Abstract
Breeding crops with ideal root system architecture for efficient absorption of phosphorus is an important strategy to reduce the use of phosphate fertilizers. To investigate genetic variants leading to changes in root system architecture, 405 oilseed rape cultivars were genotyped with a 60K Brassica Infinium SNP array in low and high P environments. A total of 285 single-nucleotide polymorphisms were associated with root system architecture traits at varying phosphorus levels. Nine single-nucleotide polymorphisms corroborate a previous linkage analysis of root system architecture quantitative trait loci in the BnaTNDH population. One peak single-nucleotide polymorphism region on A3 was associated with all root system architecture traits and co-localized with a quantitative trait locus for primary root length at low phosphorus. Two more single-nucleotide polymorphism peaks on A5 for root dry weight at low phosphorus were detected in both growth systems and co-localized with a quantitative trait locus for the same trait. The candidate genes identified on A3 form a haplotype ‘BnA3Hap’, that will be important for understanding the phosphorus/root system interaction and for the incorporation into Brassica napus breeding programs.
Keywords: oilseed rape, root system architecture, single-nucleotide polymorphism, genome-wide association study, BnA3Hap
1. Introduction
The demand for oilseed rape (Canola; Brassica napus L.) as a food and fuel crop has increased global production to 24.6 Mt yr−1 in 2013 (FAO 2013; http://faostat.fao.org/). Phosphorus (P) is critical in the production of oilseed crops, and with worldwide demand for P fertilizer rising,1 breeding P use efficiency into crops can be an effective strategy. Root system architecture (RSA) plays an important role in P utilization. This is due to low soil P mobility resulting in heteromorphic spatial variation within the soil, but also from variation in bio-availability due to varying soil pH, microbial activity, and colloid chemistry.2
Improved RSA traits have been identified in Brassica which deliver greater resistance to P starvation.3 For example, under low P in B. napus3 and B. oleracea4 primary roots become shorter and lateral roots increase in number and length. In A. thaliana, quantitative trait locus (QTL) LPR1 (Low Phosphate Root 1) is mapped to a 36-kb region on chromosome 1 in a recombinant inbred line (RIL) population.5 In rice (Oryza sativa), a major QTL for P uptake (Pup1) is mapped to a 150-kb region on chromosome 12 in an F3 near-isogenic line (NIL) population.6 The gene conferring the phenotype (PSTOL1) is attributed to this region, and is a protein kinase in this QTL region that acts as an enhancer of early root growth, thereby enabling plants to acquire more phosphorus and other nutrients.7 In B. napus, a total of 62 significant QTL for total root length, root surface area, root volume, total dry weight and plant P uptake at two P levels were detected in three independent experiments with a F10 RIL population under low and high P conditions. Two QTL clusters, uq.C3a and uq.C3b were identified as specifically for root traits and P uptake under LP stress.8 A total of 131 QTL are identified for RSA traits under low and normal phosphate supply using a ‘pouch and wick’ system and an agar based system. QTLs for root and shoot biomass co-localized on chromosome A3, and a QTL for lateral root emergence colocalized with chromosomes A4/C4 and C8/C9 in a doubled haploid population (BnaTNDH).3,9
Genome-wide association studies (GWAS) have been successfully applied to the genetic dissection of complex traits in crops, such as drought resistance in Oryza sativa10,11 and flowering time and leaf architecture in Zea mays.12,13 Compared to QTL mapping, GWAS can achieve a higher resolution of the underlying genetic loci and does not require a mapping population.14 A total of 40 SNP peaks were detected for symbiotic nitrogen fixation traits in bean (Phaseolus vulgaris).15 Six SNPs were attributed to root efficiency of P uptake in traditional rice.16 Seventy-four significant SNPs associated with P-efficiency were detected in soybean (Glycine max).17 Candidate genes for P efficiency in Arabidopsis,18 rice11 and maize19 have been identified. GWAS has also been applied to the dissection of complex traits in B. napus, e.g. flowering time,14 seed weight and seed quality,20 seed oil content,21 stem resistance to Sclerotinia sclerotiorum22 and seedling shoot ionomic traits.23
Haplotypes comprise SNPs or other markers on the same chromosome that are typically inherited together with little chance of recombination.24 To understand the effects of allelic variation on seed oil content, 324 accessions of B. napus with favourable and undesirable haplotypes at ≥ 15 loci were grouped into four classes.21 Other important haplotypes were discovered for resistance to Sclerotina stem rot in B. napus22 and P efficiency in soybean.17
In the present study, an association panel of 405 B. napus cultivars was genotyped with the 60K Brassica Infinium SNP array. RSA traits were investigated in ‘pouch and wick’ and hydroponic system experiments,9,25 under low and high P treatments. The aims of this study were (i) to determine the genetic diversity and RSA of the population, (ii) to investigate the genetics of RSA under high and low P, and (iii) to predict candidate genes associated with the genetic loci to (iv) reveal a favourable haplotype for breeding P-efficient B. napus.
2. Materials and methods
2.1. Plant material
The association panel comprised 405 B. napus cultivars and inbred lines, including 342 semi-winter, 34 spring, 26 winter and 3 unknown types, collected from major breeding centers across China.21 370 lines originated in China, 20 from Europe, 5 from Canada, 4 from Australia, 4 from Korea and 2 from Japan (Supplementary Table S1).
2.2. Phenotyping root system architecture
The root system architecture (RSA) of the panel was screened at low P (LP: 5 μmol l−1 P) and normal P (NP: 250 μmol l−1 P) using a modified standard Hoagland’s solution in 2013-14. The seeds were sterilized using 70% (v/v) ethanol and NaOCl (2.5% active chlorine), and then germinated on gauze with pure water for three days. The seedlings were transplanted to a hydroponic growth tank and grown for the first week with a 25% standard Hoagland’s solution, and then with a 50% standard Hoagland’s solution followed by a 100% standard Hoagland’s solution for a further five days. After 12 days the primary root length (PRL) was measured by a ruler. Seedling samples were dried at 80 °C for seven days for dry weight measurements (shoot dry weight − SDW; root dry weight − RDW). The panel was also screened at LP (LP: 0 μmol l−1 P) using a modified Hoagland’s solution whereby the KH2PO4 was replaced with KCl. The seedlings were grown using a ‘pouch and wick’ system,26 adapted by Thomas.25 Briefly, this method comprises growth pouches composed of germination paper down with the seedling growing between sheets of black polythene, the pouch is hung in a tank and suspended in nutrient solution, and each tank can accommodate 96 pouches (192 seedlings, one on each side of the paper). For each line, 16 seeds were sown across eight independent replicates distributed across four different aluminium frames. The nutrient solution was changed every 3 days. The RSA of samples were imaged at 14 days as previously.25 The RSA including PRL (primary root length), LRN (lateral root number), MLRL (mean lateral root length = LRL/LRN), LRL (lateral root length), TRL (total root length = PRL + LRL) and LRD (lateral root density = LRN/PRL) was measured using RootReader 2D (http://www.plantmineralnutrition.net/rr2d.php). Shoot and root samples were dried at 80 °C and dry weights (shoot, SDW; root, RDW) were determined (Supplementary Fig. S1).
2.3. Data analysis
Restricted maximum likelihood (REML) procedures were used to estimate line means and sources of variation in root traits.9,25 Means were estimated using the [Genotype] term as a fixed factor, retaining [(Replicate/Side of paper/Tray/Frame/Room)] as random factors.3 Random terms and no defined fixed factor were used to estimate sources of variation. Statistical analysis was completed using GenStat15th Edition (VSN International, Oxford, UK).
2.4. Genotyping of the association panel and in silico mapping of SNPs
Four young leaves from different individuals of each variety were planted in Wuhan and were sampled. DNA was extracted using a modified CTAB method27 and then adjusted to 50 ng μl−1. The 60K Brassica Infinium SNP array was employed and the SNP data were analyzed using Illumina BeadStudio (Illumina Inc. San Diego, California, USA).14,20,21 The SNPs with call frequencies > 0.8 or minor allele frequencies (MAF) > 0.05 and homozygous genotype frequency > 0 were selected for association mapping analysis.20 The physical position of the SNPs was identified by aligning the sequence of a 50 bp SNP probe attached to each SNP with the genome sequences of B. napus28 using local BLASTn (BLAST: Basic Local Alignment Search Tool, http://blast.ncbi.nlm.nih.gov/Blast.cgi). If the SNP probe matched two or more locations in the reference genome, the SNPs were regarded as non-specific markers and abandoned.20
2.5. Population structure, relative kinship and linkage disequilibrium analysis of the association panel
The population structure was estimated using the Bayesian Markov chain Monte Carlo model (MCMC) in STRUCTURE V. 2.3.4.29,30 Each K value (the number of real populations) was obtained with five independent runs with a putative number of populations set from 1 to 10, based on a model for admixture and correlated allele frequencies. The length of the burn-in period and number of MCMC replications after burn-in were each set to 100,000. Based on the rate of change in log probability of the data (LnP(D)) and an adhoc statistic ΔK between successive K values, the true K value was determined.3 The Q matrix, which integrates the cluster membership coefficient matrices of replicate runs from STRUCTURE, was determined using CLUMPP (http://www.stanford.edu/group/rosenberglab/clumpp.html). Genetic distances among varieties were calculated by PowerMarker version 3.25 (http://statgen.ncsu.edu/powermarker/downloads.htm). Based on the analysis of molecular variance, AMOVA, the subgroup differentiation, pairwise FST, was measured using ArlequinV. 3.5.2.2 (http://cmpg.unibe.ch/software/arlequin35/). After double centering, distance matrices were used to obtain ‘eigenvectors’ in NTSYS V. 2.2 (http://www.exetersoftware.com/cat/ntsyspc/ntsyspc.html). The relative kinship matrix was calculated by SPAGeDi (http://ebe.ulb.ac.be/ebe/SPAGeDi.html) when all negative kinship values between two individuals were set to 0. The polymorphism information content (PIC) and gene diversity of the SNP markers were estimated using PowerMarker version 3.25 (http://statgen.ncsu.edu/powermarker/). The parameter r2 between each pair of markers was used to estimate the linkage disequilibrium (LD) using TASSEL V. 4.0.14
2.6. Genome-wide association analysis of RSA traits
Population structure and kinship were used to correct for spurious association created by genotype-phenotype covariance. GLM (generalized linear model) and mixed linear model (MLM) calculations were performed in TASSEL to determine the best model for association analyses. The GLM included a naïve model without controlling for population structure, and the Q model which controlled for population structure using the Q matrix to identify populations. The MLM included the K model which assesses inter-individual relative kinship using the kinship matrix obtained in SPAGeDi software, and the Q + K model which controlled for population structure with the Q matrix, and kinship with the kinship matrix. Quantile–quantile plots were created with a negative log10 (P) value of the expected P-value from the genotype–phenotype association and the expected P-value from the assumption that no association exists between genotype and phenotype. The threshold of significance was set to P < 5.15 × 10−5 (1/total SNPs used).22
2.7. Differential expression genes underlying the QTL and significant SNP loci
The B. napus cultivar ‘Eyou Changjia’ were planted at normal P (250 μmol l−1) for ten days, and then half of the plants were transferred to a nutrition solution with zero P (0 μmol l−1) for five days. The shoot (S) and root (R) tissues of three plants at normal P and zero P were separated and immediately placed in liquid nitrogen and stored at 80 °C. The sequencing libraries of twelve RNA samples: S1_LP, S2_LP, S3_LP, S1_NP, S2_NP, S3_NP, R1_LP, R2_LP, R3_LP, R1_NP, R2_NP and R3_NP, were generated using the Illumina RNA Library Prep Kit and sequenced on an Illumina Hiseq 2000 platform with 100-bp paired-end reads. The B. napus reference genome was constructed using bowtie 2 (Broad Institute, Cambridge, MA, USA). Sequencing reads were aligned to the B. napus reference genome with bowtie2 and then assembled using TopHat 2.0.0 and Cufflinks.22 Gene expression was estimated using FPKM (fragments per kilobase of exon per million mapped fragments). Differentially expressed genes (DEGs) between two samples were identified with Cuffdiff, based on criteria False Discovery Rate (FDR) < 0.01 and |log2 (FPKM_LP/FPKM_NP)|>2 (Ding, unpublished data).
The QTLs for root traits previously detected in the Tapidor x Ningyou7 doubled haploid (BnaTNDH) population9 were explored for colocalization with SNP marker associations in the present study. The SNP markers flanking co-located QTL and SNP regions on chromosomes A3 were mapped onto the genome of cultivar Darmor-bzh, respectively. The intervals of a significant locus ranged from loci minus LD to loci plus LD. Genes/transcription factors which were located in the homologous physical regions of the reference map and previously identified as DEGs (differentially expressed genes) responding to high and low P in the B. napus cultivar ‘Eyou changjia’, were selected as putative candidate genes: BnaA03g05800D, BnaA03g05820D, BnaA03g05830D, BnaA03g05850D, BnaA05g22460D, BnaA05g22070D.
2.8. Expression profile of putative candidate genes in P-efficient and P-inefficient B. napus cultivars
The P efficiency of the varieties was evaluated using a P efficiency coefficient of the ratio of SDW under LP/SDW under HP.31 Two P-efficient (121 and 246) and two P-inefficient (292 and 453) varieties were selected for gene expression analysis of putative candidate genes. Plants were grown as above in the hydroponic system under a modified standard Hoagland’s solution for 10 days, and then transplanted to zero P for a further 5 days. Leaves and root were sampled and immediately snap-frozen in liquid nitrogen and stored at −80 °C for downstream RNA extraction. Total cDNA was synthesized from 1 μg RNA with RevertAid First Stand cDNA Synthesis kit (Cat No: K1621, Thermo Scientific, Lithuania, EU). Primers are listed in Supplementary Table S2. The final reaction mixture for RT-PCR contained 10 μl SSo Advanced SYBR Green Supermix (Bio-Rad), 2.0 μl cDNA, 1 μl primer and 7 μl distilled water. Real-time PCR was carried out in triplicate on each sample using Bio-Rad CFX Real-Time PCR System.22 Relative expression levels were evaluated using the 2-△△CT method. Actin was used as an internal control for normalization. The relative expression of genes was normalized to the level of a reference gene BnPAP17 in the P- efficient cultivar (246) at low P.
2.9. Geographical distribution of B. napus haplotypes
There were 21 significant SNPs for the RSA traits at LP, which formed a genetic cluster on A3 within a target QTL region. TASSEL version 4.0 was used to analyze the sliding window of LD on chromosome A3. The SNPs located in the LD intervals in the A3 cluster region were employed to identify the haplotype blocks in the accessions. The geographical distribution of haplotypes was predicted through a joint analysis of correlations between haplotypes and RSA traits, and origin of the panel varieties.
3 Results
3.1. Phenotypic variation in root system architecture at low and normal P
Extensive phenotypic variation was observed in all traits under the two growth systems (Supplementary Fig. S1). For example, in the hydroponic system (HS) under normal P (NP) in 2013, mean values ranged as follows: SDW 0.086–0.834 g, RDW 0.0080–0.081 g, R/S ratio 0.023–0.213 and PRL 13.73–37.67 cm; and under low P (LP), SDW 0.063–0.213 g, RDW 0.022–0.065 g, R/S ratio 0.198–0.502 and PRL 23.02–42.05 cm. In the HS the RDW, R/S ratio and PRL were all greater at LP as compared with NP in both experimental years (HSI; HSII) (Table 1), and the coefficient of variation (CV) at NP was higher. In the ‘pouch and wick’ system (PS) experiment at LP, the panel phenotypes are shown in Table 2. All traits showed continuous variation with an approximate normal distribution (Supplementary Fig. S1).
Table 1.
Mean, standard deviation (SD), minimum(min), maximum (max) and Coefficient of Variation (CV, %) of the shoot dry weight (SDW, g), root dry weight (RDW, g), R/S ratio and primary root length (PRL) in the OSR panel, in the hydroponic system experiments in 2013 (HSI) and 2014 (HSII), under low phosphorus (LP) and normal phosphorus (NP) treatments
| Trait | P Level | Mean ± SD |
Min |
Max |
CV (%) |
||||
|---|---|---|---|---|---|---|---|---|---|
| 2013 | 2014 | 2013 | 2014 | 2013 | 2014 | 2013 | 2014 | ||
| SDW (g) | LP | 0.129b± 0.026 | 0.105ab± 0.028 | 0.063 | 0.030 | 0.213 | 0.209 | 20.0 | 26.3 |
| NP | 0.357a± 0.136 | 0.372a± 0.144 | 0.086 | 0.069 | 0.834 | 1.266 | 38.1 | 38.8 | |
| RDW (g) | LP | 0.041a± 0.008 | 0.032a± 0.008 | 0.022 | 0.012 | 0.065 | 0.0623 | 19.7 | 25.1 |
| NP | 0.032ab± 0.012 | 0.030ab± 0.014 | 0.008 | 0.006 | 0.081 | 0.147 | 36.4 | 41.9 | |
| R/S ratio | LP | 0.322a± 0.049 | 0.312a± 0.055 | 0.198 | 0.178 | 0.502 | 0.499 | 15.2 | 17.5 |
| NP | 0.096b± 0.031 | 0.091c± 0.028 | 0.023 | 0.038 | 0.213 | 0.249 | 32.2 | 31.1 | |
| PRL (cm) | LP | 31.00a± 2.55 | 32.40a± 3.33 | 23.02 | 21.69 | 42.05 | 41.78 | 8.2 | 10.38 |
| NP | 25.8ab± 3.75 | 24.2ab± 2.85 | 13.73 | 15.61 | 37.67 | 37.36 | 14.5 | 11.8 | |
Values with different letters show significant differences at P<0.05.
Table 2.
Mean, standard deviation (SD), minimum (min), maximum (max) and coefficient of variation (CV, %) of the shoot dry weight (SDW, g), root dry weight (RDW, g), R/S ratio, primary root length (PRL), lateral root length (LRL), total root length (TRL), lateral root density (LRD), lateral root number (LRN) and mean lateral root length (MLRL) in the OSR panel, in the ‘pouch and wick’ system (PS) experiment, under a low phosphorus (LP) treatment
| Trait | Mean ± SD | Min | Max | CV (%) |
|---|---|---|---|---|
| SDW (g) | 0.006±0.002 | 0.002 | 0.011 | 28.4 |
| RDW (g) | 0.002±0.001 | 0.001 | 0.005 | 31.3 |
| R/S ratio | 0.322±0.068 | 0.177 | 0.585 | 21.2 |
| PRL (cm) | 12.80±2.70 | 4.50 | 22.70 | 21.2 |
| LRL (cm) | 34.50±9.63 | 10.51 | 71.16 | 27.9 |
| TRL (cm) | 47.30±11.70 | 18.45 | 90.49 | 24.7 |
| LRD (cm/cm2) | 1.30±0.210 | 0.53 | 2.17 | 16.3 |
| LRN (N) | 16.4±4.3 | 5.0 | 33.5 | 26.4 |
| MLRL (cm) | 0.65±0.050 | 0.46 | 0.81 | 7.2 |
3.2. LD decay, population structure and relative kinship
The 60K Brassica Infinium SNP array contains 52,157 SNPs. A total of 30,976 matched to a unique location in the reference genome of cultivar darmor-bzh and 19,397 high-quality SNPs with MAF > 0.05 and call frequencies of SNPs < 0.8 (Supplementary Table S3) were selected to assess the population structure, relative kinship, LD, and to conduct association analyses. SNP number on each chromosome ranged from 482 on C9 to 2131 on C4. The density of SNPs on each chromosome ranged from 98.31 kb/SNP on C9 to 19.23 kb/SNP on A10 (Table 3). LD decay on each chromosome ranged from 8250 kb on C1 to 125 kb on A3, A4 and A6 when r2 was 0.2. SNP density in the LD region ranged from 4 SNP/LDchrA2 on A2 to 347 SNP/LDchrC1 on C1 (Table 3). The LD decay on the A genome, C genome and the whole genome was 1065 kb, 250 kb and 1100 kb, respectively (Fig. 1b). SNP density on chromosomes and in LD regions on the A genome were 27 SNP/kb and 35 SNP/LDA, respectively, on the C genome were 36.97 SNP/kb and 10 SNP/LDC, respectively and on the whole genome were 33 SNP/kb and 18 SNP/LDC, respectively (Table 3).
Table 3.
SNP number, LD decay (physical distance on the genome when the value of r2 is 0.2) and SNP density each chromosome in the OSR panel
| Chromosome | No. of SNPs | LD decay (kb) | SNP density per chromosome (SNP/kb) | SNP density per LD region (SNP/LDchr) |
|---|---|---|---|---|
| A1 | 815 | 150 | 28 | 5 |
| A2 | 555 | 165 | 45 | 4 |
| A3 | 1224 | 125 | 24 | 5 |
| A4 | 837 | 125 | 23 | 5 |
| A5 | 886 | 200 | 25 | 8 |
| A6 | 898 | 125 | 27 | 5 |
| A7 | 1051 | 130 | 23 | 6 |
| A8 | 565 | 3300 | 33 | 100 |
| A9 | 916 | 600 | 37 | 16 |
| A10 | 895 | 475 | 19 | 25 |
| C1 | 1626 | 8250 | 24 | 347 |
| C2 | 1586 | 7125 | 29 | 250 |
| C3 | 1754 | 400 | 35 | 12 |
| C4 | 2131 | 6000 | 23 | 262 |
| C5 | 505 | 575 | 84 | 7 |
| C6 | 675 | 840 | 55 | 15 |
| C7 | 1016 | 775 | 39 | 20 |
| C8 | 980 | 1800 | 38 | 47 |
| C9 | 482 | 1350 | 98 | 14 |
| A genome | 8642 | 250 | 27 | 35 |
| C genome | 10755 | 1100 | 37 | 10 |
| Whole genome | 19397 | 1065 | 33 | 18 |
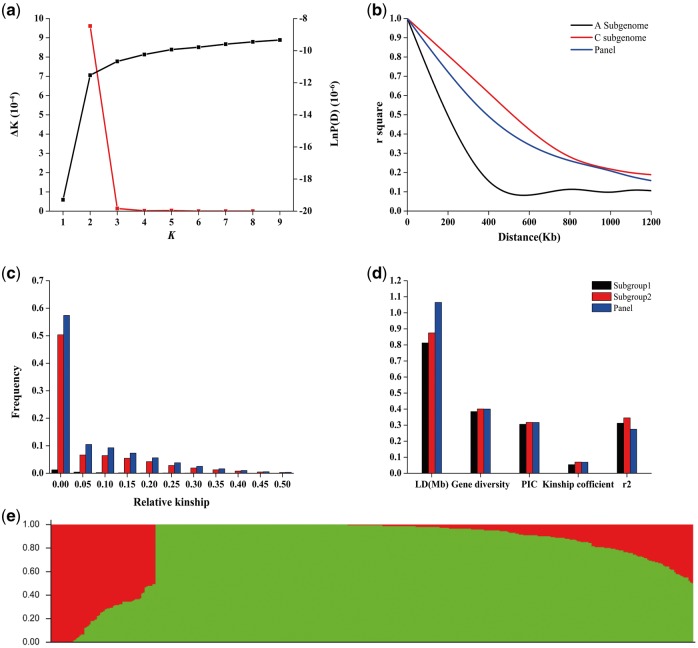
Figure 1.
(a) The rate of change in log probability (LnP(D)) and adhoc statistic ΔK (delta K) of population structure in the 405 B. napus population association panel. (b) Linkage disequilibrium (LD) of the B. napus panel and A, C subgenomes. (c) The relative kinship of the B. napus panel and two subgroups. (d) LD, gene diversity, polymorphism information content (PIC) and kinship coefficient of the B. napus panel and two subgroups. (e) Two subgroups of the B. napus panel when K equals 2.
The population structure of the B. napus panel was assessed. The K values increased continuously with no obvious inflexion points of LnP(K) values (Fig. 1a). The ΔK values indicated that the rate of change adhoc statistic was equal to 2, therefore the population could be divided into two clusters; Subgroup1 and Subgroup2 (Fig. 1e). The FST of Subgroup1 and Subgroup2 was 0.027 (P < 0.05). A total of 66 lines were in Subgroup1; 56, 3, 2, 2, 2 and 1 line(s) originated from China, Korea, Australia, Denmark, Germany and Sweden, respectively. A total of 339 lines were in Subgroup 2; 314, 2, 1, 2, 5, 9, 2, 2, 1 and 1 line originated from China, Japan, Korea, Australia, Canada, Germany, France, Sweden, Poland and Denmark, respectively (Supplementary Table S1). The LD decay distance in Subgroup2 (875 kb) was longer than in Subgroup1 (812.5 kb) and shorter than in the whole panel (1065 kb). Gene diversity of Subgroup2 (0.4010) was higher than in Subgroup1 (0.3847). The PIC in Subgroup2 (0.3175) was higher than in Subgroup1 (0.3057) (Fig. 1d). The Kinship coefficient of Subgroup2 (0.0697) was higher than Subgroup1 (0.0540). The r2 in Subgroup 2 (0.3452) was higher than in Subgroup1 (0.3122) (Fig. 1d).
The relative kinships of the association panel were evaluated. The lines with kinship coefficients < 0.05 accounted for 67.9, 69.5 and 70.6%, and the lines with kinship coefficients equal to 0 accounted for 57.4, 51.3 and 62.4% in the whole panel, Subgroup 1 and Subgroup 2, respectively (Fig 1d), indicating that the majority of the accessions have a weak relationship with one another (Fig. 1c).
3.3. Genome wide association mapping of RSA
The Naïve model, Q model, K model and Q + K model were used for association mapping. The deviations of observed values from the expected values were shown in the QQ plots (Supplementary Fig. S2) and were applied to select the most suitable model for each trait in the different experiments. Significant associations between SNPs and traits were identified by scanning the genome with a P < 5.15 × 10−5 threshold. In the hydroponic experiment I (HSI) there were 41 significant SNPs at LP and 9 at NP. Under LP; 7 SNPs for PRL were detected with the Q model; 4 SNPs for RDW were detected with the Q + K model; 6 SNPs for R/S ratio and 24 SNPs for SDW were detected with the Q model. Under NP; 1 SNP for PRL, 2 for RDW, 3 for R/S ratio and 3 for SDW were detected with the Q model. In the hydroponic experiment II (HSII), there were 31 significant SNPs at LP and 58 at NP. Under LP; 6 SNPs for PRL were detected with the Q + K model; 7 SNPs for RDW were detected with the naïve model; 6 SNPs for R/S ratio were detected with Q model and 12 SNPs for SDW detected with the Q model. At NP; 9 SNPs for PRL, 2 SNPs for R/S ratio and 37 SNPs for SDW were detected with the Q model; and 10 SNPs for RDW detected with the naïve model (Supplementary Table S4). In the ‘pouch and wick’ system (PS) 165 significant SNPs were identified at LP; 28 SNPs for PRL, 33 for RDW, 22 for R/S ratio, 32 for TRL, 5 for LRD, 31 for LRL and 7 for MLRL were detected with the Q model and 3 SNPs for SDW and 4 SNPs for LRN were detected with the Q + K model (Supplementary Fig. S3 and Table S4).
3.4. Co-locating SNPs and QTLs
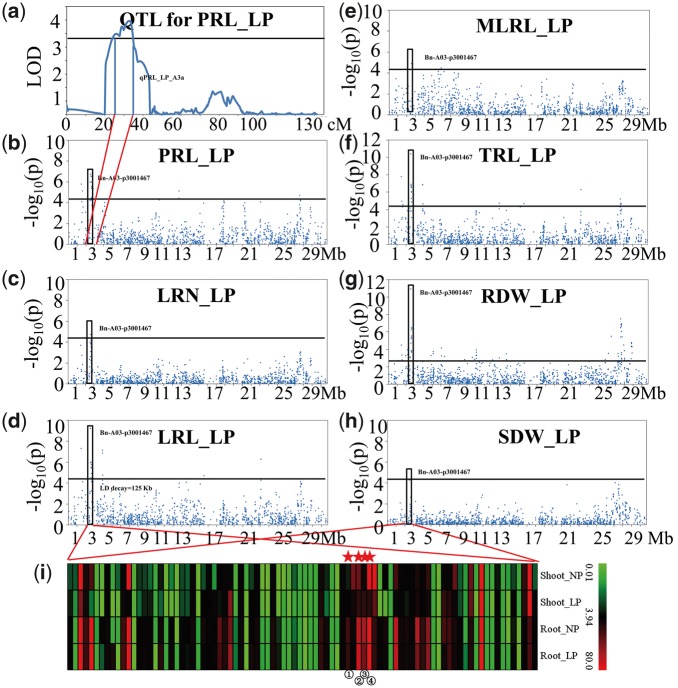
Nine significant SNPs detected with GWAS analyses co-localized with the intervals of the QTLs for the same traits in a previous linkage analyses of the BnaTNDH population,9 including one SNP on A3 for PRL, two SNPs on A5 for RDW (Supplementary Fig. S4a–c), three SNPs on A7 and C4 for SDW, and one SNP on A3 for TRL detected at LP; two SNPs on A3 for SDW detected at NP (Table 4). Therefore, the co-localized loci were detected for the same traits under the same P treatment in the two studies. The two nearly significant SNP markers on A5; Bn-scaff_16893_1-p69538 and Bn-A05-p18820057 at 16.76–17.25 Mb for RDW at LP, were detected in both systems employed in this study (Supplementary Fig. S4b and c). Moreover, there was a significant SNP marker Bn-A03-p3001467 at 2.49–2.72 Mb on A3 detected for all root traits measured in the PS experiment at LP (Fig. 2b–h). Bn-A03-p3001467 could explain phenotypic variation (PVE) of PRL (9.2%), LRN (6.0%), LRL (12.4%), MLRL (6.3%), TRL (13.8%), RDW (14.5%) and SDW (5.9%) at LP (Table 3), and thus was the main locus responding to P deficiency.
Table 4.
SNPs detected by GWAS in the OSR panel; chromosomes (Chr.); physical position (Pos. Mb); P value (significance); phenotypic variation explained (PVE %) and QTLs detected in the BnaTN DH mapping population9; LOD score (significance); marker interval; confidence interval (CI cM). co-located for root system architecture (RSA) traits, under low P (LP) and normal P (NP) treatments
| Significant SNPs |
Significant QTLs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait/treatment | SNP | Chr. | Pos. (Mb) | P value | PVE (%) | Experiment | QTLs | LOD score | Marker interval | Cl (cM) |
| PRL_LP | Bn-A03-p3001467 | A3 | 2.56 | 1.91E-07 | 9.20 | 2014PS | qPRL_LP_A3a | 3.24 | Bn-A03-p2491346/Bn-A03-p4473205 | 28.8-43.5 |
| RDW_LP | Bn-A05-p18820057 | A5 | 17.08 | 1.35E-05 | 5.63 | 2014PS | qRDW_LP_A5a | 3.77 | Bn-A09-p24520612/Bn-A05-p18958083 | 54.5-63.0 |
| RDW_LP | Bn-scaff_16893_1-p69538 | A5 | 6.91 | 1.28E-04 | 4.79 | 2014HS | qRDW_LP_A5a | 3.77 | Bn-A09-p24520612/Bn-A05-p18958083 | 54.5-63.0 |
| SDW_LP | Bn-A07-p11068231 | A7 | 12.31 | 2.07E-05 | 5.94 | 2013HS | qSDW_LP_A7a | 5.87 | Bn-A07-p9849702/Bn-A02-p126006 | 31.7-39.2 |
| SDW_LP | Bn-scaff_16394_1-p124017 | C4 | 31.52 | 3.31E-05 | 5.74 | 2013HS | qSDW_LP_C4a | 3.10 | Bn-scaff_17180_1-p59560/Bn-scaff_15798_1-p699979 | 60.6-70.3 |
| SDW_LP | Bn-A07-p10557165 | A7 | 11.74 | 7.01E-05 | 5.28 | 2014HS | qSDW_LP_A7a | 5.87 | Bn-A07-p9849702/Bn-A07-p10891272 | 31.7-39.2 |
| SDW_NP | Bn-A03-p7688578 | A3 | 6.99 | 1.02E-05 | 6.48 | 2014HS | qSDW_HP_A3b | 6.00 | Bn-A03-p7420835/Bn-A03-p7814324 | 48.9-51.2 |
| SDW_NP | Bn-A04-p18962378 | A4 | 19.01 | 5.25E-05 | 5.52 | 2014HS | qSDW_HP_A4a | 4.62 | Bn-scaff_18903_1-p856363/Bn-scaff_27676_1-p191772 | 78.0-81.6 |
| TRL_LP | Bn-A03-p4408895 | A3 | 3.94 | 1.38E-07 | 7.90 | 2014PS | qTRL_LP_A3a | 3.93 | Bn-A03-p4473205/Bn-A03-p5045095 | 45.4-46.9 |
SDW, shoot dry weight; RDW, root dry weight; PRL, primary root length; TRL, total root length.
Figure 2.
Colocalized locus on chromosome A3 for root traits of B. napus at LP. (a) QTL detected for PRL in the BnaTNDH population linkage analysis (Zhang et al. 2016). (b) peak SNP for PRL. (c) peak SNP for LRN. (d) peak SNP for LRL. (e) peak SNP for MLRL. (f) peak SNP for TRL. (g) peak SNP for RDW. (h) peak SNP for SDW detected with GWAS in the ‘pouch and wick’ (PS) experiment. i) expression profile detected with transcriptome sequencing of candidate genes located in the QTL linkage disequilibrium intervals; ①BnaA03g05800D, ②BnaA03g05820D, ③BnaA03g05830D and ④BnaA03g05850D. primary root length (PRL); lateral root number (LRN); lateral root length (LRL); mean lateral root length (MLRL); total root length (TRL).
3.5. Gene expression profile of candidate genes
A total of 88 candidate genes were within the QTL confidence interval on A3 (Fig. 2i and Supplementary Table S5); BnaA03g05800D, BnaA03g05820D, BnaA03g05830D and BnaA03g05850D were located in the LD decay region of the peak SNP Bn-A03-p3001467 at 2.49–2.72 Mb, and formed the haplotype blocks on A3 (BnA3Hap). In the cultivar ‘Eyou changjia’, the relative expression of BnaA03g05800D and BnaA03g05830D increased, and BnaA03g05820D and BnaA03g05850D decreased, under P-deficiency in the shoot. In the root, the relative expression of BnaA03g05820D increased, and the BnaA03g05800D and BnaA03g05830D decreased, under P-deficiency (Fig. 2i).
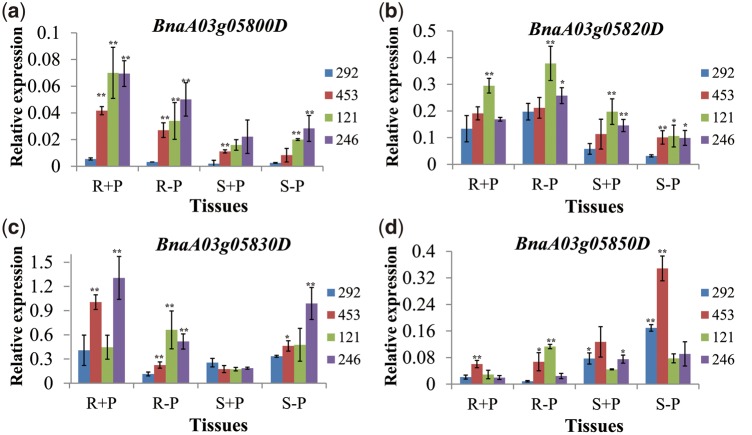
Two P-efficient varieties- 121 and 246 and two P-inefficient varieties- 292 and 453 were selected to measure gene expression of BnaA03g05800D, BnaA03g05820D, BnaA03g05830D and BnaA03g05850D by RT-qPCR. In comparison to P-inefficient varieties, the P-efficient varieties had a higher relative expression of BnPAP17 (BnaA05g22460D) in both root and shoot at LP. The relative expression of BnRACK1C (BnaA05g22070D) (Supplementary Fig. S5) was higher in the root at both P levels but lower in shoot at NP. BnaA03g05800D expression was higher in the root and shoot at both P levels, the relative expression of BnaA03g05820D was higher in the shoot at NP and in the root at LP, the relative expression of BnaA03g05830D was higher in the root at LP, and the relative expression of BnaA03g05850D was lower in shoot at LP (Fig. 3). This provides further evidence that the four putative genes were closely associated with P efficiency.
Figure 3.
Relative expression of differentially expressed genes (DEGs) in the BnA3Hap haplotype region located on chromosomes A3. (a) BnaA03g05800D. (b) BnaA03g05820D. (c) BnaA03g05830D. (d) BnaA03g05850D *P < 0.05; **P < 0.01. Root at NP (R + P); Root at LP (R-P); Shoot at NP (S + P); Shoot at LP (S-P). P –efficient- 121 and 246 and –inefficient varieties- 292 and 453.
3.6 BnA3Hap haplotypes associated RSA traits and geographical distribution
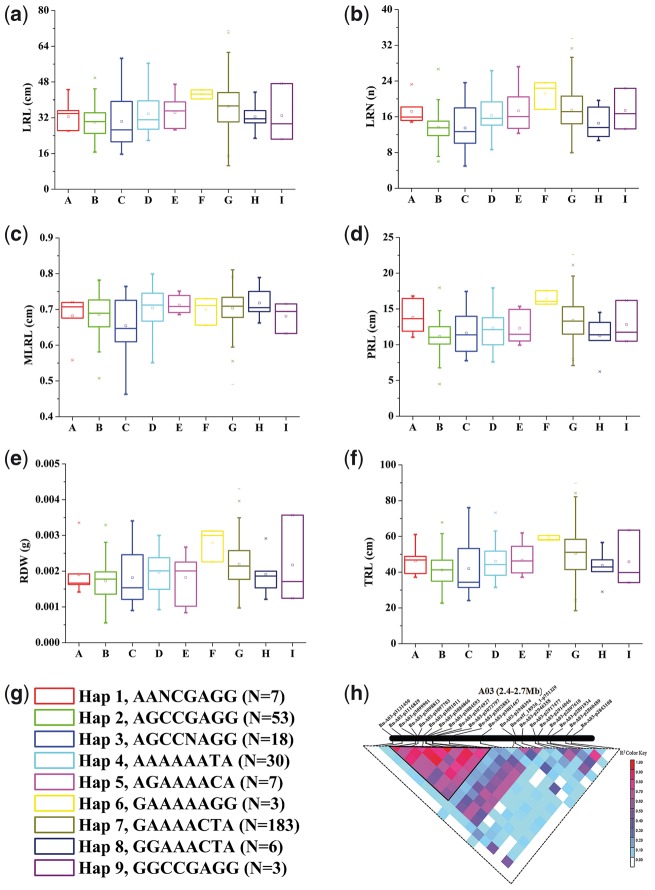
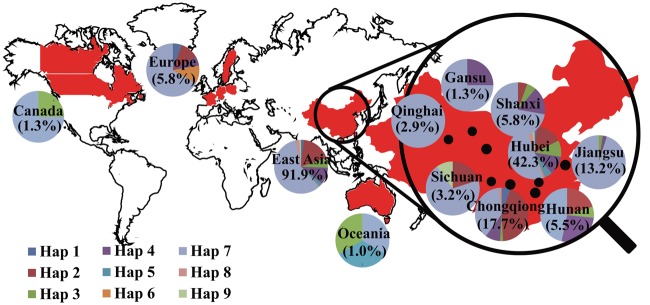
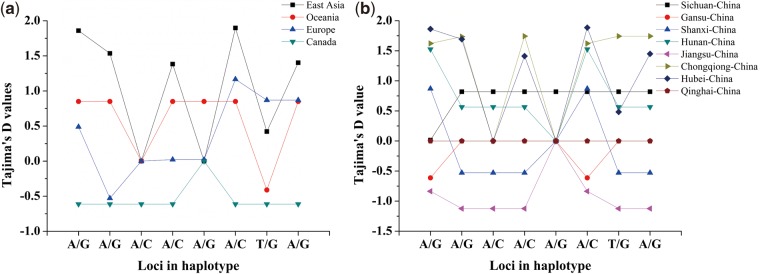
There were 21 SNPs located in the QTL confidence intervals on A3 (Fig. 4h). Eight successive SNPs located near SNP Bn-A03-p3052882 at 25.6 kb and the SNP Bn-A03-p3001467 at 75.2 kb formed nine haplotypes (Fig. 4g), designated ‘BnA3Hap haplotypes’. The varieties with haplotype 6 (GAAAAAGG) had higher average values in all the investigated RSA traits than those with the other haplotypes (Fig. 4a–f); two of the three varieties with haplotype 6 originated from European countries. Whereas, the varieties with haplotype 2 (AGCCGAGG) had lower average values in all of the investigated RSA traits than those with the other haplotypes (Fig. 5). Fifty-one of the 53 varieties with haplotype 2 originated from China. Haplotype 7 (GAAAACTA) was the most common haplotype (n = 183) and the RSA parameters of Haplotype 7 varieties were intermediate to haplotypes 6 and 2 varieties, respectively (Fig. 4a–f). The geographic distribution of the varieties was East Asia (285), Oceania (3), Europe (18) and North America (Canada) (4). Moreover, in China, the varieties distributed in Sichuan (10), Gansu (4), Shanxi (18), Hunan (17), Jiangsu (41), Chongqing (55), Hubei (131) and Qinghai (9) provinces (Fig. 5). Tajima’s D test of each SNP loci of BnA3Hap in B. napus indicated varieties from Gansu, Shanxi and Jiangsu provinces of China and Canada showed haplotype- BnA3Hap selection (Fig. 6).
Figure 4.
The haplotypes associated with RSA (root system architecture) traits in the ‘pouch and wick’ system (PS) at low P (LP). (a). Lateral root length (LRL). (b) Lateral root number (LRN). (c) Mean lateral root length (MLRL). (d) Primary root length (PRL). (e) Root dry weight (RDW). (f) Total root length (TRL). (g) BnA3Hap haplotypes. (h) The BnA3Hap haplotypes on A3 (darker colour trilateral).
Figure 5.
The geographical distribution of the BnA3Hap haplotypes in the B. napus panel. The percentage of the varieties in a region accounted for in the panel is shown in the circle.
Figure 6.
The Tajima’s D values of geographical distribution in BnA3Hap haplotypes.
The alignment of BnaA03g05800D, BnaA03g05820D, BnaA03g05830D and BnaA03g05850D in B. napus was identical to that of their orthologous genes in B. rapa, however, they differed from orthologs genes in A. thaliana. One gene was located on chromosome 3, and three were located on chromosome 5 of A. thaliana, suggesting hybridization between B. rapa and B. oleracea, while the BnA3Hap was conserved in the A genome (Supplementary Fig. S6).
4 Discussion
4.1. Genetic diversity, relative kinship and LD
In previous B. napus association panels, two subgroups were identified in 192 B. napus accessions32 and 523 B. napus accessions,21 but three subgroups in 472 B. napus accessions.20 In the present study, 405 varieties of the panel of Liu21 were used and similarly divided into two subgroups (Fig. 1). The LD, gene diversity, PIC, and relative kinship were slightly different between Subgroup1 and Subgroup2 (Fig. 1). As compared with European B. napus, the Chinese B. napus accessions studied had a more diverse genetic background (Supplementary Table S1), due to greater recombination in the A subgenome introduced through inter-specific hybridizations between B. napus and B. rapa. The LD in the C genome is greater than in the A genome (Table 3), but lower in PIC and gene diversity, indicating that the C genome may contribute more valuable alleles to adapt to different growth environments. In addition, the LD decay observed here- 1.06 Mb was different from previous B. napus panels, which observed 0.5–1 cM in 192 inbred lines,32 2.0 Mb in 472 inbred lines20 and 6.5 Mb in 523 inbred lines,14 and larger than that of other species, e.g. 100 kb–1 Mb in rice,33 1–100 kb in maize12 and 250 kb in Arabidopsis.34 The comparatively high resolution provided by association mapping is dependent upon the LD of genome.14 The low LD detected in this study indicated that this B. napus panel is suitable for association analysis and has the potential to identify SNPs in a narrow intervals equivalent to the distance of LD decay of 1.06 Mb. In addition, the number of SNPs, the length of LD decay, and the SNP density on each chromosome were different in this study (Table 3), similar to the studies with 52314 and 47220 accessions.
In this study, we did not investigate the association between the population structure and the breeding history, as the cultivars in this panel lack breeding information history. The subpopulations in a panel of 70 tomato lines corresponded to historical patterns of breeding conducted for specific production environments.35 However, the population structure in a 472 accessions Brassica napus population showed only weak association with breeding history.36
Structural variation of this B. napus panel was evaluated by gsrc, an R package for genome structure rearrangement calling.37 The C subgenome had more deletions and duplications than the A subgenome (Supplementary Fig. S7a). The number of CNVs in the region of ‘BnA3Hap’ was particularly low and therefore they have been not included in our association model. Genome structure rearrangements are a common phenomenon in allopolyploid species, which have been shown to affect phenotypic traits, and therefore it is of interest to monitor them on a genome wide scale in the future to detect further QTL. Subgenome C contains many CNVs and requires a deeper analysis to validate these and account for them in the association. For some regions (e.g. A10) CNVs were detected in the majority of lines in the panel and indicate a systematic effect.
4.2. Candidate genes identified for RSA
In this study, only 9 out of 285 SNPs colocalized with previously identified QTLs for RSA traits, which can be attributed to three reasons. First, as compared with QTL mapping, GWAS can identify relatively higher resolution genetic loci from underlying traits because natural populations incorporate ancient recombination events. Second, RSA traits in the BnaTNDH population were screened at low and high P with a ‘pouch and wick’9 and an agar culture systems,3 respectively. RSA traits of GWAS population were screened using a solution culture system at both low and high P and a ‘pouch and wick’ system at low P. The growth conditions influenced genotypic ranking and the range of variability.38 Third, uncorrelated data may be the result different environmental parameters between experiments.
The response of RSA traits to low Pi availability in B. napus has been well characterized.3,8 With low P availability, typically a reduction in the development of the primary roots occur,4,8,9 and increases the number and length of lateral roots are observed.3,35LPR2 (At1g71040) was shown to regulate primary root development in Arabidopsis under low Pi availability. It is a paralogue of Arabidopsis LPR1 (At1g23010), which encodes a multicopper oxidase protein, which are essential for the reducing root growth observed when root tips contact low-Pi media.5,39 The RACK1 orthologous gene in Arabidopsis regulates lateral root formation by changing the sensitivity of the plant to gibberellin, brassinosteroid, abscisic acid, and auxin.40
The genes associated with significant SNPs for root traits in the loci of target QTL were predicted to be the candidate genes for P efficiency. BnaA03g05800D, BnaA03g05820D, BnaA03g05830D and BnaA03g05850D were predicted to be closely associated with P efficiency, which were also located in the confidence interval of the QTL for primary root length at low P as well as the LD decay region of the peak SNP Bn-A03-p3001467 at 2.49–2.72 Mb on A3 (Fig. 2). All genes had significantly different expression in the P-efficient cultivar ‘Eyou Changjia’ between low P and high P (Fig. 2). The differential expression of the putative candidate genes in response to low-P stress was also confirmed in the P-efficient and P-inefficient accessions (Fig. 4). BnaA03g05800D in B. napus is orthologous to AT5G16010 in Arabidopsis, which encodes a 3-oxo-5-alpha-steroid 4-dehydrogenase family protein that moves between various organs under normal or nutrient-limiting conditions.41BnaA03g05820D is orthologous to AT3G02530 encoding a TCP family transcription factor.42BnaA03g05830D is homologous to AT5G16110, which regulates lateral root development.43 Along with MEX1, the AT5G16150- plastidic GLC translocator (PGLCT) is essential for the growth and development of starch degradation products from chloroplasts and starch-mediated pathway for photoassimilate export in Arabidopsis.
BnaA03g05800D, BnaA03g05820D, BnaA03g05830D and BnaA03g05850D had differential SNPs within the 5’ UTR or introns; the A/G alleles of lead SNP Bn-A03-p3073797 were located in the introns of BnaA03g5800D, A/C alleles of lead SNP Bn-A03-p3084593 were located in the introns of BnaA03g5820D, A/G alleles of lead SNP Bn-A03-p3091011 were located in the introns of BnaA03g5830D, and the A/G alleles of lead SNP Bn-A03-p3097703 were located in introns of BnaA03g5850D. These polymorphisms may cause differential gene expression between P-efficient and P-inefficient accessions (Figs. 3 and 4h). The characterization of the genetic elements associated with these traits could therefore provide elite alleles for the breeding of crops adapted to low P.
4.3. BnA3Hap haplotypes
Breeders have matched functionally different P-tolerance alleles to specific environments during selection. Oilseed rape was selected as a crop 300–400 years ago.36 In this study, the SNPs within candidate genes (BnaA03g5800D, BnaA03g5820D, BnaA03g5830D and BnaA03g5850D) on A3 formed a haplotype ‘BnA3Hap’, contributes to B. napus tolerance to low P stress (Fig. 4g and h). BnA3Hap haplotypes were detected at low P under the ‘pouch and wick’ system (Fig. 4a–f), and verified under the hydroponic system for two years. In addition, the Tajima’s D values of BnA3Hap haplotypes in B. napus varieties were strongly associated with the distribution of the agronomic P surpluses in soil44 (Figs. 5 and 6). Compared to ancestor species (B. rapa and A. thaliana), the order of the four orthologous genes of BnA3Hap haplotypes in A3 in B. napus showed collinearity with that in B. rapa, and were slightly different to that in A. thaliana (Supplementary Fig. S6), which indicates that this haplotype contributed to six RSA traits at LP (Fig. 4a–f) that were conserved during B. napus breeding. The discovery of the optimal haplotype of BnA3Hap can now enable the accurate selection of B. napus with higher P efficiencies and improve our understanding of the molecular mechanisms underlying P efficiency in plants.
Supplementary data
Supplementary data are available at DNARES Online.
Conflict of interest
None declared.
Funding
The authors acknowledge the financial support from the National Nature Science Foundation of China (Grant No. 31471933), the UK Biotechnology and Biological Sciences Research Council (BBSRC) (Crop Improvement Research Club, CIRC, Project BB/J019631/1; International Partnering Award BB/J020443/1), New Century Excellent Talents in University of Ministry of Education of China (Grant No. NCET-13-0809), Natural and Fundamental Research Funds for the Central Universities of China (Grant No. 2014PY020; 2662015PY105).
Supplementary Material
References
- 1. Kochian L.V. 2012, Plant nutrition, rooting for more phosphorus. Nature, 488, 466–7. [DOI] [PubMed] [Google Scholar]
- 2. Lynch J.P. 2015, Root phenes that reduce the metabolic costs of soil exploration, opportunities for 21st century agriculture. Plant Cell Environ., 38, 1775–84. [DOI] [PubMed] [Google Scholar]
- 3. Shi L., Shi T., Broadley M.R., et al. 2013, High-throughput root phenotyping screens identify genetic loci associated with root architectural traits in Brassica napus under contrasting phosphate availabilities. Ann. Bot-London., 112,381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hammond J.P., Broadley M.R., White P.J., et al. 2009, Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. J. Exp. Bot., 60, 1953–68. [DOI] [PubMed] [Google Scholar]
- 5. Svistoonoff S., Creff A., Reymond M., et al. 2007, Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet., 39, 792–6. [DOI] [PubMed] [Google Scholar]
- 6. Wissuwa M., Wegner J., Ae N., Yano M.. 2002, Substitution mapping of Pup1, a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet., 105, 890–7. [DOI] [PubMed] [Google Scholar]
- 7. Gamuyao R., Chin J.H., Pariasca-Tanaka J., et al. 2012, The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature, 488, 535–9. [DOI] [PubMed] [Google Scholar]
- 8. Yang M., Ding G., Shi L., Feng J., Xu F., Meng J.. 2010, Quantitative trait loci for root morphology in response to low phosphorus stress in Brassica napus. Theor. Appl. Genet., 121, 181–93. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Thomas C.L., Xiang J., et al. 2016. QTL meta-analysis of root traits in Brassica napus under contrasting phosphorus supply in two growth systems. Sci. Rep., 6, 33113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang X., Wei X., Sang T., et al. 2010, Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet., 42, 961–7. [DOI] [PubMed] [Google Scholar]
- 11. Courtois B., Audebert A., Dardou A., et al. 2013, Genome-wide association mapping of root traits in a japonica rice panel. PLoS One, 8, e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Remington D.L., Thornsberry J.M., Matsuoka Y., et al. 2001, Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA, 98, 11479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian F., Bradbury P.J., Brown P.J., et al. 2011, Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet., 43, 159–62. [DOI] [PubMed] [Google Scholar]
- 14. Xu L., Hu K., Zhang Z., et al. 2016, Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res., 23, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamfwa K., Cichy K.A., Kelly J.D.. 2015. Genome-wide association analysis of symbiotic nitrogen fixation in common bean. Theor. Appl. Genet., 128, 1999–2017. [DOI] [PubMed] [Google Scholar]
- 16. Mori A., Fukuda T., Vejchasarn P., Nestler J., Pariasca-Tanaka J., Wissuwa M.. 2016, The role of root size versus root efficiency in phosphorus acquisition in rice. J. Exp. Bot., 67, 1179–89. [DOI] [PubMed] [Google Scholar]
- 17. Zhang D., Song H., Cheng H., et al. 2014, The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet., 10, 229–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosas U., Cibrian-Jaramillo A., Ristova D., et al. 2013, Integration of responses within and across arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc. Natl. Acad. Sci. USA, 110, 15133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Wang H., Liu S., et al. , Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet., 48, 1233–41. [DOI] [PubMed] [Google Scholar]
- 20. Li F., Chen B., Xu K., et al. 2014, Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res., 21, 355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu S., Fan C., Li J., et al. 2016, A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet., 129, 1203–15. [DOI] [PubMed] [Google Scholar]
- 22. Wei L., Jian H., Lu K., et al. 2016, Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J., 14, 1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bus A., Korber N., Parkin I.A., et al. 2014, Species- and genome-wide dissection of the shoot ionome in Brassica napus and its relationship to seedling development. Front. Plant Sci., 5, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cox C.B., Moore P.D., Ladle R.. 2016, Biogeography: An Ecological and Evolutionary Approach. Wiley-Blackwell., ISBN 978-1-118-96858-1, pp.106. [Google Scholar]
- 25. Thomas C.L., Graham N.S., Hayden R., et al. 2016, High-throughput phenotyping HTP, identifies seedling root traits linked to variation in seed yield and nutrient capture in field-grown oilseed rape (Brassica napus L.). Ann. Bot-London., 118, 655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atkinson J.A., Wingen L.U., Griffiths M., et al. 2015, Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot., 66, 2283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu P., Lv Z., Zhang X., et al. 2013, Identification of molecular markers linked to trilocular gene (mc1) in Brassica juncea L. Mol. Breeding, 33, 425–34. [Google Scholar]
- 28. Chalhoub B., Denoeud F., Liu S., et al. 2014, Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345, 950–3. [DOI] [PubMed] [Google Scholar]
- 29. Pritchard J.K., Stephens M., Rosenberg N.A., Donnelly P.. 2000, Association mapping in structured populations. Am. J. Hum. Genet., 67, 170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evanno G., Regnaut S., Goudet J.. 2005, Detecting the number of clusters of individuals using the software STRUCTURE, a simulation study. Mol. Ecol., 14, 2611–20. [DOI] [PubMed] [Google Scholar]
- 31. Duan H., Shi L., Ye X., Wang Y., Xu F.. 2009, Identification of Phosphorous Efficient Germplasm in Oilseed Rape. J. Plant Nutr., 32, 1148–63. [Google Scholar]
- 32. Xiao Y., Cai D., Yang W., et al. 2012, Genetic structure and linkage disequilibrium pattern of a rapeseed Brassica napus L., association mapping panel revealed by microsatellites. Theor. Appl. Genet., 125, 437–47. [DOI] [PubMed] [Google Scholar]
- 33. Zhao K., Tung C.W., Eizenga G.C., et al. 2011, Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun., 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nordborg M., Borevitz J.O., Bergelson J., et al. 2002, The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet., 30, 190–3. [DOI] [PubMed] [Google Scholar]
- 35. Sim S.C., Robbins M.D., Van Deynze A.,et al. 2011, Population structure and genetic differentiation associated with breeding history and selection in tomato (Solanum lycopersicum L.). Heredity, 106, 927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang N., Li F., Chen B., et al. 2014, Genome-wide investigation of genetic changes during modern breeding of Brassica napus. Theor. Appl. Genet., 127, 1817–29. [DOI] [PubMed] [Google Scholar]
- 37. Grandke F., Snowdon R., Samans B.. 2017. gsrc-an R package for genome structure rearrangement calling. Bioinformatics, 33, 545–6. [DOI] [PubMed] [Google Scholar]
- 38. Nestler J., Wissuwa M.. 2016, Superior root hair formation confers root efficiency in some, but not all, rice genotypes upon p deficiency. Front. Plant Sci., 7, e75997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reymond M., Svistoonoff S., Loudet O., Nussaume L., Desnos T.. 2006, Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ., 29, 115–25. [DOI] [PubMed] [Google Scholar]
- 40. Guo J., Wang J., Xi L., Huang W.D., Liang J., Chen J.G.. 2009, RACK1 is a negative regulator of ABA responses in Arabidopsis. J. Exp. Bot., 60, 3819–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thieme C.J., Rojas-Triana M., Stecyk E., et al. 2015, Endogenous arabidopsis messenger RNAs transported to distant tissues. Nat. Plants, 1. [DOI] [PubMed] [Google Scholar]
- 42. Jin J., Zhang H., Kong L., Gao G., Luo J.. 2014, PlantTFDB 3.0, a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res., 42, 1182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laskowski M., Biller S., Stanley K., Kajstura T., Prusty R.. 2006, Expression profiling of auxin-treated Arabidopsis roots, toward a molecular analysis of lateral root emergence. Plant Cell Physiol., 47, 788–92. [DOI] [PubMed] [Google Scholar]
- 44. MacDonald G.K., Bennett E.M., Potter P.A., Ramankutty N.. 2011, Agronomic phosphorus imbalances across the world's croplands. Proc. Natl. Acad. Sci. USA, 108, 3086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.