Abstract
Transcription comprises a highly regulated sequence of intrinsically stochastic processes, resulting in bursts of transcription intermitted by quiescence. In transcription activation or repression, a transcription factor binds dynamically to DNA, with a residence time unique to each factor. Whether the DNA residence time is important in the transcription process is unclear. Here, we designed a series of transcription repressors differing in their DNA residence time by utilizing the modular DNA binding domain of transcription activator-like effectors (TALEs) and varying the number of nucleotide-recognizing repeat domains. We characterized the DNA residence times of our repressors in living cells using single molecule tracking. The residence times depended non-linearly on the number of repeat domains and differed by more than a factor of six. The factors provoked a residence time-dependent decrease in transcript level of the glucocorticoid receptor-activated gene SGK1. Down regulation of transcription was due to a lower burst frequency in the presence of long binding repressors and is in accordance with a model of competitive inhibition of endogenous activator binding. Our single molecule experiments reveal transcription factor DNA residence time as a regulatory factor controlling transcription repression and establish TALE-DNA binding domains as tools for the temporal dissection of transcription regulation.
INTRODUCTION
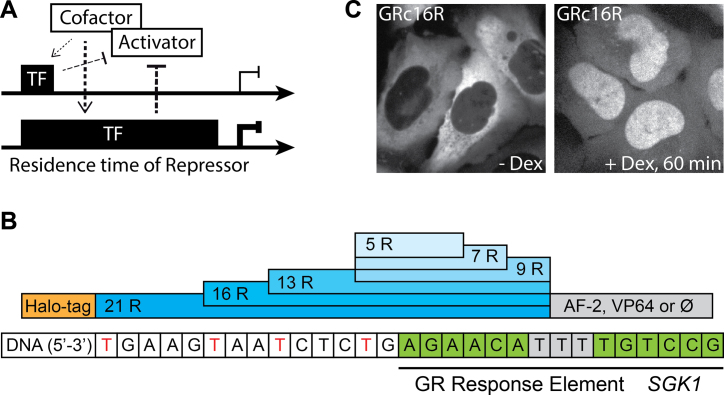
Transcription factor (TF) binding to DNA is central to transcription or repression of a gene, as it is the first step in a cascade of events resulting in assembly or inhibition of the transcription complex (1). Repressive TFs binding to DNA either inhibit transcription by competitively blocking the binding sites of activators or the transcription machinery, by quenching activator function or by recruiting repressively acting co-factors (2–4). While TF binding to DNA was initially pictured to be persistent throughout the transcription process, it is now established for many TFs that DNA residence time is short (5,6). The typical time a TF spends bound specifically to DNA ranges from seconds to minutes (7–10). Intriguingly, for each TF, the DNA residence time is well defined and unique. Moreover, it can widely differ for factors binding to very similar target sequences, for example seen for the steroid receptors glucocorticoid receptor (GR) (9,11,12), estrogen receptor (9) and androgen receptor (12,13). Thus, the question arises whether the DNA residence time of a TF might have regulatory functions in transcription activation or repression. A longer bound transcription repressor for example might be more efficient in recruiting co-factors or blocking other factors from accessing DNA (Figure 1A).
Figure 1.
Design of TALE-TF constructs. (A) Potential regulatory effect of transcription factor (TF) DNA residence time on gene transcription. Long DNA binding of a transcription repressor might lead to enhanced repression efficiency compared to short DNA binding due to more efficient competitive inhibition of an activator or recruitment of co-factors. (B) Domain structure of the TALE-based TFs. After a N-terminal HaloTag varying numbers of TALE DBD repeat domains recognizing the GR response element of SGK1 are inserted, followed by a C-terminal domain (AF-2 of GR or VP64) or no domain (⊘). (C) U2-OS cells expressing GRc16R labeled with TMR in the absence (left panel) and presence (right panel) of 1 μM Dex.
The effect of differing TF affinity to different target sequences on the transcription of the respective gene has been studied for steroid receptors (14,15), the transcription activator GCN4 (16) and transcription activator-like effectors (TALEs) (17). These experiments point to a high correlation between TF affinity and transcription levels. Analogously, in a repressive system, it was found that only the high affinity state of a repressor complex completely repressed transcription while partial dissociation gave rise to low transcription (18). A similar relationship between TF affinity and transcription outcome was suggested by experiments on the effect of organizational features of promoters on gene expression (19). However, measurements of binding energies or affinities do not yield association and dissociation rate constants of a TF. While it is commonly assumed that changes in affinity are due to altered dissociation rate constants, this has not been directly tested and varying association rate constants have not been excluded. In addition, knowledge of affinities enables calculations of occupation probabilities, but kinetic information is necessary for a full mechanistic insight into cellular processes. Moreover, the results of in vitro measurements of affinity are often not directly transferable to the living cell, where TF binding might be influenced by additional factors. Examples are the GR (20) and the TATA-binding protein (21), whose binding to DNA are influenced by chaperones. Consistently, the in vitro DNA residence time of GR was observed to be much longer compared to the value observed in vivo (9,11,22). Other influences, such as concentration-dependent facilitated dissociation (23,24), are difficult to correctly reconstitute in vitro. Resolving the kinetic aspects of the influence of TF–DNA interactions on gene transcription thus asks for DNA residence time measurements in the living cell.
A correlation between the DNA residence time of a TF and transcription output has been observed in competition chromatin immunoprecipitation assays for Rap1 in yeast (7). Long Rap1 residence on DNA occurred predominantly at promoters and was coupled to transcription activation, while fast Rap1 turnover on non-promoter sequences entailed low transcription. In their study, Lickwar et al. compared the DNA residence time of Rap1 for different genes. Thus, while suggesting a role for DNA residence time on transcription regulation, causality cannot be inferred, and other gene-specific regulatory traits might be dominating. For GR, fluorescence recovery after photobleaching (FRAP) experiments of several mutants revealed a correlation between longer recovery times and higher transcriptional activity (25). Similarly, different GR ligands exhibited differing FRAP recovery times (26,27). Again, these studies could not exclude mutant or ligand-specific regulatory traits, and left the role of DNA residence time on activation or repression an open question.
Here, we systematically investigated the effect of TF DNA residence time on transcription repression. As model system we chose the endogenous GR-activated gene, SGK1. We used the modular DNA binding domain (DBD) of transcription activator-like effectors (TALEs) to create a series of TFs (TALE-TFs), all binding to the same target sequence and fused to the same regulatory domain, but differing in the time they spend bound to DNA. The TALE DBD consists of a sequence of similar repeat domains differing by two amino acid residues that lend specificity towards a certain DNA base (28). We characterized the DNA residence time of TALE-TFs in living cells using single-molecule fluorescence microscopy and achieved different DNA residence times by altering the number of repeat domains. We observed higher repression if the TALE-TF bound longer to DNA, and could trace the mechanism of TALE-TF induced SGK1 repression back to competitive DNA binding between TALE-TFs and endogenous GR.
MATERIALS AND METHODS
Cloning of TALE-TF constructs and generation of stable cell lines
We assembled TALE-TFs using the Golden Gate TALEN and TAL Effector Kit2.0 (Addgene kit # 1000000024 (29)) and generated cell lines stably expressing TALE-TF constructs by Puromycin selection. For details see Supplementary Information.
Sample preparation
Before induction of the GRcXR, we grew cells on 35 mm glass bottom μ-dishes (Ibidi)(smFISH imaging) or glass-bottom dishes (Delta T, Bioptechs)(single molecule imaging) for at least 3 days in Dulbecco's modified Eagle's medium (DMEM) medium without phenol red and with 10% of charcoal-stripped fetal bovine serum. to achieve uninduced conditions for GR and SGK1. Subsequently, we treated cells with either 0.4% ethanol as vehicle or 1 μM of Dex for 3 h. We labeled GRcXR with HaloTag-TMR ligand or SiR fluorescent ligand according to the HaloTag protocol (Promega), ensuring that equal numbers of single molecules were visible in each sample for single molecule tracking. We performed smFISH imaging in Fluorobrite DMEM and single molecule imaging in phenol red free Opti-MEM at 35°C.
Detection of SGK1 mRNA with smFISH
For smFISH experiments, we labeled TALE-TFs with equal concentrations of HaloTag-TMR ligand and performed smFISH according to a modified Stellaris RNA-FISH protocol (Biosearch Technologies). For details see Supplementary Information.
We imaged individual SGK1 mRNA molecules throughout cells on a custom built spinning disk microscope (Supplementary Information) by taking z-stacks with a step size of 500 nm. We analyzed smFISH images with the Matlab toolbox FISH-quant (30) to obtain the number of mRNA molecules per cell, the number of nascent transcripts per transcription site (burst size) and the fraction of cells with at least one transcription site (burst frequency). Outlines of cells expressing TALE-TFs were identified using membrane staining. We adjusted the detection settings for each sample individually to allow for accurate detection of single mRNA molecules.
Single molecule fluorescence microscopy and data analysis
The single molecule fluorescence microscope was custom built around a commercial microscope body (see Supplementary Information). We fixed the camera integration time to 50 ms and varied laser dark times between different time-lapse conditions. Data analysis was performed in MATLAB following the procedures published in (9). To extract DNA residence times of TALE-TFs, we implemented a global fitting approach to histograms of fluorescent ‘on’ times as described in (9) in Python 2.7.9. For details see Supplementary Information.
RESULTS
Design of artificial transcription factors with varying length of the DNA binding domain
We utilized the DBD of TALEs to construct artificial TFs and investigated the role of DNA residence time on transcription repression. The modular structure of the TALE DBD bears several characteristics important for our design (Figure 1B): first, the TF can be targeted to any DNA sequence that starts with a thymine (28), and second the number of nucleotides addressed by the binding domain can be altered easily (29,31). In addition, we designed the artificial TFs to be ligand-inducible (32), to enable a quantitative comparison of transcript levels in presence and absence of TFs in the same cellular background. Therefore, we chose the hormone-inducible C-terminal domain AF-2 of the GR, which translocates GR to the nucleus upon ligand binding (Figure 1B) (33). As target sequence, we chose the glucocorticoid response element (GRE) in the promoter region of isoform 1 of the serum and glucocorticoid-regulated kinase 1 (SGK1) gene (Figure 1B). We used Golden Gate cloning to construct TALE DBDs comprising 5, 7, 9, 13, 16 and 21 repeat domains (29) (Figure 1B and Supplementary Table S1). Deviating from the original counting scheme (28), we included the N-terminal thymine and the last half repeat in the number of repeat domains to arrive at an integer value of DNA-interacting residues. We confirmed efficient recruitment and preferred binding of these TALE DBDs to SGK1 by ChIP (Supplementary Figure S1 and Supplementary Information). All artificial TFs were fused N-terminally to a HaloTag for fluorescent labeling (Figure 1B). To control for the influence of AF-2, which acts as activating domain in wild-type GR (33), we cloned some artificial TFs without AF-2 or exchanged it with the VP64 domain. These constructs were targeted to the nucleus by a SV40 nuclear localization signal. We will refer to the complete artificial TFs including HaloTag, X repeat domains and the GR AF-2 activation domain as GRcXR, VPXR for the factors with VP64 domain and XR for factors without regulatory domain (Supplementary Table S1).
To ensure low-level expression of our artificial TFs across cells, we created stable U2-OS cell lines expressing the TALE-TFs by antibiotic selection, isolated colonies arising from single cells and sorted these cells by flow cytometry. For GRcXR, only colonies exhibiting a clear cytoplasmic localization before and a nuclear localization after induction with the ligand Dexamethasone (Dex) were considered (Figure 1C). The selected cell lines showed only moderate variations in GRcXR concentrations (Supplementary Figure S2). We performed experiments with induced cells three hours after addition of Dex, when all constructs showed nuclear localization. In contrast to GRcXR, VPXR and XR were localized permanently in the nucleus, without the need for ligand induction.
DNA residence time of GRcXR is a non-linear function of the number of repeat domains
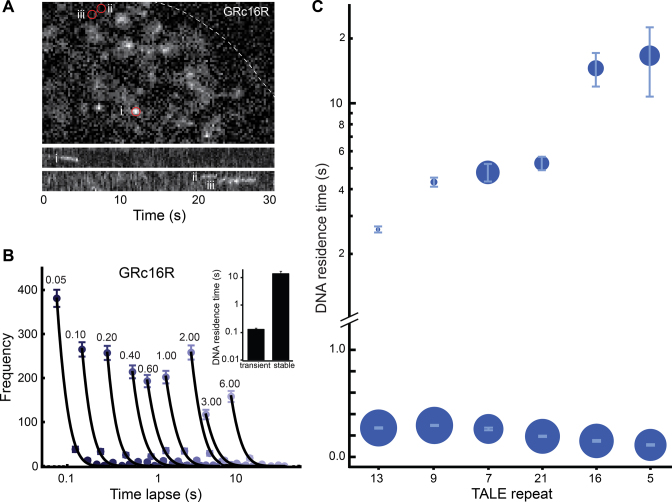
We quantified the DNA residence time of GRcXR by single molecule imaging of individual GRcXR labeled with an organic dye (9). Cells were plated on a temperature-controlled glass bottom dish and excited with an inclined laser beam on a custom built microscope. We could clearly resolve single molecules and observed short and long binding events, as exemplified for GRc16R (Figure 2A and Supplementary Movie S1). To quantify the DNA residence time of GRcXR, we employed time-lapse illumination with different dark times after a constant time period of laser excitation. This enabled us to separate the dissociation rate constants of GRcXR from the photobleaching rate constant of the dye molecules (9).
Figure 2.
DNA residence times of GRcXR are dependent on the number of TALE repeat domains. (A) Upper panel: fluorescence image of TMR-labeled GRc16R in a U2-OS cell nucleus (dashed line) upon induction with 1 μM Dex and 561 nm laser excitation at 50 ms camera integration time. The image is taken from Movie S1. Lower panels: kymographs of regions (i)–(iii) highlighted by circles in the upper panel. (B) Histograms of fluorescent ‘on’ times of TMR-labeled GRc16R at different time-lapse conditions (n = 442 (0.05 s); n = 313 (0.10 s); n = 315 (0.20 s); n = 260 (0.40 s); n = 248 (0.60 s); n = 255 (1.00 s); n = 306 (2.00 s); n = 142 (3.00 s); n = 183 (6.00 s)). Numbers denote the time-lapse time in s. Lines are a global fit by an exponential decay model with two off-rate constants (Equation (14) in Supplementary Information). Inset: DNA residence times of GRc16R extracted from the fit. Error bars denote s.d. (C) DNA residence times of GRcXR. The symbol area is proportional to the frequency of molecules entering a binding mode with the corresponding DNA residence time. GRcXR constructs are ordered by increasing DNA residence time of the long binding population. Error bars denote s.d.
We compiled the distribution times during which GRcXR was fluorescent (fluorescent ‘on’ times) at different time-lapse conditions and extracted the dissociation rate constants using an exponential decay model (9). Two dissociation rate constants corresponding to two distinct populations of DNA residence times best described the distribution of fluorescent ‘on’ times for GRc16R (Figure 2B, Equation 14 in Supplementary Information). Similarly, all GRcXR exhibited a high frequency of transient interactions with DNA with a residence time in the range of 0.14–0.28 s, and infrequent long interactions with a residence time between 2.6 s (GRc13R) and 16.4 s (GRc5R) (Figure 2C, Supplementary Figure S3 and Table S2). We note that all binding events observed in the nucleus enter our analysis, thus our measurements represent an average residence time of GRcXR on chromatin. GRc21R bound to DNA on average for (0.18 ± 0.01) s and (5.20 ± 0.37) s. These values are highly comparable to recently reported in vitro DNA residence times of (0.67 ± 0.09) s and (4.40 ± 2.24) s of a TALE construct comprising 21.5 repeat domains (34). Wild-type GR binds to DNA for ∼2 s (9,11), comparable to the GRcXR constructs with shorter residence times.
A biphasic kinetic behavior similar to our observations has been observed previously for various other TFs and was ascribed to the presence of unspecific and specific DNA sequences for the TF (10,11,35,36). This interpretation is further supported by two-color experiments in which only those TFs that co-localized with RNA polymerase II exhibited stable DNA associations (37), measurements on the DNA residence time of a bacterial factor in a mammalian cell (38) and investigations in yeast (39), all reporting unspecific binding times in a range similar to the transient interaction times of GRcXR. Thus, our data support the notion that we observe GRcXR molecules interacting transiently with unspecific DNA elements and a fraction of GRcXR molecules that are bound to their specific target sequences on DNA.
While the GRcXR constructs together provide a series of TFs with continuously increasing DNA residence times, we found that the relationship between the number of repeat domains and the DNA residence time was not linear, with maxima around 16 repeat domains and at lower repeat domain numbers (Supplementary Figure S4). Such a relationship might result from the helical conformation a TALE DBD assumes when bound to DNA (40,41). Elucidating the functional dependency between the length of the TALE DBD and DNA residence time is an important future task.
Transcription repression by GRcXR increases with DNA residence time
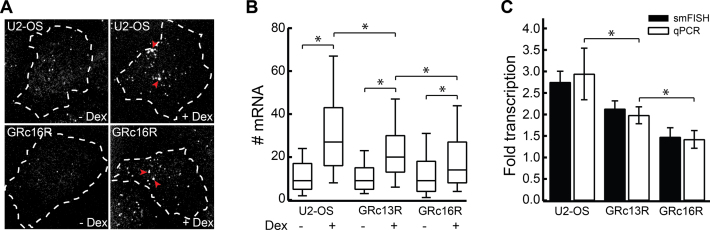
SGK1 transcription is activated by GR in the presence of hormone, but can be stimulated, too, by serum and phenol red (42,43). U2-OS cells have been reported to express GR (44), and we verified GR expression in our U2-OS cells by Western Blot, using HeLa and MCF-7 cells as positive control (Supplementary Figure S5 and Supplementary Information). As expected, after addition of 1 μM Dex, U2-OS cells grown in charcoal stripped FBS and phenol red free medium exhibited SGK1 expression enhanced by a factor of 2.7 compared to Dex-free conditions, as quantified by single molecule fluorescence in situ hybridization (smFISH) and quantitative polymerase chain reaction (qPCR) (Figure 3A–C).
Figure 3.
GRc13R and GRc16R repress SGK1 transcription. (A) smFISH of SGK1 mRNA in wild-type U2-OS cells (upper panels) and U2-OS cells expressing GRc16R (lower panels) in the absence (left panels) and presence (right panels) of 1 μM Dex. Dashed lines indicate the nuclear membrane. Arrowheads highlight loci of nascent transcription. The contrast was adjusted individually for each panel. (B) mRNA distributions in wild-type U2-OS cells and U2-OS cells expressing GRc13R and GRc16R in absence and presence of 1 μM Dex. (*) indicates a significant difference according to the Wilcoxon–Mann–Whitney two-sample rank test, two tailed, P < 0.05. (C) Fold change of transcription calculated as ratio between the mean number of mRNA molecules upon Dex induction and the mean number of mRNA molecules in absence of Dex for wild-type U2-OS cells (308/304 cells) and U2-OS cells expressing GRc13R (308/294 cells) and (GRc16R (270/211 cells), or from qPCR experiments (triplicates on at least three biological replicates). (*) indicates a significant difference according to Student's t-test, two-tailed, P < 0.05. Error bars denote s.e.m. (smFISH) or s.d. (qPCR).
We compared the transcription level of SGK1 determined with smFISH experiments in U2-OS cells stably expressing GRc13R and GRc16R with wild-type U2-OS cells, in the presence of 1 μM Dex (Figure 3A–C). The presence of both constructs significantly shifted the population-wide distribution of SGK1 transcripts to lower values (Figure 3B and C). Notably, repression of SGK1 transcription in the presence of long binding GRc16R was significantly stronger than repression by short binding GRc13R.
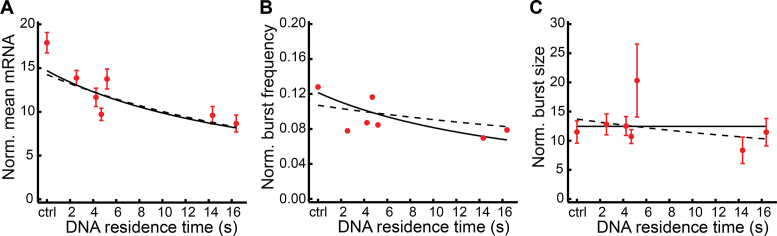
Similar to GRc13R and GRc16R, all GRcXR constructs exhibited reduced Dex-activated SGK1 transcription compared to wild-type U2-OS cells in smFISH experiments (Figure 4A). Notably, the decrease in the mean SGK1 transcription level was more pronounced, the longer the DNA residence time of GRcXR was (linear correlation R2 = 0.63). We confirmed the decrease in mean SGK1 transcription by qPCR (Figure 3C; Supplementary Figure S6 and Supplementary Information).
Figure 4.
Repression of SGK1 transcription increases with GRcXR DNA residence time. (A) Normalized mean number of SGK1 mRNA molecules as function of DNA residence time in wild-type U2-OS cells as control (ctrl) and U2-OS cells expressing GRcXR (spheres, 308/304, 308/294, 296/316, 267/274, 167/157, 270/211 and 290/283 cells). Error bars denote s.e.m. (B) Normalized burst frequency of SGK1 transcription as function of GRcXR DNA residence time. Cell numbers are as in (A). (C) Normalized burst size of SGK1 transcription as function of GRcXR DNA residence time (70/19, 51/28, 53/29, 69/34, 22/7, 31/12 and 42/20 nascent sites). Error bars denote s.e.m. Lines are calculated based on a model in which the on-rate constant of a two-state gene transcription model is a function of GRcXR DNA residence time (continuous lines) or in which both the on-rate constant and the transcription rate constant are a function of GRcXR DNA residence time (dashed lines).
To test whether the effect on repression was due to the regulatory domain AF-2 that we used for controlled nuclear translocation, we compared the fold change of SGK1 transcription of cells expressing GRc16R with those expressing a TALE-TF with the VP64 domain (VP16R) or lacking a regulatory domain (16R) (Supplementary Figure S7A). All constructs exhibited comparable repression. We further tested whether the cell line selection process or the presence of other parts of the fusion protein affected SGK1 transcription by exchanging the DBD in GRc16R with one targeting the forkhead box L1 (FoxL1) promoter region or a random sequence (Supplementary Figure S7B and Table S1). Both control constructs did not alter the transcription level of SGK1 compared to wild-type U2-OS cells.
Since the SGK1 sequence targeted by our GRcXR constructs contains the consensus GR half-site, it is conceivable that GRcXR exhibits at least some affinity towards other GREs. We tested whether GRcXR affected other GR genes containing this half-site by comparing the fold change of cells expressing GRc16R with wild-type U2-OS cells for several GR genes (Supplementary Figure S8). As expected from partial binding to related GREs, we observed a tendency toward transcription repression for GR genes activated in our conditions.
Overall, our experiments revealed an increase in SGK1 repression with increasing DNA residence time of GRcXR.
Transcription repression by GRcXR is due to competitive inhibition of the endogenous activator
In unstimulated U2-OS cells as well as in cells activated by Dex, we could identify up to two sites of SGK1 transcription by the accumulation of smFISH probes, corresponding to the two alleles of SGK1 (Figure 3A). This observation is consistent with the occurrence of a burst of transcription frequently observed for many genes (45,46). The measured number of transcription sites can be used as readout of burst frequency (47). As expected, the fraction of wild-type U2-OS cells exhibiting a transcription site was considerably higher, by a factor 3.6, in activated cells. In the presence of Dex, the fraction of active alleles (as proxy for burst frequency) in cells expressing GRcXR was smaller, the longer the DNA residence time of GRcXR was (linear correlation R2 = 0.40) (Figure 4B). In contrast, we found that the mean number of SGK1 transcripts within a burst (burst size) did not change in wild-type U2-OS cells upon Dex addition (fold change 0.9 ± 0.2), and was mostly independent of the DNA residence time of GRcXR in cells expressing these factors (linear correlation R2 = 0.10) (Figure 4C). We note that quantification of mRNA transcripts at a transcription site by smFISH might underestimate the real burst size as RNA molecules might leave the transcription site while transcription is still active (47). Our further analysis, however, was not affected by this potential misestimate, since activated and non-activated burst sizes were similar and we used the ratio of both burst sizes (Supplementary Information).
Transcription of a gene switching between periods of high and low activity is frequently described by a two-state model in which DNA is transcribed only in the active state of the gene (48). Stochastic and dynamic TF binding to DNA provides a possible explanation for such a bursting behavior (49). To reveal to which extend GRcXR affected the kinetic parameters of the two-state model, we calculated the rate of gene activation, kon, the off-rate of the gene, koff and the rate of gene transcription, ktx (Figure 5), from the measured values of mean SGK1 mRNA, burst frequency and burst size (Supplementary Information). Using these extracted rate constants, simulated mRNA distributions closely reproduced the measured distributions if we corrected the measured burst sizes by a small factor to account for deviations from the real burst sizes (Supplementary Figure S9 and Supplementary Information).
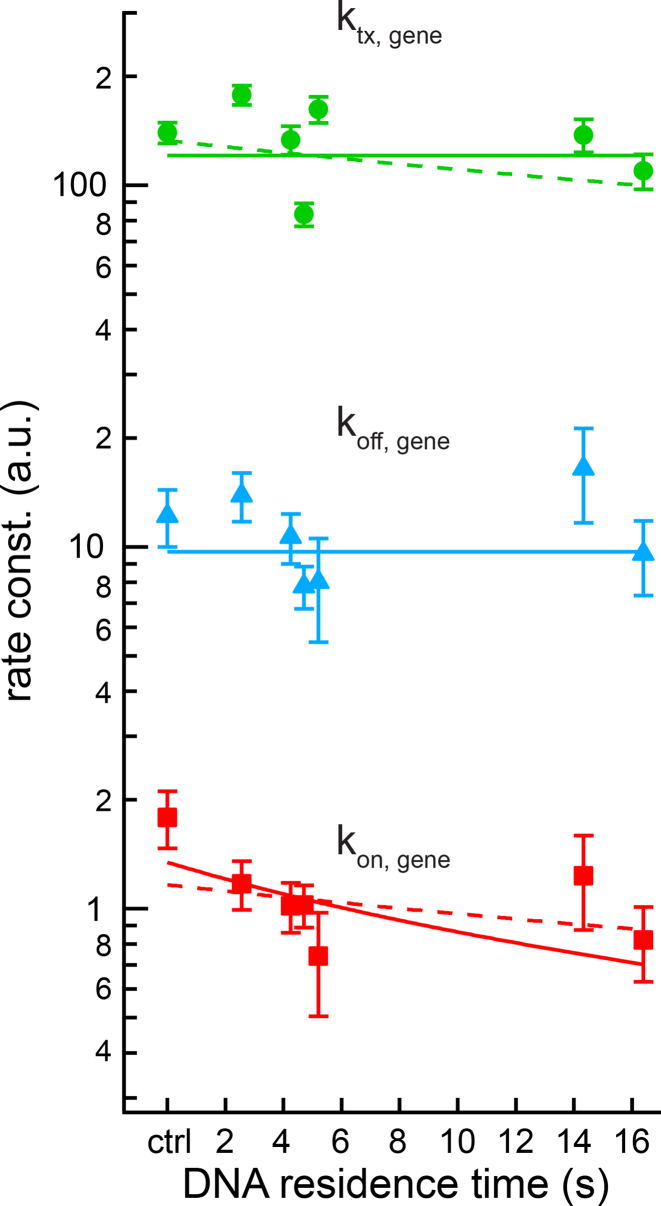
Figure 5.
Rate constants of the two-state model of SGK1 transcription. On-rate constant (squares), off-rate constant (triangles) and transcription rate constant (spheres) were calculated from the measured values of mean SGK1 mRNA, burst frequency and burst size (Figure 4A–C), refer to one allele, are normalized to the SGK1 degradation rate constant and are plotted as function of GRcXR DNA residence time (ctrl denotes wild-type U2-OS cells without GRcXR expression) (Supplementary Information). Error bars denote s.e.m. Lines are fits based on a model in which the on-rate constant is a function of GRcXR DNA residence time (Equation (12) in Supplementary Information, reduced 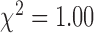 ) and off-rate and transcription rate are constant with respect to GRcXR DNA residence time (continuous lines) or a global fit in which both the on-rate constant and the transcription rate constant are a function of GRcXR DNA residence time (Equations (12) and (13) in Supplementary Information, reduced
) and off-rate and transcription rate are constant with respect to GRcXR DNA residence time (continuous lines) or a global fit in which both the on-rate constant and the transcription rate constant are a function of GRcXR DNA residence time (Equations (12) and (13) in Supplementary Information, reduced 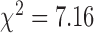 ) and the off-rate is constant with respect to GRcXR DNA residence time (dashed line).
) and the off-rate is constant with respect to GRcXR DNA residence time (dashed line).
We found that kon was decreasing with the DNA residence time of GRcXR (linear correlation R2 = 0.20), while koff was independent of this parameter (linear correlation R2 = 0.03). The transcription rate ktx showed only a small dependency on the residence time (linear correlation R2 = 0.10). Accordingly, we modeled kon with a function decreasing proportionately to the fraction of time that GRcXR was bound to DNA (Equation (12) in Supplementary Information), and ktx and koff as independent of DNA residence time (Figure 5). Such a model is equivalent to a model of competitive inhibition of the endogenous activating factor GR by GRcXR binding to the GRE (Figure 6 and Supplementary Information). Simulations of this extended two-state model suggest that the association and dissociation rate constants of TALE-TFs are much faster than the transitions between the on and off states of the gene (Supplementary Figure S10). Consistent with competitive inhibition, ChIP-qPCR revealed that occupancy of endogenous GR to SGK1 was reduced in the presence of VPXR, at comparable levels of GR expression (Supplementary Figure S11A), the more the longer the DNA residence time of the inhibiting factor was (Supplementary Figure S11B and Supplementary Information).
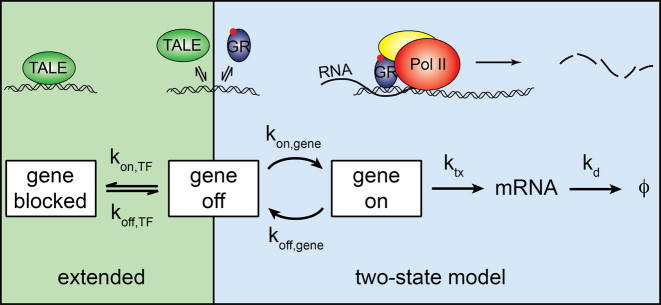
Figure 6.
Model of SGK1 transcription repression by TALE-TF constructs. TALE-TF binds to and unbinds from the GRE of SGK1 with rate constants 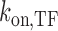 and
and 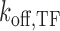 competitive to endogenous GR. If TALE-TF is bound, it prevents gene activation by competitive inhibition of GR DNA binding, and the gene assumes a blocked state (left shade) in addition to the ‘off’- and ‘on’-states of the two-state model of gene transcription (right shade).
competitive to endogenous GR. If TALE-TF is bound, it prevents gene activation by competitive inhibition of GR DNA binding, and the gene assumes a blocked state (left shade) in addition to the ‘off’- and ‘on’-states of the two-state model of gene transcription (right shade). 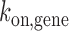 : rate constant of SGK1 activation,
: rate constant of SGK1 activation, 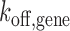 : rate constant of SGK1 deactivation,
: rate constant of SGK1 deactivation, 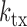 : transcription rate constant of SGK1,
: transcription rate constant of SGK1, 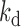 : degradation rate constant of SGK1. Cartoons illustrate the different states of the extended two-state model.
: degradation rate constant of SGK1. Cartoons illustrate the different states of the extended two-state model.
We used the functions describing kon, ktx and koff to calculate the mean SGK1 mRNA (Figure 4A) as well as the burst frequency (Figure 4B) and burst size (Figure 4C) as function of the DNA residence time of GRcXR. All three directly measured datasets were well reproduced by the model of competitive inhibition of GR by GRcXR. An alternative model, in which also the rate constant of transcription varied with the DNA residence time of GRcXR (Equation (13) in Supplementary Information), did not improve the description of the mean number of SGK1 mRNA (Figure 4A).
Overall, our data and quantitative model suggest that GRcXR are competitive inhibitors to GR and repress GR dependent on their DNA residence time.
DISCUSSION
We studied the influence of DNA residence time of a transcription repressor on transcription using SGK1 as model gene and found that this temporal parameter greatly affected the transcription outcome. Exchange or removal of the regulatory domain confirmed the repressive function of our TALE-TF constructs and pointed to a mechanism of competitive inhibition of the endogenous activator. Consistently, a model of competitive inhibition based on the measured kinetic parameters could well reproduce the dependency of repression strength with increasing DNA residence time. We can exclude a repressive effect of long TALE domains due to blocking of promoter regions adjacent to the GRE, as both GRc5R and GRc16R exhibited very similar repression strengths. Compared to DNA residence time, the contribution of nuclear concentration on repression strength plays a minor role in our experiments, since these two constructs differed most in their nuclear concentration but had similar residence times. At first sight our observations contradict a previous study that did not observe competition between GR and ER at the MMTV array (50). However, we note that the residence times of most of our TALE-TFs are longer than those of GR and ER, and thus might have a higher capability to compete off GR from SGK1. Overall, we infer that repression of SGK1 is a function of TALE-TF DNA residence time and consistent with a mechanism of competitive inhibition of endogenous GR by our TALE-TF constructs. The DNA residence time of a TF thus has a regulatory function in transcription repression.
The DNA residence time-dependent repression of our TALE-TFs is able to explain the repressive function of many bacterial (51–54), archaeal (55–57) and eukaryotic (58–62) repression factors suggested to act through competitive inhibition of activating TFs or the transcription machinery (3,4), and the DNA-binding-mediated repression of artificial repressors (63,64). For such a repression by competitive inhibition, our model predicts that complete repression will be achieved for very long DNA residence times. While in eukaryotes repression of protein-coding genes seems to depend mainly on recruiting corepressors, repression of polymerase I and III—mediated transcription often relies on direct inhibition, reminiscent of prokaryotic repression (4). Yet it is very likely that both co-factor-dependent repression involving silencing domains as well as transcription activation are similarly influenced by the DNA residence time of regulatory TFs. Co-factors recruited to a silencing or activating regulatory domain of a TF will differently sense the presence of this domain dependent on the TF DNA residence time. If the TF exchanges fast compared to the on-rate constant of a co-factor, the co-factor will sense an average probability of TF presence, but it will directly follow TF occupancy on DNA if the TF exchanges slowly.
GRcXR mainly affects the on-rate of gene transcription in the two-state model. Thus, in reverse, the GR activating SGK1 transcription should also affect the on-rate of transcription. This is in contrast to observations in bacteria, where gene activation was achieved by reducing the off-rate of the gene (65), and experiments on the bursting of genes in eukaryotes that pointed to an effect of TF affinity on the rate constant of transcription (45). A recent study on the oligomerization state of GR suggests a novel mechanism of GR action by tetramerization, possibly achieved by association of two GR dimers bound to different GREs, thereby inducing a DNA loop (66). In such a model GR binding will influence the on-rate of the gene, and inhibition of GR binding by GRcXR will result in reduction of this on-rate, consistent with our observations. However, the two-state model is a highly simplified model of transcription regulation, and the rate constants of this model comprise a multitude of successive and parallel regulatory processes (67). Thus, care has to be taken when interpreting the rate constants of the two-state model.
The range of 5–21 repeat domains we explored was sufficient to create TFs differing by a factor of 6 in their DNA residence time. This time was a non-linear function of the number of repeat domains. Notably, we measured a peak in DNA residence time between 16 and 18 repeat domains, the range of naturally occurring and of most artificial TALE constructs designed to activate or repress gene transcription (28,31,32,68–70). Crystal structures of TALE DBDs revealed that repeat domains comprising 34 amino acids closely follow the major groove of B-form DNA (40,41). Thus, an increase in binding affinity with increasing length of the TALE DBD would be expected. This is in contrast to our observation of a short DNA residence time for GRc21R. Yet, our measurements are consistent with in vitro experiments (34), a publication reporting a higher contribution of N-terminal repeats to binding affinity compared to C-terminal repeats (17), and the observation that longer TALEs are more tolerant of DNA sequence mismatches (71). It might be that TALE DBDs longer than 16–18 repeat domains start to exhibit a saturation in their affinity to DNA, analogous to the finite melting temperature of long DNA sequences.
Analogous to our TALE-TFs, the effect on transcription of other artificial TALE constructs most likely will depend on the DNA residence times of these constructs. Since TALEs that recognize different target sequences while comprising the same number of repeat domains often achieve very different transcription fold-changes (31,69,70), we anticipate that the DNA residence time of a TALE will be dependent on repeat domain composition such as the choice of the variable di-residue (72), in addition to the number of repeat domains. A systematic evaluation of the effect of repeat domain composition on the DNA residence time of a TALE will be necessary to identify predictive rules. When designing transcription-modulating TALE-TFs, attention should also be paid towards possible side effects on other genes resulting from partial sequence homology, as seen by the effect of GRc16R on GR genes other than SGK1. Overall, our results suggest that including the DNA residence time in the design process of a TALE, in addition to the rules of TALE structure (17,29,70–73) and target sequence position (74) will help to identify the most active factors.
In addition to the dissociation rate constant of a TF, gene transcription may be regulated by its association rate constant (15). However, while modulating the association rate constant of a TF via changes in TF concentration will globally affect the activity of this TF, modulating the dissociation rate constant respective DNA residence time of a TF instead allows for a gene-specific regulation. The cell may achieve changes in DNA residence time through small changes in the base pair sequence of the target region capable of altering TF affinity (15). In addition, co-factors or enzymes are able to affect the DNA residence time of a TF (20,21), and concentration-dependent changes of TF DNA residence time have been reported (24,75). Overall, global concentration-dependent effects combined with sequence-specific strategies in altering the TF DNA residence time will allow for tailored fine-tuning of genetic networks in the cell. Besides dissecting the kinetics of such transcription networks, our approach of controlling the DNA residence time of a protein using TALE DNA binding domains should also be widely applicable to investigate the influence of the time parameter in other chromatin-related processes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anja Palmer for support with single molecule microscopy. SiR dye was kindly provided by K. Johnsson (Ecole Polytechnique Fédérale de Lausanne).
Author contributions: J.C.M.G. conceived the study; M.R. constructed the single molecule microscope; K.C., A.P.P. and L.S. designed experiments; K.C., A.P.P. and L.S. performed experiments; L.E.T., A.P.P. and N.H.U. performed ChIP-qPCR experiments, K.C., A.P.P., L.S. and M.R. analyzed data; J.H. and J.C.M.G. modeled data; J.C.M.G. supervised the study and contributed to experimental design and data analysis; J.C.M.G., K.C. and A.P.P. wrote the manuscript with input from all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [GE 2631/1–1 to J.C.M.G., Emmy Noether Programme UH 275 1/1 to N.H.U.]; European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme [637987 ChromArch to J.C.M.G., 638573 SILENCE to N.H.U.]; German Academic Scholarship Foundation (to M.R.). Funding for open access charge: Deutsche Forschungsgemeinschaft; ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Voss T.C., Hager G.L.. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2013; 15:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levine M., Manley J.L.. Transcriptional repression of eukaryotic promoters. Cell. 1989; 59:405–408. [DOI] [PubMed] [Google Scholar]
- 3. Gaston K., Jayaraman P.S.. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 2003; 60:721–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Payankaulam S., Li L.M., Arnosti D.N.. Transcriptional repression: conserved and evolved features. Curr. Biol. 2010; 20:R764–R771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hager G.L., McNally J.G., Misteli T.. Transcription dynamics. Mol. Cell. 2009; 35:741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mueller F., Stasevich T.J., Mazza D., McNally J.G.. Quantifying transcription factor kinetics: at work or at play. Crit. Rev. Biochem. Mol. Biol. 2013; 48:492–514. [DOI] [PubMed] [Google Scholar]
- 7. Lickwar C.R., Mueller F., Hanlon S.E., McNally J.G., Lieb J.D.. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012; 484:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazza D., Abernathy A., Golob N., Morisaki T., McNally J.G.. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012; 40:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebhardt J.C.M., Suter D.M., Roy R., Zhao Z.W., Chapman A.R., Basu S., Maniatis T., Xie X.S.. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013; 10:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Zhang Z., Li L., Chen B.-C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014; 156:1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Groeneweg F.L., van Royen M.E., Fenz S., Keizer V.I.P., Geverts B., Prins J., de Kloet E.R., Houtsmuller A.B., Schmidt T.S., Schaaf M.J.M.. Quantitation of glucocorticoid receptor DNA-binding dynamics by single-molecule microscopy and FRAP. PLoS One. 2014; 9:e90532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nenseth H.Z., Dezitter X., Tesikova M., Mueller F., Klokk T.I., Hager G.L., Saatcioglu F.. Distinctly different dynamics and kinetics of two steroid receptors at the same response elements in living cells. PLoS One. 2015; 9:e105204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Royen M.E., van Cappellen W.A., Geverts B., Schmidt T., Houtsmuller A.B., Schaaf M.J.M.. Androgen receptor complexes probe DNA for recognition sequences by short random interactions. J. Cell Sci. 2014; 127:1406–1416. [DOI] [PubMed] [Google Scholar]
- 14. Connaghan-Jones K.D., Heneghan A.F., Miura M.T., Bain D.L.. Thermodynamic analysis of progesterone receptor-promoter interactions reveals a molecular model for isoform-specific function. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2187–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bain D.L., Yang Q., Connaghan K.D., Robblee J.P., Miura M.T., Degala G.D., Lambert J.R., Maluf N.K.. Glucocorticoid receptor–DNA interactions: binding energetics are the primary determinant of sequence-specific transcriptional activity. J. Mol. Biol. 2012; 422:18–32. [DOI] [PubMed] [Google Scholar]
- 16. Nutiu R., Friedman R.C., Luo S., Khrebtukova I., Silva D., Li R., Zhang L., Schroth G.P., Burge C.B.. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011; 29:659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meckler J.F., Bhakta M.S., Kim M.S., Ovadia R., Habrian C.H., Zykovich A., Yu A., Lockwood S.H., Morbitzer R., Elsaesser J. et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 2013; 41:4118–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi P.J., Cai L., Frieda K., Xie X.S.. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008; 322:442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharon E., Kalma Y., Sharp A., Raveh-Sadka T., Levo M., Zeevi D., Keren L., Yakhini Z., Weinberger A., Segal E.. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012; 30:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stavreva D.A., Muller W.G., Hager G.L., Smith C.L., McNally J.G.. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 2004; 24:2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Auble D.T., Wang D., Post K.W., Hahn S.. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol. 1997; 17:4842–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lieberman B.A., Nordeen S.K.. DNA intersegment transfer, how steroid receptors search for A target site. J. Biol. Chem. 1997; 272:1061–1068. [DOI] [PubMed] [Google Scholar]
- 23. Graham J.S., Johnson R.C., Marko J.F.. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011; 39:2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sing C.E., Olvera de la Cruz M., Marko J.F.. Multiple-binding-site mechanism explains concentration-dependent unbinding rates of DNA-binding proteins. Nucleic Acids Res. 2014; 42:3783–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kino T., Liou S.H., Charmandari E., Chrousos G.P.. Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function. Mol. Med. 2004; 10:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schaaf M., Cidlowski J.A.. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol. Cell. Biol. 2003; 23:1922–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schaaf M., Lewis-Tuffin L.J., Cidlowski J.A.. Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol. Endocrinol. 2005; 19:1501–1515. [DOI] [PubMed] [Google Scholar]
- 28. Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U.. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009; 326:1509–1512. [DOI] [PubMed] [Google Scholar]
- 29. Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F.. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011; 39:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., Zimmer C.. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013; 10:277–278. [DOI] [PubMed] [Google Scholar]
- 31. Zhang F., Cong L., Lodato S., Kosuri S., Church G.M.. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature. 2011; 29:149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mercer A.C., Gaj T., Sirk S.J., Lamb B.M., Barbas C.F.I.. Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. Acs Synth. Biol. 2014; 3:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicolaides N.C., Galata Z., Kino T., Chrousos G.P., Charmandari E.. The human glucocorticoid receptor: Molecular basis of biologic function. Steroids. 2010; 75:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cuculis L., Abil Z., Zhao H., Schroeder C.M.. Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nat. Commun. 2015; 6:7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sugo N., Morimatsu M., Arai Y., Kousoku Y., Ohkuni A., Nomura T., Yanagida T., Yamamoto N.. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci. Rep. 2015; 5:10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., El Beheiry M., Masson J.-B., Dahan M. et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015; 350:823–826. [DOI] [PubMed] [Google Scholar]
- 37. Morisaki T., ller W.G.M.U., Golob N., Mazza D., McNally J.G.. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014; 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caccianini L., Normanno D., Izeddin I., Dahan M.. Single molecule study of non-specific binding kinetics of LacI in mammalian cells. Faraday Discuss. 2015; 184:393–400. [DOI] [PubMed] [Google Scholar]
- 39. Ball D.A., Mehta G.D., Salomon-Kent R., Mazza D., Morisaki T., Mueller F., McNally J.G., Karpova T.S.. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 2016; 44:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.K., Shi Y., Yan N.. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012; 335:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mak A.N.S., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L.. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012; 335:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Webster M.K., Goya L., GE Y., Maiyar A.C., Firestone G.L.. Characterization of Sgk, a novel member of the serine threonine protein-kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol. Cell. Biol. 1993; 13:2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berthois Y., Katzenellenbogen J.A., Katzenellenbogen B.S.. Phenol red in tissue-culture media is a weak estrogen—implications concerning the study of estrogen-responsive cells in culture. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:2496–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kay P., Schlossmacher G., Matthews L., Sommer P., Singh D., White A., Ray D.. Loss of glucocorticoid receptor expression by DNA methylation prevents glucocorticoid induced apoptosis in human small cell lung cancer cells. PLoS One. 2011; 6:e24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suter D.M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F.. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011; 332:472–474. [DOI] [PubMed] [Google Scholar]
- 46. Lionnet T., Singer R.H.. Transcription goes digital. EMBO Rep. 2012; 13:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senecal A., Munsky B., Proux F., Ly N., Braye F.E., Zimmer C., Mueller F., Darzacq X.. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014; 8:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peccoud J., Ycart B.. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 1995; 48:222–234. [Google Scholar]
- 49. Coulon A., Chow C.C., Singer R.H., Larson D.R.. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat. Rev. Genet. 2013; 14:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Voss T.C., Schiltz R.L., Sung M.-H., Yen P.M., Stamatoyannopoulos J.A., Biddie S.C., Johnson T.A., Miranda T.B., John S., Hager G.L.. Dynamic exchange at regulatory elements during chromatin remodelingunderlies assisted loading mechanism. Cell. 2011; 146:544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis M. The lac repressor. C R Biol. 2005; 328:521–548. [DOI] [PubMed] [Google Scholar]
- 52. Hawley D.K., Johnson A.D., McClure W.R.. Functional and physical characterization of transcription initiation complexes in the bacteriophage lambda OR region. J. Biol. Chem. 1985; 260:8618–8626. [PubMed] [Google Scholar]
- 53. Bertrand-Burggraf E., Hurstel S., Daune M., Schnarr M.. Promoter properties and negative regulation of the Uvra gene by the Lexa repressor and its amino-terminal DNA-binding domain. J. Mol. Biol. 1987; 193:293–302. [DOI] [PubMed] [Google Scholar]
- 54. Ramos J.L., Martinez-Bueno M., Molina-Henares A.J., Teran W., Watanabe K., Zhang X.D., Gallegos M.T., Brennan R., Tobes R.. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005; 69:326–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bell S.D., Jackson S.P.. Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem. 2000; 275:31624–31629. [DOI] [PubMed] [Google Scholar]
- 56. Vierke G., Engelmann A., Hebbeln C., Thomm M.. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 2003; 278:18–26. [DOI] [PubMed] [Google Scholar]
- 57. Lie T.J., Wood G.E., Leigh J.A.. Regulation of nif expression in Methanococcus maripaludis—roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 2005; 280:5236–5241. [DOI] [PubMed] [Google Scholar]
- 58. Jaynes J.B., O’Farrell P.H.. Activation and repression of transcription by homeodomain-containing proteins that bind a common site. Nature. 1988; 336:744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Glass C.K., Holloway J.M., Devary O.V., Rosenfeld M.G.. The thyroid-hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid-hormone and estrogen response elements. Cell. 1988; 54:313–323. [DOI] [PubMed] [Google Scholar]
- 60. Kageyama R., Pastan I.. Molecular-cloning and characterization of a human DNA-binding factor that represses transcription. Cell. 1989; 59:815–825. [DOI] [PubMed] [Google Scholar]
- 61. Schüle R., Umesono K., Mangelsdorf D.J., Bolado J., Pike J.W., Evans R.M.. Jun-Fos and receptors for vitamin-A and vitamin-D recognize a common response element in the human osteocalcin gene. Cell. 1990; 61:497–504. [DOI] [PubMed] [Google Scholar]
- 62. Austin D., Biggin M.D.. A domain of the even-skipped protein represses transcription by preventing TFIID binding to a promoter—repression by cooperative blocking. Mol. Cell. Biol. 1995; 15:4683–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Werner J., Gossen M.. Modes of TAL effector-mediated repression. Nucleic Acids Res. 2014; 42:13061–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Botta S., Marrocco E., de Prisco N., Curion F., Renda M., Sofia M., Lupo M., Carissimo A., Bacci M.L., Gesualdo C. et al. Rhodopsin targeted transcriptional silencing by DNA-binding. Elife. 2016; 5:e12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. So L.-H., Ghosh A., Zong C., Sepulveda L.A., Segev R., Golding I.. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011; 43:554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Presman D.M., Ganguly S., Schiltz R.L., Johnson T.A., Karpova T.S., Hager G.L.. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8236–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Corrigan A.M., Tunnacliffe E., Cannon D., Chubb J.R.. A continuum model of transcriptional bursting. Elife. 2016; 5:e13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schornack S., Meyer A., Römer P., Jordan T., Lahaye T.. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006; 163:256–272. [DOI] [PubMed] [Google Scholar]
- 69. Cong L., Zhou R., Kuo Y.-C., Cunniff M., Zhang F.. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012; 3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Garg A., Lohmueller J.J., Silver P.A., Armel T.Z.. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012; 40:7584–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guilinger J.P., Pattanayak V., Reyon D., Tsai S.Q., Sander J.D., Joung J.K.. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA cleavage specificity. Nat. Methods. 2014; 11:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Streubel J., Blücher C., Landgraf A., Boch J.. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012; 30:593–595. [DOI] [PubMed] [Google Scholar]
- 73. Lin Y., Fine E.J., Zheng Z., Antico C.J., Voit R.A., Porteus M.H., Cradick T.J., Bao G.. SAPTA: a new design tool for improving TALE nuclease activity. Nucleic Acids Res. 2014; 42:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Uhde-Stone C., Cheung E., Lu B.. TALE activators regulate gene expression in a position- and strand-dependent manner in mammalian cells. Biochem. Biophys. Res. Commun. 2014; 443:1189–1194. [DOI] [PubMed] [Google Scholar]
- 75. Chen T.-Y., Santiago A.G., Jung W., Krzemiński Ł., Yang F., Martell D.J., Helmann J.D., Chen P.. Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat. Commun. 2015; 6:7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.