Abstract
In prokaryotes, RNA polymerase and ribosomes can bind concurrently to the same RNA transcript, leading to the functional coupling of transcription and translation. The interactions between RNA polymerase and ribosomes are crucial for the coordination of transcription with translation. Here, we report that RNA polymerase directly binds ribosomes and isolated large and small ribosomal subunits. RNA polymerase and ribosomes form a one-to-one complex with a micromolar dissociation constant. The formation of the complex is modulated by the conformational and functional states of RNA polymerase and the ribosome. The binding interface on the large ribosomal subunit is buried by the small subunit during protein synthesis, whereas that on the small subunit remains solvent-accessible. The RNA polymerase binding site on the ribosome includes that of the isolated small ribosomal subunit. This direct interaction between RNA polymerase and ribosomes may contribute to the coupling of transcription to translation.
INTRODUCTION
In eubacteria, transcription and translation occur in close spatial and temporal proximity, allowing the processes to couple. In Escherichia coli, most proteins are translated while their genes are still being transcribed (1). The inhibition of translation results in the genome-wide stalling of transcription (2). Stalled RNA polymerases act as a barrier for the DNA replication machinery, jeopardizing the processivity of replication, and with it, the integrity of the genome (3,4).
Functional interactions between RNA polymerase and the ribosome have been demonstrated for polycistronic operons (5–7). For example, a nonsense mutation in an upstream gene attenuates the transcription of downstream genes (8,9). Premature translation termination causes ribosomes to dissociate from nascent RNA (10,11). Unhindered by ribosomes, transcription termination factor rho proceeds along the nascent RNA to RNA polymerase, where it induces transcription termination (12–14).
The functional interaction between ribosomes and RNA polymerase is also exploited in the regulation of gene expression, as exemplified by the regulation of the trp operon. During tryptophan starvation, ribosomes translating the operon's leader peptide stall at the two consecutive tryptophan codons. This stalling prevents the nascent RNA from forming a short stem–loop that acts as an intrinsic transcription termination signal; without the stem–loop, RNA polymerase transcribes the downstream genes of the operon, which are necessary for tryptophan synthesis (5–7).
In some cases, the functional coupling of transcription and translation is thought to be promoted by physically connecting the ribosome to RNA polymerase via a small protein, such as transcription factor NusG or its paralog, RfaH (15,16). Both factors consist of two domains, the N- and C-terminal domains. The N-terminal domain binds directly to RNA polymerase (17,18), while the C-terminal domain binds to ribosomal protein S10 on a surface accessible on the ribosome (15,16). These results point to NusG and RfaH as a physical link between RNA polymerase and ribosomes (15,16).
However, biophysical considerations of NusG’s interactions with RNA polymerase and ribosomes point to a significant contribution of factor-independent interactions in the physical linking of RNA polymerase and ribosomes. Because the two domains of NusG are structurally independent of each other (19), formation of the NusG-link between RNA polymerase and ribosome can be broken down into two thermodynamically separate events: the binding of NusG’s N-terminal domain to the RNA polymerase and the binding of NusG’s C-terminal domain to the ribosome. During exponential growth, the segregation of the nucleoid from the cytoplasm (20–22) increases the local concentration of RNA polymerase and NusG in the nucleoid (1–2 μM (20,21,23)) beyond the NusG dissociation constant for RNA polymerase (0.15 μM (24)), resulting in near-saturation of the NusG binding to RNA polymerase. On the other hand, the same segregation also limits the local concentration of ribosomes in the nucleoid (2–8 μM (22,25)) to as much as an order of magnitude below NusG’s dissociation constant for ribosomes (50 μM (15)), implying that only a small fraction of ribosomes is engaged by NusG. These data suggest that only a modest amount of the ternary complex of RNA polymerase, NusG, and ribosomes accumulates under conditions of transcription–translation coupling, raising the question of whether additional mechanisms of coupling ribosomes to RNA polymerase exist.
We hypothesize that direct interactions between RNA polymerase and ribosome may contribute to the coupling of RNA polymerase and ribosomes during transcription–translation coupling. Early electron microscopy as well as recent functional studies demonstrate that ribosomes can directly contact RNA polymerase by translating all of the nascent mRNA being synthesized (26,27). Such proximity may be stabilized by one of several ribosomal proteins that directly bind to RNA polymerase and moonlight as transcription factors (e.g., ribosomal protein S4, which binds to RNA polymerase and inhibits the premature termination of ribosomal RNA transcription (28)).
The present work tests the hypothesis of direct physical interactions between RNA polymerase and the ribosome. By applying biophysical methods and chemical crosslinking in combination with mass spectrometry, we demonstrate that these interactions occur. We expect that these direct interactions between RNA polymerase and the ribosome play an important role in the coupling of translation to transcription.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli MRE 600.rif was a kind gift from Dr. Knud Nierhaus, and pVS10 (T7P-α–β–β′-His6–ω), pIA900 (T7P-α-β-β′-TEV-His10-ω) and pIA1127 (T7P-His6-TEV-σ70[1–613]) were from Dr. Irina Artismovitch. Chemically competent T7 Express and BL21(DE3) E. coli were from New England Biolabs (Ipswich, MA, USA).
RNA polymerase and ribosome preparation
The purification of E. coli RNA polymerase and ribosomes followed published protocols (29–32) with minor modifications that were designed to reduce the co-purification of RNAs and other factors. Briefly, RNA polymerase captured on a Ni2+ affinity column was washed with two column volumes of 1.8 M NaCl before elution (33); ribosomes from the lysate were pelleted through a high-salt sucrose cushion (34). To remove ribosome-bound mRNAs, ribosomes were dissociated into subunits, and the purified subunits were re-associated into vacant ribosomes following a published protocol (32).
RNA polymerase•ribosome complex formation
In a standard reaction, 2.5 μM RNA polymerase and 2.5 μM ribosomes were incubated for 15 min at 37°C in buffer A (20 mM HEPES–KOH pH 7.5, 20 mM Mg(OAc)2 and 30 mM KCl), followed by 5 min of incubation at 4°C and a 10-min centrifugation at 19 000 × g at 4°C.
Separation of free RNA polymerase from ribosome–bound RNA polymerase
Mixtures of RNA polymerase and ribosomes were analyzed by rate zonal centrifugation or by gel filtration. For rate zonal centrifugation, samples were loaded on a 10–40% sucrose or 10–40% glycerol gradient in buffer A and centrifuged in an SW32.1 Ti rotor for 18 h at 24 000 rpm at 4°C. After centrifugation, the gradients were collected in 13 fractions starting from the bottom. The protein content of each fraction was precipitated with trichloroacetic acid (TCA) and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
For gel filtration, samples were loaded onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in buffer A with the KCl concentrations indicated in the text. The collected elution fractions were TCA-precipitated and analyzed by SDS-PAGE.
To probe the interactions between RNA polymerase and ribosomes, complex formation was performed in the presence of 20 μM bovine serum albumin (BSA), a 14-nucleotide-long RNA (rGrArGrUrCrUrGrCrGrGrCrGrArU) at a 5 μM concentration, 10 mM sodium phosphate pH 7.5, or additional KCl.
Separation of free ribosomes from RNA polymerase–bound ribosomes
RNA polymerase, ribosomes and mixtures of both were loaded onto Ni Sepharose High Performance spin columns and washed several times with increasing concentrations of imidazole before step-eluting the complex. Flowthrough, wash, and elution fractions were collected, TCA-precipitated, and analyzed by SDS-PAGE.
Capturing ribosomes with RNA polymerase immobilized on an affinity matrix
A Ni Sepharose High Performance column was loaded with saturating amounts of His-tagged RNA polymerase before loading purified ribosomes. The column was washed with increasing concentrations of imidazole before step-eluting the complex. Fractions from all steps were TCA-precipitated and analyzed by SDS-PAGE.
Determining the RNA polymerase–ribosome binding curves
To generate binding curves, various concentrations of ribosomes and RNA polymerase were incubated in buffer A with different concentration of KCl as indicated in the text. The content of each sample was analyzed by sucrose gradient centrifugation followed by SDS-PAGE and staining with colloidal Coomassie (35). The stained gels were imaged with a ChemiDoc™ Touch Gel Imaging System (Bio-Rad), and the β/β′ bands of each digital gel image were quantified using ImageJ 1.46 (36). The fraction of bound RNA polymerase was determined by subtracting the concentration-adjusted profile of the free RNA polymerase from that of the overall RNA polymerase profile.
The experimental data were modeled assuming that the RNA polymerase binds to one binding site on the ribosome in the presence of a dimer-monomer equilibrium of RNA polymerase. The affinities of RNA polymerase for ribosomes were estimated by nonlinear least-square fitting of the partition function
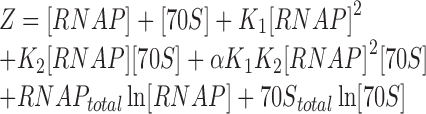 |
to the experimentally determined fraction of bound RNA polymerase using the ‘Equilibrium Expert’ add-in for Microsoft Excel® (37). [RNAP] and [70S] are the concentrations of free RNA polymerase and ribosomes, respectively; RNAPtotal and 70Stotal represent the total concentrations of RNA polymerase and ribosomes, respectively; K1 and K2 are the association constants for RNA polymerase dimer formation and RNA polymerase•ribosome complex formation, respectively; and α is the cooperativity factor for the binding of an RNA polymerase dimer to a ribosome. An average core enzyme•ribosome dissociation constant and its pooled standard deviation were calculated from the dissociation constants determined at 55 and 250 mM KCl according to (38) and yielded ∼0.93 ± 0.21 μM. To calculate the holoenzyme•ribosome dissociation constant in the presence of a dimer–monomer equilibrium of the holoenzyme, we assumed a dissociation constant of 10 μM (39,40) and a cooperativity factor (α) of one. To calculate the complex formation in the absence of RNA polymerase dimerization (i.e. holoenzyme), K1 was set to zero. The fraction of bound RNA polymerase was set to 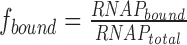 for titrating the complex with increasing amounts of ribosomes and
for titrating the complex with increasing amounts of ribosomes and 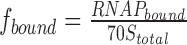 for titration with increasing amounts of RNA polymerase.
for titration with increasing amounts of RNA polymerase.
Preparation of NusG and sigma factor σ70
C-terminally His6-tagged NusG was purified by Ni2+ affinity chromatography. Sigma factor σ70 was purified using a protein construct with an N-terminal His6 tag and TEV protease site. After the initial capture of the protein on a Ni-NTA column, the protein was washed with two column volumes of 1 M NaCl before eluting with an imidazole gradient. The purified His6-tagged factor was digested with TEV protease, and the free sigma factor was then separated from its cleaved His-tag and the TEV protease by passing it over a Ni-NTA column.
Formation of functional RNA polymerase complexes
For preparation of holoenzyme, stoichiometric amounts of σ70 and core RNA polymerase were incubated (41). The transcription elongation complex was prepared following (42), in which first a 14-nucleotide oligoribonucleotide (rGrArGrUrCrUrGrCrGrGrCrGrArU) and non-template DNA (GCGATTCAGACAGG) are annealed to the template DNA strand (CCTGTCTGAATCGCTATCGCCGC) to form a DNA:RNA hybrid scaffold before incubating with core RNA polymerase.
Formation of tRNA–bound ribosome complexes
For preparation of tRNA–bound ribosomes, vacant ribosomes were incubated with an mRNA containing a Shine-Dalgarno sequence and codons for Pro and Phe (rArArArGrGrArArArUrArArArArCrCrArUrUrC), followed by sequential incubation with E. coli UGG tRNAPro isoacceptor and yeast tRNAPhe (43,44). The UGG tRNAPro isoacceptor was transcribed in vitro, purified by gel electrophoresis, and N1 methylated at G37 with TrmD and AdoMet (45).
Chemical crosslinking of RNA polymerase•ribosome complexes
The RNA polymerase•ribosome complexes from E. coli were incubated with 5 mM 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide (EDC) and 5 mM sulfo-N-hydroxysuccinimide (sulfo-NHS) at room temperature for 30 min before quenching the crosslinking reaction with 50 mM Tris–HCl. The protein content of the crosslinked sample was analyzed by SDS-PAGE with either a discontinuous 6–10% Tris-glycine or a 6–9% Tris-acetate gradient gel. The compositions of the SDS gel bands that occurred only in the presence of RNA polymerase and ribosomes were further analyzed by western blot or excised and stored at 4°C for further analysis by liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Sample preparation for LC–MS/MS
Excised SDS-PAGE bands were washed overnight before reducing and alkylating the captured protein with dithiothreitol and iodoacetamide, respectively. The alkylated proteins were subjected to tryptic digestion, followed by the extraction and desalting of the peptide fragments. The lyophilized peptides were resuspended in 0.1% formic acid and immediately analyzed by LC–MS/MS.
LC–MS/MS for protein identification and quantification
On-line LC–MS/MS analysis was performed on an LTQ-Orbitrap Velos mass spectrometer coupled with an EASY-nLC II HPLC system and a nanoelectrospray ionization source (Thermo, San Jose, CA, USA). Sample injection, enrichment, desalting, and HPLC separation were conducted automatically. The HPLC was equipped with an in-house-packed ReproSil-Pur C18-AQ column. The peptides were separated using a linear gradient of 2–40% acetonitrile in 0.1% formic acid at a flow rate of 230 nl/min and electrosprayed (spray voltage: 1.8 kV) into the mass spectrometer operating in the positive-ion mode. Data-dependent acquisition was enabled, and the twenty most abundant ions found in the full-scan (m/z 300–1500 at a resolution of 60 000 at m/z 400) MS exceeding a threshold of 1000 counts were selected for collision-induced dissociation to generate the MS/MS.
LC–MS/MS data analysis
Proteins were identified and quantified using MaxQuant software (version 1.5.3.8) (46) to search raw LC–MS/MS data against the E. coli database downloaded from Uniprot (47), which contains 4306 protein entries and additional entries for known contaminants. The fixed modification option was set to include cysteine carboamidomethylation, and the maximum number of missed cleavages for trypsin was set to two per peptide. The tolerance levels in mass accuracy for MS and MS/MS were set to 4.5 ppm and 0.6 Da, respectively. The false positive rate was set to 1%. For each protein, the spectral index (SI) was calculated as the sum of the ion intensities of all the tryptic peptides detected throughout the LC–MS/MS analysis of a sample (48):
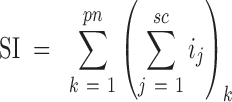 |
where ij is the ion intensity of the jth spectrum of peptide fragment k summed over all spectra sc for all tryptic peptides of the protein of interest, pn. The SI of each protein was weighted based on the length of the amino acid sequence of the protein:
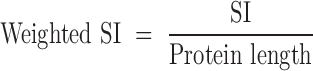 |
The weighted SI values for all identified proteins were normalized to the highest weighed SI value within the sample (48). Proteins were only considered enriched in the crosslink if they were present in all three biological replicates of the crosslinked band. We excluded proteins as potential RNA polymerase–ribosome interaction partners when they were present at the same relative mobility of the crosslinked species in either the crosslinked RNA polymerase or crosslinked ribosome sample and their SIs exceeded two-thirds of the SI observed for the crosslinked RNA polymerase–ribosome sample. The SIs of the remaining proteins were normalized to that of the protein with the highest index in each replicate. The average normalized SI values were calculated. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE17 partner repository (49) with the data set identifier PX006717.
RESULTS
E. coli RNA polymerase and ribosomes form a complex in vitro
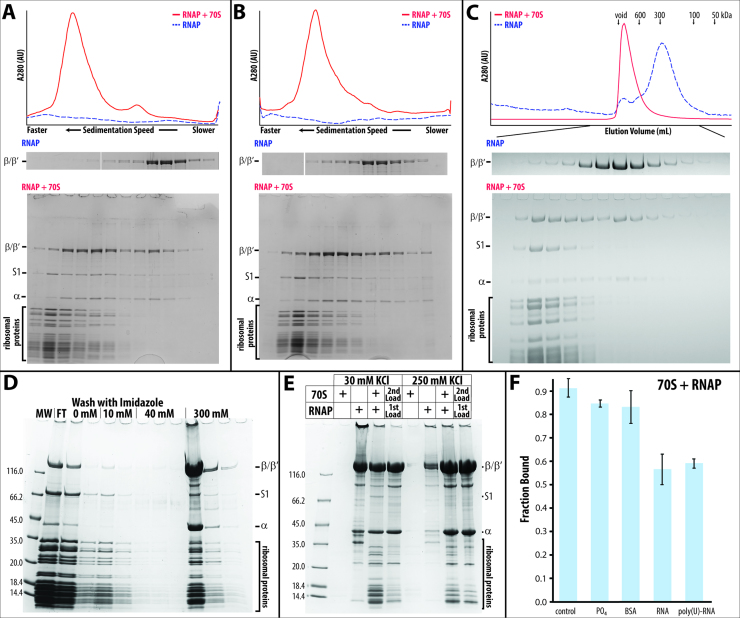
Eighty percent of RNA polymerase co-migrates with ribosomes when a micromolar mixture of stoichiometric amounts of RNA polymerase (core enzyme, consisting of α2ββ′ω subunits) and vacant ribosomes (lacking bound mRNA and tRNAs) from E. coli are separated by sucrose density gradient centrifugation (Figure 1A). The presence of sucrose or glycerol in the density gradient does not influence the extent of complex formation (compare Figure 1A with B). Similar levels of complex formation are detected when the mixture is separated by size exclusion chromatography (Figure 1C). Density gradient centrifugation and size exclusion chromatography probe the hydrodynamic parameters of the complex, which are dominated by the sedimentation coefficient and the size of the ribosome, preventing the separation of free ribosomes from bound ribosomes.
Figure 1.
Isolating RNA polymerase•ribosome complexes using different methods: (A) Sucrose gradient centrifugation. The top panel displays the sedimentation profiles of RNA polymerase alone (dashed blue line) and a stoichiometric mixture of RNA polymerase and ribosomes (solid red line) recorded at 280 nm. The two bottom panels display the SDS-polyacrylamide gel electrophoresis (SDS-PAGE) result of each of the sucrose gradient fractions. The top panel shows the β/β′ region of the SDS-PAGE result of the gradient centrifugation of RNA polymerase (RNAP) alone, while the bottom presents the full gel of a mixture of RNA polymerase and ribosomes (RNAP + 70S, marker lane removed for clarity). (B) Glycerol gradient centrifugation. The panels are the same as in A. During ultracentrifugation, the complex of RNA polymerase and ribosome constantly re-equilibrates, causing the bound RNA polymerase to trail the ribosome in A and B. (C) Size exclusion chromatography. The top panel shows the elution profiles of a mixture of RNA polymerase and ribosomes (solid red line) and of RNA polymerase alone (dashed blue line; for comparison, the absorption is increased by 160-fold) from a 10/30 Superdex 200 column. (D and E) Capturing His-tagged RNA polymerase•ribosome complexes on a Ni-sepharose spin column. (D) SDS-PAGE results of all fractions, i.e. flowthrough (FT), washes with 0, 10, and 40 mM imidazole, and elution with 300 mM imidazole. (E) SDS-PAGE results of the first 300 mM imidazole elution step from Ni2+ affinity binding experiments. Various amounts of RNA polymerase and ribosomes are either loaded together or sequentially—first RNA polymerase (‘1st Load’), followed by a stoichiometric amount of ribosomes (‘2nd Load’). These experiments are performed in the presence of 30 mM KCl and 250 mM KCl. (F) RNA polymerase binding to ribosomes in the presence of 10 mM phosphate buffer (PO4), 20 μM bovine serum albumin (BSA), a 14-nucleotide long RNA (RNA) at 5 μM, or 80 μg of poly(U)-RNA.
To separate free ribosomes from bound ribosomes, we captured the complex on a Ni2+ affinity matrix via a C-terminal poly(His)-tag on the β′ subunit of the RNA polymerase, either by pre-forming the RNA polymerase•ribosome complex or by first immobilizing the core enzyme on the matrix and then capturing vacant ribosomes from solution (Figure 1D). Although only an estimated 10% of the applied RNA polymerase–ribosome complex is captured by the Ni2+ affinity matrix, none of the ribosomes are captured in the absence of RNA polymerase (compare 70S alone and RNAP + 70S in Figure 1E).
Although all employed methods exploit different molecular principles, they all allude to a high yield of RNA polymerase•ribosome complex formation at micromolar concentrations of RNA polymerase and ribosomes, pointing to an apparent dissociation constant in the one to two micromolar range.
Non-specific competitors do not significantly impair complex formation
While RNA polymerase consists only of protein subunits, two-thirds of the mass of the ribosome consists of RNA, implying that protein–protein and/or RNA-protein interactions may contribute to complex formation. To exclude non-specific interactions between RNA polymerase and ribosomes, we formed the complex in the presence of several potential non-specific competitors for RNA-protein and protein–protein interactions, such as phosphate buffer, a short RNA, random-sized poly(U) RNA, or BSA. The extent of complex formation is not significantly impaired by the presence of either an 8-fold excess of BSA or by 10 mM phosphate buffer at pH 7.4 (Figure 1F). At higher phosphate buffer concentrations, ribosomes dissociate into subunits, i.e. in the presence of 10 mM phosphate buffer, none of the ribosomes dissociate, whereas with 100 mM phosphate buffer, approximately 40 % of the ribosomes dissociate into subunits. The presence of poly(U) RNA or a 14-nucleotide-long RNA reduces the extent of complex formation by approximately one-third. A fraction of RNA polymerase, ribosomes, and the RNA polymerase•ribosome complex appears to bind poly(U), causing it to sediment faster during sucrose gradient centrifugation. The nominal effect of these non-specific competitors for binding argues in favor of a specific interaction between the RNA polymerase core enzyme and the vacant ribosome. Thus, we hypothesize that the interactions between RNA polymerase and the ribosome are direct and specific.
Stoichiometry and dissociation constants of the RNA polymerase•ribosome complex
Specific interactions between macromolecules cause the formation of defined stoichiometric complexes. Depending on the ionic conditions, the RNA polymerase core enzyme exists in an equilibrium of multiple oligomeric states (39,50–52). To distinguish between the binding of an oligomer and the binding of multiple monomers, titration experiments were performed at salt concentrations that favor either the oligomeric or monomeric state of the RNA polymerase.
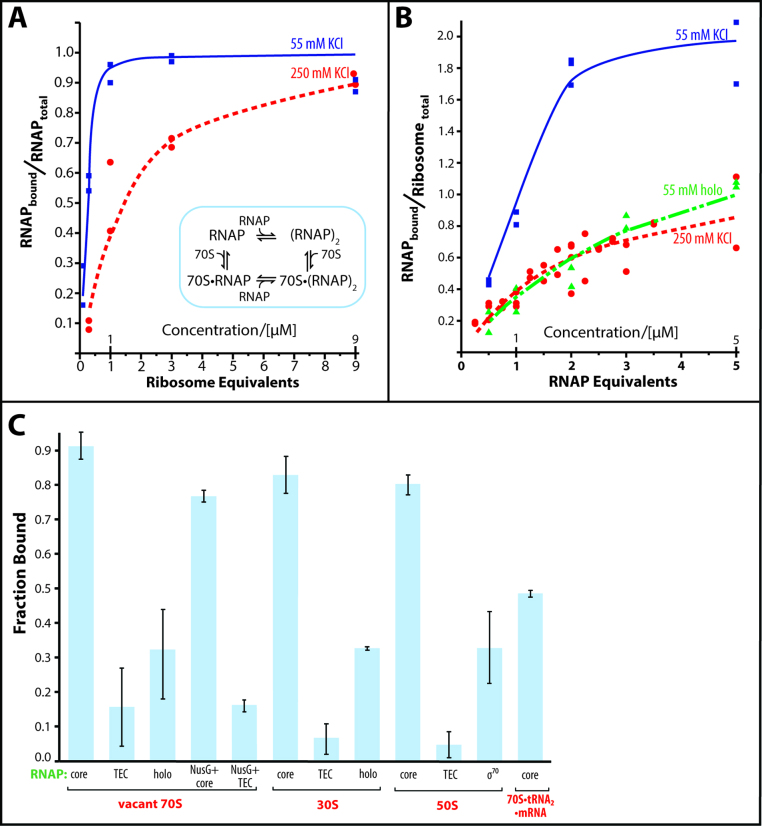
The titration of the core enzyme with vacant ribosomes in 55 mM KCl saturates at a one-to-one stoichiometry, while two equivalents of RNA polymerase bind to one equivalent of ribosomes under saturating conditions (Figure 2A and B, 55 mM KCl). At 250 mM KCl, a one-to-one complex is formed between the RNA polymerase and ribosomes at a saturating concentration of RNA polymerase (Figure 2B, 250 mM KCl). The similar sedimentation coefficients of ribosomes and of RNA polymerase•ribosome complexes indicate that only one ribosome is bound in each of the observed complexes. Ribosome dimers would sediment much faster than monomers, i.e. 100 Svedbergs versus 70, respectively (53–55).
Figure 2.
Titration of RNA polymerase with ribosomes and vice versa. (A) Binding of ribosomes to RNA polymerase. RNA polymerase (1 μM) is incubated with increasing concentrations of ribosomes in the presence of 55 mM KCl (blue squares) or 250 mM KCl (red circles). The inset displays the proposed binding model of complex formation. (B) Binding of RNA polymerase to ribosomes. Ribosomes (1 μM) are incubated with increasing concentrations of RNA polymerase core enzyme (blue squares) or holoenzyme (green triangles) in the presence of 55 mM KCl or 250 mM KCl (red circles, core enzyme only). The lines connecting the data in A and B are the binding curves calculated as described in the Materials and Methods section of the text. For the holoenzyme, the simulated binding curve in the presence of dimer–monomer equilibrium is shown. (C) Influence of the functional state of the RNA polymerase and of the ribosome on the RNA polymerase•ribosome complex formation. The complex formation of ribosomes (70S), small subunits (30S), large subunits (50S) and tRNA-bound ribosomes (70S•tRNA2•mRNA) with RNA polymerase core enzyme (core), transcription elongation complex (TEC), holoenzyme (holo), core enzyme and transcription elongation complex in the presence of NusG (NusG + core and NusG + TEC) was analyzed by sucrose gradient centrifugation.
We were able to model the formation of the RNA polymerase•ribosome complex assuming the presence of one binding site on the vacant ribosome and a dimer–monomer equilibrium of the core enzyme (with estimated dissociation constants of 0.02 μM and 0.2 mM for 55 and 250 mM KCl, respectively, based on (51), inset in Figure 2A). This model yields a nearly identical RNA polymerase•ribosome dissociation constant of 0.93 ± 0.21 μM. However, upon transitioning from 55 to 250 mM KCl, the presence of ribosomes ceases to skew the RNA polymerase towards dimer formation, as reflected by a drop in the cooperativity factor, α, from 50 to 1. The significantly lower ratio of ribosomes to RNA polymerase captured on the Ni-affinity matrix at 250 mM KCl compared to 30 mM KCl supports the results from sucrose gradient centrifugation (compare experiments at 30 mM with 250 mM KCl in Figure 1E).
The observed dissociation constant for the RNA polymerase•ribosome complex likely constitutes an upper limit, as the presence of a nascent RNA that connects both interaction partners will serve to further significantly reduce the dissociation constant. Nevertheless, this upper limit of the dissociation constant of the RNA polymerase•ribosome complex is well within in a physiologically relevant range, when compared with values for other processes that regulate RNA polymerase or ribosome activity (i.e. 0.9 μM for RNA polymerase binding to ribosomes versus 0.1 μM for transcription factor NusA binding to RNA polymerase (41) or 0.2 μM for EF-G (56) and 0.5 μM for EF-Tu•GTP•Phe-tRNAPhe (57) binding to ribosomes).
Characterization of the interaction between RNA polymerase and the ribosome
The RNA polymerase adopts multiple functional states in the course of transcribing a gene. Therefore, in addition to testing the core enzyme, we also tested RNA polymerase with bound sigma factor σ70 (holoenzyme) and RNA polymerase with a bound DNA:RNA scaffold (transcription elongation complex, TEC) as examples of the initiation and elongation states, respectively. The sigma factor, as well as the radioactively labeled RNA of the DNA:RNA scaffold, co-migrates with the RNA polymerase–ribosome complex (Supplementary Figure S2A and B), indicating that actively transcribing RNA polymerase may also participate in complex formation. Both states display reduced affinity for the ribosome, albeit to different extents—31% of the holoenzyme and 15% of the TEC bind to the ribosome versus 90% of the core enzyme (Figure 2C).
Nonlinear regression to best fit the measured binding of vacant ribosomes to holoenzyme results in a computed dissociation constant of 1.4 ± 0.2 μM; for this fit, we assumed the presence of dimer-monomer equilibrium for the holoenzyme, as predicted under our experimental conditions (39,40). Assuming the presence of only monomeric holoenzyme reduces the quality of the non-linear fit, yet yields a dissociation constant of 1.2 ± 0.2 μM, which is consistent with a weaker ribosome binding of the holoenzyme than of the core enzyme (Kd of core enzyme complex 0.9 ± 0.2 μM).
Like RNA polymerase, ribosomes adopt multiple functional states when translating a gene. Therefore, in addition to testing the vacant ribosomes, we tested tRNA-bound ribosomes. The tRNA-bound ribosomes display a weaker affinity for the core RNA polymerase (Figure 2C). The modulation of the dissociation constant by the functional states of the RNA polymerase and ribosome may indicate that certain combinations of functional states permit tight binding, possibly synchronizing transcription and translation during transcription–translation coupling.
The ribosome consists of a small and a large ribosomal subunit. To identify the contribution of each subunit to the binding of the RNA polymerase, we investigated the interaction of the RNA polymerase with each subunit individually. The RNA polymerase core enzyme interacts with both subunits (Figure 2C), and the non-specific competitors have similar effects on the complex formation as on the RNA polymerase•ribosome complex formation (Supplementary Figure S3). These results suggest that either each ribosomal subunit interacts with a different part of the RNA polymerase or that one RNA polymerase binding site is blocked upon subunit association. To distinguish between these two possibilities, we identified the RNA polymerase binding interfaces on the ribosome and on each of its subunits using chemical crosslinking.
Chemical crosslinking of RNA polymerase and ribosomes
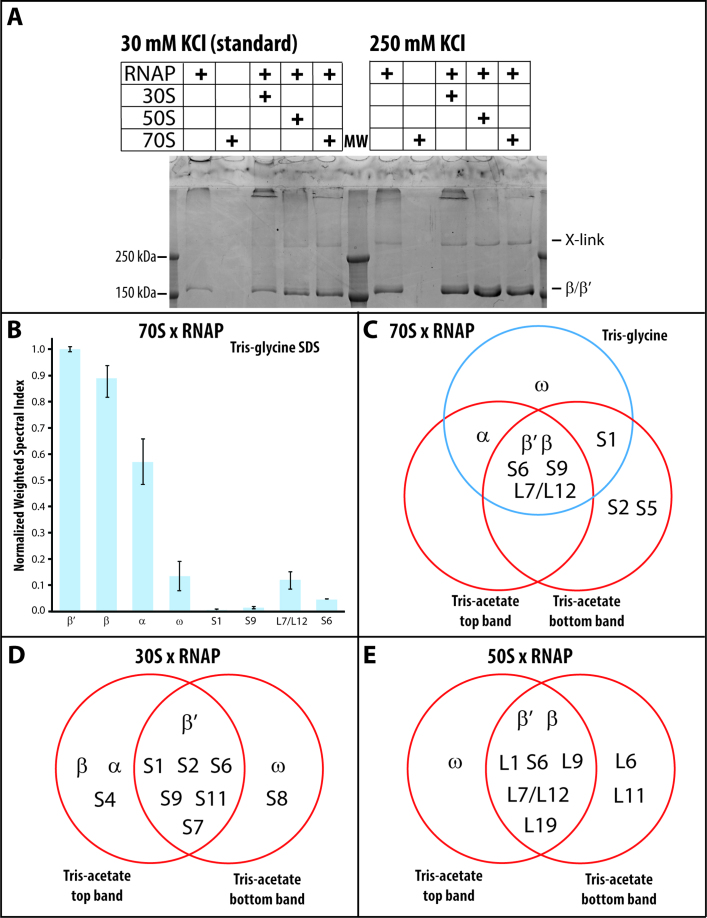
In the presence of sulfo-NHS, the zero crosslinker 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide (EDC) produces a ribosome-dependent crosslink of the core enzyme (Figure 3A, 30 mM KCl). The effect of non-specific competitors on the crosslinking efficiency mirrors the effect of these competitors on RNA polymerase•ribosome complex formation (Supplementary Figure S4A). In addition, the crosslinking efficiency correlates with the affinity of the RNA polymerase for the ribosome. The holoenzyme and TEC, which have lower affinity for ribosomes, display no increase in crosslink formation in the presence of ribosome (Supplementary Figure S4A inset).
Figure 3.
EDC crosslinking of RNA polymerase–ribosome complexes. (A) EDC crosslinking of RNA polymerase in the presence of the small ribosomal subunit (30S), large ribosomal subunit (50S), and ribosome (70S) with 30 mM and 250 mM KCl. (B) Normalized weighted spectral index of LC–MS/MS analysis of the Tris-glycine SDS-PAGE purified RNA polymerase–ribosome crosslink. (C) Venn diagram of crosslinked proteins after exposing a mixture of RNA polymerase and ribosomes to EDC and isolating the unique crosslinked species by 6–10% Tris-glycine (single band) and by 6–9% Tris-acetate SDS-PAGE (two bands). (D and E) Venn diagram of the proteins in the two species isolated from 6–9% Tris-acetate PAGE, which are specific to the EDC crosslinking of RNA polymerase in the presence of the small and the large ribosomal subunits.
However, an LC–MS/MS analysis of the SDS-PAGE-purified crosslink reveals a significant enrichment of the RNA polymerase subunits α, β, and β′ in the crosslink (Figure 3A). The presence of the RNA polymerase subunits in the crosslink was confirmed by western blot analysis (Supplementary Figure S4B). Therefore, we conclude that upon ribosome binding, the core enzyme adopts a conformation that promotes EDC-induced crosslinking within the polymerase. A similar crosslink is produced with the core enzyme alone in the presence of 250 mM KCl (Figure 3A, 250 mM KCl). RNA polymerase can be coaxed into different functional states by manipulating the solvent conditions (58,59). The similarity of the two crosslinks may point to a similar conformation of the RNA polymerase when bound to the ribosome and in the presence of 250 mM KCl.
EDC induces a limited number of crosslinks between RNA polymerase and the ribosome
MS analysis of the crosslink identified several ribosomal proteins that co-migrate with the intramolecularly crosslinked polymerase. These ribosomal proteins are less abundant than the α, β, or β′ subunits in the crosslinked band (Figure 3B), implying that only a fraction experienced an additional crosslinking event to a ribosomal protein. Except for minuscule amounts of β and β′ subunits, no other protein can be identified at the relative mobility of the crosslinked species in the absence of the chemical crosslinker (Supplementary Table S1). The crosslinked species resolve into at least three bands on Tris-acetate SDS gels (Supplementary Figure S5). LC–MS/MS analysis of two of these bands confirms that the single crosslinked species on our standard Tris-glycine gel indeed contains multiple components. Common to all three analyzed crosslinks are β, β′, S6, S9 and L7/L12 (Figure 3C; L7 is the acetylated form of L12. The ratio of L7 to L12 varies with the cell's growth phase (60)). The observed crosslinks place the ribosome-bound RNA polymerase on the cytosolic site of the small ribosomal subunit, covering the mRNA exit and entry sites (Figure 4C). The restricted number of ribosomal proteins in the crosslinks further hints at a defined arrangement between the RNA polymerase and the ribosome, which also supports our observation of a stoichiometric complex between RNA polymerase and ribosomes in solution.
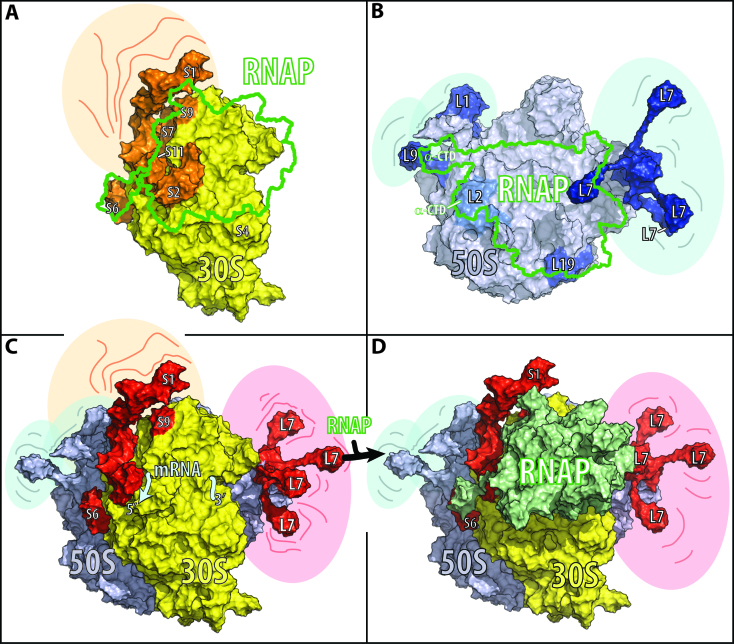
Figure 4.
Models for RNA polymerase binding to the small ribosomal subunit, the large ribosomal subunit, and the ribosome. Ribosomal proteins crosslinked to RNA polymerase (RNAP, in green) are indicated in orange for the small ribosomal subunit (30S, in yellow), in dark blue for the large ribosomal subunit (50S, in light blue), and red for the whole ribosome (70S). The model of the full-length RNA polymerase is based on the cryo-EM structure of E. coli RNA polymerase (PDB: 5UPC (89)) and the NMR structure of the C-terminal domain of the α subunit (PDB: 2MAX (90)). The models of the ribosome and its subunits are based on the cryo-EM structure of E. coli ribosomes (PDB: 4V6Q (91)). To complete the ribosome model, the L1 stalk is modeled based on the crystal structure of the L1 stalk (PDB: 1U63 (92)), the L7/L12 stalk is based on the NMR structure of L10•(L7/L12)4 (PDB 1RQU (93)), the C-terminal residues of ribosomal protein S6 are modeled according to the full-length S6 in the cryo-EM structure of the ribosome (PDB: 4V6P (91)), and ribosomal protein S1 is modeled based on the crystal structure of domain I in complex with ribosomal protein S2 (PDB 4TOI (94)) and the NMR structures of domains 4 and 6 (PDB 2KHI and 2KHJ (95)). The relative position of the RNA polymerase on the small ribosomal subunit and ribosome is restrained by the identified crosslinks as well as by the assumption that the nascent RNA between the RNA polymerase and ribosome has the shortest length during transcription–translation coupling. The shaded areas surrounding different components of the ribosome in A–D indicate spatial flexibility. On the small ribosomal subunit the flexible region involves ribosomal protein S1; on the large ribosomal subunit it involves the L1 stalk, the L7 stalk, and of the ribosomal protein L9; and on the ribosome the L1 stalk, the L7 stalk, and the ribosomal proteins S1 and L9. (A) The small ribosomal subunit viewed from the cytosolic site with crosslinked ribosomal proteins S6, S2, S11, S7, S9 and S1 in orange. The green boundary outlines the RNA polymerase position on the small ribosomal subunit. (B) The large subunit viewed from the ribosomal subunit interface. In dark blue are the crosslinked proteins L1, L9 and L19, and in blue is L2, a known binding partner of the RNA polymerase α-subunit. (C) Ribosome with identified crosslinked proteins S6, S9 and L7 in red. (D) Model of RNA polymerase–ribosome interactions based on the chemical crosslinks identified in this study. The figures were prepared in PyMOL (Schrödinger, LLC) using a solvent radius of 5 Å.
Crosslinks between RNA polymerase and ribosomal subunits overlap with those between RNA polymerase and the ribosome
The same crosslinked species are present on a Tris-acetate gel when the RNA polymerase is crosslinked to the small and to the large ribosomal subunits. LC–MS/MS analysis of two crosslinked species of the small and of the large ribosomal subunit indicates that the ribosomal proteins have a similar abundance relative to that of the RNA polymerase subunit, as they do when the RNA polymerase is crosslinked to the ribosome. Common to the two RNA polymerase-small subunit crosslinks are the RNA polymerase β′ subunit and the small ribosomal subunit proteins S1, S2, S6, S9, S11 and S7 (Figure 3D). All of the identified ribosomal proteins cluster next to the mRNA exit site on the small ribosomal subunit (Figure 4A).
Common to the two RNA polymerase-large ribosomal subunit crosslinks are the β and β′ RNA polymerase subunits and the ribosomal proteins L1, S6, L9, L7/L12 and L19 (Figure 3E). The proteins of the large ribosomal subunit are clustered at the tRNA entry (L7/L12) and exit sites (L1 and L9). L19 is the only ribosomal protein on the interface between the small and large ribosomal subunits (Figure 4B), which, upon association of these subunits, is buried within the ribosome. The presence of the S6 crosslink in experiments with the 50S subunit arises from cross-contamination of our large ribosomal subunits with a small amount of 30S subunits (<15 mol%). The identified crosslinks position the RNA polymerase on the ribosomal subunit interface of the large subunit, indicating that only the RNA polymerase interface of the small ribosomal subunit contributes to RNA polymerase binding to the ribosome.
DISCUSSION
In E. coli, the translation rate is the same as the transcription rate (27). The first ribosome trailing the transcribing RNA polymerase directly assists the polymerase during elongation (27) and suppresses transcription termination within coding regions (61–63). Recent ribosome profiling studies show that translational elongation speed is not uniform (64,65). Prolonged pausing of translation decouples RNA synthesis from protein synthesis, causing the premature termination of transcription (66). Prolonged pausing of transcription turns the RNA polymerase into a roadblock for the leading ribosome. Any barrier encountered by the translating ribosome promotes the loss of its reading frame (67–70), which, in turn, results in the premature termination of translation. Premature translation termination causes premature rho-dependent and rho-independent transcription termination (14,61,66). Consequently, the synchronization of transcription and translation is essential for gene expression in eubacteria.
The direct interaction between the RNA polymerase and ribosome results in stoichiometric complex formation. The strength of the interaction is modulated by the functional state of the RNA polymerase and the ribosome (Figure 2C), with the vacant ribosome binding more tightly to the core polymerase and holoenzyme than to the transcription elongation complex. Although this analysis was performed with stable functional states, the results suggest that the different states the RNA polymerase and ribosome adopt during transcription and translation will modulate the interaction between them. For instance, the binding of DNA to the RNA polymerase restricts the number of conformations the polymerase can sample, while binding of σ70 does not affect the conformational dynamics of the polymerase (71). Therefore, when bound to the ribosome, the core and holoenzyme may adopt a conformation that is not attainable by the transcription elongation complex, but enables a better interaction with the ribosome, thus explaining the observed difference in the binding affinities.
The spatial arrangement of the polymerase and ribosome is reflected in the proteins that are crosslinked within the complex and allows us to triangulate the location of the RNA polymerase on the surface of the subunits and the ribosome. The interface between the polymerase and the large subunit is located on the face that binds the small subunit, which, upon association with the small subunit, is buried within the ribosome (Figure 4B and C). Modeling the binding of RNA polymerase on the large ribosomal subunit places the RNA polymerase in close proximity to the ribosomal protein L2, which is known to bind the α subunit of RNA polymerase (72). The set of proteins crosslinked in the complex with ribosomes overlaps with that of the small ribosomal subunit complex (Figure 3C and D), clustering around the mRNA exit site of the ribosome (Figure 4A and C), hinting at the possible coordination between the transcription and translation initiation of the nascent RNA.
During the initial phase of translation initiation, the mRNA binds to a ‘standby’ site on the ribosome that encompasses the entire mRNA exit site (73). The binding of translation initiation factors repositions the mRNA on the small ribosomal subunit, permitting translation initiation to progress (74). In vivo, transcription pauses near the promoter (23,75). This pausing of the RNA polymerase seems to allow the ribosome to initiate translation and catch up with the polymerase. The ‘standby’ position of the mRNA may interfere with the interaction of the RNA polymerase with the mRNA exit site, thus coordinating the accommodation of the nascent RNA and the subsequent initiation of translation with the resumption of transcription. During eukaryotic translation initiation, initiation factor eIF3 also engages with the mRNA in the exit site while it is bound to both the mRNA entry and exit sites, thereby coordinating mRNA accommodation with the downstream steps of translation initiation (76,77).
Common to the identified RNA polymerase interfaces on the ribosome and small ribosomal subunit are ribosomal proteins S6 and S9; additionally, S1 is present in all but one of the identified crosslinked species (Figure 3C). Ribosomal protein S1 binds to RNA polymerase near the exit site of the nascent RNA (78,79) and promotes the recycling of the RNA polymerase after transcription termination (80). Similar to the C-terminal domains of ribosomal protein S1, the two to six glutamic acid residues of the C-terminal tail of S6 extend away from the ribosome, reaching into the surrounding solution. This local accumulation of glutamic acid residues may form the same salt bridges with the RNA polymerase as free glutamate at high cellular concentrations, when it releases the RNA polymerase stalled at the osmY promoter DNA (81). Although ribosomal protein S9 is farther from the mRNA exit site, its long C-terminal tail reaches through the head of the subunit to the mRNA channel. In the mRNA channel, the tail stabilizes the tRNA–mRNA interactions at the P-site (82), contributing to the fidelity of translation initiation and the maintenance of the reading frame (83) (Figure 4A and D).
The potential mechanistic implications of the observed interaction of the RNA polymerase with the large ribosomal subunit are difficult to reconcile with our current understanding of the coupling of transcription and translation. However, this interaction might hint at the potential coordination of ribosomal RNA transcription and ribosome assembly during ribosome biogenesis. Ribosome assembly factors can, like RNA polymerase, bind to mature ribosome particles in vitro and in vivo (84–87).
In summary, our binding studies suggest that during transcription–translation coupling, the RNA polymerase binds to the cytosolic site of the small ribosomal subunit, extending from the mRNA exit to the mRNA entry site. This binding allows the polymerase to monitor the translation rate of the ribosome while providing it with more nascent RNA. The coordination of transcription and translation may be conferred via the interaction with ribosomal protein S9.
While revising our manuscript, Cramer, Landick, and colleagues published a 9 Å cryo-EM structure of the RNA polymerase•ribosome complex (88). Although the stoichiometry and the ribosomal interface components of our model agree well with that of the EM structure, the relative orientation of the polymerase and ribosome are distinct. The EM structure places the polymerase more towards the mRNA entry site of the ribosome, with the nascent RNA exit site of the polymerase in closer contact with the mRNA entry site of the ribosome and the RNA polymerase rotated more towards the cytosol. Further studies will be required to understand the origins of these differences.
Our biochemical study, as well as the recently published EM structure of the RNA polymerase•ribosome complex (88), demonstrates the existence of a direct interaction between RNA polymerase and ribosomes, and points to its functional relevance during transcription–translation coupling. However, the here determined equilibrium constants only reflect the strength of the interaction between two the molecules, but not the time that interaction will persist in a dynamic setting, such as during on-going transcription and translation. Consequently, detailed kinetic studies will be needed to fully understand the feedback between the RNA polymerase and ribosome during transcription–translation coupling. The mechanistic insights derived from such studies will add to this new paradigm of how gene expression is controlled.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Carlos Rodriguez and Gabriela Sanchez for technical assistance during sample preparation, Dr. Jacques Barbet for unwavering technical support with the ‘Equilibrium Expert’ add-in for Microsoft Excel®, Dr. Whitney Yin for critical discussion, Weili Miao for assistance in analyzing the LC–MS/MS data, and Tim Rowsell, Timothy Tam, and Kacey Kilpatrick for assisting in the preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R21 ES025392 to Y.W.]; University of California, Riverside (to G.B. and Y.W.). Funding for open access charge: University of California, Riverside.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bakshi S., Choi H., Weisshaar J.C.. The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 2015; 6:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Y., Mooney R., Grass J., Sivaramakrishnan P., Herman C., Landick R., Wang J.. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol. Cell. 2014; 53:766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dutta D., Shatalin K., Epshtein V., Gottesman M.E., Nudler E.. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011; 146:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev.: MMBR. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yanofsky C., Ito J.. Nonsense codons and polarity in the tryptophan operon. J. Mol. Biol. 1966; 21:313–334. [DOI] [PubMed] [Google Scholar]
- 6. Landick R., Carey J., Yanofsky C.. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:4663–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yanofsky C. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 1999; 182:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franklin N.C., Luria S.E.. Transduction by bacteriophage P-1 and the properties of the lac genetic region in E. coli and S. dysenteriae. Virology. 1961; 15:299–311. [DOI] [PubMed] [Google Scholar]
- 9. Jacob F., Monod J.. Frisch L. Cellular Regulatory Mechanisms. 1961; XXVI, NY: The Biological Laboratory, Cold Spring Harbor, L.I; 193–211. [Google Scholar]
- 10. Kaempfer R. Nomura M, Tissieres A, Lengyel P. Ribosomes. 1974; NY: Cold Spring Harbor Laboratory; 679–704. [Google Scholar]
- 11. Baggett N.E., Zhang Y., Gross C.A.. Global analysis of translation termination in E. coli. PLoS Genet. 2017; 13:e1006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richardson J.P., Grimley C., Lowery C.. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:1725–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adhya S., Gottesman M., de Crombrugghe B., Court D.. Losick R, Chamberlin M. RNA Polymerase. 1976; NY: Cold Spring Harbor Laboratory; 719–730. [Google Scholar]
- 14. Adhya S., Gottesman M.. Control of transcription termination. Annu. Rev. Biochem. 1978; 47:967–996. [DOI] [PubMed] [Google Scholar]
- 15. Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rösch P.. A NusE:NusG complex links transcription and translation. Science. 2010; 328:501–504. [DOI] [PubMed] [Google Scholar]
- 16. Burmann B., Knauer S., Sevostyanova A., Schweimer K., Mooney R., Landick R., Artsimovitch I., Rösch P.. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012; 150:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drogemuller J., Strauss M., Schweimer K., Jurk M., Rosch P., Knauer S.H.. Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy. Scientific Rep. 2015; 5:16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belogurov G.A., Sevostyanova A., Svetlov V., Artsimovitch I.. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol. 2010; 76:286–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mooney R.A., Schweimer K., Rösch P., Gottesman M., Landick R.. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009; 391:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endesfelder U., Finan K., Holden S.J., Cook P.R., Kapanidis A.N., Heilemann M.. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys. J. 2013; 105:172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bratton B.P., Mooney R.A., Weisshaar J.C.. Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli. J. Bacteriol. 2011; 193:5138–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bakshi S., Siryaporn A., Goulian M., Weisshaar J.. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012; 85:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mooney R., Davis S., Peters J., Rowland J., Ansari A., Landick R.. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009; 33:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turtola M., Belogurov G.A.. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. eLife. 2016; 5:e18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanamrad A., Persson F., Lundius E.G., Fange D., Gynna A.H., Elf J.. Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11413–11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller O., Hamkalo B., Thomas C.. Visualization of bacterial genes in action. Science. 1970; 169:392–395. [DOI] [PubMed] [Google Scholar]
- 27. Proshkin S., Rahmouni A.R., Mironov A., Nudler E.. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010; 328:504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torres M., Condon C., Balada J.M., Squires C., Squires C.L.. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001; 20:3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nedialkov Y.A., Opron K., Assaf F., Artsimovitch I., Kireeva M.L., Kashlev M., Cukier R.I., Nudler E., Burton Z.F.. The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim. Biophys. Acta. 2013; 1829:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Artsimovitch I., Svetlov V., Murakami K.S., Landick R.. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lilneage-specific sequence insertions. J. Biol. Chem. 2003; 278:12344–12355. [DOI] [PubMed] [Google Scholar]
- 31. Fong B.A., Gillies A.R., Ghazi I., LeRoy G., Lee K.C., Westblade L.F., Wood D.W.. Purification of Escherichia coli RNA polymerase using a self-cleaving elastin-like polypeptide tag. Protein Sci. 2010; 19:1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blaha G., Stelzl U., Spahn C., Agrawal R., Frank J., Nierhaus K.. Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 2000; 317:292–309. [DOI] [PubMed] [Google Scholar]
- 33. Martin R.G., Gillette W.K., Martin N.I., Rosner J.L.. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 2002; 43:355–370. [DOI] [PubMed] [Google Scholar]
- 34. Robertson J.M., Paulsen H., Wintermeyer W.. Pre-steady-state kinetic studies on ribosomal translocation. Methods Enzymol. 1988; 164:581–597. [DOI] [PubMed] [Google Scholar]
- 35. Westermeier R. Sensitive, quantitative, and fast modifications for Coomassie Blue staining of polyacrylamide gels. Proteomics. 2006; 6(Suppl. 2):61–64. [DOI] [PubMed] [Google Scholar]
- 36. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raguin O., Gruaz-Guyon A., Barbet J.. Equilibrium expert: an add-in to Microsoft Excel for multiple binding equilibrium simulations and parameter estimations. Anal. Biochem. 2002; 310:1–14. [DOI] [PubMed] [Google Scholar]
- 38. Box G.E.P., Hunter J.S., Hunter W.G.. Statistics for Experimenters: Design, Innovation, and Discovery. 2005; 2nd ednHoboken: Wiley-Interscience. [Google Scholar]
- 39. Shaner S.L., Piatt D.M., Wensley C.G., Yu H., Burgess R.R., Record M.T. Jr. Aggregation equilibria of Escherichia coli RNA polymerase: evidence for anion-linked conformational transitions in the protomers of core and holoenzyme. Biochemistry. 1982; 21:5539–5551. [DOI] [PubMed] [Google Scholar]
- 40. Berg D., Chamberlin M.. Physical studies on ribonucleic acid polymerase from Escherichia coli B. Biochemistry. 1970; 9:5055–5064. [DOI] [PubMed] [Google Scholar]
- 41. Gill S., Weitzel S., von Hippel P.. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 1991; 220:307–324. [DOI] [PubMed] [Google Scholar]
- 42. Kashkina E., Anikin M., Tahirov T.H., Kochetkov S.N., Vassylyev D.G., Temiakov D.. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 2005; 34:4036–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schilling-Bartetzko S., Franceschi F., Sternbach H., Nierhaus K.H.. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J. Biol. Chem. 1992; 267:4693–4702. [PubMed] [Google Scholar]
- 44. Holschuh K., Gassen H.G.. Mechanism of translocation. Binding equilibria between the ribosome, mRNA analogues, and cognate tRNAs. J. Biol. Chem. 1982; 257:1987–1992. [PubMed] [Google Scholar]
- 45. Christian T., Evilia C., Williams S., Hou Y.M.. Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 2004; 339:707–719. [DOI] [PubMed] [Google Scholar]
- 46. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 47. Alpi E., Griss J., da Silva A.W., Bely B., Antunes R., Zellner H., Rios D., O’Donovan C., Vizcaino J.A., Martin M.J.. Analysis of the tryptic search space in UniProt databases. Proteomics. 2015; 15:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Griffin N.M., Yu J., Long F., Oh P., Shore S., Li Y., Koziol J.A., Schnitzer J.E.. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 2010; 28:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stevens A., Emery A.J. Jr, Sternberger N.. Sedimentation properties of E. coli RNA polymerase and its complexes with polyuridylic acid. Biochem. Biophys. Res. Commun. 1966; 24:929–936. [DOI] [PubMed] [Google Scholar]
- 51. Harris S.J., Williams R.C. Jr, Lee J.C.. Self-association of Escherichia coli DNA-dependent RNA polymerase core enzyme. Biochemistry. 1995; 34:8752–8762. [DOI] [PubMed] [Google Scholar]
- 52. Kansara S.G., Sukhodolets M.V.. Oligomerization of the E. coli core RNA polymerase: formation of (alpha2betabeta’omega)2-DNA complexes and regulation of the oligomerization by auxiliary subunits. PLoS One. 2011; 6:e18990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stoffler G., Hasenbank R., Lutgehaus M., Maschler R., Morrison C.A., Zeichhardt H., Garrett R.A.. The accessibility of proteins of the Escherichia coli 30S ribosomal subunit to antibody binding. Mol. Gen. Genet.: MGG. 1973; 127:89–110. [DOI] [PubMed] [Google Scholar]
- 54. Morrison C.A., Tischendorf G., Stoffler G., Garrett R.A.. Accessibility of proteins in 50S ribosomal subunits of Escherichia coli to antibodies: an ultracentrifugation study. Mol. Gen. Genet.: MGG. 1977; 151:245–252. [DOI] [PubMed] [Google Scholar]
- 55. Kato T., Yoshida H., Miyata T., Maki Y., Wada A., Namba K.. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010; 18:719–724. [DOI] [PubMed] [Google Scholar]
- 56. Katunin V.I., Savelsbergh A., Rodnina M.V., Wintermeyer W.. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002; 41:12806–12812. [DOI] [PubMed] [Google Scholar]
- 57. Maracci C., Peske F., Dannies E., Pohl C., Rodnina M.V.. Ribosome-induced tuning of GTP hydrolysis by a translational GTPase. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:14418–14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ruff E.F., Kontur W.S., Record M.T. Jr. Using solutes and kinetics to probe large conformational changes in the steps of transcription initiation. Methods Mol. Biol. 2015; 1276:241–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ruff E.F., Record M.T. Jr, Artsimovitch I.. Initial events in bacterial transcription initiation. Biomolecules. 2015; 5:1035–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gordiyenko Y., Deroo S., Zhou M., Videler H., Robinson C.V.. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 2008; 380:404–414. [DOI] [PubMed] [Google Scholar]
- 61. Li R., Zhang Q., Li J., Shi H.. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator. Nucleic Acids Res. 2016; 44:2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lesnik E.A., Sampath R., Levene H.B., Henderson T.J., McNeil J.A., Ecker D.J.. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001; 29:3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Smit M.H., Verlaan P.W., van Duin J., Pleij C.W.. In vivo dynamics of intracistronic transcriptional polarity. J. Mol. Biol. 2009; 385:733–747. [DOI] [PubMed] [Google Scholar]
- 64. Mohammad F., Woolstenhulme C.J., Green R., Buskirk A.R.. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016; 14:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Elgamal S., Artsimovitch I., Ibba M.. Maintenance of transcription–translation coupling by elongation factor P. mBio. 2016; 7:doi:10.1128/mBio.01373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yan S., Wen J.D., Bustamante C., Tinoco I. Jr. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015; 160:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen J., Petrov A., Johansson M., Tsai A., O’Leary S.E., Puglisi J.D.. Dynamic pathways of -1 translational frameshifting. Nature. 2014; 512:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim H.K., Liu F., Fei J., Bustamante C., Gonzalez R.L. Jr, Tinoco I. Jr. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Caliskan N., Katunin V.I., Belardinelli R., Peske F., Rodnina M.V.. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014; 157:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chakraborty A., Wang D., Ebright Y.W., Korlann Y., Kortkhonjia E., Kim T., Chowdhury S., Wigneshweraraj S., Irschik H., Jansen R. et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012; 337:591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rippa V., Cirulli C., Di Palo B., Doti N., Amoresano A., Duilio A.. The ribosomal protein L2 interacts with the RNA polymerase alpha subunit and acts as a transcription modulator in Escherichia coli. J. Bacteriol. 2010; 192:1882–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yusupova G., Jenner L., Rees B., Moras D., Yusupov M.. Structural basis for messenger RNA movement on the ribosome. Nature. 2006; 444:391–394. [DOI] [PubMed] [Google Scholar]
- 74. Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell. Mol. Life Sci. 2015; 72:4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S.. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014; 344:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Aylett C.H., Boehringer D., Erzberger J.P., Schaefer T., Ban N.. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat. Struct. Mol. Biol. 2015; 22:269–271. [DOI] [PubMed] [Google Scholar]
- 77. Aitken C.E., Beznoskova P., Vlckova V., Chiu W.L., Zhou F., Valasek L.S., Hinnebusch A.G., Lorsch J.R.. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife. 2016; 5:e20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sukhodolets M.V., Garges S.. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003; 42:8022–8034. [DOI] [PubMed] [Google Scholar]
- 79. Liu B., Zuo Y., Steitz T.A.. Structural basis for transcription reactivation by RapA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2006–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sukhodolets M.V., Garges S., Adhya S.. Ribosomal protein S1 promotes transcriptional cycling. RNA. 2006; 12:1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee S.J., Gralla J.D.. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004; 14:153–162. [DOI] [PubMed] [Google Scholar]
- 82. Selmer M., Dunham C., Murphy F., Weixlbaumer A., Petry S., Kelley A., Weir J., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 83. Arora S., Bhamidimarri S.P., Weber M.H., Varshney U.. Role of the ribosomal P-site elements of m(2)G966, m(5)C967, and the S9 C-terminal tail in maintenance of the reading frame during translational elongation in Escherichia coli. J. Bacteriol. 2013; 195:3524–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sharma M.R., Barat C., Wilson D.N., Booth T.M., Kawazoe M., Hori-Takemoto C., Shirouzu M., Yokoyama S., Fucini P., Agrawal R.K.. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol. Cell. 2005; 18:319–329. [DOI] [PubMed] [Google Scholar]
- 85. Gibbs M.R., Moon K.M., Chen M., Balakrishnan R., Foster L.J., Fredrick K.. Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Qin Y., Polacek N., Vesper O., Staub E., Einfeldt E., Wilson D.N., Nierhaus K.H.. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006; 127:721–733. [DOI] [PubMed] [Google Scholar]
- 87. Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008; 14:381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kohler R., Mooney R.A., Mills D.J., Landick R., Cramer P.. Architecture of a transcribing-translating expressome. Science. 2017; 356:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kang J.Y., Olinares P.D., Chen J., Campbell E.A., Mustaev A., Chait B.T., Gottesman M.E., Darst S.A.. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLife. 2017; 6:e25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Borin B.N., Tang W., Krezel A.M.. Helicobacter pylori RNA polymerase alpha-subunit C-terminal domain shows features unique to varepsilon-proteobacteria and binds NikR/DNA complexes. Protein Sci. 2014; 23:454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Agirrezabala X., Liao H.Y., Schreiner E., Fu J., Ortiz-Meoz R.F., Schulten K., Green R., Frank J.. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6094–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nevskaya N., Tishchenko S., Gabdoulkhakov A., Nikonova E., Nikonov O., Nikulin A., Platonova O., Garber M., Nikonov S., Piendl W.. Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA. Nucleic Acids Res. 2005; 33:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bocharov E.V., Sobol A.G., Pavlov K.V., Korzhnev D.M., Jaravine V.A., Gudkov A.T., Arseniev A.S.. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 2004; 279:17697–17706. [DOI] [PubMed] [Google Scholar]
- 94. Byrgazov K., Manoharadas S., Kaberdina A.C., Vesper O., Moll I.. Direct interaction of the N-terminal domain of ribosomal protein S1 with protein S2 in Escherichia coli. PLoS ONE. 2012; 7:e32702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Salah P., Bisaglia M., Aliprandi P., Uzan M., Sizun C., Bontems F.. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 2009; 37:5578–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.