Abstract
Background and Aims Polyploidy and hybridization are important factors for generating diversity in plants. The species-rich dog roses (Rosa sect. Caninae) originated by allopolyploidy and are characterized by unbalanced meiosis producing polyploid egg cells (usually 4x) and haploid sperm cells (1x). In extant natural stands species hybridize spontaneously, but the extent of natural hybridization is unknown. The aim of the study was to document the frequency of reciprocal hybridization between the subsections Rubigineae and Caninae with special reference to the contribution of unreduced egg cells (5x) producing 6x offspring after fertilization with reduced (1x) sperm cells. We tested whether hybrids arose by independent multiple events or via a single or few incidences followed by a subsequent spread of hybrids.
Methods Population genetics of 45 mixed stands of dog roses across central and south-eastern Europe were analysed using microsatellite markers and flow cytometry. Hybrids were recognized by the presence of diagnostic alleles and multivariate statistics were used to display the relationships between parental species and hybrids.
Key Results Among plants classified to subsect. Rubigineae, 32 % hybridogenic individuals were detected but only 8 % hybrids were found in plants assigned to subsect. Caninae. This bias between reciprocal crossings was accompanied by a higher ploidy level in Rubigineae hybrids, which originated more frequently by unreduced egg cells. Genetic patterns of hybrids were strongly geographically structured, supporting their independent origin.
Conclusions The biased crossing barriers between subsections are explained by the facilitated production of unreduced gametes in subsect. Rubigineae. Unreduced egg cells probably provide the highly homologous chromosome sets required for correct chromosome pairing in hybrids. Furthermore, the higher frequency of Rubigineae hybrids is probably influenced by abundance effects because the plants of subsect. Caninae are much more abundant and thus provide large quantities of pollen. Hybrids are formed spontaneously, leading to highly diverse mixed stands, which are insufficiently characterized by the actual taxonomy.
Keywords: Microsatellites, asymmetrical crossing barriers, meiosis, anorthoploidy, hybridization, polytopic origin, Rosa, Rosa micrantha, Rosa agrestis, Rosa canina, Caninae, Rubigineae
INTRODUCTION
In angiosperms, polyploidy is a nearly ubiquitous phenomenon and is often observed in extant lineages (Soltis et al., 2014), and whole-genome duplications have also occurred several times during the phylogeny of angiosperms (Jiao et al., 2011). However, it is still debated whether polyploidy acts as an evolutionary driving force enhancing speciation or whether it has no major impact on the long-term evolutionary success of a lineage (Stebbins, 1950, 1971; Otto and Whitton, 2000; Van de Peer, 2011; Heslop-Harrison, 2012). Recent polyploidization events have very often been accompanied by hybridization and are then termed allopolyploidy, which is considered a major pathway of sympatric speciation in plants (Soltis and Soltis, 2009). Since genome doubling in hybrids facilitates correct bivalent formation during meiosis, it could provide a loophole from hybrid sterility (Rieseberg and Willis, 2007). The most prominent process of polyploid formation is the production of unreduced gametes (Ramsey and Schemske, 1998), which are more often developed in hybrids with susceptible meiosis (Mason and Pires, 2015).
The northern hemisphere woody genus Rosa serves as a good example for studying the evolutionary effects of polyploidy and hybridization because most lineages contain allopolyploid taxa (Wissemann and Ritz, 2005; Joly and Bruneau, 2006, 2007; Joly et al., 2006; Fougère-Danezan et al., 2015). This is especially true for the polyploid sect. Caninae (dog roses), which evolved by multiple hybridization events (Wissemann, 1999, 2000; Ritz et al., 2005). Genomes involved refer to diploid progenitors related to at least two different sections of Rosa and to a so-called Protocaninae genome, which has not been found in extant diploid roses (Ritz et al., 2005). Dog roses are tetra- to hexa- (rarely hepta- and octo-)ploid but pentaploid cytotypes are most frequent (2n = 5x = 35) (Klášterská, 1969; Klásterská and Natarajan, 1974; Končalová and Klášterský, 1978; Małecka and Popek, 1982, 1984; Pachl, 2011). Despite their predominant somatic odd ploidy, dog roses reproduce sexually due to the unique canina meiosis (Täckholm, 1920, 1922; Blackburn and Harrison, 1921; Blackburn, 1925). During this meiosis, only two sets of chromosomes form bivalents: one set of bivalent-forming chromosomes is transmitted by the haploid pollen grain (1n = 1x = 7) and the other is transmitted together with all sets of univalent-forming chromosomes by the egg cell. The number of univalent-forming chromosome sets depends on the somatic ploidy level, e.g. pentaploids (2n = 5x = 35) have tetraploid egg cells (1n = 4x = 28) with three univalent-forming chromosome sets. Molecular studies revealed that chromosome pairing is not random, because always the same genetically very similar chromosome sets form bivalents during meiosis (Nybom et al., 2004, 2006; Ritz and Wissemann, 2011). Dog roses are distributed from Europe to West Asia and comprise ∼60 species (Henker, 2000). Due to the above-mentioned allopolyploid constitution, skewed maternal inheritance and ongoing hybridization, the taxonomy of sect. Caninae is notoriously difficult. Dog roses are conventionally divided into six subsections; the three larger of these (Caninae, Rubigineae and Vestitae) are unambiguously differentiated and each contains several less clear-cut microspecies (Henker, 2000). During this study we focus on subsects. Caninae and Rubigineae, which are morphologically as well as genetically clearly separated from each other (De Cock et al., 2008; Koopman et al., 2008; De Riek et al., 2013).
Apart from the hybridogenic origin of the section, numerous cases of hybridization involving extant dog rose species have been observed in single populations (Schanzer and Vagina, 2007; Schanzer and Kutlunina, 2010; Ritz and Wissemann, 2011; Kellner et al., 2012; Herklotz and Ritz, 2014). Using microsatellite and morphological data, Ritz and Wissemann (2003, 2011) and Herklotz and Ritz (2014) demonstrated that both Rosa micrantha and Rosa agrestis (subsect. Rubigineae) originated by hybridization between a maternal parent from subsect. Rubigineae and a paternal parent from subsect. Caninae. In both cases hybrids were mainly hexaploid, although they originated from pentaploid parents, because unreduced egg cells were involved. We hypothesized that the establishment of hybrids is facilitated by unreduced gametes since they provide the two highly homologous chromosome sets needed for correct bivalent formation during canina meiosis.
Since the above-mentioned observations of spontaneous hybridization between subsections Rubigineae and Caninae have remained anecdotal, the aim of the present study was to investigate the extent to which hybridogenic individuals occur in mixed stands of the two parental subsections across a wide geographic range by analysing ploidy levels and microsatellites. In particular, we wanted to answer the following questions: (1) Does the number of hybridogenic individuals vary between the two parental subsections (reciprocal crossings: maternal Rubigineae × paternal Caninae versus maternal Caninae × paternal Rubigineae) and are these hybrids more frequently formed by unreduced gametes? (2) Do hybrids originate independently and multiple times in mixed stands (polytopic origin) or are they related to one or few hybridogenic ancestors that subsequently spread across the area (monotopic origin)?
MATERIALS AND METHODS
Study species
Subsection Caninae is widely distributed in Europe, occurring in hedgerows and forest edges on various soils, and is characterized by glabrous or hairy leaves and pedicels with no or odourless glands (Henker, 2000, 2011). Subsection Rubigineae is found in more thermophilic habitats on base-rich soils (Henker, 2000); the leaflets are pubescent and bear numerous glands spreading a fruity scent. Within subsect. Rubigineae a group of morphotypes with wide-angled, roundish leaflet bases and glandular pedicels are summarized as Rosa rubiginosa agg. (Christ, 1873; Henker, 2000). In contrast, the Rosa elliptica agg. is characterized by leaflets with acute-angled, cuneate bases and non-glandular pedicels. Within each subsection or aggregate, several microspecies are differentiated based on sets of correlated characters emphasizing fruit morphology (Christ, 1873; Henker, 2000). For the purpose of the present study we focus on subsections and aggregates.
Plant material
We collected 811 samples of Rosa at 45 stands in central and south-eastern Europe in August to September 2012 and 2013 (Fig. 2; Supplementary Data Table S1). Additionally, two stands from the Ukraine and one individual from Azerbaijan were included. Subsection Caninae and subsect. Rubigineae co-occurred at 36 stands. We sampled (4–) 15–20 (−32) individuals per stand (on average ten individuals of subsect. Caninae and seven of subsect. Rubigineae). We aimed for a balanced sampling between the number of individuals of both subsections and tried to sample all microspecies at a stand. Species belonging to other taxonomic groups of Rosa were occasionally found but were excluded from further analysis (29 individuals in total; Table S1). For each individual the geographic position was determined as WGS84 coordinates using a GPS device; leaf material was dried in silica gel and a herbarium specimen was deposited in the Herbarium Senckenbergianum Görlitz, Germany (GLM) (Table S1). Identification of rose species followed Henker (2000, 2011).
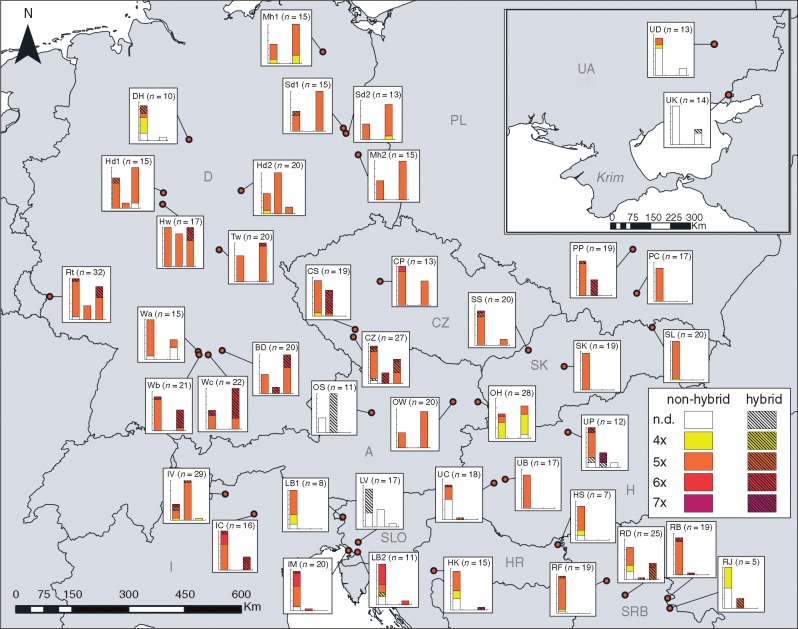
Fig. 2.
Distribution of cytotypes and hybrids of subsect. Caninae and Rubigineae in the study area. Study sites are abbreviated according to Table S1. The first bar plot represents the cytotypes in subsect. Caninae, the second bar plot cytotypes in the R. elliptica agg. and the third bar plot cytotypes in the R. rubiginosa agg. of subsect. Rubigineae. Localities with fewer than five samples are not shown (As, SE, HP, OB, UV; see Table S2). A, Austria; CZ, Czech Republic; D, Germany; HR, Croatia; H, Hungary; I, Italy; PL, Poland; SK, Slovakia; SLO, Slovenia; SRB, Serbia; UA, Ukraine.
Flow cytometry
Ploidy levels were determined by flow cytometry from silica-dried leaflets. Rosa arvensis (2n = 2x = 14) grown in the garden of the Senckenberg Museum of Natural History Görlitz was analysed simultaneously as an internal standard in each sample. The exact chromosome number of this calibration standard was determined with traditional cytological methods (Supplementary Data Fig. S1). Leaf material was chopped with a sharp razor blade in nucleus extraction buffer according to Pfosser et al. (1995) or woody plant buffer (Loureiro et al., 2007). Both buffers were modified with 10 g L−1 polyvinylpyrrolidone K30 (Yokoya et al., 2000) and 200 mm d(−)-mannitol (Doležel et al., 2007). The lysates were filtered through nylon gauze (30 µm) and stained with Otto II buffer including 4 µg mL−1 4′, 6-diamidino-2-phenylindole. In case of the extraction buffer of Pfosser et al. (1995) we added 2 µl mL−1 β-mercaptoethanol to the Otto II buffer. Fluorescence intensity was measured with a CyFlow Ploidy Analyzer (Partec, Münster, Germany) equipped with a UV LED (365 nm). Each sample was measured at least twice with a minimum of 3000 particles and a mean coefficient of variation <6 %. Primary data were analysed with the software Cyflogic v. 1.2.1 (Cyflo Ltd, Finland). Ploidy levels were calculated from the ratio of fluorescence intensity between the sample and the internal calibration standard.
Microsatellites
DNA was extracted from silica-gel-dried leaflets according to Dumolin et al. (1995) and deposited in the Senckenberg DNA bank (http://sesam.senckenberg.de/). We amplified nine microsatellite loci (RhEO506, RhD201, RhB303, RhAB73, RhP507, RhP50, RhO517, RhD206, RhP518) with primers developed for Rosa hybrida (Esselink et al., 2003). For three of them we used the M13 ‘poor man’s’ labelling technique according to Schuelke (2000). The non-M13-labelled amplifications were carried out in 11 µL of reaction mixture containing 30 ng of template DNA, 1×Y-Reaction Buffer (Peqlab, Erlangen, Germany), 2 mm MgCl2, 200 µm dNTPs, 0·07 µm forward primer, 0·36 µm reverse primer and 0·05 U Taq DNA polymerase (Peqlab, Erlangen, Germany). Forward primers were directly labelled with fluorochromes (6-FAM, VIC, PET, NED). The PCRs were performed in an Eppendorf Mastercycler EP S (Eppendorf, Hamburg, Germany) programmed for 180 s at 94 °C followed by 32 cycles of 30 s at 94 °C, 30 s at 53 °C and 45 s at 72 °C and a final extension for 5 min at 72 °C. For the nested amplification with M13-labelled primers we followed Schuelke (2000). Fragment analysis was performed using an ABI3730 automated sequencer (Life Technology, Darmstadt, Germany) and the size standard LIZ-500 (Life Technology) at the Senckenberg Biodiversity and Climate Research Centre (BIK-F) in Frankfurt am Main (Germany). Scoring of fragments was done with the software Peak Scanner v. 1.0 (Thermo Fisher Scientific). The length of M13-labelled fragments was reduced by 18 bp according to the M13 sequence length.
Identification of hybrids
Since canina meiosis violates the assumptions for hybrid identification used by common software applications, we identified hybrids according to the following premises derived from previous studies (Nybom et al., 2004, 2006; Ritz and Wissemann, 2011; Herklotz and Ritz, 2014). Dog roses often contain fewer alleles per locus than expected from their ploidy level (e.g. at maximum four different alleles in pentaploids). Thus, at least one allele has two identical copies, which are presumably located on the highly homologous bivalent-forming chromosome sets. Pentaploid and hexaploid hybrids (the latter were derived from unreduced egg cells) may have five different alleles at a locus when the pollen parent contributed an allele not present in the egg cell.
For hybrid identification, all individuals were first assigned to one of the two subsections by morphology. An individual assigned to a subsection represents either a potential maternal parent or a hybrid derived from the maternal parent of this subsection and a paternal parent of the other. Henceforward, we refer to hybrids arising from maternal parents of subsect. Caninae as Caninae hybrids and hybrids derived from the subsect. Rubigineae maternal parent as Rubigineae hybrids. Second, we determined diagnostic alleles of the respective paternal subsection according to the following premises. (1) A candidate allele must be more frequent in samples of the maternal subsection with five or more alleles per locus (at at least one of nine investigated loci) compared with samples of the same subsection containing a maximum of four alleles per locus. (2) After passing the first criterion, alleles were considered diagnostic if their relative frequency was at least 5-fold lower in samples of the maternal subsection with a maximum of four alleles per locus than their relative frequency in individuals of the paternal subsection with a maximum of four alleles per locus (1/5 = pmat4/ppat4). The relative frequencies for each subsection were calculated by dividing the frequency of the respective allele in samples with a maximum of four alleles per locus by the total number of individuals with a maximum of four alleles per locus to account for the higher number of investigated plants and the higher allelic diversity in subsect. Caninae compared with Rubigineae. Third, all investigated plants of a subsection (without taking the number of alleles per locus into account) were screened for the presence of diagnostic alleles. Plants with at least three diagnostic alleles across all loci were considered to be hybrids. Additionally, we performed a series of analyses changing the threshold for comparing the relative frequencies of the two subsections from one to nine to test its quantitative influence on hybrid identification.
Statistical analysis
We treated the microsatellite data as allelic phenotypes (presence/absence of alleles). Samples containing missing values at more than one locus were excluded. Thus, the final data set contained 2·1% missing values and was reduced to 742 samples. These data were transferred into Bruvo distances (Bruvo et al., 2004) using the POLYSAT package (Clark and Jasieniuk, 2011) running under the R environment (R Core Team, 2015). Since microsatellite genotypes are mostly unknown in polyploids, Bruvo distances assume ambiguous allele copy numbers in partial heterozygotes and take mutational distances into account by including repeat themes of the microsatellites. Principal coordinate analysis (PCoA) based on square-rooted Bruvo distances was computed in R using the VEGAN package version 2.3.5 (Oksanen et al., 2015). To analyse molecular variation at several hierarchical levels (subsections, aggregates, species, ploidy levels), analysis of molecular variance (AMOVA) was performed using the POPPR package (Kamvar et al., 2014, 2015). Percentages of polymorphic alleles were computed with GenAlEx 6.5 (Peakall and Smouse, 2006, 2012). To test matrix correlations between geographic distances (WGS84 coordinates transformed into Euclidean distances) and genetic distances, Mantel tests with 9999 permutations were computed with the ADE-4 package (Dray et al., 2007), but samples from outlier locations (Azerbaijan, Ukraine) were excluded.
Due to the above-mentioned challenges of canina meiosis, traditional methods for analysing hybrid origin cannot be applied. Thus, we used a multiple response permutation procedure (MRPP) based on Bruvo distances with 999 permutations implemented in the VEGAN package. If hybrids originated independently in mixed stands, genetic distances between hybrids within a locality should be smaller than between randomly selected hybrids from ‘pseudo-localities’ generated by permutations. In the case of a single hybridogenic origin, hybrids differ slightly among localities. Localities with a single hybrid plant were excluded from the analysis.
RESULTS
Species composition
We analysed five species of the subsect. Caninae [Rosa canina (316 individuals), R. subcanina (44), R. dumalis Bechst. (3), R. corymbifera (87), R. subcollina (10)] and five species of subsect. Rubigineae [R. inodora (25), R. agrestis (63) of the R. elliptica agg. and R. rubiginosa (25), R. gremlii (87), R. micrantha (82) of the R. rubiginosa agg.] at our study sites (in total 742 individuals) (Supplementary Data Table S2). However, except for R. rubiginosa and a few individuals of R. dumalis we did not find the so called D-type microspecies of either subsection (Henker, 2000; R. elliptica, R. caesia).
Microsatellite alleles
Alleles ranged from 89 to 365 bp in length (Table 1). The highest number of alleles (40) was found at locus RhP50 and the lowest (eight) at locus RhO517. Individuals contained between one and seven alleles per locus. We detected 22 diagnostic alleles across all loci for each subsection (Table S2). The number of diagnostic alleles per individual counted across all loci ranged from zero to four for subsect. Caninae and from one to seven for subsect. Rubigineae. In most individuals we found at most one diagnostic allele per locus. In subsect. Caninae a few individuals contained two diagnostic alleles per locus (one 4x, eight 6x and six 6x individuals; Table S2). In subsect. Rubigineae only three heptaploid individuals contained a maximum of two diagnostic alleles per locus.
Table 1.
Characteristics of microsatellite loci and the number of diagnostic alleles per locus used for hybrid identification
| Range of allele length (bp) | No. of detected alleles | No. of alleles per individual | No. of diagnostic alleles | ||
|---|---|---|---|---|---|
| Locus | subsect. Caninae | subsect. Rubigineae | |||
| RhEO506 | 188–251 | 21 | 2–6 | 3 | 4 |
| RhD201 | 169–231 | 22 | 1–5 | 4 | 3 |
| RhB303 | 100–146 | 16 | 1–4 | 0 | 2 |
| RhAB73 | 155–191 | 19 | 1–3 | 4 | 1 |
| RhP507 | 89–203 | 18 | 2–6 | 2 | 2 |
| RhP50 | 225–364 | 40 | 3–7 | 2 | 7 |
| RhO517 | 163–270 | 8 | 1–2 | 0 | 1 |
| RhD206 | 185–365 | 31 | 3–6 | 3 | 1 |
| RhP518 | 123–183 | 16 | 1–4 | 4 | 1 |
| Total | 22 | 22 | |||
Ploidy levels and hybrids
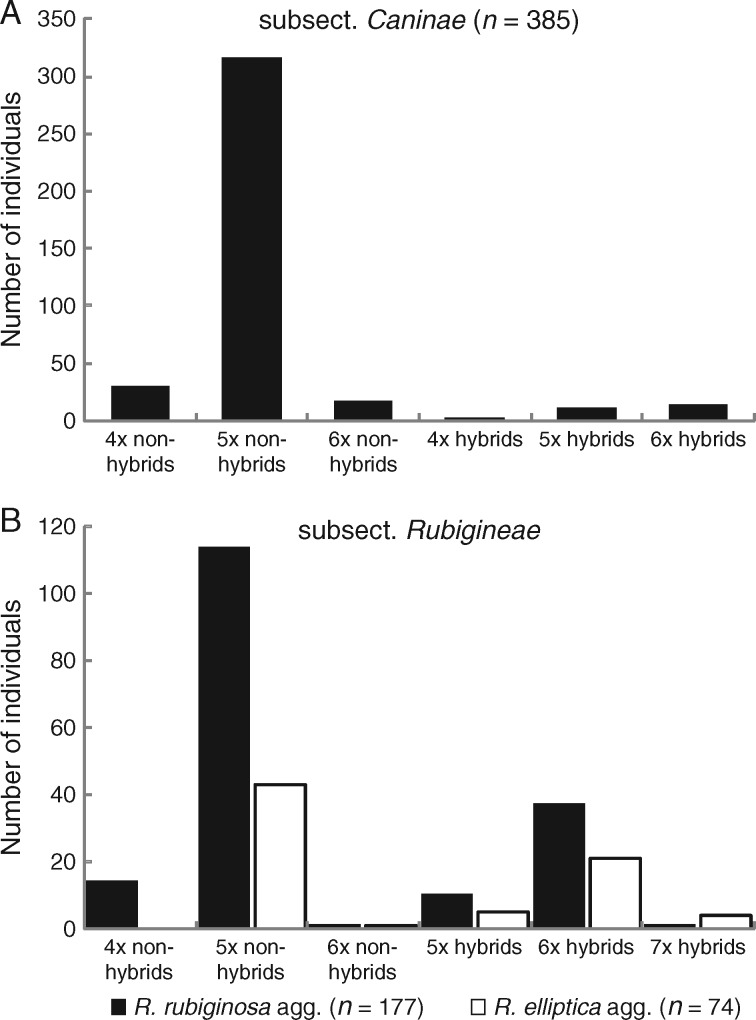
Ploidy levels ranged from 4x to 7x (Fig. 1, Table 2; Table S2). However, we were not able to determine ploidy levels in 75 individuals of subsect. Caninae and 31 individuals of subsect. Rubigineae due to withered leaf samples. Pentaploids were most frequent (71 % Caninae, 64 % R. rubiginosa agg., 55 % R. elliptica agg.); tetraploids were rare (7 % in each of subsect. Caninae and R. rubiginosa agg.) and missing in the R. elliptica agg. Hexaploids occurred more frequently in subsect. Rubigineae (20 % in R. rubiginosa agg. and 25 % in R. elliptica agg.) compared with subsect. Caninae (6 %). We detected one heptaploid individual in R. rubiginosa agg. and four heptaploids in the R. elliptica agg.
Fig. 1.
Proportions of different cytotypes in hybrids and non-hybrids of subsect. Caninae (A) and in the two aggregates [R. rubiginosa agg. (black) and R. elliptica agg. (white)] of subsect. Rubigineae (B).
Table 2.
Number of plant samples per subsection and aggregate, hybrid status and ploidy levels
| Taxonomic affiliation | Number of plants (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Non-hybrids | Hybrids | Ploidy level | |||||
| n.d.a | 4x | 5x | 6x | 7x | ||||
| Subsect. Caninae | 460 | 424 (92) | 36 (8) | 75 (16) | 31 (7) | 325 (71) | 29 (6) | |
| Subsect. Rubigineae | 282 | 192 (68) | 90 (32) | 31 (11) | 14 (5) | 172 (61) | 60 (21) | 5 (2) |
| R. elliptica agg. | 88 | 49 (56) | 39 (44) | 14 (16) | 48 (55) | 22 (25) | 4 (4·5) | |
| R. rubiginosa agg. | 194 | 143 (74) | 51 (26) | 17 (9) | 14 (7) | 124 (64) | 38 (20) | 1 (0·5) |
Not determined.
In sum, 45 % (128 individuals) of subsect. Rubigineae and 24 % (110 individuals) of subsect. Caninae met the first precondition for harbouring candidate alleles (five or more alleles at one or more loci) without taking relative allele frequencies into account. Low thresholds for pmat4/ppat4 turned out to be unrealistic because the number of diagnostic alleles per individual and locus exceeded expectations from canina meiosis (one or, in case of pollen grains that were not fully reduced, two diagnostic alleles per individual and locus; Supplementary Data Fig. S2). However, the overall trend across different thresholds remained the same: subsect. Caninae contained fewer hybrids compared with subsect. Rubigineae (Fig. S2). Applying a threshold of 1/5 for pmat4/ppat4, we identified 126 hybrids (17 %) in total. Thirty-six of these hybrids were found in subsect. Caninae (8 % of subsect. Canine; Fig. 1, Table 2). Hybrids were much more frequent in subsect. Rubigineae, containing 90 hybrids [32 %: 39 in R. elliptica agg. (44 %) and 51 in R. rubiginosa agg. (26 %)].
Non-hybridogenic dog roses were mostly pentaploid (Fig. 1), while the majority of hybrids were hexaploid (57 %). Except for two individuals (IM8 and LB11; Table S2), all hexa- and heptaploids of subsect. Rubigineae were identified as hybrids. Within R. rubiginosa agg. we detected ten pentaploid hybrids, 37 hexaploid hybrids and one heptaploid hybrid (Fig. 1). The R. elliptica agg. contained five pentaploid, 21 hexaploid and four heptaploid hybrids. In subsect. Caninae one tetraploid, ten pentaploids and 13 hexaploids were identified as hybrids.
Pentaploid cytotypes were recorded at every mixed stand (Fig. 2). The highest diversity of ploidy levels (4x–6x) was found at the mixed stands OH and IV in Austria and Italy, respectively. The 45 tetraploid individuals were found in northern Germany, in Austria or irregularly dispersed in the south of the study area. In southern Germany and the Czech Republic hexaploid Rubigineae were frequent, but they were also recorded from eastern Poland, northern Italy and the western Balkan states. We found numerous hybrids close to the Alps, in Hungary (UP), Serbia (RD, RB) and southern Poland (PP). In sum, there was no apparent geographical pattern of cytotypes, hybrids and species.
Genetic structure within subsections and aggregates
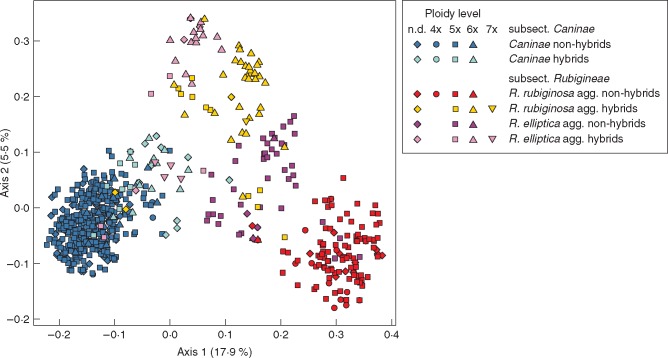
In the PCoA all individuals of subsect. Caninae were clearly separated from subsect. Rubigineae (Fig. 3). All individuals of subsect. Caninae were densely clustered and we did not detect any structure when analysing data from subsect. Caninae separately (Supplementary Data Fig. S3). The ordination differentiated between the non-hybrids of the R. rubiginosa agg. and the R. elliptica agg. of subsect. Rubigineae. Most of the 6x Rubigineae hybrids were separated from the Rubigineae non-hybrids along the second axis, which, however, explained only 5·5 % of the variation (Fig. 3). The pentaploid and hexaploid Caninae hybrids were intermingled with Caninae-non-hybrids (Fig. 3). The 6x hybrids of the R. rubiginosa agg. and the R. elliptica agg. were close to each other. Heptaploid hybrids of the R. elliptica agg. were closer to subsect. Caninae.
Fig. 3.
PCoA of 742 samples based on 177 alleles of nine microsatellite loci transformed into square-rooted Bruvo distances.
Analysis of molecular variance attributed 26 % of the variation to subsections (Table 3). The largest share of variance (55 %) was partitioned to individuals within localities. Within subsect. Caninae only 1 % of the variation was within microspecies, while 79 % of the variation was found between individuals within localities. Aggregates in subsect. Rubigineae explained 19 % of the variance; the remaining variance was attributed to localities (40 %) and individuals within localities (41 %). Differences between microspecies within Rubigineae aggregates hardly captured any variance. Ploidy level within subsect. Rubigineae explained 20 % of the variation, which was not surprising because almost all 6x individuals were assigned as hybrids containing alleles of subsect. Caninae.
Table 3.
Distribution of molecular variance between subsections, aggregates, microspecies and cytotypes
| Source of variation | d.f. | Sum of squares | Mean of squares | Percentage of variance | ΦST | P-value |
|---|---|---|---|---|---|---|
| AMOVA on subsections (all samples) | ||||||
| Between subsections | 1 | 21·60 | 21·60 | 26 | 0·25 | 0.001 |
| Between localities within subsections | 81 | 43·12 | 0·53 | 19 | 0·26 | 0·001 |
| Within localities | 659 | 84·66 | 0·12 | 55 | 0·45 | 0·001 |
| AMOVA on species of subsect. Caninae | ||||||
| Between species | 4 | 1·31 | 0·33 | 1 | 0·01 | 0·015 |
| Between localities within species | 105 | 29·65 | 0·28 | 20 | 0·20 | 0·001 |
| Within localities | 350 | 48·47 | 0·14 | 79 | 0·21 | 0·001 |
| AMOVA on aggregates of subsect. Rubigineae | ||||||
| Between aggregates | 1 | 5·20 | 5·20 | 19 | 0·18 | 0·001 |
| Between localities within aggregate | 47 | 24·14 | 0·51 | 40 | 0·50 | 0·001 |
| Within localities | 233 | 18·37 | 0·09 | 41 | 0·59 | 0·001 |
| AMOVA on ploidy levels of subsect. Rubigineae | ||||||
| Between ploidy levels | 4 | 7·82 | 1·96 | 20 | 0·20 | 0·001 |
| Between localities within ploidy level | 51 | 23·55 | 0·46 | 42 | 0·52 | 0·001 |
| Within localities | 226 | 16·34 | 0·07 | 38 | 0·62 | 0·001 |
| AMOVA on species of R. rubiginosa agg. | ||||||
| Between species | 2 | 2·35 | 1·18 | 9 | 0·09 | 0·003 |
| Between localities within species | 54 | 16·90 | 0·31 | 52 | 0·57 | 0·001 |
| Within localities | 137 | 7·88 | 0·06 | 39 | 0·61 | 0·001 |
| AMOVA on species of R. elliptica agg. | ||||||
| Between species | 1 | 0·54 | 0·54 | −3 | −0·02 | 0·608 |
| Between localities within species | 23 | 10·07 | 0·44 | 62 | 0·48 | 0·001 |
| Within localities | 63 | 4·61 | 0·07 | 41 | 0·47 | 0·001 |
d.f., degrees of freedom.
The percentage of polymorphic alleles was ∼3-fold lower in subsect. Rubigineae (R. rubiginosa agg. 11·5 %, R. elliptica agg. 10·3 %) compared with subsect. Caninae (28·5 %). Genetic and geographic distances were only very weakly correlated in non-hybridogenic individuals of subsect. Caninae, but moderately correlated in subsect. Rubigineae (Table 4).
Table 4.
Standardized Mantel correlations between geographic and genetic distances
| Subsection | Aggregates | Non-hybrids | Hybrids |
|---|---|---|---|
| Caninae | 0·09*** | 0·01* | |
| Rubigineae | |||
| R. elliptica agg. | 0·41*** | 0·40*** | |
| R. rubiginosa agg. | 0·31*** | 0·50*** |
P < 0·05;
P < 0·001.
Monotypic versus polytopic origin of hybrids
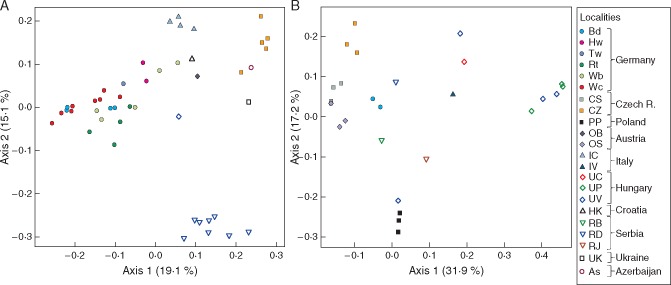
Since we detected many more hybrids in subsect. Rubigineae (Fig. 1) and Caninae hybrids occurred often as single individuals per stand (Fig. 2), we focused the analysis on Rubigineae hybrids. In the PCoAs, hybrids clustered according to their geographic origin (Fig. 4A, B). Hybrids from the Serbian population (RD) were clearly separated along the second axis; hybrids from German populations were separated along the first axis and some of them were densely clustered (Bd, Rt, Wb, Wc). Accordingly, in the R. elliptica agg., hybrids originating from the same locality clustered together and the Hungarian (UV, UP) hybrids were separated from the rest (Fig. 4B).
Fig. 4.
PCoA of hybrids from the R. rubiginosa agg. (A, n = 51) and the R. elliptica agg. (B, n = 40) based on 177 alleles of nine microsatellite loci converted into square-rooted Bruvo distances. Colours and symbols indicate localities (see also Table S1).
The AMOVA (Table 5) covering variation between and within localities attributed the major part of the variance to differences between localities for hybrids of the R. rubiginosa agg. (58 %, P < 0·001) and for hybrids of the R. elliptica agg. (68 %, P < 0·001).
Table 5.
Distribution of molecular variance among Rubigineae hybrids between and within localities
| Source of variation | d.f. | Sum of squares | Mean of squares | Percentage of variance | P-value |
|---|---|---|---|---|---|
| AMOVA on hybrids of R. rubiginosa agg. | |||||
| Between localities | 7 | 3·19 | 0·46 | 58 | 0·001 |
| Within localities | 37 | 1·95 | 0·05 | 42 | |
| AMOVA on hybrids of R. elliptica agg. | |||||
| Between localities | 6 | 2·54 | 0·42 | 68 | 0·001 |
| Within localities | 27 | 1·02 | 0·04 | 32 | |
d.f., degrees of freedom.
The Mantel tests detected a slightly higher correlation between geographic and genetic distances in hybrids compared with non-hybrids in the R. rubiginosa agg. but no differences were observed between hybrids and non-hybrids of the R. elliptica agg. (Table 4).
The MRPP for the hybrids of the R. rubiginosa agg. grouped by localities resulted in a chance-corrected within-group agreement of A = 0·56 (P < 0·001). An even stronger agreement was detected for hybrids of the R. elliptica agg. (A = 0·74; P < 0·001). In the case of agreement of A = 1 all hybrids are identical within a locality, and in the case of A = 0 the heterogeneity of hybrids within a locality is equal to the heterogeneity of randomly selected hybrids across all localities (represented by permutations of pseudo-localities). Relatively high values imply a close relationship of hybrids within a locality and thus support the ‘multiple origins’ hypothesis.
DISCUSSION
Genetic characterization of intersectional hybrids
Genetic data supported a clear distinction between subsections and aggregates (Fig. 3, Table 3), which is in agreement with previous studies using AFLP markers (De Cock et al., 2008; De Riek et al., 2013). Within these groups genetic variance was mainly attributed to localities, individuals or cytotypes, but morphology-defined microspecies were not reflected (Table 3), which will be discussed in a separate study (V. Herklotz and C. M. Ritz, unpubl. res.).
In ∼80 % of all pentaploids and hexaploids we detected a maximum of four or five different microsatellite alleles per locus, respectively (Table S2). This result matches previous observations that dog rose species often contain at least one allele with two identical copies, which are assumed to be located on the highly homologous bivalent-forming chromosomes (Nybom et al., 2004,2006; Ritz and Wissemann, 2011; Ritz et al., 2011). In contrast, we found 33 tetraploids with four and 12 tetraploids with five alleles in at least one locus (Table S2). We suspect that, at least in the latter case, ploidy level estimation failed and/or aneuploids occurred. Twenty-two percent of the pentaploids (109 individuals) had five alleles in at least one locus but we identified only 15 % of these plants (16 individuals) as hybrids (Table S2). In contrast, hexaploid Rubigineae with five or six alleles were all classified as hybrids (Table S2).
We set rather high thresholds for the recognition of diagnostic alleles (Fig. S2), possibly leading to the underestimation of hybrids. We chose this conservative threshold to minimize random effects of allele frequencies, mutations of microsatellite alleles and PCR artefacts. However, irrespective of the height of threshold, subsect. Caninae contained fewer hybrids compared with subsect. Rubigineae (Fig. S2). The number of alleles diagnostic for subsect. Caninae could be underestimated because the percentage of polymorphic alleles was 3-fold higher in subsect. Caninae and the percentage of variance within individuals per stand was twice as high compared with subsect. Rubigineae (Table 3), a fact that we tried to compensate for by computing relative allele frequencies. Another reason for unidentified diagnostic alleles might be hybridization with other non-investigated rose species. Such hybridization events are probably more frequent if the widespread subsect. Caninae is involved because most interspecific hybrids recorded in Great Britain originated from at least one parental species of subsect. Caninae (Graham and Primavesi, 1993; Stace et al., 2015). We found only occasionally other rose species at the mixed stands, yet wild roses are pollinated by bees and bumblebees flying distances of up to 1·5 km (Walther-Hellwig and Frankl, 2000; Osborne et al., 2008; Zurbuchen et al., 2010).
Apart from a few exceptions, we detected one diagnostic allele per locus and individual in hybrids. This is in line with expectations of canina meiosis because the diagnostic alleles were transmitted by haploid pollen grains. Interestingly, three of five 7x hybrids carried two diagnostic alleles at one locus, implying that they arose by a merger of an unreduced 5x egg cell of subsect. Rubigineae and an incompletely reduced 2x pollen grain of subsect. Caninae. Alternatively, they could be derived from unreduced egg cells of a 6x Rubigineae hybrid backcrossing with a fully reduced pollen grain (1x) of subsect. Caninae. Unfortunately, our study could not cover the potential effects of backcrossing. Since the pollen parent transmits only one genome to the F1 and the segregation of genomes during meiosis in the F1 is unknown, the paternal genome could be either transmitted to the egg cell or the pollen grain produced by F1 hybrids. In the latter case, the hybrid egg cell would be genetically identical to an egg cell of a non-hybrid, preventing the origin of backcrossing lineages.
Asymmetrical hybridization towards hexaploid Rubigineae hybrids
Although the strong bias towards Rubigineae hybrids (Table 2; Fig. 1) might be partly influenced by methodological shortcomings of hybrid identification, there are two additional arguments supporting our results. First, the Caninae microspecies R. caesia and R. dumalis were absent or very rare at the mixed stands (Table S1). A previous study showed that they were morphologically identical to artificial Caninae hybrids (Ritz and Wissemann, 2003). Based on genetic analyses, Herklotz and Ritz (2014) demonstrated that individuals of these species constituted Caninae hybrids in a single population in Eastern Germany. However, R. dumalis is obviously not exclusively hybridogenic (Table S2; Ritz and Wissemann, 2011).
Second, and more important, the majority of hybrids were hexaploid, whereas non-hybrids were mostly pentaploid and the proportion of hexaploids varied considerably between both subsections (Fig. 1). Thus, hybrid formation occurs more frequently in subsect. Rubiginae, involving unreduced gametes. Since 6x Rubigineae hybrids contained a maximum of one diagnostic allele per locus, they originated from unreduced (5x) egg cells and haploid (1x) pollen grains. In contrast, Nybom et al. (2006) reported hexaploid synthetic dog rose hybrids that arose either from unreduced egg cells or unreduced (2x) pollen grains, and unreduced gametes of both sexes also gave rise to spontaneous Cardamine hybrids (Mandáková et al., 2013).
Asymmetrical crossing barriers have been already documented in wild roses (Kellner et al., 2012) and in many other plant genera, and are caused by either pre- or postzygotic isolation mechanisms (Tiffin et al., 2001). The biased hybrid formation is not likely to be caused by intrinsic prezygotic barriers leading to unidirectional mating, because both subsections are self-compatible (Wissemann and Hellwig, 1997) and their pollen viability does not differ (Herklotz and Ritz, 2014). Flowering times are largely overlapping, but the most frequent microspecies of subsect. Caninae start to flower early and bloom for a long period (Henker, 2000). In addition, subsect. Caninae is more abundant (Kurtto et al., 2004). Both facts lead to a surplus of Caninae pollen available for potential hybridizations. Such frequency-dependent effects were also shown in hybrids of Rosa (Kellner et al., 2012) and Morus (Burgess et al., 2005).
Furthermore, the coincidence of hexaploidy and hybridization suggests either a facilitated origin of unreduced egg cells in subsect. Rubigineae or a non-reciprocal selective advantage for these hybrids. We hypothesized that the unreduced egg cells provide the two highly homologous chromosome sets required for correct bivalent formation in these hybrids (Ritz and Wissemann, 2011; Herklotz and Ritz, 2014). Unreduced gametes are considered to be the primary mechanism for polyploidization (Harlan, 1975; Mason and Pires, 2015) and are more frequently developed in hybrids with low fertility (Ramsey and Schemske, 1998). All dog roses are allopolyploids (Wissemann and Hellwig, 1998; Wissemann, 2000; Ritz et al., 2005). Studies on microsatellite markers revealed close similarities among the presumed bivalent-forming genome and larger differences among the univalent-forming genomes, whose composition might differ between subsections (Nybom et al., 2004, 2006; Zhang et al., 2013). However, recent phylogenies based on chloroplast markers placed subsect. Caninae and subsect. Rubigineae into different clades, suggesting a polyphyletic origin of dog roses and canina meioisis (Wissemann and Ritz, 2005; Bruneau et al., 2007; Fougère-Danezan et al., 2015). Possibly, the meiosis in subsect. Rubigineae is more prone to the production of unreduced gametes. However, referring to the above-mentioned correlation between hybrid fertility and unreduced gametes, differences in seed set between subsections were not observed (Herklotz and Ritz, 2014). Unreduced gamete formation is also triggered by environmental stress (Ramsey and Schemske, 1998; De Storme and Geelen, 2013), but we lack ecological data on growing conditions at the mixed stands. The biased hybridization could also be affected by asymmetrical gene dosage (Osborn et al., 2003) or plastome–genome incompatibilities (Greiner et al., 2008). Data on differential hybrid fitness are scarce: pollen was less viable in Caninae hybrids compared with Rubigineae hybrids but ploidy was not taken into account (Herklotz and Ritz, 2014). In a comparable study, Werlemark (2000) detected lower male fitness of hybrids compared with their parents, but the direction of crossing had no influence on pollen viability. In contrast, seed set of pentaploid artificial crossings between Rubigineae seed parents and Caninae pollen parents was lower than vice versa (Wissemann and Hellwig 1997; Werlemark, 2000), but the surviving offspring in the first-mentioned study were pentaploid (Ritz and Wissemann, 2011) and those in the second study were of unknown ploidy.
Polytopic origin of hybrids
Given that hybrids were mostly hexaploids (Fig. 1, Table 2) and that species and cytotypes co-occurred and were not geographically clustered (Fig. 2), a multiple origin of hybrids is likely. In support of this hypothesis, microsatellite data grouped hybrids according to their locality (Fig. 4), genetic distances of hybrids were more strongly correlated with geographic distances compared with non-hybrids in the extensively sampled R. rubiginosa agg. (Table 4), and the largest part of the genetic variance in hybrids was attributed to locality (Table 5). Furthermore, the MRPP suggested high genetic similarity among hybrids of the same locality compared with randomly chosen hybrids. Unfortunately, an extensive screening of 11 chloroplast markers revealed no variation within sequences of the respective subsections (Fiedler, 2015). High levels of gene flow between rose species within localities were also reported by De Cock et al. (2008). Multiple origins of allopolyploids appear to be more common (reviewed in Soltis and Soltis, 2000; Weiss-Schneeweiss et al., 2013) than single origins reported for e.g. Aster amellus (Münzbergová et al., 2013), Helianthus paradoxus (Welch and Rieseberg, 2002) and Spartina anglica (Raybould et al., 1991). Interestingly, hybridogenic species of Onosma, characterized also by an unbalanced meiosis, originated multiple times (Kolarčik et al., 2014). Further research is needed to investigate whether these polytopic dog rose hybrids represent distinct evolutionary entities that are differentiated from their parents by ecological features (e.g. occupying certain niches) or by reproductive barriers preventing backcrossing (e.g. apomixes or assortative mating among hexaploids).
In conclusion, natural dog rose stands consist of a mixture of hybridogenic and non-hybridogenic individuals, whose genetic relatedness is inadequately reflected by the current taxonomic system (Henker, 2000). Hybrids evolve independently and apparently do not disperse across larger areas. Hybridization occurs much more frequently if the rarer subsect. Rubigineae serves as seed parent and the frequent subsect. Caninae as pollen parent. Furthermore, hybridization is often accompanied by unreduced Rubigineae egg cells probably providing the homologous chromosome sets for bivalent formation. However, the meiotic behaviour and fertility of hexaploid hybrids have not been studied and should be the subject of future investigations.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: mitotic metaphase plate of R. arvensis. Figure S2: different thresholds of relative allele frequencies for assigning diagnostic alleles. Figure S3: PCoA based on samples of subsect. Caninae. Table S1: taxonomic affiliation and collection information. Table S2: information on microsatellite alleles, hybrid status and ploidy levels.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank I. Schanzer (Main Botanical Garden, Russian Academy of Sciences, Russian Federation, Moscow) and V. Kerenyi-Nagy (Szent István University, Gödöllő, Hungary) for providing plant samples. We thank the following botanists for useful information on suitable mixed stands and for their assistance in the field: B. and J. Adler, Z. Dajdok, P. Erzberger, T. Gregor, Z. Hroudová, N. Jogan, Z. Kącki, G. Kleesadl, H. Korsch, S. Kühnel, M. Lepší, P. Mair, A. Mohr, A. Mrkvicka, R. Paulič, F. Prosser, J. Pusch, H. Reichert, W. Rottensteiner, H. Sonnenberg, F. Starlinger and H. Wolf. Help with statistics was given by K. Wesche (Senckenberg Museum of Natural History, Görlitz), and J. Paule (Senckenberg Research Institute and Natural History Museum Frankfurt am Main, Germany) gave valuable advice on flow cytometry. J. Lunerová and A. Kovařík (Department of Molecular Epigenetics, Institute of Biophysics, Brno, Czech Academy of Sciences) helped with cytological studies in R. arvensis and comments on the manuscript. We acknowledge S. Kovac and M. Olđa (‘Vojvodinasume’, ŠG ‘Banat’, Pančevo, Serbia), the federal state government of Lower Austria, the Untere Naturschutzbehörde des Landkreises Mecklenburgische Seenplatte and the Bistum Hildesheim (Germany) sampling permissions. The DNA Bank of the Senckenberg Research Institute and R. Kohli (Autohaus Klische, Görlitz, Germany) kindly provided financial and logistical support, respectively. Primers of the microsatellite loci were provided by R. Smulders (Stichting Wageningen Research, Research Institute Wageningen, Plant Research, Business Unit Plant Breeding, the Netherlands). We are indebted to M. Schwager, D. Altmann, S. Dorf, A. Smolka (Senckenberg Museum of Natural History, Görlitz) and to the staff of the Senckenberg BIK-F laboratory (Frankfurt am Main, Germany) for their great technical support in the laboratory, and we cordially thank R. Christian and P. Gebauer (Senckenberg Museum of Natural History, Görlitz) for their help with the herbarium specimens. We thank K. Wesche, J. Wesenberg (Senckenberg Museum of Natural History) and the editor and the anonymous referees for their very thoughtful comments and improvements on the manuscript.
LITERATURE CITED
- Blackburn K. 1925. Chromosomes and classification in the genus Rosa. American Naturalist 59: 200–205. [Google Scholar]
- Blackburn KB, Harrison JWH.. 1921. The status of the British rose forms as determined by their cytological behaviour. Annals of Botany 35: 159–188. [Google Scholar]
- Bruneau A, Starr JR, Joly S.. 2007. Phylogenetic relationships in the genus Rosa: new evidence from chloroplast DNA sequences and an appraisal of current knowledge. Systematic Botany 32: 366–378. [Google Scholar]
- Bruvo R, Michiels NK, D’Souza TG, Schulenburg H.. 2004. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molecular Ecology 13: 2101–2106. [DOI] [PubMed] [Google Scholar]
- Burgess KS, Morgan M, Deverno L, Husband BC.. 2005. Asymmetrical introgression between two Morus species (M. alba, M. rubra) that differ in abundance. Molecular Ecology 14: 3471–3483. [DOI] [PubMed] [Google Scholar]
- Christ H. 1873. Die Rosen der Schweiz mit Berücksichtigung der umliegenden Gebiete Mittel- und Süd-Europas: ein monographischer Versuch. Basel: H. Georg. [Google Scholar]
- Clark LV, Jasieniuk M.. 2011. POLYSAT: an R package for polyploid microsatellite analysis. Molecular Ecology Resources 11: 562–566. [DOI] [PubMed] [Google Scholar]
- De Cock K, Vander Mijnsbrugge K, Breyne P, Van Bockstaele E, Van Slycken J.. 2008. Morphological and AFLP-based differentiation within the taxonomical complex section Caninae (subgenus Rosa). Annals of Botany 102: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim: John Wiley & Sons. [Google Scholar]
- Dray S, Dufour AB, Chessel D.. 2007. The ade4 package-II: two-table and K-table methods. R News 7: 47–52. [Google Scholar]
- Dumolin S, Demesure B, Petit RJ.. 1995. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Esselink GD, Smulders MJM, Vosman B.. 2003. Identification of cut rose (Rosa hybrida) and rootstock varieties using robust sequence tagged microsatellite site markers. Theoretical and Applied Genetics 106: 277–286. [DOI] [PubMed] [Google Scholar]
- Fiedler S. 2015. Molekulargenetische Untersuchungen an Weinrosen (Rosa sect. Caninae subsect. Rubigineae) und ihren Hybriden anhand von Chloroplasten-DNA und ribosomaler DNA. Masters Thesis, International Graduate School (IHI) Zittau, Germany.
- Fougère-Danezan M, Joly S, Bruneau A, Gao X-F, Zhang L-B.. 2015. Phylogeny and biogeography of wild roses with specific attention to polyploids. Annals of Botany 115: 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham G, Primavesi A.. 1993. Roses of Great Britain and Ireland. BSBI Handbook 7. London: Botanical Society of the British Isles. [Google Scholar]
- Greiner S, Wang X, Herrmann RG, et al. 2008. The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Molecular Biology and Evolution 25: 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR. 1975. On Ö. Winge and a prayer: the origins of polyploidy. Botanical Review 41: 361–390. [Google Scholar]
- Henker H. 2000. Rosa L In: Conert HJ, Jäger EJ, Kadereit JW, Schultze-Motel W, Wagenitz G, Weber HE, eds. Gustav Hegi: Illustrierte Flora von Mitteleuropa. Berlin: Parey Buchverlag, 1–108. [Google Scholar]
- Henker H. 2011. Rosa L In: Jäger EJ, ed. Rothmaler – Exkursionsflora von Deutschland. Grundband. Heidelberg: Springer Spektrum, 444–453. [Google Scholar]
- Herklotz V, Ritz CM.. 2014. Spontane Hybridisierung von Hundsrosen (Rosa L. sect. Caninae (DC). Ser.) an einem natürlichen Vorkommen in der Oberlausitz (Sachsen, Deutschland). Peckiana 9: 119–131. [Google Scholar]
- Heslop-Harrison J. 2012. Genome evolution: extinction, continuation or explosion? Current Opinion in Plant Biology 15: 115–121. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Joly S, Bruneau A.. 2006. Incorporating allelic variation for reconstructing the evolutionary history of organisms from multiple genes: an example from Rosa in North America. Systematic Biology 55: 623–636. [DOI] [PubMed] [Google Scholar]
- Joly S, Bruneau A.. 2007. Delimiting species boundaries in Rosa sect. Cinnamomeae (Rosaceae) in Eastern North America. Systematic Botany 32: 819–836. [Google Scholar]
- Joly S, Starr JR, Lewis WH, Bruneau A.. 2006. Polyploid and hybrid evolution in roses east of the Rocky Mountains. American Journal of Botany 93: 412–425. [DOI] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grünwald NJ.. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Brooks JC, Grünwald NJ.. 2015. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Frontiers in Genetics 6: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner A, Ritz CM, Wissemann V.. 2012. Hybridization with invasive Rosa rugosa threatens the genetic integrity of native Rosa mollis. Botanical Journal of the Linnean Society 170: 472–484. [Google Scholar]
- Klášterská I. 1969. Cytology and some chromosome numbers of Czechoslovak roses I. Folia Geobotanica 4: 175–189. [Google Scholar]
- Klásterská I, Natarajan AT.. 1974. Cytological studies of the genus Rosa with special reference to the section Caninae. Hereditas 76: 97–108. [DOI] [PubMed] [Google Scholar]
- Kolarčik V, Zozomová-Lihová J, Ducár E, Mártonfi P.. 2014. Evolutionary significance of hybridization in Onosma (Boraginaceae): analyses of stabilized hemisexual odd polyploids and recent sterile hybrids. Biological Journal of the Linnean Society 112: 89–107. [Google Scholar]
- Končalová MN, Klášterský I.. 1978. Cytology and chromosome numbers of some Czechoslovak roses III. Folia Geobotanica et Phytotaxonomica 13: 67–93. [Google Scholar]
- Koopman WJM, Wissemann V, De Cock K, et al. 2008. AFLP markers as a tool to reconstruct complex relationships: a case study in Rosa (Rosaceae). American Journal of Botany 95: 353–366. [DOI] [PubMed] [Google Scholar]
- Kurtto A, Lampine R, Junikka L. eds. 2004. Atlas Flora Europaeae. Distribution of vascular plants in Europe. 13. Rosaceae (Spiraea to Fragaria, excl. Rubus). Helsinki: Committee for Mapping the Flora of Europe and Societas Biologica Fennica Vanamo. [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C.. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małecka J, Popek R.. 1982. Karyological studies in the Polish representatives of the genus Rosa L. I. Acta Biologica Cracoviensia, Series Botanica 24: 79–90. [Google Scholar]
- Małecka J, Popek R.. 1984. Karyological studies in the Polish representatives of the genus Rosa L.: II. Acta Biologica Cracoviensia, Series Botanica 26: 43–54. [Google Scholar]
- Mandáková T, Kovařík A, Zozomová-Lihová J, et al. 2013. The more the merrier: recent hybridization and polyploidy in Cardamine. The Plant Cell 25: 3280–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AS, Pires JC.. 2015. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends in Genetics 31: 5–10. [DOI] [PubMed] [Google Scholar]
- Münzbergová Z, Surinová M, Castro S.. 2013. Absence of gene flow between diploids and hexaploids of Aster amellus at multiple spatial scales. Heredity 110: 123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H, Esselink GD, Werlemark G, Vosman B.. 2004. Microsatellite DNA marker inheritance indicates preferential pairing between two highly homologous genomes in polyploid and hemisexual dog-roses, Rosa L. Sect. Caninae DC. Heredity 92: 139–150. [DOI] [PubMed] [Google Scholar]
- Nybom H, Esselink GD, Werlemark G, Leus L, Vosman B.. 2006. Unique genomic configuration revealed by microsatellite DNA in polyploid dogroses, Rosa sect. Caninae. Journal of Evolutionary Biology 19: 635–648. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2015. vegan: Community Ecology Package. R package version 2.3.5. https://cran.r-project.org/src/contrib/Archive/vegan/ (last accessed 16 June 2016).
- Osborn TC, Chris Pires J, Birchler JA, et al. 2003. Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Osborne JL, Martin AP, Carreck NL, et al. 2008. Bumblebee flight distances in relation to the forage landscape. Journal of Animal Ecology 77: 406–415. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Pachl Š. 2011. Variablita botanických druhů rodu Rosa L., a možosti jejich využití v krajinářské tvorbê. PhD Thesis, Slovak University of Agriculture, Nitra, Slovakia.
- Peakall R, Smouse PE.. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE.. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y. 2011. A mystery unveiled. Genome Biology 12: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfosser M, Amon A, Lelley T, Heberle-Bors E.. 1995. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat-rye addition lines. Cytometry 21: 387–3893. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: A language and environment for statistical computing .Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- Raybould AF, Gray AJ, Lawrence MJ, Marshall DF.. 1991. The evolution of Spartina anglica CE Hubbard (Gramineae): origin and genetic variability. Biological Journal of the Linnean Society 43: 111–126. [Google Scholar]
- De Riek J, De Cock K, Smulders MJM, Nybom H.. 2013. AFLP-based population structure analysis as a means to validate the complex taxonomy of dogroses (Rosa section Caninae). Molecular Phylogenetics and Evolution 67: 547–559. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH.. 2007. Plant speciation. Science 317: 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz CM, Wissemann V.. 2003. Male correlated non-matroclinal character inheritance in reciprocal hybrids of Rosa section Caninae (DC.) Ser. (Rosaceae). Plant Systematics and Evolution 241: 213–221. [Google Scholar]
- Ritz CM, Wissemann V.. 2011. Microsatellite analyses of artificial and spontaneous dogrose hybrids reveal the hybridogenic origin of Rosa micrantha by the contribution of unreduced gametes. Journal of Heredity 102: 217–227. [DOI] [PubMed] [Google Scholar]
- Ritz CM, Schmuths H, Wissemann V.. 2005. Evolution by reticulation: European dogroses originated by multiple hybridization across the genus Rosa. Journal of Heredity 96: 4–14. [DOI] [PubMed] [Google Scholar]
- Ritz CM, Köhnen I, Groth M, Theissen G, Wissemann V.. 2011. To be or not to be the odd one out – allele-specific transcription in pentaploid dogroses (Rosa L. sect. Caninae (DC.) Ser). BMC Plant Biology 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanzer IA, Kutlunina NA.. 2010. Interspecific hybridization in wild roses (Rosa L. sect. Caninae DC.). Biology Bulletin 37: 564–573. [PubMed] [Google Scholar]
- Schanzer I, Vagina AV.. 2007. ISSR (inter simple sequence repeat) markers reveal natural intersectional hybridization in wild roses [Rosa L., sect. Caninae (DC.) Ser. and sect. Cinnamomeae (DC.) Ser.]. Wulfenia 14: 1–14. [Google Scholar]
- Schuelke M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology 18: 233–234. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2000. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences of the USA 97: 7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2009. The role of hybridization in plant speciation. Annual Review of Plant Biology 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS.. 2014. The polyploidy revolution then … and now: Stebbins revisited. American Journal of Botany 101: 1057–1078. [DOI] [PubMed] [Google Scholar]
- Stace CA, Preston CD, Pearman DA.. 2015. Hybrid flora of the British Isles. Bristol: Botanical Society of Britain and Ireland. [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold. [Google Scholar]
- De Storme N, Geelen D.. 2013. Sexual polyploidization in plants – cytological mechanisms and molecular regulation. New Phytologist 198: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Täckholm G. 1920. On the cytology of the genus Rosa: a preliminary note. Svensk Botanisk Tidskrift 14: 300–311. [Google Scholar]
- Täckholm G. 1922. Zytologische Studien über die Gattung Rosa. Acta Horti Bergiani 7: 97–381. [Google Scholar]
- Tiffin P, Olson MS, Moyle LC.. 2001. Asymmetrical crossing barriers in angiosperms. Proceedings of the Royal Society of London B: Biological Sciences 268: 861–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther-Hellwig K, Frankl R.. 2000. Foraging habitats and foraging distances of bumblebees, Bombus spp. (Hym., Apidae), in an agricultural landscape. Journal of Applied Entomology 124: 299–306. [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang T-S, Schneeweiss GM.. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch ME, Rieseberg LH.. 2002. Patterns of genetic variation suggest a single, ancient origin for the diploid hybrid species Helianthus paradoxus. Evolution 56: 2126–2137. [DOI] [PubMed] [Google Scholar]
- Werlemark G. 2000. Evidence of apomixis in hemisexual dogroses, Rosa sect. Caninae. Sexual Plant Reproduction. 12: 353–359. [Google Scholar]
- Wissemann V. 1999. Genetic constitution of Rosa sect. Caninae (R. canina, R. jundzilli) and sect. Gallicanae (R. gallica). Journal of Applied Botany 73: 191–196. [Google Scholar]
- Wissemann V. 2000. Molekulargenetische und morphologisch-anatomische Untersuchungen zur Evolution und Genomzusammensetzung von Wildrosen der Sektion Caninae (DC.) Ser. Botanische Jahrbücher für Systematik 122: 357–429. [Google Scholar]
- Wissemann V, Hellwig FH.. 1997. Reproduction and hybridisation in the genus Rosa, section Caninae (Ser.) Rehd. Botanica Acta 110: 251–256. [Google Scholar]
- Wissemann V, Hellwig FH.. 1998. Ist die Pollenqualitat in der Gattung Rosa, Sektion Caninae Indikator fur eine hybridogene Entstehung dieser Sektion? Acta Rhodologica 1: 43–50. [Google Scholar]
- Wissemann V, Ritz CM.. 2005. The genus Rosa (Rosoideae, Rosaceae) revisited: molecular analysis of nrITS-1 and atpB-rbcL intergenic spacer (IGS) versus conventional taxonomy. Botanical Journal of the Linnean Society 147: 275–290. [Google Scholar]
- Yokoya K, Roberts A, Mottley J.. 2000. Nuclear DNA amounts in roses. Annals of Botany 85: 557–561. [Google Scholar]
- Zhang J, Esselink GD, Che D, Fougere-Danezan M, Arens P, Smulders MJM.. 2013. The diploid origins of allopolyploid rose species studied using single nucleotide polymorphism haplotypes flanking a microsatellite repeat. Journal of Horticultural Science & Biotechnology 88: 85–92. [Google Scholar]
- Zurbuchen A, Landert L, Klaiber J, Müller A, Hein S, Dorn S.. 2010. Maximum foraging ranges in solitary bees: only few individuals have the capability to cover long foraging distances. Biological Conservation 143: 669–676. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.