Abstract
Two of the many goals of synthetic biology are synthesizing large biochemical systems and simplifying their assembly. While several genes have been assembled together by modular idempotent cloning, it is unclear if such simplified strategies scale to very large constructs for expression and purification of whole pathways. Here we synthesize from oligodeoxyribonucleotides a completely de-novo-designed, 58-kb multigene DNA. This BioBrick plasmid insert encodes 30 of the 31 translation factors of the PURE translation system, each His-tagged and in separate transcription cistrons. Dividing the insert between three high-copy expression plasmids enables the bulk purification of the aminoacyl-tRNA synthetases and translation factors necessary for affordable, scalable reconstitution of an in vitro transcription and translation system, PURE 3.0.
INTRODUCTION
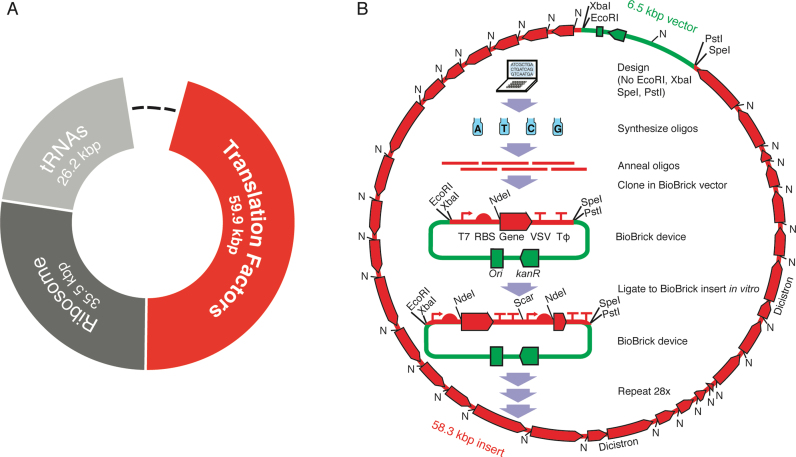
Two of the many goals of synthetic biology are making the design and construction of biological systems easier (1–12) and synthesizing a minimal cell (13–17). An important milestone towards both goals was synthesis of a 1080-kb near-minimal bacterial genome from oligodeoxyribonucleotides and transplantation into a host cell (17). This advance used nature's genome design, the order of the DNA parts needed to be pre-determined (for homologous recombinations in yeast and one Gibson assembly), and took 200 person-years of work at a cost of $40 million (18). The top-down approach to minimization from here involves exhaustive combinatorial knock-out experiments that are very informative about cellular principles (19) though applications are less obvious. Our complementary long-term approach to investigating core cellular principles is bottom-up theoretical design of an Escherichia coli (E. coli)-based minimal proteinaceous cell and its genome and stepwise modular construction (14). Along the way, construction of potentially functional pieces (e.g. Figure 1A) of this theoretical genome have been proposed to have utility for biopolymer syntheses (14). The main progress to date was synthesis of a 15-kb DNA encoding all 21 small ribosomal subunit proteins (part of the dark grey piece in Figure 1A), although expression of only a few genes was demonstrated (7). Here, we focus on the piece encoding the translation factor module (red in Figure 1A).
Figure 1.
De novo design of a DNA sequence encoding translation, and method for synthesis of its fragment that encodes translation factors. Synthesized sequences are in red; vector sequences are in green. (A) Modular design of a theoretical DNA sequence encoding all macromolecules necessary and sufficient for protein synthesis (a ‘translatome’ of 121.6 kb) based upon biochemistry central to E. coli. Module sizes are updated from (14) to include recently discovered enzymes (Supplementary Table S1). Dashes represent non-translation sequences of undefined size. The ‘translation factors’ module (red) encodes all aminoacyl-tRNA synthetases and translation factors required for protein synthesis. (B) BioBrick construction of the translation factors module shown in red in (A), but omitting EF-Tu. The final bacterial artificial chromosome (BAC) is shown surrounding the standardized workflow. NdeI sites (N), T7 RNA polymerase promoters (↱), ribosome binding sites (RBS), vesicular stomatitis virus (VSV) and TΦ terminators (⊤⊤) are denoted.
Reconstitution in vitro of the simplest, best-characterized, protein-synthesis system, that from E. coli, requires 31 translation factors (33 protein subunits) (20–22) in addition to the ribosome, mRNA and tRNAs (a subset of Supplementary Table S1). Synthesis of this translation factor module is attractive for two reasons. Firstly, it is the largest module (some 60 kb; red in Figure 1A) of our proposed minimal genome. Secondly, preparation of the 31 translation factors is the most labor-intensive aspect of assembling a purified translation system because, unlike ribosomes and tRNAs, they cannot be purified in bulk from cells. Factor preparation was simplified by separate plasmid cloning of each as an independent cistron with a His tag to give the PURE system (Protein synthesis Using Recombinant Elements (21)). Nevertheless, over-expressing translation factors from 31 different strains was so time consuming, difficult to scale up, and expensive (see Table 1 of (18)) that we proposed simplification by encoding the system on just a few DNAs instead of 31 (23). One approach was insertion of His tags directly into these genes on the native E. coli chromosome, but despite considerable effort, the number of necessary strains could not be reduced below 10 and this ‘PURE 2.0’ was only 11% as active as PURE (8). Here we use the BioBrick method (1,24) to combine these 30 His-tagged cistrons (deliberately omitting EF-Tu) onto just three plasmids for inexpensive reconstitution of ‘PURE 3.0,’ then combine these into a single circular translation factors module (Figure 1B). This establishes the feasibility, flexibility and cost-effectiveness of completely de novo design and assembly of DNA at the systems level.
MATERIALS AND METHODS
Design and synthesis of cistrons
Sequences for the 32 translation factor proteins were compiled from the NCBI gene database specific to E. coli str. K-12 substr. MG1655 (25). Restriction sites for EcoRI, XbaI, SpeI, PstI and NdeI among others were destroyed by silent point mutations, taking into account codon usage (see Supplementary Methods and Supplementary Table S2). Additionally, His6 tags were encoded for each translation factor (21). All cistrons contained a single gene except the two-subunit proteins GlyRS and PheRS, which were maintained as two-gene cistrons as found in wild type E. coli. Each gene (flanked by a VSV terminator) was synthesized from oligodeoxyribonucleotides, cloned and sequence-verified by GenScript (NJ, USA; total cost was $36492 in 2008), then subcloned into a BioBrick vector after a standard T7 promoter/lac operator/RBS (Figure 1B). See Supplementary Methods for more details.
Assembly of pLD plasmid constructs (Figure 2B and Supplementary Figure S8)
Figure 2.
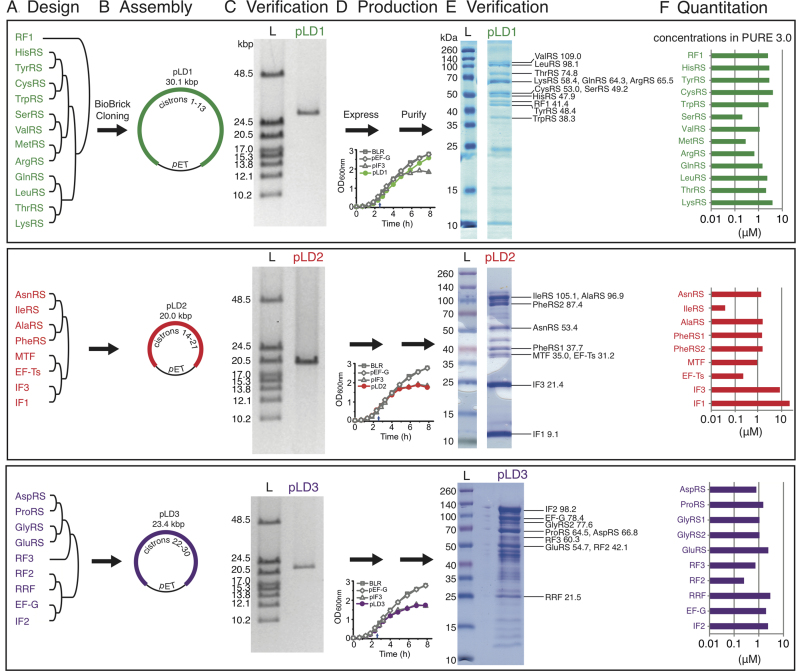
Design, synthesis and assembly of 30 cistrons into three plasmids for simplified construction of a purified translation system (PURE 3.0). (A) Pre-designed groups of BioBrick genes. Final assembled gene orders are listed from 5′-3′, top-bottom. (B) Pairwise BioBrick assembly into three high copy plasmids. (C) Size verification of plasmids by linearization and pulsed field gel electrophoresis (L, ladder). Further digests are in Supplementary Figure S2. (D) Representative growth curves of duplicate cultures before and after induction at 155 min (small arrows) with 0.5 mM IPTG. Controls (gray) are no plasmid (BLR), pET-EF-G and pET-IF3. (E) Representative patterns of nickel-column-purified proteins from over-expression of each of the three pLD plasmids, as assayed by SDS-PAGE and Coomassie blue staining (L, pre-stained ladder). Plasmid-encoded proteins were assigned (labels on right) based on mass spectrometry of excised bands (Supplementary Table S3). Calculated MWs of encoded proteins in kDa are shown. (F) Approximate relative concentrations of nickel-purified proteins from each plasmid, as estimated from gel densitometries of the gels in (E) with comigrations resolved by mass spectrometry (Supplementary Table S3). Concentrations are from Supplementary Table S5 and are plotted here on a log scale in the order of the genes on the plasmids.
Each BioBrick assembly step was pairwise in digestion and ligation. To verify each assembly, plasmid DNA was digested with NdeI and PstI, or XbaI digestion for linearization, before agarose gel analysis. BioBrick assembly was iterated to generate three large pET-based constructs (pLD1, pLD2 and pLD3). As binary cloning at this size range is only limited by in vivo amplification (2 days per cloning cycle), parallel assembly of these constructs was quick. For example, pLD2 was assembled within 8 days, with only a few hours of bench time needed each day. See Supplementary Methods for more details.
Expression and purification of translation factors
BLR (F−ompT hsdSB(rB− mB−) gal dcm (DE3) Δ(srl-recA)306::Tn10 (TetR)) E. coli cells (Merck, Germany), a recombination-deficient T7 RNA polymerase expression strain, were transformed with each construct of interest using electroporation. Proteins were expressed (Figure 2D) at 37°C with 1 mM (IPTG) for 5 h in LB. Purification of the proteins was done on a Ni-NTA (Qiagen) column in HEPES buffer. Proteins were stored in a HEPES/glycerol buffer. Total protein yields from pLD1, pLD2 and pLD3 were 32, 12 and 37 mg/l of TB, respectively. Detailed expression, purification and product characterization protocols can be found in Supplementary Methods.
Mass spectrometry of proteins
Nickel column eluate (pLD1 proteins) or slices from 12% acrylamide SDS gels (pLD1-3 in Figure 2E) were digested with trypsin and subjected to tandem mass spectrometry for analysis of proteins on a 7.4 T LTQ-FTICR mass spectrometer (ThermoFischer Scientific, Bremen Germany) connected to an Agilent 1100 nanoflow system (Agilent Technologies, Waldbronn, Germany). The peptide separations were performed on 15-cm fused silica emitters (75-μm inner diameter, 375-μm outer diameter) packed with reversed-phase, fully end-capped Reprosil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Matching tryptic peptides to proteins was accomplished by using the MASCOT search engine (version 2.2.2, Matrix Science, UK) against the UniProt_SwissProt database. The search parameters were set to Taxonomy: E. coli, enzyme: trypsin, fixed modifications: carbamidomethyl (C), variable modifications: oxidation (M) and deamidated (NQ), peptide tolerance: 0.03 Da, MS/MS tolerance: 0.8 Da and maximum two missed cleavage sites. Sometimes MASCOT scores include duplicates of the same protein present in the database containing all E. coli strains. Proteins were only considered to be positively matched (Supplementary Table S3) if they passed the more stringent MudPIT MASCOT ion scoring (only peptides identified with a peptide ion score P ≤ 0.05 were included in the search) and at least one peptide passing the required bold red criteria. The output from each sample (gel slice or eluate) is a list of proteins ranked according to their protein score where a higher number equals a higher degree of certainty of the identity of the protein. Each protein score is calculated as the sum of the peptide ion scores, where a higher peptide ion score describes a higher similarity between the experimental spectrum and the theoretical spectrum. This unbiased approach provides a powerful way of identifying virtually any protein. However, due to sequence homology between proteins, the top scoring protein may not always correspond to the correct protein. Thus, the obtained protein score should be regarded as probabilistic and the correct protein may end up among the top candidates. In making assignments for gel bands (Figure 2E), most weight was given to top scores but the mass spectrometer detected multiple proteins in every band, detection is only semi-quantitative (26), portions of the gels were not excised, and not all assignments are perfect with respect to predicted mobilities.
Kinetics (Figure 3A and B and Supplementary Figure S5)
Figure 3.
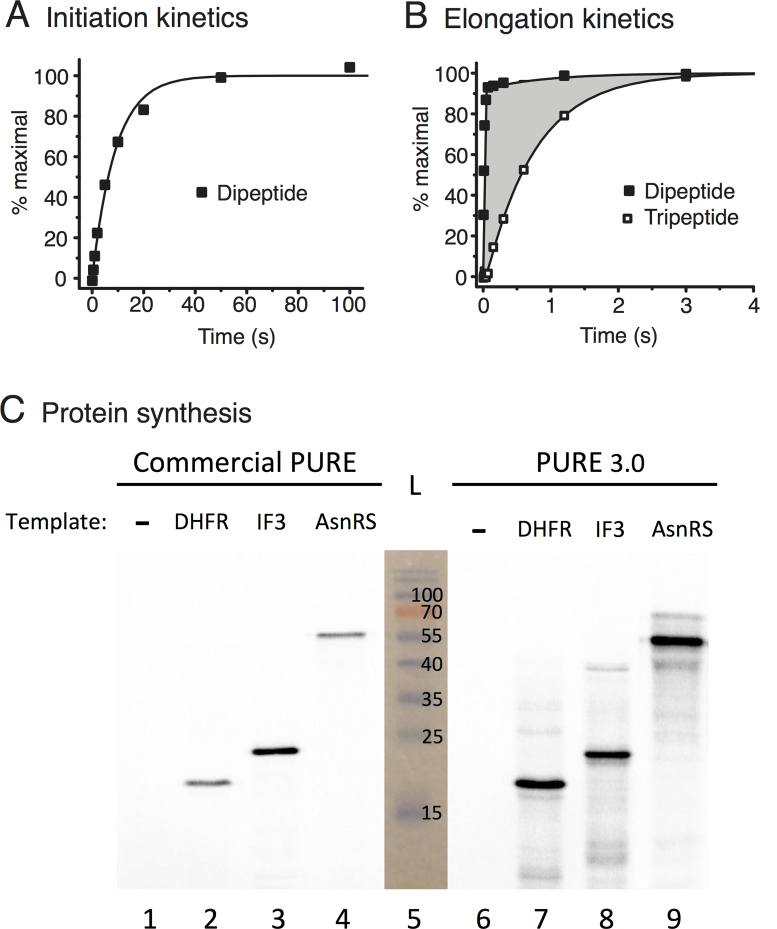
Assay of purified proteins from pLD plasmids for kinetics of initiation, di- and tripeptide syntheses and for synthesis of full-length proteins (see Materials and Methods). (A) Rate-limiting splitting of vacant 70S ribosomes (initiation) monitored by time course of subsequent non-rate-limiting fMet-Phe dipeptide bond formation using proteins encoded by pLD2 and pLD3. (B) Elongation efficiency measured by templating synthesis of fMet-Phe-Phe tripeptide and monitoring formation of both dipeptide and tripeptide products using proteins encoded by pLD2 and pLD3. The shaded area is a measure of translocation mean time. (C) Comparison of full-length protein yields of DHFR, His6-IF3 and His6-AsnRS (predicted 18.0, 21.4 and 53.4 kDa, respectively) templated by uncut plasmids in a commercial PURE system versus our PURE 3.0 containing proteins encoded by pLD1, pLD2 and pLD3. Products were quantitated by incorporation of 35S-Met, separation by SDS-PAGE and phosphoimaging of the gel. A photograph of the pre-stained ladder (L) in lane 5 is superimposed, and the results are reproducible.
All kinetics experiments were done at 37°C in 1× polymix buffer pH 7.5 supplemented with 1 mM ATP, 1 mM GTP, 10 mM PEP, 20 μg/ml pyruvate kinase and 2 μg/ml myokinase for energy regeneration. The mRNAs encoding fMet-Phe-Ala-Stop (mMFA) and fMet-Phe-Phe-Stop (mMFF) were prepared by in vitro transcription with T7 RNA polymerase and purified on an oligo-dT column as described (27). Both of the mRNAs had the same strong Shine-Dalgarno sequence (uaaggaggu) upstream of the start codon. The mRNA sequences were as follows (with sense codons xxx):

To demonstrate activities of PURE 3.0 components in translation initiation kinetics, the total average time for the splitting of tight bound vacant 70S ribosomes and the first initiation event was measured by mixing vacant 70S ribosomes with the other components necessary for dipeptide synthesis. Although the product measured was synthesis of dipeptide fMet-Phe, the average reaction time in assays performed this way is dominated by the splitting of the tight bound vacant 70S ribosomes and the first initiation steps as these are much slower compared with dipeptide bond formation per se (28,29). Two mixes were prepared and preincubated at 37°C for 15 min. A ribosomal mix contained 3.0 μM tight bound 70S ribosomes. A factor mix contained 20 μM EF-Tu, 6 μM tRNAPhe, 0.4 mM Phe, 0.4 μM PheRS, 2.9 mg/ml of pLD2 proteins and 3.6 mg/ml of pLD3 proteins (respectively 1.6 and 1.9× concs in Figure 2F). After 15 min preincubation, mMFA was added to the factor mix to 6 μM and 3H-fMet-tRNAfMet to 2 μM. Then equal volumes of the two mixes were rapidly mixed in a temperature-controlled quench flow apparatus (RQF-3; KinTeck Corp.) and the reaction was quenched with 0.5 M KOH at the indicated times. Sample treatment and data analysis were as described (30). The fraction of dipeptide peak out of total signal was plotted against time. The reaction average time was estimated by fitting the data to a single exponential equation with Origin 7.5 (OriginLab Corp.). The control experiment done in parallel for comparison (Supplementary Figure S5A) used purified components that were well characterized in such kinetics assays (29,30): the ribosomal mix was the same (3.0 μM tight bound 70S ribosomes) while the factor mix contained 4 μM EF-G, 3 μM EF-Ts, 31.6 μM IF1, 3.6 μM IF2, 23.2 μM IF3, 13.2 μM RF2, 8.4 μM RRF and 6.2 μM RF3 (instead of pLD2 and pLD3 proteins; relative concentrations were based roughly on Figure 2F) in addition to the same 20 μM EF-Tu, 6 μM tRNAPhe, 0.4 mM Phe and 0.4 μM PheRS.
To demonstrate activities of PURE 3.0 components in translation elongation kinetics, the average time for the first translocation event was measured when fMet-Phe-Phe tripeptide was synthesized from pre-initiated 70S ribosomes as described (31). Two mixes were prepared and preincubated for 15 min at 37°C. A ternary complex mix contained 20 μM EF-Tu, 1 μM EF-Ts, 8.0 μM tRNAPhe, 0.4 mM Phe, and 0.4 μM PheRS. To form pre-initiated 70S ribosomes, a ribosomal mix contained 3.0 μM tight bound 70S ribosomes, 5 μM mMFF, 1.6 μM 3H-fMet-tRNAfMet, 2.9 mg/ml of pLD2 proteins and 3.6 mg/ml of pLD3 proteins. Equal volumes of the two mixes were rapidly mixed in the quench flow apparatus and quenched by 0.5 M KOH at the indicated times. Sample treatment was done as described (30) and the average time for translocation was estimated as described (31). The control experiment done in parallel for comparison (Supplementary Figure S5B) used purified components that were well characterized in such kinetics assays: the factor mix was the same (20 μM EF-Tu, 1 μM EF-Ts, 8.0 μM tRNAPhe, 0.4 mM Phe, and 0.4 μM PheRS) while the ribosomal mix contained 4 μM EF-G, 3 μM EF-Ts, 31.6 μM IF1, 3.6 μM IF2, 23.2 μM IF3, 13.2 μM RF2, 8.4 μM RRF and 6.2 μM RF3 (instead of pLD2 and pLD3 proteins; relative concentrations were based roughly on Figure 2F) in addition to the same 3.0 μM tight bound 70S ribosomes, 5 μM mMFF and 1.6 μM 3H-fMet-tRNAfMet. Figure 3A,B and Supplementary Figure S5 show representative plots of duplicate experiments performed on different days. Kinetic parameters are reported as mean ± propagated standard deviation in the Supplementary Figure S5 legend.
In vitro transcription and translation using PURE 3.0 (Figure 3C)
Component preparation is detailed in Supplementary Methods. Briefly, ribosomes were purified using zonal centrifugation from MRE600 E. coli cells. T7 RNA polymerase was purified on Ni-NTA (32), and EF-Tu was prepared as reported (33). All other reagents were purchased. The PURE 3.0 translation reaction was adapted from the CytoMim and original PURE systems (21,34). Optimization experiments revealed large stimulations by switching to phosphocreatine energy and by switching template from mRNA to DNA, then adjusting concentrations of DNA template, NTPs and Mg2+. Changing the relative amounts of bulk pLD-encoded products revealed flexibility, as both the ratio in Figure 2F and Supplementary Tables S5 and S8 and the ratio in the protocol detailed below for Figure 3C synthesized full length protein products. The reconstitution of PURE 3.0 was reproducible by different researchers.
The reactions in Figure 3C were set up as follows. On ice, 4.3 μl Stock Buffer (425 mM HEPES pH 7.6, 950 mM K(Glu), 110 mM Mg(Glu)2, 15 mM spermidine), 0.5 μl 1M Mg(Glu)2 and 3.4 μl water were combined with 1.9 μl 100 mM DTE, 0.9 μl of 50 mM each of 18 amino acids (without Met and Cys), 0.9 μl of 50 mM Cys, 0.7 μl of 20 mM Met, 1.1 μl of 20 mM 10-CHO-THF, 1.8 μl each of 100 mM Na ATP, CTP, GTP and UTP, 0.9 μl 1M phosphocreatine di(tris) and 0.6 μl of 4 A260/μl total E. coli tRNA. Then 1.1 μl energy enzymes mix (containing myokinase, creatine phosphokinase, pyrophosphatase, nucleoside diphosphate kinase) and 1.1 μl 40 U/μl RiboLock (ThermoFischer Scientific) were added. Then 1.6 μl of 17 mg/ml LD1 proteins, 6.4 μl of 5 mg/ml LD2 proteins, and 1.3 μl of 23 mg/ml LD3 proteins were added (final reaction bulk protein concentrations: LD1, 0.59 mg/ml; LD2, 0.70 mg/ml; LD3, 0.65 mg/ml). Then 0.5 μl of 90 μM T7 RNA polymerase was added along with 5.2 μl of 21 μM ribosomes. The pH was adjusted to 7.6 with about 0.5 μl of 2 M KOH with the components being kept on ice. Next 2.3 μl of 530 μM EF-Tu and 1.1 μl 35S-Met (10 μCi/μl, 1175 Ci/mmol) were added. Other components of the system listed in Supplementary Table S5 (e.g. calcium, putrescine, BME, DTE, buffering agents, magnesium) were carried over from the LD protein purifications and from the T7 RNA polymerase, ribosome and EF-Tu preparations in polymix. Then 9.5 μl of this 43.5 μl reaction mix was added to 0.5 μl of water (negative control) or 50 ng uncut plasmid DNA of T7-promoter-controlled dhfr (NEB) or His6-IF3 or His6-AsnRS (Supplementary Figure S1A) in 0.5 μl of water. Reactions were incubated for 2 h at 37°C before freezing at –80°C or quenching by boiling in SDS loading buffer.
For positive controls, the PUREfrex commercial PURE translation kit (GeneFrontier, Chiba, Japan) was used with the same specific activity of35S-Met and same concentrations of DHFR, IF3 and AsnRS plasmids. In each of four tubes, 0.5 μl 100 ng/μl template or water was added to 5 μl Solution I, 0.5 μl Solution II and 1 μl Solution III with 2.6 μl ddH2O and 0.4 μl 35S-Met (10 μCi/μl, 1175 Ci/mmol) to give 10 μl reactions containing 0.5 mM Met. Reactions were incubated at 37°C for 2 h before freezing at –80°C.
One μl from each reaction was added to 8 μl final 1× SDS loading buffer and samples were boiled for 5 minutes before loading on a 15% SDS-PAGE gel (7.5 cm × 0.75 mm) and run in SDS running buffer until the Bromophenol blue dye was at the bottom. The gel was removed to fixing solution (10% acetic acid, 40% methanol, 3% glycerol) for 15 min. The gel was then dried at 80°C on Whatman 3MM chromatography paper under vacuum and exposed to a phosphoimaging screen for 16 h before scanning. The intensities of the bands (Figure 3C) were measured by software Quantity One (BioRad) and yields quantitated by comparison with known amounts of [35S]-methionine, taking into account the number of methionines in each protein and measuring approximate fold quenching by the gel. The PURE 3.0 yields calculated for two replicates of DHFR, IF3 and AsnRS syntheses performed on different days were 12.9 ± 0.1, 7.1 ± 0.7 and 6.2 ± 0.4 pmol/μl (means ± standard deviations), respectively. These are comparable with the yield of DHFR of 8 pmol/μl specified by the manufacturer of the commercial PURE kit.
Assembly of pTFM1 (Figure 4A–C and Supplementary Figure S8)
Figure 4.
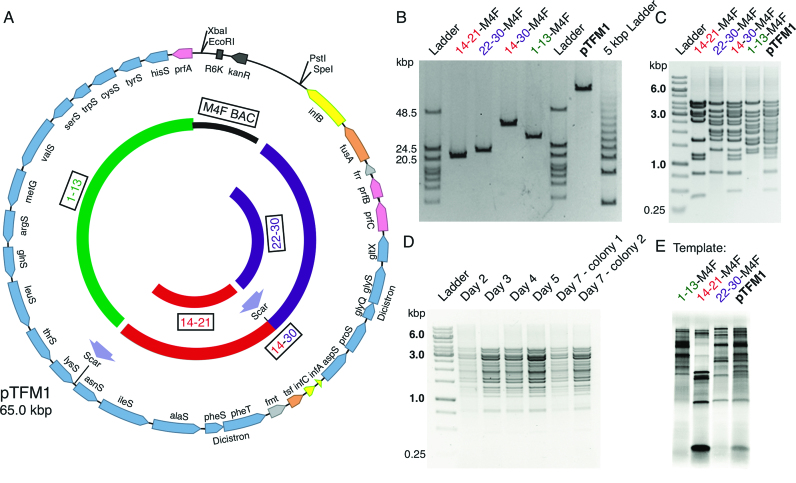
Construction of a 30-cistron translation factor module. (A) Plasmid map of final assembled Translation Factor Module (pTFM1). Linearized low copy number M4F-14-21 (containing cistrons 14–21; red arc) and 22–30 insert (purple arc) were ligated to generate a 14–30 cassette (red-purple arc). This was ligated to M4F-1-13 (black and green arc) to obtain 30 cistrons (117 basic parts) on a single plasmid (pTFM1). BioBrick restriction sites and ligation scars are indicated. Genes are labeled and shown to scale: synthetases are blue; initiation factors are yellow; elongation factors are orange, release factors are pink, and other factors are light grey. (B) Pulsed field gel analysis of linearized intermediate plasmids and final assembled pTFM1. Predicted sizes from L-R are 21.4, 24.8, 39.8, 31.8 and 65.0 kb. The 5 kbp Ladder (right) has increments of 4.9 kb from 9.8 kb. (C) Agarose gel of NdeI+PstI-digested intermediate plasmids and final pTFM1 showing DNA fragments containing each cistron with its 3′ intergenic region (further digests in Supplementary Figure S6). (D) Agarose gel of NdeI+PstI-digested pTFM1 DNA isolated from each of 7 days of culturing in E. coli. (E) In vitro protein expressions from uncut BAC constructs 1–13, 14–21, 22–30 and pTFM1 using a commercial PURE system, 35S-Met and gradient SDS-PAGE. Similar patterns are seen for in vitro synthesis from pTFM1 using a commercial S30 E. coli extract (Supplementary Figure S7).
Inserts from pLD1, pLD2 and pLD3 were ligated into an engineered BAC (pM4F) containing an R6K origin of replication with subsequent cloning and propagation of DNA done in Pir+ (F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG pir+(DHFR)) E. coli from Epicentre (WI, USA). Binary cloning continued between cistrons 22–30 (insert) and M4F-14-21, followed by ligating 14–30 (insert) to M4F-1-13 to yield the final pTFM1, and the method was reproducible. Recombination during cloning was limited by ensuring high DNA quality (using fresh DNA purifications and minimizing UV exposure, freeze-thaw cycles and pipetting) and increasing ligation efficiency (prolonged, thermal-cycled ligations in fresh T4 DNA ligase buffer). Positive clones were verified by NdeI and PstI digestion (Figure 4C), XbaI linearization (Figure 4B) and DNA sequencing of the entire pTFM1. See Supplementary Methods for details.
RESULTS
Our strategy, developed mindful of a number of challenges predicted for de novo design and characterization of large systems-level expression modules, included:
inducible, orthogonal expression from bacteriophage T7 RNA polymerase promoters to minimize toxicity during cellular amplification and to enable over-expression;
a transcription promoter and terminator for each translation factor cistron to minimize gene proximity effects;
amplification in recombination-deficient E. coli to minimize recombination between non-coding repeated sequences;
modular BioBrick assembly in vitro (based on the restriction enzymes EcoRI, XbaI, SpeI and PstI) (1) to minimize cost and eliminate pre-customized arrangements of genes; and
His-tagging for easy copurification.
Other predicted challenges (incorrect published coding sequences (35), immutable BioBrick restriction sites (24), inefficient termination (36) and uneven expression and purification from RBS/His-tag/N-terminal coding region fusions (37)) were left for trouble-shooting if, and when, they appeared.
EF-Tu was omitted from the constructs because it is required at much higher levels than any other translation factor. The remaining 30 cistrons (Figure 2A) were prepared commercially from synthetic oligodeoxyribonucleotides with all BioBrick restriction endonuclease sites mutated to synonymous codons (Supplementary Table S2). BioBrick sites were removed previously from pET-24a plasmid (23) and here from a BAC plasmid by site-directed mutagenesis (Supplementary Figure S1). Each synthetic gene flanked by the VSV class II T7 RNA polymerase terminator was then ligated into the BioBrick pET vector after the standard pET T7 RNA polymerase promoter/lac operator/strong RBS (designed for maximal expression). Assemblies were performed pairwise in parallel (Figure 1B); all but the final two used the high-copy-number BioBrick pET vector to facilitate eventual protein over-expression.
Initial beta testing of our design strategy was performed on assemblies of five genes (23,38) and many more (not shown). This synthetic biology approach revealed unexpected massive gene-proximity effects (23), leading to the astonishing conclusion that the well-known class II T7 terminators (which we selected because of their tiny sizes) only functioned in vitro, not in vivo (38). To address this, we synthesized a BioBrick-compatible 119-nucleotide classic TΦ class I terminator, demonstrated efficient termination in vivo (38) and ligated it to all of our 30 BioBrick cistrons (Figures 1B and 2A).
We then assembled larger numbers of the cistrons into groups (Figure 2A) defined by relative amounts of proteins needed for the PURE translation system (21) to enable some adjustment of final relative protein levels (pLD1 groups lowest amounts needed, pLD2 groups highest and pLD3 groups intermediate). Surprisingly, despite the circular topology of the multigene constructs, there were some restrictions in gene order. The ThrRS gene could only be cloned in a highly restricted position at, or near, the 3′ end of the gene clusters. This was apparently due to the open reading frame containing two internal promoters for E. coli RNA polymerase (39) causing toxic expression of downstream genes (perhaps mischarging by an over-expressed aminoacyl-tRNA synthetase (40)) because mutation of these promoters (Supplementary Table S2) relieved the positional constraint. We thus incorporated these internal promoter mutations into the assembly of pLD1 and larger constructs. The RF1 gene exhibited the opposite constraint, only being clonable upstream of gene clusters. Although we were unable to determine the specific reason for this, our finding below that several of the most highly expressed cistrons were located at the downstream ends of the clusters (Figure 2F) may be pertinent. Recognition and solving of order-related cloning problems was facilitated by the BioBrick method because any gene arrangement can be attempted in parallel without pre-customization.
Next, we pushed the limits of insert size in the high-copy-number pET vector to minimize the number of plasmids needed for over-expression of the PURE system (Figure 2B and Supplementary Figure S8). Remarkably, an insert as big as 33 kb (cloning the red insert downstream from the purple insert of Figure 2B) was clonable (yielding pLD4; Supplementary Figure S3). Analyses were facilitated by our synthetic DNA design that incorporated an NdeI site immediately upstream of every cistron while replacing all other cistronic NdeI sites with synonymous codons (Figure 1B). Given the combination of huge insert size, many genes each containing a 254-base intergenic repeat, and high plasmid copy number, it was important to test stability over a number of generations. Indeed the largest pET construct (pLD4) proved highly unstable (Supplementary Figure S3). However, the next three largest constructs (Figure 2B and C) were sufficiently stable, both during amplification in E. coli NEB10β and during induction of protein over-expression in recombination-deficient E. coli BLR (Merck, Germany) even though the concentration of lac repressor had not been increased to match the increased number of lac operators (Supplementary Figure S4; note that prolonged cultures showed some instability of pLD1). The cell growth curves before and after over-expression were similar to controls (Figure 2D).
Despite the genetic complexity of the three constructs in Figure 2B, purification of all 32 encoded protein products was enabled by just three protein over-expressions and three elutions from nickel columns, as verified by the following data specific to each of the 32 proteins. Gel bands stained by Coomassie blue were visible reproducibly at positions corresponding to the expected migrations of all encoded proteins (Figure 2E) and the presence in these bands of 30 of the 32 encoded proteins was confirmed by mass spectrometry of excised bands (Supplementary Table S3 and its legend; protein assignments are labelled on the right of the gels in Figure 2E). Of these assignments, only SerRS had low mass spectrometry scores for both the excised band and the eluate, so SerRS was confirmed by measuring tRNA charging with serine (Supplementary Table S4). Regarding the two of the 32 encoded protein subunits not detected in excised bands: MetRS (77.0 kDa) was detected in mass spectrometry of the eluate (Supplementary Table S3) and confirmed by tRNA charging (Supplementary Table S4) while GlyRS1 (34.8 kDa; the non-His-tagged subunit of the 2 GlyRS subunits) was also detected by tRNA charging (as were all of the 20 encoded aminoacyl-tRNA synthetases; Supplementary Table S4). Other bands were minor and, based on the top mass spectrometry scores of excised bands, corresponded to EF-Tu (43.3 kDa; the most abundant protein in E. coli) or FKBP-type peptidyl prolyl cis-trans isomerase (20.9 kDa; a well-known nickel-binding contaminant) or fragments of encoded proteins (Supplementary Table S3).
Relative ratios of nickel-purified proteins from each plasmid were estimated from densitometric scanning of the gel lanes in Figure 2E, with individual protein ratios within comigrations (or eluate in the case of MetRS) being estimated by mass spectrometry (Supplementary Table S3). The ratios (Figure 2F) are considered as rough approximations because, in making band assignments, most weight was given to top MASCOT scores but the mass spectrometer detected multiple proteins in every band, mass spectrometry is only semi-quantitative (26), and not all assignments are perfect with respect to theoretically-predicted mobilities of the His-tagged proteins. Mindful of these limitations, we concluded that the higher-yielding proteins tended to come from the downstream ends of the three inserts and tended to be small, yet expression of most of the proteins was relatively balanced. Non-identical yields were expected based on the variation in coding sequences because these sequences lie just downstream from the standardized RBS (expected to affect the secondary structure of the RBS and thus the efficiency of initiation of protein synthesis) and lie directly adjacent to the His tag (expected to affect binding and elution from nickel (8)). Given our estimated ratios (Figure 2F) and that the lowest-yielding protein, IleRS, had a tRNA-charging activity comparable with six other aminoacyl-tRNA synthetases (Supplementary Table S4), we deemed the relative yields sufficient to proceed with translation assays using our database-derived, codon-altered over-expressed proteins.
The pooled nickel eluates of pLD2 and pLD3 exhibited fast kinetics of initiation, di- and tripeptide syntheses (Figure 3A and B) that were comparable with controls using individually-purified proteins (Supplementary Figure S5), demonstrating high activities of initiation factors and elongation factors. The relative amounts of the extracts from the three constructs and also other components of the PURE translation system were then optimized to enable protein synthesis as efficiently as in a commercial PURE system (Figure 3C; see Materials and Methods for optimization and protein synthesis yields).
To test if BioBrick cloning and 30 repeated sequences are compatible with assemblies at the 30-cistron scale, we attempted ligating the three inserts from pET vectors (Figure 2B) into our low-copy-number BioBrick BAC (Figure 4A and Supplementary Figure S6). The cloning was successful (Figure 4B and C) and reproducible, but non-trivial, perhaps due to the higher ratio of internal DNA bases to DNA ends during ligation of such large fragments because thermo-cycled ligations were helpful. The complete 64 979 bp DNA sequence of the final pTFM1 plasmid was verified by real-time single-molecule sequencing (Pacific Biosciences RSII system at SciLifeLab, Uppsala), which also validates the correctness of the three ‘parent’ inserts from the pLD1, pLD2 and pLD3 plasmids. Importantly, the resulting BAC plasmid was stable through 150 generations (Figure 4D) despite the repeated sequences.
Finally, we tested if a completely de-novo-designed 30-cistron plasmid can act as a template for transcription and translation, as did the three smaller, parent, PURE 3.0 plasmids. Using PURE and crude S30 translation systems, the in vitro expression patterns obtained matched that expected by combining the expression patterns of the three pLD clones encoding PURE 3.0 (Figure 4E and Supplementary Figure S7, respectively). However, useful translation activity is yet to be obtained from nickel-purified proteins expressed in vivo from this single plasmid because protein expression yields from a low-copy BAC are much lower than from high copy pET plasmids and the PURE system requires high concentrations of protein factors. Given this high concentration requirement of factors, it should also be noted that break-even expression of all PURE factors from PURE (using any number of plasmids) remains an important challenge for future work. Thus, for preparation of PURE translation system factors, we recommend in vivo expression from our three pLD plasmids.
DISCUSSION
Our characterizations of the three PURE 3.0 plasmids, their protein over-expression products, and the translation systems prepared from them demonstrate that they are ready for free distribution to the community. They enable production of an affordable, scalable PURE system for protein synthesis and unnatural amino acid incorporation under defined conditions in the absence of cellular barriers and significant nuclease and protease activities (Figure 3C). Though our PURE 3.0 system shows activity similar to the commercially-available PURE system, it is much cheaper to produce. With respect to translation factors, it only requires fermentation and purification from 4 translation factor clones instead of 31 (a factor of 8×), and this allows easy scale up (mostly by adding more LB). Ribosomes and tRNAs can readily be prepared in bulk or purchased. Energy usage is an important factor also, and the costs of those reagents and enzymes are significant at larger scales. For a rough comparison at the 25 ml scale, we estimate a cost of consumable reagents and enzymes of $0.033 per μl of reaction (Supplementary Table S8, exclusive of equipment, labor and overhead) compared with the cost of a commercial PURE system of $0.69 per μl of reaction. Notably, as with the original PURE system (21), the PURE 3.0 proteins are His tagged, thus allowing reverse purification of protein products synthesized without His tags by nickel chromatography.
We have also demonstrated that simple BioBrick cloning is a practical route for assembling large DNAs. This was certainly not a foregone conclusion, given that different DNA assembly methods work best at different scales (41). Success required removing two cryptic promoters, using recombination-deficient strains, making a BioBrick BAC vector and handling the large DNAs very carefully. Relative expression levels of our individual genes or extra added ones potentially could be adjusted rationally by ligating different promoter/RBS sequences into a gene's unique NdeI site at its start codon (standardized T7 promoter mutants (35) and RBSs are available). In addition, our BioBrick constructs fill important gaps in the Registry of Biological Parts, a rapidly expanding BioBrick collection containing the vast majority of standardized synthetic biology parts. Regarding concerns about the reliability of deposited parts (42), the inflexibility of using a standard (43) and the feasibility of making large multigene constructs from them, the number of parts to synthesize and characterize is finite (some 4300 genes in E. coli (44)) and we have now demonstrated that recombination and large-scale removal of BioBrick sites are not prohibitive. And if there are any essential BioBrick sites, they could always be added last by an alternative method.
Multi-gene to systems-scale assemblies are enabling a new era in designer biology. For example, our 30-cistron construct and strategy can be employed in projects working towards synthesizing self-replication. A comparison with alternative strategies for potentially designing and building orthogonal systems at the 30-gene scale is warranted. The 16-gene cluster for nitrogen fixation was refactored and assembled by GoldenGate cloning. This involved removal of illegal restriction sites, pre-determined gene orders, polymerase chain reactions (PCR), high throughput assembly of more than two parts per reaction and optimization by functional screening of 484 constructs (9). More complicated cloning strategies have been used for refactoring a 6-gene polyketide cluster (45). Bottom-up synthesis of multigene constructs have also been applied to engineer information-gathering functionality into organisms, demonstrated by the 14-gene multi-plasmid ‘photocopying’ E. coli, which was generated across four plasmids and a genomic modification (46). To avoid homologous recombination, multiple terminators were used. Another powerful approach for multigene assembly is Gibson assembly, but it requires pre-specified gene order and PCR, the latter of which can be problematic with repeated sequences and for fidelity (47). Yeast recombination (17,48) has also become an enabling technology for large genetic systems assembly, enabling parallel synthesis of, for example, a synthetic minimal Mycoplasma genome (19), 6.5 synthetic yeast chromosomes totalling approximately 4 Mb built in 10–60 kb fragments (10,12,49–53) and 55 recoded 50 kb genetic fragments towards generating a 57-codon E. coli genome (54). Importantly, design criteria introduced in these synthetic systems, such as codon reduction and incorporation of loxP sites, are compatible with the BioBrick approach here. Yet, yeast assembly mandates PCR-based pre-specified gene orders and by its nature is incompatible with the multiple repeats we needed due to lack of T7 terminator variants. So BioBrick cloning at the systems level has some advantages in terms of compatibility with multiple repeats and the largest library of standardized parts, fidelity, simplicity, affordability and eliminating pre-determined gene orders (which we found can determine viability). This modular cistron approach is thus ready for systems-level constructions, as shown by going from completely de novo design to de novo synthesis to functional characterization of a 30-cistron system across three plasmids, to assembly of a single 30-cistron BAC.
DATA AVAILABILITY
The plasmids and strains described here are available upon request. The DNA sequence for pTFM1 has been deposited in GenBank under accession number KT347311.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tom Knight and Barry Canton for advice and technical assistance during the pilot stage that included pointing out two internal promoter sequences within the ThrRS gene, and Chris Anderson for the BAC vector pBACr-NESd. We also thank Raymond Fowler for technical assistance, Olga Pettersson and Ignus Bunikis of Uppsala SciLifeLab for DNA sequencing, Marek Kwiatkowski for assistance with sequence analysis and the anonymous referees of NAR for comments on the manuscript in October 2015.
Footnotes
Present addresses:
Tyson R. Shepherd, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Liping Du, Center for Quantitative Science, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Samudyata, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Stockholm 171 77, Sweden.
Jinfan Wang, Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Marcus O.D. Sjödin, Swedish Toxicology Sciences Research Center (Swetox), Södertälje 151 36, Sweden.
Magnus Wetterhall, GE Healthcare Bio-Sciences, Life Sciences, Uppsala 751 84, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-AI0727453 to A.C.F.]; American Cancer Society [RSG07-238-01 to A.C.F.]; Swedish Research Council [NT 2011-5787, NT 2016-1 to A.C.F.]; Japanese Exploratory Research for Advanced Technology (ERATO) [to A.C.F. and T.Y.]. Funding for open access charge: Swedish Research Council [2016-1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Knight T. Idempotent Vector Design for Standard Assembly of Biobricks. 2003; Cambridge: MIT; http://hdl.handle.net/1721.1/21168. [Google Scholar]
- 2. Endy D. Foundations for engineering biology. Nature. 2005; 438:449–453. [DOI] [PubMed] [Google Scholar]
- 3. Kahl L.J., Endy D.. A survey of enabling technologies in synthetic biology. J. Biol. Eng. 2013; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Way J.C., Collins J.J., Keasling J.D., Silver P.A.. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014; 157:151–161. [DOI] [PubMed] [Google Scholar]
- 5. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson J.C., Dueber J.E., Leguia M., Wu G.C., Goler J.A., Arkin A.P., Keasling J.D.. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 2010; 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tian J., Gong H., Sheng N., Zhou X., Gulari E., Gao X., Church G.. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004; 432:1050–1054. [DOI] [PubMed] [Google Scholar]
- 8. Wang H.H., Huang P.Y., Xu G., Haas W., Marblestone A., Li J., Gygi S.P., Forster A.C., Jewett M.C., Church G.M.. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth. Biol. 2012; 1:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smanski M.J., Bhatia S., Zhao D., Park Y., L B.A.W., Giannoukos G., Ciulla D., Busby M., Calderon J., Nicol R. et al. . Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014; 32:1241–1249. [DOI] [PubMed] [Google Scholar]
- 10. Annaluru N., Muller H., Mitchell L.A., Ramalingam S., Stracquadanio G., Richardson S.M., Dymond J.S., Kuang Z., Scheifele L.Z., Cooper E.M. et al. . Total synthesis of a functional designer eukaryotic chromosome. Science. 2014; 344:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liljeruhm J., Gullberg E., Forster A.C.. Synthetic Biology: A Lab Manual. 2014; Singapore: World Scientific Publishing Company. [Google Scholar]
- 12. Richardson S.M., Mitchell L.A., Stracquadanio G., Yang K., Dymond J.S., DiCarlo J.E., Lee D., Huang C.L., Chandrasegaran S., Cai Y. et al. . Design of a synthetic yeast genome. Science. 2017; 355:1040–1044. [DOI] [PubMed] [Google Scholar]
- 13. Szostak J.W., Bartel D.P., Luisi P.L.. Synthesizing life. Nature. 2001; 409:387–390. [DOI] [PubMed] [Google Scholar]
- 14. Forster A.C., Church G.M.. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006; 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glass J.I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M., Hutchison C.A. 3rd, Smith H.O., Venter J.C.. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kita H., Matsuura T., Sunami T., Hosoda K., Ichihashi N., Tsukada K., Urabe I., Yomo T.. Replication of genetic information with self-encoded replicase in liposomes. ChemBioChem. 2008; 9:2403–2410. [DOI] [PubMed] [Google Scholar]
- 17. Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M. et al. . Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010; 329:52–56. [DOI] [PubMed] [Google Scholar]
- 18. Jewett M.C., Forster A.C.. Update on designing and building minimal cells. Curr. Opin. Biotechnol. 2010; 21:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hutchison C.A. 3rd, Chuang R.Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L. et al. . Design and synthesis of a minimal bacterial genome. Science. 2016; 351:aad6253. [DOI] [PubMed] [Google Scholar]
- 20. Kung H.F., Redfield B., Treadwell B.V., Eskin B., Spears C., Weissbach H.. DNA-directed in vitro synthesis of beta-galactosidase. Studies with purified factors. J. Biol. Chem. 1977; 252:6889–6894. [PubMed] [Google Scholar]
- 21. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T.. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001; 19:751–755. [DOI] [PubMed] [Google Scholar]
- 22. Kazuta Y., Adachi J., Matsuura T., Ono N., Mori H., Yomo T.. Comprehensive analysis of the effects of Escherichia coli ORFs on protein translation reaction. Mol. Cell. Proteomics. 2008; 7:1530–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du L., Gao R., Forster A.C.. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase. Biotechnol. Bioeng. 2009; 104:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shetty R.P., Endy D., Knight T.F. Jr. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008; 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B.L., Mori H. et al. . Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006; 2, doi:10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M.. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005; 4:1265–1272. [DOI] [PubMed] [Google Scholar]
- 27. Pavlov M.Y., Ehrenberg M.. Rate of translation of natural mRNAs in an optimized in vitro system. Arch. Biochem. Biophys. 1996; 328:9–16. [DOI] [PubMed] [Google Scholar]
- 28. Johansson M., Bouakaz E., Lovmar M., Ehrenberg M.. The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell. 2008; 30:589–598. [DOI] [PubMed] [Google Scholar]
- 29. Pavlov M.Y., Antoun A., Lovmar M., Ehrenberg M.. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 2008; 27:1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J., Kwiatkowski M., Pavlov M.Y., Ehrenberg M., Forster A.C.. Peptide formation by N-methyl amino acids in translation is hastened by higher pH and tRNA(Pro). ACS Chem. Biol. 2014; 9:1303–1311. [DOI] [PubMed] [Google Scholar]
- 31. Mandava C.S., Peisker K., Ederth J., Kumar R., Ge X., Szaflarski W., Sanyal S.. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012; 40:2054–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He B., Rong M., Lyakhov D., Gartenstein H., Diaz G., Castagna R., McAllister W.T., Durbin R.K.. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expression Purif. 1997; 9:142–151. [DOI] [PubMed] [Google Scholar]
- 33. Ehrenberg M., Bilgin N., Kurland C.G.. Spedding G. Ribosomes and Protein Synthesis: A Practical Approach. 1990; Oxford: IRL Press; 101–108. [Google Scholar]
- 34. Jewett M.C., Fritz B.R., Timmerman L.E., Church G.M.. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 2013; 9:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Temme K., Zhao D., Voigt C.A.. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:7085–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Studier F.W., Moffatt B.A.. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986; 189:113–130. [DOI] [PubMed] [Google Scholar]
- 37. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. . Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 38. Du L., Villarreal S., Forster A.C.. Multigene expression in vivo: supremacy of large versus small terminators for T7 RNA polymerase. Biotechnol. Bioeng. 2012; 109:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wertheimer S.J., Klotsky R.A., Schwartz I.. Transcriptional patterns for the thrS-infC-rplT operon of Escherichia coli. Gene. 1988; 63:309–320. [DOI] [PubMed] [Google Scholar]
- 40. Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., Soll D.. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988; 242:1548–1551. [DOI] [PubMed] [Google Scholar]
- 41. Casini A., Storch M., Baldwin G.S., Ellis T.. Bricks and blueprints: methods and standards for DNA assembly. Nat. Rev. Mol. Cell Biol. 2015; 16:568–576. [DOI] [PubMed] [Google Scholar]
- 42. Vilanova C., Porcar M.. iGEM 2.0–refoundations for engineering biology. Nat. Biotechnol. 2014; 32:420–424. [DOI] [PubMed] [Google Scholar]
- 43. Alnahhas R.N., Slater B., Huang Y., Mortensen C., Monk J.W., Okasheh Y., Howard M.D., Gottel N.R., Hammerling M.J., Barrick J.E.. The case for decoupling assembly and submission standards to maintain a more flexible registry of biological parts. J. Biol. Eng. 2014; 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Horiuchi T., Keseler I.M., Kosuge T. et al. . Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006; 34:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osswald C., Zipf G., Schmidt G., Maier J., Bernauer H.S., Muller R., Wenzel S.C.. Modular construction of a functional artificial epothilone polyketide pathway. ACS Synth. Biol. 2014; 3:759–772. [DOI] [PubMed] [Google Scholar]
- 46. Fernandez-Rodriguez J., Moser F., Song M., Voigt C.A.. Engineering RGB color vision into Escherichia coli. Nat. Chem. Biol. 2017; 13:706–708. [DOI] [PubMed] [Google Scholar]
- 47. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 48. Muller H., Annaluru N., Schwerzmann J.W., Richardson S.M., Dymond J.S., Cooper E.M., Bader J.S., Boeke J.D., Chandrasegaran S.. Assembling large DNA segments in yeast. Methods Mol. Biol. 2012; 852:133–150. [DOI] [PubMed] [Google Scholar]
- 49. Mitchell L.A., Wang A., Stracquadanio G., Kuang Z., Wang X., Yang K., Richardson S., Martin J.A., Zhao Y., Walker R. et al. . Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017; 355, doi:10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 50. Shen Y., Wang Y., Chen T., Gao F., Gong J., Abramczyk D., Walker R., Zhao H., Chen S., Liu W. et al. . Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017; 355, doi:10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu Y., Li B.Z., Zhao M., Mitchell L.A., Xie Z.X., Lin Q.H., Wang X., Xiao W.H., Wang Y., Zhou X. et al. . Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017; 355, doi:10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie Z.X., Li B.Z., Mitchell L.A., Wu Y., Qi X., Jin Z., Jia B., Wang X., Zeng B.X., Liu H.M. et al. . “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017; 355, doi:10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 53. Zhang W., Zhao G., Luo Z., Lin Y., Wang L., Guo Y., Wang A., Jiang S., Jiang Q., Gong J. et al. . Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017; 355, doi:10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 54. Ostrov N., Landon M., Guell M., Kuznetsov G., Teramoto J., Cervantes N., Zhou M., Singh K., Napolitano M.G., Moosburner M. et al. . Design, synthesis, and testing toward a 57-codon genome. Science. 2016; 353:819–822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plasmids and strains described here are available upon request. The DNA sequence for pTFM1 has been deposited in GenBank under accession number KT347311.