Abstract
Small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), play a variety of important regulatory roles in many eukaryotes. Their small size has made it challenging to study them directly in live cells. Here we describe an RNA-based fluorescent sensor for small RNA detection both in vitro and in vivo, adaptable for any small RNA. It utilizes an sxRNA switch for detection of miRNA–mRNA interactions combined with a fluorophore-binding sequence ‘Spinach’, a GFP-like RNA aptamer for which the RNA–fluorophore complex exhibits strong and consistent fluorescence under an excitation wavelength. Two example sensors, FASTmiR171 and FASTmiR122, can rapidly detect and quantify the levels of miR171 and miR122 in vitro. The sensors can determine relative levels of miRNAs in total RNA extracts with sensitivity similar to small RNA sequencing and northern blots. FASTmiR sensors were also used to estimate the copy number range of miRNAs in total RNA extracts. To localize and analyze the spatial distribution of small RNAs in live, single cells, tandem copies of FASTmiR122 were expressed in different cell lines. FASTmiR122 was able to quantitatively detect the differences in miR122 levels in Huh7 and HEK293T cells demonstrating its potential for tracking miRNA expression and localization in vivo.
INTRODUCTION
Small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), are 21–24 nt in size, and play a variety of important regulatory roles including developmental regulation, activation of pathogen defenses and epigenetic regulation in both plants and animals (1–4).
Plant small RNAs are produced as RNA duplexes from longer RNA precursors cut by endonuclease activity of DICER-LIKE proteins (2). Thereafter, one strand of a small RNA duplex is incorporated into the RNA-induced silencing complex (RISC) that includes Argonaute (AGO). In animals, mature miRNAs are generated from precursor molecules (pre-miRNAs) by the action of two RNase III-type endonucleases, Drosha and Dicer. Following Dicer cleavage, the resulting RNA is loaded onto an AGO protein to generate the RISC (5). Small RNAs in the RISC target complementary sequences, directing endonucleolytic cleavage (6), translational repression/activation (7), DNA methylation (8) or heterochromatin formation (9). However, tools for the detection and localization of mature small RNAs are laborious, static, low in resolution and prone to artefacts. Current methods include RNA gel blots (northerns), DNA-nanosilver clusters (DNA/AgNC), in situ hybridization and fluorescent-protein-based sensors (10–12). The DNA/AgNC fluorescent sensor detects miRNAs in vitro by a decrease in fluorescence rather than an increase in fluorescence (12). Both northern blots and DNA/AgNC are inappropriate for analyses of subcellular localization. In situ hybridization can precisely localize small RNAs in plant and animal tissues (10,13,14), but requires fixed tissue, paraffin-embedding and many subsequent steps. Fluorescent–protein-based small RNA sensors have been developed to indirectly detect miRNAs, such as miR156 and miR171, by measuring changes in fluorescence of GFP protein resulting from decreases in GFP transcripts targeted by miRNAs; but this approach has limited spatial and temporal resolution (11,15). Moreover, none of these approaches can monitor live-cell subcellular dynamics or transiently expressed miRNAs.
Genetically encoded fluorescent tagging of RNAs is possible by fusion of a target RNA with a fluorescent RNA aptamer, Spinach, that exhibits strong and consistent fluorescence upon binding of a fluorophore like DFHBI (3,5-difluoro-4-hydroxybenzylidene imidazolinone) (16). The Spinach structure is divided into paired regions 1, 2, 3 (P1–3) and junctions between them, J1–2 and J2–3 (17,18), as shown in Figure 1A. DFHBI can fluoresce when it binds between the top platform of the G-quadruplex and a Hoogsteen-paired U and A of the base triple of J2–3 (highlighted in blue in Figure 1A) of Spinach (17,18). Cellular metabolite sensors based on Spinach could detect a variety of small molecules in vitro and in Escherichia coli (19,20). Recently, Aw et al. described a Spinach-based Pandan sensor to detect miRNAs in vitro. Pandan is a circular bimolecular sensor with a destabilized G-quadruplex and base triplet required for DFHBI binding (21). Binding of miRNA re-forms the DFHBI-binding pocket of the sensor which leads to increased fluorescence in vitro (21).
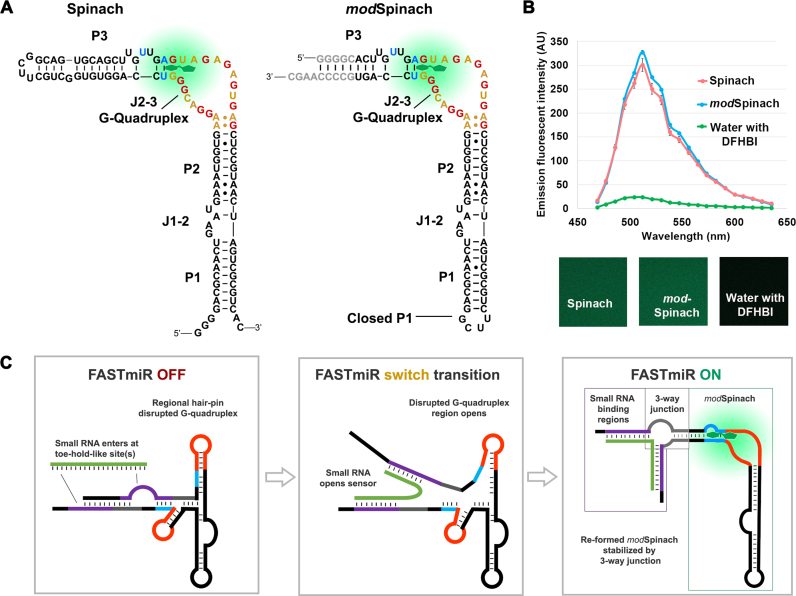
Figure 1.
Modification of original Spinach and sensor design. (A) Structure of the original Spinach and the modified Spinach (modSpinach) drawn according to Huang et al. (18). There are three paired regions, P1, P2 and P3. The two junctions between the paired regions are J1–2 and J2–3 (17). The G’s from the G-quadruplex are highlighted in red. A’s and U’s from the same region are highlighted in gold. DFHBI (green) binds between the G’s comprising the top G-quartet and the Hoogsteen-paired (blue) U and A of the base triple of J2–3. To create modSpinach, P1 was closed and P3 was opened and replaced with the 24–2 min Spinach sequence. The bases colored gray were used to stabilize the modSpinach, but not used in the sensor designs. The crucial G-quadruplex and base triple required for DFHBI binding were unaltered in the modSpinach. (B) Fluorescence emission spectra of modSpinach was nearly identical to the original Spinach and was obtained from spectral confocal micrographs (lower panels). The water control shows the background level of free, unbound DFHBI. (C) Overview diagram of the FASTmiR sensor design. The FASTmiR sensor relies on a sxRNA switch that has both an OFF-state and ON-state. The DFHBI binding regions consisting of the G-guadruplex (orange) and the base triple (blue) are disrupted in the OFF-state. The small RNA binding regions (purple) form toe-hold-like site(s) (unpaired) that allow the small RNA target (green) to initiate binding. This potentially destabilizes the OFF-state which allows the FASTmiR sensor to switch (middle panel) to the ON-state (right panel). Once in the ON-state, the DFHBI binding pocket reforms and then fluoresces. The switch may happen also happen independently of the small RNA, but the ON-state is only stabilized when bound by the target small RNA via a three-way junction (3WJ). The ON-state also depicts the three modular components of FASTmiR sensors: the small RNA binding regions, the base of the 3WJ (gray) and the modSpinach.
Here we describe a variation of Spinach that functions as a fluorescent sensor of small RNAs both in vitro and in vivo. This sensor can directly monitor in vitro the levels of miRNAs or other small RNAs, and quantify miRNA from RNA extracted from tissue. We also modified our sensor for live-cell, real-time imaging of small RNAs. We describe the difficulties of detecting Spinach in vivo that can be partially overcome by generating a tandemly repeated version of the FASTmiR sensor stabilized by three-way junctions (3WJs).
MATERIALS AND METHODS
Modification of the original Spinach and design of small RNA sensor
Based on the original Spinach structure 24–2 and 24–2 min, the uucg tetra loop was picked for sxRNA switch reinforcement. The loop structure of 24–2 min was used in this loop. We then closed the original open loop with the same uucg tetra loop sequence. Sequences reverse complement to the small RNAs were added to the sxRNA switch for the small RNA sensors. Secondary structure prediction was performed using the RNAfold and RNAcofold software from the Vienna RNA package (22).
The modified Spinach (modSpinach) sequence is: GCACTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTCGCGTCTTCGGACGCAACTGAATGAAATGGTGAAGGACGGGTCCAGTGCCCC
Sequences for small RNA sensors are as follows:
psRS2 (FASTmiR122): CAAACACCATTGTCAAAAGGGTACTTGTTGAGTAGAGTGTGAGCTCCGTA ACTAGTCGCGTCTTCGGACGCAACTGAATGAAATGGTGAAGGACGGGTCCAGTGCCTACACTCCA
psRS5 (FASTmiR171): GATATTGGCAAAGGGTACTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGT CGCGTCTTCGGACGCAACTGAATGAAATGGTGAAGGACGGGTCCAGTGCCTAGCGGCTCAATCA
6xFASTmiR122
GGGCAAACACCATTGTCAAAAGGGTACTTG TTGAGTAGAGTGTGAGCTCCGTAACTAGTTAC ATCTTCGGATGTAACTGAATGAAATGGTGAAG GACGGGTCCAGTGCCTACACTCCAACTAGTGG GCAAACACCATTGTCAAAAGGGTACTTGTTGA GTAGAGTGTGAGCTCCGTAACTAGTTACATCTTC GGATGTAACTGAATGAAATGGTGAAGGACGGG TCCAGTGCCTACACTCCAACTAGTGGGCAAAC ACCATTGTCAAAAGGGTACTTGTTGAGTAGAG TGTGAGCTCCGTAACTAGTTACATCTTCGGATGT AACTGAATGAAATGGTGAAGGACGGGTCCAGT GCCTACACTCCAACTAGTGGGCAAACACCATT GTCAAAAGGGTACTTGTTGAGTAGAGTGTGAG CTCCGTAACTAGTTACATCTTCGGATGTAACTGA ATGAAATGGTGAAGGACGGGTCCAGTGCCTAC ACTCCAACTAGTGGGCAAACACCATTGTCAAA AGGGTACTTGTTGAGTAGAGTGTGAGCTCCGT AACTAGTTACATCTTCGGATGTAACTGAATGA AATGGTGAAGGACGGGTCCAGTGCCTACACTC CAACTAGTGGGCAAACACCATTGTCAAAAGGG TACTTGTTGAGTAGAGTGTGAGCTCCGTAACT AGTTACATCTTCGGATGTAACTGAATGAAATG GTGAAGGACGGGTCCAGTGCCTACACTCCA
>hsa-miR-122–5p MIMAT0000421
UGGAGUGUGACAAUGGUGUUUG
>ath-miR171a MIMAT0000202
UGAUUGAGCCGCGCCAAUAUC
Construct design
For in vitro assays, modSpinach, FASTmiR122 and FASTmiR171 constructs were synthesized with GenScript (Piscataway, NJ, USA) (vector pUC57-Kan). The FASTmiR171_Design5 and FASTmiR122_Design5 constructs were made using Q5® Site-Directed Mutagenesis Kit from NEB (New England BioLabs, Inc.) using primer pairs (23 and 24, 25 and 26) according to manufacturer's instructions.
For expression of Spinach in plants, the Spinach sequence was polymerase chain reaction (PCR) amplified using primers 1 and 2 (Supplementary Table S1) using the original Spinach vector from the Jaffrey Lab (pAV-5S-Spinach). The positive control and the small RNA sensor sequences were synthesized with GenScript (Piscataway, NJ, USA) with the att sites corresponding with Gateway vector pGWB461 (P35S-TagRFP-attR1-Cmr-ccdB-attR2-TNOS). A stop codon was placed before the sensor sequence to prevent translation of the sensor. Then the sensors were introduced into pGWB461 using Gateway® LR Clonase II Enzyme mix (Thermo Fisher Scientific).
For expression of Spinach and FASTmiR in mammalian cells, the positive control and the small RNA sensor sequences were synthesized with GenScript, and each piece was amplified with BamHI and XbaI sites using primers 9 and 10 (for the original Spinach), 11 and 12 (for the modified Spinach), 13 and 14 (for FASTmiR122), 15 and 16 (for FASTmiR171), 21 and 22 (for Spinach2), with all primer sequences listed in Table S1. PCR products were digested with BamHI and XbaI and ligated to the mammalian expression vector pcDNA3.1(+) from Thermo Fisher Scientific.
Generation of FASTmiR for in vitro assays
DNA was fused with T7 promoter and amplified using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Scientific). The PCR products were transcribed in vitro using MEGAshortscriptTM T7 transcription kit (Thermo Fisher Scientific) according to the manufacturer's instructions. RNA was purified by phenol:chloroform extraction and alcohol precipitation.
All RNAs were tagged with the T7 promoter and transcribed in vitro using MEGAshortscriptTM T7 Kit (ThermoFisher Scientific, Inc), and dissolved in high-salt buffer (10 mM Tris 100 mM NaCl, and 1 mM MgCl2). The same amount of each RNA (5 μM) or water control was heated to 95°C for 10 min and slowly cooled down to room temperature. DFHBI (400 μM) was incubated with each sample for 15 min prior to imaging. For all in vitro experiments, we used RNase-free, HPLC purified small RNA synthesized by IDT (Integrated DNA Technologies).
Linear dose-response curve measurements
Concentrations of 0, 25, 50 and 75 μM miR122 or miR171 were incubated with their respective sensors (FASTmiR122 and FASTmiR171) in μ-Plate Angiogenesis 96 well plate (ibidi, GmbH). Fluorescence intensity was measured using Zeiss LSM 710 confocal microscope with 458 nm excitation and 500–550 nm band pass GaAsP detector and EC Plan-Neofluar 40x/1.3 Oil DIC lens. The laser power was kept at 65% and pinhole was wide open to collect all the light. Reflected light was collected at 420–480 nm to mark the bottom of each well. Then we collect the Spinach signal 20 μm above the bottom of the each well into the solution using a 505–550 nm band-pass filter.
miRNA detection in total RNA
Total RNA from Arabidopsis was isolated using TRI Reagent (Molecular Research Center) according to the manufacturer's instructions with an additional 48 h at −80°C during the precipitation step in isopropanol. RNA was quantified using a Qubit® 2.0 Fluorometer RNA BR (Broad-Range) Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
For the FASTmiR sensor detecting miR171 (Figure 3C and D), 1 μg of FASTmiR171 and 1 ug flower/leaf total RNA extract were incubated at room temperature for 30 min with 400 μM DFHBI. Images were taken with Zeiss LSM 710 confocal with 458 nm excitation and 500–550 nm band pass GaAsP detector and EC Plan-Neofluar 40x/1.3 Oil DIC lens. We used 2′ O-methyl small RNA from TriLink Biotechnologies to build the standard curve in vitro. To check whether FASTmiR171 was able to detect changes and was specific to miR171 in a background of total RNA, the reverse complement of miR171 (designated as miR171-rc, with the sequence GATATTGGCGCGGCTCAATCA [MW = 5613.6 g/mole], underlined nucleotides are LNAs) was added to total RNA from Arabidopsis inflorescences, prior to incubation (final concentration either 20, 30 or 40 pM) at room temperature. LNA locked probes from Exiqon were used for higher sensitivity, specificity and higher melting temperatures. A total of 500 μM of miR171 was added to leaf RNA to assess whether FASTmiR171 could detect an increase of miR171 levels in the total RNA extract. For FASTmiR122 sensor detecting miR122 in total Huh7 RNA extraction, the reverse complement of miR122 (miR122-rc, with the sequence CAAACACCATTGTCACACTCCA (MW = 6830.5 g/mole, TM = 91°C), underlined nucleotides are LNAs) was added prior to folding (final concentration 600–900 pM) and incubated at 95°C for 10 min. FASTmiR122 sensor was added when the mixture cooled down to room temperature, and then incubate at 75°C for 5 min for the sensor to bind residual RNA. DFHBI was added after the solution was cooled to room temperature and incubated for 15 min before imaging.
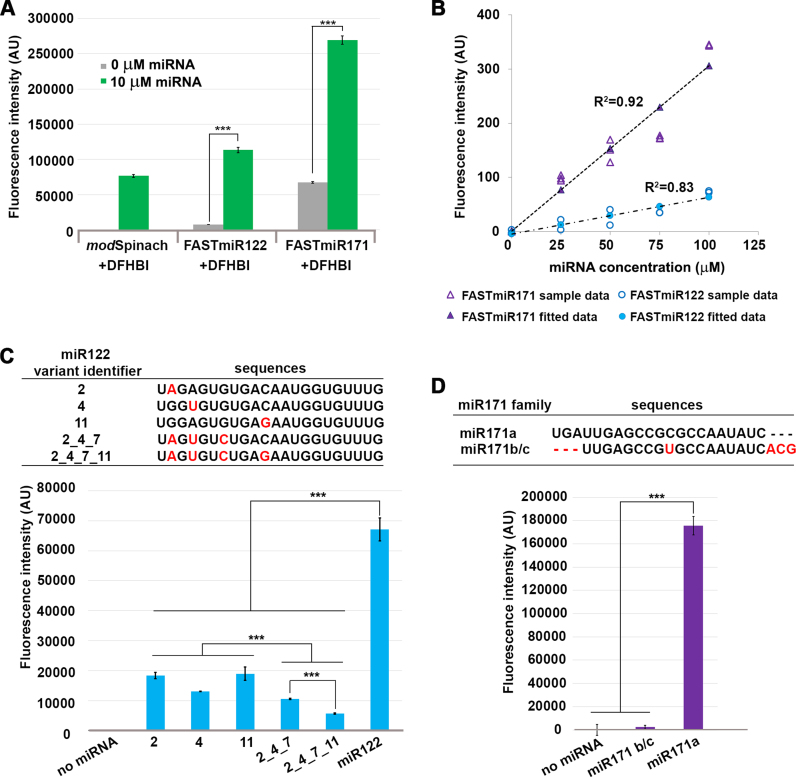
Figure 3.
Characterization of FASTmiR122 and FASTmiR171 sensors in vitro. (A) Fluorescence intensity of FASTmiR122 and FASTmiR171 with 0 or 10 uM of cognate miRNAs. FASTmiR122 with miR122 show fluorescence equivalent to the modSpinach positive control. FASTmiR171 with miR171 showed an increased fluorescence compared to the 0 mM miR171 negative control, but less than the modSpinach positive control. Fluorescence values were normalized by subtracting background DFHBI fluorescence in the water control (number of considered samples, n = 4). (B) Dose-response curves of FASTmiR122 and FASTmiR171. Fluorescence intensity increased linearly with increasing concentrations of miRNAs (0, 25, 50, 75 and 100 μM miR122 (or miR171). Fluorescence values were normalized to the sensor alone background DFHBI fluorescence. (C and D) Specificity of the FASTmiR122 and FASTmiR171 sensors to their cognate miRNAs. FASTmiR122 showed significantly higher fluorescence when incubating with miR122, compared to miR122 variant with 1–4 point mutations. FASTmiR171 showed significantly higher fluorescence when incubated with its specific miR171 target, miR171a compared to its naturally occurring family member miR171b/c or no miRNA. Fluorescence values were normalized by subtracting background DFHBI fluorescence in the water control (n = 4). Significance codes: ‘***’ for <0.001, ‘**’ for <0.01, ‘*’ for < 0.05.
Expression of Spinach in plants
Arabidopsis ecotype Col-0 was grown under a 20 h day/4 hr night cycle, at 24°C. Mature leaves and flower buds were collected 5–6 weeks after planting. Sensors were transiently expressed in planta by Agrobacterium-mediated transient expression in leaves (‘Agroinfiltration’). Agrobacterium strain GV2260 containing each sensor vector was cultured for 2 days on LB plates, then resuspended in infiltration medium containing 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone, and induced at 28°C for 2 h. Four-week-old Nicotiana benthamiana plant grown under 24-h light was used for infiltration. All vectors were coinfiltrated with GV2260 Agrobacterium containing P19. Leaves were harvested 5 days post-infiltration for imaging.
Mammalian cell transfection
HEK293 or HEK293T cells were transfected using Nucleofector™ (Lonza), Lipofectamine 3000 (Thermo Fisher Scientific) or FuGeneHD (Promega) according to manufacturer's instructions (eGFP was used as a positive control). Huh7 cells were transfected using Lipofectamine 3000. For Nucleofector™ transfection, 2 μg DNA was used for each transfection. For FuGeneHD and Lipofectamine 3000 transfection, a 4-well Nunc™ chamber slide (Thermo Fisher Scientific) was coated with 300 μl of poly-L-lysine (PLL) overnight at 4°C. The next morning, the chambers were washed once with ddH2O and UV sterilized for 5 min. The chambers were further coated with 50 μl of rat collagen-I and laminin (1:100) for 2 h at 37°C. After removing the coating solution by washing 2× with 1 ml ddH2O, cells were plated at 80 000 cells/ml. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) + 10% Fetal Bovine Serum (FBS) and switched to DMEM + 10% FBS without dye (Thermo Fisher Scientific, catalog # 31053036) before imaging. HEK293T cells were transfected with 0.4 μg of DNA the next day (25–40% confluent) with FuGeneHD (Promega, catalog #E2311). For each transfection, 0.6 μg DNA and 1.65 μl FuGeneHD solution was used. Huh7 cells were transfected with 0.625 μg of DNA. Imaging experiments were performed 24 h after transfection. Lentivirus expression of miR122 (a gift from Alexander Ploss, Princeton University) (23) in HEK293T cells was performed as previously described by adding HEK293T 333 μl of lentivirus stock (4 μg/ml polybrene, 20 mM HEPES). HEK293TCells were imaged by confocal microscopy at 0, 1 and 5 h post inoculation. Cells lines were authenticated via Biomolecular Core Laboratory (Nemours/A. I. duPont Hospital for Children).
Fluorescence microscopy and live cell imaging of Spinach
Live plant imaging (Supplementary Figure S3A) was performed using a Zeiss LSM 780 confocal microscope (Carl Zeiss) with a 40x/1.2 C-Apochromat water immersion objective lens. The 458 nm laser line and emission filter 464–560 nm was used to detect Spinach and the Spinach-based sensors. The 561 nm laser line and emission filter 569–648 nm was used to detect the tagRFP signal. To collect the spectra, emitted light was collected from 469–635 nm under lambda mode (Supplementary Figure S3). Linear spectral unmixing of images was conducted using the Zen 2010 software (Carl Zeiss).HEK293 (Supplementary Figure S5 and 6) cells were imaged using the Zeiss LSM 710 confocal microscope and an EC Plan-Neofluar 40x/1.3 Oil DIC lens. Samples were excited with 30% 458 nm laser. Spinach signal was collected using a Bi-GaAsP (BiG) detector using 500–550 band pass filter. For imaging Spinach, 30 min prior to experiment, we replaced the HEK293 cell media with pre-warmed imaging media (DMEM with no phenol red or vitamins and supplemented with 25 mM HEPES pH 7.4, 5 mM MgSO4 and 5 μg/ml Hoechst 33342). In addition, 80 μM DFHBI or DFHBI-1T (diluted from 40 mM stock in DMSO) was added to the cells. For imaging 6× tandem repeats, images were taken using a Zeiss LSM 880 confocal microscope (Carl Zeiss) with EC Plan-Neofluar 40×/1.30 Oil DIC objective, pinhole 99 μm, under 488 nm excitation. Live-cell images were processed using median function (3 × 3 kernel size) in Zen 2010 software (Carl Zeiss).
Fluorescent in situ hybridizations
HEK293T or Huh7 cells were grown directly onto PLL coated coverslips in DMEM + 10% FBS, washed once in 1× phosphate buffered saline (PBS) (100 mM Na2HPO4, 20 mM KH2PO4, 137 mM NaCl, 27 mM KCl, pH 7.4) and fixed for 10 min at room temperature in 4% paraformaldehyde, 1× PBS. After two washes in 1× PBS, cells were permeabilized by treatment with 70% ethanol 2 h at 4°C. A 20 min protease (protease from Streptomyces griseus, Sigma-Aldrich, cas # 9036–06-0) treatment in TE buffer (final concentration 65 μg/ml) was followed by 0.2% glycine treatment in 1× PBS 2 min, 1× PBS for 2 min, 95% ethanol for 1 min and 100% ethanol for 1 min. Cells were then hybridized overnight at 55°C in 100 μl of a mixture containing 10% dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNAse-free bovine serum albumin, 40 μg E. coli tRNA, 2×Saline-Sodium Citrate buffer (SSC), 50% formamide, 30 ng of the miR122 probe or a scrambled probe (see below). After hybridization, cells were washed twice for 30 min at the appropriate stringency: 0.5× SSC, 55°C and rinsed twice in Tris Buffered Saline (TBS). Digoxigenin-labeled probes were detected with sheep anti-digoxigenin antibodies (1/500), and then with donkey anti-sheep antibodies conjugated to AlexaFluor647 (1/1000). Slides are incubated overnight at 4°C with primary antibody, then washed in washing buffer three times for 20 min at room temperature. Slides were incubated overnight at 4°C with secondary antibody, then washed in washing buffer three times for 20 min at room temperature. For final mounting, samples were washed in 1× TBS, and mounted in slow-fade gold with DAPI (Thermo Fisher Scientific).
Probe sequence: AAACACCATTGTCACACTCCA/3Dig_N/.
Scrambled probe sequence: GTGTAACACGTCTATACGCCCA/3Dig_N/.
Statistical and computational analysis
Significances were calculated using a standard t-test. Significance codes indicate the following: ‘***’ for <0.001, ‘**’ for <0.01, ‘*’ for < 0.05. Dose-response curves were fitted using simple linear regression using all the sample data collected. P-value for each slope were calculated using ANOVA. HEK 293 and Huh7 libraries were all acquired from GenBank's Gene Expression Omnibus (GEO). The accession IDs for the HEK 293 cells are GSE46254, GSE43666 and the sample GSM1536635. The accession ID for the one Huh7 library is GSM1908181. SRA files for each sample were downloaded and converted into FASTQ format where they had their Illumina adapters removed using Trimmomatic v0.32 (24). These reads were then mapped to the human genome assembly hg19 with bowtie v1.1.2 (25,26). We then manually searched the mapped reads for the presence of hsa-miR122 in each of these libraries and quantified the counts of each small RNA. hsa-miR122 was identified in three libraries, and those abundances were then normalized to reads per 30 million.
RESULTS
Spinach-based small RNA sensor design
To construct this sensor, we first modified the Spinach sequence to open the paired stem-loop region (P3). P3 is adjacent to the critical base triple of junction J2–3 and the G-quadruplex region where DFHBI binds and also corresponds to the same stem loop that was destabilized previously to form a Spinach-based sensor for cellular metabolites (19). We replaced P3 with the 24–2 min Spinach stem (16) and closed the remaining open loop with the UUCG tetraloop to form a modSpinach (Figure 1A) (27). Our modSpinach had nearly identical fluorescent intensity as the original Spinach, with the same excitation (458 nm) and emission (∼520 nm) peaks, detected by laser scanning confocal microscopy (Figure 1B).
Next, we designed the sensor out of three modular components, the RNA binding regions, the base of a 3WJ and modSpinach (Figure 1C). Only the small RNA binding regions need to be changed for different small RNA targets; the junction and modSpinach components remained unchanged. Rather than simply destabilizing the open stem (19), we employed an sxRNA (structurally interacting RNA) (28) switch designed to have a discrete OFF-state in the absence of a targeted small RNA and an ON-state that forms a stable 3WJ (28) between the two small RNA binding paired regions and the opened P3 region of the modSpinach sequence (Figure 1C) (28,29). The non-paired bulge at the base of the 3WJ provides sufficient flexibility to include the entire sequence of the small RNA target. In comparison, the Pandan sensor requires two sequence-specific, unpaired nucleotides at the split position (21). The small RNA complementary binding sequence was split into two parts near the 5′ and 3′ ends of the sensor. The choice of the split position impacts the predicted OFF-state folding (see next paragraph and ‘Discussion’ section). The ON-state reforms the base triple and G-quadruplex so that DFHBI can bind between to create fluorescent sensor. We name the sensors FASTmiR, for Fluorescent Aptamer Sensor for Tracking microRNAs. To adapt this design to any small RNA only the small RNA binding region needs to be changed according to the target small RNA sequence. Below we describe how our design was implemented for two different miRNAs.
Design of FASTmiR122 and FASTmiR171 sensors
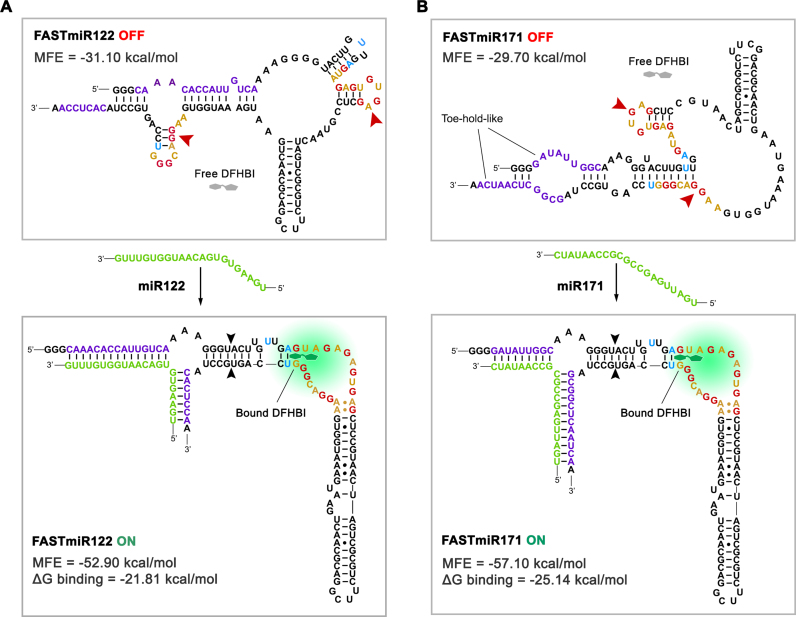
We used our sensor design to create FASTmiR sensors to human miR122 and Arabidopsis miR171 (Figure 2). The critical step in the design process is deciding on the split position of the two small RNA binding regions of the sensor near the 3′ and 5′ ends. Changing this position greatly affects the switching between the OFF- and ON-state. We examined five different split position designs along the miRNAs for either FASTmiR122 or FASTmiR171 (Supplementary Figure S1) and examined three criteria: (i) a minimum free energy (MFE) sensor bound to its target small RNA should be lower than the sensor alone; (ii) disruption of the G-quadruplex; and (iii) a toe-hold-like binding region (30) that allows the small RNA to open up the OFF-state. All of the five different split positions resulted in a negative ΔG MFE and a disrupted G-quadruplex, but only three different designs had a toe-hold-like structure in the OFF-state, suggesting this is the most important criterion (Supplementary Figure S1B). Designs with predicted toe-hold-like structures were selected and used to generate the final FASTmiR122 and FASTmiR171 sensors (Figure 2). We also examined FASTmiRs lacking toe-hold-like structures to verify their importance and found that there was almost no change in FASTmiR122 (Design5) or FASTmiR171 (Design5) fluorescence when incubated with their target miRNAs (Supplementary Figure S1C and D). We hypothesize that without the toe-hold-like structures the FASTmiRs are locked in the OFF-state. Furthermore, FASTmiR171 with miR171 showed higher fluorescence than FASTmiR122 with miR122 and had two rather than one toe-hold-like structures. When designing a new sensor, these criteria should be used as a guide, but multiple split positions may need to be examined since these are based only on a prediction of the MFE structure of the OFF-state. The ON-state is known, since the crystal structure of Spinach has been described (17). Any given RNA sequence, such as the OFF-state sensor, will have an MFE structure prediction as well as a host of sub-optimal energy structures (31).
Figure 2.
OFF- and ON-states of FASTmiR122 and FASTmiR171. (A) The predicted structure of the FASTmiR122 OFF-state and the ON-state with the miR122 target. The OFF-state contains a disrupted base triple and G-quadruplex that forms two small hairpin structures (arrowheads). Free DFHBI (gray) cannot bind and then fluoresce. The miR122 (green) binds and opens the structure, resulting in a re-formed modSpinach (right of black arrowheads) that can bind DFHBI and then fluorescence. The FASTmiR122 ON-state bound by miR122 is favored thermodynamically (MFE = −52.90 kcal/mol) compared to the unbound OFF-state (MFE = −31.10 kcal/mol. ΔG binding =−21.81 kcal/mol). (B) The predicted structure of the FASTmiR171 OFF-state and the ON-state with the miR171 target. The G-quadruplex of the OFF-state is disrupted by one hairpin structure and one paired region (arrowheads). The OFF sequence has two toe-hold-like regions where the miR171 (green) can enter, bind and open the OFF-state structure. The FASTmiR sensor then switches to the ON-state that has a reformed modSpinach (right of arrowhead) that can fluoresce. The FASTmiR171 ON-state bound by miR171 is favored thermodynamically (MFE = 57.10 kcal/mol) compared to the unbound OFF-state (MFE =−29.7 kcal/mol. ΔG binding= −25.14 kcal/mol). The OFF-states in A and B are drawn based on predicted structure from Vienna RNA secondary structure server. MFE and ΔG binding were calculated using Vienna RNAfold and RNAcofold (version 2.1.1). The ON-states in A and B are drawn according to Huang et al. (18).
FASTmiR sensors can specifically detect cognate miRNAs and can linearly detect miRNA levels in vitro
Without miRNA binding, the sensors remained at their lowest free-energy structural status and yielded only background fluorescence after incubation with the fluorophore DFHBI (Figure 3A). Incubation of FASTmiR122 with miR122, and FASTmiR171 with miR171, resulted in high fluorescence compared to the control (Figure 3A). To assess the sensor's effectiveness for in vitro assays, we tested whether a linear increase in miRNA resulted in a linear increase in sensor fluorescence. Concentration series from 0 to 100 μM of miR122 or miR171 were incubated with a fixed concentration of their respective sensors. Both sensors generated linear increases in fluorescence with increasing concentrations of miRNA (Figure 3B). This assay was conducted in a 96-well plate on a standard laser-scanning confocal microscope, showing that FASTmiR sensors can be used for large scale, rapid detection of miRNAs. Next, we tested whether FASTmiR sensors can recognize their cognate miRNAs. The seed region is important for binding to mRNA target for animal miRNAs (32,33). So we generated four types of mutations to miR122: (i) 1 nt mutation in the seed region (variants 2 and 4); (ii) 1 nt mutation in the non-seed region (variant 11); (iii) 3 nt mutations in the seed region (variant 2_4_7); and (iv) 4 nt mutations including the seed region (variant 2_4_7_11) (Figure 3C). The combination of FASTmiR122 with miR122 showed higher levels of fluorescence compared with miR122 variants that have 1, 3 or 4 point mutations (Figure 3C). miR122 variants with only one mutation showed significantly higher fluorescence than three or four mutations. miR122 variant 2_4_7_11 gave the least fluorescence, almost 12-fold different from the original miR122. And mutations in the seed region (2 and 4) showed lower fluorescence than a mutation in non-seed region (11) (4 versus 11 is significant). There are no closely related miRNAs to miR122; however, there are three miR171 family members (34). miR171a is unique and although miR171b and miR171c have different precursors, their mature miRNA sequence is identical. Our FASTmiR171 specifically detected miR171a and showed a 75-fold higher level of fluorescence when incubated with miR171a compared to miR171b/c (Figure 3D).
FASTmiR sensors can specifically detect cognate miRNAs in RNA extracts
Next, we examined if FASTmiR sensors can distinguish cognate miRNA in a complex RNA mixture. Here we took advantage of our knowledge of miRNA distribution in Arabidopsis; miR171 is enriched in Arabidopsis flowers and has very little or no expression in leaves (15). We examined if FASTmiR171 can directly quantify miR171 levels in Arabidopsis total leaf and flower RNA extracts, applied as an in vitro assay. FASTmiR171 was able to detect specific miRNA levels in the total RNA background. After incubation at room temperature, we observed significantly higher sensor fluorescence with total RNA extracts from flowers (Figure 4A, red) compared with total RNA extracts from leaf tissue (Figure 4A, green). We further tested whether this increase of fluorescence was due to binding of miR171 to the FASTmiR171 by adding miR171 to the leaf extract. Artificial introduction of miR171 into leaf total RNA extracts increased FASTmiR171 fluorescence (Figure 4B). The added miR171 was unmodified and not LNA, suggesting FASTmiR171 can specifically detect endogenous miR171.
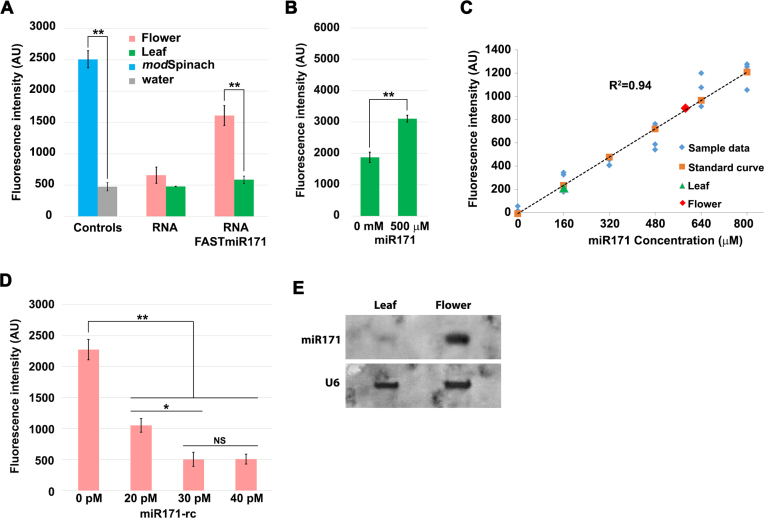
Figure 4.
FASTmiR171 can directly measure miR171 concentration in Arabidopsis total RNA extracts. (A) FASTmiR171 detected endogenous miR171 levels in a complex mixture of RNA in total RNA extracted from Arabidopsis flowers or leaves. FASTmiR171 sensor incubated with total flower RNA extracts showed significantly higher fluorescence intensity (pink) compared with FASTmiR171 incubated with leaf total RNA extracts (green) (n = 4). (B) FASTmiR171 sensor detected the addition of miR171 to total leaf extracts. Fluorescence increased significantly after adding synthetic miR171 into total leaf RNA extracts. (C) In vitro quantification of small RNAs with a FASTmiR sensor. A standard curve of FASTmiR171 fluorescence was generated with different concentrations of miR171 (0, 160, 320, 480, 640 and 800 μM) (n = 3). Fluorescence values were normalized to the sensor alone background DFHBI fluorescence. Linear regression P-value: 2.82134E-11. (D) Depleting the miR171 pool with an LNA reverse-complemented miR171 (miR171-rc) decreased FASTmiR171 fluorescence. A total of 20, 30 or 40 pM miR171-rc was first added to total flower RNA extracts to quench the available miR171 (n = 3). After the addition of FASTmiR171, there was a reduced amount of fluorescence in the sample pre-treated with all concentrations of miR171-rc, but no significant difference between 30 and 40 pM miR171-rc. Fluorescence values were normalized to the background fluorescence of flower RNA alone with DFHBI. (E) Northern blot of 20 μg of total RNA extracts from Arabidopsis flowers and leaves. Similar to FASTmir171, northern blotting detected a higher level of miR171 in total flower RNA extracts compared to total leaf RNA extracts. The digoxigenin-labeled miR171-rc (same as in D) was used as the probe. U6 was used as loading control. Significance codes: ‘***’ for <0.001, ‘**’ for <0.01, ‘*’ for < 0.05. ‘NS’ for no significance.
miR122 is highly expressed in hepatic cells and qRT-PCR shows that the relative miR122 level is much higher in the hepatic Huh7 cell line compared to the HEK293T cell line (35). We extracted total RNA from Huh7 and HEK293T cells and incubated them with our FASTmiR122 sensor. The FASTmiR122 sensor was able to detect a significantly higher amount of miR122 in Huh7 total RNA extract, compared with HEK293T RNA extract or total RNA extracted from Arabidopsis flowers, which was used as a control containing no miR122 (Supplementary Figure S2A).
FASTmiR sensors can more rapidly measure the range and estimate the copy number of miRNAs compared to other methods
To approximate the relative concentration of miR171 in leaf and flower RNA extracts, we created a standard curve using a concentration series of 0–800 μM of miR171 and FASTmiR171 (Figure 4C). Based on the standard curve, we were able to calculate the relative leaf and flower miR171 concentrations. Our FASTmiR171 determined that the flower RNA extracts had 3.6-fold higher miR171 compared to leaf RNA extracts. The miR171 concentrations were verified by small RNA sequencing. The normalized sRNA read count of miR171 was ∼4.3-fold higher in flowers (1826 transcripts per 10M, TP10M; GSM893112) compared to leaves (422 TP10M, GSM984196), which is comparable to the 3.6-fold higher measurement via our sensor.
To measure absolute concentrations of miR171 we used a more specific and sensitive FASTmiR assay that included the reverse complement LNA-locked miR171 (miR171-rc). We pre-incubated 1 μg of total flower RNA extract with picomolar concentration of miR171-rc. This method of quantification has two advantages. First, the LNAs of the miR171 can quench the pool of miR171 in the RNA extracts, due to the much higher affinity of LNAs (36,37) and FASTmiR171 cannot out compete the LNA miR171-rc. Second, this approach is more specific because the reduction in FASTmiR171 fluorescence by the LNA miR171-rc is dependent on the specificity of the LNA miR171-rc and not the specificity of the FASTmiR171 sensor. This is advantageous because LNA probes in general show high levels of specificity (36,37), overcoming any potential non-specific detection by FASTmiR sensors. We found that 20, 30 and 40 pM concentrations of miR171-rc all reduced the fluorescence of 1 μg of FASTmiR sensor (Figure 4D). We saw no significant difference in fluorescence between 30 and 40 pM of miR171-rc, suggesting 30 pM sufficiently titrated out all of the miR171. Therefore, in 10 pg of total flower RNA extract there were an estimated 1204–1807 copies of miR171. It has been estimated that 10 pg of total RNA can be extracted from an individual mammalian cell (38); therefore, we can use the same method to estimate the copy number of miR122 per individual Huh7 cell. Total RNA extracts from Huh7 cells were incubated with 600, 700, 800 and 900 pM of miR122-rc (Supplementary Figure S2B). We estimated between 33 723 and 38 541 copies of miR122 in 10 pg RNA, with the calculation performed as in Supplementary Table S2), which is equivalent to a single Huh7 cell (39). This range is higher but in concordance with previously reported copy number of miR122 in Huh7 cells (∼16 000 copies) (35,38–40). We were also able to detect significantly higher fluorescence in Arabidopsis leaf RNA extract with addition of 800 pM miR122. Then 800 pM addition of miR122-rc was able to quench the miR122 pool (Supplementary Figure S2C).
We also compared our detected fold difference with northern blots of the same RNA extract (Figure 4E). Quantification of miRNAs using FASTmiR sensors is faster than sequencing or northern blots; FASTmiR sensors require only RNA extraction, in vitro transcribed sensor and a fluorescence detector. Another advantage of our method is that once the sensor is made, a large number of samples can be easily tested using 96-well plates and a plate-reader or an automated confocal microscope.
Spinach-based fluorescent aptamers are difficult to detect in vivo in plants and mammalian cell lines
Live-cell imaging with Spinach is challenging due to the low fluorescence of the RNA aptamer, as well as quick dissociation of fluorophore DFHBI (∼100 ms) upon excitation (41). Spinach fluorescence was not detectable when transiently over-expressed in N. benthamiana leaves by Agrobacterium infiltration (Supplementary Figure S3A). The expression was verified using a tagRFP reporter and by reverse transcriptase polymerase chain reaction (RT-PCR) (Supplementary Figure S3B). We hypothesized that the cell wall may interfere with DFHBI uptake; we tested this by using plant cell protoplasts, in which the cell wall has been enzymatically removed. We were able to detect background levels of DFHBI in the protoplasts; however, we failed to detected reliable signal above that background DFHBI fluorescence in cells expressing Spinach (Supplementary Figure S3C and D).
Next, we examined Spinach and modSpinach in mammalian cells. First, we tested both constructs in Chinese Hamster Ovary (CHO) cells; however, we failed to detect any reliable signal above the DHFBI background fluorescence. We confirmed the expression by RT-PCR (data not shown). We next tested the exact protocol and constructs used by the Jaffrey laboratory (42), but we were unable to detect an increase in fluorescence in cells transfected with Spinach, Spinach2 or modSpinach compared to the negative controls in HEK293 or HEH293T cells (Supplementary Figures S4 and 5). Unlike previous reports on Spinach (16), sucrose stress treatment did not induce an increase in signal. Spectral analysis of the non-transfected controls showed signal in granules surrounding HEK293 cells was likely background DFHBI signal (Supplementary Figure S5B).
Tandemly repeated FASTmiR sensors can localize and monitor changes of small RNAs in vivo
Previous studies suggested that Spinach2 might be superior for live-cell imaging and tandem copies might be required for detection in live cells (42,43). Tandemly repeated Spinach2 applied in a multiplex CRISPR detection system in live cells showed enhanced fluorescence (43). Similarly, a tandemly repeated Spinach consisting of 8–64 repeats was required to detect RNA in E. coli (44). Therefore, to improve FASTmiR sensors for live-cell imaging, we made the sensor with Spinach2 and generated a 6× tandem repeat of the sensor sequence (6×FASTmiR122) and 6×modSpinach2 (Supplementary Figure S6). We omitted the previously-used tRNA stabilizer (16), as it would result in a large amount of additional sequence in a 6× tandem array and has been shown to induce cleavage, promoting faster degradation (45). In vitro experiments showed that 6×FASTmiR122 is still responsive specifically to miR122, but the overall fluorescence of 6×FASTmiR122 in vitro is very low (Supplementary Figure S7).
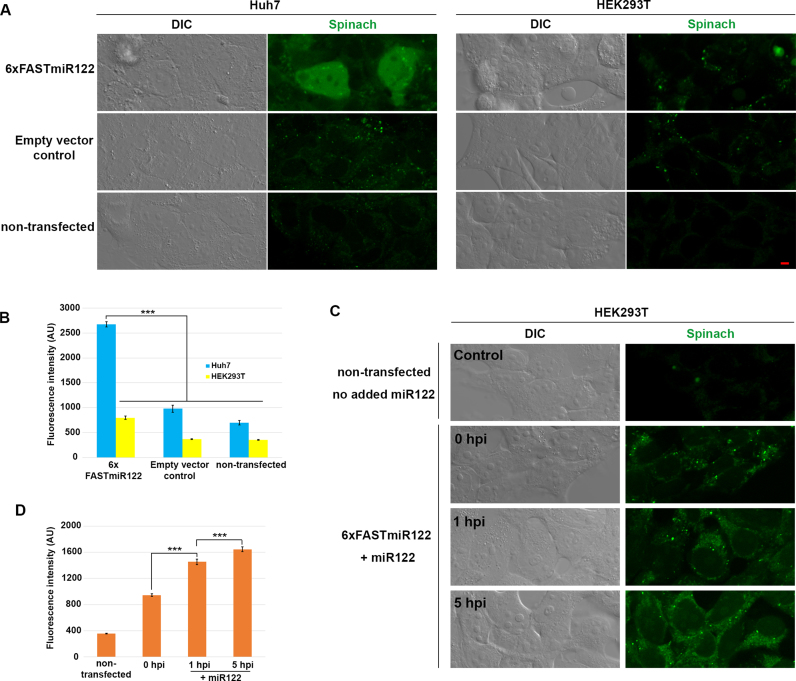
To determine if our FASTmiR122 sensor can detect differences in miR122 in vivo, we compared the miR122-enriched Huh7 cells and the miR122-depleted HEK293T cells. Huh7 cells transfected with 6×FASTmiR122 construct showed significantly higher fluorescence (Figure 5A and B) compared to HEK293T cells transfected with the same vector. Non-transfected cells or cells transfected with empty vector showed similar background level fluorescence. miR122 mainly localized in the nucleus and cytosol in Huh7 cells. We hypothesize that the 3WJ between our sensor and the miRNA stabilizes the DFHBI-1T binding pocket (45). Previous reports show that binding of miRNAs with mRNA by splitting two non-contiguous regions may be used to reinforce the base of stem-loop motif (28,46). RNA nanoparticles with a 3WJ core have been shown to be resistant to 8M urea and stable in serum even at extremely low concentration both in vitro and in vivo (47).
Figure 5.
FASTmiR122 sensor localized miR122 and detects changes of miR122 in live mammalian cells. (A) The 6xFASTmiR122 sensor detected higher level of miR122 in nuclei and the cytosol of Huh7 cells (green, row 1). The empty vector (row 2) and non-transfected control (row 3) showed similar levels of background fluorescence. (B) Quantification of fluorescence in nuclei showed substantially stronger fluorescence intensity in Huh7 cells transfected with 6xFASTmiR122 sensors compared to the controls (n = 13). (C) HEK293T cells were transfected with the 6xFASTmiR122 sensor and miR122 was expressed using a lentivirus delivery system. Fluorescence increased 1 hpi (hours post infection) and 5 hpi compared with 0 hpi and cells without the 6xFASTmiR122 sensor (non-transfected control). (D) Quantification of nucleus-localized signal for miR122-expressing HEK293T cells (n = 15). Scale bars = 5 μM for all images. Error bars show standard error. Significance codes: ‘***’ for <0.001, ‘**’ for <0.01, ‘*’ for < 0.05, ‘NS’ for no significance (n = 10).
We analyzed several published small RNA libraries (GSM1067869, GSM1067867 and GSM1908181), and verified the difference of miR122 in HEK293 and Huh7 cells (Supplementary Table S3). We also performed fluorescent in situ hybridization with a miR122 probe and verified the localization of miR122 signal detected with our 6×FASTmiR122 sensor in Huh7 cells (Supplementary Figure S8A). We confirmed the expression of 6×FASTmiR122 RNA with RT-PCR using RNA from the cells shown in Figure 5 (Supplementary Figure S8B).
In order to further test if 6×FASTmiR122 sensor can track miR122 abundance differences in live cells, we increased miR122 using a lentivirus delivery system (23). The FASTmiR122 sensor was first transfected in HEK293T cells and then miR122 was expressed for 0, 1 or 5 h. We observed a gradual increase in FASTmiR122 fluorescence following delivery and expression of miR122. After 1 and 5 h of miR122 expression, HEK293T cells showed significantly higher FASTmiR122 fluorescence in nuclei (including nucleoli) and the cytosol, similar to Huh7 cells (Figure 5A and C).
DISCUSSION
In summary, we have demonstrated a fluorescent, RNA aptamer-based small RNA sensor that can rapidly quantify small RNAs in vitro and directly detect and localize small RNAs in vivo. Our sensors can be used to detect their cognate small RNAs in total RNA extracts, and could be used for rapid, large-scale screening and quantification of candidate miRNAs. Furthermore, we demonstrated that a FASTmiR sensor can be used in vivo to localize specific miRNAs.
Similar to DNA/AgNC miRNA sensors (48), our method is faster and easier than traditional miRNA quantification methods. The northern blot assay requires a long, labor-intensive protocol handling radioactive probes (radiolabeled) or expensive DIG-labeled LNA probes. Quantitative reverse transcription PCR (qRT-PCR) is sensitive and specific, but only medium-throughput with respect to the number of samples that can be processed per day, and it relies on reverse transcription of miRNA to cDNA, which is prone to error (49). Small RNA sequencing is high-throughput, but requires substantial computational support and is costly (49). Quantification of miRNAs using FASTmiR sensors is fast and low-cost, which only requires RNA extraction, an in vitro transcribed sensor sequence and a confocal microscope. We chose confocal microscopy as our detection method because it was readily available to our research group and because most researchers have access to a modern confocal microscope. An alternative detection method is a sensitive fluorescence microplate reader and others have shown that this method can detect Spinach-based sensors (21). In either case, once the sensor has been designed and tested, a large quantity of samples can be easily measured using a 96-well plate format. We have used automated confocal microscopy to measure 96-well plates, but a fluorescence microplate reader may be more suitable for large-scale studies.
FASTmiR detection of small RNAs does not require chemical synthesis and uses standard nucleic acid laboratory techniques. FASTmiR directly detects small RNAs rather than using amplification or indirect detection, enabling absolute quantification. We were able to estimate that 10 pg of total flower RNA extract has between 1204 and 1807 copies of miR171. The precision of that measurement probably can be improved using additional concentrations of miR171-rc. Furthermore, it could be used to estimate the average absolute concentrations of miRNAs in cells. This approach has been used previously for DNA/AgNC miRNA sensors (48), and here we used a similar approach to estimate 33 723–38 541 copies of miR122 per Huh7 cell. Our estimation is similar but higher than the previously reported copy number of miR122 in a Huh7 (∼16 000 copies), which was estimated by an RNase protection assay (39). The discrepancy may be caused by differences in either the probe; our method utilizes LNA (locked nucleic acid) probes, which provide more affinity and specificity to target small RNAs compared to 32P-labeled RNA probes (37). Alternatively, the difference in copy number could also be due to differences in the cell growth phase when the materials were harvested, differences in culture conditions, or differences in RNA extraction efficiency.
Both the FASTmiR122 and FASTmiR171 easily detected their targets, suggesting that the basic design of FASTmiR sensors is robust and can be modified to specifically detect other small RNAs. The only sequence that needs to be changed for a given small RNA is the binding site regions. We used the Vienna RNA package to search for an OFF-state that forms small loops within the critical sequences comprising the G-quadruplex, and then miRNA binding changes the conformation to an ON-state in which the fluorophore is bound. The findings by Aw et al., show that Spinach-based sensors can detect a wide variety of small RNAs. Our design has advantages relative to the Aw et al. Spinach-based sensor design (21); FASTmiR sensors have a clear OFF-state, rather than an unstable or loose structure state and a clear ON-state stabilized by a 3WJ that locks in the binding of small RNA. The circularly permuted Pandan sensor with closed ends is not amenable to tandem repetition, a feature critical for RNA-based sensors in vivo (21,43,44). As a result, Pandan sensors may be more sensitive for in vitro assays, but a single FASTmiR sensor design can be used for both in vitro detection of miRNAs in total RNA extracts and in vivo detection of miRNAs in live cells. Although an RNA-based FASTmiR sensor is required for in vivo detection, a DNA-based fluorescent aptamer sensor may be more stable for in vitro assays.
Our extensive experimentation with Spinach-based RNA aptamers suggests that their fluorescence is difficult to detect in live cells and are challenging to use for live-cell imaging in mammalian and plant cells. We overcame those challenges for our sensor by combining various approaches to successfully detect the signal from Spinach. These approaches include using the improved Spinach variant, Spinach2, tandem duplications and a stabilizing 3WJ (43,44,50). Our sensor can also be used to detect miR122 levels and changes in miR122 levels, with regular imaging techniques, when the miRNA is highly enriched, e.g. Huh7 cells and HEK293T cells infected with lentivirus expressing miR122. Spinach-based sensors clearly require stabilization structures, such as a tRNA or F30 stabilizer (16,45). A functional RNA-based sensor in live cells may require additional stabilization or protection to prevent decay. Shechner et al. showed that fusion of Spinach2 to a deactivated Cas9 (dCas9)-loaded guide RNA, resulted in detectable fluorescence in the nucleus of live cells (43). Our sensor relies on a 3WJ that has been shown previously to stabilize RNAs (28).
Even though DFHBI can penetrate mammalian cell membranes, the concentration used in live-cell detection is very high (μM) compared to other live cell dyes (nM). Another challenge was that DFHBI tended to accumulate and fluoresce in dying or dead cells, resulting in misleading results. This is true in plant cells as well; however, we discovered that DFHBI could not successfully pass an intact plant cell wall; even an esterified version of DFHBI (ACO-DFHBI) that we synthesized and tested did not increase its permeability in plant cells (data not shown). Despite these challenges, our data suggests that our FASTmiR sensor can detect small RNAs and changes in small RNA levels in live cells, and should be easy to adapt to miRNAs of interest.
Future advances in fluorescent RNA aptamers will make new FASTmiR sensors even more robust for additional live-cell applications. This could include better stabilization and novel fluorescent RNA aptamers with higher fluorescence in vivo. As advances are made, exciting applications of fluorescent RNA aptamer sensors have the potential to revolutionize our ability to visualize the location and dynamics of small RNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alexander Ploss from Princeton University for Huh7 cells and the miR122-lentivirus delivery system, Jongyoun Baik at the Delaware Biotechnology Institute for CHO and HEK cell transfections, Yuchen Zhang in the Department of Chemistry at the University of Delaware for DFHBI and synthesis of its derivatives, Jenn Holbrook for her assistance with cell line authentication, and members of the Meyers and Caplan labs for support and helpful discussions. We also thank the Jaffrey lab for Spinach constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Science Foundation (NSF) via an ‘EAGER’ Award [1252939 to B.C.M., J.L.C.]; NSF Plant Genome Research Program [1339229 to B.C.M.]; National Institutes of Health [1R41GM110877, 1R41GM097811 to S.A.T., F.D., Z.E.W., in part]; NIH-NIGMS [P20 GM103446, S10 OD016361]; NSF [EPSCoR IIA-1301765]. Funding for open access charge: NSF IOS Award 1339229.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005; 579:5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones-Rhoades M.W., Bartel D.P., Bartel B.. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006; 57:19–53. [DOI] [PubMed] [Google Scholar]
- 3. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 4. Simon S.A., Meyers B.C.. Small RNA-mediated epigenetic modifications in plants. Curr. Opin. Plant Biol. 2011; 14:148–155. [DOI] [PubMed] [Google Scholar]
- 5. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S. et al. . The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003; 425:415–419. [DOI] [PubMed] [Google Scholar]
- 6. Llave C., Xie Z., Kasschau K.D., Carrington J.C.. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002; 297:2053–2056. [DOI] [PubMed] [Google Scholar]
- 7. Djuranovic S., Nahvi A., Green R.. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012; 336:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E.. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006; 38:721–725. [DOI] [PubMed] [Google Scholar]
- 9. Ekwall K. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell. 2004; 13:304–305. [DOI] [PubMed] [Google Scholar]
- 10. Javelle M., Timmermans M.C.P.. In situ localization of small RNAs in plants by using LNA probes. Nat. Protoc. 2012; 7:533–541. [DOI] [PubMed] [Google Scholar]
- 11. Nodine M.D., Bartel D.P.. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010; 24:2678–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang S.W., Vosch T.. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal. Chem. 2011; 83:6935–6939. [DOI] [PubMed] [Google Scholar]
- 13. Rozhkov N.V., Zelentsova E.S., Shostak N.G., Evgen’ev M.B.. Expression of Drosophila virilis retroelements and role of small RNAs in their intrastrain transposition. PLoS One. 2011; 6:e21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bak M., Silahtaroglu A., Moller M., Christensen M., Rath M.F., Skryabin B., Tommerup N., Kauppinen S.. MicroRNA expression in the adult mouse central nervous system. RNA. 2008; 14:432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O.. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004; 18:2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferre-D’Amare A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R.. Fluorescence imaging of cellular metabolites with RNA. Science. 2012; 335:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. You M., Litke J.L., Jaffrey S.R.. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. PNAS. 2015; 112:E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aw S.S., Tang M.X., Teo Y.N., Cohen S.M.. A conformation-induced fluorescence method for microRNA detection. Nucleic Acids Res. 2016; 44:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003; 31:3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vogt A., Scull M.A., Friling T., Horwitz J.A., Donovan B.M., Dorner M., Gerold G., Labitt R.N., Rice C.M., Ploss A.. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology. 2013; 444:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J.. Improved placement of multi-mapping small RNAs. G3. 2016; 6:2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C., Nikonov S., Dumas P.. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000; 304:35–42. [DOI] [PubMed] [Google Scholar]
- 28. Doyle F., Tenenbaum S.A.. Trans-regulation of RNA-binding protein motifs by microRNA. Front. Genet. 2014; 5:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doyle F., Lapsia S., Spadaro S., Wurz Z.E., Bhaduri-McIntosh S., Tenenbaum S.A.. Engineering Structurally Interacting RNA (sxRNA). Sci. Rep. 2017; 7:45393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Green A.A., Silver P.A., Collins J.J., Yin P.. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014; 159:925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zuker M., Jaeger J.A., Turner D.H.. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991; 19:2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narbus C.M., Israelow B., Sourisseau M., Michta M.L., Hopcraft S.E., Zeiner G.M., Evans M.J.. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J. Virol. 2011; 85:12087–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mullany L.E., Herrick J.S., Wolff R.K., Slattery M.L.. MicroRNA seed region length impact on target messenger RNA expression and survival in colorectal cancer. PLoS One. 2016; 11:e0154177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bari A., Orazova S., Ivashchenko A.. miR156- and miR171-binding sites in the protein-coding sequences of several plant genes. Biomed. Res. Int. 2013; 2013:307145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fukuhara T., Kambara H., Shiokawa M., Ono C., Katoh H., Morita E., Okuzaki D., Maehara Y., Koike K., Matsuura Y.. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with Hepatitis C Virus. J. Virol. 2012; 86:7918–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Braasch D.A., Corey D.R.. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001; 8:1–7. [DOI] [PubMed] [Google Scholar]
- 37. Vester B., Wengel J.. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004; 43:13233–13241. [DOI] [PubMed] [Google Scholar]
- 38. Bissels U., Wild S., Tomiuk S., Holste A., Hafner M., Tuschl T., Bosio A.. Absolute quantification of microRNAs by using a universal reference. RNA. 2009; 15:2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. et al. . miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 40. Oliveira K.G., Malta F.M., Nastri A.C., Widman A., Faria P.L., Santana R.A., Alves V.A., Carrilho F.J., Pinho J.R.. Increased hepatic expression of miRNA-122 in patients infected with HCV genotype 3. Med. Microbiol. Immunol. 2016; 205:111–117. [DOI] [PubMed] [Google Scholar]
- 41. Han K.Y., Leslie B.J., Fei J.Y., Zhang J.C., Ha T.. Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 2013; 135:19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strack R.L., Disney M.D., Jaffrey S.R.. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013; 10:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shechner D.M., Hacisuleyman E., Younger S.T., Rinn J.L.. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods. 2015; 12:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang J.C., Fei J.Y., Leslie B.J., Han K.Y., Kuhlman T.E., Ha T.. Tandem Spinach array for mRNA imaging in living bacterial cells. Sci. Rep. 2015; 5:17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filonov G.S., Kam C.W., Song W.J., Jaffrey S.R.. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 2015; 22:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lescoute A., Westhof E.. Topology of three-way junctions in folded RNAs. RNA. 2006; 12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shu D., Shu Y., Haque F., Abdelmawla S., Guo P.. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011; 6:658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah P., Choi S.W., Kim H.J., Cho S.K., Bhang Y.J., Ryu M.Y., Thulstrup P.W., Bjerrum M.J., Yang S.W.. Locking-to-unlocking system is an efficient strategy to design DNA/silver nanoclusters (AgNCs) probe for human miRNAs. Nucleic Acids Res. 2016; 44:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pritchard C.C., Cheng H.H., Tewari M.. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012; 13:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guet D., Burns L.T., Maji S., Boulanger J., Hersen P., Wente S.R., Salamero J., Dargemont C.. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat. Commun. 2015; 6:8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.