Google Scholar]
- 51. Jin Q., Yu L.-R., Wang L., Zhang Z., Kasper L.H., Lee J.-E., Wang C., Brindle P.K., Dent S.Y.R., Ge K.. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011; 30:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chakroun I., Yang D., Girgis J., Gunasekharan A., Phenix H., Kærn M., Blais A.. Genome-wide association between Six4, MyoD, and the histone demethylase Utx during myogenesis. FASEB J. 2015; 29:4738–4755. [DOI] [PubMed] [Google Scholar]
- 53. Eto I. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: implication in phorbol ester-induced tumour promotion. Cell Prolif. 2000; 33:167–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eeckhoute J., Carroll J.S., Geistlinger T.R., Torres-Arzayus M.I., Brown M.. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006; 20:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. . Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Faralli H., Dilworth F.J.. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp. Funct. Genomics. 2012; 2012:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Halevy O., Novitch B.G., Spicer D.B., Skapek S.X., Rhee J., Hannon G.J., Beach D., Lassar A.B.. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995; 267:1018–1021. [DOI] [PubMed] [Google Scholar]
- 58. Weintraub H., Tapscott S.J., Davis R.L., Thayer M.J., Adam M.A., Lassar A.B., Miller A.D.. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conerly M.L., Yao Z., Zhong J.W., Groudine M., Tapscott S.J.. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation article distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev. Cell. 2016; 36:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O’Dwyer N.J., Neilson P.D., Nash J.. Mechanisms of muscle growth related to muscle contracture in cerebral palsy. Dev. Med. Child Neurol. 1989; 31:543–547. [DOI] [PubMed] [Google Scholar]
- 61. Meryon E. On granular and fatty degeneration of the voluntary muscles. Med. Chir. Trans. 1852; 35:73–84. [PMC free article] [PubMed] [Google Scholar]
- 62. Carpenter S., Karpati G.. Duchenne muscular dystrophy: plasma membrane loss initiates muscle cell necrosis unless it is repaired. Brain. 1979; 102:147–161. [DOI] [PubMed] [Google Scholar]
- 63. Webster C., Silberstein L., Hays A.P., Blau H.M.. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988; 52:503–513. [DOI] [PubMed] [Google Scholar]
- 64. Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P.A.. Peroxisome proliferator-activated receptor controls muscle development and oxydative capability. FASEB J. 2003; 17:2299–2301. [DOI] [PubMed] [Google Scholar]
- 65. Wang Y.-X.Y., Zhang C.C.-L., Yu R.R.T., Cho H.K., Nelson M.M.C., Bayuga-Ocampo C.R., Ham J., Kang H., Evans R.M., Abou M. et al. . Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2004; 2:e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gaudel C., Schwartz C., Giordano C., Abumrad N.A., Grimaldi P.A.. Pharmacological activation of PPARβ promotes rapid and calcineurin-dependent fiber remodeling and angiogenesis in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008; 295:E297–E304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq and ChIP-seq data from this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, assigned as SuperSeries GSE94561. This SuperSeries is composed of two SubSeries assigned as GSE94560 (RNA-seq data) and GSE94558 (ChIP-seq data), respectively.