Abstract
Poly ADP-ribose polymerases (PARPs) catalyze massive protein poly ADP-ribosylation (PARylation) within seconds after the induction of DNA single- or double-strand breaks. PARylation occurs at or near the sites of DNA damage and promotes the recruitment of DNA repair factors via their poly ADP-ribose (PAR) binding domains. Several novel PAR-binding domains have been recently identified. Here, we summarize these and other recent findings suggesting that PARylation may be the critical event that mediates the first wave of the DNA damage response. We also discuss the potential for functional crosstalk with other DNA damage-induced post-translational modifications.
INTRODUCTION
The genetic information stored in the DNA is prone to damage by environmental and internal hazards such as ultraviolet light, mutagenic chemicals, ionizing radiation and reactive oxygen species (1–3). Exposure to these genotoxic stresses induces various types of DNA lesions, including DNA single-strand breaks (SSBs) and double-strand breaks (DSBs). Without repair, accumulated lesions can drastically alter the genome. Fortunately, cells have evolved a sophisticated DNA damage response system to repair these DNA lesions and maintain genomic stability.
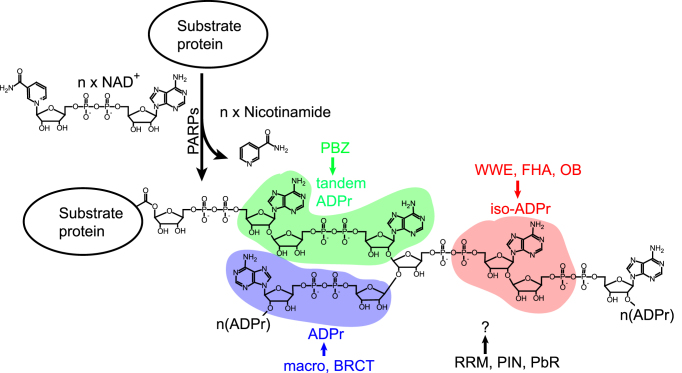
After induction of DNA damage, the immediate reaction of a cell is to detect various DNA lesions using DNA damage sensors. These sensors are abundant in the nucleus for damage surveillance and consequent activation of the repair process. In the presence of DNA damage, these sensors initiate signals to recruit DNA damage repair (DDR) factors and activate other relevant biological processes, such as cell cycle arrest, to facilitate the repair process (4). Accumulated evidence indicates that PARP1, the founding member of PARP family, recognizes both SSBs and DSBs. PARP1 transfers the ADP-ribose moiety of NAD+ to the side chains of asparagine, aspartic acid, glutamic acid, arginine, lysine, serine and cysteine residues on its substrates (5–16). The ribose sugar adjacent to the adenine side is linked to the next ADP-ribose residue through glycosidic bonds to form a linear PAR chain containing up to 200 ADP-ribose residues (17) (Figure 1). Branched ADP-ribose chains are also generated by α(1″′-2″)-ADP-ribose linkage (18) (Figure 1). These PAR chains form a platform to recruit DNA repair proteins via their PAR-binding domains (Figure 1). Together, these properties allow PARP1 to function as an important DNA damage sensor for both SSBs and DSBs (20).
Figure 1.
Schematic representation of PAR synthesis. PARPs hydrolyze nicotinamide from NAD+ and covalently link the remaining ADP-ribose moieties to their substrates, forming linear or branched PAR chains. Different PAR readers recognize distinct units of the PAR chain. The PBZ motif recognizes tandem ADP-ribose. The WWE, FHA and OB domains recognize iso-ADP-ribose. The macro and BRCT domains recognize ADP-ribose. The recognition units of the RRM, PIN and PbR domains need to be identified. ADPr: ADP-ribose; iso-ADPr: iso-ADP-ribose.
With the identification of new proteins as ‘readers’ of protein PARylation, the PARylation-dependent early DNA damage response has emerged as an important aspect of the complex repair process in response to new DNA lesions. This review highlights the biological function of PARylation in response to DNA damage, focusing on the idea that PARylation may serve as an early signal to initiate the DNA damage response. Interestingly, PARylation is a transient response to DNA damage that is also controlled by dePARylation. Thus, the role of protein dePARylation, an equally important process for DDR, will also be discussed. Moreover, this review will explore the functional interactions between PARylation and other signals generated after DNA damage, including phosphorylation and ubiquitination.
PARP1 ACTS AS A SENSOR FOR BOTH SSBS AND DSBS
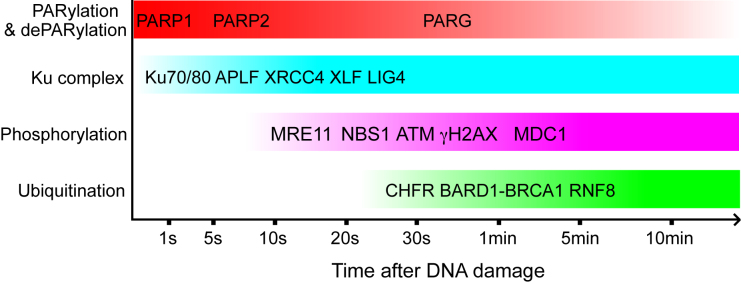
PARylation is a unique post-translational modification synthesized in response to DNA damage, which acts as a crucial signal after SSB or DSB induction. The basal level of PARylation is very low in cells under normal conditions (21), indicating that most PARPs are in the inactive state. However, PARP1, which catalyzes about 90% of DNA damage-induced PARylation (22), is strongly activated after binding to SSBs and DSBs. PARP1 is one of the most abundant nuclear polypeptides, with an estimated 1–2 million molecules per nucleus (23,24). On average, one PARP1 molecule scans approximately 10 nucleosomes of chromatin (25). This scanning function enables PARP1 to quickly detect DNA damage. In laser micro-irradiation experiments, PARP1 is recruited to DNA damage sites with a t1/2 of only ∼1.6 s (Figure 2) (26).
Figure 2.
Schematic representation of the temporal dynamics of DDR proteins recruited to the DNA damage sites in response to laser micro-irradiation.
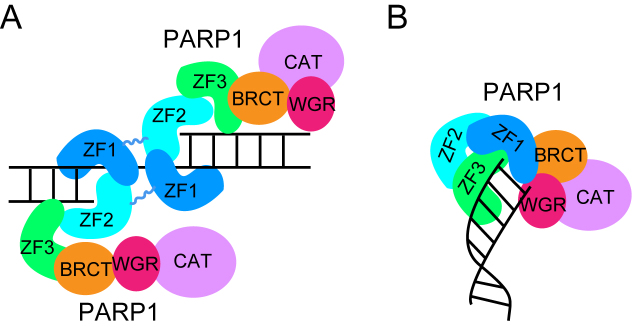
PARP1 contains multiple domains, including three N-terminal zinc finger motifs (ZF1-ZF3), a BRCT domain, a WGR domain and a C-terminal catalytic domain. As shown in Figure 3A, once an SSB is generated, ZF2 stacks onto the 3′ terminal base pair via its hydrophobic platform. The ZF1 motif from a second PARP1 molecule dimerizes with the ZF2, and leaves an open channel to accommodate the undamaged nucleotide. This dimerization of ZF1 and ZF2 may then lead to an additional intramolecular conformational change and activation of the catalytic domain of PARP1 (27).
Figure 3.
PARP1 is a sensor for both SSBs (A) and DSBs (B). The domains within one PARP1 molecule are colored differently. ZF1-3, N-terminal zinc finger motif 1-3; BRCT, BRCT domain; WGR, WGR domain; CAT, C-terminal catalytic domain.
In contrast to SSBs, PARP1 recognizes DSBs by a slightly different mechanism. For each end generated from a DSB, the ZF1 domain recognizes the terminal base pair, whereas the ZF3 and the WGR domains contact each side of the backbone of the DNA helix and bind the minor and major grooves to stabilize the interaction (28). The WGR domain stacks to the 5′-terminus of the DNA end but the ZF2 domain is dispensable for DSB recognition (Figure 3B). This interaction with one DSB end triggers intramolecular conformational changes within the catalytic domain and activates PARylation (28). Together, these structural studies suggest that PARP1 can sense both SSBs and DSBs.
In addition to PARylation, accumulation of a phosphorylated form of histone H2AX, known as γH2AX, is one of the earliest events that occur at DSBs. H2AX differs from canonical H2A in the C-terminal tail, which contains an evolutionarily conserved serine motif in the H2AX variant. Phosphorylation at this serine residue occurs within one minute of IR treatment (Figure 2) (29,30). The accumulation of γH2AX is important for retaining numerous DDR factors at DSBs (31,32). However, in H2AX-deficient cells, most DDR factors can still transiently relocate to the sites of DSBs (31), indicating that γH2AX is not essential for their early recruitment. Moreover, the genetic deficiency in H2AX has modest effects on DSB repair in vivo (31–33). These results suggest that although H2AX phosphorylation is important, the precise role of γH2AX in DSB repair remains elusive.
Ataxia-telangiectasia mutated (ATM) is the major kinase responsible for H2AX phosphorylation in response to DSB induction (34–37). Accumulated evidence suggests that the MRN complex, which consists of MRE11, RAD50 and NBS1, is essential for ATM activation (38). Among these three subunits, RAD50 recognizes naked DNA ends and holds them in close proximity to each other (39,40). MRE11 is an endo- and exo-nuclease that processes DNA ends prior to religation for resection-dependent nonhomologous end joining (NHEJ) or prior to more extensive resection by other nucleases for homologous recombination (HR) (41). NBS1 mediates protein-protein interactions within the complex (42–44). The ability of the MRN complex to bind directly to the DNA ends suggests that it could be a DSB sensor (45–47). However, in laser micro-irradiation experiments, the t1/2 for MRN recruitment is ∼13 s (Figure 2) (26), which is almost 10 times slower than the t1/2 for PARP1 recruitment. Of note, laser micro-irradiation generates mixtures of different DNA lesions, including both SSBs and DSBs. Thus, DNA damage sensors on both SSBs and DSBs will be recruited to lesions generated by laser micro-irradiation. Nevertheless, in spite of certain limitations like the presence of heterogeneous mixture of various types of DNA lesions, micro-irradiation in the UV range is the most efficient method for experimental induction of DNA damage that allows measurement in the narrow time scale of few seconds (48). The relatively slow recruitment of the MRN complex suggests that this complex may serve as a secondary sensor for DSB repair. Consistently, the early recruitment of MER11 and NBS1 is mediated by PARP1 (26,49). Moreover, our recent findings indicate that NBS1 recognizes DNA damage-induced PARylation and mediates the early recruitment of the MRN complex (50). Because DNA damage-induced PARylation is mainly catalyzed by PARP1, one hypothesis is that PARP1 acts upstream of the MRN complex and γH2AX in response to DSBs. Alternatively, structural analysis indicates that the N-terminus of PARP1 favors binding to the blunt ends of DSBs (28), whereas MRE11, the catalytic subunit of the MRN complex, recognizes 5′-overhang DNA (51). Thus, it is also possible that PARP1 and the MRN complex preferentially sense different types of DSBs.
Similar to PARP1, the Ku complex, consisting of Ku70 and Ku80 subunits, also relocates to DSBs within few seconds after DNA damage and binds directly to DSBs with high affinity (Figure 2) (52–54). The Ku complex is also highly abundant, with ∼500 000 molecules per cell (55). Thus, in addition to PARP1, the Ku complex is another important DSB sensor. However, the mechanism of crosstalk between the Ku complex and PARP1 is still unclear. As DSBs can be repaired through HR, alternative NHEJ (a-NHEJ) or canonical NHEJ (c-NHEJ) pathways (56–58), it is possible that PARP1 and the Ku complex mediate DSB repair pathway choice separately through distinct mechanisms. Binding of the Ku complex to DSBs recruits and activates the DNA-PK catalytic subunit, which facilitates c-NHEJ (59). The Ku complex also interacts with numerous other c-NHEJ factors, including XRCC4 (53,60,61), APLF (54), XLF (62) and LIG4 (60). In contrast, the binding of PARP1 promotes a-NHEJ and HR repair (63–68). Thus, the competition between PARP1 and the Ku complex at DSBs may play an important role in determining the repair pathway. Moreover, these two DNA damage sensors may have complementary roles in activation of the DNA damage response. Loss of either sensor in mammals only generates minor DSB repair defects (69–73). Interestingly, double knockout of the mouse Parp1 and Xrcc5 (Ku80) genes causes early embryonic lethality (74,75), consistent with the idea that PARP1 and Ku have complementary functions. In addition, it is also possible that PARP1 and the Ku complex function together at certain steps of DSB repair. It is interesting to note that Ku70 in Dictyostelium discoideum even has a PAR-binding motif (76). Also, PARylation retains the Ku complex at DSBs for efficient NHEJ in Dictyostelium discoideum (77). Moreover, in mammals, the Ku complex is ADP-ribosylated after DNA damage induction, although the function of this ADP-ribosylation is unclear (9,16,78). Thus, future studies on the functional interaction between PARP1 and the Ku complex may reveal novel molecular mechanisms of NHEJ.
CONTRIBUTION OF PARP2 AND PARP3 TO DNA DAMAGE SIGNALING
In addition to PARP1, PARP2 also participates in the DNA damage response (79–81). Unlike PARP1, PARP2 does not contain N-terminal zinc finger motifs. However, the N-terminal region and the adjacent WGR domain of PARP2 act together to bind the ends of nucleic acid strands; this binding activates the C-terminal catalytic domain for the synthesis of PAR (82). Thus, the activation mode of PARP2 is similar to that of PARP1. Recent genetic evidence shows that Parp1- or Parp2-knockout mice has very mild DDR defects, whereas the double knockout of Parp1 and Parp2 arrests embryonic development at gastrulation (83), suggesting that PARP1 and PARP2 have overlapping functions. Although PARP2 is recruited to the sites of laser-induced DNA damage slower than PARP1, it persists comparatively longer (Figure 2) (84). Thus, it is likely that PARP1 and PARP2 play slightly different roles in the DNA damage response.
Recent studies show that PARP3 also plays an important role in DDR (85–87). PARP3 has an N-terminal WGR domain and a C-terminal catalytic domain, but lacks other domains implicated in DNA binding. However, PARP3 can be activated in response to DSBs, especially by DNA breaks with 5′-phosphoryl ends (82,85,88). It is possible that the conserved WGR domain facilitates DSB recognition (89). The enzymatic activity of PARP3 is still controversial. Rulten et al. reported that PARP3 could generate PAR, although it was very short compared to that generated by PARP1 (85). However, Loseva et al. and Vyas et al. found that PARP3 catalyzed only mono ADP-ribosylation (MARylation) in vitro (88,90). PARP3 interacts with the Ku complex, DNA-PKcs, LIG4 and APLF (85,87,91), which are key factors in the c-NHEJ pathway. Also, Ku complex can be ADP-ribosylated by PARP3 (78), which is then recognized by the PAR-binding zinc finger (PBZ) motif of APLF, and thus promotes the recruitment of XRCC4 and LIG4 to enable c-NHEJ (85,87).
Specific substrates of PARP1, PARP2 and PARP3 were identified using engineered PARP mutants that only use NAD+ analogs for PARylation (78,92,93). Although most of the substrates modified by each PARP are distinct, they can be categorized into multiple common ontological groups, such as transcription, RNA processing, chromatin organization and DNA damage repair (78). Moreover, the preferred ADP-ribosylation motifs around the ADP-ribosylation site (here indicated as E*) were also identified for each PARP. For instance, PARP1 prefers E*P, E*XP and E*E sites; PARP2 prefers EE* and GXXXXXE* sites, and PARP3 prefers K or R residues within ±8 residues of ADP-ribosylation sites (78).
Because each ADP-ribose residue contains two negatively charged phosphate groups, PARylation imparts a substantial amount of negative charge to the sites of DNA damage (19). As DNA itself is also negatively charged, PARylation may loosen the higher-order structure of chromatin due to charge-charge repulsion, which would facilitate access to the DNA repair machinery. Additionally, as PARP1 is a major substrate for PARylation (94), automodified PARP1 could disassociate from the damaged sites because of its negative charge (95,96). Histones, such as H1, H2A and H2B, are also important substrates of PARP1 (16,97,98). PARylation at the sites of DNA damage would also enhance the accessibility of large protein complexes assembled during the DDR process. Recent evidence also suggests that, in addition to chromatin remodeling, PARylation functions as a signal for recruitment of numerous DNA damage response factors to the sites of DNA damage (50,66,76,99–105). Over the past few years, several types of PAR-binding modules have been identified, including the PBZ motif; the macro, WWE, BRCT, FHA, OB-fold, RRM, and PIN domains; and PAR-binding regulatory (PbR) motif (Table 1 and Figure 1). With live cell imaging, we and other researchers have shown that the interactions between these modules and PAR mediate their recruitment to the DNA damage sites within 20–30 seconds after DNA damage (50,66,105,106). These DNA damage response factors play key roles in multiple repair mechanisms, including base excision repair (BER), SSB repair, and DSB repair. Consistently, previous analyses showed a similar role for PARPs in these repair pathways (106–108). Apart from their functions in DNA damage response, the ADP-ribosylation readers play important roles in other biological processes, such as DNA replication, cell cycle regulation, chromatin remodeling, RNA metabolism and protein turnover (109).
Table 1. Summary of PAR-binding modules.
| Module name | Recognition unit | Protein(s) | Reference(s) |
|---|---|---|---|
| PAR-binding zinc finger (PBZ) | Tandem ADP-ribose | APLF, CHFR | (76,161,172) |
| Macro domain | ADP-ribose | macroH2A1.1, ARTD7, ARTD8, ARTD9, macroD1, macroD2, macroD3, ALC1, TARG1, PARG | (108,173) |
| WWE domain | iso-ADP-ribose | RNF146, HUWE1, ULF, Deltex1, Deltex2, Deltex4, ARTD11 | (174,175) |
| BRCT domain | ADP-ribose | BARD1, LIG4, NBS1, XRCC1, ECT2 | (50,66,176) |
| FHA domain | iso-ADP-ribose | PNKP, APTX | (50) |
| OB-fold domain | iso-ADP-ribose | SSB1, CTC1, MEIOB, SSB2 | (105) |
| RRM domain | NONO | (177) | |
| PIN domain | EXO1, GEN1, SMG5 | (106) | |
| PAR-binding regulatory (PbR) motif | Chk1 | (178) |
Although PARPs catalyze massive PARylation at the sites of DNA damage, the half-life of PAR is only a few minutes (110,111). Thus, PARylation may act as the first wave of signaling, to transiently activate DDR. Because the process of DDR continues until the lesions are fixed, especially for DSB repair, it is possible that PARylation mediates the priming of DDR by recruiting the DNA repair machinery to the region near DNA lesions through their PAR-binding modules. For example, the BRCT domain of NBS1 recognizes DNA damage-induced PAR and mediates the early recruitment of the MRN complex to DSBs within 20 seconds (50). The early recruitment of the MRN complex may prime early ATM activation and ATM-dependent cell cycle arrest. Collectively, the first wave of PAR signaling after DNA damage recruits PAR readers to the sites of DNA damage and induces DNA damage response by activating cell cycle checkpoints and DDR.
Although the cellular concentration of NAD+ is roughly 0.3–1 mM (112–114), PARPs are able to use abundant NAD+ to synthesize massive amounts of PAR in a very short period of time in response to DNA damage. It has been reported that when wild-type fibroblasts are treated with 0.5 mM Methylnitronitrosoguanidine (MNNG), the NAD+ level drops 80% within 15 minutes and is no longer detectable by 30 minutes (115). Similar results were also reported when cells were treated with lower concentrations of MNNG or other DNA damage-inducing agents (116–120). Thus, DNA damage-induced PARylation may have much more far-reaching effects on cellular physiology than other post-translational modifications in cells.
These massive amounts of PAR likely provide abundant docking sites for DNA damage response factors with distinct PAR-binding modules. However, how DNA damage response factors are loaded to DNA lesions remains elusive. One possibility is that these DNA damage response factors are loaded sequentially via specific PARylation motifs. Alternatively, PARylation facilitates the recruitment of these DNA damage response factors to the proximity of DNA lesions. With dePARylation and other post-translational modifications, specific DDR machineries are retained at different types of lesions for DNA repair.
DEPARYLATION IS ALSO REQUIRED FOR DDR
Although extensive and rapid PARylation recruits DNA damage response factors to the vicinity of DNA lesions, PARylation is removed quickly. This rapid dePARylation can perhaps prevent the trapping of other factors involved in the first wave of the DNA damage response. Otherwise, trapping these DNA damage repair factor may block access of other downstream repair factors. Therefore, the timely and orderly degradation of PAR by dePARylation enzymes is the next necessary step towards DDR.
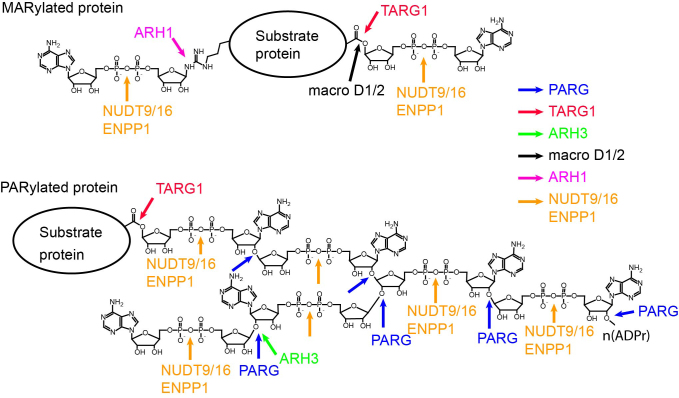
To date, six human dePARylation enzymes have been identified, including poly ADP-ribose glycohydrolase (PARG), TARG1, ARH3, NUDT9 and NUDT16 and ENPP1 (Table 2, Figure 4). PARG contains a macro domain and it has exo- and endo-glycohydrolysis activity to hydrolyze the glycosidic bond between ADP-ribose units in PAR chain and release free ADP-ribose residues (121–124). TARG1 is also a macro domain-containing protein. However, TARG1 cannot hydrolyze PAR to ADP-ribose; rather, it removes the whole PAR chain from the glutamate residue on the PARylated proteins (125). TARG1 can also hydrolyze the glutamate-ADP-ribose bond and release the terminal ADP-ribose unit from MARylated proteins (125). Similar to PARG, ARH3 has exo-glycohydrolysis activity to digest PAR chains and release free ADP-ribose, although the ARH3 activity is much lower than PARG activity (126). NUDT9 and NUDT16 have nucleoside diphosphate-linked moiety X (Nudix) domains, which cleave pyrophosphate bonds, and release iso-ADP-ribose and AMP from PAR chains or AMP from MARylated proteins (127,128). It is possible that Nudix pyrophosphatases collaborate with other dePARylation enzymes to further digest ADP-ribose. ENPP1 is a newly discovered pyrophosphatase lacking a Nudix domain; it can digest PAR chains and release iso-ADP-ribose from PAR chains or AMP from MARylated proteins (129).
Table 2. Summary of deADP-ribosylation enzymes.
| Name | Synonyms | Subcellular location | Key domain | Substrates and activity | Reference(s) |
|---|---|---|---|---|---|
| PARG | Nucleus, cytoplasm, mitochondria | Macro domain | PAR chain, exo- and endo-glycohydrolysis to produce ADP-ribose and short PAR chain. | (179,180) | |
| TARG1 | C6orf130 OARD1 | Nucleus, cytoplasm | Macro domain | Mono ADP-ribosylated protein, cleaving the bond between acidic residues and ADP-ribose; poly ADP-ribosylated protein, releasing the whole PAR chain from protein; deacylation of O-acetyl-ADP-ribose, O-propionyl-ADP-ribose, and O-butyryl-ADP-ribose to produce ADP-ribose and acetate, propionate, and butyrate, respectively. | (125,130,181) |
| MacroD1 | LRP16 | Nucleus | Macro domain | Mono ADP-ribosylated protein, cleaving the bond between acidic residues and ADP-ribose; deacetylation of O-acetyl-ADP-ribose. | (130) |
| MacroD2 | C20orf133 | Nucleus, cytoplasm | Macro domain | Mono ADP-ribosylated protein, cleaving the bond between acidic residues and ADP-ribose; deacetylation of O-acetyl-ADP-ribose. | (130) |
| ADPRH | ARH1 | Nucleus, cytoplasm | Mono ADP-ribose-arginine protein, cleaving the N-glycosidic bond of ADP-ribose attached to an Arg residue of a protein to produce free ADP-ribose and unmodified protein. | (131) | |
| ADPRHL2 | ARH3 | Nucleus, cytoplasm, mitochondria | Deacetylation of O-acetyl-ADP-ribose; PAR chain, exo-glycohydrolysis to produce ADP-ribose. | (126,182,183) | |
| NUDT9 | Cytoplasm | Nudix hydrolase | Cleaving ADP-ribose and IDP-ribose to form the corresponding nucleoside 5′-monophosphates and ribose 5-phosphate; cleaving O-acetyl-ADP-ribose to form AMP and acetylated ribose 5′-phosphate; low activity to digest PAR by cleaving the pyrophosphate bonds. | (127,128,184) | |
| NUDT16 | Nucleus, cytoplasm | Nudix hydrolase | Cleaving m7G or m227G caps from U8 snoRNA or mRNA and leaving a 5′-monophosphate-RNA; Poly/mono ADP-ribosylated protein, cleaving the pyrophosphate bonds of ADP-ribose. | (128,185) | |
| ENPP1 | Extracellular, lysosome, plasma membrane | Cleaving the phosphodiester bonds in (d)NTP, (d)NDP, NAD, ADP-ribose, FAD, diadenosine polyphosphates, UDP sugars, PAR chains and mono ADP-ribosylated proteins. | (129) |
Figure 4.
Schematic representation of the deADP-ribosylation process. The cutting sites of each enzyme are shown by the arrows with indicated colors. ADPr: ADP-ribose.
Recent studies have also found enzymes that remove MAR groups from proteins. These enzymes include macro D1, macro D2, ARH1 and the aforementioned TARG1, NUDT9, NUDT16 and ENPP1 (Table 2, Figure 4). Macro D1 and macro D2 are two macro domain-containing enzymes that can release ADP-ribose from ADP-ribosylated acidic residues (130). ARH1 is the only hydrolase that specifically removes MAR from arginine residues (131).
Among these deADP-ribosylation enzymes, the most potent in the context of DDR is PARG (121–124). The early recruitment of PARG to the sites of DNA damage is mediated by the interaction of its macro domain with the PAR signal (132), but PARG recruitment occurs more slowly than recruitment of PARP1, with a t1/2 of about 50 seconds (Figure 2) (133). PARG is stably retained by proliferating cell nuclear antigen (PCNA) (133). As expected, reduction or loss of PARG expression causes a significant delay in PAR degradation and extends the half-life of PAR at DNA lesions (16,134,135). Instead of facilitating DDR, prolongation of PARylation impairs both SSB and DSB repair, and thereby inducing apoptosis (136–138). Moreover, loss of the Parg gene induces early embryo lethality at the gastrulation stage (136), which phenocopies the Parp1 and Parp2 double-knockout mouse (83). The genetic similarity further indicates that dePARylation is essential for DDR.
Besides PARG, other dePARylation enzymes are also recruited to the sites of DNA damage, suggesting that these enzymes are involved in the DNA damage response and may act sequentially following ADP-ribosylation (125,133,139). In particular, TARG1 is not only recruited to DNA lesions, but germline mutation of TARG1 induces progressive neurodegenerative disorders (125), a phenomenon often observed when BER is impaired. Thus, these findings indicate that TARG1 participates in BER. However, the specificity of these dePARylation enzymes towards the different substrates remains elusive.
The ADP-ribose released by PAR glycohydrolases may not be merely a byproduct of the reaction, but also an important second messenger accounting for Ca2+ influx and caspase activation (140). Moreover, a recent study shows that ADP-ribose released from dePARylation enzymes can be digested further by NUDT5, another Nudix domain family member, to form AMP and ribose phosphate. The released AMP moiety can be used to synthesize ATP for chromatin remodeling during DDR (141). Finally, ADP-ribose can also be recycled to form NAD+ to maintain the cellular level of NAD+, which is a co-enzyme required for numerous biochemical reactions (142). Collectively, the digested ADP-ribose monomer may be involved in many physiological relevant processes in response to DNA damage.
FUNCTIONAL INTERACTION BETWEEN DNA DAMAGE-INDUCED PARYLATION AND PHOSPHORYLATION
Besides PARylation, other post-translational modifications, like DSB-induced phosphorylation and ubiquitination, are also induced by DNA damage and function as signals to mediate the recruitment of DNA damage machinery (143,144). The phosphorylation cascade in response to DNA damage is initiated by a group of well-documented PI3-like kinases, including ATM, ATM- and Rad3-related (ATR), and/or DNA-dependent protein kinase (DNA-PKcs) (145–147). These kinases are mainly activated in response to DSBs, and some redundancy of these kinases in DSB-induced phosphorylation events has been shown. Among these kinases, ATM is considered the primary inducer of the phosphorylation cascade in response to DSBs (148). The rapid accumulation and activation of the ATM kinase cascade results in Ser or Thr phosphorylation of several hundreds of proteins, including effectors of the DNA damage response such as BRCA1, CHK2 and p53 (146), which further activate cell cycle checkpoints and DDR.
Phosphorylation of H2AX is observed about 1 minute after DNA damage (29,30), a time scale that is considerably slower than the PARP1 recruitment (Figure 2). It is possible that the PI3-like kinase-induced phosphorylation cascade is the second wave of signaling during the DNA damage response. As mentioned earlier, PARylation can prime the activation of the ATM-dependent phosphorylation cascade via recruitment of the MRN complex (50). In our previous study, we showed that the BRCT domain of NBS1, one of the components of the MRN complex, recognizes PARP1-dependent PARylation at DNA lesions, which recruits and activates ATM (50). PARylation may thus serve as the first wave of signaling at DNA lesions, thereby facilitating ATM-dependent phosphorylation as a second wave of signaling.
One of the prominent phosphorylation targets of ATM is H2AX. Following DNA damage, ATM-induced γH2AX can be observed within one minute in the range of ∼1 kb of the chromatin flanking the DNA lesion. γH2AX can spread up to ∼500 kb of the flanking chromatin regions in a few hours, and is important for anchoring numerous DDR factors surrounding DNA lesions (31,32). Interestingly, PARylation, the first wave of signals, may negatively regulate this H2AX phosphorylation event. Mass spectrometry analysis indicates that H2AX is quickly PARylated at E141 after DNA damage (16). Because PARylation at E141 brings a large amount of negative charge close to Ser139, it is possible that PARylation of E141 suppresses the phosphorylation of Ser139, which may subsequently delay the recruitment of DDR factors or destabilize some DDR factors at DNA lesions. However, because the PARylation on H2AX is also quickly removed by dePARylation enzymes, the Ser139 motif can be re-exposed to PI3-like kinases. Such transient delay of H2AX phosphorylation adds another layer of regulation for the recruitment of DDR factors. The wave of PARylation mediates the recruitment of numerous DDR complexes, whereas dePARylation and subsequent phosphorylation of H2AX may selectively stabilize certain DDR factors at DNA lesions to fulfill the repair function.
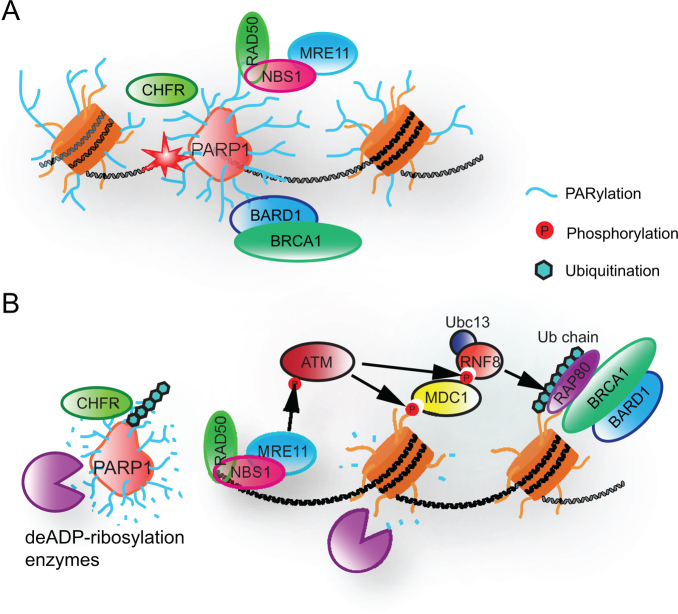
One typical example of two-stage recruitment is the recruitment of the BRCA1/BARD1 complex, a key complex for DSB repair. The recruitment of this complex involves both PARylation and the γH2AX-dependent pathway. Both BRCA1 and BARD1 have N-terminal Ring domains, through which they bind to each another (149). They also have C-terminal BRCT domains. The BRCT domain of BRCA1 is a phospho-Ser binding domain (150,151), whereas the BRCT domain of BARD1 is a PAR-binding domain (66). After the induction of DSBs, BARD1 recognizes the PAR signal at the sites of DNA damage through its BRCT domain, which mediates the quick mobilization of BRCA1 to DNA lesions, thereby recruiting a functional BRCA1/BARD1 complex at the sites of DNA damage (Figure 5A). After the dePARylation, retention of the BRCA1/BARD1 complex is mediated through a γH2AX-dependent pathway (31,66) (Figure 5B). The BRCT domain of BRCA1 has a phospho-serine binding domain through which it recognizes a phospho-Ser motif in the RAP80 complex, which is anchored at the DNA lesions via a γH2AX-dependent pathway (152–155). Thus, both PARylation and phosphorylation play important roles in recruiting and retaining the BRCA1 complex at DSBs for lesion repair. The synergistic effects of PARylation and phosphorylation on DSB repair are further supported by mouse genetic studies. In vivo studies show that mice lacking the Parp1, H2ax, or Atm genes have minor DNA repair defects (33,69,70,72,156), whereas the loss of both Parp1 and H2ax (or Atm) leads to early embryonic lethality at the gastrulation stage (157,158). Again, this embryonic lethal phenotype of the Parp1 and H2ax (or Atm) double-mutants is similar to that of HR-deficient mice, including the Brca1 knockout mouse, which is consistent with the notion that both PAR and γH2AX are required for the stable recruitment of BRCA1 (159).
Figure 5.
Functional interactions between DNA damage-induced PARylation and other post-translational modifications. (A) PARP1-mediated PARylation facilitates the early recruitment of DNA damage factors (e.g. NBS1, BARD1 and CHFR). (B) In response to PARylation, other post-translational modifications (e.g. phosphorylation and ubiquitination) stabilize the DDR machinery.
However, PARylation may not always function together with phosphorylation. The Ser139 motif in H2AX is a typical phosphorylation motif for the PI3-like kinases, as the consensus phosphorylation motif is S/T-Q-D/E (146). Interestingly, PARylation is usually observed at aspartic acid and/or glutamic acid residues on the substrates (9,16). Because the consensus motif of the PI3-like kinases contains aspartic acid and/or glutamic acid at the +2 position, these motifs can also be potentially PARylated. Additionally, competition between the phosphorylated and PARylated states can regulate the subsequent choice of repair pathways. Thus, PARylation can also act as a competitor for DNA damage-induced phosphorylation at certain loci.
FUNCTIONAL INTERACTION BETWEEN DNA DAMAGE-INDUCED PARYLATION AND UBIQUITINATION
Ubiquitination is another significant post-translational modification linked with DDR. Similar to phosphorylated proteins, ubiquitinated proteins begin to accumulate at the sites of DNA damage several minutes after DSB damage (160,161), indicating that ubiquitination can also serve as a secondary wave of signaling in response to DNA damage. Both H2A and H2AX have been identified to be ubiquitinated primarily at the K13/15 and K119/120 residues on their tails (162–164), and ubiquitination at K13/15 is induced by DNA damage. Interestingly, recent evidence shows that lysine residues can also be PARylated by PARP1 (5,12). Thus, PARylation may also compete with ubiquitination to modify histones at DNA lesions. Although it is unclear if H2A and H2AX are PARylated at K13/15 in response to DNA damage, PARylation on any adjacent residue could be sufficient to suppress DNA damage-induced ubiquitination. However, further studies are needed to elucidate if PARylation and ubiquitination act in concert during sequential steps of DDR.
In addition, PARylation can prime the ubiquitination at DNA lesions. Two ubiquitin E3 ligases, CHFR and RNF146, are recruited to DNA lesions by PARylation (76,161,165). The C-terminus of CHFR harbors a PBZ motif that recognizes PAR chains (76,161). Once DNA damage occurs, CHFR is quickly recruited to the sites of DNA damage via the PBZ motif and initiates ubiquitination (Figure 5A). However, dePARylation mediates quick release of CHFR from DNA lesions. Thus, the role of CHFR-dependent ubiquitination may be regulated during DDR. Moreover, our studies have shown that CHFR-dependent ubiquitination may have overlapping function with RNF8-dependent ubiquitination during DDR (166). In addition to CHFR, RNF146 contains a WWE domain that also interacts with PAR (165). Although RNF146 is a cytoplasmic protein, it relocates to the nucleus and is enriched at DNA lesions in response to genotoxic stress. The enrichment of RNF146 at DNA lesions is mediated by DNA damage-induced PARylation. However, the substrates of RNF146 at the sites of DNA damage remain elusive. Nevertheless, these studies on CHFR and RNF146 further support a model wherein PARylation, the first wave of DNA damage signaling, activates ubiquitination, the later wave of signaling at DNA lesions.
CONCLUDING REMARKS
PARylation is a dynamically regulated post-translational modification, which plays a versatile role in the early steps of DDR. It creates the first wave of DNA damage response through recruitment of numerous DNA damage response factors to the regions near DNA lesions. PARylation also regulates other late post-translational modifications, such as phosphorylation and ubiquitination, at the sites of DNA damage. The additive effect of these post-translational modifications leads to stable retention of the DNA repair machinery. PARylation also affects chromatin remodeling directly through the negative charge in each ADP-ribose unit. The coupling of PARylation and dePARylation can also provide a ready and abundant source of energy for DDR.
However, there are still a lot of interesting questions about the synthesis, recognition and degradation of PAR, which need to be addressed to fully understand the function of PARylation in DNA damage response. The biological function of branched PAR chains remains unclear. With the advancements in proteomics, more site-specific PARylation events can be studied, shedding light on their potential roles in the subsequent steps of DNA repair. Additionally, the roles of the dePARylation enzymes other than PARG are yet to be deciphered. It is possible that different dePARylation enzymes could be directed at specific removal of different PARylation targets or activated in other physiological conditions. Besides DDR, PARylation also participates in many other biological processes, which have been comprehensively summarized in other reviews (167–169). Studies of the molecular mechanism of other PARylation-dependent cellular functions may also provide novel clues to understanding the first wave of DNA damage response.
Another interesting feature of PARPs is that only four out of 16 PARP family enzymes catalyze PARylation, whereas others catalyze MARylation. To date, several lines of evidence suggest that similar to PARylation, MARylation plays very important roles in DDR (89,170). In particular, PARP3 is recruited to the sites of DNA damage and facilitates chromatin remodeling (85,87). PARP3-dependent MARylation is important for both DSB and SSB repair (85,91). Similarly, PARP10, another mono ADP-ribosyltransferase is also involved in DDR (171). In particular, PARP10 has tandem ubiquitin-interacting motifs, which may recognize ubiquitin signals at the sites of DNA damage. Future characterization of these mono ADP-ribosyltransferases may reveal the versatile role of ADP-ribosylation in the DNA damage response.
ACKNOWLEDGEMENTS
We thank suggestions and proofreading of the manuscript by Drs Jeremy Stark, Shan Zha and Nancy Linford.
FUNDING
National Institutes of Health [CA132755, CA130899, CA187209 to X.Y.]. X.Y. is a recipient of Era of Hope Scholar Award from the Department of Defense and Research Scholar Award from Leukemia and Lymphoma Society. Funding for open access charge: National Institutes of Health [CA132755, CA130899, CA187209].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. [DOI] [PubMed] [Google Scholar]
- 2. Lindahl T., Barnes D.E.. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000; 65:127–133. [DOI] [PubMed] [Google Scholar]
- 3. Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M.. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006; 160:1–40. [DOI] [PubMed] [Google Scholar]
- 4. Zhou B.B., Elledge S.J.. The DNA damage response: putting checkpoints in perspective. Nature. 2000; 408:433–439. [DOI] [PubMed] [Google Scholar]
- 5. Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O.. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009; 37:3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daniels C.M., Ong S.E., Leung A.K.. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J. Proteome Res. 2014; 13:3510–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geipel U., Just I., Schering B., Haas D., Aktories K.. ADP-ribosylation of actin causes increase in the rate of ATP exchange and inhibition of ATP hydrolysis. Eur. J. Biochem. 1989; 179:229–232. [DOI] [PubMed] [Google Scholar]
- 8. Hoshino S., Kikkawa S., Takahashi K., Itoh H., Kaziro Y., Kawasaki H., Suzuki K., Katada T., Ui M.. Identification of sites for alkylation by N-ethylmaleimide and pertussis toxin-catalyzed ADP-ribosylation on GTP-binding proteins. FEBS Lett. 1990; 276:227–231. [DOI] [PubMed] [Google Scholar]
- 9. Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L.. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013; 52:272–285. [DOI] [PubMed] [Google Scholar]
- 10. Just I., Wollenberg P., Moss J., Aktories K.. Cysteine-specific ADP-ribosylation of actin. Eur. J. Biochem. 1994; 221:1047–1054. [DOI] [PubMed] [Google Scholar]
- 11. Manning D.R., Fraser B.A., Kahn R.A., Gilman A.G.. ADP-ribosylation of transducin by islet-activation protein. Identification of asparagine as the site of ADP-ribosylation. J. Biol. Chem. 1984; 259:749–756. [PubMed] [Google Scholar]
- 12. Messner S., Altmeyer M., Zhao H., Pozivil A., Roschitzki B., Gehrig P., Rutishauser D., Huang D., Caflisch A., Hottiger M.O.. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010; 38:6350–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moss J., Stanley S.J.. Amino acid-specific ADP-ribosylation. Identification of an arginine-dependent ADP-ribosyltransferase in rat liver. J. Biol. Chem. 1981; 256:7830–7833. [PubMed] [Google Scholar]
- 14. Ogata N., Ueda K., Kagamiyama H., Hayaishi O.. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 1980; 255:7616–7620. [PubMed] [Google Scholar]
- 15. Sekine A., Fujiwara M., Narumiya S.. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 1989; 264:8602–8605. [PubMed] [Google Scholar]
- 16. Zhang Y., Wang J., Ding M., Yu Y.. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013; 10:981–984. [DOI] [PubMed] [Google Scholar]
- 17. Alvarez-Gonzalez R., Jacobson M.K.. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987; 26:3218–3224. [DOI] [PubMed] [Google Scholar]
- 18. Miwa M., Saikawa N., Yamaizumi Z., Nishimura S., Sugimura T.. Structure of poly(adenosine diphosphate ribose): identification of 2′-[1″-ribosyl-2″-(or 3″'-)(1″′-ribosyl)]adenosine-5′,5″,5″′-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P.. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ame J.C., Spenlehauer C., de Murcia G.. The PARP superfamily. Bioessays. 2004; 26:882–893. [DOI] [PubMed] [Google Scholar]
- 21. Ji Y., Tulin A.V.. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 2010; 20:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shieh W.M., Ame J.C., Wilson M.V., Wang Z.Q., Koh D.W., Jacobson M.K., Jacobson E.L.. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998; 273:30069–30072. [DOI] [PubMed] [Google Scholar]
- 23. Ludwig A., Behnke B., Holtlund J., Hilz H.. Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates. An analysis of the in vivo status in mammalian species and in lower eukaryotes. J. Biol. Chem. 1988; 263:6993–6999. [PubMed] [Google Scholar]
- 24. Yamanaka H., Penning C.A., Willis E.H., Wasson D.B., Carson D.A.. Characterization of human poly(ADP-ribose) polymerase with autoantibodies. J. Biol. Chem. 1988; 263:3879–3883. [PubMed] [Google Scholar]
- 25. Lautier D., Lagueux J., Thibodeau J., Menard L., Poirier G.G.. Molecular and biochemical features of poly (ADP-ribose) metabolism. Mol. Cell. Biochem. 1993; 122:171–193. [DOI] [PubMed] [Google Scholar]
- 26. Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J., Poirier G.G.. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008; 283:1197–1208. [DOI] [PubMed] [Google Scholar]
- 27. Ali A.A., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W.. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012; 19:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science (New York, N.Y.). 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rogakou E.P., Boon C., Redon C., Bonner W.M.. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 31. Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A.. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003; 5:675–679. [DOI] [PubMed] [Google Scholar]
- 32. Yuan J., Chen J.. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 2010; 285:1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J. et al. Genomic instability in mice lacking histone H2AX. Science (New York, N.Y.). 2002; 296:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J.. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001; 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- 35. Girard P.M., Riballo E., Begg A.C., Waugh A., Jeggo P.A.. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene. 2002; 21:4191–4199. [DOI] [PubMed] [Google Scholar]
- 36. Stiff T., O’Driscoll M., Rief N., Iwabuchi K., Lobrich M., Jeggo P.A.. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004; 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- 37. Stucki M., Jackson S.P.. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.). 2006; 5:534–543. [DOI] [PubMed] [Google Scholar]
- 38. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y.. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003; 22:5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Jager M., van Noort J., van Gent D.C., Dekker C., Kanaar R., Wyman C.. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001; 8:1129–1135. [DOI] [PubMed] [Google Scholar]
- 40. Moreno-Herrero F., de Jager M., Dekker N.H., Kanaar R., Wyman C., Dekker C.. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005; 437:440–443. [DOI] [PubMed] [Google Scholar]
- 41. Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F. et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014; 53:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A.. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001; 105:473–485. [DOI] [PubMed] [Google Scholar]
- 43. Paull T.T., Gellert M.. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999; 13:1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trujillo K.M., Sung P.. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 2001; 276:35458–35464. [DOI] [PubMed] [Google Scholar]
- 45. Petrini J.H., Stracker T.H.. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003; 13:458–462. [DOI] [PubMed] [Google Scholar]
- 46. Mirzoeva O.K., Petrini J.H.. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 2001; 21:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mirzoeva O.K., Petrini J.H.. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res.: MCR. 2003; 1:207–218. [PubMed] [Google Scholar]
- 48. Kong X., Mohanty S.K., Stephens J., Heale J.T., Gomez-Godinez V., Shi L.Z., Kim J.S., Yokomori K., Berns M.W.. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009; 37:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haince J.F., Kozlov S., Dawson V.L., Dawson T.M., Hendzel M.J., Lavin M.F., Poirier G.G.. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007; 282:16441–16453. [DOI] [PubMed] [Google Scholar]
- 50. Li M., Lu L.Y., Yang C.Y., Wang S., Yu X.. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013; 27:1752–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Jager M., Wyman C., van Gent D.C., Kanaar R.. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002; 30:4425–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koike M., Yutoku Y., Koike A.. Nuclear localization of mouse Ku70 in interphase cells and focus formation of mouse Ku70 at DNA damage sites immediately after irradiation. J. Vet. Med. Sci./Jap. Soc. Vet. Sci. 2015; 77:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M. et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:18597–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grundy G.J., Rulten S.L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., Oliver A., Caldecott K.W.. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013; 32:112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mimori T., Hardin J.A., Steitz J.A.. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 1986; 261:2274–2278. [PubMed] [Google Scholar]
- 56. Hopfner K.P., Putnam C.D., Tainer J.A.. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 2002; 12:115–122. [DOI] [PubMed] [Google Scholar]
- 57. West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 2003; 4:435–445. [DOI] [PubMed] [Google Scholar]
- 58. Lieber M.R., Ma Y., Pannicke U., Schwarz K.. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003; 4:712–720. [DOI] [PubMed] [Google Scholar]
- 59. Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J.. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007; 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Costantini S., Woodbine L., Andreoli L., Jeggo P.A., Vindigni A.. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst.). 2007; 6:712–722. [DOI] [PubMed] [Google Scholar]
- 61. Nick McElhinny S.A., Snowden C.M., McCarville J., Ramsden D.A.. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000; 20:2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yano K., Morotomi-Yano K., Wang S.Y., Uematsu N., Lee K.J., Asaithamby A., Weterings E., Chen D.J.. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008; 9:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang M., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G.. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006; 34:6170–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mansour W.Y., Rhein T., Dahm-Daphi J.. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010; 38:6065–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T.. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009; 28:2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li M., Yu X.. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013; 23:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang F., Shi J., Bian C., Yu X.. Poly(ADP-ribose) mediates the BRCA2-dependent early DNA damage response. Cell Rep. 2015; 13:678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F.. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014; 329:18–25. [DOI] [PubMed] [Google Scholar]
- 69. de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Masutani M., Suzuki H., Kamada N., Watanabe M., Ueda O., Nozaki T., Jishage K., Watanabe T., Sugimoto T., Nakagama H. et al. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2301–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C.. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996; 382:551–555. [DOI] [PubMed] [Google Scholar]
- 72. Wang Z.Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., Wagner E.F.. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995; 9:509–520. [DOI] [PubMed] [Google Scholar]
- 73. Zhu C., Bogue M.A., Lim D.S., Hasty P., Roth D.B.. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996; 86:379–389. [DOI] [PubMed] [Google Scholar]
- 74. Henrie M.S., Kurimasa A., Burma S., Menissier-de Murcia J., de Murcia G., Li G.C., Chen D.J.. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair (Amst.). 2003; 2:151–158. [DOI] [PubMed] [Google Scholar]
- 75. Tong W.M., Cortes U., Hande M.P., Ohgaki H., Cavalli L.R., Lansdorp P.M., Haddad B.R., Wang Z.Q.. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 2002; 62:6990–6996. [PubMed] [Google Scholar]
- 76. Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C.. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008; 451:81–85. [DOI] [PubMed] [Google Scholar]
- 77. Couto C.A., Wang H.Y., Green J.C., Kiely R., Siddaway R., Borer C., Pears C.J., Lakin N.D.. PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J. Cell Biol. 2011; 194:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H., Schwede F., Yu Y., Kraus W.L.. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science (New York, N.Y.). 2016; 353:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ame J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Hoger T., Menissier-de Murcia J., de Murcia G.. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999; 274:17860–17868. [DOI] [PubMed] [Google Scholar]
- 80. Schreiber V., Ame J.C., Dolle P., Schultz I., Rinaldi B., Fraulob V., Menissier-de Murcia J., de Murcia G.. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002; 277:23028–23036. [DOI] [PubMed] [Google Scholar]
- 81. Huber A., Bai P., de Murcia J.M., de Murcia G.. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst.). 2004; 3:1103–1108. [DOI] [PubMed] [Google Scholar]
- 82. Langelier M.F., Riccio A.A., Pascal J.M.. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014; 42:7762–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Menissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Ame J.C., Dierich A., LeMeur M. et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003; 22:2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mortusewicz O., Ame J.C., Schreiber V., Leonhardt H.. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007; 35:7665–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rulten S.L., Fisher A.E., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W.. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011; 41:33–45. [DOI] [PubMed] [Google Scholar]
- 86. Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F. et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fenton A.L., Shirodkar P., Macrae C.J., Meng L., Koch C.A.. The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 2013; 41:4080–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T., Ahel I., Chang P.. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014; 5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Grundy G.J., Polo L.M., Zeng Z., Rulten S.L., Hoch N.C., Paomephan P., Xu Y., Sweet S.M., Thorne A.W., Oliver A.W. et al. PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B(Glu2). Nat. Commun. 2016; 7:12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Loseva O., Jemth A.S., Bryant H.E., Schuler H., Lehtio L., Karlberg T., Helleday T.. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010; 285:8054–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rouleau M., McDonald D., Gagne P., Ouellet M.E., Droit A., Hunter J.M., Dutertre S., Prigent C., Hendzel M.J., Poirier G.G.. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 2007; 100:385–401. [DOI] [PubMed] [Google Scholar]
- 92. Jiang H., Kim J.H., Frizzell K.M., Kraus W.L., Lin H.. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J. Am. Chem. Soc. 2010; 132:9363–9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Carter-O’Connell I., Jin H., Morgan R.K., David L.L., Cohen M.S.. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. J. Am. Chem. Soc. 2014; 136:5201–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ogata N., Ueda K., Kawaichi M., Hayaishi O.. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 1981; 256:4135–4137. [PubMed] [Google Scholar]
- 95. Ferro A.M., Olivera B.M.. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J. Biol. Chem. 1982; 257:7808–7813. [PubMed] [Google Scholar]
- 96. Zahradka P., Ebisuzaki K.. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 1982; 127:579–585. [PubMed] [Google Scholar]
- 97. Rouleau M., Aubin R.A., Poirier G.G.. Poly(ADP-ribosyl)ated chromatin domains: access granted. J. Cell Sci. 2004; 117:815–825. [DOI] [PubMed] [Google Scholar]
- 98. Krishnakumar R., Gamble M.J., Frizzell K.M., Berrocal J.G., Kininis M., Kraus W.L.. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science (New York, N.Y.). 2008; 319:819–821. [DOI] [PubMed] [Google Scholar]
- 99. Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science (New York, N.Y.). 2009; 325:1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A.. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9129–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Luo X., Kraus W.L.. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012; 26:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G.. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998; 18:3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Okano S., Lan L., Caldecott K.W., Mori T., Yasui A.. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003; 23:3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang F., Chen Y., Li M., Yu X.. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7278–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang F., Shi J., Chen S.H., Bian C., Yu X.. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li N., Chen J.. ADP-ribosylation: activation, recognition, and removal. Mol. Cells. 2014; 37:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu C., Yu X.. ADP-ribosyltransferases and poly ADP-ribosylation. Curr. Protein Pept. Sci. 2015; 16:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Teloni F., Altmeyer M.. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016; 44:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Alvarez-Gonzalez R., Althaus F.R.. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mut. Res. 1989; 218:67–74. [DOI] [PubMed] [Google Scholar]
- 111. Bernardi R., Rossi L., Poirier G.G., Scovassi A.I.. Analysis of poly(ADP-ribose) glycohydrolase activity in nuclear extracts from mammalian cells. Biochim. Biophys. Acta. 1997; 1338:60–68. [DOI] [PubMed] [Google Scholar]
- 112. Yamada K., Hara N., Shibata T., Osago H., Tsuchiya M.. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal. Biochem. 2006; 352:282–285. [DOI] [PubMed] [Google Scholar]
- 113. Yang H., Yang T., Baur J.A., Perez E., Matsui T., Carmona J.J., Lamming D.W., Souza-Pinto N.C., Bohr V.A., Rosenzweig A. et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007; 130:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Belenky P., Racette F.G., Bogan K.L., McClure J.M., Smith J.S., Brenner C.. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell. 2007; 129:473–484. [DOI] [PubMed] [Google Scholar]
- 115. Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J., Poirier G.G., Dawson T.M., Dawson V.L.. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science (New York, N.Y.). 2002; 297:259–263. [DOI] [PubMed] [Google Scholar]
- 116. Sims J.L., Berger S.J., Berger N.A.. Poly(ADP-ribose) Polymerase inhibitors preserve nicotinamide adenine dinucleotide and adenosine 5′-triphosphate pools in DNA-damaged cells: mechanism of stimulation of unscheduled DNA synthesis. Biochemistry. 1983; 22:5188–5194. [DOI] [PubMed] [Google Scholar]
- 117. Alano C.C., Garnier P., Ying W., Higashi Y., Kauppinen T.M., Swanson R.A.. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J. Neurosci. 2010; 30:2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ying W., Alano C.C., Garnier P., Swanson R.A.. NAD+ as a metabolic link between DNA damage and cell death. J. Neurosci. Res. 2005; 79:216–223. [DOI] [PubMed] [Google Scholar]
- 119. Zhang F., Xie R., Munoz F.M., Lau S.S., Monks T.J.. PARP-1 hyperactivation and reciprocal elevations in intracellular Ca2+ during ROS-induced nonapoptotic cell death. Toxicol. Sci. 2014; 140:118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Coppola S., Nosseri C., Maresca V., Ghibelli L.. Different basal NAD levels determine opposite effects of poly(ADP-ribosyl)polymerase inhibitors on H2O2-induced apoptosis. Exp. Cell Res. 1995; 221:462–469. [DOI] [PubMed] [Google Scholar]
- 121. Bonicalzi M.E., Haince J.F., Droit A., Poirier G.G.. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when. Cell. Mol. Life Sci. 2005; 62:739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. D’Amours D., Desnoyers S., D'Silva I., Poirier G.G.. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999; 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 123. Gagne J.P., Hendzel M.J., Droit A., Poirier G.G.. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr. Opin. Cell Biol. 2006; 18:145–151. [DOI] [PubMed] [Google Scholar]
- 124. Rouleau M., Patel A., Hendzel M.J., Kaufmann S.H., Poirier G.G.. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010; 10:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sharifi R., Morra R., Appel C.D., Tallis M., Chioza B., Jankevicius G., Simpson M.A., Matic I., Ozkan E., Golia B. et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013; 32:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Oka S., Kato J., Moss J.. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006; 281:705–713. [DOI] [PubMed] [Google Scholar]
- 127. Lin S., Gasmi L., Xie Y., Ying K., Gu S., Wang Z., Jin H., Chao Y., Wu C., Zhou Z. et al. Cloning, expression and characterisation of a human Nudix hydrolase specific for adenosine 5′-diphosphoribose (ADP-ribose). Biochim. Biophys. Acta. 2002; 1594:127–135. [DOI] [PubMed] [Google Scholar]
- 128. Palazzo L., Thomas B., Jemth A.S., Colby T., Leidecker O., Feijs K.L., Zaja R., Loseva O., Puigvert J.C., Matic I. et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015; 468:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Palazzo L., Daniels C.M., Nettleship J.E., Rahman N., McPherson R.L., Ong S.E., Kato K., Nureki O., Leung A.K., Ahel I.. ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 2016; 283:3371–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rosenthal F., Feijs K.L., Frugier E., Bonalli M., Forst A.H., Imhof R., Winkler H.C., Fischer D., Caflisch A., Hassa P.O. et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013; 20:502–507. [DOI] [PubMed] [Google Scholar]
- 131. Takada T., Iida K., Moss J.. Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J. Biol. Chem. 1993; 268:17837–17843. [PubMed] [Google Scholar]
- 132. Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I.. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011; 477:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mortusewicz O., Fouquerel E., Ame J.C., Leonhardt H., Schreiber V.. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011; 39:5045–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cortes U., Tong W.M., Coyle D.L., Meyer-Ficca M.L., Meyer R.G., Petrilli V., Herceg Z., Jacobson E.L., Jacobson M.K., Wang Z.Q.. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 2004; 24:7163–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Min W., Cortes U., Herceg Z., Tong W.M., Wang Z.Q.. Deletion of the nuclear isoform of poly(ADP-ribose) glycohydrolase (PARG) reveals its function in DNA repair, genomic stability and tumorigenesis. Carcinogenesis. 2010; 31:2058–2065. [DOI] [PubMed] [Google Scholar]
- 136. Koh D.W., Lawler A.M., Poitras M.F., Sasaki M., Wattler S., Nehls M.C., Stoger T., Poirier G.G., Dawson V.L., Dawson T.M.. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17699–17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Fisher A.E., Hochegger H., Takeda S., Caldecott K.W.. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007; 27:5597–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Shirai H., Poetsch A.R., Gunji A., Maeda D., Fujimori H., Fujihara H., Yoshida T., Ogino H., Masutani M.. PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis. 2013; 4:e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., Ladurner A.G.. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013; 20:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Blenn C., Wyrsch P., Bader J., Bollhalder M., Althaus F.R.. Poly(ADP-ribose)glycohydrolase is an upstream regulator of Ca2+ fluxes in oxidative cell death. Cell. Mol. Life Sci. 2011; 68:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wright R.H., Lioutas A., Le Dily F., Soronellas D., Pohl A., Bonet J., Nacht A.S., Samino S., Font-Mateu J., Vicent G.P. et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science (New York, N.Y.). 2016; 352:1221–1225. [DOI] [PubMed] [Google Scholar]
- 142. Hassa P.O., Haenni S.S., Elser M., Hottiger M.O.. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going. Microbiol. Mol. Biol. Rev.: MMBR. 2006; 70:789–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Brown J.S., Jackson S.P.. Ubiquitylation, neddylation and the DNA damage response. Open Biol. 2015; 5:150018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. van Attikum H., Gasser S.M.. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009; 19:207–217. [DOI] [PubMed] [Google Scholar]
- 145. Falck J., Coates J., Jackson S.P.. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434:605–611. [DOI] [PubMed] [Google Scholar]
- 146. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science (New York, N.Y.). 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 147. Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003; 3:155–168. [DOI] [PubMed] [Google Scholar]
- 148. Shiloh Y., Ziv Y.. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013; 14:197–210. [PubMed] [Google Scholar]
- 149. Wu L.C., Wang Z.W., Tsan J.T., Spillman M.A., Phung A., Xu X.L., Yang M.C., Hwang L.Y., Bowcock A.M., Baer R.. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996; 14:430–440. [DOI] [PubMed] [Google Scholar]
- 150. Manke I.A., Lowery D.M., Nguyen A., Yaffe M.B.. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science (New York, N.Y.). 2003; 302:636–639. [DOI] [PubMed] [Google Scholar]
- 151. Yu X., Chini C.C., He M., Mer G., Chen J.. The BRCT domain is a phospho-protein binding domain. Science (New York, N.Y.). 2003; 302:639–642. [DOI] [PubMed] [Google Scholar]
- 152. Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J.. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science (New York, N.Y.). 2007; 316:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kim H., Chen J., Yu X.. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science (New York, N.Y.). 2007; 316:1202–1205. [DOI] [PubMed] [Google Scholar]
- 154. Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A.. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science (New York, N.Y.). 2007; 316:1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F., Jetten A.M.. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007; 67:6647–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D. et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996; 86:159–171. [DOI] [PubMed] [Google Scholar]
- 157. Menisser-de Murcia J., Mark M., Wendling O., Wynshaw-Boris A., de Murcia G.. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol. 2001; 21:1828–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Orsburn B., Escudero B., Prakash M., Gesheva S., Liu G., Huso D.L., Franco S.. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Mol. Cell. Biol. 2010; 30:2341–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang X., Liu L., Montagna C., Ried T., Deng C.X.. Haploinsufficiency of Parp1 accelerates Brca1-associated centrosome amplification, telomere shortening, genetic instability, apoptosis, and embryonic lethality. Cell Death Differ. 2007; 14:924–931. [DOI] [PubMed] [Google Scholar]
- 160. Ma T., Keller J.A., Yu X.. RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta Biochim. Biophys. Sin. (Shanghai). 2011; 43:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Liu C., Wu J., Paudyal S.C., You Z., Yu X.. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013; 41:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K.. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 163. Goldknopf I.L., Busch H.. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hunt L.T., Dayhoff M.O.. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem. Biophys. Res. Commun. 1977; 74:650–655. [DOI] [PubMed] [Google Scholar]
- 165. Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G. et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wu J., Chen Y., Lu L.Y., Wu Y., Paulsen M.T., Ljungman M., Ferguson D.O., Yu X.. Chfr and RNF8 synergistically regulate ATM activation. Nat. Struct. Mol. Biol. 2011; 18:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Hottiger M.O. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 2015; 84:227–263. [DOI] [PubMed] [Google Scholar]
- 168. Abd Elmageed Z.Y., Naura A.S., Errami Y., Zerfaoui M.. The poly(ADP-ribose) polymerases (PARPs): new roles in intracellular transport. Cell. Signal. 2012; 24:1–8. [DOI] [PubMed] [Google Scholar]
- 169. Virag L., Robaszkiewicz A., Rodriguez-Vargas J.M., Oliver F.J.. Poly(ADP-ribose) signaling in cell death. Mol. Aspects Med. 2013; 34:1153–1167. [DOI] [PubMed] [Google Scholar]
- 170. Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V.. SIRT6 promotes DNA repair under stress by activating PARP1. Science (New York, N.Y.). 2011; 332:1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Nicolae C.M., Aho E.R., Vlahos A.H., Choe K.N., De S., Karras G.I., Moldovan G.L.. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J. Biol. Chem. 2014; 289:13627–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Eustermann S., Brockmann C., Mehrotra P.V., Yang J.C., Loakes D., West S.C., Ahel I., Neuhaus D.. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat. Struct. Mol. Biol. 2010; 17:241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Han W., Li X., Fu X.. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mut. Res. 2011; 727:86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001; 26:273–275. [DOI] [PubMed] [Google Scholar]
- 175. Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W.. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012; 26:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Li M., Bian C., Yu X.. Poly(ADP-ribosyl)ation is recognized by ECT2 during mitosis. Cell Cycle. 2014; 13:2944–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y.. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012; 40:10287–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Min W., Bruhn C., Grigaravicius P., Zhou Z.W., Li F., Kruger A., Siddeek B., Greulich K.O., Popp O., Meisezahl C. et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013; 4:2993. [DOI] [PubMed] [Google Scholar]
- 179. Braun S.A., Panzeter P.L., Collinge M.A., Althaus F.R.. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur. J. Biochem. 1994; 220:369–375. [DOI] [PubMed] [Google Scholar]
- 180. Miwa M., Tanaka M., Matsushima T., Sugimura T.. Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose). J. Biol. Chem. 1974; 249:3475–3482. [PubMed] [Google Scholar]
- 181. Peterson F.C., Chen D., Lytle B.L., Rossi M.N., Ahel I., Denu J.M., Volkman B.F.. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 2011; 286:35955–35965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ono T., Kasamatsu A., Oka S., Moss J.. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:16687–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Niere M., Kernstock S., Koch-Nolte F., Ziegler M.. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell. Biol. 2008; 28:814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Rafty L.A., Schmidt M.T., Perraud A.L., Scharenberg A.M., Denu J.M.. Analysis of O-acetyl-ADP-ribose as a target for Nudix ADP-ribose hydrolases. J. Biol. Chem. 2002; 277:47114–47122. [DOI] [PubMed] [Google Scholar]
- 185. Ghosh T., Peterson B., Tomasevic N., Peculis B.A.. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell. 2004; 13:817–828. [DOI] [PubMed] [Google Scholar]