Abstract
Although epidermal keratinocyte development and differentiation proceeds in similar fashion between humans and mice, evolutionary pressures have also wrought significant species-specific physiological differences. These differences between species could arise in part, by the rewiring of regulatory network due to changes in the global targets of lineage-specific transcriptional master regulators such as p63. Here we have performed a systematic and comparative analysis of the p63 target gene network within the integrated framework of the transcriptomic and epigenomic landscape of mouse and human keratinocytes. We determined that there exists a core set of ∼1600 genomic regions distributed among enhancers and super-enhancers, which are conserved and occupied by p63 in keratinocytes from both species. Notably, these DNA segments are typified by consensus p63 binding motifs under purifying selection and are associated with genes involved in key keratinocyte and skin-centric biological processes. However, the majority of the p63-bound mouse target regions consist of either murine-specific DNA elements that are not alignable to the human genome or exhibit no p63 binding in the orthologous syntenic regions, typifying an occupancy lost subset. Our results suggest that these evolutionarily divergent regions have undergone significant turnover of p63 binding sites and are associated with an underlying inactive and inaccessible chromatin state, indicative of their selective functional activity in the transcriptional regulatory network in mouse but not human. Furthermore, we demonstrate that this selective targeting of genes by p63 correlates with subtle, but measurable transcriptional differences in mouse and human keratinocytes that converges on major metabolic processes, which often exhibit species-specific trends. Collectively our study offers possible molecular explanation for the observable phenotypic differences between the mouse and human skin and broadly informs on the prevailing principles that govern the tug-of-war between evolutionary forces of rigidity and plasticity over transcriptional regulatory programs.
INTRODUCTION
In mammals, the skin offers protection against dehydration and trauma and acts as body's first line of defense against a whole range of environmental onslaughts like extremities of temperature and microbial infections. The keratinocyte-enriched stratified epidermis constitutes the outer layer of the skin, which along with appendages such as the hair follicles, sebaceous and sweat glands, contribute to its function as a mechanical and immunological barrier (1). A better understanding of the molecular underpinnings of the development and differentiation program of keratinocytes is thus critically important from both a basic science perspective and clinical and translational point of view. Not surprisingly, a versatile toolkit of traditional biochemical and genetics-based strategies aided by more recently developed next generation sequencing (NGS)-based genomics approaches has propelled exciting research in the field of cutaneous biology. In this context, mouse models have served as an invaluable workhorse for keratinocyte and skin-centric studies, however the relevance of findings from such rodent studies to human skin physiology and disorders has often been overlooked and underappreciated. This is an important consideration given the fact that despite the similarities in the underlying anatomy and physiology, there also exist key differences in skin structure and function between mouse versus human skin (2–4)—a finding that can be attributed to ∼70 million years of evolution separating the two organisms. Indeed, the mouse epidermis is thinner (<25 μm) and hence less efficient as a structural barrier in comparison to the human epidermis which is 6–10 layers and >100 μm thick (4). Additional differences include the mouse skin being more densely populated with hair follicles and being occupied by distinct subtypes of immune cells (5). Importantly, these species-specific structural differences are also accompanied by distinct physiological attributes that for example, include faster turnover and metabolic rates for mouse keratinocytes compared to their human counterparts (6). The underlying molecular basis for such differences however is not well understood.
Given the widespread use and reliance on transgenic animals for modeling various facets of human biology and disease, it is important to elucidate both the evolutionarily conserved and divergent biological pathways between mouse and human. Toward this end, in the recent years, driven by the power of NGS, high-resolution annotation of genomes has provided new insights into the evolution of gene regulatory mechanisms as a potential driver of phenotypic variations between mouse and human (7). While these studies clearly illustrate that gene regulation machinery and networks are conserved in mouse and human, the similarities appear only to be surface-deep. Indeed, an emerging and rather surprising finding is that orthologous transcription factors (TFs) target enhancer regulatory networks that undergo extensive re-wiring between species (8–14). For example, studies in human and mouse embryonic stem cells have revealed that lineage-specific TFs such as OCT4 and NANOG undergo more dramatic changes in genome-wide binding profiles (only 3–5% of the TF binding sites are conserved) compared to ubiquitously expressed factors like CTCF (∼50% binding sites conserved) (15). This altered transcriptional circuitry of pluripotent stem cells have been attributed in part due to emergence of species-specific transposable elements. Additional mechanisms that contribute to generation of new regulatory modules include co-option of existing enhancers or de-novo generation of regulatory elements (10). Importantly, this turnover of TF binding sites, especially of master regulatory factors can also influence the underlying chromatin architecture, ultimately leading to differential functional output (16). Given the proven differences in the species-specific regulatory landscape, it is thus reasonable to posit that the epidermal development and differentiation program is also subjected to such evolutionary forces. Furthermore, it is expected that such differences would be more apparent for TFs that play a cardinal role in the skin developmental pathway such as p63.
p63 is a p53-like conserved TF that is perched at the apex of the transcriptional regulatory network in keratinocytes and other epithelial cells, where it regulates almost every major signaling pathway (17–19). In both mouse and human skin, p63 is highly expressed in the basal keratinocytes, where it regulates myriad cellular processes including proliferation, differentiation, stratification and adhesion (20–22). Mice lacking p63 exhibit profound developmental defects including catastrophic failure in the formation of stratified epithelial structures such as the skin and limb malformations (23,24). The diverse biological functions of p63 are carried out by the products of a structurally complex gene, which generates several isoforms anchored by the longer TAp63 or N-terminally truncated ΔNp63 proteins (17). The ΔNp63 isoforms serve as the predominant functional isoform in most epithelial cells including keratinocytes as revealed by studies with isoform-specific mouse knockouts and transcriptomic analysis (25–29). Over the years, molecular understanding of p63 function has been greatly aided by chromatin immunoprecipitation (ChIP) experiments performed in both primary and immortalized human keratinocytes, which has led to the identification of a relatively large number of genome-wide targets of p63 (30–34). Further follow-up studies on the intricate interplay between the underlying chromatin architecture and p63 binding in the genomic context have also revealed a possible deterministic role of p63 in recruiting active histone modifications at its target sites in human keratinocytes (35–37). However, despite the widespread use of mouse models for genetic and biochemical studies on p63, similar information about its target repertoire is lacking in this species, particularly in the context of keratinocytes.
Here we address the functional role of p63 by generating global p63 binding datasets and major epigenetic profiles in mouse keratinocytes. Furthermore, by integrating our mouse-based studies with available human datasets, we have probed into the evolutionary forces that have shaped common and species-specific p63 regulatory network. Interestingly, we find that the p63 regulatory network has expanded by ∼3 times during the 70 million years of evolution between mouse and human with only a small core set of 1639 genomic sites conserved in p63 occupancy between species. In parallel, our analysis demonstrates that even in spite of broad conservation and underlying sequence homology, majority of the regulatory segments bound by p63 reflect a species-specific bias. We show that these differences in p63 binding are associated with a significant deviation of the p63 motif from the consensus during evolution. The substantial rewiring of the p63 regulatory network between mouse and humans is further accompanied by a changed epigenetic landscape characterized by inaccessibility and lack of an active chromatin signature. Finally, by incorporating RNA-Seq-based transcriptomic data, we infer the effects of these differences in the p63-driven gene regulatory environment on downstream gene expression. Taken together, our findings might serve as a mechanistic basis to account for the distinct physiological characteristics of mouse and human skin keratinocytes.
MATERIALS AND METHODS
Cell culture
Two independently derived mouse keratinocyte (MK1 and MK2) cell lines were utilized for the experiments described in this paper. MK1, a spontaneously immortalized mouse keratinocyte cell line (also known as UG-1) has been described previously (38,39) and was grown in a low Ca++ medium which comprised of a 3:1 mixture of Ham's F12 and Dulbecco's modified Eagle's medium supplemented with 15% chelated fetal bovine serum. MK2 cell line was commercially obtained from CeLLnTEC and grown in the serum-free CnT-07 media as recommended. According to the supplier, MK2 (referred to as MPEK-BL6 or Epidermal Keratinocyte Progenitors) were isolated from normal C57BL/6 mouse skin, have not been actively transformed and retain the ability to differentiate. Primary mouse keratinocytes (PMK) were isolated from post-natal day 1 BL6 mouse skin after separating the epidermis from dermis by Trypsin treatment and grown in CnT–Prime media (CeLLnTEC).
ChIP qPCR and ChIP-Seq
ChIP data generation and validation
MK1, MK2 and PMK cells were grown to ∼ 90% confluency and cross-linked with 1% Formaldehyde for 10 min. Cell lysates from fixed keratinocytes were sonicated with a Bioruptor to obtain sheared chromatin to an approximate length of 150–400 bp using a low sodium dodecyl sulphate shearing kit for Histones or TFs (Diagenode: C01020010/C01020013). ChIP experiments were performed with chromatin from 1–2 × 106 cells per histone mark or 1 × 107 cells for p63 using the Indirect ChIP protocol (Auto-Histone kit using protein G beads: C01010023) implemented on the IP-Star Compact Automated system (Diagenode: B03000002) or manually using the protocol from the iDeal ChIP-Seq kit for TFs (Diagenode: C01010055). ChIP for p63 was carried out using ∼3 μg each of 4A4 or p40 mouse monoclonal anti-p63 antibodies, whereas Histone ChIPs were performed using ∼2 μg each of H3K4me1 (Diagenode: C15410194), H3K4me3 (Diagenode: C15410003) and H3K27ac (Diagenode: C15410174) antibodies. After cross-linking reversal and proteinase K treatment, the immunoprecipitated and input DNAs were purified using the Qiagen MinElute kit (Cat No. 28004) and libraries prepared using ThruPLEX DNA-seq kit from Rubicon Genomics. ChIP DNA and input controls from the two MK cell-lines (one replicate each from MK1 and MK2 cell-lines) were then subjected to 50 bp single-end Next-Gen sequencing at ∼25 million reads/per sample on an Illumina HiSeq 2500. For validation of specific p63 targets, ChIP-qPCR was performed with chromatin obtained from independent ChIP experiments with 4A4 and p40 antibodies in MK2 and PMK cell-lines. Experimental details and primer sequence information is provided in Supplementary Methods.
Analysis pipeline
Raw sequencing reads from both in-house generated MK1 and MK2 (4A4, H3K4me1, H3K4me3, H3K27ac, Inputs), and human keratinocyte (two 4A4 replicates and H129 anti-p63 antibodies) and NHEK (two replicates each of H3K4me1, H3K4me3, H3K27ac and input) ChIP-Seq datasets were analyzed through an identical pipeline. Human keratinocyte p63 ChIP-Seq data (30) was downloaded from GEO GSE17611, whereas Histone ChIP-Seq, were downloaded from the ENCODE repository, available here: http://genome.ucsc.edu/ENCODE/. The raw reads were mapped to the Mus musculus genome (mm10 build) and Homo sapiens genome (hg19 build) respectively using Bowtie v1.1.1 (40) with the following parameters: m = 1 (i.e. removes all those alignments with more than one match).
Determining p63 targets using IDR
High-confidence p63 targets were generated based on IDR analysis (https://sites.google.com/site/anshulkundaje/projects/idr), which is well suited for determining optimal peak calling parameters (41). Briefly, each p63 ChIP-Seq bam file was randomly split into two parts with approximately equal number of reads to create pseudo-replicates. Then pooled data files were generated from both the actual replicates and the pseudo-replicates by using the merge function in samtools v1.1. MACS2 peak-calling software (42) was used to call peaks from all three types of files (actual replicates, pseudo-replicates and pooled bam files) under very low stringency conditions (cutoff P-value: 5e-2) using input as control. The resulting peak files were sorted by P-value and filtered to keep only the top 100 000 peaks, which were then used to run IDR (Irreproducible Discovery Rate) analysis. Finally using optimal cutoffs (as defined here: https://sites.google.com/site/anshulkundaje/projects/idr), we identified 8451 p63 ChIP-Seq targets in mouse and 25 920 p63 ChIP-Seq targets in human keratinocytes.
Aligning genomic target regions between mouse and human
The 8451 genomic segments bound by p63 in mouse were mapped to their homologous regions in human using reciprocal-best chained blastz alignments (43) from UCSC, with the target species as mouse and query species as human (mm10.hg19.rbest.chain found here http://hgdownload-test.cse.ucsc.edu/goldenPath/hg19/vsMm10/reciprocalBest/). Two different software programs were used to process these alignments and find homologous regions to mouse p63 targets: UCSC liftover tool and bnMapper (44) implemented in bx-python (More details: https://bitbucket.org/james_taylor/bx-python/wiki/bnMapper). Similar analysis was also repeated in reverse, to map the 25 920 p63 bound genomic targets in human to homologous regions in the mouse genome. We used Reciprocal-best chained blastz alignments (hg19.mm10.rbest.chain found here http://hgdownload-test.cse.ucsc.edu/goldenPath/hg19/vsMm10/reciprocalBest/) from UCSC, with the target species set as human and query species as mouse, respectively.
RNA-Seq
Data generation
Total RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research), from MK1 and MK2 and cDNA libraries for NGS were prepared using the TrueSeq RNA Sample Preparation Kit (Illumina). The samples were then 50 bp single-end sequenced at ∼25 million reads per sample on an Illumina HiSeq 2500. For these experiments, we generated two replicates per cell-line (four replicates in total from MK) to provide higher statistical power for downstream differential expression analysis.
Analysis pipeline
Raw sequencing reads from both in-house generated (MK1, MK2) and publicly available (human keratinocytes from ENCODE: NHEK cell-line) RNA-Seq datasets were analyzed through an identical pipeline. They were mapped to the Mus musculus genome (mm10 build) and Homo sapiens genome (hg19 build) respectively with TopHat2 v2.0.13, using Bowtie2 v2.2.3 as the underlying aligner (45). The software was run with default parameters using Illumina's iGenomes transcript annotation file ‘genes.gtf’ from RefSeq, available at http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn. The aligned reads were quantified with featureCounts-1.4.6 (46) and normalized using the DESeq2 median ratio method (47). The normalized reads were then used to derive transcript per million (TPM) values.
Differential expression analysis
We determined the human orthologs for the corresponding 16 134 mouse genes by using Biomart (http://www.biomart.org/). An input matrix was made of the expression estimates (in count space) of these common genes in both mouse (two replicates of both MK1 and MK2) and human (two replicates of NHEK from ENCODE/CSHL) samples. The expression level of the genes in humans was normalized by total exonic gene length (determined by featureCounts-1.4.6 (46)) using the following formula: length_normalized_expression = (expression/human_gene_length)* mouse_gene_length. This modified count matrix was then used as input for the DESeq2 (47) model to find differentially expressed genes (P-value < 0.05, and median expression > 5 TPM in either mouse or human).
Additional methods
Additional details about experiments and other bioinformatics analysis carried out in this study are provided in the Supplementary Methods section.
RESULTS
Mapping the p63 bound regulatory landscape in mouse
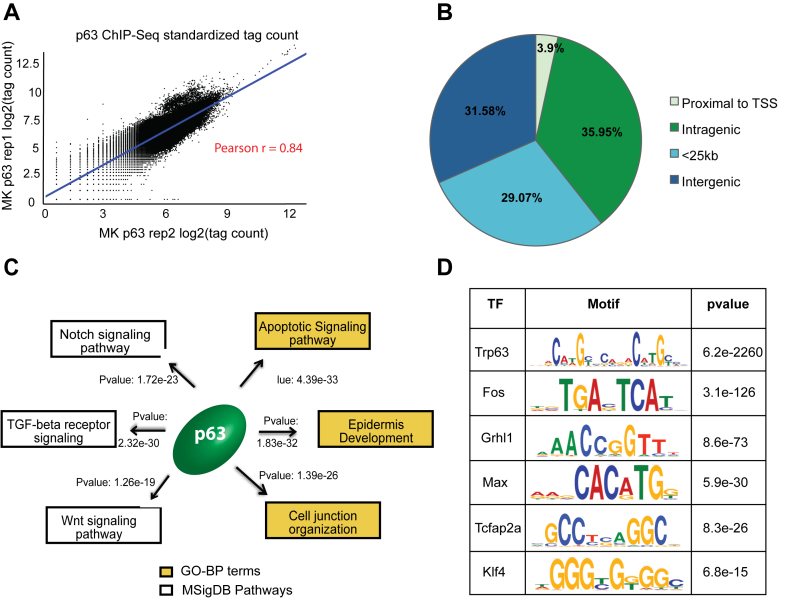
To date, the ChIP-Seq-based p63 target discoveries have been largely confined to human cell-lines that include primary keratinocytes and squamous cancer cell lines. To complement these results, here we identified global p63 transcriptional binding events in two independently established mouse keratinocyte (MK1 and MK2) cell lines by performing ChIP-Seq experiments with a pan-isoform antibody (4A4) directed against the p63 DNA-binding domain. Our ChIP-Seq datasets from the two MK cell-lines showed high level of correlation and led to the identification of 8451 high confidence p63 targets based on peak calling using the ENCODE-project approved IDR (Irreproducible Discovery Rate) method (see ‘Materials and Methods’ for details) (Figure 1A and Supplementary Table S1). To ensure the validity of the ChIP-Seq results, in parallel, we performed independent p63 ChIP studies using PMK and a different antibody, p40 targeted against ΔNp63 proteins, which are the exclusive p63 isoforms in mouse keratinocytes. Six p63 target sites chosen from the ChIP-Seq datasets showed selective enrichment of p63 binding compared to IgG control as measured by quantitative-polymerase chain reaction (Supplementary Figure S1). The results were reproducibly similar in MK and PMK cells and for both the 4A4 and p40 antibodies suggesting that our MK datasets were robust and less likely to be biased due to differences in inherent nature of the mouse keratinocytes or antibody specificity. For valid comparisons, next we performed identical re-analysis of the most comprehensive published p63 ChIP-Seq datasets from human keratinocytes (30), which generated 25,920 p63 targets (Supplementary Figure S2A and Table S2). The fact that the p63 regulatory network in mouse was only about one-third the size of the network targeted by p63 in human keratinocytes was an interesting finding and the first hint of the possible species-specific traits of the p63 targeted cistrome.
Figure 1.
Annotation and characterization of genomic target of p63 in mouse keratinocytes (MK) based on ChIP-Seq. (A) Scatterplot representing high degree of correlation between the MK1 and MK2 p63 ChIP-seq replicates based on total genomic tag density. The datasets were normalized to 20 million reads and the tag density was measured across the entire genome in 1 kb bins. (B) Distribution of p63 targets relative to RefSeq genes. The p63 binding sites are divided into 4 groups: Proximal to TSS (500bp +/− of TSS), intragenic (all introns and exons except first 500 bp), <25 kb (500 bp – 25 kb upstream of TSS or 25 kb downstream) or intergenic (the remaining genomic targets). (C) The top GO-BP terms and MSigDB pathways that are enriched for p63 targets based upon GREAT analysis. (D) TFs motifs enriched at mouse p63 target sites as determined using DREME and Tomtom implemented in MEME software suite.
We next sought to determine the distribution profile of p63 binding in the mouse genome and to examine whether the p63–genome interactions were skewed toward distal regulatory regions similar to what has been reported in human keratinocytes. For this purpose, the regulatory elements were assigned to four groups based on their distance to transcriptional start site (TSS) of genes: proximal to TSS (500 bp upstream and downstream of TSS), intragenic (all introns and exons excluding the 500 bp flanking the TSS), <25 kb (500 bp – 25 kb upstream of TSS or 25 kb downstream of gene) or intergenic (the remaining genomic targets). Our analysis revealed that only a small percentage (3.9%) of mouse p63 binding events were located close to the TSS, suggesting that the primary mode of gene regulation by p63 is mediated mostly through enhancers rather than promoters (Figure 1B).
Notably, the distribution pattern of the p63-bound genomic regions was found to be very similar between mice and human (Supplementary Figure S2B). Upon annotation to the nearest gene using Genomic Regions Enrichment of Annotation Tool (GREAT), we found that mouse p63 targets were enriched for Gene-Ontology Biological Process (GO-BP) such as epidermis development (Figure 1C) and MSigDB Pathways like Notch and Wnt signaling, similar to what has been reported in human studies (30,35). As expected, DREME (48) based motif discovery analysis showed that the canonical 20 bp bipartite p63 DNA-binding sequence was the most significantly enriched motif in the mouse p63 ChIPed regions (Figure 1D). This motif when compared using TOMTOM (49) to the de novo human p63 motif was found to be almost identical (P-value: 2.6e−25) between species (Supplementary Figure S2C). Additional motifs enriched at the p63-bound regions in mouse keratinocytes included those for TFs such as AP-1 and AP-2 that have already been shown to be p63 co-factors (32,35) in human keratinocytes (Figure 1D). Taken together, these studies suggested that even though the p63 regulatory network was significantly expanded in human keratinocytes, the overall patterns of p63–genome interactions and consequently the biological functions modulated by p63 might be mostly conserved between species. These interesting broad observations prompted us to delve deeper into both the similarities and differences of the p63 regulated cistrome between mouse and human, particularly in the chromatin context.
The keratinocyte epigenome and its reciprocal interaction with p63
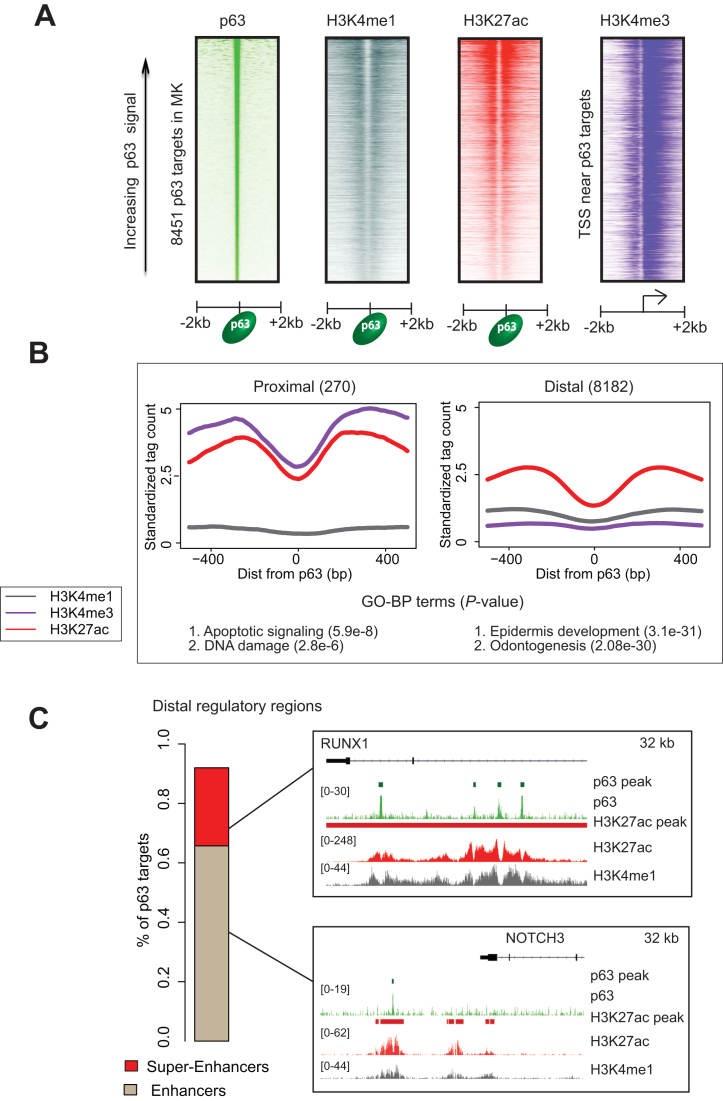
We and others have previously shown that in human keratinocytes, p63 targets are characterized by a distinctive epigenetic signature consisting of active histone but lacking repressive marks (34,35,50). Given this intimate link between p63 and chromatin, we next investigated p63’s interaction with the underlying chromatin architecture in mouse keratinocytes. We therefore performed ChIP-Seq analysis with antibodies against well-characterized histone modifications using chromatin isolated from both MK1 and MK2. To gain maximum insights into the diverse nature of regulatory regions, we chose to focus on H3K4me1, which is found on nucleosomes flanking enhancers, H3K4me3, which marks promoter regions and H3K27ac which can be utilized to distinguish active regulatory regions from inactive/poised state (51,52). Aggregate chromatin profiles of mouse keratinocytes demonstrated a distinct histone signature corresponding to the gene-proximal and distal p63-bound regulatory elements as revealed by their telltale high H3K4me3 and H3K27ac levels, respectively (Figure 2A and B). Interestingly, these two broad categories of regulatory elements also exhibited a clear dichotomy in the functional nature of the corresponding p63-regulated genes, with proximal bound elements being enriched for pathways such as DNA damage and apoptosis, whereas the distal ones representing broad developmental pathways.
Figure 2.
Histone architecture at MK p63 targets. (A) Heatmaps represent standardized read density plots of p63, H3K4me1 H3K27ac and H3K4me3 ChIP-Seq. The signal intensity for p63, H3k4me1 and H3k27ac were plotted centered over p63 binding sites while for H3K4me3 was plotted centered at TSSs proximal to p63 targets. These regions were ordered in descending order of peak strength of p63 signal (averaged across 4 kb genomic window). (B) Aggregate plots representing histone signatures at proximal and distal p63 targets. Standardized signal was plotted in a 1 kb window centered at the p63 summit. Bottom panel shows GO-BP terms enriched at the proximal and distal targets (C) Left: stacked barplot showing the distribution of mouse p63 targets between regular and super-enhancers. Right: UCSC browser shots showing NOTCH3 and RUNX1 as examples of p63 target genes regulated by enhancer and super-enhancer respectively.
For our next analysis, we focused on genome-wide H3K27ac binding profiles, which led to the identification of 20, 534 putative distal enhancer elements in mouse keratinocytes as determined by the ROSE algorithm developed by the Young lab (data not shown). A subset of these enhancers was further subtyped as super-enhancers (Supplementary Figure S3A) based on their higher levels of H3K27ac signals and a defined ‘stitch’ size of composite individual enhancers (53,54). Super-enhancers differ from typical enhancers in size, TF density and ability to induce transcription by virtue of their frequent association with lineage-specific TFs (53,54). Notably, 67% of all p63 bound regions in the mouse genome were constituents of regular enhancers as exemplified by the Notch3 gene, whereas a sizeable 26% of the p63 target sites represented super-enhancers as exemplified by the Runx1 gene (Figure 2C). Impressively, a vast majority (83% or 1024 out of the 1233) of the identified MK super-enhancers was found to bind p63 based on ChIP-Seq analysis—an observation that fits well with the potential master regulatory role of p63 (Supplementary Figure S3A and Table S3A). A similar analysis performed with available H3K27ac data from human keratinocytes revealed 1304 of the 1510 super-enhancers in this species to be bound by p63 (Supplementary Figure S3A and Table S3B), in agreement with a recent report (34). Not surprisingly, key p63 target genes like Krt5, Grhl2 and Dlx2 that are intimately associated with epidermal development and differentiation were found to be associated with regulatory regions that fall in the super-enhancer category in both the species (Supplementary Table S4A and B). However, a closer analysis also revealed key differences in the super-enhancer landscape. Strikingly we found that a relatively small 22% of the genes in mouse keratinocytes that are potentially regulated by p63 bound super-enhancers are also subjected to transcriptional regulation by super-enhancers elements in human keratinocytes. Thus, while the Trp63 gene itself is under an auto-regulatory target of super-enhancer common to both species, other p63 target genes like Src and PTPN14 were found to be subjected to control by species-specific super-enhancers (Supplementary Figure S3B). Overall these findings suggested that although p63 seemed to possess similar molecular function in both mouse and human keratinocytes, a subtle but distinct species-specific alteration in the targeting of p63 was prevalent in a genome-wide fashion, driven most likely by an underlying evolutionary pressure.
The genomic landscape of p63-bound regions undergoes extensive alterations between mouse and human
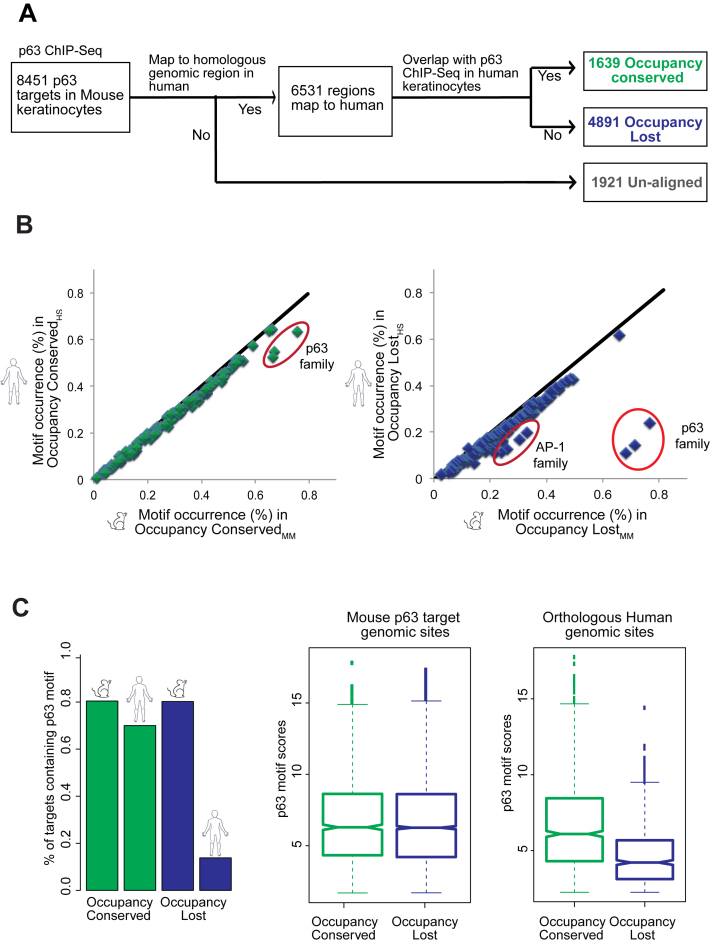
To examine the full-spectrum of the impact of evolution on the global p63 regulatory network, we next compared the p63 bound enhancer segments between mouse and human throughout the genome. To accomplish this, we first mapped the p63 binding sites in mouse to their counterpart DNA sequences in human by using best-chained Blastz pairwise alignments (43). This alignment tool is available from the UCSC browser (55) and has been used to identify homologous TF bound regions of the human and mouse genomes (44). Next, by probing for p63 occupancy at these corresponding regions of the human genome, we determined the rate of conservation/divergence of the p63-bound regulatory network between species (Figure 3A and Supplementary Table S1). Similar to what has been reported by other studies carried out for various tissue-specific TFs (9,16), we found that 22.7% of p63 targets in mouse have diverged in the corresponding DNA sequence to such a degree that they could not be aligned to the human genome. This relatively high divergence between mouse and human was not surprising given the notion that non-coding DNA elements are under less selection pressure than for e.g. protein-coding genes. We next proceeded with the 6530 aligned genomic regions and queried for the homologous human sequences that are represented among the 25 920 ChIP targets of p63 in human keratinocytes. Interestingly, we found only 25% of the aligned targets to be bound by p63 in both species. We refer to this smaller subset of 1639 mice p63 target sites as the Occupancy Conserved group in contrast to the rest of the 4891 locations that are targeted in a species-specific manner and thus denote the Occupancy Lost fraction.
Figure 3.
Comparison of p63 genomic targets in mouse to their aligned homologous regions in the human genome. (A) Flowchart depicting that p63 targets in mouse can be classified into three groups: Occupancy Conserved (genomic regions conserved both in sequence and p63 binding between mouse and human), Occupancy Lost (regions conserved in sequence but not p63 binding between species) and Unaligned (regions not conserved in sequence. (B) Scatterplots of 126 TF motif occurrence rates in p63 targets are drawn with mouse p63 targets represented on the X-axis and the homologous human regions plotted on the Y-axis. The percentage of sites containing each motif is determined by CentriMo. Left panel: represents Occupancy Conserved targets between mouse and human. Right panel: represents Occupancy Lost targets between species. (C) Left panel: a bar plot representing the percentage of aligned p63 targets in mouse and their corresponding homologous regions in human that retain the p63 motif (as determined by Patser). Right panel: all aligned p63 targets in mouse (Occupancy Conserved and Occupancy Lost) and the homologous locations in human are filtered for those that have a p63 motif. Notched box plots illustrate the scores of p63 motif strength at each of the two groups of p63 targets: Occupancy Conserved and Occupancy Lost and their homologous human sequences.
One likely mechanism that explains the lack of p63 binding in the Occupancy Lost sites in the human genome is evolutionary changes in the underlying DNA sequence that leads to disruption of p63 or adjoining cofactor TF binding motifs. To examine this facet, we utilized CentriMo (56) to identify all possible TFs motifs that are present and enriched in the mouse p63-bound genomic segments (denoted by subscript MM) and the corresponding homologous human DNA sequences (denoted by subscript HS) (Supplementary Table S5). We found 126 DNA-binding motifs, all of which occur at same or similar frequency between Occupancy ConservedMM and Occupancy ConservedHS genomic regions (Figure 3B). The p63 DNA binding motif seemed to be disrupted at some of the Occupancy ConservedHS sites but most of them retained a strong version of the motif. However, in contrast, the Occupancy LostHS genomic regions were characterized by a striking loss in occurrence of the DNA-binding motif of the p63 family and to a lesser extent of it's known partner TF family, AP-1 (Figure 3B). To confirm these results in an independent and unbiased fashion, we next utilized pattern matching software PATSER (57) to examine the occurrence of p63 motif based on an established p63 family position weight matrix (58). In agreement with the CentriMo analysis we found that ∼80% of all p63 targets in mouse posses a consensus p63 motif, which is conserved at 72% of the Occupancy ConservedHS regions (Figure 3C). In contrast the Occupancy LostHS regions either contain a motif that matched rather weakly to the consensus (15%) or not all (85%). We next also carried out a reciprocal analysis with the human p63 ChIPed targets and their homologous mouse genomic sites (Supplementary Table S2). This analysis gave similar results of majority of p63 targets in human belonging to the Occupancy Lost category and characterized by disruption of p63 motif at homologous mouse sequences (Supplementary Figure S4A and B). Hence it seemed that changes in p63 regulatory landscape between mouse and human could mostly be attributed to evolutionary loss of p63 and key associated TFs motifs.
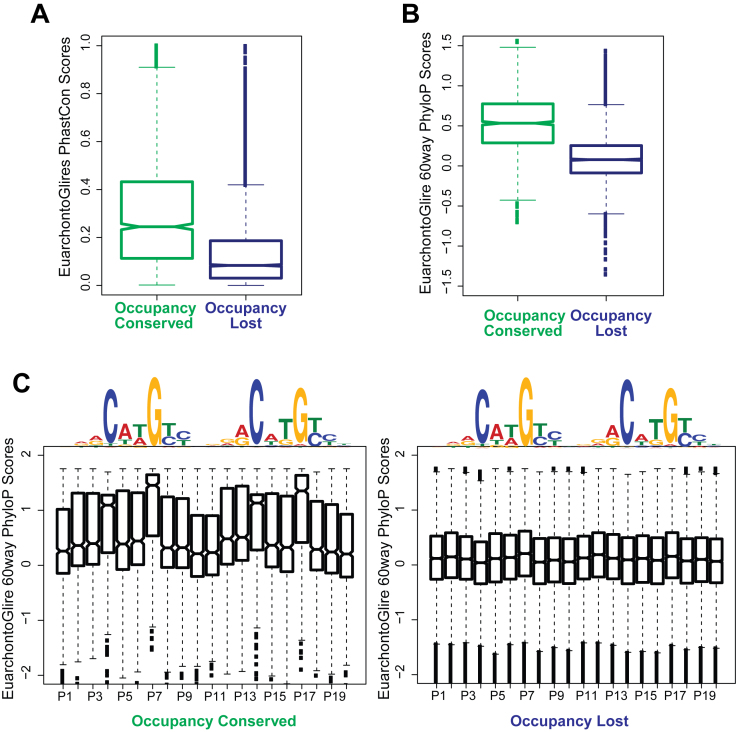
Given the dynamic nature of the presence and distribution of the p63 DNA binding motif between the mouse target regions and their human counterparts, we focused on the possible role of evolutionary forces, if any, on p63 targets. For this we used two different measures: phastCons (59) and phyloP (60) scores, both available through UCSC server (55). We limited ourselves to the EuarchontoGlires clade, as Euarchonta and Glires clades together account for both Rodentia and Primates. We first used phastCons scores and found that the resulting scores reflected moderate to high probability of the Occupancy Conserved targets being under negative selection (Figure 4A). However, as expected the Occupancy Lost targets showed low probabilities of being conserved elements, as were the mouse target sequences that could not be aligned (Figure 4A and Supplementary Figure S5A). We next posited that the p63 motif in species-specific sites could be created by accelerated evolution driven by positive selection. To test this idea, we used phyloP score, which can detect departure from neutral rate of substitution in either direction, i.e. both conservation and acceleration. We plotted phyloP scores that were averaged either over the entire 20 bp or at each individual nucleotide of the p63 consensus motif at both the Occupancy Conserved and Occupancy Lost p63 targets. These results confirmed that the consensus p63 motif, especially the crucial C and G nucleotides at 4th, 14th, 7th and 17th positions are under strong purifying selection in the Occupancy conserved targets, however we did not find any evidence of accelerated evolution at either the Occupancy Lost targets or the Unaligned sites (Figure 4B and C; Supplementary Figure S5B). Taken together our results suggest two modes of p63 binding: a core set of Occupancy Conserved regulatory elements that are co-targeted in mouse and human and subjected to negative selection but also more interestingly a larger species-specific set of regulatory elements that lose/gain the p63 motif due to neutral drift during evolution.
Figure 4.
Occupancy Conserved targets but not Occupancy Lost are under purifying selection across the EuarchontoGlires clade. (A) Conservation of the entire p63 ChIPed region: notched box plots represent average PhastCon scores for the EuarchontoGlires clade for mouse p63 targets (calculated across a 200 bp window centered at the p63 summit). The scores reflect the probability of the region being conserved. (B) Conservation of p63 motif: p63 targets belonging to both groups are filtered for only those genomic locations that have a p63 motif. For a 20 bp window centered at the p63 motif, average PhyloP scores (<0: positive selection; >0: negative selection) for the EuarchontoGlires clade is calculated at each genomic location. Notched boxplots are plotted of these PhyloP scores for both groups of p63 targets. (C) Notched box plots are plotted of PhyloP scores at p63 motif at per base pair resolution. Left panel: scores at all Occupancy Conserved genomic regions containing p63 motif. Right panel: scores at all Occupancy Lost genomic regions containing p63 motif.
Distinctive chromatin architecture at p63 bound genomic segments in mouse and human
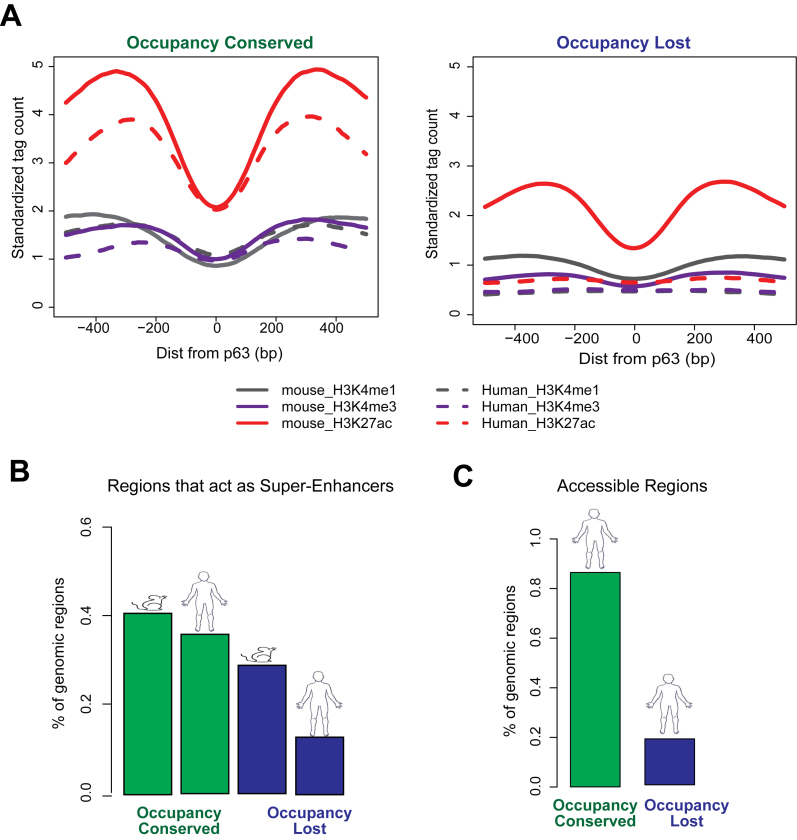
Given the established correlation of p63 binding with chromatin accessibility and presence of active histone marks, it is likely that the notable differences in the genomic distribution of p63 targets between mouse and human keratinocytes is associated with distinctive epigenetic states. To examine this, we plotted aggregate chromatin profiles at Occupancy Conserved and Occupancy Lost targets and found a strong epigenetic signature of H3K4me1, H3K4me3 and H3K27ac modifications flanking the p63 targets in mouse (Figure 5A). At the homologous human p63 binding sites, this epigenetic signature was preserved at the Occupancy ConservedHS but lost at the Occupancy LostHS genomic regions. Importantly, this trend was prevalent when examined in the reverse context also. Accordingly, we found the human p63 ChIPed sites to be characterized by active histone marks with the homologous mouse targets preserving the signature at Occupancy ConservedMM regions but not the Occupancy LostMM sites (Supplementary Figure S6). To better investigate this dichotomy, we next expanded our studies from aggregate profiles to examining histone profiles at individual p63 target locations. We investigated the super-enhancer status of p63 targets in mouse and their homologous human regions. In agreement with the previous analysis we found that that there was a preferential loss of super-enhancers at the homologous human regions in Occupancy lost sites (30% of p63 targets) relative to Occupancy Conserved sites (10% of p63 targets) (Figure 5B). Therefore, vast majority of p63 targets in mouse (Occupancy Lost) seem to lose both p63 occupancy and active chromatin architecture in human. This observation led us to posit that the Occupancy LostHS genomic regions are likely to be in an inaccessible chromatin state. Indeed, we found only 15% of the Occupancy LostHS regions to be in an open chromatin conformation based on available DNase-Seq data from human keratinocytes (61) (Figure 5C). This result was in stark contrast to the Occupancy ConservedHS regions, for which 85% were deemed to be accessible based on the same metric. These findings implied that the vast majority of genomic regions in the human that are homologous to mouse p63 targets might not play any functional role in human keratinocytes.
Figure 5.
Evolutionary differences in p63 targeting reflected in changes in epigenetic landscape. (A) Average profiles of three active histone modifications (H3K4me1, H3K4me3 and H3K27ac) plotted for a 1 kb window centered at p63 motif. The average chromatin architecture is plotted for both mouse and the corresponding homologous human regions at Occupancy Conserved and Occupancy Lost sites. (B) Bar plots showing percentage of p63 targets in mouse and their corresponding homologous regions in human that can be categorized as super-enhancers. (C) Bar plots showing percentage of the homologous human regions to both Occupancy Conserved and Occupancy Lost targets that are accessible in human keratinocytes (as defined by DNase-Seq).
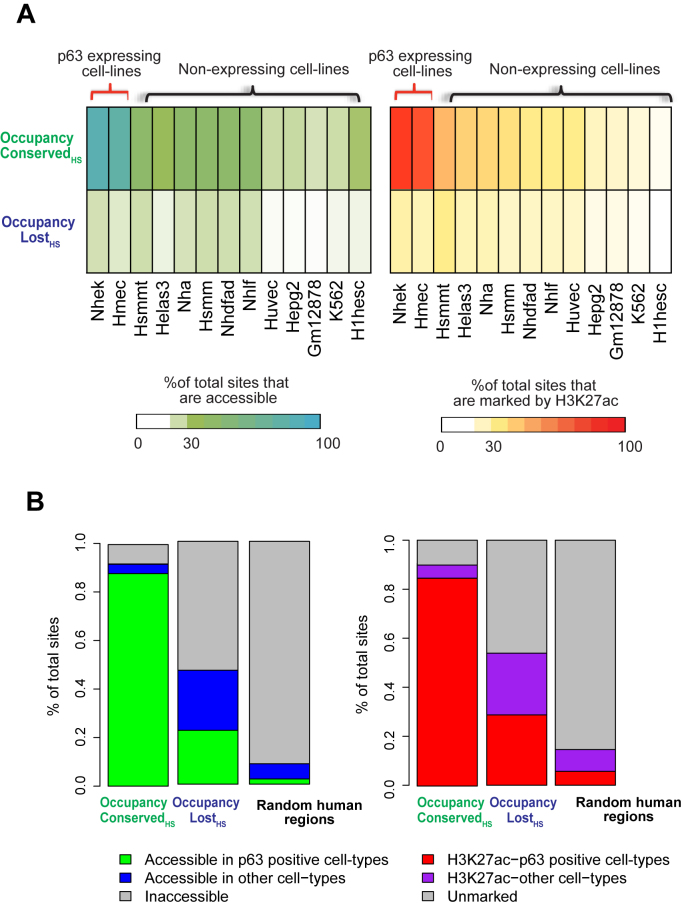
To address if the Occupancy LostHS regions serve potential regulatory roles in any other human cellular context, we next examined ENCODE-generated H3K27ac ChIP-Seq and DNase-Seq accessibility peaks from 13 human cell-lines. This included NHEK and HMEC, two cell lines with relatively high levels of p63. We found that a vast majority of Occupancy ConservedHS sites were associated with an active H3K27ac mark and open chromatin conformation in both NHEK (∼85%) and HMEC (∼78%)—an association that was distinctly less pronounced (∼15–45%) when compared to the p63 negative cell lines. In contrast only ∼10–25% of Occupancy LostHS genomic locations were deemed to be in an active state, based on the H3K27ac signal and DNase accessibility of cells, irrespective of their p63 expression levels (Figure 6A). However, when Occupancy LostHS regions of the mouse were compared for accessibility or active H3K27ac mark across an aggregate of all 13 human cell-lines, the number of genomic regions that could be categorized as functionally active in at least one cell line was closer to 50% (Figure 6B). The effects on the Occupancy ConservedHS sites upon similar comparison of all cell lines were rather modest—a finding not surprising given that a vast majority of these sites are already functional in NHEK cells. These results are especially striking and in contrast to randomly chosen genomic regions, of which only 10–15% were found to be open and marked by active H3K27ac modification across the 13 human cell-lines. To probe this further, we expanded our comparative analysis to 88 human cell-lines for which DNase-Seq data was available from the ENCODE project. In this larger dataset, we found that almost 75% of Occupancy LostHS genomic sites seemed to be in an open chromatin state in contrast to only ∼20% of random genomic regions (Supplementary Figure S7). Taken together these analyses indicate that although a vast majority of the human regulatory regions homologous to mouse p63 targets remain silent in keratinocytes, a subset of them might indeed be functional in a different cellular context. We suspect that this activity in non-keratinocyte cell lines may be driven, in part by other TFs whose motifs are conserved between mouse and human in those genomic segments, as illustrated in Figure 3B.
Figure 6.
Occupancy Lost mouse p63 targets might be functional in other contexts in human development. Occupancy Conserved and Occupancy Lost p63 targets are mapped to their homologous regions in humans. These genomic locations are overlapped with DNase-Seq and H3K27ac peaks in 13 diverse human cell-types, as generated by the ENCODE consortium. (A) Heatmaps: left panel: represents the percentage of genomic sites that overlap with DNase-Seq peaks in each individual cell-type. Right panel: represents the percentage of genomic sites that overlap with H3K27ac peaks in each individual cell-type. (B) Stacked bar plots: left panel: represents percentage of genomic sites that are accessible (at least 1 bp overlap with DNase-Seq peaks) across any cell-type, right panel: represents percentage of genomic sites that are marked by H3K27ac (at least 1 bp overlap with H3K27ac peaks) in any cell-type. Regions that are accessible or marked by H3K27ac in p63 negative cell-lines are colored separately from the rest. Random human genomic regions serve as negative control.
Influence of evolutionary plasticity of the p63 regulatory network on gene network and transcriptional output of keratinocytes
The dramatic alterations in the p63 regulatory landscape during evolution prompted us to next investigate the functional relevance of these findings, particularly as a possible contributing factor for the phenotypic differences in the epidermal morphogenesis program between mouse and human. To examine this, we first annotated p63 targets by assigning the ChIP-Seq peaks to nearest gene in mouse and human keratinocytes, which generated a list of 4870 mouse and 9304 human genes, respectively (Supplementary Tables S1 and 2). We next determined the human and mouse orthologs using the Biomart data mining tool implemented in Ensembl Genome and found that 3065 of these genes constituted a common core which is targeted by p63 in both species. Surprisingly, there was a large set of genes that represented species-specific p63 targets (1805 in mouse, 6239 in human) (Supplementary Table S6). Interestingly, most of the species-specific p63 target genes have a large share of corresponding orthologs in the other species (63.5% for mouse and 85.2% for human) but presumably in these cases the orthologs have opted out from direct regulation by p63 due to evolutionary rewiring of the enhancer landscape. These initial comparisons suggested a divergence of the p63 controlled gene network between mouse and human and hence demanded further exploration.
We hypothesized that syntenic genomic regions in vertebrates that represents gene-rich islands with conserved biological functions between mouse and human would offer a good model to compare species-specific p63 binding patterns under stringent conditions. We therefore delved into a detailed examination of an ultra-conserved micro-syntenic block, the epidermal differentiation complex (EDC) locus, which consists of a cohort of genes that play important roles in keratinocyte development and differentiation (62). The EDC locus spans ∼3 Mb on mouse 3qF1 and 3qF2.1 (and ∼1.5 Mb on human 1q21.3) and comprises of four gene families, common to both species (Supplementary Figure S8). Three of these: Filaggrin (FLG)-like, Late Cornified Envelope (LCE) and Small Proline Rich Region (SPRR) all encode for structural proteins essential for epidermal barrier function whereas the fourth: S100 genes, encodes for proteins whose altered expression is associated with impairment of barrier function. The mouse EDC loci is larger due to a 1.3 Mb gene desert insertion in the genome, (63) which importantly also lacks any p63 binding. A comparative and close-up examination of all the p63 targeted putative enhancers in the EDC locus showed that this region was enriched for common p63 target genes as expected, i.e. genes that are targeted by p63 in both mouse and human (∼50% EDC locus versus ∼32% entire genome). Interestingly, however all of the 36 p63 target genes in the EDC loci, including the common ones were found to be exclusively regulated by species-specific (Occupancy Lost or unaligned) enhancers. These studies, both genome-wide and centered on conserved genomic blocks indicated that there are some differences in the p63-targeted gene network between mouse and human, primarily at the level of the enhancer landscape.
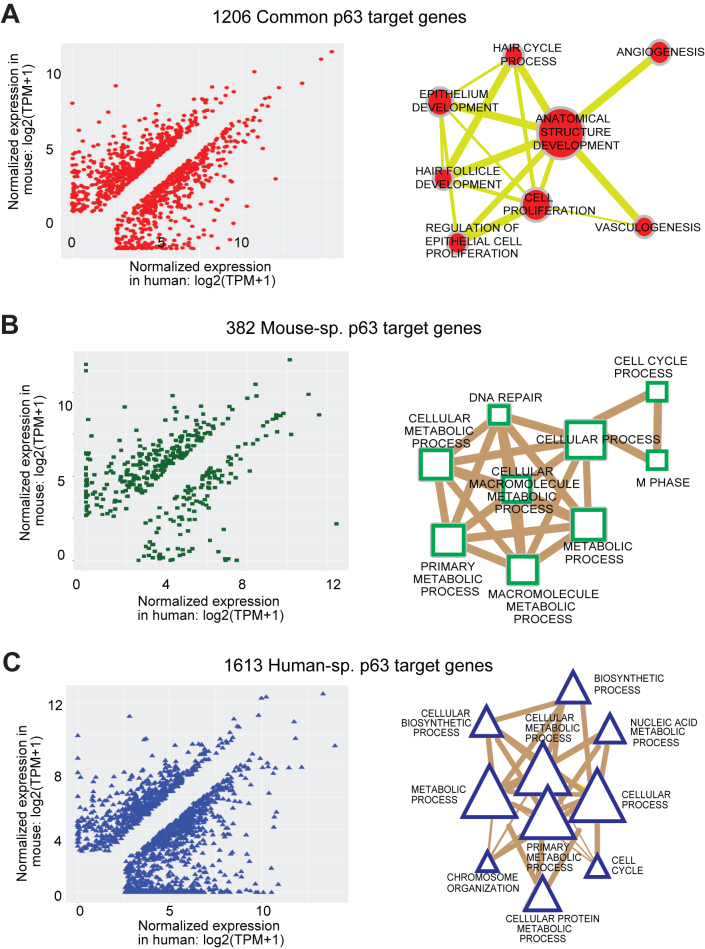
Although the p63 binding spectrum across the genome of mouse and human keratinocyte showed extensive differences, whether such disparities lead to species-specific alterations in the gene expression levels of p63 targets is biologically more relevant question. To address this, we generated the global transcriptomic map of MK1 and MK2 cells by RNA-Seq. Hierarchical clustering analysis performed with RNA-Seq data obtained from a large cohort of mouse tissues and organs (64) demonstrated that both MK1 and MK2 cell lines segregated closest to the skin, as would be expected from their epidermal origin (Supplementary Figure S9). To account for meaningful comparisons of gene expression levels between species, we first compiled a comprehensive list of 16, 134 genes in mouse with identifiable corresponding orthologs in human and standardized their expression estimates (calculated in TPM). By comparing our MK RNA-Seq to similar datasets from human keratinocytes (61), we identified 3201 differentially expressed genes between mouse and human that are targeted by p63 in either mouse and/or human. We found that one-third (1206) of these genes were modulated by p63 in both mouse and human (‘common genes’) although the majority (∼85%) of them exhibited differential usage of enhancers between species (Figure 7A). An example is Sox15, a gene that we have previously found to be co-expressed with p63 (26), which we now show is regulated by an additional mouse-specific p63 targeted enhancer that could potentially account for its differential expression between species (Supplementary Figure S10A). We also found another 382 genes to be targeted by p63 exclusively in mouse keratinocytes (‘Mouse sp. genes’) and 1613 targeted exclusively in human keratinocytes (‘Human sp. genes’) as exemplified by Prim1 and ADAM9 genes respectively (Figure 7B and C; Supplementary Figure S10B and C). Notably many mouse-specific and human-specific genes showed a propensity for higher expression in their respective species suggesting a link between gene expression levels and the changed p63-bound enhancer landscape. We next asked what are the biological processes and pathways that are modulated by these three distinct groups of genes. GO-term annotation using Cytoscape and pathway analysis by IPA revealed that common genes showed enrichment for core p63 functions like anatomical structure, epithelium development and hair cycle while both mouse and human-specific genes reflected primarily metabolic processes (Figure 7 and Supplementary Table S7). The intriguing finding that species-specific p63 target genes were enriched for the same GO-term of broad metabolic processes prompted us to perform a more targeted IPA analysis by focusing on different sub-types of metabolic processes. Interestingly, we found that the majority of the metabolic pathways were somewhat differentially targeted by p63 between mouse and human keratinocytes (Supplementary Figure S11).
Figure 7.
Genes that are differentially regulated by species-specific p63 targeting. (A) A total of 3201 differentially expressed p63 target genes between mouse and human are divided into three groups. Left panel: scatter plot showing 1206 common genes that are targeted by p63 in both mouse and human. Right panel: network representing some of the most highly enriched GO-BP terms for the common differentially expressed genes (adjusted P-value cutoff: 1e-5). The size of the node represents the number of genes that are enriched for that biological process whereas the thickness of the lines connecting the nodes represents the number of genes that are common between the two processes. (B) Left panel: scatter plot showing 382 Mouse-sp. genes that are targeted by p63 exclusively in mouse. Right panel: network representing most highly enriched GO-BP terms for the Mouse-sp. differentially expressed genes (adjusted P-value cutoff: 1e-5). (C) Left panel: scatter plot showing 1613 Human-sp. genes that are targeted by p63 exclusively in human. Right panel: network representing most highly enriched GO-BP terms for the human sp. differentially expressed genes (adjusted P-value cutoff: 1e-5).
Taken together, these data suggest that although the p63 enhancer landscape undergoes sweeping changes during evolution, these may translate to rather limited but observable downstream differences in gene expression profiles. Case in point is the canonical Wnt-β-Catenin signaling pathway—a primary target of p63 with well documented role in skin development (65–67). Although a majority of the key gene components in this pathway are regulated by p63 in both mouse and human, interestingly, there exists also several species-specific p63 target genes, such as Wnt5b, Fzd3 in human and Fzd2 in mouse (Supplementary Figure S12). Further adding to the complexity is the fact that different members of multi-family genes (Frizzled and CBP for e.g.) are targeted by p63 in mouse versus human keratinocytes (Supplementary Figure S12). These findings together highlight the possibility that although the common p63 target genes may seem broadly to impinge upon the same molecular pathways, there might be subtle but distinct variations in both enhancer utilization and resulting gene expression levels that can generate species-specific signaling outputs. This effect of differential p63 targeting on specific downstream genes and functions is even more pronounced in case of metabolic pathways, which is targeted in a species-specific manner. Indeed, as illustrated in Supplementary Figure S13, all the identified p63 target genes in the Fatty acid β oxidation I pathway are unique to only mouse or human keratinocytes. Collectively these studies reveal that both conserved and re-wired portions of the p63 regulatory networks might operate in a complex manner to maintain similar biological functions between species while contributing to observable phenotypic differences.
DISCUSSION
We have performed ChIP-Seq analysis to identify the genome-wide binding repertoire of p63 in mouse keratinocytes within the context of the underlying chromatin state. Although the human cistrome for p63 has been relatively well studied and already been reported, similar studies in mouse was lacking. Our cross-species comparison of the genome-wide patterns of p63 binding and the corresponding transcriptomic milieu in keratinocytes has revealed interesting insights into evolutionary forces that shape gene regulatory modules. In particular, we find that the p63–genome interactions have undergone significant evolutionary changes between mouse and human, and that these changes, which also feature distinctive epigenomic signatures might account for the phenotypic and anatomical differences between the two species. Indeed, we find that similar to what has been described in recent studies on cross-species analyses of other TFs (7,9,10,12–14), the majority of mouse p63 binding sites are not retained in human keratinocytes and thus fall into two discrete categories—those lacking counterpart homologous regions in the human genome and a larger subset representing the Occupancy Lost sites. In contrast, our analysis revealed that there was only a limited conservation of the p63 ChIPed elements between mouse and human as signified by the relatively lower number of Occupancy Conserved sites.
The rather striking divergence in the genomic profile of p63 binding between rodent and human led us to carefully examine the underlying molecular rules and evolutionary forces that could possibly explain how conserved p63 enhancers are retained and species-specific regulatory elements are created. Our major observation in this regard was that while the Occupancy Conserved p63 targets were subjected to purifying selection across the Euarchontoglires clade, the Occupancy Lost targets seemed to be under less evolutionary constraints. Further distinguishing the two classes of p63 targets was the finding that on average, the Occupancy Conserved sites were characterized by greater signals for both p63 (data not shown) and histone modification (Figure 5) than Occupancy Lost sites. An important point worth stressing is that for the sake of simplicity, in our primarily mouse-centric analysis, we have labeled sites that lack p63 binding in humans as Occupancy Lost. However, it is not implicit that these potential p63 target elements are actually lost in human, in the strict sense of the term. Indeed, as an alternative these elements may be gained in the mouse by divergent evolutionary forces of neutral drift creating de-novo p63 binding motifs. Either way, other possible explanations to account for such differences includes differences in the inherent topological nature, spacing and other biophysical attributes of the p63 and/or adjoining TF-binding elements present in these genomic regions. In this regard, it is important to note that the overall p63 consensus motif inferred from the in vivo ChIP data in both species was indistinguishable as would be expected from the high levels of amino acid similarity of the DNA-binding domain of p63 between mouse and human. However, in case of the Occupancy Lost sites, the observed lack of p63 binding in human keratinocytes showed strong correlation with a significant deviation in the underlying sequence from the consensus p63 motif and that of AP-1, a well-known p63 co-TF. Furthermore, mirroring the disrupted p63 binding, we also found that these genomic sites had lost marks for active chromatin architecture and remain largely inaccessible under most circumstances. These evolutionary changes, we posit are likely driven by forces of neutral drift and similar genomic studies in other species in the Euarchontoglires clade are warranted for more insights into this phenomenon. Furthermore, these interesting findings also shed light on p63’s role as a master regulator of keratinocyte function and offer additional support to the growing notion that it acts as a pioneer factor to open and unshackle regulatory elements from the normal chromatin restraints.
A natural follow-up to the aforementioned findings is the possible link between the diverse spectrums of p63-bound enhancers in keratinocytes to functional differences in gene expression between species. Indeed, recent studies have highlighted that even though the underpinnings of TFs regulatory networks are often dynamic, the targeted gene networks are typically conserved between species. We therefore took a gene-centric approach to examine if the p63 controlled regulome varied significantly between mouse and human in keeping with the species-specific differences in the interaction of p63 with the genome. Two caveats remain in our analysis, the first being the assigning of each p63-binding region to the nearest gene as a straightforward means to identify p63 targets. This supposition disregards the premise that in some measurable cases, p63-bound enhancers might rather influence expression of genes located more distally. The second limitation is that binding of a TF at any given site might not universally equate a measurable effect on downstream gene expression. Adding to the complexity is the fact that multiple TF target sites are typically associated with each target gene, as is also likely the case for p63. Thus, many p63 target genes have a multitude of p63 bound sites, of which only a subset may fall in the species-specific category. It is also conceivable that some of these sites may be inactive, play redundant roles or act in additive and/or synergistic fashion thus complicating gene-by-gene comparisons between species. Despite these limitations, we found that two distinct evolutionary mechanisms are likely in play for regulating species-specific differences in p63 target gene expression.
We identified a cohort of ‘common’ genes that despite being targeted by p63-bound enhancers in both mouse and human keratinocytes, showed differential expression based on RNA-Seq analysis (Figure 7A). For many of these genes, however the p63-bound enhancer landscape was found to undergo varying degree of reshuffling and rearrangements that included appearance (or disappearance) of enhancers exclusively in one species. Whether such species-specific binding events of p63 in the context of a common gene could be (i) driver of the differential expression of the gene, and (ii) possibly contribute to phenotypic differences like thicker skin in humans or increased hair follicle density in mouse remains outstanding questions worthy of future investigation. Interestingly, these common set of genes overwhelmingly represented key aspects of developmental pathways such as epithelium development, proliferation and hair cycle, cellular processes in which the functional role of p63 is well established (68). The comparison of mouse versus human cistrome also revealed a whole new class of genes (382 and 1613 genes targeted exclusively by p63 in mouse and human, respectively) that serve in the p63-controlled network in a species-specific manner (Figure 7). Interestingly, these species-specific p63 target genes are predominantly associated with major metabolic processes in both human and mouse (Supplementary Figure S11). These findings lead us to a tempting speculation that the evolution of species-specific p63 targeting to distinct set of metabolic genes perhaps is an adaptive response to the unique cellular needs and demands of mouse and human keratinocytes.
It is worth noting that some aspects of the observed interspecies differences in the genome-wide targeting of p63 could result from intrinsic differences in the keratinocyte cell culture models, batch effects and reflect different experimental setups. We have attempted to minimize such effects by employing datasets obtained using the same anti-p63 antibodies for ChIP-Seq experiments, ensuring concordance between duplicate experiments, applying robust statistical analysis and importantly using epigenomic marks to corroborate p63 binding. However, for future studies, the state of DNA methylation and activity of modifying enzymes such as DNA methyltransferases needs to be taken into account to get a complete and comprehensive picture—a case in point is recent studies on Dnmt3a, which can interact with p63 in primary human keratinocytes and can functionally interact at enhancers (69). Overall, despite the different interspecies models and experimental conditions, our experiments clearly demonstrate that there is indeed a high percentage of shared p63 target genes even though many of the binding sites in some enhancers are turned over during evolution. This turnover, we posit is well tolerated and does not lead to major alterations in the corresponding gene expression levels possibly because of built-in redundancy, as noted above. It is important to stress that our study does not imply that the majority of species-specific p63 targeting events have functional implications. Although some species-unique p63 binding regions and associated species-unique target genes could offer selection pressure that might drive phenotypic differences between mice and humans, we suspect that such differences in most cases are modest due to evolutionary tinkering (70). In sum, our study reinforces the notion that cross-species conservation and retention is not effective in identifying TF-bound sites across species, and sets the stage for the challenges that lie ahead in experimentally discerning the functional consequences of species-specific rewiring of regulatory elements.
DATA AVAILABILITY
All of the ChIP-Seq and RNA-Seq datasets generated in this study have been deposited in GEO repository under accession number: GSE86902.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Marc Halfon for helpful suggestions and advice and Kirsten Smalley for assistance with mouse keratinocyte experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NY State Department of Health [C026714 to M.J.B., in part]. Funding for open access charge: Department of Biochemistry, SUNY at Buffalo.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fuchs E. Scratching the surface of skin development. Nature. 2007; 445:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fuchs E. Epithelial skin biology: three decades of developmental biology, a hundred questions answered and a thousand new ones to address. Curr. Top. Dev. Biol. 2016; 116:357–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khavari P.A. Modelling cancer in human skin tissue. Nat. Rev. Cancer. 2006; 6:270–280. [DOI] [PubMed] [Google Scholar]
- 4. Pasparakis M., Haase I., Nestle F.O.. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014; 14:289–301. [DOI] [PubMed] [Google Scholar]
- 5. Mestas J., Hughes C.C.. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004; 172:2731–2738. [DOI] [PubMed] [Google Scholar]
- 6. Demetrius L. Of mice and men. When it comes to studying ageing and the means to slow it down, mice are not just small humans. EMBO Rep. 2005; 6:S39–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D. et al. . A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014; 515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ballester B., Medina-Rivera A., Schmidt D., Gonzalez-Porta M., Carlucci M., Chen X., Chessman K., Faure A.J., Funnell A.P., Goncalves A. et al. . Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014; 3:e02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt D., Wilson M.D., Ballester B., Schwalie P.C., Brown G.D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C.P., Mackay S. et al. . Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010; 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M., Park T.J., Deaville R., Erichsen J.T., Jasinska A.J. et al. . Enhancer evolution across 20 mammalian species. Cell. 2015; 160:554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villar D., Flicek P., Odom D.T.. Evolution of transcription factor binding in metazoans - mechanisms and functional implications. Nat. Rev. Genet. 2014; 15:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odom D.T., Dowell R.D., Jacobsen E.S., Gordon W., Danford T.W., MacIsaac K.D., Rolfe P.A., Conboy C.M., Gifford D.K., Fraenkel E.. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007; 39:730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rockowitz S., Zheng D.. Significant expansion of the REST/NRSF cistrome in human versus mouse embryonic stem cells: potential implications for neural development. Nucleic Acids Res. 2015; 43:5730–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jubb A.W., Young R.S.. Enhancer turnover is associated with a divergent transcriptional response to glucocorticoid in mouse and human macrophages. J. Immunol. 2016; 196:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G.. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010; 42:631–634. [DOI] [PubMed] [Google Scholar]
- 16. Ulirsch J.C., Lacy J.N., An X., Mohandas N., Mikkelsen T.S., Sankaran V.G.. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet. 2014; 10:e1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dotsch V., Andrews N.C., Caput D., McKeon F.. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998; 2:305–316. [DOI] [PubMed] [Google Scholar]
- 18. Botchkarev V.A., Flores E.R.. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 2014; 4:a015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Candi E., Amelio I., Agostini M., Melino G.. MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 2015; 22:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truong A.B., Kretz M., Ridky T.W., Kimmel R., Khavari P.A.. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006; 20:3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crum C.P., McKeon F.D.. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu. Rev. Pathol. 2010; 5:349–371. [DOI] [PubMed] [Google Scholar]
- 22. Senoo M., Pinto F., Crum C.P., McKeon F.. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007; 129:523–536. [DOI] [PubMed] [Google Scholar]
- 23. Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A.. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999; 398:708–713. [DOI] [PubMed] [Google Scholar]
- 24. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C. et al. . p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999; 398:714–718. [DOI] [PubMed] [Google Scholar]
- 25. Romano R.A., Smalley K., Magraw C., Serna V.A., Kurita T., Raghavan S., Sinha S.. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012; 139:772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sethi I., Romano R.A., Gluck C., Smalley K., Vojtesek B., Buck M.J., Sinha S.. A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genomics. 2015; 16:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rizzo J.M., Romano R.A., Bard J., Sinha S.. RNA-seq studies reveal new insights into p63 and the transcriptomic landscape of the mouse skin. J. Invest. Dermatol. 2015; 135:629–632. [DOI] [PubMed] [Google Scholar]
- 28. Chakravarti D., Su X., Cho M.S., Bui N.H., Coarfa C., Venkatanarayan A., Benham A.L., Flores Gonzalez R.E., Alana J., Xiao W. et al. . Induced multipotency in adult keratinocytes through down-regulation of DeltaNp63 or DGCR8. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E572–E581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pignon J.C., Grisanzio C., Geng Y., Song J., Shivdasani R.A., Signoretti S.. p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8105–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L. et al. . Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010; 6:e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vigano M.A., Lamartine J., Testoni B., Merico D., Alotto D., Castagnoli C., Robert A., Candi E., Melino G., Gidrol X. et al. . New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 2006; 25:5105–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDade S.S., Henry A.E., Pivato G.P., Kozarewa I., Mitsopoulos C., Fenwick K., Assiotis I., Hakas J., Zvelebil M., Orr N. et al. . Genome-wide analysis of p63 binding sites identifies AP-2 factors as co-regulators of epidermal differentiation. Nucleic Acids res. 2012; 40:7190–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zarnegar B.J., Webster D.E., Lopez-Pajares V., Vander Stoep Hunt B., Qu K., Yan K.J., Berk D.R., Sen G.L., Khavari P.A.. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am. J. Hum. Genet. 2012; 91:435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kouwenhoven E.N., Oti M., Niehues H., van Heeringen S.J., Schalkwijk J., Stunnenberg H.G., van Bokhoven H., Zhou H.. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015; 16:863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sethi I., Sinha S., Buck M.J.. Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genomics. 2014; 15:1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bao X., Rubin A.J., Qu K., Zhang J., Giresi P.G., Chang H.Y., Khavari P.A.. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015; 16:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cavazza A., Miccio A., Romano O., Petiti L., Malagoli Tagliazucchi G., Peano C., Severgnini M., Rizzi E., De Bellis G., Bicciato S. et al. . Dynamic transcriptional and epigenetic regulation of human epidermal keratinocyte differentiation. Stem Cell Rep. 2016; 6:618–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romano R.A., Birkaya B., Sinha S.. Defining the regulatory elements in the proximal promoter of DeltaNp63 in keratinocytes: potential roles for Sp1/Sp3, NF-Y, and p63. J. Invest. Dermatol. 2006; 126:1469–1479. [DOI] [PubMed] [Google Scholar]
- 39. Gat U., DasGupta R., Degenstein L., Fuchs E.. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998; 95:605–614. [DOI] [PubMed] [Google Scholar]
- 40. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Myers R.M., Stamatoyannopoulos J., Snyder M., Dunham I., Hardison R.C., Bernstein B.E., Gingeras T.R., Kent W.J., Birney E., Wold B. et al. . A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011; 9:e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng J., Liu T., Zhang Y.. Using MACS to identify peaks from ChIP-Seq data. Curr. Protoc. Bioinformatics. 2011; doi:10.1002/0471250953.bi0214s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W.. Human-mouse alignments with BLASTZ. Genome Res. 2003; 13:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Denas O., Sandstrom R., Cheng Y., Beal K., Herrero J., Hardison R.C., Taylor J.. Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution. BMC Genomics. 2015; 16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 47. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011; 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kouwenhoven E.N., van Bokhoven H., Zhou H.. Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim. Biophys. Acta. 2015; 1849:590–600. [DOI] [PubMed] [Google Scholar]
- 51. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rivera C.M., Ren B.. Mapping human epigenomes. Cell. 2013; 155:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G. et al. . Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013; 24:777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B. et al. . The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013; 41:D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bailey T.L., Machanick P.. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012; 40:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hertz G.Z., Stormo G.D.. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999; 15:563–577. [DOI] [PubMed] [Google Scholar]
- 58. Perez C.A., Ott J., Mays D.J., Pietenpol J.A.. p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene. 2007; 26:7363–7370. [DOI] [PubMed] [Google Scholar]
- 59. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. et al. . Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G. et al. . ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013; 41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Henry J., Toulza E., Hsu C.Y., Pellerin L., Balica S., Mazereeuw-Hautier J., Paul C., Serre G., Jonca N., Simon M.. Update on the epidermal differentiation complex. Front. Biosci. 2012; 17:1517–1532. [DOI] [PubMed] [Google Scholar]
- 63. de Guzman Strong C., Conlan S., Deming C.B., Cheng J., Sears K.E., Segre J.A.. A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis. Hum. Mol. Genet. 2010; 19:1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gluck C., Min S., Oyelakin A., Smalley K., Sinha S., Romano R.A.. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genomics. 2016; 17:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lim X., Nusse R.. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb. Perspect. Biol. 2013; 5:a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu N., Rollin J., Masse I., Lamartine J., Gidrol X.. p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network. J. Biol. Chem. 2012; 287:5627–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Romano R.A., Smalley K., Liu S., Sinha S.. Abnormal hair follicle development and altered cell fate of follicular keratinocytes in transgenic mice expressing DeltaNp63alpha. Development. 2010; 137:1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Melino G., Memmi E.M., Pelicci P.G., Bernassola F.. Maintaining epithelial stemness with p63. Sci. Signal. 2015; 8:re9. [DOI] [PubMed] [Google Scholar]
- 69. Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J.I., Matallanas D., Von Kriegsheim A. et al. . Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016; 19:491–501. [DOI] [PubMed] [Google Scholar]
- 70. Kratochwil C.F., Meyer A.. Evolution: tinkering within gene regulatory landscapes. Curr. Biol. 2015; 25:R285–R288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the ChIP-Seq and RNA-Seq datasets generated in this study have been deposited in GEO repository under accession number: GSE86902.