Abstract
The birth of novel genes, including their cell-specific transcriptional control, is a major source of evolutionary innovation. The lens-preferred proteins, crystallins (vertebrates: α- and β/γ-crystallins), provide a gateway to study eye evolution. Diversity of crystallins was thought to originate from convergent evolution through multiple, independent formation of Pax6/PaxB-binding sites within the promoters of genes able to act as crystallins. Here, we propose that αB-crystallin arose from a duplication of small heat shock protein (Hspb1-like) gene accompanied by Pax6-site and heat shock element (HSE) formation, followed by another duplication to generate the αA-crystallin gene in which HSE was converted into another Pax6-binding site. The founding β/γ-crystallin gene arose from the ancestral Hspb1-like gene promoter inserted into a Ca2+-binding protein coding region, early in the cephalochordate/tunicate lineage. Likewise, an ancestral aldehyde dehydrogenase (Aldh) gene, through multiple gene duplications, expanded into a multigene family, with specific genes expressed in invertebrate lenses (Ω-crystallin/Aldh1a9) and both vertebrate lenses (η-crystallin/Aldh1a7 and Aldh3a1) and corneas (Aldh3a1). Collectively, the present data reconstruct the evolution of diverse crystallin gene families.
Keywords: aldehyde dehydrogenase, crystallin, eye evolution, heat shock responsive element, lens, Pax6, small heat shock protein
Introduction
The birth of novel genes with potentially new functions is thought to be a major contributor to evolution (Kaessmann 2010). Evolution from ancestral proteins is a process by which mutation and selection processes generate new materials for either improved functions and/or formation of novel biological structures, that is, morphological evolution (Carroll 2008). It has been shown that the total number of protein-coding genes in genomes of diverse model metazoan organisms is similar. However, besides differences in protein content and sequences, the underlying functional differences of the encoded information are determined by differences in the wiring of the gene regulatory networks (GRNs) that control both formation and function of individual tissues and organs (Davidson and Erwin 2006; Wagner 2007; Carroll 2008; Davidson, 2010). Although a number of important insights into the molecular evolution have been achieved since the dawn of DNA sequencing, particularly regarding coding sequences, evolution of regulatory sequences is not well understood (Wray etal. 2003).
The key principles of metazoan evolution include gene duplication to generate novel genes (Conant and Wolfe 2008), changes in gene control mechanisms (Piatigorsky and Wistow 1991; Wray etal. 2003; Wagner 2007; Carroll 2008), and activity of mobile genetic elements (Kazazian 2004). Simulation of processes required to establish novel cis-acting sites through local point mutations (Stone and Wray 2001; Wagner etal. 2007; Behrens and Vingron 2010), identification of prospective cis-sites in DNA-repetitive sequences (Zhou etal. 2002; Zemojtel etal. 2009), and comparisons of homologous regulatory sequences in related species (Lowe etal. 2011) indicate that new cis-sites can be generated on microevolutionary timescales (Stone and Wray 2001; Stern and Orgogozo 2008; Behrens and Vingron 2010).
Eye evolution exemplifies a multitude of challenges to test evolutionary theories; at the same level, the eye provides a considerable number of molecular insights to directly link tissue morphogenesis with conserved and/or novel gene functions (Lamb etal. 2007; Nilsson 2009, 2013; Oakley and Speiser 2015). Reconstruction of key processes in eye evolution has now become possible (Gehring and Ikeo 1999; Gehring 2004; Carroll 2008; Vopalensky and Kozmik 2009; Gehring 2012; Vopalensky etal. 2012). To simplify analysis of rapidly evolving eye complexity, four discrete stages have been proposed (Nilsson 2009, 2013). Nondirectional photoreception (stage I) requires only a few photosensitive cells. Directional photoreception (stage II) requires a combination of pigmented cells that shield one or multiple photoreceptors forming the eyespot. Low-resolution vision (stage III), achieved by pit/cup eyes, does not employ any focusing optics. Finally, high-resolution vision (stage IV) also added an ocular lens as a focusing apparatus to generate the camera-like eye (Jonasova and Kozmik 2008). The ocular lens is a cellular structure mostly composed of highly elongated lens fiber cells, the crystallin-producing cellular factories (Piatigorsky 1981; Bassnett etal. 2011). At present, camera-like eyes (supplementary fig. S1, Supplementary Material online) are found in some jellyfish (Cnidarians), a range of invertebrates including scallops, squid and octopus (Mollusks), and in vertebrates starting during the Cambrian revolution (525 Ma, e.g., lampreys, supplementary fig. S1, Supplementary Material online). The fossil record shows that primitive eyes existed prior to this period and were further improved to provide high-resolution vision during the Cambrian period (Land and Nilsoon 2002). Thus, understanding lens evolution is intimately linked with studies of its principal structural proteins, the crystallins as well their encoding genes (Piatigorsky and Wistow 1991; Piatigorsky 2007).
Crystallins are water-soluble proteins that are highly expressed in lens fiber cells (Wistow and Piatigorsky 1988); however, there is no apparent similarity between crystallins of vertebrates and invertebrates. In vertebrates, crystallins are represented by over a dozen proteins that are classified by their elution profiles in gel filtration as α-, β- and γ-crystallins (Piatigorsky 1981). It has been proposed earlier that either αB- or βB2-crystallin could be the “original” vertebrate crystallin as both of them are expressed outside of the eye (Wistow 1993). The pair of αA- and αB-crystallins are structurally and functionally related to small heat shock proteins, called Hspbs (Ingolia and Craig 1982). Accordingly, the mammalian αB-crystallin transcriptional regulatory regions contain multiple heat shock responsive elements (HSEs) (Klemenz etal. 1991; Somasundaram and Bhat 2000, 2004). Hspbs form a family of ten vertebrate genes (Kappe etal. 2003; Franck etal. 2004; Wistow 2012). Vertebrate lenses also contain a variable number of proteins from the β/γ-crystallin superfamily that is composed of at least 14 members (Kappe etal. 2010). This β/γ-crystallin superfamily also includes Ca2+-binding proteins encoded by microbial genomes (Kappe etal. 2010; Srivastava etal. 2014).
In contrast, different abundant proteins were found in invertebrate lenses, including S-crystallins (squid and octopus), Ω-crystallins (scallop and squid), and J-crystallins (jellyfish) (supplementary fig. S1, Supplementary Material online). These crystallins are identical to basic metabolic enzymes, such as aldehyde dehydrogenases (Aldhs, Ω-crystallins) and glutathione-S-transferases (S-crystallins) (Piatigorsky 2007; Marchitti etal. 2008; Chen etal. 2013; Vasiliou etal. 2013). In particular, both scallop (Placopecten magellanicus) and squid (Omnastrephes sloani pacificus) express Aldh1a9 as their Ω-crystallin (Tomarev and Piatigorsky 1996). In addition, other crystallins were identified in jellyfish (Tripedalia cystophora) with more distant similarities to known proteins, including ADP-ribohydroxylase (J1-crystallins) and saponins (J3-crystallin) (Piatigorsky and Kozmik 2004). In the absence of any common denominator between vertebrate and invertebrate crystallins, the remarkable diversity of crystallins was thought to originate from their ability to accumulate to high concentrations while maintaining their solubility through the processes of convergent evolution.
To explain the origin of crystallin genes, a concept of “gene recruitment” was proposed (Piatigorsky and Wistow 1991). The hallmark of this idea is that gene-duplication is not essential to establish any novel function; only the transcriptional regulation of a gene needs to change to gain neofunction. Thus, an acquisition of lens-preferred (“lens-specific”) mechanism(s) of gene expression allows quantitatively significant increases in expression of these proteins in lens, while maintaining appropriate levels and timing of expression in other tissues/organs (Piatigorsky and Wistow 1991). Despite the differences among vertebrate and invertebrate crystallins in the origins of their protein-coding sequences, a series of studies of their transcriptional control revealed a remarkable similarity in the use of an evolutionarily conserved DNA-binding transcription factor Pax6 (Callaerts etal. 1997; Chow and Lang 2001; Piatigorsky 2007). However, none of the earlier studies have examined contribution of divergent and convergent evolutionary processes to explain how the lens-specific GRNs have evolved.
Transcriptional studies of vertebrate α-, β-, and γ-crystallins have shown Pax6, c-Maf, Hsf4, Prox1, and Sox1 as their key DNA-binding factors (supplementary fig. S2, Supplementary Material online) (Cvekl and Duncan 2007; Cvekl etal. 2015). Both jellyfish J2- and J3-crystallins are regulated by PaxB (Kozmik etal. 2003), and scallop Ω-crystallin/Aldh1a9 is directly regulated by Pax6 (Carosa etal. 2002). PaxB is a Pax gene that lost a major part of its homeodomain following gene duplications of the ancestral Pax gene formed by proto-Pax transposase inserted into a homeobox-containing genomic region about 1 billion years ago (Breitling and Gerber 2000). Taken together, several lines of evidence support the model of a conversion of a preexisting gene into a crystallin-coding gene via acquisition of Pax6/PaxB-binding sites (Cvekl and Piatigorsky 1996; Kozmik etal. 2003; Kozmik etal. 2008).
Key questions for understanding the origins of individual crystallin genes are 1) how lens regulatory mechanisms evolved in specific gene(s) and 2) why proteins encoded by these genes evolved successfully as lens crystallins. Here we propose that Pax6-binding sites could evolve from preexisting cis-sites governing stress-response. Multiple sequence alignments and phylogenetic tree analyses of Hspb/Cryab/Cryaa genes demonstrate that an ancestral Hspb1-like gene evolved into a pair of αΑ- and αΒ-crystallin paralogs. Our data also show that the ancestral β/γ-crystallin could have been formed in the cephalochordate/tunicate lineage as a gene fusion in which the Hspb1-like gene provided a promoter region inserted 5′ to the coding region of a Ca2+-binding protein. Finally, notable similarities between multiple invertebrate crystallin promoters raise the possibility that they evolved from an ancestral Aldh gene promoter.
Materials and Methods
Sequence Data Retrieval
The amino acid and nucleotide coding sequences were obtained from sequenced genomes available from the UCSC Genome Browser and from the Department of Energy (DOE) Joint Genome Institute.
Annotation of Crystallin Promoters
The following “consensus” binding site sequences were used: HSE, 5′-GAANNTTCNNGAA-3′ (Fujimoto etal. 2008), P6CON (Epstein etal. 1994), and Pax6 ChIP-seq (Sun etal. 2015).
Multiple Sequence Alignments
Multiple sequence alignments with DNA and protein sequences were conducted using T-coffee (Notredame etal. 2000), MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar 2004), and MAFFT (version 7) (Katoh and Standley 2013). The phylogenetic tree analysis was conducted with maximum likelihood method and bootstrap with 1,000 calculations using MEGA7 (Kumar etal. 2016). All the alignments and phylogenetic analysis used default setting unless otherwise stated.
Expression of Cryaa, Cryab, Hspb1, Hspb2, and Hspb6 in Mouse Heart and Lens
Mouse embryonic E16.5 lenses (n = 8) and newborn and hearts (n = 2) were obtained from CD-1 mice (Charles River Laboratories). Dissected tissues were incubated overnight in in 35 mm dishes at 37 °C in 5% CO2 in 2 ml of DMEM (10% FBS). To treat the tissues with heat shock, the media was replaced with DMEM (10%FBS) at 42 °C and the dishes incubated for 1 h at 42 °C in 5% CO2. The tissues were then returned to 37 °C media and allowed to recover for 0.5, 1, and 4 h at 37 °C in 5% CO2. Total RNAs were prepared using the RNeasy Mini Kit (QIAGEN, Valencia, CA), followed by quantitative RT-PCR analysis as described elsewhere (Xie and Cvekl 2011). Ct values were normalized using GAPDH and fold changes of gene expression were calculated relative to untreated tissues. Primer sequences used for quantitative RT-PCR were: Cryaa forward 5′-GAGATTCACGGCAAACACAA-3′; Cryaa reverse 5′-ACATTGGAAGGCAGACGGTA-3′; Cryab forward 5′-TCTCTCCGGAGGAACTCAAA-3′; Cryab reverse 5′-TCCGGTACTTCCTGTGGAAC-3′; Hspb1 forward 5′-CCGGAAATACACGCTCCCTC-3′; Hspb1 reverse 5′-ATGGTGATCTCCGCTGACTG-3′; Hspb2 forward 5′-CCGAGTACGAATTTGCCAACC-3′; Hspb2 reverse 5′-CCCGAGGCCGAACATAGTAG-3′; Hspb6 forward 5′-TGTCCACGGACTCTGGGTAT-3′; Hspb6 reverse 5′-TGAATCCGTGTTCATCCGGG-3′; GAPDH forward 5′-CCAATGTGTCCGTCGTGGATCT-3′; GAPDH reverse 5′-GTTGAAGTCGCAGGAGACAACC-3′.
Transcriptional Regulation by Putative Pax6-Binding Sites Identified in B. floridae
Six copies of candidate Pax6-binding sites (WT1 and WT2), their mutants (M1 and M2), and “optimized” sites (O1 and O2), were synthesized by GeneScript (Piscataway, NJ) and cloned into a E4 TATA-luc minimal promoter followed by transient cotransfection experiments as we described earlier (Chauhan etal. 2004a, 2004b). The cells used included mouse lens epithelial cell line αTN4-1 and embryonic carcinoma cells P19 as used previously to evaluate transactivation by Pax6 (Xie and Cvekl 2011; Chauhan etal. 2004a).
Results
A Pax6 Consensus-Binding Site Resembles Recognition Motifs Recognized by Stress-Induced TFs
Pax6 binds to DNA through a combinatorial use of paired domain (PD) and homeodomain (HD) (fig. 1A) that by footprinting occupies 16–20 bp long sites (Epstein etal. 1994; Xu etal. 1999; Yang and Cvekl 2005) and contains variants of a 14 bp consensus sequence, P6CON (Epstein etal. 1994), which is highly similar to the recently established invivo 15 bp Pax6 consensus sequence (Sun etal. 2015). Evolution of such sequences for Pax6-binding could originate from random mutations and/or through a smaller number of nucleotide changes using preexisting cis-sites.
Fig. 1.
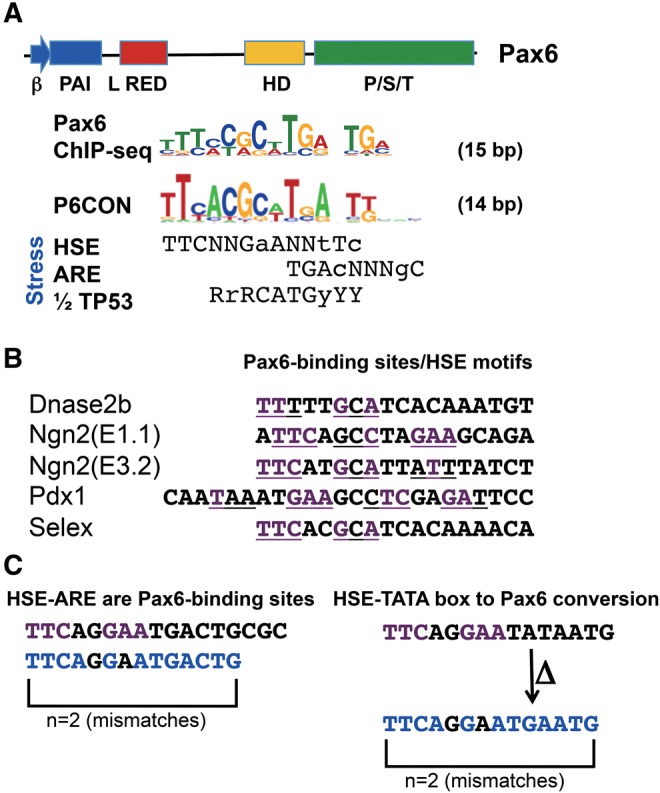
—Comparison of Pax6 consensus binding site with three stress-response sites. (A) Alignments of invivo Pax6 consensus (Sun etal. 2015) and P6CON (Epstein etal. 1994) with stress-response elements, including canonical trimeric HSE, ARE, and half of p53 (1/2 TP53) sites. Five highly interactive DNA-binding subdomains are found in Pax6: β−turn, PAI, RED, linker (L), and HD (Xu etal. 1999). (B) Identification of optimal HSEs (underlined, conserved nucleotide, and purple) in a set of four validated Pax6-binding sites, and in multiple “optimal” Pax6-binding sites generated by Selex (Xie and Cvekl 2011). (C) A model for the immediate utilization and simple conversion of a HSE-ARE and HSE-TATA box into a Pax6-binding site without any nucleotide change (left panel) or via a single nucleotide substitutions (arrow) coupled with a single nucleotide deletion (Δ) (right panel), respectively.
Here, we hypothesize that a Pax6-binding site could have rapidly emerge from related cis-regulatory regions in various promoters and enhancers of genes during the course of eye evolution. Given the link between vertebrate crystallins and stress-response, we evaluated a mutual relationship between HSEs, antioxidant response elements (AREs), p53-binding sites, and both the invivo Pax6 consensus sequence and Pax6 paired domain consensus, P6CON (fig. 1A). This “optimal” Pax6 site could be indeed aligned with these stress-response elements (fig. 1A). Among a group of ∼60 validated Pax6-binding sites (Xie and Cvekl 2011), an appreciable number of them indeed preserve high sequence similarity to HSE motifs (fig. 1B). To illustrate minimal nucleotide changes required to generate a Pax6-binding site, we show two plausible model “seed” sequences. First, a HSE-ARE tandem is already a Pax6-binding site with two nucleotide mismatches (fig. 1C). Second, a region comprised of a single HSE and a canonical TATA-box would require only one nucleotide substitution (A to G) and a single nucleotide deletion (ΔT) to convert this array of cis-elements into a Pax6-binding site (fig. 1C). This scenario of Pax6 site emergence is conceivably more parsimonious than the alternative that a random 14-bp sequence was converted to Pax6 site. In addition, a consensus ARE, identified in promoters of squid crystallins (Tomarev etal. 1994) partially overlaps with the “optimal” Pax6-binding site (fig. 1A). Considering stress regulation at the level of transcription by p53 (Lohrum and Vousden 1999; Danilova etal. 2008), coupled with recent identification of p53-binding sites in distal regions of mouse Cryaa and Cryba1 genes (Ji etal. 2013), we also noticed similarities between a Pax6-binding site and a half of the p53-decameric sequence (fig. 1A). These sequence comparisons suggest that an individual gene can be recruited into a Pax6-dependent gene regulation through a small number of nucleotide changes within the preexisting cis-elements, in agreement with a recent systematic analysis reporting that binding sites of a number of transcription factors differ indeed by a few nucleotides (Payne and Wagner 2014).
Gene Duplication and Transcriptional Regulation of Hspb1, Hspb2, Hspb6, and Cryab/Hspb5
To reconstruct molecular evolution underlying the birth of crystallin genes in vertebrate genomes, it is necessary to identify an ancestral gene to track changes in transcriptional mechanisms that proceeded towards the evolution of high lens-preferred gene expression. A group of three mammalian proteins, including Hspb1, Hspb6, and Hspb2, is highly related to both α-crystallins (Kappe etal. 2003; Franck etal. 2004; Basha etal. 2012). However, a precise evolutionary history of this group of genes is still not clear from the protein analysis, even if exon/intron boundaries and other features are considered (Franck etal. 2004).
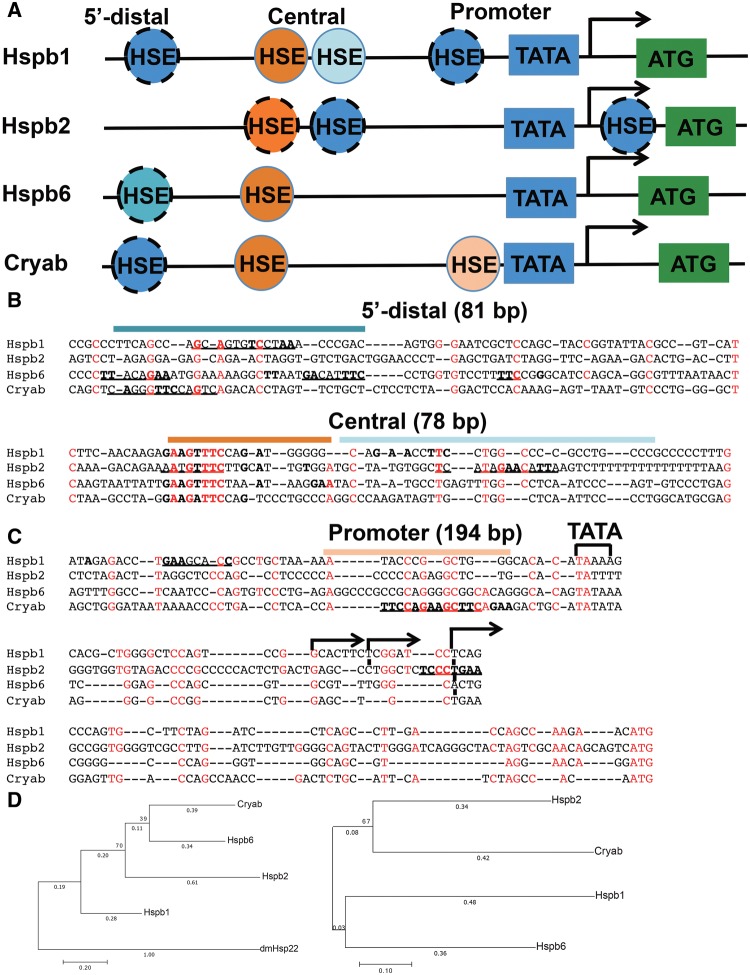
To resolve this issue, we hypothesized that the analysis of transcriptional regulatory sequences of Hspb1, Hspb2, Hspb6, and Cryab could provide valuable clues to explain the process that led to the evolution of lens-preferred expression of an ancestral Cryab gene. Previous studies identified multiple HSEs in human and mouse HSPB1/Hspb1 promoters (Gaestel etal. 1993; Oesterreich etal. 1996). In contrast, gene regulation of Hspb6 and Hspb2 genes is poorly understood (Hough etal. 2002; Li etal. 2007; Swamynathan and Piatigorsky 2007). Initial studies of αB-crystallin gene regulation identified distal HSE in the muscle-lung-heart (MLH) enhancer (−427/−254) and proximal HSE adjacent to the TATA-box (fig. 2A;Klemenz etal. 1991; Somasundaram and Bhat 2004; Jing etal. 2013). To align regulatory sites of Hspb1, Hspb2, Hspb6, and Cryab, we first identified HSEs (fig. 2A). These HSEs were classified as “optimal” (at least two perfect consensus trinucleotides; fig. 2A, solid circles) and “imperfect” (a sequence with only one consensus trinucleotide and/or with at least three scattered “core” nucleotides (Fujimoto etal. 2008; fig. 2A, dashed circles). All four genes examined showed comparable arrangements of multiple HSEs in their 5′-flanking regions (fig. 2A). At the nucleotide level, these promoter regions show notable similarities (fig. 2B and C), including the 5′-distal 81 bp region (fig. 2B), a central 78 bp region with two perfect HSE trinucleotides in Hspb1, Hspb6, and Cryab genes (fig. 2B), and a 194 bp long promoter region (fig. 2C). The central region (fig. 2B) harbors a characteristic HSE motif, 5′-GAAGTTTC-3′. Phylogenetic analysis of Hspb1, Hspb2, Hspb6, and αB-crystallin proteins suggests that Hspb6 and Cryab could be derived from a putative Hspb1-like ancestor gene, which could also give rise to the Hspb2 gene as supported by reasonably good bootstrapping values (fig. 2D). Since both HSE and Pax6-site evolved from the proximal promoter region (fig. 2C), we call this region the “HSE/Pax6 precursor binding region.” Additional comparison of their regulatory DNA sequences provided further support to the putative evolutionary events. A duplication of an ancestral Hspb produced two pairs, Hspb2/Cryab (two genes in head-to-tail orientation) and Hspb1/Hspb6 (currently on two different chromosomes; figs. 2D and 3A). Note that similar a conclusion can be drawn if additional protein and DNA sequences, including human, mouse, and naked mole rat Hspbs, are analyzed (supplementary fig. S3, Supplementary Material online). Although some degree of uncertainty remains in the sequential evolutionary events, taken together, these analyses uncover both evolutionary conserved and seed sequences for novel transcriptional control elements in the 5′-flanking promoter regions among this family of vertebrate Hspb genes.
Fig. 2.
—Sequence similarities in noncoding regions of mouse Hspb1, Hspb2, Hspb6, and Cryab genes. (A) Diagrammatic representation of promoter regions of mouse Hspb1, Hspb2, Hspb6, and Cryab genes. Optimal” (“imperfect”) HSEs are indicated by full (dashed) circles, respectively (see text for details). The regions are partitioned into the 5′-distal, central, and promoter. (B) Sequence alignment of “distal” (81 bp) and “central” (78 bp) regions containing multiple HSEs using T-coffee. Conserved nucleotides in the 5′-GAANNTTC-3′ motifs (horizontal bars) are shown in bold. 100%-conserved nucleotides are shown in red. “Imperfect” HSEs are underlined. (C) Sequence alignment of the “proximal” promoter region (194 bps), including the TATA-box (horizontal bracket) and start site of transcription (+1, hooked arrows) using T-coffee. A “weak” trimeric HSE is in both Hspb1 and Hspb2 promoters, though at different locations. (D) Phylogenetic analysis of the protein sequences (MAFFT for alignment with E-INS-i for multiple conserved regions and long gaps and leave gappy regions, others as default) and regulatory regions (T-coffee for alignment) of the mouse Hspb1, Hspb2, Hspb6, and Cryab. Drosophila melanogaster small heat shock protein 22 (dmHsp22) was also included in the protein analysis as the outgroup. The clustering was carried out in the software MEGA7 using Maximum likelihood method and bootstrap with 1,000 calculations (Kumar etal. 2016). Additional alignments including human, mouse, and naked mole rat sequences are shown in supplementary figure S3, Supplementary Material online.
Comparative Analysis of Mouse αA- and αB-Crystallin Promoters
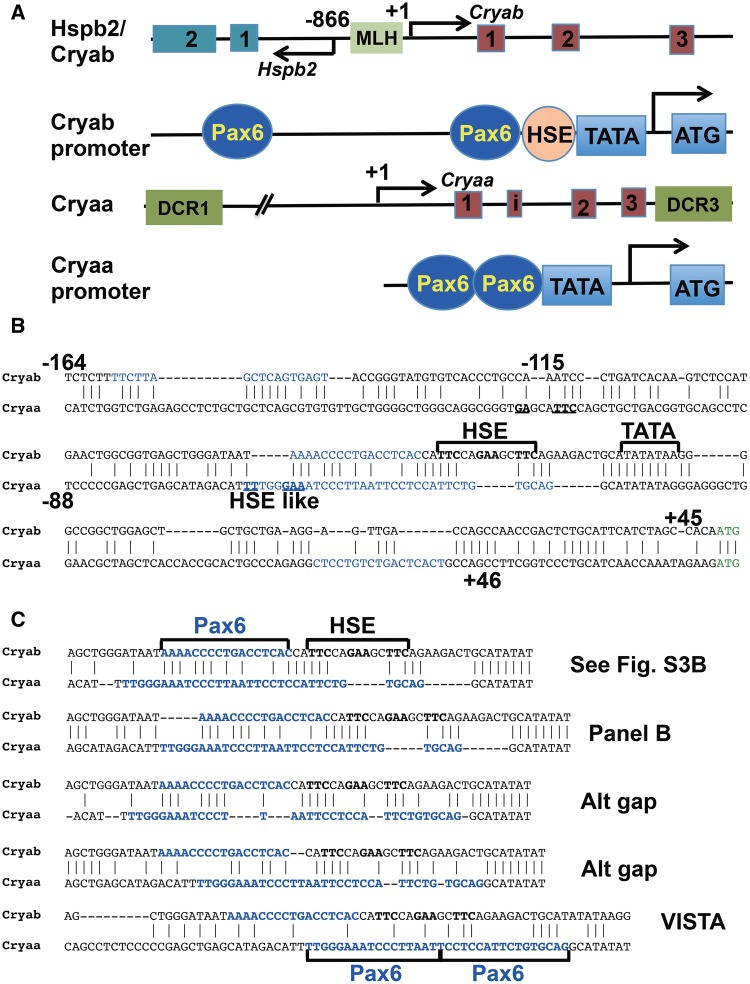
Duplication of the ancestral αB-crystallin into the αA-crystallin-like gene (supplementary fig. S3, Supplementary Material online) most likely occurred in the early vertebrates as Cryaa homologues are not found in the sea urchin, amphioxus, sea squirt, and lamprey genomes (data not shown); however, it is not known how long of the Cryab-containing sequence was duplicated to generate the earliest precursor of Cryaa gene. Using the VISTA alignment, we found sequence similarities over 9 kb of 5′-sequences of mouse Cryab/Cryaa loci that include the Hspb2-Cryab loci (fig. 3A andsupplementary fig. S4A, Supplementary Material online) as well as the distal lens-specific enhancer (DCR1) (Yang etal. 2006a) in the Cryaa locus (fig. 3A and supplementary fig. S4A, Supplementary Material online). Importantly, similar findings are obtained from analysis of evolutionary lower elephant shark and zebrafish genomes (supplementary fig. S4B and C, Supplementary Material online). In summary, these studies show that the Cryaa gene originated from a duplication of over 9 kb of genomic DNA (5′-sequences and coding regions) including the Hspb2-Cryab head-to-tail paired genes that occurred in the early fish ancestor.
Fig. 3.
—Evolution of αA-crystallin via gene duplication of the inverted tandem Hspb2/αB-crystallin and nucleotide changes in its 5′-proximal promoter region. (A) Schematic representation of mouse Hspb2/Cryab and Cryaa loci including their promoter regions. The HSE and Pax6-binding sites are shown as circles and ovals, respectively. (B) Pair-wise alignment of mouse lens-specific Cryab and Cryaa promoters using T-coffee. Both αB-crystallin promoter fragments (−162/+45) and (−115/+45) shown here are lens-specific (Gopal-Srivastava etal. 1996). The αA-crystallin promoter (−88/+46) is also lens-specific (Wawrousek etal. 1990). Conserved nucleotides in the 5′-GAANNTTC-3′ HSE motifs are shown in bold. Pax6-binding sites, blue; first ATG codon, green; “imperfect” HSE in the Cryaa promoter, HSE-like. C) Five alignments (Alt I–V) of the 5′-promoter proximal regions of mouse Cryab × Cryaa. Alt I, see alignment in supplementary figure S5, Supplementary Material online; Alt II, see alignment in panel B; Alt III and IV, manually adjusted/gaps changed; Alt V, a result of 9 kb pair-wise alignment of mouse, elephant shark, and zebrafish Cryab × Cryaa loci (supplementary fig. S4A–C, Supplementary Material online, respectively).
Defined by transgenic studies, the shortest lens-specific αA-crystallin promoter (fig. 3A) resides within the region of −88/+46 (Wawrousek etal. 1990). Similarly, the minimal αB-crystallin promoter resides within an −115/+44 fragment (fig. 3A) (Gopal-Srivastava etal. 1996). Lens-specific expression of both αA- and αB-crystallin promoters is driven by Pax6 and c-Maf (fig. 3A and supplementary fig. S2, Supplementary Material online) (Yang etal. 2004; Yang and Cvekl 2005) and Hsf4 binding to the Cryab promoter (Somasundaram and Bhat 2004). A comparison of the two promoters indicates HSE to Pax6 replacement in the αA-crystallin promoter supported by a pair-wise alignment of mouse Cryab and Cryaa promoters (fig. 3B). The alignment between these regulatory elements remains nearly intact even if corresponding Hspb1, Hspb2, and Hspb6 sequences are added (compare fig. 3C and supplementary fig. S5, Supplementary Material online). However, subtle nuances in the Cryab × Cryaa pair-wise alignment exist, depending on method used and gap penalty values (fig. 3C andsupplementary fig. S5, Supplementary Material online), as alignments of short DNA regulatory regions remain challenging (Wray etal. 2003; Meireles-Filho and Stark 2009). Nevertheless, all these alignments support the idea that the evolution of the TATA-proximal Pax6-binding site in the αA-crystallin promoter was aided by the “preexisting” HSE region.
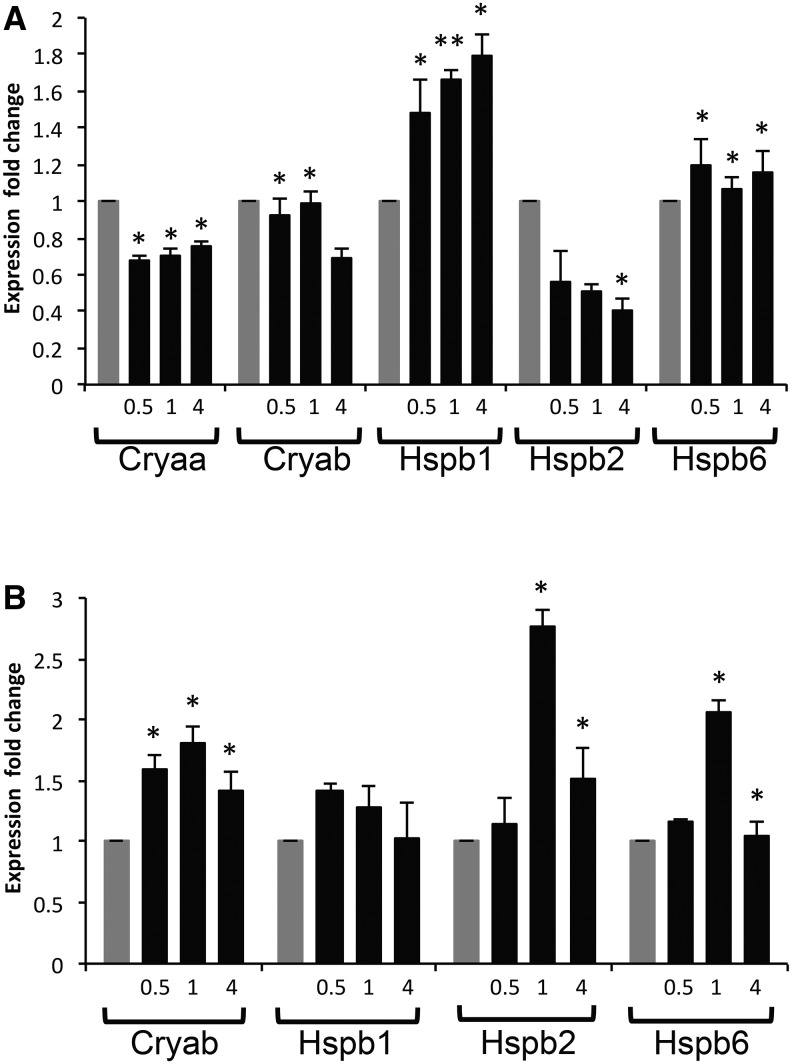
The differences such as the number, structure and location of HSEs among the Hspb/crystallin promoters could elicit different regulation of these genes under heat shock conditions (Jing etal. 2013). For example, the cell-specific environment of the lens provides Pax6 and Hsf4; whereas these proteins are not significantly expressed in the heart (Walther and Gruss 1991; Fujimoto etal. 2004), and heat inducible Hsf1 and Hsf2 might show different levels of expression in these tissues especially in early development (Somasundaram and Bhat 2004). We evaluated response of Cryab, Cryaa, Hspb1, Hspb2, and Hspb6 following the 42 °C/1 h heat shock treatments in isolated mouse embryonic lens and heart. In E16.5 lens, expression of Hspb1 and Hspb6 was induced during the entire time window examined (0.5–4 h). In contrast, no induction of expression of Cryaa, Cryab, or Hspb2 was found (fig. 4A). In newborn heart, Hspb6 and Hspb2 transcripts were most abundant 1 h following the stress treatment and expression of Cryab was also increased following the heat shock (fig. 4B). Expression of Cryaa was so low in heart that it was not possible to accurately observe the effect of heat shock on expression. Thus, although the HSE-like sequences exist in the Cryaa promoter (fig. 3B), they are not functional. While no heat induction of Cryab in lens may look surprising in light of Cryab heat-inducibility in the heart, expression of Hsf1 and Hsf2 may be too low at this stage of lens morphogenesis (Somasundaram and Bhat 2000, 2004). We conclude that unique tissue-specific heat shock responses of individual Cryab, Cryaa, Hspb1, Hspb2, and Hspb6 genes reflect pressure on DNA to retain the HSEs while lens environment evolved mechanisms utilizing lens-specific factors, such as Hsf4 and Pax6 (Cvekl and Ashery-Padan 2014).
Fig. 4.
—Regulation of Cryaa, Cryab, Hspb1, Hspb2, and Hspb6 by heat shock is tissue-specific. (A) E16.5 lenses and (B) P1 hearts were subjected to heat shock treatment at 42 °C for 1 h. Dissected tissues were allowed to recover at 37 °C for 0.5, 1, and 4 h. RT-PCR analysis of the genes was performed using total RNAs isolated from the tissues and fold changes of expression were calculated relative to untreated controls (grey bars). Expression of Cryaa in heart tissues was at too low levels to measure accurately and was therefore omitted. Biological repeats were performed and similar trends observed. P values were calculated using the Student’s t test (* ≈ P < 0.05, ** ≈ P < 0.001).
A Possible Origin of the Ciona intestinalis β/γ-Crystallin Gene from an Hspb1 Fragment Inserted 5′-of the β/γ-Coding Sequences
Multiple microbial genomes encode proteins that can serve as the founding members of the β/γ-crystallin superfamily (Mishra etal. 2014). Two precursors of vertebrate β/γ-crystallin genes have been identified previously in lower invertebrates: geodin and β/γ-crystallin-like gene. In sea sponge Geodia cydonium, geodin is an intron-less gene encoding a 163 amino acid protein (supplementary fig. S6, Supplementary Material online), the most ancient member of the β/γ-crystallin superfamily in metazoans (Di Maro etal. 2002; Giancola etal. 2005; Srivastava etal. 2014). The tunicate Ciona intestinalis genome harbors an ancient β/γ-crystallin-like gene with multiple exons (Shimeld etal. 2005). Its current open reading frame (ORF) lacks as much as 108 amino acid residues from the putative N-terminal region compared with the mouse βB2- and γA-crystallins (supplementary fig. S6, Supplementary Material online). Analysis of sea urchin (Strongylocentrotus purpuratus) and amphioxus (Branchyostoma floridae) genomes revealed a novel β/γ-crystallin-like ORF in the amphioxus (Bf Crybg) (supplementary fig. S6, Supplementary Material online). These analyses led us to hypothesize that the amphioxus Bf Crybg gene (see fig. 6B) is an ancestor form of the sea squirt β/γ-crystallin-like gene and that the vertebrate β/γ-crystallins evolved from this precursor via gene duplication including their 5′-regulatory regions.
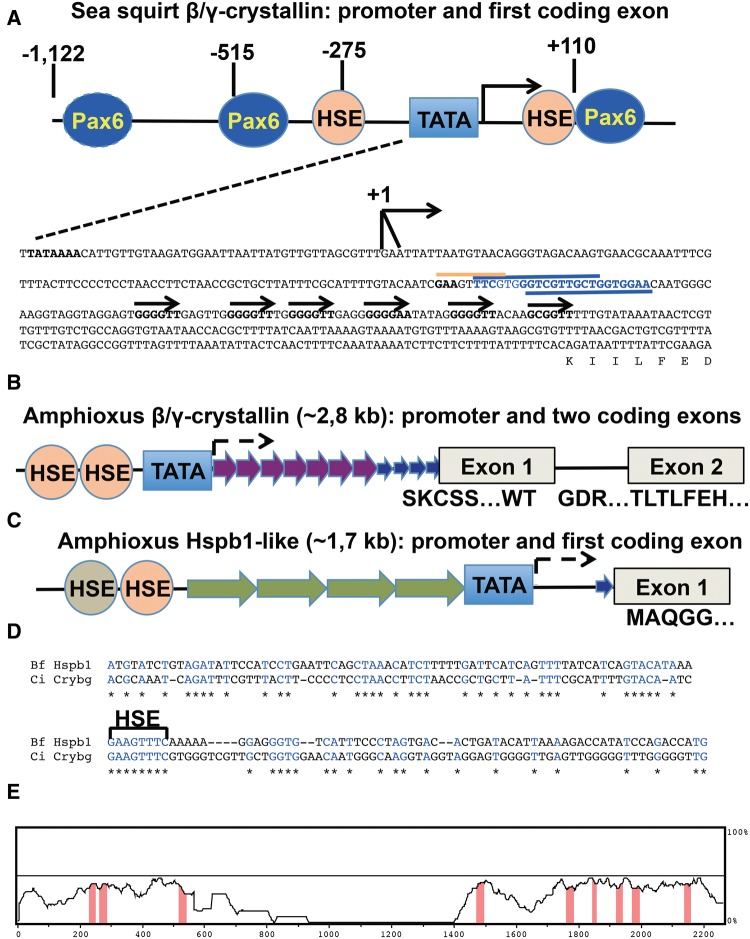
Fig. 6.
—Origin of β/γ-crystallin gene family. (A) Genomic organization of Ciona β/γ-crystallin-like gene and its 1.2 kb promoter (the numbering of nucleotides is from Shimeld etal. 2005). The TATA-box (blue rectangle), start sites of transcription (+1, hooked arrows), putative HSE (beige circles) and Pax6-binding (blue ovals) sites, downstream HSE and Pax6 sites, horizontal lines (beige, and blue, respectively), six short repetitive sequences (horizontal arrows, consensus 5′-GGGGTT-3′) are shown. (B) Genomic organization of B. floridae β/γ-crystallin-like gene and its putative promoter. The 16 bp and 4 bp repetitive sequences are shown by purple and blue arrows. (C) Genomic organization of B. floridae Hspb1-like gene and its putative promoter. Four repetitive sequences (each ∼ 200 bp) are shown as green arrows. (D) The pair-wise alignment centered around the conserved HSEs by T-coffee. (E) The alignment between a Ciona β/γ-crystallin promoter and an upstream region of the B. floridae Hspb1-like gene by LAGAN (Berezikov etal. 2004).
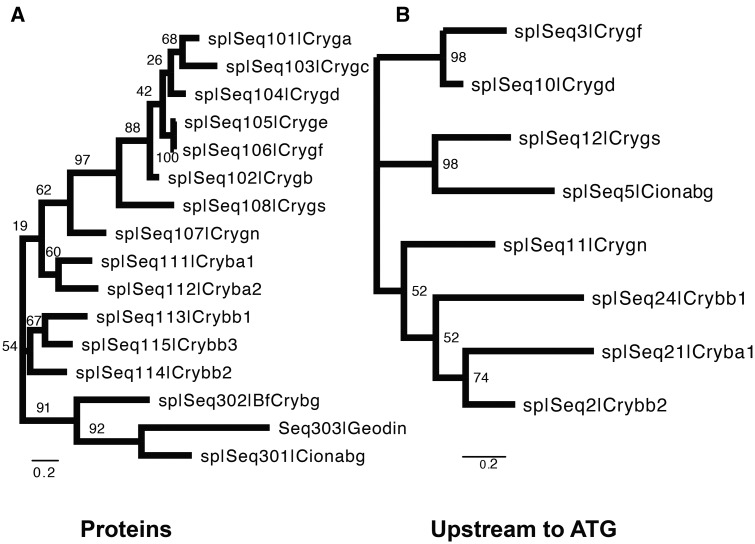
To test this hypothesis, we aligned 16 proteins, including all 13 mouse β/γ-crystallins, plus amphioxus Crybg (BfCrybg), sea squirt Crybg (Cionabg), and geodin as an obvious “outlier.” The alignments were conducted by T-coffee. As shown in figure 5A, a separation of the β- and γ-crystallin subfamily is clear, whereas both invertebrate Crybg and geodin were placed into a separate clade. Although these alignments support common evolutionary origin of coding sequences among the β/γ-crystallins as proposed earlier (Kappe etal. 2010), it remains possible that their promoters and regulatory elements formed separately. We thus examined their 5′-regions prior and including the first ATG codon (Cryba1, Crybb1, Crybb2, Crygd, Crygf, Crygs, and Crygn lengths between 414 to 504 bp; Ciona Crybg, 537 bp). Notably, the phylogenetic tree (fig. 5B) generated with DNA regulatory regions—ATG, differed from the tree based on protein sequences since Ciona Crybg was placed in a clade with Crygs. Nevertheless; at the nucleotide level, multiple sequence alignment of eight promoter regions, including seven β/γ-crystallins and Ciona Crybg, revealed a number of conserved blocks as well as gaps (supplementary fig. S7, Supplementary Material online). Taken together, the current data support the idea that the ancestor of the sea squirt Crybg likely represents the founding member of the β/γ-crystallin family, including their promoters.
Fig. 5.
—Phylograms of crystallin proteins and their 5′-promoter regions. (A) Tree constructed from protein coding sequences without the distance correction. (B) Tree constructed from promoter sequences up to the starting ATG codon. Both phylogenetic trees are constructed using T-coffee for alignment and maximum likelihood method with 1,000 bootstrap calculations with MEGA7.
A series of Ciona Crybg promoter fragments (−1, 122/+110 to −275/+110) have been studied in sea squirt larvaes and transgenic frogs (Shimeld etal. 2005; Chen etal. 2014); however, the origin of this promoter and its possible regulation by Pax6 and HSEs identified here (fig. 6A) was not discussed. The transition from amphioxus into sea squirt β/γ-crystallin-like gene could happen by various mechanisms such as trans-insertion of a novel genomic fragment 5′-of the β/γ-like ORF, or cis-deletion of an internal genomic fragment, a combination of these processes, and by cumulative nucleotide modifications of the promoter region. We first considered an “insertional” model from “scratch” as proposed elsewhere (Kaessmann 2010) and searched for the possible “donor” sequence. As the putative inserted sequence has a conserved 8 bp HSE (5′GAAGTTTC-3′) similar to what is found in mouse Hspb1 and Hspb6 promoters (fig. 2B), an intriguing possibility arose that evolution of both α- and β/γ-crystallin promoters could be linked together. The amphioxus β/γ-crystallin and Hspb1-like genes are shown in figure 6B and C, respectively. We found that amphioxus Hspb1-like gene promoter also harbors a conserved 8 bp HSE, (5′GAAGTTTC-3′) (fig. 6C and D). A conservation of this HSE across evolutionary diverse genomes is evident from multiple sequence alignments of five Hspb1-related genes, including D. melanogaster Hsp23, mouse Hspb1, mouse Hspb6, C. intestinalis Crybg, and B. floridae Hspb1-like gene, demonstrating conservation of this sequence over a region of ∼150 bps (supplementary fig. S8, Supplementary Material online). Importantly, the “conserved” 5′-GAAGTTTC-3′ HSE in the sea squirt Crybg and amphioxus Hspb1 genomic fragments were precisely aligned if five Hspb1 genomic fragments were used (see supplementary fig. S8, Supplementary Material online). Likewise, their pair-wise alignment, including 146 and 149 bp with centrally positioned HSEs, matched these HSEs (fig. 6D). Finally, a pair-wise alignment between functionally defined C. intestinalis β/γ-crystallin promoter (Shimeld etal. 2005) and the 5′-flanking region of the B. floridae Hspb1-like gene confirmed similarities between these sequences over a span of 2 kb (fig. 6E). These findings indicate that a common cephalochordate/tunicate ancestor Hspb1-like gene provided a genomic fragment that was inserted 5′- of the β/γ-crystallin-like ORF to produce the ancestral β/γ-crystallin gene.
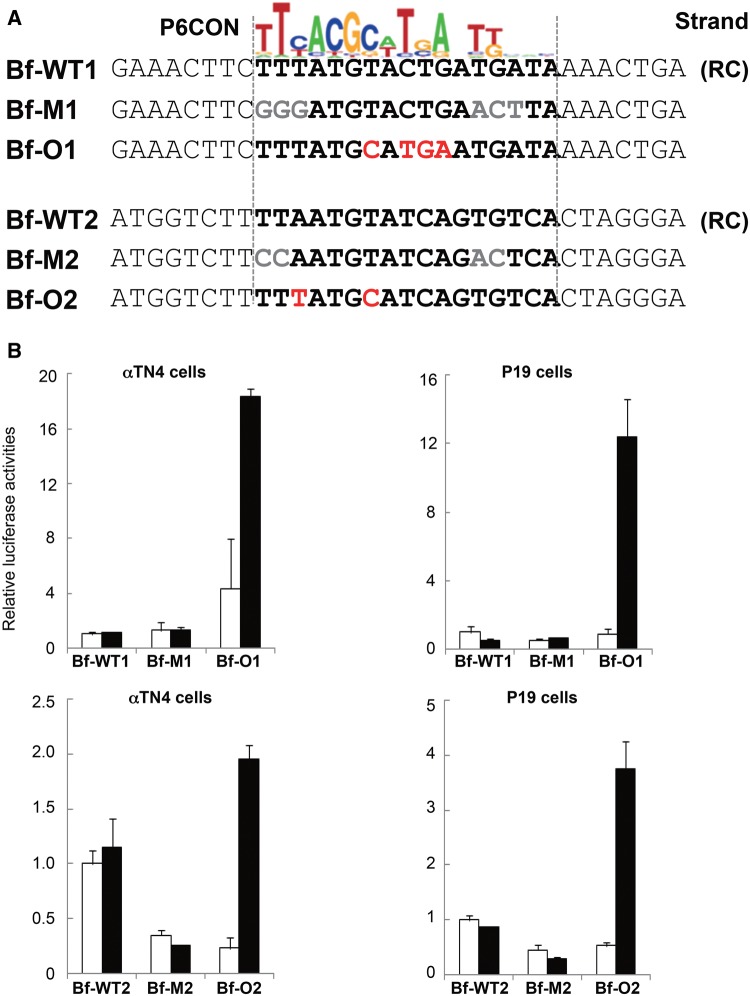
The possible origin of Pax6-binding sites from HSE-rich sequences described above was experimentally tested using a specific region of the B. floridae Hspb1-like gene. Using a 14 bp Pax6 consensus binding site (P6CON), we identified two putative Pax6-like sequences, Bf-WT1 and Bf WT2, separated by the 5′-GAAGTTTC-3′ HSE (fig. 7A). Six copies of wild type (Bf-WT1 and Bf WT2), their point mutants (Bf-M1 and Bf-M2), and optimized Pax6-binding sites (Bf-O1 and Bf-O2), were inserted 5′-of the E4 TATA-box in a luciferase reporter gene pGL3 as we described elsewhere (Chauhan etal. 2004a). Transient cotransfections with Pax6 cDNA were conducted in both lens (expressing Pax6) and nonlens (no Pax6 expression) cultured cells (fig. 7B). The results showed that reporter genes driven by putative and mutated Pax6-binding sites were not activated by Pax6. In contrast, four individual nucleotide changes towards the “optimal′ Pax6-binding sites, Bf-O1 and Bf-O2, produced a pair of reporters activated by Pax6 in both cell culture systems. Taken together, these experiments showed that a small number of nucleotide changes is sufficient to produce functional Pax6-binding sites from regions rich in HSE-like motifs.
Fig. 7.
—Functional evaluation of the HSE to Pax6-binding site conversion model within the B. floridae Hspb1-like gene. (A) Identification of HSEs and two putative Pax6-binding sites. The binding sites were found on the reverse complement (RC) strand. (B) Relative luciferase expression driven by six copies of the wild type (Bf-WT1 and Bf WT2), their mutants (Bf-M1 and Bf-M2), and optimized Pax6-binding sites (Bf-O1 and Bf-O2) in lens (αTN4) and embryonic carcinoma (P19) cells.
Invertebrate Crystallin Promoters: Connections to Aldhs?
Camera-like eyes are also found in multiple evolutionarily distant invertebrates and Ω-, S-, and J-crystallins have been identified in their lenses (Tomarev and Piatigorsky 1996; Piatigorsky and Kozmik 2004; Jonasova and Kozmik 2008). The overarching hypothesis is that invertebrate Ω-crystallin genes evolved by gene duplication from an ancestral Aldh gene. Aldh genes encode enzymes critical for retinoic acid biosynthesis, a key ligand employed by retinoic acid activated nuclear receptors (Cvekl and Wang 2009; Kumar etal. 2012). Importantly, these proteins protect cells from UV light damage and oxidative stress (Chen etal. 2013; Lassen etal. 2008). In scallop, the lens-cornea form continuous tissues and both highly express Ω-crystallin/Aldh1a9 (Piatigorsky etal. 2000). In addition to the major S-crystallins, the Ω-crystallin is also expressed in squid lenses (Tomarev and Piatigorsky 1996). Aldh1a1/Raldh1, Aldh1a2/Raldh2, and Aldh1a3/Raldh3 are expressed in the mammalian lens, neuroretina, and retinal pigmented epithelium (RPE) (Cvekl and Wang 2009; Kumar etal. 2012) and Aldh1a3 is a Pax6-regulated gene (Suzuki etal. 2000). Furthermore, Aldh3a1 is a major component of the mouse cornea with a low lens expression (Lassen etal. 2007; Chen etal. 2013, 2016), and is also directly regulated by Pax6 (Davis etal. 2008). In elephant shrew (a small mammal), η-crystallin is encoded by an Aldh1a7 gene (Graham etal. 1996). Multiple sequence alignments of Ω-crystallin/Aldh1a9, η-crystallin/Aldh1a7, Aldh3a1, Aldh1a1, Aldh1a2, and Aldh1a3 proteins (supplementary fig. S9A, Supplementary Material online) reveal extensive amino acid similarities supporting their common evolutionary origin. We next aligned their promoters, experimentally defined for mouse Aldh3a1 (Davis etal. 2008) and scallop Ω-crystallin/Aldh1a9 (Carosa etal. 2002), and deduced from genome annotation for mouse Aldh1a1, Aldh1a2, Aldh1a3, and Aldh1a7. Many blocks of conservation were identified in the alignment of two evolutionarily distant genomes (mouse and scallop) whose common ancestor is at least 650 million years old (supplementary fig. S9B, Supplementary Material online). We therefore wondered whether it is possible to reconstruct various aspects of invertebrate crystallin gene recruitment through the analysis of Ω-crystallin/Aldh1a9 promoter formation by considering a single origin of the invertebrate crystallin promoter and convergent evolution assisted by the preexisting cis-regulatory sites.
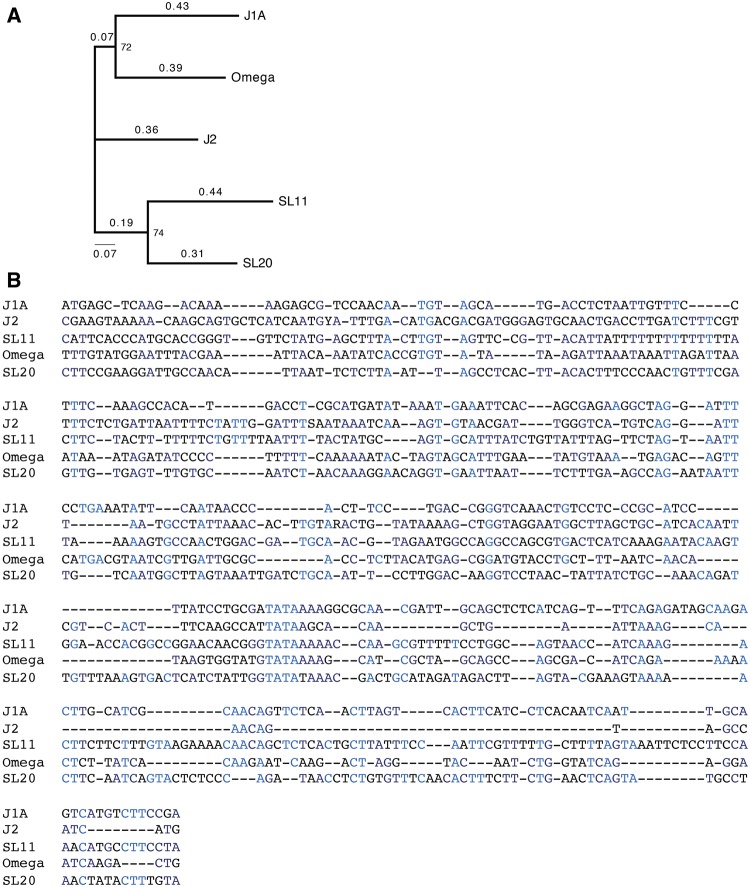
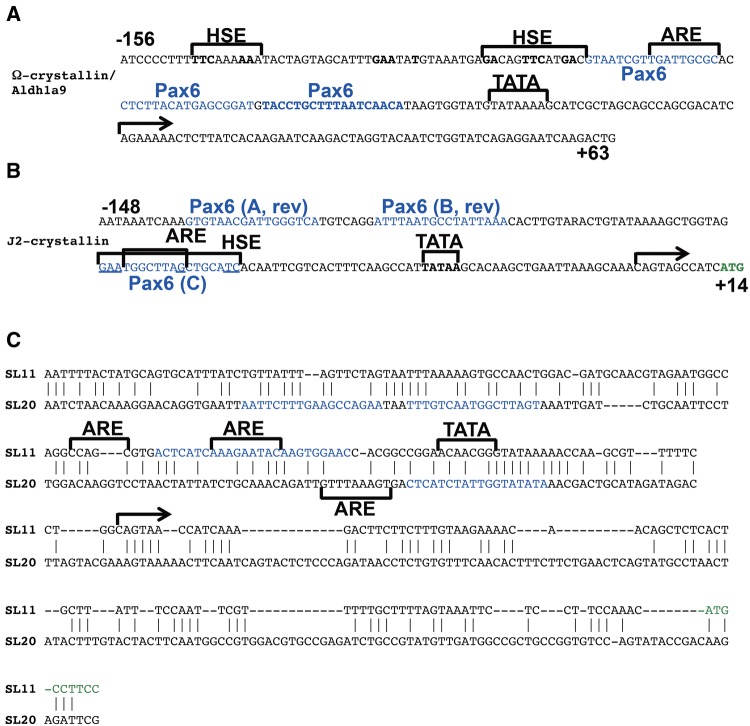
To test the first model, we aligned five known scallop Ω-, jellyfish J1A- and J2-, and squid SL11 and SL20 S-crystallin promoters (fig. 8). This alignment shows that both jellyfish J1A- and J2-crystallin promoters form a group with Ω-crystallin/Aldh1a9, whereas SL11 and SL20 form their own clade (fig. 8A). Multiple blocks of conservation combined with gap patterns therefore support the intriguing possibility of the common ancestral origin of these promoters. To probe the model of preexisting sites, we focused on the idea that the invertebrate Pax6 sites also evolved from HSEs and/or AREs. In sea scallop (Placopecten magellanicus) Ω-crystallin/Aldh1a9 promoter, three Pax6-binding sites were identified and characterized previously (fig. 9A) (Carosa etal. 2002). The distal Pax6-site overlaps with an ARE (fig. 9A). The cubozoan jellyfish (Tripedalia cystophora) J2-crystallin promoter fragment −148/+10 is comprised of three functional PaxB-binding sites (fig. 9B, Kozmik etal. 2008). Site C overlaps ARE, while two putative HSE motifs are located in the adjacent 5′-flanking region. The J3-crystallin promoter (Kozmik etal. 2003) contains a pair of PaxB/Pax6-binding sites though its complete promoter sequence remains to be reported. Finally, two squid (Ommastrephes sloani) crystallin promoters have been characterized using vertebrate lens nuclear extracts (Tomarev etal. 1994). The SL11 and SL20 crystallin promoters are similar (46% identity across 204 bp). In the SL11-crystallin promoter, two putative Pax6-binding sites overlap with AREs (fig. 9C). In the SL20-crystallin promoter, a putative Pax6-binding site also partially overlaps with an ARE. The Pax6-binding sites (n = 16, eight experimentally validated) identified here were aligned with a Pax6 consensus site (P6CON, Epstein etal. 1994; Xie and Cvekl 2011) and Pax6 “optimal” binding site (Sun etal. 2015) to demonstrate that many of these sites contain five or more mismatches (supplementary fig. S10, Supplementary Material online). Thus, it is possible that Pax6/PaxB-binding sites in invertebrate crystallin promoters could also originate from AREs in the ancestral crystallin-precursor genes by the process of convergent evolution.
Fig. 8.
—Multiple sequence alignments of five invertebrate crystallin promoters. (A) Phylogram of the promoter sequences: Scallop Ω-crystallin/Aldh1a9, squid SL11 and SL20 S-crystallin promoters, jellyfish J1A and J2-crystallin promoters. The phylogram employed sequences from the 5′-end throughout the TATA-box using alignments in panel B with maximum likelihood method and 1,000 bootstrap calculation. (B) Multiple sequence alignments of the invertebrate crystallin promoters. The alignment shown was generated from two blocks based on the use of TATA-box and experimentally defined transcriptional start sites (Tomarev etal. 1994; Carosa etal. 2002; Kozmik etal. 2008) using T-coffee. The 5′-box included all sequences up to the TATA. The downstream box started from the TATA-box and included the first ATG sequences in J1A, J2, and SL11 sequences. 100% nucleotide conservation, light blue; at least 60% conservation, dark blue. The clustering was carried out by T-coffee.
Fig. 9.
—Analysis of invertebrate Ω-, J2-, SL11, and SL20-crystallin promoters. (A) Scallop Ω-crystallin promoter. (B) Jellyfish J2-crystallin promoter. (C) Squid S-crystallin SL11 and SL20 promoters. Pax6-binding sites (blue font), ARE and TATA-box (horizontal brackets), HSEs (bold and underlined), start sites of transcription (horizontal arrows), coding sequences (green font).
Discussion
Reconstruction of an evolutionary history of any family of genes encoding proteins that determine a unique cell type-specific property, such as lens transparency, is aided by a) following general principles of evolution applied to formation of novel cell types/tissues/organs (Arendt etal. 2016), b) analysis of functionally annotated genomic DNA regions from related and distant organisms (Park etal. 2016), and c) detailed knowledge on the molecular mechanisms that govern tissue-specific gene expression (Davidson and Erwin 2006; Sun and Kim 2011; Ettensohn 2013). The present study was aimed to examine origin of lens-specific Pax6-binding sites, identify ancestral genes from which diverse families of crystallins evolved, and search for one or more unifying principles of crystallin gene recruitment throughout metazoan evolution.
The current studies were conducted with the aim to shed new light into the genome evolutionary changes and their consequences for pathways underlying eye evolution in both vertebrate and invertebrate organisms (Gehring and Ikeo 1999; Lamb 2013; Nilsson 2013). Thus, we aimed to address the potential use of parallel and convergent evolutionary steps and/or their combinations. The present data shed new light onto ocular lens evolution through the identification of complementary evolutionary mechanisms, including gene duplication, formation of Pax6-binding sites from preexisting cis-elements, and insertion-mediated de novo gene formation.
Through convergent evolution, any Pax6-binding site could evolve from nonfunctional and/or preexisting cis-sites near active transcriptional control regions. The complementary hypothesis states that an initial formation of Pax6-dependent regulatory region occurred through the processes described above but only in one or very few genes. If expression of the regulated gene provides a selective advantage for the cell expressing it, the gene could be “recruited” with an opportunity to maintain the original function and/or produce novel function(s) (Piatigorsky 2006). In case of α-crystallins, the original function could be similar to what other small heat shock proteins do, that is, to protect proteins from missfolding and inhibit programmed cell death (Arrigo and Simon 2010). The present data show that Pax6-binding sites in α-crystallin genes could indeed emerge from proximal promoter sequences, called here the “HSE/Pax6 precursor binding region”, found in the ancestral Hspb1/6 genes. The conservation of distal HSEs makes this region available for evolutionary changes towards formation of either additional HSEs like in αB-crystallin promoter, and subsequently, by destroying the nonessential HSE in the αA-crystallin promoter. It is important to state that the current Hspb1, Hspb6, Cryab, and Cryaa promoter sequences do not provide any definitive insight into the putative nucleotide sequence of the ancestral gene Hspb1-like gene. Similarly, multiple HSE-like motifs found in both amphioxus and sea squirt β/γ-crystallin-like genes could provide specific “seed” sequences for the emergence of new Pax6-binding sites. The β/γ-crystallin family of genes expanded by gene duplication while retaining and evolving their 5′-flanking sequences. From a purely evolutionarily perspective, it is easier to envision a common origin from a single “improbable” event compared with many parallel processes by convergent evolution within a family of duplicated genes (Gehring 2004, 2012).
An interesting question is how consensus Pax6-binding sites are related to the optimal binding sites of all known transcription factors as any of these sites has a potential to give rise to a new site recognized by different factors and change gene regulatory mechanisms. Comparisons of optimal binding sites have an internal limitation as it has been proposed that promoter/enhancer mediated transcriptional regulation offers better opportunities if the individual regulatory sites are “suboptimal” (Crocker etal. 2015; Jolma etal. 2015). An unbiased analysis using the TOMTOM motif comparison software (http://meme-suite.org/tools/tomtom) to compare the Pax6 motif with all vertebrate TF motifs in the Jaspar database (http://jaspar.genereg.net/) identified binding motifs for glial cells missing homolog 1 (GCM1) and T-box transcription factors (e.g., Tbr1) that use DNA-binding domains distinct from the helix-turn-helix domains forming the Pax6 paired domain, that is, metal chelating and Tbox DNA-binding domain, respectively. This analysis illustrates that there are many routes how preexisting cis-sites can be modified towards Pax6-binding sites and the actual mechanisms ultimately selected by evolution depends on the transcription factors available and gene product function(s).
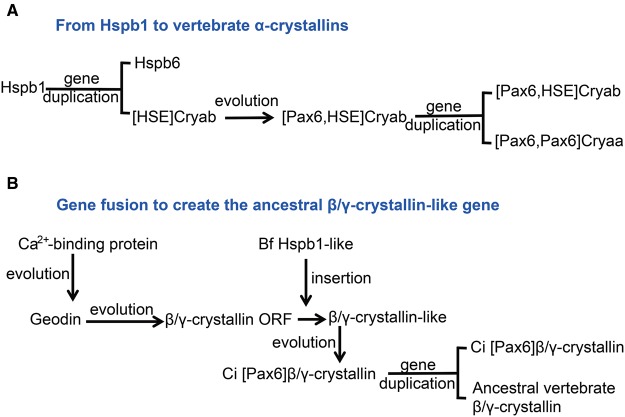
Our studies suggest that multiple invertebrate crystallins originated from an ancestral Aldh gene, a pair of vertebrate α-crystallins originated from an Hspb1 gene, and 14 vertebrate β/γ-crystallins emerged from a single β/γ-crystallin-like gene (see fig. 10). The common denominator between Aldh and Hspb genes is their stress-inducibility and inhibition of apoptosis by the encoded proteins. Small heat shock proteins are multifunctional proteins used to protect cells from heat shock and other types of stress (Arrigo and Simon 2010). Mouse Aldh3a1 possesses both antiapoptotic function and stress inducibility (Choudhary etal. 2005; Lassen etal. 2007) likely mediated by at least two HSEs and an ARE predicted here in the Ω-crystallin/Aldh1a9 promoter.
Fig. 10.
—Summary of evolution model of multigene α- and β/γ-crystallin families. (A) The process of gene duplication originating from an ancestral Hspb1 gene to produce a pair of Cryab and Cryaa genes. (B) The process of gene fusion to create the ancestral β/γ-crystallin-like gene. Tentative acquisition of HSE and Pax6 sites is shown in brackets.
The recruitment of small heat shock proteins, other stress-related proteins, and ubiquitous enzymes to function as lens crystallins has been extensively discussed earlier mostly through their ability to support lens transparency and its refractive power (Piatigorsky 2006). It is obvious that any crystallin protein needs to be water soluble at a very high concentration, and selection pressures had to choose for the right proteins. Since evolution works by improvement of something that already works, it is unlikely that the issue of water-solubility and concentration would have any impact at the beginning of this process. It has been proposed that elongation of lens fiber cells requires elaborate changes in the cellular cytoskeleton as well as changes in the osmoregulation that would preferentially select small heat shock proteins (Wistow 1993). Indeed, both αB-crystallin and small heat shock proteins interact with actin and have roles in cytoskeleton assembly (Xi etal. 2006; Ghosh etal. 2007; Singh etal. 2007) and play several noncanonical roles in lens and nonlens cells (Gangalum and Bhat 2009; Gangalum etal. 2011). It is also possible that β/γ-crystallins originally served as UV light filters (Hibbert etal. 2015). We propose here that the reason to initially select small heat shock proteins is their strong antiapoptotic function (Arrigo and Simon 2010; Basha etal. 2012) coupled with their compatibility with the cytoskeleton remodeling during cellular elongation (Wistow 1993). The hallmark of lens differentiation is an organized degradation of organelles inside of the lens fiber cells to prevent light scattering. Degradation of organelles combined with preservation of cells is a unique property of the lens, and thus, a reasonable initial force to improve lens function. However, there is an internal conflict between the degradation of organelles that leads to apoptosis in other cell types, and the necessity to make organelle-free fiber cells as a structural foundation of the bulk of the lens. The elevation of expression of an ancestral crystallin/small heat shock protein would provide the antiapoptotic cellular environment to preserve the cells while undergoing demise of the mitochondria, endoplasmatic reticulum, Golgi apparatus, and finally nuclei. The αA-crystallin indeed protects lens epithelial cells from apoptosis during mitosis (Xi etal. 2003b). Loss of both αA- and αB-crystallins in double-knockout mouse lens resulted in a caspase 3/6-dependent lens fiber cell disintegration (Morozov and Wawrousek 2006). These studies demonstrate that a specific antiapoptotic role of α-crystallins is essential for the lens viability. Interestingly, specific mutations in both α- and γ-crystallins inhibit the denucleation process (Sandilands etal. 2002; Graw etal. 2004; Gupta etal. 2011) further supporting the link between shsps and denucleation of lens fiber cells. Finally, it is possible that the Pax6/α-crystallin regulatory mechanism evolved in ancestral neuronal cells prior to its subsequent deployment in the ancestral lens fibers. This inference is supported by recent studies demonstrating that Pax6 regulates expression of αA-, βB2-, and other crystallins in a subset of olfactory bulb neurons, and that the key role of αA-crystallin in these cells is inhibition of apoptosis (Ninkovic etal. 2010).
Likewise, what are the selective advantages to recruit Aldhs as lens crystallins? In the scallop eye, Ω-crystallin/Aldh1a9 is expressed both in the cornea/lens joint structure (Piatigorsky etal. 2000). The Ω-crystallin/Aldh1a9 is both a major crystallin in scallop and a minor crystallin in squid lens (Tomarev and Piatigorsky 1996). In the present mammalian eye, Aldh3a1 is the major corneal crystallin (Lassen etal. 2007) and protects cells from UV light damage (Chen etal. 2013). Thus, Aldh-based proteins were successfully employed by lens and cornea in evolutionarily very distant organisms (Jonasova and Kozmik 2008). The rodent Aldh1a3 and Aldh3a1 (Suzuki etal. 2000; Davis etal. 2008) and scallop Ω-crystallin/Aldh1a9 (Carosa etal. 2002) are all Pax6-regulated genes. Although we do not know if the ancestral “prototype” Aldh gene was also regulated by Pax6/PaxB, the direct link between Pax6/PaxB and Aldh fits to the “intercalary evolution” hypothesis to explain both initiating and universal roles of Pax6/PaxB in eye formation and evolution (Gehring and Ikeo 1999) as Aldh1a1, Aldh1a2, and Aldh1a3 are important enzymes in the metabolism of retinoic acid, a key morphogen that controls eye development (Cvekl and Wang 2009; Kumar etal. 2012). Our data also support an intriguing possibility that Aldh gene recruitment could be accompanied by duplication of their promoter sequences and their insertion in other parts of the ancestral genomes to give rise to the J- and S-crystallin promoters. Taken together, prior to lens evolution, Aldh gene(s) were already important for the formation of the most primitive eyes, and were available to expand their roles as lens and corneal structural proteins.
Conclusions
In summary, the foundation for the present study employs detailed knowledge of lens-specific regulatory mechanisms of both vertebrate and invertebrate crystallin genes through their transcriptional control by Pax6 and more ancestral PaxB in jellyfish. Lens morphogenesis can be unified by the use of small heat shock proteins for the formation of vertebrate lens and suggest that the nonvertebrate crystallins could follow the same strategy via antiapoptotic and UV-filtering activities of Aldhs. Although our data on the origin of Pax6-regulation of vertebrate crystallins support the model of HSE-like to Pax6 conversion followed by gene duplication, the origin of invertebrate crystallin promoters is less clear and awaits for complete DNA sequences of the invertebrate genomes of interest. Finally, it is possible to envision experimental testing of the current models as it is now feasible to screen large promoter libraries using massively parallel reporter assays (Inoue and Ahituv 2015).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
Designed research: A.C. and D.Z. Performed experiments: A.C., Y.Z., R.S.M., Q.X., and D.Z. Data analysis and interpretation: A.C., Y.Z., R.S.M., Q.X., X.G., and D.Z. Wrote the manuscript: A.C. and D.Z.
Supplementary Material
Acknowledgments
We thank Drs Joram Piatigorsky, Claude Desplan, and Jack Lenz for critical comments and Dr Walter J. Gehring for initial encouragement. We are grateful to Dr Sebastian Shimeld for the DNA sequence of the Ciona β/γ-crystallin-like promoter. This work was supported by NIH grants R01 EY012200 (A.C.), and EY014237 (A.C.), and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences.
Literature Cited
- Arendt D, et al. 2016. The origin and evolution of cell types. Nat Rev Genet. 17(12):744–757. [DOI] [PubMed] [Google Scholar]
- Arrigo AP, Simon S.. 2010. Expression and functions of heat shock proteins in the normal and pathological mammalian eye. Curr Mol Med. 10(9):776–793. [DOI] [PubMed] [Google Scholar]
- Basha E, O'Neill H, Vierling E.. 2012. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 37(3):106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Shi Y, Vrensen GF.. 2011. Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci. 366(1568):1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Vingron M.. 2010. Studying the evolution of promoter sequences: a waiting time problem. J Comput Biol. 17(12):1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, Plasterk RHA, Cuppen E.. 2004. CONREAL: Conserved regulatory elements anchored alignment algorithm for indentification of transcription factor binding sites by phylogenetic footprinting. Genome Res. 14(1):170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Gerber JK.. 2000. Origin of the paired domain. Dev Genes Evol. 210(12):644–650. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ.. 1997. PAX-6 in development and evolution. Annu Rev Neurosci. 20:483–532. [DOI] [PubMed] [Google Scholar]
- Carosa E, Kozmik Z, Rall JE, Piatigorsky J.. 2002. Structure and expression of the scallop Ω-crystallin gene. Evidence for convergent evolution of promoter sequences. J Biol Chem. 277(1):656–664. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134(1):24–36. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Yang Y, Cveklova K, Cvekl A.. 2004a. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 32:1696–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Yang Y, Cveklova K, Cvekl A.. 2004b. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 45(2):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WC, et al. 2014. Dissection of a Ciona regulatory element reveals complexity of cross-species enhancer activity. Dev Biol. 390(2):261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Thompson DC, Koppaka V, Jester JV, Vasiliou V.. 2013. Ocular aldehyde dehydrogenases: protection against ultraviolet damage and maintenance of transparency for vision. Prog Retin Eye Res. 33:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. 2016. Corneal haze phenotype in Aldh3a1-null mice: invivo confocal microscopy and tissue imaging mass spectrometry. Chem Biol Interact. doi:10.1016/j.cbi.2016.12.017. [DOI] [PubMed] [Google Scholar]
- Chow RL, Lang RA.. 2001. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 17:255–296. [DOI] [PubMed] [Google Scholar]
- Choudhary S, et al. 2005. Role of aldehyde dehydrogenase isozymes in the defense of rat lens and human lens epithelial cells against oxidative stress. Invest Ophthalmol Vis Sci. 46(1):259–267. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9(12):938–950. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R.. 2014. The cellular and molecular mechanisms of vertebrate lens development. Development 141(23):4432–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Duncan MK.. 2007. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 26(6):555–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Piatigorsky J.. 1996. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays 18(8):621–630. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Wang WL.. 2009. Retinoic acid signaling in mammalian eye development. Exp Eye Res. 89(3):280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, McGreal R, Liu W.. 2015. Lens development and crystallin gene expression. Prog Mol Biol Transl Sci. 134:129–167. [DOI] [PubMed] [Google Scholar]
- Crocker J, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160(1–2):191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S.. 2008. p53 family in development. Mech Dev. 125(11–12):919–931. [DOI] [PubMed] [Google Scholar]
- Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468(7326):911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH.. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311(5762):796–800. [DOI] [PubMed] [Google Scholar]
- Davis J, Davis D, Norman B, Piatigorsky J.. 2008. Gene expression of the mouse corneal crystallin Aldh3a1: activation by Pax6, Oct1, and p300. Invest Ophthalmol Vis Sci. 49(5):1814–1826. [DOI] [PubMed] [Google Scholar]
- Di Maro A, Pizzo E, Cubellis MV, D'Alessio G.. 2002. An intron-less βγ-crystallin-type gene from the sponge Geodia cydonium. Gene 299(1–2):79–82. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high througput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R.. 1994. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 269(11):8355–8361. [PubMed] [Google Scholar]
- Ettensohn CA. 2013. Encoding anatomy: developmental gene regulatory networks and morphogenesis. Genesis 51(6):383–409. [DOI] [PubMed] [Google Scholar]
- Franck E, et al. 2004. Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol. 59(6):792–805. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, et al. 2004. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 23(21):4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, et al. 2008. Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. J Biol Chem. 283(44):29961–29970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M, Gotthardt R, Muller T.. 1993. Structure and organisation of a murine gene encoding small heat-shock protein Hsp25. Gene 128(2):279–283. [DOI] [PubMed] [Google Scholar]
- Gangalum RK, Atanasov IC, Zhou ZH, Bhat SP.. 2011. AlphaB-crystallin is found in detergent-resistant membrane microdomains and is secreted via exosomes from human retinal pigment epithelial cells. J Biol Chem. 286(5):3261–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangalum RK, Bhat SP.. 2009. AlphaB-crystallin: a Golgi-associated membrane protein in the developing ocular lens. Invest Ophthalmol Vis Sci. 50(7):3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue F, Ahituv N.. 2015. Decoding enhancers using massively parallel reporter assays. Genomics 106(3):159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ. 2004. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol. 48(8–9):707–717. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. 2012. The animal body plan, the prototypic body segment, and eye evolution. Evol Dev. 14(1):34–46. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K.. 1999. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 15(9):371–377. [DOI] [PubMed] [Google Scholar]
- Ghosh JG, Houck SA, Clark JI, Egli M.. 2007. Interactive domains in the molecular chaperone human αB crystallin modulate microtubule assembly and disassembly. PLoS One 2(6):e498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancola C, Pizzo E, Di Maro A, Cubellis MV, D'Alessio G.. 2005. Preparation and characterization of geodin. A βγ-crystallin-type protein from a sponge. FEBS J. 272(4):1023–1035. [DOI] [PubMed] [Google Scholar]
- Gopal-Srivastava R, Cvekl A, Piatigorsky J.. 1996. Pax-6 and αB-crystallin/small heat shock protein gene regulation in the murine lens. Interaction with the lens-specific regions, LSR1 and LSR2. J Biol Chem. 271(38):23029–23036. [DOI] [PubMed] [Google Scholar]
- Graham C, Hodin J, Wistow G.. 1996. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of η-crystallin. J Biol Chem. 271(26):15623–15628. [DOI] [PubMed] [Google Scholar]
- Graw J, et al. 2004. Genetic and allelic heterogeneity of Cryg mutations in eight distinct forms of dominant cataract in the mouse. Invest Ophthalmol Vis Sci. 45(4):1202–1213. [DOI] [PubMed] [Google Scholar]
- Gupta R, Asomugha CO, Srivastava OP.. 2011. The common modification in αA-crystallin in the lens, N101D, is associated with increased opacity in a mouse model. J Biol Chem. 286(13):11579–11592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert SA, et al. 2015. A potential role for endogenous proteins as sacrificial sunscreens and antioxidants in human tissues. Redox Biol. 5:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough RB, et al. 2002. Adaptive evolution of small heat shock protein/αB-crystallin promoter activity of the blind subterranean mole rat, Spalax ehrenbergi. Proc Natl Acad Sci U S A. 99(12):8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia TD, Craig EA.. 1982. Four small Drosophila heat shock proteins are related to each other and to mammalian α-crystallin. Proc Natl Acad Sci U S A. 79(7):2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WK, et al. 2013. p53 directly regulates αA- and βA3/A1-crystallin genes to modulate lens differentiation. Curr Mol Med. 13(6):968–978. [DOI] [PubMed] [Google Scholar]
- Jing Z, Gangalum RK, Lee JZ, Mock D, Bhat SP.. 2013. Cell-type-dependent access of HSF1 and HSF4 to alphaB-crystallin promoter during heat shock. Cell Stress Chaperones 18(3):377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, et al. 2015. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527(7578):384–388. [DOI] [PubMed] [Google Scholar]
- Jonasova K, Kozmik Z.. 2008. Eye evolution: lens and cornea as an upgrade of animal visual system. Semin Cell Dev Biol. 19(2):71–81. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe G, et al. 2003. The human genome encodes 10 α-crystallin-related small heat shock proteins: HspB1-10. Cell Stress Chaperones 8(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe G, Purkiss AG, van Genesen ST, Slingsby C, Lubsen NH.. 2010. Explosive expansion of βγ-crystallin genes in the ancestral vertebrate. J Mol Evol. 71(3):219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, et al. 1999. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 274(27):19254–19260. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303(5664):1626–1632. [DOI] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH.. 1999. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 96(7):3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R, Fröhli E, Steiger RH, Schäfer R, Aoyama A.. 1991. αB-crystallin is a small heat shock protein. Proc Natl Acad Sci U S A. 88(9):3652–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z, et al. 2003. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell 5(5):773–785. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, et al. 2008. Cubozoan crystallins: evidence for convergent evolution of pax regulatory sequences. Evol Dev. 10(1):52–61. [DOI] [PubMed] [Google Scholar]
- Kumar S, Sandell LL, Trainor PA, Koentgen F, Duester G.. 2012. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim Biophys Acta 1821(1):198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TD. 2013. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 36:52–119. [DOI] [PubMed] [Google Scholar]
- Lamb TD, Collin SP, Pugh EN Jr.. 2007. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci. 8(12): 960–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land MF, Nilsson D-E.. 2002. Animal eyes Cambridge, UK: Oxford University Press. [Google Scholar]
- Lassen N, et al. 2007. Multiple and additive functions of ALDH3A1 and ALDH1A1. Cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 282(35):25668–25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Black WJ, Estey T, Vasiliou V.. 2008. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin Cell Dev Biol. 19(2):100–112. [DOI] [PubMed] [Google Scholar]
- Li Y, Hough RB, Piatigorsky J.. 2007. Tissue-specific activity of the blind mole rat and the two nucleotide-mutated mouse αB-crystallin promoter in transgenic mice. Proc Natl Acad Sci U S A. 104(8):2608–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Vousden KH.. 1999. Regulation and activation of p53 and its family members. Cell Death Differ. 6(12):1162–1168. [DOI] [PubMed] [Google Scholar]
- Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR, Kingsley DM, Lindblad-Toh K, Haussler D.. 2011. Three periods of regulatory innovation during vertebrate evolution. Science 333(6045):1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V.. 2008. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 4(6):697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles-Filho AC, Stark A.. 2009. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev. 19(6):565–570. [DOI] [PubMed] [Google Scholar]
- Mishra A, Krishnan B, Srivastava SS, Sharma Y.. 2014. Microbial βγ-crystallins. Prog Biophys Mol Biol. 115(1):42–51. [DOI] [PubMed] [Google Scholar]
- Morozov V, Wawrousek EF.. 2006. Caspase-dependent secondary lens fiber cell disintegration in αA-/αB-crystallin double-knockout mice. Development 133(5):813–821. [DOI] [PubMed] [Google Scholar]
- Nilsson DE. 2009. The evolution of eyes and visually guided behaviour. Philos Trans R Soc Lond B Biol Sci. 364(1531):2833–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson DE. 2013. Eye evolution and its functional basis. Vis Neurosci. 30(1–2):5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J, et al. 2010. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin αA. Neuron 68(4):682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J.. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302(1):205–217. [DOI] [PubMed] [Google Scholar]
- Oakley TH, Speiser DI.. 2015. How complexity originates: the evolution of animal eyes. Annu Rev Ecol Evol Syst. 46(1):237–260. [Google Scholar]
- Oesterreich S, Hickey E, Weber LA, Fuqua SA.. 1996. Basal regulatory promoter elements of the hsp27 gene in human breast cancer cells. Biochem Biophys Res Commun. 222(1):155–163. [DOI] [PubMed] [Google Scholar]
- Park Y, Nnamani MC, Maziarz J, Wagner GP.. 2016. Cis-regulatory evolution of forkhead box O1 (FOXO1), a terminal selector gene for decidual stromal cell identity. Mol Biol Evol. 33(12):3161–3169. [DOI] [PubMed] [Google Scholar]
- Payne JL, Wagner A.. 2014. The robustness and evolvability of transcription factor binding sites. Science 343(6173):875–877. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. 1981. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation 19(3):134–153. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. 2006. Evolutionary genetics: seeing the light: the role of inherited developmental cascades in the origins of vertebrate lenses and their crystallins. Heredity (Edinb) 96(4):275–277. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. 2007. Gene sharing and evolution: the diversity of protein function. Cambridge, (MA: ): Harvard University Press. [Google Scholar]
- Piatigorsky J, Kozmik Z.. 2004. Cubozoan jellyfish: an Evo/Devo model for eyes and other sensory systems. Int J Dev Biol. 48(8–9):719–729. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, Wistow G.. 1991. The recruitment of crystallins: new functions precede gene duplication. Science 252(5009):1078–1079. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, et al. 2000. Ω-Crystallin of the scallop lens. A dimeric aldehyde dehydrogenase class 1/2 enzyme-crystallin. J Biol Chem. 275(52):41064–41073. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS.. 2000. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127(2):307–317. [DOI] [PubMed] [Google Scholar]
- Sandilands A, et al. 2002. Altered aggregation properties of mutant γ-crystallins cause inherited cataract. EMBO J. 21(22):6005–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, et al. 2005. Urochordate βγ-crystallin and the evolutionary origin of the vertebrate eye lens. Curr Biol. 15(18):1684–1689. [DOI] [PubMed] [Google Scholar]
- Singh BN, Rao KS, Ramakrishna T, Rangaraj N, Rao CM.. 2007. Association of αB-crystallin, a small heat shock protein, with actin: role in modulating actin filament dynamics invivo. J Mol Biol. 366(3):756–767. [DOI] [PubMed] [Google Scholar]
- Somasundaram T, Bhat SP.. 2000. Canonical heat shock element in the αB-crystallin gene show tissue-specific and developmentally controlled interactions with heat shock factor. J Biol Chem. 275(22):17154–17159. [DOI] [PubMed] [Google Scholar]
- Somasundaram T, Bhat SP.. 2004. Developmentally dictated expression of heat shock factors: exclusive expression of HSF4 in the postnatal lens and its specific interaction with αB-crystallin heat shock promoter. J Biol Chem. 279(43):44497–44503. [DOI] [PubMed] [Google Scholar]
- Srivastava SS, Mishra A, Krishnana B, Sharma Y.. 2014. Ca2+-binding motif of βγ-crystallins. J Biol Chem. 289(16):10958–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V.. 2008. The loci of evolution: how predictable is genetic evolution?. Evolution 62(9):2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JR, Wray GA.. 2001. Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol. 18(9):1764–1770. [DOI] [PubMed] [Google Scholar]
- Sun MG, Kim PM.. 2011. Evolution of biological interaction networks: from models to real data. Genome Biol. 12(12):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. 2015. Identification of invivo DNA-binding mechnaisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 43(14):6827–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, et al. 2000. Identification of RALDH-3, a novel retinaldehyde dehydrogenase, expressed in the ventral region of the retina. Mech Dev. 98(1–2):37–50. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK, Piatigorsky J.. 2007. Regulation of the mouse αB-crystallin and MKBP/HspB2 promoter activities by shared and gene specific intergenic elements: the importance of context dependency. Int J Dev Biol. 51(8):689–700. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Duncan MK, Roth HJ, Cvekl A, Piatigorsky J.. 1994. Convergent evolution of crystallin gene regulation in squid and chicken: the AP-1/ARE connection. J Mol Evol. 39(2):134–143. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Piatigorsky J.. 1996. Lens crystallins of invertebrates–diversity and recruitment from detoxification enzymes and novel proteins. Eur J Biochem. 235(3):449–465. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y.. 2013. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem Biol Interact. 202(1–3):2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vopalensky P, Kozmik Z.. 2009. Eye evolution: common use and independent recruitment of genetic components. Philos Trans R Soc Lond B Biol Sci. 364(1531):2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vopalensky P, et al. 2012. Molecular analysis of the amphioxus frontal eye unravels the evolutionary origin of the retina and pigment cells of the vertebrate eye. Proc Natl Acad Sci U S A. 109(38):15383–15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. 2007. The developmental genetics of homology. Nat Rev Genet. 8(6):473–479. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Otto W, Lynch V, Stadler PF.. 2007. A stochastic model for the evolution of transcription factor binding site abundance. J Theor Biol. 247(3):544–553. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P.. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113(4):1435–1449. [DOI] [PubMed] [Google Scholar]
- Wawrousek EF, Chepelinsky AB, McDermott JB, Piatigorsky J.. 1990. Regulation of the murine αA-crystallin promoter in transgenic mice. Dev Biol. 137(1):68–76. [DOI] [PubMed] [Google Scholar]
- Wistow G. 1993. Lens crystallins: gene recruitment and evolutionary dynamism. Trends Biochem Sci. 18(8):301–306. [DOI] [PubMed] [Google Scholar]
- Wistow G. 2012. The human crystallin gene families. Hum Genomics 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow GJ, Piatigorsky J.. 1988. Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem. 57:479–504. [DOI] [PubMed] [Google Scholar]
- Wray GA, et al. 2003. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 20(9):1377–1419. [DOI] [PubMed] [Google Scholar]
- Xi JH, Bai F, Andley UP.. 2003. Reduced survival of lens epithelial cells in the αA-crystallin-knockout mouse. J Cell Sci. 116(Pt 6):1073–1085. [DOI] [PubMed] [Google Scholar]
- Xi JH, Bai F, McGaha R, Andley UP.. 2006. crystallin expression affects microtubule assembly and prevents their aggregation. FASEB J. 20(7):846–857. [DOI] [PubMed] [Google Scholar]
- Xie Q, Cvekl A.. 2011. The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. J Biol Chem. 286(50):43259–43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, et al. 1999. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 13(10):1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chauhan BK, Cveklova K, Cvekl A.. 2004. Transcriptional regulation of mouse αB- and γF-crystallin genes in lens: opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J Mol Biol. 344(2):351–368. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cvekl A.. 2005. Tissue-specific regulation of the mouse αA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. J Mol Biol. 351(3):453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. 2006. Regulation of αA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 25(10):2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Kielbasa SM, Arndt PF, Chung HR, Vingron M.. 2009. Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet. 25(2):63–66. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Zheng JB, Gu X, Saunders GF, Yung WK.. 2002. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 12(11):1716–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.