Abstract
The general transcription factor IIH (TFIIH) is a multi-protein complex and its 10 subunits are engaged in an intricate protein–protein interaction network critical for the regulation of its transcription and DNA repair activities that are so far little understood on a molecular level. In this study, we focused on the p44 and the p34 subunits, which are central for the structural integrity of core-TFIIH. We solved crystal structures of a complex formed by the p34 N-terminal vWA and p44 C-terminal zinc binding domains from Chaetomium thermophilum and from Homo sapiens. Intriguingly, our functional analyses clearly revealed the presence of a second interface located in the C-terminal zinc binding region of p34, which can rescue a disrupted interaction between the p34 vWA and the p44 RING domain. In addition, we demonstrate that the C-terminal zinc binding domain of p34 assumes a central role with respect to the stability and function of TFIIH. Our data reveal a redundant interaction network within core-TFIIH, which may serve to minimize the susceptibility to mutational impairment. This provides first insights why so far no mutations in the p34 or p44 TFIIH-core subunits have been identified that would lead to the hallmark nucleotide excision repair syndromes xeroderma pigmentosum or trichothiodystrophy.
INTRODUCTION
The general transcription factor IIH (TFIIH) has been identified as a common factor involved in both transcription and nucleotide excision repair (NER) processes. TFIIH is highly conserved among eukaryotes and is composed of ten subunits, three of which possess enzymatic activities (Figure 1A). The transcription factor can be divided into two main functional sub-complexes: core-TFIIH comprising the XPB helicase, p62, p52, p44, p34, p8/TTD-A and the heterotrimeric Cdk-activating-kinase (CAK) composed of Cdk7, cyclin H and MAT1. The XPD helicase bridges core-TFIIH to CAK. The CAK complex plays an important role in transcription by phosphorylating the C-terminal domain of RNA polymerase II (RNA pol II) as well as several transcription regulators such as the nuclear hormone receptor RAR or PPAR (1). XPB and XPD possess adenosine triphosphate (ATP)-dependent translocase and helicase activities, respectively. XPB is involved in promoter opening during transcription initiation, whereas XPD is responsible for strand separation around the DNA lesion during NER.
Figure 1.
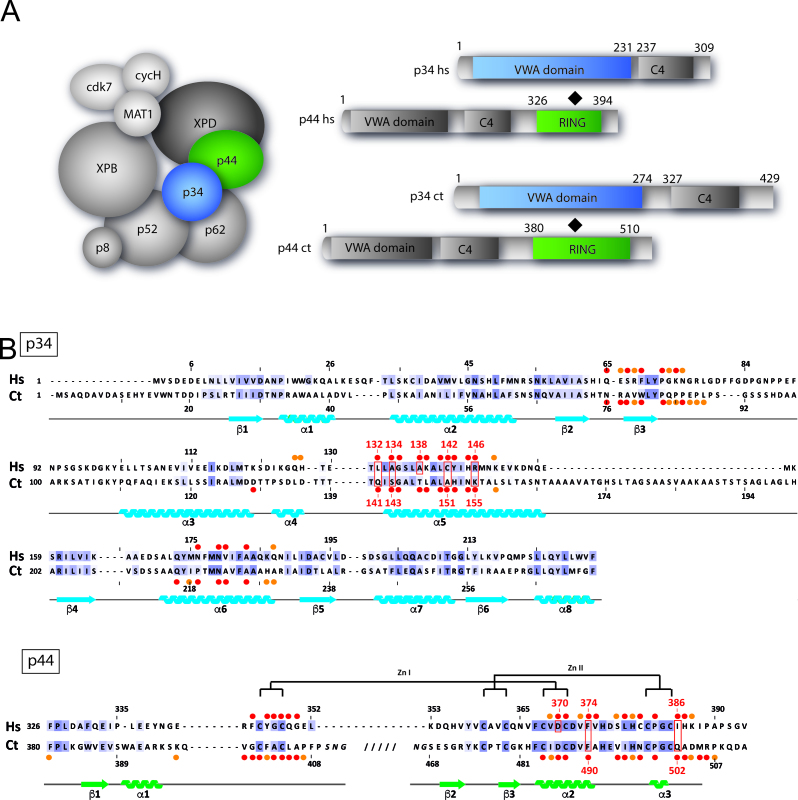
Organization of TFIIH and characterization of a minimal p34/p44 complex. (A) Composition of TFIIH and domain organization of the p34 and p44 TFIIH subunits: the Homo sapiens (hs)/Chaetomium thermophilum (ct) nomenclature of each subunit is indicated. The different structural domains of the p34/p44 subunits are represented in blue, gray and green shades, respectively. The interaction between the two subunits as observed in the structures is indicated for each protein and framed by a black diamond. (B) Structure based alignment of H. sapiens (hs) and C. thermophilum (ct) p34 and p44. Secondary structure elements are indicated below the sequence, with arrows representing β-strands and coils for α-helices. Red (largely buried) and orange circles (partially buried) indicate interface residues as defined by PDBsum. Residues that were mutated are boxed and numbered. Conserved residues are color coded (light purple for moderate conservation and dark purple for highly conserved residues (threshold 30% and 50% identify computed from a multiple sequence alignment of the human and ct sequences with orthologs from Danio rerio, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae and Arabidopsis thaliana). The highly conserved Zn knuckles of the p44 subunits are shown.
Mutations in XPB, XPD and the p8/TTD-A subunits of TFIIH are associated with autosomal recessive disorders such as xeroderma pigmentosum (XP), sometimes combined with Cockayne syndrome (XP/CS), or trichothiodystrophy (TTD) (2,3). Many of the known mutations that cause the aforementioned syndromes do not directly affect the activity of XPB and XPD but rather the interactions of these subunits with their regulatory partners. No further disease associated mutations in other TFIIH subunits have been identified in humans so far. Yeast genetics has shown that the p44/SSL1 and XPB/SSL2 subunits are stem loop suppressors, essential for translation initiation (4,5). In drosophila, mutations in p62 and p52 lead to photosensitivity and XP/TTD like phenotypes (6,7).
The intricate network of interactions between the 10 subunits of yeast or human TFIIH that governs its assembly and activity has been analyzed by several groups (8–11). For human TFIIH it was shown that XPB and XPD interact tightly with the p52 and p44 core-TFIIH subunits, respectively (12–14). p52 is physically associated with p8/TTD-A (15) to regulate the ATPase and translocase activities of XPB (16), whereas p44 stimulates the ATPase and helicase activities of XPD (12,17). The trimeric CAK complex formed between cdk7, cyclin H and MAT1 is required for optimal kinase activity of cdk7 (18). CAK interacts with XPB and XPD and has been shown to inhibit XPD’s helicase activity in vitro (19,20).
So far, low resolution EM reconstructions of the entire yeast and human TFIIH have been determined as well as crystal and nuclear magnetic resonance (NMR) structures of isolated subunits and/or functional domains (Supplementary Table S1). These include the crystal structures of the N-terminal von Willebrand Factor A (vWA) like domain of p34 from the eukaryotic fungus Chaetomium thermophilum (ct) (21), the N-terminal domain of yeast p44/SSL1 (22) and the NMR structure of the C-terminal RING finger of human p44 (23,24). However, with the exception of the crystal structure of the interaction domain of p52/Tfb2 in complex with p8/TTD-A/Tfb5, little is known regarding the interaction between core-TFIIH subunits and no high resolution data on protein interfaces (15) between TFIIH subunits are available.
Here, we analyzed the interaction between the p34 and p44 subunits (Figure 1A) and solved the crystal structure of the N-terminal vWA domain of p34 in complex with the RING domain of p44. Surprisingly, interface variants only mildly affected the association between the full length proteins and did not impinge on TFIIH activities due to the presence of an additional interface involving the C4 domain of p34. Further analyses revealed that the C4 domain of p34 does not only rescue the interaction between p34 and p44 but also seems to be necessary to maintain additional core-TFIIH interactions.
MATERIALS AND METHODS
Protein expression, purification and crystallization of p34 vWA/p44 RING complexes
The p34hs(1–233)/p44hs(321–395) and p34ct(1–277)/p44ct(386–584) complexes were co-expressed in Escherichia coli BL21(DE3) and purified using affinity followed by size exclusion chromatography (SEC). Details are provided in the Supplementary Materials and Methods section. Crystals of the human complex grew in hanging drops at 20°C from a 0.1 mM Tris–HCl or HEPES (pH 7.0 to 8.5) and 20–40% (w/v) PEG 4000 solution containing 5 mg/ml or 2.5 mg/ml of protein. The ct p34/p44 minimal complex I (MC I) was crystallized at 20°C in 15% (w/v) PEG 20 000 and 100 mM MES pH 6.5, whereas the ct p34/p44 minimal complex II (MC II) was crystallized at 20°C in 5–10% (w/v) PEG 4000, 20–33% (v/v) MPD and 100 mM HEPES pH 7.0–7.5. Crystals of the ct p34/p44 minimal complexes I and II were crystallized at concentrations of 5–10 mg/ml using the vapor diffusion method in sitting drops.
Structure determination and analysis
X-ray diffraction data were collected at the SOLEIL Proxima1 beamlines and the European Synchrotron Radiation Facility and processed using XDS (25). The structures of the p34ct/p44ct complex were solved by molecular replacement using the p34ct vWA domain (PDB ID: 4PN7) as a search model. Phases permitted the building of the p44ct RING domain using the human p44 NMR structure (PDB ID: 1Z60) as a guide. The structure of the p34hs/p44hs complex was solved by molecular replacement using the same strategy. We used the intrinsic Zn anomalous scattering signal of the two zinc ions in the p44 RING domain to confirm the solution. Models were adjusted and extended in Coot (26) and refined with Phenix (27) or Refmac (28). Figures containing structures were generated using UCSF Chimera (29) or PyMOL (30). Details are provided in the Supplementary Materials and Methods section.
Construction of baculoviruses, production and purification
Procedures used for insect cell manipulation, generation of recombinant baculoviruses and production of multiprotein complexes are described in (31). To produce wild-type and variant p34hs/p44hs complexes, viruses expressing p34hs in fusion with an N-terminal twin-strep tag under the control of a p10 promoter and with p44hs under the control of a PH promoter were generated. These viruses were co-infected with the virus Vcore Δp34p44, expressing the core-TFIIH subunits His-XPB, p62, p52 and p8 but missing p34 and p44 to reconstitute core-TFIIH variants. Details of the purification procedures are provided in the Supplementary Materials and Methods section.
Characterization of the protein complexes
The interaction between p34ct(1–277) or p34ct full length and p44ct(368–534), which can both be expressed and purified as isolated proteins, was investigated using SEC and isothermal titration calorimetry (ITC). Pull down experiments followed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis with Coomassie staining or western blot detection or high resolution LabChip electrophoresis analysis were used to analyze the p34hs/p44hs complexes. Protein thermal stability was measured by label-free fluorimetric analysis. Details are provided in the Supplementary Materials and Methods section.
Transcription and dual-incision assays
In vitro transcription and dual-incision assays were performed as previously described (19): Run-off transcription assays were performed using reaction mixes containing the adenovirus major late promoter sequence (AdMLP) EcoRI–SalI DNA template (50 ng), TFIIB (15 ng), TFIIE (160 ng), TFIIF (500 ng of the phenyl fraction, (32)), TBP (30 ng), endogenous RNA Pol II (10 μg of the 1 M DEAE fraction) and TFIIH was replaced by a mixture of purified wild-type and variant core-IIH6, XPD (100 ng) and CAK (100 ng). NER dual-incision assays were performed using a plasmid with a single 1,3-intrastrand d(GpTpG) (50 ng) in a buffer containing 50 mM HEPES-KOH (pH 7.8), 5 mM MgCl2, 1 mM Dithiothreitol (DTT), 0.3 mM ethylenediaminetetraacetic acid, 10% (v/v) glycerol, 2.5 μg bovine serum albumin, 50 mM KCl and 2 mM ATP. Reaction mixtures (10 μl) containing XPG (5 ng), XPF/ERCC1 (15 ng), XPC/hHR23B (10 ng), RPA (50 ng), XPA (25 ng) and XPD (100 ng) and increasing amounts of purified wild-type or variant core-IIH6 (see figure legends) were incubated at 30°C for 90 min and stopped by rapid freezing. About 6 ng of an oligonucleotide complementary to the excised DNA fragment containing four extra G nucleotides at the 5′ end was annealed to the excision products. Sequenase (0.1 units) and 1 μCi of [α-32P]dCTP were used to add four radiolabeled C nucleotides to excision products that were separated on a denaturing 14% (w/v) polyacrylamide gel and visualized by autoradiography. Concentrations of core-IIH6 wild-type and variants were estimated using quantitative western blot analysis with a monoclonal antibody directed against XPB (1B3).
Automated Hydrogen–Deuterium eXchange coupled to Mass Spectrometry (HDX-MS)
Hydrogen–Deuterium eXchange coupled to Mass Spectrometry (HDX-MS) experiments were performed on a Synapt-G2 HDMS (Waters Scientific, Manchester, UK) coupled to a Twin HTS PAL dispensing and labelling robot (LEAP Technologies, Carborro, NC, USA) via a NanoAcquity system with HDX technology (Waters, Manchester, UK). Briefly, 5 μl of protein at 31.5 μM were diluted in 95 μl of protonated (for peptide mapping) or deuterated buffer (20 mM Tris pD 7.4, 50 mM NaCl) and incubated at 20°C for 0, 0.5, 1, 5 and 10 min. A total of 95 μl were then transferred to vials containing 15 μl of pre-cooled quenching/digestion solution (0.8 mg/ml pepsin in 500 mM glycine pH 2.6). After 2 min of digestion, 105 μl were injected into a 100 μl loop. Peptides were desalted for 2 min on a C18 pre-column (Acquity UPLC BEH 1.7 μM, VANGUARD) and separated on a C18 column (Acquity UPLC BEH 1.7 μM, 1.0 × 100 mm) by a linear gradient (2% to 40% (v/v) acetonitrile in 7 min). Experiments were run in triplicates and protonated buffer was injected between each sample to wash the column and avoid cross-over contamination.
Peptide identification was performed with the ProteinLynx Global SERVER (PLGS, Waters, Manchester, UK) based on the MSE data acquired on the non-deuterated samples. Peptides were filtered in DynamX 3.0 with the following parameters: peptides identified in at least 3 acquisitions, 0.2 fragments per amino-acid, intensity threshold 1000. Structural representation of the differential uptake was performed using UCSF Chimera (29).
RESULTS
Structure of the minimal p34/p44 complex
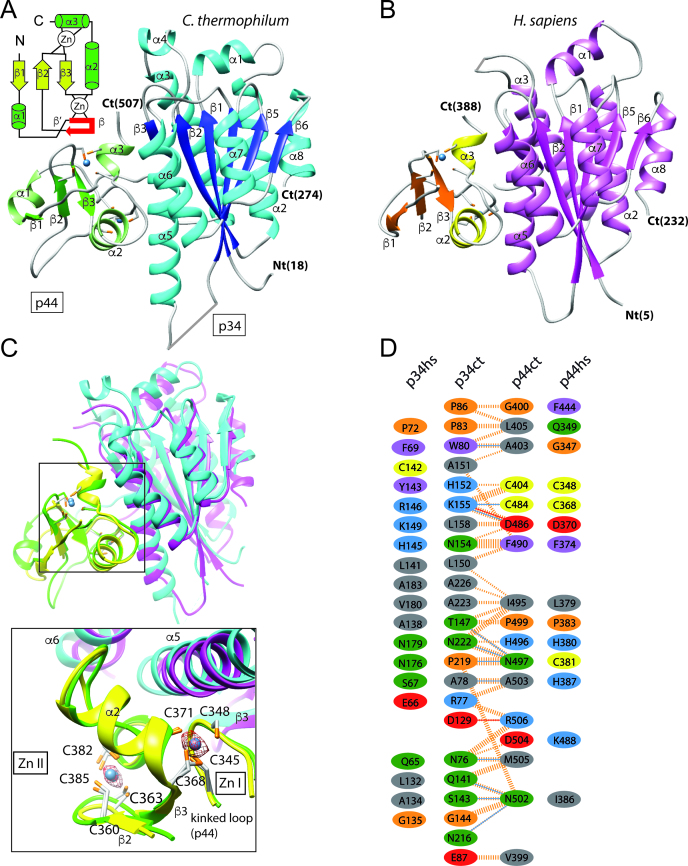
Based on previous studies (33), we pursued the structural characterization of the human p34/p44 minimal complex (p34hs(1–233) and p44hs(321–395)) and the corresponding complex from the fungal model organism Chaetomium thermophilum (p34ct(1–277) and p44ct(368–534)). The p34hs/ct and p44hs/ct sequences share 28% and 40% sequence identity, respectively (Figure 1B). The human and the fungal p34/p44 complexes are structurally similar (Figure 2A–D) and can be superimposed with a root mean square (rms) deviation of 1.5 Å using 236 equivalent Cα-atoms (182 and 54 residues for p34 and p44, respectively), indicating a high structural conservation of the p34/p44 complex. Their secondary structure elements are almost identical except for two helices (α4 of p34ct and α1 of p44ct) that are only present in the fungal complex. Data collection and refinement statistics can be found in Table 1. A detailed description of the structure solution process is available in the Supplementary Materials and Methods section.
Figure 2.
Overall structure of the p34/p44 complex. (A and B) Ribbon representation of the ct and hs p34/p44 complexes. The topology of the p44 RING domain is shown on the upper left side of panel A. The kinked loop between helix α1 and strand β2 is shown in red. (C) Overall superposition of the human (in magenta and yellow for p34 and p44, respectively) and fungal (in blue and green for p34 and p44, respectively) p34/p44 complex structures. Close-up view of the interactions between the two proteins. The blue spheres represent the Zn ions. The anomalous electron density map from the human complex calculated using the phases of the p34 molecule is contoured at 3σ. (D) Schematic diagram of the p34 vWA/p44 RING interface. The fungal interface was analyzed with PDBsum: interacting residues are colored by residue type (blue positive (H,K,R); red negative (D,E); green neutral (S,T,N,Q); gray aliphatic (A,V,L,I,M); magenta aromatic (F,Y,W); orange (P,G); yellow (C) cysteine) and interactions are indicated by colored lines (blue and red lines for potential H-bonds and salt bridges, dashed orange lines for non-bonded contacts). Structurally similar residues (identified from a geometry-based alignment of the human and fungal interfaces) are represented using the same color code.
Table 1. Data collection and refinement statistics.
| Data collection | p34ct/p44ct MC (I) | p34ct/p44ct MC (II) | p34hs/p44hs * |
| Space Group | P 63 2 2 | P 63 2 2 | I 21 21 21 |
| Cell Dimensions | |||
| a, b, c [Å] | 138.21, 138.21, 95.24 | 147.06, 147.06, 87.60 | 81.41, 120.39, 128.67 |
| α, β, γ [°] | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution [Å] | 45.24 – 3.7 (4.05 – 3.70) | 48.14 – 2.2 (2.27 – 2.20) | 40.7 – 3.4 (3.58 – 3.40) |
| Wavelength [Å] | 0.8726 | 0.97625 | 1.2826 |
| Unique Reflections | 6,120 (1,418) | 28,834 (2,439) | 9014 (1306) |
| I/σI | 9.5 (1.4) | 19.6 (1.0) | 10.5 (2.0) |
| I/σI ≥ 2.0 [Å] ** | 3.97 | 2.36 | 3.40 |
| CC (1/2) | 0.998 (0.601) | 0.999 (0.700) | 0.998 (0.820) |
| Rmerge [%] | 20.0 (142.5) | 12.1 (454.8) | 13.3 (145.7) |
| Completeness [%] | 99.8 (100.0) | 100.0 (100.0) | 99.4 (96.4) |
| Redundancy | 8.1 (8.4) | 18.5 (14.8) | 13.1 (13.1) |
| SigAno | 0.8 (0.6) | ||
| Refinement | p34ct/p44ct MC (I) | p34ct/p44ct MC (II) | p34hs/p44hs * |
| Resolution [Å] | 45.2 – 3.7 | 41.4 – 2.2 | 40.7 – 3.4 |
| Unique Reflections | 5828 | 28 436 | 9014 |
| Number of Atoms | 2088 | 2,211 | 3896 |
| Rwork (Rfree) [%] | 19.6 (24.6) | 21.0 (22.7) | 24.0 (29.0) |
| Mean B-Factor [Å2] | 152.7 | 73.88 | 146.2 |
| Bond Lengths [Å] | 0.011 | 0.003 | 0.003 |
| Bond Angles [°] | 1.610 | 0.728 | 0.720 |
| Ramachandran Statistics (%) | |||
| Favored | 93.56 | 96.83 | 95.00 |
| Allowed | 4.92 | 3.17 | 4.35 |
| Outliers | 1.52 | 0.0 | 0.65 |
Values in parentheses refer to the highest resolution shell. * Friedel mates were averaged when calculating reflection statistics. ** Resolution [Å] at which I/σI ≥ 2.0
Ramachandran analyses were performed using MolProbity.
In both complexes, p34 adopts a vWA like architecture as it was previously described by Schmitt et al. (21) (Figure 2A and B). The p44ct/hs C4C4 RING finger displays a compact α/β global fold and binds two zinc ions with an interweaved zinc-binding motif (Figure 2A–C). Interestingly, the p44ct/hs C4C4 RING fingers present significant structural homology to other RING finger domains. The best matches were obtained with the RFN38 and Ark2C E3-RING domains (pdb entries 5D0M chain B and 4V3L chain C) that can be superimposed with the human p44-RING with rms deviations of 1.9 Å and 2.5 Å for 50 and 46 equivalent Cα-atoms, respectively (Supplementary Figure S1, left panel). Furthermore, this similarity is extended toward their interactions with other proteins, since both zinc-binding sites of the p44-RING and the E3-RING domain participate in the recognition of the partner molecule (Supplementary Figure S1, middle and right panel).
Analysis of the p34 vWA/p44 RING interface
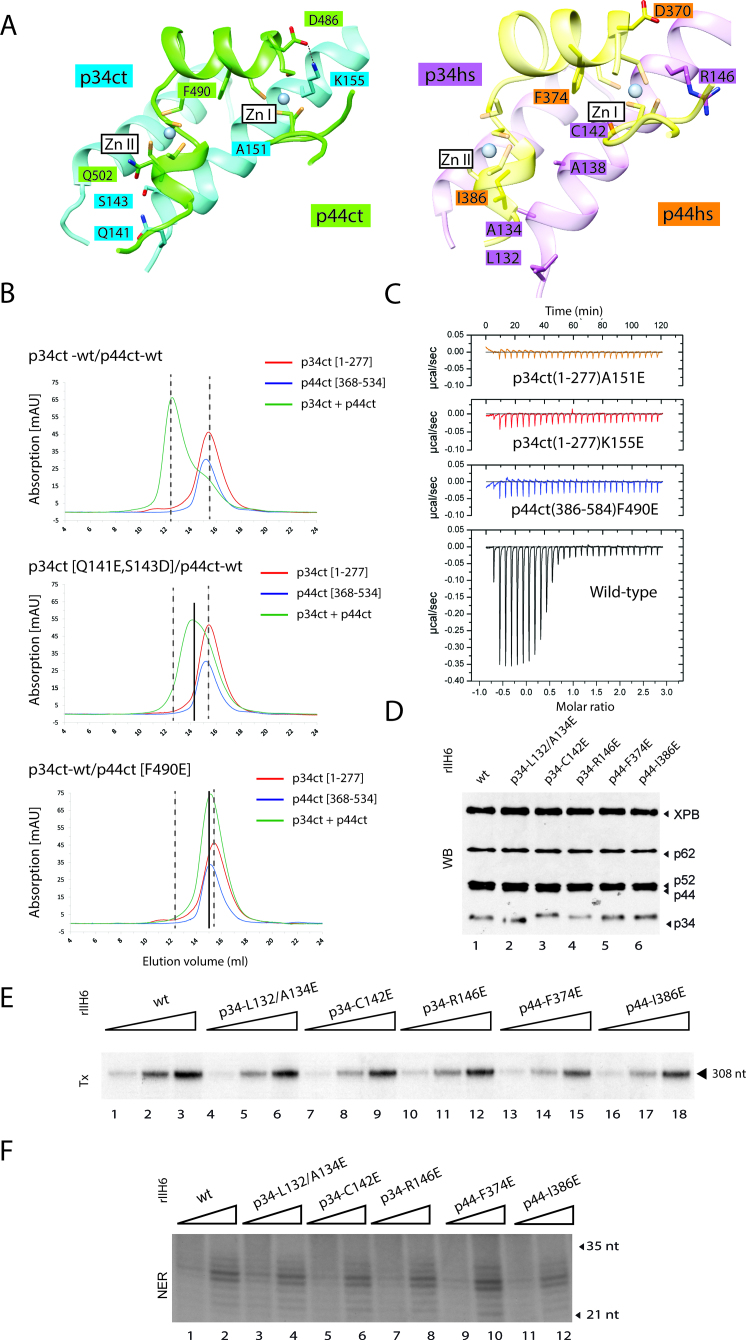
The p34ct/hs-p44ct/hs interactions are governed by strand β3 and helices α5 and α6 of p34 and structural elements flanking the cross-braced zinc-binding motif of p44 including the kinked loop between helix α1 and strand β2 as well as helices α2 and α3 (see Figure 2C). In line with the overall spatial conservation of the complexes, a geometry-based comparison of the interfaces (34) from both species provided a structural alignment of 39 residues (IS-score of 0.53, P-value 0.44 10−11 and rmsd 1.6 Å). Within these aligned residues, 28% are identical, a value that is comparable to that obtained when all surface residues are considered (28% and 34% sequence identity for p34 and p44, respectively). A total of 11 residues share a hydrophobic character while 8 are polar or charged in both structures, resulting in a complex mixture of small hydrophobic patches and polar interactions scattered over the entire interface. Conserved hydrophobic residues contributing to the interface include F69, P72, L141 and A183 from p34hs and L379, F374 and P383 from p44hs, corresponding to W80, P83, L150 and A226 from p34ct and I495, F490 and P499 from p44ct (Figure 2D). However, some interactions also differ between the two complexes, for example, the orientation of the positively charged side chain of R146 in p34hs, corresponding to K155 in p34ct that forms a salt bridge with D386 from p44ct (Figure 3A, left panel). R146 from p34hs points into a different direction and therefore, does not participate in similar interactions.
Figure 3.
Mutational analysis of the p34/p44 interface. (A) Detailed view of the contact area for the fungal and the human complex: selected side chains of interface residues are shown. Residues 76–85, 131–151 and 213–231 of p34ct (blue) and 398–407 and 484–506 of p44ct (green) of the fungal complex are represented in the left panel. Residues 65–74, 131–151 and 172–188 of p34hs (magenta) and 343–351 and 368–506 of p44hs (yellow) are represented in the right panel. (B) SEC of p34ct(1–277) with p44ct(368–534) wild-type or variants. p34ct(1–277) (red) and p44ct(368–534) (blue) were analyzed separately and after mixing them in a 1:1 stoichiometry (green) prior to SEC. (C) Quantification of the interaction between wild-type or variant p34ct(1–277) and p44ct(368–534) by ITC. The thermodynamic parameters for the association of the wild-type protein domains are: KD = 11 nM, n = 0.71, ΔH = −25 kcal/mol and ΔS = −45 cal/mol/degree. (D) Core-TFIIH wild-type or variant were co-expressed in insect cells and purified using immobilized metal affinity chromatography (IMAC) followed by immuno-precipitation using an anti-p44 antibody. Equal amounts of purified rIIH6 were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis followed by western blot (WB) using antibodies directed against XPB, p62, p52, p44 and p34. (E) Basal transcription activity. Increasing amounts of purified wild-type and variant rIIH6 (20, 40 and 80 ng) were added to an in vitro reconstituted transcription system lacking core-TFIIH. Transcripts were analyzed by electrophoresis followed by autoradiography. The length of the corresponding transcript is indicated on the right. (F) In vitro dual-incision assay. Increasing amounts of immune purified wild-type and variant rIIH6 (60 and 160 ng) were added to an incision/excision assay using recombinant NER factors, and the reaction was analyzed by electrophoresis followed by autoradiography. Quantification of the basal transcription and of the double incision assays are presented in Supplementary Figure S5C.
Mutational analysis of the p34/p44 interface
To further dissect the interaction of both complexes, we generated mutations in the interface of the p34hs/ct-p44hs/ct complexes: L132Ehs/Q141Ect, A134Ehs/S143Dct, C142Ehs/A151Ect and R146Ehs/K155Ect of p34 and D370Ahs, F374Ehs/F490Ect and I386Ehs/Q502Ect of p44 (Figures 1B and 3A, Table 2). Circular dichroism spectroscopy of the fungal variants showed that they all display similar spectra as the wild-type proteins indicating that the overall fold has been maintained (Supplementary Figure S2).
Table 2. Human (Hs) and Chaetomium thermophilum (Ct) p34/p44 variants and their location.
| Variant in Hs p34 | Variant in Ct p34 | Structure element |
| L132E | Q141E | helix α5 |
| A134E | S143D | helix α5 |
| A138E | helix α5 | |
| C142E | A151E | helix α5 |
| R146E | K155E | helix α5 |
| Variant in Hs p44 | Variant in Ct p44 | Structure element |
| D370A | helix α2 | |
| F374E | F490E | helix α2 |
| I386E | Q502E | helix α3 |
Analysis by SEC revealed that equimolar ratios of wild-type p34ct(1–277) and p44ct(368–534) form a stable complex as demonstrated by a clear shift in the elution volume compared to the individual components (Figure 3B, upper panel). The p44ct(368–534)-Q502E variant does not prevent complex formation when combined with WT p34ct(1–277) (Supplementary Figure S3A, second panel). The double p34ct(1–277)-Q141E/S143D variant that most likely disrupts the hydrogen bonding network between helices α5 of p34ct and α3 of p44ct (Figure 3A) shows a weakened interaction with wild-type p44ct(368–534) as illustrated by the observation of a shifted peak for the complex (Figure 3B, middle panel), whereas the three single mutations A151E and K155E of p34ct and F490E of p44ct clearly disrupt the interaction between p34ct(1–277) and p44ct(368–534) (Figure 3B, lower panel and Supplementary Figure S3A, third and fourth panels). The p34ct(1–277)-A151E and p44ct(368–534)-F490E single mutations most likely weaken the van der Waals contacts in close proximity to the Zn I binding site, while the p34ct(1–277)-K155E variant disrupts the electrostatic interaction with D486 of p44ct (Figure 3A).
Additional characterization by ITC shows that the p34ct(1–277) and p44ct(368–534) fragments associate with a KD of 11 nM and a 1:1 stoichiometry (Figure 3C and Supplementary Figure S3B). However, no interaction was observed when the p34ct(1–277)-A151E or the p34ct(1–277)-K155E variant was titrated into p44ct(368–534). Similarly, the p44ct(368–534)-F490E variant also abolished binding to p34ct(1–277) underlining the key roles of the p34ct amino acids A151 and K155 as well as F490 from p44ct toward the stabilization of the interaction between p34ct(1–277) and p44ct(368–534) (Figure 3C) as observed in the crystal structure. Similar conclusions were obtained when we introduced the R146E mutation (K155E in p34ct) in p34hs(1–233) and the F374E mutation (F490E in p44ct) in p44hs(321–395) (Supplementary Figure S4).
We next transferred mutations that impair the interaction between the two minimal domains into the full length p34hs and p44hs proteins. Full length p34hs-L132E/A134E, p34hs-C142E, p34hs-R146E and p44hs-F374E or p44hs-I386E variants were co-expressed with the remaining core-TFIIH subunits, and the purified recombinant complexes (rIIH6) were tested in in vitro transcription and DNA repair assays. Interestingly, while variants p34hs-R146E (K155E in p34ct), p44hs-F374E (F490E in p44ct) and p34hs-C142E (A151E in p34ct) impaired the formation of the p34/p44 minimal complex (Figure 3B and C, Supplementary Figures S3 and 4), western blot analysis of the rIIH6 complexes showed that none of these mutations affected the rIIH6 composition or stoichiometry (Figure 3D). Moreover, when the core-TFIIH variants were added to an in vitro transcription assay containing in addition to the AdMLP template, RNA polymerase II, TFIIB, TFIIE, TFIIF, TBP transcription factors as well as XPD and CAK, RNA synthesis was performed almost as accurately as observed for the wild-type proteins (Figure 3E and Supplementary Figure S5C). Similarly, these core-TFIIH variants were able to remove the damaged oligonucleotide as well as wild-type core-TFIIH (Figure 3F and Supplementary Figure S5C), when added to an in vitro nucleotide excision repair (NER) assay containing XPC/Hr23B, XPA, RPA, XPF/ERCC1, XPG in addition to the XPD subunit of TFIIH as well as a cisplatinated damaged plasmid.
The p34 C-terminal zinc-binding domain interacts with the p44 RING domain
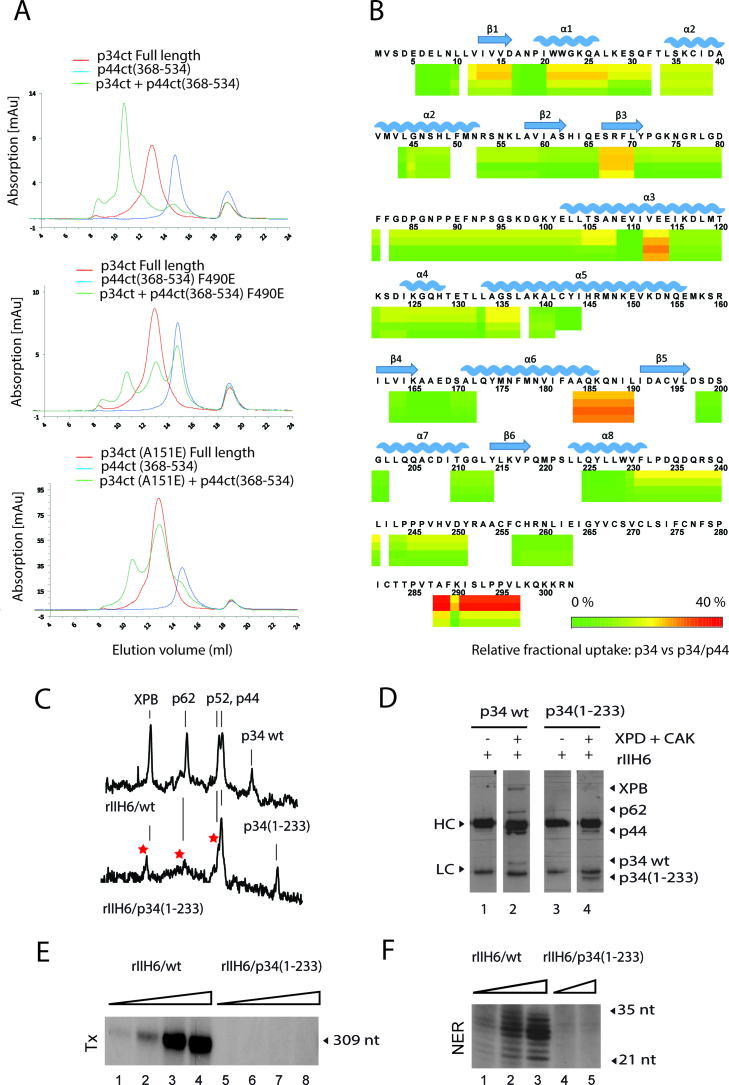
Due to this disparity between lack of interaction as observed for the minimal complexes and no disruption of core-TFIIH, we speculated that additional p34/p44 interacting domains could circumvent the impaired interactions between the p34 vWA domain and the p44 RING finger domain. To investigate this further, SEC experiments originally pursued with the A151E and the F490E variants of the minimal complex domains (Figure 3B and Supplementary Figure S3A) were now performed with full length p34ct. Contrary to what we observed with the truncated version of p34ct, full length p34ct and p44ct(368–534)-F490E as well as p34ct-A151E and p44ct(368–534) led to, albeit highly reduced, complex formation (Figure 4A). These results are in line with pull down data on the human complexes where no effect could be observed for all variants except for the p34hs-C142E variant that showed a diminished interaction (Supplementary Figure S5A and B), suggesting that the C-terminal C4 domain of p34 can compensate for mutations in the p34 vWA or in the p44 RING domain by providing an additional interaction site for p44.
Figure 4.
Association between p34 and p44 also involves the C-terminal domain of p34. (A) SEC of the p34ct full length protein with p44ct(368–534) wild-type or p44ct(368–534)-F490E and p34ct-A151E with p44ct(368–534) wild-type. p34ct full length (red) and p44ct(368–534) (blue) were analyzed separately and mixed in a 1:1 stoichiometry (green) prior to SEC. (B) HDX-MS differential heatmap of p34hs versus p34hs/p44hs. Hydrogen–Deuterium eXchange coupled to mass spectrometry experiments, in which the deuterium exchange labeling of p34hs in the presence and absence of p44hs was compared. Data could be measured only for the highlighted areas as depicted in the sequence coverage presented in Supplementary Figure S6B. (C) Densitometry analysis of a Coomassie stained gel of rIIH6/p34wt and rIIH6/p34(1–233) purified using IMAC followed by immuno-precipitation using an anti-p44 antibody. The gel as well as a western blot analysis of the complexes are presented in the supplementary data. (D) Extracts from SF9 cells co-infected by viruses expressing core-TFIIH wild-type (rIIH6/p34 wt) or mutant (rIIH6/p34(1–233)) were incubated with XPD plus CAK, immuno-precipitated using an antibody directed against cdk7 and immobilized proteins were analyzed using specific antibodies. Heavy (HC) and light (LC) chains of the anti-cdk7 antibody are indicated. (E) and(F) Complexes purified in (C) were analyzed for their transcription and NER activities as described in Figure 3.
To further validate the importance of the C4 domain of p34 for the p34/p44 interaction, we deleted this domain and compared the stability of the p34hs(1–233)/p44hs complex with that of full length p34hs/p44hs. Deletion of the C-terminal region of p34hs does not prevent association between p34hs and p44hs (Supplementary Figure S6A, left panel). The thermal stability of the mutant complex is, however, affected. Analysis of the intrinsic tryptophan fluorescence changes upon thermal unfolding yielded a clear melting curve with a single transition indicative of a melting temperature close to 50°C for the wild-type complex, but an altered profile when the p34 C4 domain was deleted (Supplementary Figure S6A, right panel) emphasizing the fortification of the p34/p44 interaction by this domain.
We also performed HDX-MS experiments, in which we compared deuterium exchange labeling of p34hs in the presence and absence of p44hs (Figure 4B). Association of p34hs with p44hs caused an appreciable reduction of backbone amide deuterium uptake on several p34hs peptides. When mapped onto the p34 structure, the deuterium exchange nicely validates the crystallographically observed interface (Supplementary Figure S6B and C). In addition to the known interface a significant reduction in exchange was observed in one region of the C4 domain (residues 288–297), strongly supporting our biochemical observations that the C-terminus of p34 provides an additional interface for the p34/p44 interaction. To further analyze the observed effects, we generated a model of the p34hs C4 domain (Supplementary Figure S7). According to this model the potential region of interaction starts just after a highly conserved zinc binding motif and is extended to the solvent accessible surface, clearly supporting the possibility of an interaction interface. However, the sequence conservation between p34hs and p34ct in that region is not very high (Supplementary Figure S7).
Since the p34 C4 domain constitutes an additional interface for the interaction with p44, we decided to conduct experiments to probe its importance on the integrity of core-TFIIH and its impact on transcription and repair. Pull down experiments with extracts of insects cells co-infected with baculoviruses expressing p34hs(1–233) together with the other core-TFIIH subunits showed that an anti-p44 antibody permits the isolation of a complex with levels of p34 almost comparable to those observed for the wild-type complex, verifying that the redundant network between p34 and p44 ensures an interaction between the two proteins. These complexes, however, showed highly reduced levels of p62, p52 and XPB (Figure 4C and Supplementary Figure S8A), indicating an additional more central role of the p34 C4 domain in the architecture of core-TFIIH. For further characterization, lysates containing core-TFIIH were mixed with XPD and CAK and in vitro reconstituted complexes were immobilized using an antibody directed against the cdk7 subunit of CAK. In agreement with the results described above, the p34 deletion mutant but not XPB or p62 were detected (Figure 4D and Supplementary Figure S8B). These results not only confirm the central role of the p34 C4 domain in the architecture of core-TFIIH but also show that the p34 C-terminal deletion is not compensated by interactions mediated by CAK and/or XPD. Consequently, the rIIH6/p34(1–233) complex is unable to support RNA synthesis and to function in DNA repair when incorporated into in vitro transcription and NER assays (Figure 4E and F).
DISCUSSION
High-resolution structural data on the molecular interactions within TFIIH are indispensable for our understanding of the functional networks within this complex. Here, we focused on the association between the p34 and p44 subunits of core-TFIIH. The p44 subunit was described for both its ubiquitin ligase activity and its role as a regulator of XPD’s helicase activity (12,35). It interacts with XPD and anchors the CAK via XPD to core-TFIIH on the one hand, whereas its interaction with the p34 subunit of core-TFIIH is crucial for TFIIH core integrity and function on the other hand (13). We solved crystal structures of the p34 vWA domain in complex with the p44 RING domain and analyzed the p34/p44 interaction using a combination of biophysical and biochemical approaches. A closer analysis of the structure indicates that the p44 RING domain displays significant structural homology to other ubiquitin ligase E3 RING domains as observed in the RFN38 and Ark2C proteins, which is consistent with the E3 ligase activity described for the yeast SSL1/p44 TFIIH subunit (35). Moreover, as observed in E3-RING/E2 ubiquitin ligase interfaces, structural elements flanking the zinc-binding motif of the p44 RING also govern the recognition of the p34 subunit in an E3-RING/E2 like fashion, indicating a similar recognition mechanism for the p34/p44 complex (Supplementary Figure S1). However, the fact that this interface is required for the formation of core-TFIIIH interactions renders a p44 E3 ligase function within the TFIIH context unlikely.
We identified a set of amino acids that are essential for the interaction within the minimal complex (Figure 3A–C; Supplementary Figures S3 and 4). However, when these residues were mutated in the full length human p34 and p44 proteins, we discovered that most of them did not interfere with complex stability (Supplementary Figure S5A and B). Furthermore, disrupting this interaction in the context of the entire TFIIH impinges neither on its composition nor on its transcription or NER activities (Figures 3D–F and 5) suggesting the presence of additional binding site(s) that compensate for the impairment in complex formation between the vWA domain of p34 and the C4C4 RING finger of p44.
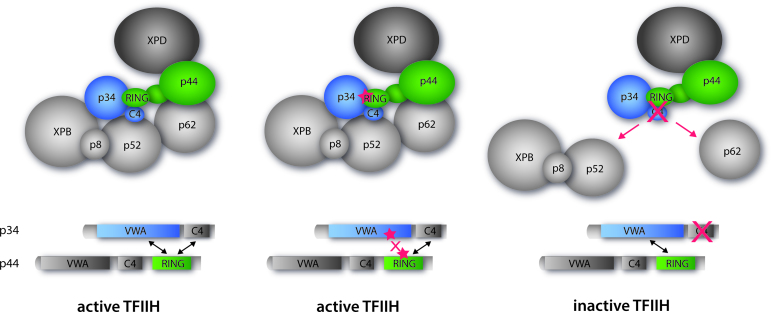
Figure 5.
Cartoon representation depicting the summary of our observations with the same coloring scheme as used in Figure 1 showing core-TFIIH with XPD. Left panel: wild-type active TFIIH. Middle panel: TFIIH mutated within the minimal p34/p44 interface (structurally characterized in this study) maintaining the active complex. Right panel: TFIIH with the C4 domain of p34 missing. This p34 variant still interacts with p44 but other vital interactions are missing leading to an inactive TFIIH.
In search for an additional interface, we identified the C4 domain of p34 to be involved in the association with the p44 RING domain and to further stabilize the association between the two proteins (Figure 4A). This observation is strongly supported by our HDX-MS experiments, where the C4 domain of p34 shows the lowest exchange of interacting interfaces (Figure 4B). The interaction network of the p44 RING domain with two interfaces within p34 is also highly important for the integrity of core-TFIIH, which is demonstrated by our pull down experiments. The use of p34hs(1–233) lacking the C-terminal domain led to almost no reduction in p44 interaction whilst other TFIIH components such as p52, p62 and XPB clearly show weaker binding profiles (Figures 4C–D and 5). This demonstrates that the loss of the p34 C4 domain cannot directly be compared with the effect of point mutations in the p34 vWA/p44 RING interface as it has a more dramatic impact on the structure and function of TFIIH. However, the robustness of the p34/p44 interaction in the absence of the p34 C-terminal domain clearly indicates that both interfaces of p34 need to be disrupted to affect the p34/p44 association. In addition, our data show that the C-terminal domain of p34 not only interacts with p44 but also stabilizes interactions with p62 and p52. The reduction of XPB is most likely a secondary effect due to the loss of the p52 interaction. Consequently, the deletion of the C-terminal p34 domain affects both the stability of human TFIIH and its NER and transcription activities (Figure 4E and F). Intriguingly, yeast genetics demonstrated that the deletion of the C4 domain was lethal (10). How the C-terminal domain of p34 exactly interacts with other TFIIH partners requires further investigation in future studies.
Our studies provide evidence of an intricate network within TFIIH that governs the structure and function of the 10 subunits complex with p34 and p44 being positioned at the very core of this network (Figure 5). p44 interacts through its N-terminal vWA domain with XPD; the latter interacts through its ARCH domain with the central region of MAT1 (14,19) to anchor the regulatory CAK to core-TFIIH. p44 further interacts with p62 (36). p34, apart from interacting with p44, might also interact with p52 as previously suggested (11) and as indicated by our data (Figure 4C and D) suggesting that the p34/p44 pair could act as a seed to assemble the remaining TFIIH core subunits (Figure 5). This hypothesis is further supported by the fact that both proteins contain vWA domains, which are known to be able to perform multiple protein–protein interactions (37,38) and the presence of two interfaces to safeguard the p34/p44 interaction.
The strong and redundant multipoint interaction between p34 and p44 suggests the presence of a failsafe mechanism for a pair of proteins that is central to TFIIH stability and function, explaining why individual point mutations in either subunit have so far not been associated with a disease phenotype. Most disease-associated mutations have been identified in XPB, XPD and the p8/TTDA subunits of TFIIH (3,39). Mutations in XPB or XPD disturb either their catalytic activity and/or their regulation by p52 and p44, respectively (1). Due to the importance of the p34/p44 pair within TFIIH in acting as a core seed and mediator for other TFIIH subunits, a disruption by multiple mutations may not be tolerated.
DATA AVAILABILITY
Coordinates and structure factors for the p34 vWA/p44 RING structures have been deposited in the Protein Data Bank under accession codes 5NUS, 5OBZ and 5O85.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sebastien Fribourg and Anass Jawhari for the efforts in the early stages of the project, Alexandre Ourjoumtsev and Dino Moras for fruitful advice and constructive discussions. Simon Pichard is acknowledged for excellent technical help, Nathalie Troffer-Charlier and Isabelle Kolb-Cheynel (IGBMC Baculovirus Facility) for production of recombinant proteins in insect cells. We also thank the members of SOLEIL Proxima1 beamlines and the European Synchrotron Radiation Facility–European Molecular Biology Laboratory joint Structural Biology groups for the use of beamline facilities and for help during X-ray data collection.
Footnotes
Present addresses:
Laura Radu, Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, CB2 1GA Cambridge, UK.
Julien Marcoux, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [KI-562/7–1]; Agence Nationale de la Recherche [ANR-12-BSV8–0015-01, ANR-12-BSV8.001–01]; Institut National du Cancer [INCA_9378]; Association pour la Recherche sur le Cancer [ARC SL120120304592]; la ligue contre le cancer [CCIR-GE 2013]; la ligue contre le cancer Post-doctoral Fellowship (to L.R.); French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05–01]; Proteomic French Infrastructure (ProFI) [ANR-10-INSB-08-03]; Instruct as part of the European Strategy Forum on Research Infrastructures (ESFRI); German Excellence Initiative to the Graduate School of Life Sciences; University of Würzburg PhD Fellowship (to E.S.). Funding for open access caharge: German Research Foundation [KI-562/7–1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Compe E., Egly J.M.. Nucleotide excision repair and transcriptional regulation: TFIIH and beyond. Annu. Rev. Biochem. 2016; 85:265–290. [DOI] [PubMed] [Google Scholar]
- 2. Cleaver J.E. Splitting hairs–discovery of a new DNA repair and transcription factor for the human disease trichothiodystrophy. DNA Repair (Amst). 2005; 4:285–287. [DOI] [PubMed] [Google Scholar]
- 3. Lehmann A.R., Norris P.G.. DNA repair and cancer: speculations based on studies with xeroderma pigmentosum, Cockayne's syndrome and trichothiodystrophy. Carcinogenesis. 1989; 10:1353–1356. [DOI] [PubMed] [Google Scholar]
- 4. Yoon H., Miller S.P., Pabich E.K., Donahue T.F.. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992; 6:2463–2477. [DOI] [PubMed] [Google Scholar]
- 5. Gulyas K.D., Donahue T.F.. SSL2, a suppressor of a stem-loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992; 69:1031–1042. [DOI] [PubMed] [Google Scholar]
- 6. Castro J., Merino C., Zurita M.. Molecular characterization and developmental expression of the TFIIH factor p62 gene from Drosophila melanogaster: effects on the UV light sensitivity of a p62 mutant fly. DNA Repair (Amst). 2002; 1:359–368. [DOI] [PubMed] [Google Scholar]
- 7. Fregoso M., Laine J.P., Aguilar-Fuentes J., Mocquet V., Reynaud E., Coin F., Egly J.M., Zurita M.. DNA repair and transcriptional deficiencies caused by mutations in the Drosophila p52 subunit of TFIIH generate developmental defects and chromosome fragility. Mol. Cell. Biol. 2007; 27:3640–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iyer N., Reagan M.S., Wu K.J., Canagarajah B., Friedberg E.C.. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996; 35:2157–2167. [DOI] [PubMed] [Google Scholar]
- 9. Feaver W.J., Huang W., Gileadi O., Myers L., Gustafsson C.M., Kornberg R.D., Friedberg E.C.. Subunit interactions in yeast transcription/repair factor TFIIH. Requirement for Tfb3 subunit in nucleotide excision repair. J. Biol. Chem. 2000; 275:5941–5946. [DOI] [PubMed] [Google Scholar]
- 10. Warfield L., Luo J., Ranish J., Hahn S.. Function of Conserved Topological Regions within the Saccharomyces cerevisiae Basal Transcription Factor TFIIH. Mol. Cell. Biol. 2016; 36:2464–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo J., Cimermancic P., Viswanath S., Ebmeier C.C., Kim B., Dehecq M., Raman V., Greenberg C.H., Pellarin R., Sali A. et al. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol. Cell. 2015; 59:794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coin F., Marinoni J.C., Rodolfo C., Fribourg S., Pedrini AM., Egly J.M.. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 1998; 2:184–188. [DOI] [PubMed] [Google Scholar]
- 13. Coin F., Oksenych V., Egly J.M.. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell. 2007; 26:245–256. [DOI] [PubMed] [Google Scholar]
- 14. Busso D., Keriel A., Sandrock B., Poterszman A., Gileadi O., Egly J.M.. Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J. Biol. Chem. 2000; 275:22815–22823. [DOI] [PubMed] [Google Scholar]
- 15. Kainov D.E., Vitorino M., Cavarelli J., Poterszman A., Egly J.M.. Structural basis for group A trichothiodystrophy. Nat. Struct. Mol. Biol. 2008; 15:980–984. [DOI] [PubMed] [Google Scholar]
- 16. Coin F., Proietti De Santis L., Nardo T., Zlobinskaya O., Stefanini M., Egly J.M.. p8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell. 2006; 21:215–226. [DOI] [PubMed] [Google Scholar]
- 17. Kuper J., Braun C., Elias A., Michels G., Sauer F., Schmitt D.R., Poterszman A., Egly J.M., Kisker C.. In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol. 2014; 12:e1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roy R., Adamczewski J.P., Seroz T., Vermeulen W., Tassan J.P., Schaeffer L., Nigg E.A., Hoeijmakers J.H., Egly J.M.. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994; 79:1093–1101. [DOI] [PubMed] [Google Scholar]
- 19. Abdulrahman W., Iltis I., Radu L., Braun C., Maglott-Roth A., Giraudon C., Egly J.M., Poterszman A.. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E633–E642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riedl T., Hanaoka F., Egly J.M.. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003; 22:5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmitt D.R., Kuper J., Elias A., Kisker C.. The structure of the TFIIH p34 subunit reveals a von Willebrand factor A like fold. PLoS One. 2014; 9:e102389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J.S., Saint-Andre C., Lim H.S., Hwang C.S., Egly J.M., Cho Y.. Crystal structure of the Rad3/XPD regulatory domain of Ssl1/p44. J. Biol. Chem. 2015; 290:8321–8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fribourg S., Kellenberger E., Rogniaux H., Poterszman A., Van Dorsselaer A., Thierry J.C., Egly J.M., Moras D., Kieffer B.. Structural characterization of the cysteine-rich domain of TFIIH p44 subunit. J. Biol. Chem. 2000; 275:31963–31971. [DOI] [PubMed] [Google Scholar]
- 24. Kellenberger E., Dominguez C., Fribourg S., Wasielewski E., Moras D., Poterszman A., Boelens R., Kieffer B.. Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions. J. Biol. Chem. 2005; 280:20785–20792. [DOI] [PubMed] [Google Scholar]
- 25. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 30. Schrodinger, LLC The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- 31. Abdulrahman W., Radu L., Garzoni F., Kolesnikova O., Gupta K., Berger I., Poterszman A.. The production of multi-protein complexes in insect cells using the baculovirus expression system. Methods Mol. Biol. 2015; 1261:91–114. [DOI] [PubMed] [Google Scholar]
- 32. Gerard M., Fischer L., Moncollin V., Chipoulet J.M., Chambon P., Egly J.M.. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem. 1991; 266:20940–20945. [PubMed] [Google Scholar]
- 33. Fribourg S., Romier C., Werten S., Gangloff Y.G., Poterszman A., Moras D.. Dissecting the interaction network of multiprotein complexes by pairwise coexpression of subunits in E. coli. J. Mol. Biol. 2001; 306:363–373. [DOI] [PubMed] [Google Scholar]
- 34. Gao M., Skolnick J.. iAlign: a method for the structural comparison of protein-protein interfaces. Bioinformatics. 2010; 26:2259–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takagi Y., Masuda C.A., Chang W.H., Komori H., Wang D., Hunter T., Joazeiro C.A., Kornberg R.D.. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol. Cell. 2005; 18:237–243. [DOI] [PubMed] [Google Scholar]
- 36. Tremeau-Bravard A., Perez C., Egly J.M.. A role of the C-terminal part of p44 in the promoter escape activity of transcription factor IIH. J. Biol. Chem. 2001; 276:27693–27697. [DOI] [PubMed] [Google Scholar]
- 37. Fukuda K., Doggett T., Laurenzi I.J., Liddington R.C., Diacovo T.G.. The snake venom protein botrocetin acts as a biological brace to promote dysfunctional platelet aggregation. Nat. Struct. Mol. Biol. 2005; 12:152–159. [DOI] [PubMed] [Google Scholar]
- 38. Maita N., Nishio K., Nishimoto E., Matsui T., Shikamoto Y., Morita T., Sadler J.E., Mizuno H.. Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin: insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins. J. Biol. Chem. 2003; 278:37777–37781. [DOI] [PubMed] [Google Scholar]
- 39. Giglia-Mari G., Coin F., Ranish J.A., Hoogstraten D., Theil A., Wijgers N., Jaspers N.G., Raams A., Argentini M., van der Spek P.J. et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004; 36:714–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the p34 vWA/p44 RING structures have been deposited in the Protein Data Bank under accession codes 5NUS, 5OBZ and 5O85.