Abstract
Chromatin recruitment of effector proteins involved in gene regulation depends on multivalent interaction with histone post-translational modifications (PTMs) and structural features of the chromatin fiber. Due to the complex interactions involved, it is currently not understood how effectors dynamically sample the chromatin landscape. Here, we dissect the dynamic chromatin interactions of a family of multivalent effectors, heterochromatin protein 1 (HP1) proteins, using single-molecule fluorescence imaging and computational modeling. We show that the three human HP1 isoforms are recruited and retained on chromatin by a dynamic exchange between histone PTM and DNA bound states. These interactions depend on local chromatin structure, the HP1 isoforms as well as on PTMs on HP1 itself. Of the HP1 isoforms, HP1α exhibits the longest residence times and fastest binding rates due to DNA interactions in addition to PTM binding. HP1α phosphorylation further increases chromatin retention through strengthening of multivalency while reducing DNA binding. As DNA binding in combination with specific PTM recognition is found in many chromatin effectors, we propose a general dynamic capture mechanism for effector recruitment. Multiple weak protein and DNA interactions result in a multivalent interaction network that targets effectors to a specific chromatin modification state, where their activity is required.
INTRODUCTION
Chromatin, the nucleoprotein complex organizing eukaryotic DNA, is composed of arrays of nucleosomes. Each nucleosome contains two of each histone H2A, H2B, H3 and H4 that form a protein core around which 147 base pairs (bp) of DNA are wrapped. Connected by 30–70 bp of linker DNA, arrays of nucleosomes form higher-order structures, such as compact chromatin fibers (1). Chromatin exists in different functional states characterized by its structure, the presence of protein factors as well as patterns of histone post-translational modifications (PTMs) and DNA modifications (2–4). Dynamic multivalent interactions between the chromatin fiber and effector proteins, which carry PTM-specific reader domains, play a key role in regulating chromatin function in biological processes such as gene expression (5), whereas misregulation of these interactions may result in cancer (6). The physicochemical principles of these dynamic and specific interactions are not well understood.
HP1 family proteins function as fundamental components of heterochromatin involved in gene regulation including suppression of latent viruses and transposable elements in the genome (7). Heterochromatin denotes compact and transcriptionally repressed chromatin domains characterized by H3 di- and tri-methylated at lysine 9 (H3K9me2/3), the absence of histone acetylation and the presence of linker histone H1 (8). H3K9me2/3 serve as binding sites for HP1, which accumulates at heterochromatin loci and induces gene silencing (9). In mammals, HP1 proteins include three isoforms: HP1α, HP1β and HP1γ (9) with different sub-nuclear localization and function (10–12): Both HP1α and HP1β associate with heterochromatin, while HP1γ has also been shown to localize in euchromatin. HP1 proteins are similar in structural architecture: they contain a chromodomain (CD), which specifically interacts with H3K9me2/3 with affinities around 1–10 μM (13), a flexible hinge region (HR) that interacts with nucleic acids as well as a dimerization domain (chromoshadow domain, CSD) (14). Dimeric HP1 can bind to chromatin in a multivalent fashion by engaging two H3K9me3 sites, thereby cross-bridging nucleosomes (15,16), and reducing access for the transcription machinery (17,18). In spite of their structural role, HP1 proteins are in rapid exchange in living cells (19–21), forming locally dynamic compartments that are nevertheless stable over time. Such local dynamic behavior is indeed important, enabling the cell to rapidly react when local access to chromatin is required (22). Intriguingly, this organization of spatial chromatin states has been found to involve phase-separation behavior, dependent on multivalent interactions (23,24). Due to the complexity of multivalent effector–chromatin interactions, it is however not well understood how HP1 proteins are dynamically recruited and retained at their target sites and how they contribute to establishing the heterochromatin state. It is thus of importance to determine how the local arrangement (chromatin conformation) and nature of binding sites on the chromatin fibers (histone PTMs or other structural features) control dynamic HP1 association.
To unravel the complex interaction modes between multivalent effectors and modified chromatin fibers, an integration of detailed experimental and theoretical methods is required. We have recently developed a single-molecule total internal reflection fluorescence (smTIRF) imaging approach to directly observe the chromatin interaction kinetics of individual HP1α molecules (25). A major population of HP1α molecules exhibited rapid binding and dissociation dynamics in the millisecond timescale, whereas a smaller population remained bound for seconds due to multivalent interactions. We showed that multivalency prolonged HP1α chromatin interactions and increased the binding rate. Finally, we observed decreased HP1α residence times when H3K9 methylation was reduced across the chromatin fiber. This finding revealed that individual binding events consist of multiple rounds of dissociation and rapid local rebinding (25). Based on these observations, we hypothesize that HP1 proteins not only bind to H3K9me3, but that they engage in a dynamic interaction network on the chromatin fiber, probing a structurally and chemically complex interaction surface. To detect these dynamic interactions has however presented a challenge as the temporal and spatial resolution of single-molecule experiments is limited. Instead, kinetic modeling is required to reveal the sequence of elementary reaction steps from their effects on the apparent effector–chromatin interaction kinetics.
Here, we thus combined direct detection of HP1 dynamics on the single-molecule scale with a large-scale kinetic model of the HP1-chromatin interaction network. This enabled us to extract pathways of chromatin binding for the individual isoforms HP1α, β and γ. Using synthetic chromatin fibers we systematically altered chromatin structure and found that HP1α residence times are dependent on chromatin conformation, with a preference for compact fibers. Comparing binding dynamics of all three HP1 isoforms revealed that charge-based DNA interactions critically increase the binding rate and residence time. Indeed, transient DNA interactions are involved in shuttling HP1 proteins between individual longer lived mono- and multivalent PTM-bound states. Together, these findings demonstrate an important and general role for multivalent PTM- and DNA-dependent interactions for the recruitment and retention of HP1 proteins on modified chromatin.
MATERIALS AND METHODS
Production of DNA sequences
About 12-mer repeats of the 601 nucleosome positioning sequence (26), separated by either 30 bp linker (a177) or 50 bp linker (a197), flanked by EcoRV sites and a unique BsaI restriction site at the 3′ end were cloned into plasmids and purified from bacterial culture by size exclusion chromatography followed by a phenol/chloroform extraction. The plasmids were then subjected to a restriction digest and the 12 × 601 sequences were purified by polyethylene glycol (PEG) precipitation. For immobilization and visualization, an oligonucleotide (5′-ph-CAGCTAGTCTGCT-(amine-linker)-CAGATATCGTCG-3′-Biotin) was labeled using Atto647N-NHS ester according to the manufacturer's (AttoTec) protocol. The labeled oligonucleotide was then annealed to its complementary DNA strand (5′-CGACGATATCTGAGCAGACTA-3′) and ligated to the 12 × 601 array DNA with T4 DNA ligase for 1 h at room temperature. The excess dsDNA was removed by purification with QIAquick spin columns (Qiagen), and the final 12 × 601 DNA was finally purified by ethanol precipitation (Supplementary Figure S1).
Synthesis of H3K9me3
H3K9me3 was produced as described in ref. (25). Briefly, the modified histone peptide H3(1–14)K9me3-NHNH2 (carrying a C-terminal hydrazide) was synthesized by solid phase peptide synthesis (SPPS). The truncated protein H3(Δ1–14)A15C was recombinantly expressed as N-terminal fusion to small ubiquitin like modifier (SUMO), the N-terminal SUMO was cleaved by SUMO protease and the protein purified by RP-HPLC. For ligation, in a typical reaction 3 μmol H3(1–14)K9me3-NHNH2 was dissolved in ligation buffer (200 mM phosphate pH 3, 6 M GdmCl) at −10°C. NaNO2 was added dropwise to a final concentration of 15 mM and incubated at −20°C for 20 min. H3(Δ1–14)A15C was dissolved in mercaptophenyl acetic acid (MPAA) ligation buffer (200 mM phosphate pH 8, 6 M GdmCl, 300 mM MPAA) and added to the peptide, followed by adjustment of the pH to 7.5. The ligation was left to proceed until completion (as observed by RP-HPLC). The product (H3K9me3A15C) was purified by semi-preparative RP-HPLC. For desulfurization, H3K9me3A15C was dissolved in tris(2-carboxyethyl)phosphine (TCEP) desulfurization buffer (200 M phosphate pH 6.5, 6 M GdmCl, 250 mM TCEP). Glutathione (40 mM) and a radical initiator, VA-044 (20 mM), were added, followed by a readjustment of the pH to 6.5. The reaction mixture was incubated at 42°C until completion (verified by RP-HPLC and ESI-MS). The final product, H3K9me3, was purified by semi-preparative RP-HPLC (Supplementary Figure S2, calculated mass: 15251 Da, observed mass: 15248 Da).
Synthesis of H4 acetyl lysine analog
Preparation of H4KS16ac was performed following ref. (27). H4 carrying the point mutation K16C was expressed and purified as described (25). (Supplementary Figure S2, Calculated mass = 11211 Da, observed mass = 11211 Da). For the site-specific installation of the acetyl-lysine analog, H4K16C was dissolved in 0.2 M acetate buffer, pH 4 to a final concentration of 1 mM, followed by the addition of 50 mM N-vinylacetamide, 5 mM VA-044 and 15 mM glutathione. The reaction was incubated at 45°C for 2 h. Once the product was confirmed by analytical RP-HPLC and ESI-MS, the protein was purified by semipreparative RP-HPLC (Supplementary Figure S2, Calculated mass: 11296 Da, observed mass: 11293 Da).
Histone octamer refolding
Core histones H2A, H2B, H3 and H4 (or modified variants, see above) were dissolved at 1 mg/ml in unfolding buffer (20 mM Tris–HCl pH 7.5, 6 M GdmCl) and combined in equimolar ratios (with a 10% excess of H2A and H2B). To refold into octamers the mixture was dialyzed against refolding buffer (10 mM Tris–HCl pH 7.5, 2 M NaCl, 1 mM ehylenediaminetetraacetic acid (EDTA)). The refolded octamers were purified by size exclusion chromatography with a S200 10/300 GL column (Supplementary Figure S3). Eluted octamers were concentrated to 20–60 μM, supplemented with glycerol to 50% and stored at −20°C.
Chromatin reconstitution
Chromatin arrays or mononucleosomes were reconstituted at a concentration of around 1 μM per mononucleosome (100 pmol). Array DNA (a177 or a197) or nucleosome DNA was mixed with 1 equivalent (per 601 site) of histone octamer in reconstitution buffer (10 mM Tris pH 7.5, 10 mM KCl, 0.1 mM EDTA) containing 2 M NaCl. For chromatin arrays, 0.5 molar equivalents (eq.) of MMTV buffer DNA was added to prevent oversaturation. In the case of reconstituted chromatin containing linker histone, 0.5, 1 or 1.5 equivalents H1.1 were also added to the DNA/octamer mixture. The reactions were gradually dialyzed from high salt buffer (10 mM Tris pH 7.5, 1.4 M KCl, 0.1 mM EDTA) to reconstitution buffer over 12 h with a two-channel peristaltic pump. After the dialysis the reconstituted chromatin arrays were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis (PAGE) in 0.5x Tris-Borate-EDTA (TBE) running buffer or on a 0.6% agarose gel following ScaI restriction digest (Supplementary Figure S4).
Preparation of HP1 proteins
HP1 proteins were expressed and labeled as described in ref. (25) using streamlined expressed protein ligation for labeling (28). Briefly, for labeling, a short tripeptide (Thz-G2-C3-CONH2, Thz denotes thiazolidine) was synthesized by SPPS. For a typical labeling reaction, 1.5 mg of the purified peptide was labeled with Atto532-iodoacetamide, followed by quenching of the reaction by TCEP. Thiazolidine opening was achieved by treatment with 0.5 M methoxylamine at pH 5. The labeled tripeptide was finally purified by semipreprative RP-HPLC.
For expression of the HP1 proteins, the genes for HP1α, HP1β and HP1γ were fused to the NpuN split-intein sequence (28) at their C-termini, followed by a hexahistidine tag. HP1α and HP1β were expressed in Escherichia coli BL21 DE3 cells, whereas HP1γ was expressed in E. coli Rosetta DE3 cells. Expression was induced with 0.25 mM isopropyl-β-D-thiogalactopyranosid (IPTG) overnight at 18°C. Cells were harvested and lysed in lysis buffer (25 mM phosphate pH 7.8, 50 mM NaCl, 5 mM imidazole and protease inhibitors). The proteins were purified over a Ni-affinity resin, followed by anion exchange chromatography (1 ml HiTrap Q FF, GE Healthcare) from low (25 mM phosphate pH 7.8, 50 mM NaCl) to high salt buffer (25 mM phosphate pH 7.8, 1000 mM NaCl). A total of 500 μl of the expressed HP1-NpuN fusion constructs at a concentration of 50–100 μM were each applied to a column of 125 μl SulfoLink resin slurry, containing immobilized NpuC peptide (28). After 5 min incubation, the column was drained, followed by washes with wash buffer (100 mM phosphate pH 7.2, 1 mM EDTA, 1 mM TCEP) containing high (500 mM NaCl), intermediate (300 mM NaCl) and low salt (150 mM NaCl). About 1 mM of tripeptide in labeling buffer (100 mM phosphate pH 7.8, 50 mM MPAA, 200 mM MESNA, 150 mM NaCl, 10 mM TCEP, 1 mM EDTA) was added to the column and incubated for 16 h at room temperature. The column was drained and the eluate was collected. The column was further washed using elution buffer (100 mM phosphate pH 7.2, 200 mM MESNA, 150 mM NaCl, 10 mM TCEP, 1 mM EDTA). Elution fractions were pooled and the protein was purified by size exclusion chromatography (Superdex S200 10/300GL) in gel filtration buffer (50 mM Tris pH 7.2, 150 mM NaCl, 1 mM dithiothreitol (DTT)). Fractions containing purified labeled HP1 were pooled and concentrated, glycerol was added to 30%, frozen aliquots were stored at −80°C. (Supplementary Figure S5, HP1α: Calculated mass: 23200 Da, observed mass: 23202 Da, HP1β: Calculated mass: 22390 Da, observed mass: 22394 Da, HP1γ: Calculated mass: 21786 Da, observed mass: 21789 Da).
Histone H1
Human histone H1.1 expression was induced with 0.5 mM IPTG for 4 h in BL21 DE3 pLysS cells in LB media supplemented with 100 μg/ml ampicillin and 35 μg/ml chloramphenicol. Cells were lysed by sonication in lysis buffer (20 mM Tris–HCl pH 8, 200 mM NaCl, 1 mM EDTA, 2% Triton, 1 mM PMSF, 1 mM β-mercaptoethanol, protease inhibitors). H1.1 was purified by cation exchange (HiTrap SP HP 5 ml column) from low (20 mM Tris–HCl pH 8, 200 mM NaCl, 1 mM EDTA) to high salt concentration buffer (20 mM Tris–HCl pH 8, 1 M NaCl, 1 mM EDTA). Eluted H1.1 was further purified by size exclusion chromatography using a S75 10/300 GL column in gel filtration buffer (50 mM phosphate pH 7, 150 mM NaCl). Collected fractions containing the protein were pooled, concentrated, mixed with glycerol (to 10%), flash frozen and stored at −80°C (Supplementary Figure S7, Calculated mass: 21711 Da, observed mass: 21714 Da). For fluorescent labeling, a cysteine residue was added to the C-terminus of H1.1 by site-directed mutagenesis. After cation exchange, the protein was labeled with Atto532-iodoacetamide overnight at 4°C. The reaction was quenched with TCEP and purification was achieved by size exclusion chromatography (50 mM phosphate pH 7, 150 mM NaCl). The purified protein was mixed with glycerol (to 10%), flash frozen and stored at −80°C (Supplementary Figure S7, Calculated mass: 22582 Da, observed mass: 22587 Da).
Scanning force microscopy imaging of chromatin arrays
After reconstitution, chromatin fibers with or without H1.1 were dialyzed against 1 mM EDTA, pH 8 for 6 h. Nucleosomal arrays were then dialyzed against 1 mM EDTA, 0.1% glutaraldehyde, pH 7.7 for 6 h and finally dialyzed overnight against 1 mM EDTA, pH 7.6. All dialysis steps were performed at 4°C. A total of 2 μl of purified nucleosomal arrays was pipetted onto freshly cleaved mica and 5 μl water was added. The sample was left to settle for 5–10 min before the liquid was blown off with a nitrogen gas stream, leaving the dried chromatin arrays on the atomically flat mica. Atomic force microscope images were acquired in tapping mode in air with cantilevers at a nominal spring constant of 42 N/m and resonance frequency of 320 kHz. Typically, images were recorded at a resolution of 1024 × 512 pixel and at a line rate of 4 Hz. Data processing was done using standard SPM image processing software.
Phosphorylated HP1α
For in vitro kinase reactions ref. (29) was followed with modifications. 2.5 μg of recombinant HP1α was mixed with 100 U of casein kinase II (CK2), 100 μM adenosine triphosphate (ATP) spiked with 10 μCi [γ-32P]-ATP in kinase reaction buffer (20 mM Tris–HCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.01% Brj35, pH 7.5) in a total volume of 20 μl. The reaction was incubated at 37°C, 3 h. To stop the reaction, 5 μl loading buffer was added and the reaction mixture was analyzed by 12% sodium dodecyl sulfate (SDS)-PAGE followed by autoradiography detection. To determine the number of phosphorylation sites in each reaction, the gel was dried and the bands corresponding the protein were cut out and their radioactivity was measured by calibrated scintillation counting. For smTIRF measurements ATP was removed by dialysis.
Microscale thermophoresis titrations
Measurements were carried out on a MonoLith NT.115 (Nanotemper). H3K9me3 1–14 peptide was diluted into HEPES buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% Tween, 1 mM DTT) and serial dilutions were prepared from 1 to 100 μM with a final volume of 10 μl. 250 nM of labeled protein was then added to each peptide dilution to a final volume of 20 μl. The mixtures were then centrifuged 1 min, 15 000 rpm and loaded onto premium-coated capillaries. A cap scan was performed at 20% detection light intensity to check that each capillary contained the same protein concentration. Thermophoresis was measured at 40 or 60% infrared heating-laser power. Data were normalized to the start and end point, averaged and fitted with a quadratic binding equation.
Electrophoretic mobility shift assays
Three different concentrations of each HP1 protein (0, 3.5 and 7 μM) were incubated with 0.2 pmol of 193-bp DNA containing the 601 sequence and a Cy5 label in EMSA buffer (20 mM Tris–HCl, 50 mM NaCl, 1 mM DTT, 0.1 mg/ml bovine serum albumin) in a total volume of 10 μl. The reactions mixtures were incubated 15 min at 37°C and terminated by adding 3 μl 25% sucrose. The samples were then separated on a non-denaturing 5% polyacrylamide gel in 0.5x TBE running buffer.
Single-molecule TIRF imaging
Single molecule measurements were performed as described previously in ref. (25). Briefly, Glass coverslips (40 × 24 mm) and microscopy slides (76 × 26 mm) containing drilled holes were cleaned by sonication in 10% alconox, acetone and ethanol and finally incubated at least 1 h in a mixture of concentrated sulfuric acid to 30% hydrogen peroxide (3:1). The coverslips and slides were then submerged in acetone containing 2% (3-aminopropyl)triethoxysilane (APTES) for silanization. The slides and coverslips were dried and strips of double-sided tape were sandwiched between them to form four channels. The glass coverslips were passivated with a solution of 100 mg/ml mPEG(5000)-succinimidyl carbonate containing 1% biotin-mPEG-succinimidyl carbonate for 3 h. Once passivated, the channels were washed with water and buffer T50 (10 mM Tris, 50 mM KCl). For chromatin immobilization, 0.2 mg/ml neutravidin solution was incubated for 5 min, followed by extensive washes with T50 buffer. Then, 500 pM chromatin arrays in T50 buffer were injected into the neutravidin treated flow chamber for 5 min, followed by a wash with T50 and imaging buffer (50 mM HEPES, 130 mM KCl, 10% glycerol, 2 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox), 0.005% Tween-20, 3.2% glucose, glucose oxidase/catalase enzymatic oxygen removal system). Chromatin coverage was observed in the TIRF microscope (Nikon Ti-E) by fluorescent emission in the far-red channel upon excitation by a 640-nm laser line. Dynamic experiments were initiated by influx of 1–5 nM Atto532-labeled HP1 in imaging buffer. All smTIRF experiments were performed at room temperature (22°C). HP1 dynamics were observed with an EMCCD camera (Andor iXon) in the yellow/orange channel using a 530 nm laser line for excitation at 20 W/cm2. Ten thousand frames at 20 Hz rate were acquired over a 25 × 50 μm area at a resolution of 160 nm/pixel. Every 200 frames, an image of the chromatin positions in the far-red channel was recorded for drift correction.
Data analysis
For each chromatin fiber, an individual trace was extracted using a custom-made semi-automated Matlab (Mathworks) script, as described in ref. (25). This was carried out by a baseline correction and a drift correction from the chromatin images taken every 200 frames. A peak-finding algorithm (using the far-red images) was employed to detect individual chromatin array positions. Fluorescence intensity traces for each chromatin position were obtained by integrating over a circle of 2 pixel radius. Individual HP1 fluorescence peaks were included based on point-spread-function (PSF) and distance cut-offs. To ensure that only single-molecules are analyzed, peaks exhibiting step-wise bleaching kinetics were excluded from the analysis. Kinetics were extracted from fluorescence traces using a semi-automated thresholding algorithm. Cumulative histograms were constructed from dark and bright intervals and fitted to mono or bi-exponential functions.
H1.1 residence time determination
To determine the residence time of H1.1, chromatin fibers containing Atto532-labeled H1.1 were reconstituted. Single molecule measurements were performed as follows: Chromatin fibers were imaged with the 647 nm laser over 100 ms for localization, subsequently Atto532-labeled H1.1 was imaged over 2000 s with 100 ms exposure time. The delay between each excitation was increased stepwise up to 1000 ms to avoid contributions from photobleaching. The traces from 50 different arrays were then averaged and fitted with a bi-exponential function.
H1.1 occupancy determination
To determine the occupancy of H1.1 on chromatin arrays, Atto532-labeled H1.1 was imaged for 60 s with high laser intensity. Stepwise photobleaching was observed and analyzed from the recorded traces using a step-detection algorithm (30).
Mammalian cell culture
NIH/3T3 mouse fibroblasts were cultured in 75 cm2 tissue culture flasks in DMEM/F-12 supplemented with 10% Newborn calf serum. For transfection, cells were seeded onto 25-mm round coverslips in 6-well plates. At 70–90% level of confluency the cells were transfected 4 h with 4 μg of plasmid DNA per well using Lipofectamine. The cells were imaged the following day.
Fluorescence recovery after photobleaching assays
Microscopy was performed using an inverted LSM 700 confocal microscope (Zeiss) and a Plan apochromat × 63/1.4 NA objective. Fluorescence recovery after photobleaching (FRAP) bleaching and time-series images were acquired in 150 × 150 pixels with a pixel size of 0.1 μm, 16-bit grayscale depth and a pixel dwell time of 1.58 μs with the pinhole set at 201 μm. A circular spot of 14 pixels (1.4 μm) in diameter was used for bleaching. Five pre-bleach images were acquired before five iterations of a bleaching pulse at 100% laser power used and movies were acquired for 42 s. Photobleaching during the time-series was corrected using the intensity in the bleach region relative to the entire acquisition region. The time-intensity acquisitions were normalized to the pre-bleach intensity and the first image after the bleach pulse. Results were averaged over 17–30 individual FRAP curves for unmodified HP1α and mutants proteins.
RESULTS
HP1α residence times are sensitive to chromatin structure
We set out to probe whether effector proteins interact mainly with individual PTMs or if they integrate the global chromatin state through multivalent interactions, dynamic rebinding or additional interaction modes. In the latter case, the local density of modifications and their spatial arrangement is critical for PTM binding within chromatin (31). Chromatin can adopt different structures, including open extended conformations as well as a compact double helical fibers (1) (Figure 1A). Local structure depends on the ionic strength, the length of linker DNA connecting nucleosomes, inter-nucleosomal interactions and chromatin-associated structural proteins such as H1 (1,32,33). All these features are potentially being probed by multivalent effectors such as HP1 family proteins (34). We thus decided to determine the influence of chromatin fiber conformation on the binding and retention dynamics of the human HP1α isoform.
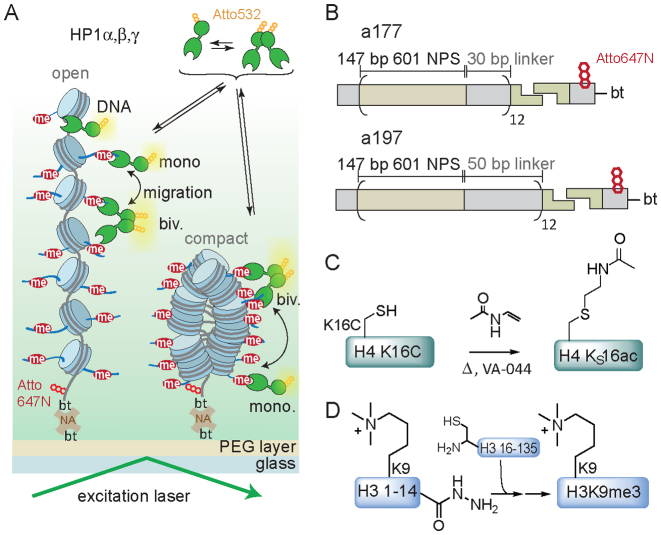
Figure 1.
Detection of effector–chromatin interactions on the single-molecule level. (A) Schematic representation of the smTIRF imaging experimental setup showing HP1 interacting with open and compact chromatin fibers. DNA: DNA interactions, mono.: monovalent PTM binding, biv.: bivalent PTM binding, bt: biotin, NA: neutravidin. (B) Structure of the chromatin DNA constructs used: a tandem array of 12 × 601 nucleosome positioning sequences with a 177 bp NRL (a177) or DNA with a 197 bp NRL (a197), ligated to an oligonucleotide containing biotin for immobilization and a dye for localization. (C) Synthesis scheme of H4KS16ac using radical based thiol-ene addition. (D) Synthesis of H3K9me3 by traceless EPL, involving in situ conversion of the peptide hydrazide to a thioester followed by EPL and desulfurization.
First, we investigated the effect of expanding chromatin structure (i.e. increasing the distance between individual nucleosomes) on HP1α binding dynamics. To this end, we designed two different chromatin DNA constructs, based on a 12-mer array of the 601 nucleosome positioning sequence (26): we employed a DNA construct with a short nucleosome repeat length (NRL) of 177 bp (a177), where individual nucleosomes are separated by 30 bp DNA linkers, as utilized in earlier experiments (25). In a second design, we increased the NRL to 197 bp (a197), resulting in 50 bp DNA linkers between nucleosomes (Figure 1B). Both DNA constructs were recombinantly produced, coupled to a fluorescently labeled (Atto647N) and biotinylated oligonucleotide anchor (Figure 1B and Supplementary Figure S1) and employed for chromatin assembly.
To further ensure an open, extended chromatin state we introduced H4 acetylated at K16 (H4K16ac), which disrupts a key internucleosomal interaction (35). Previous experiments using hydrodynamic and spectroscopic methods established that incorporation of H4K16ac disrupts local and higher-order chromatin fiber decompaction (36–38). A close mimic of this PTM was synthesized using a radical-mediated thiol-ene approach from H4 carrying a lysine 16 to cysteine mutation to yield the modified histone (H4KS16ac) (27) (Figure 1C and Supplementary Figure S2). Finally, we employed traceless expressed protein ligation (39–41) to produce H3K9me3 (Figure 1D and Supplementary Figure S2). Subsequently, octamers were refolded using recombinant as well as chemically modified human histones (Supplementary Figure S3). We then reconstituted a set of chromatin fibers on both DNA templates (a177 or a9197), containing H3K9me3 and either unmodified H4 or H4KS16ac (Supplementary Figure S4).
We proceeded to measure HP1α binding dynamics by smTIRF imaging (Figure 1A). Chromatin fibers were immobilized in PEG-passivated flow chambers and imaged in the far-red channel. We then injected HP1α, labeled with Atto532 at its C-terminus (Supplementary Figure S5), and observed transient binding interactions by single-molecule colocalization detection (Figure 2A). From the recorded movies fluorescence emission traces were extracted for each chromatin position. The kinetic traces showed transiently bound HP1α proteins as peaks in fluorescence emission (Figure 2B). Cumulative lifetime-histograms were constructed from the detected interaction events and analyzed. HP1α exhibited bi-exponential dissociation kinetics for all chromatin architectures (Figure 2C), while the association kinetics were well described with single-exponential kinetics (Figure 2D and Table 1). In the absence of H4KS16ac, the HP1α residence time was not significantly different between a177 and a197 fibers, exhibiting a dominant dissociation process (87–95% of the total amplitude) associated with a short time constant τoff,1 = 250 ± 30 ms (a177) and 240 ± 10 ms (a197), whereas a second slower process, resulting from multivalent interactions, exhibited time constants of τoff,2 = 2.3 ± 1.2 and 2.5 ± 0.3 s for the two fiber architectures (Figure 2E and Table 1). The similar behavior of HP1α indicates that the local density of PTMs is comparable in compact chromatin fibers with a NRL of 177 or 197 bp. This is in agreement with cryo-electron microscopy studies of chromatin fibers of different linker length (42).
Figure 2.
Open chromatin reduces HP1α retention. (A) Representative microscopy images showing the localization of the chromatin fibers (left panel) and individual HP1α molecules bound to chromatin (right panel). The scale bar is 5 μm. (B) Characteristic time trace (blue) of HP1α binding dynamics to a single chromatin fiber and fitted with a step function (red). Each intensity peak represents one binding event. (C) Dissociation kinetics: cumulative histograms of HP1α dwell times of 100 traces fitted with a double-exponential function. (D) Association kinetics: cumulative histogram of times between each binding event of the same 100 traces, fitted with a mono-exponential function. (E) Chromatin decompaction by H4KS16ac reduces τoff,1 and τoff,2 in both a177 and a197 fibers. Numbers indicate % amplitude. Error bars: standard deviation (s.d.), n = 7–16 replicates, *P < 0.05, Student's t-test. For the fit values, see Table 1.
Table 1. Fit results from smTIRF measurements of HP1α, β and γ chromatin interactions.
| Chromatin | NRL | Effector | τ off,1 (s) | % A1 | τ off,2 (s) | % A2 | k on (M−1s−1) × 106 | n |
|---|---|---|---|---|---|---|---|---|
| H3K9me3* | 177 | HP1α | 0.25 ± 0.03 | 87 ± 7 | 2.26 ± 1.22 | 13 ± 7 | 3.64 ± 1.56 | 16 |
| H3K9me3 and H4K16ac | 177 | HP1α | 0.19 ± 0.02 | 92 ± 4 | 1.92 ± 1.45 | 8 ± 4 | 2.76 ± 1.58 | 10 |
| H3K9me3 0.5eq H1.1 | 177 | HP1α | 0.22 ± 0.01 | 95 ± 2 | 2.90 ± 0.60 | 5 ± 2 | 3.34 ± 2.46 | 4 |
| H3K9me3 1eq H1.1 | 177 | HP1α | 0.19 ± 0.02 | 95 ± 3 | 3.68 ± 0.72 | 5 ± 3 | 2.84 ± 1.19 | 4 |
| H3K9me3 | 197 | HP1α | 0.24 ± 0.03 | 90 ± 7 | 2.49 ± 0.89 | 10 ± 7 | 2.51 ± 0.82 | 12 |
| H3K9me3 and H4K16ac | 197 | HP1α | 0.16 ± 0.02 | 95 ± 5 | 2.00 ± 0.83 | 5 ± 5 | 2.25 ± 0.44 | 7 |
| H3K9me3 0.5eq H1.1 | 197 | HP1α | 0.24 ± 0.05 | 92 ± 5 | 3.61 ± 0.84 | 8 ± 5 | 3.84 ± 1.37 | 7 |
| H3K9me3 1eq H1.1 | 197 | HP1α | 0.23 ± 0.03 | 92 ± 3 | 3.63 ± 1.18 | 8 ± 3 | 3.23 ± 1.10 | 9 |
| H3K9me3 (195 mM salt) | 177 | HP1α | 0.19 ± 0.03 | 89 ± 4 | 3.32 ± 0.61 | 11 ± 4 | 1.10 ± 0.19 | 5 |
| H3K9me3 (260 mM salt) | 177 | HP1α | 0.14 ± 0.01 | 86 ± 4 | 2.97 ± 0.71 | 14 ± 4 | 0.36 ± 0.03 | 3 |
| H3K9me3 | 177 | HP1β | 0.19 ± 0.03 | 95 ± 2 | 3.60 ± 1.13 | 5 ± 2 | 0.67 ± 0.19 | 12 |
| H3K9me3 | 177 | HP1γ | 0.17 ± 0.02 | 93 ± 3 | 2.64 ± 0.48 | 7 ± 3 | 0.89 ± 0.23 | 6 |
| H3K9me3 | 177 | pHP1α | 0.26 ± 0.02 | 90 ± 2 | 5.60 ± 0.84 | 10 ± 2 | 1.73 ± 0.35 | 5 |
n denotes the number of independent experiments, each contributing >100 kinetic traces.
* values taken from ref. (25).
Chromatin fibers containing H4KS16ac adopt an open chromatin state (36–38). In this chromatin context, the HP1α residence times were accordingly reduced by 25–35% for τoff,1 and by a similar degree for τoff,2 (Figure 2E). Importantly, this effect was more pronounced for a197 than for a177 fibers, indicating that the lower density of binding sites reduced the probability of HP1α to rebind after dissociation. To ensure that the effect of H4KS16ac originates from chromatin structure, we tested HP1α binding dynamics in mononucleosomes (Supplementary Figure S6). In the absence of a full chromatin fiber, no significant difference was observed between nucleosomes containing both H3K9me3 and H4KS16ac or only H3K9me3. Together, these measurements thus indicate that HP1α retention is sensitive to chromatin fiber conformation: Chromatin decompaction lowers the local H3K9me3 density, reducing both multivalent binding and the probability of re-binding of transiently dissociated HP1α, thereby shortening the HP1α residence time.
Linker histone incorporation prolongs HP1α binding
Secondly, we investigated HP1α interaction dynamics in compact chromatin, where the local density of H3K9me3 marks is increased. Chromatin fibers are efficiently compacted by linker histone H1 incorporation (42,43), and heterochromatin regions contain near stoichiometric levels of linker histone H1 (44,45). We thus expressed and purified human H1.1 (Supplementary Figure S7) and incorporated it into chromatin fibers at different stoichiometries (Figure 3A). Indeed, atomic force microscopy showed that H1.1 incorporation resulted in a transition from an extended beads-on-a-string conformation to compact chromatin fibers (Figure 3B). To ensure stable H1.1 incorporation under single-molecule conditions, we further prepared chromatin fibers containing H2A labeled with Alexa Fluor 488 at position 110 and H1.1 labeled with Atto532 at its C-terminus (Supplementary Figure S7). We then confirmed the colocalization of H1.1, H2A as well as chromatin DNA on the single-molecule level (Figure 3C). Moreover we quantified H1.1 incorporation into chromatin fibers by counting stepwise photobleaching (Figure 3D). This analysis revealed that in chromatin fibers with 50 bp linker DNA (a197) a 1:1 stoichiometry of H1.1 to nucleosome was established (Figure 3D and Supplementary Figure S7). In contrast, for a177 only one H1.1 molecule was incorporated per di-nucleosome, in agreement with earlier reports (43) (Supplementary Figure S7). For H1 binding to nucleosomes, residence times of up to 25 min were reported in vitro (46). We thus determined the residence time of H1.1 under our experimental conditions, and observed that the majority of H1.1 molecules (70%) remained bound over a 2000 s observation period, whereas a smaller subset (30%) dissociated with a 313 s time constant (Figure 3E). While these residence times are sufficiently long compared to our experimental timescale (∼10 min), we ensured full H1.1 saturation by performing the measurements in the presence of 100 pM H1.1 added to the measurement buffer.
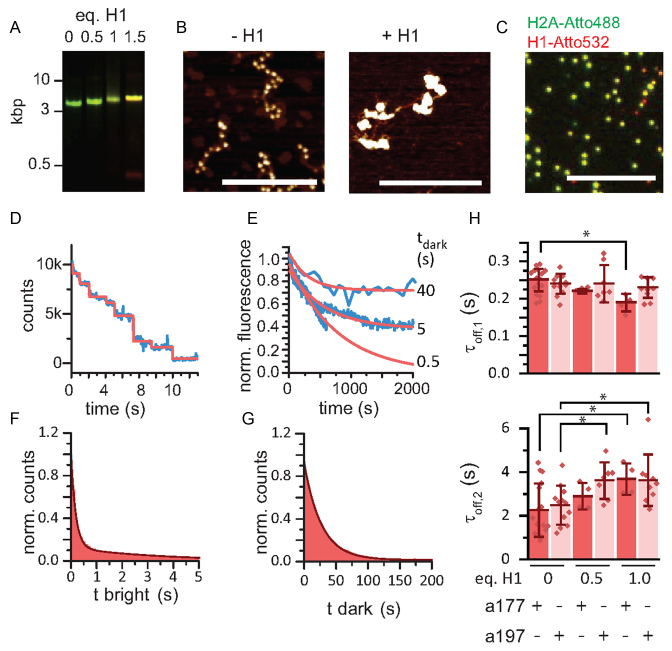
Figure 3.
H1.1 compacted chromatin prolongs HP1α binding. (A) Analysis of reconstituted chromatin containing the indicated equivalents of H1.1 (per nucleosome) by agarose gel electrophoresis (0.6%), green: DNA(Atto647N), red: H1(Atto532). (B) AFM imaging of 12-mer chromatin fibers (a197) with and without H1.1. Scale bar: 400 nm. (C) Single molecule analysis of H1.1 incorporation into chromatin fibers. Green: H2A(Atto488), red: H1(Atto532). Scale bar: 10 μm. (D) Stepwise bleaching of fibers containing H1(Atto532). (E) Determination of the residence time of H1(Atto532): >100 chromatin fibers are imaged using stroboscopic imaging with the indicated dark times (tdark = 0.5, 5, 40 s). For the longest tdark, only H1.1 dissociation is limiting showing 70% of molecules remaining bound >2000 s, whereas 30% dissociate with τ = 313 s. (F) Dissociation kinetics: cumulative histograms of HP1α dwell times of 100 traces fitted with a double exponential function. (G) Association kinetics: cumulative histogram of times between each binding event of the same 100 traces, fitted with a mono-exponential function. (H) Dependence of τoff,1 and τoff,2 on H1.1 incorporation for both a177 and a197 chromatin fibers. Error bars: s.d., n = 4–16 replicates, *P < 0.05 versus fibers of the same NRL without H1.1, Student's t-test. For the fit values, see Table 1.
Having established suitable experimental conditions, we proceeded to measure HP1α dissociation (Figure 3F) and association kinetics (Figure 3G) with chromatin fibers that were compacted by H1.1. For a177 fibers we observed a decrease in τoff,1 with H1.1 incorporation, which was concomitant with an increase in τoff,2 (Table 1, Figure 3H). Similarly, in a197 fibers τoff,2 was increased from 2.49 ± 0.26 s to 3.63 ± 0.39 s, however without a change in τoff,1 (Table 1). To ensure that the increase in HP1α residence time arises from changes in chromatin structure as opposed to direct H1.1 interactions we further performed binding experiment with H3K9me3 and H1.1 containing mononucleosomes as well as with H1.1 compacted chromatin fibers lacking the H3K9me3 mark (Supplementary Figure S6). No binding to chromatin fibers was observed for HP1α in the absence of H3K9me3, even when H1.1 was present. Similarly, H1.1 incorporation in H3K9me3-modified mononucleosomes did not significantly alter the binding dynamics. These results, in particular for a197 fibers, thus show that chromatin compaction by H1.1 induces a prolongation of the HP1α residence time. Several factors may contribute to altered kinetics: In both array contexts, chromatin compaction increases H3K9me3 density, facilitating multivalent interactions. These findings are in agreement with equilibrium experiments of HP1 binding in compact chromatin (34,47). The stabilizing effect may however be mitigated by H1.1 leading to steric restriction (45) and changes in H3 tail mobility (48). Moreover, the basic, unstructured tail of H1 interacts with linker DNA (49), thereby shielding DNA charges and reducing electrostatic interactions, which might be important for HP1α recruitment and retention.
DNA interactions modulate HP1 binding
Thus, to explore the relationship between DNA interactions and PTM recognition by HP1 proteins in a chromatin context, we decided to perform a comparative study of the three mammalian HP1 isoforms. HP1α, β and γ differ mainly in the unstructured HR as well as in the N- and C-terminal domains (NT, CT) (Figure 4A and Supplementary Figure S5). Together, these sequence differences result in altered DNA interactions, mediated by a stretch of basic amino acids in the HR: HP1β and HP1γ have been reported to exhibit significantly reduced DNA binding compared to HP1α (29). In order to investigate their chromatin binding dynamics, we thus recombinantly expressed and fluorescently labeled all isoforms (Supplementary Figure S5). Subsequently, we confirmed that HP1α interacts with DNA by performing electrophoretic mobility shift assays (EMSA), whereas DNA binding was found to be greatly reduced for HP1β and HP1γ (Figure 4B). To confirm that the increased DNA interactions in HP1α originate from the hinge-region, we further performed EMSA experiments with a mutant protein, where six lysines/arginine residues, localized in the hinge, were converted to alanine (hinge-mutant) (50). Indeed, this protein did not show detectable DNA binding (Supplementary Figure S9). In addition to differences in DNA binding, HP1 isoforms also exhibit varying affinities for their target histone PTM. The affinity for H3K9me3 was increased for both HP1β (KPTM = 1.4 ± 0.3 μM) and HP1γ (KPTM = 4.1± 1.0 μM) as compared to HP1α (KPTM = 11.6 ± 1.2 μM) when measuring binding to a modified histone peptide (51) (Supplementary Figure S7). We then measured the binding kinetics of all HP1 isoforms to H3K9-methylated chromatin fibers. Surprisingly, we found that the major residence time, τoff,1, for both HP1β and HP1γ was reduced compared to HP1α, indicating weaker chromatin binding in spite of their higher affinity for H3K9me3. The binding rate constants, kon, were significantly lowered compared to HP1α, for HP1β by a factor of 5.8 and for HP1γ by a factor of 3.9. Only the slow dissociation process in HP1β exhibited an increased time constant (τoff,2) compared to the α-isoform, indicating the presence of a strongly bound molecular subpopulation (Figure 4C).
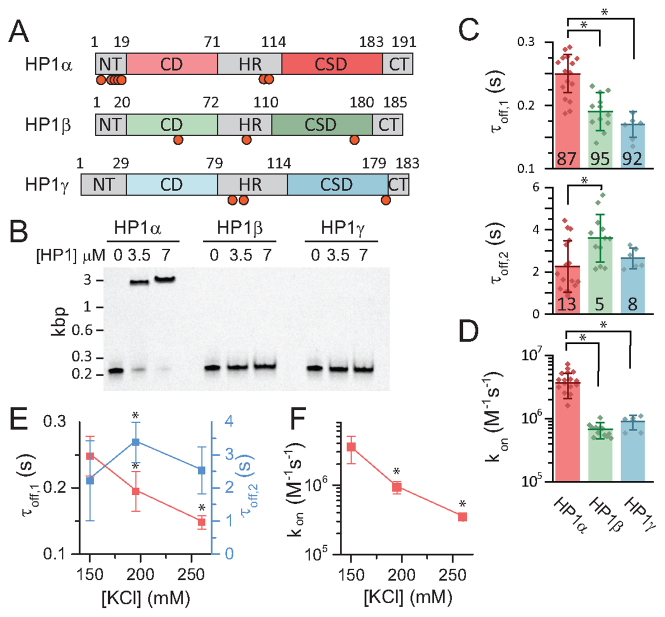
Figure 4.
HP1 isoforms show distinct chromatin binding dynamics. (A) Scheme of the three different mammalian HP1 isoforms. The CDs and CSDs are similar in structure, however they differ in the unstructured regions. All HP1 variants can be phosphorylated: HP1α (T8, S11, S12, S13, S14, S93, S95/S97), HP1β (T51, S89, S175), HP1γ (S93, S95, S176), refs. (22,50,55). The red circles indicate phosphorylation sites. (B) Electrophoretic mobility shift assay (EMSA) for DNA binding of HP1α, HP1β and HP1γ. Binding reactions with equal amount of DNA and indicated HP1 concentrations are analyzed by polyacrylamide gel electrophoresis and imaged by fluorescence detection of Cy5-labeled DNA. (C) Comparison of dissociation time constants (τoff,1 and τoff,2) and (D) association rate constants (kon) obtained from smTIRF experiments for HP1α, HP1β and HP1γ. (E) Salt dependence of τoff,1 and τoff,2 for HP1α. (F) Salt dependence of kon for HP1α. (C–F) Error bars: s.d., n = 4–16 replicates, *P < 0.05, Student's t-test for the fit values, see Table 1.
Based on these observations, we hypothesized that DNA interactions could be the origin of the fast binding kinetics of HP1α and its prolonged residence time on chromatin. We therefore tested if shielding charge–charge interactions by increasing the ionic strength of the solution might reduce fast binding and chromatin retention of HP1α. Indeed, increasing the ionic strength from 130 to 260 mM KCl resulted in both a decrease in HP1α residence time as well as a 10-fold reduction in the binding rate constant (Figure 4E and F). Of note, we cannot exclude that secondary salt-effects contribute to the observed changes in interaction kinetics, e.g. changes in H3 tail dynamics or alterations in local chromatin structure. These effects, in accordance with our own results (Figures 2 and 3) are however most likely smaller in magnitude compared to direct charge-based DNA interactions. Together, these experiments thus reveal that DNA interactions play an important role for HP1 recruitment to chromatin.
Kinetic modeling of the HP1-chromatin interaction network
To dissect the detailed mechanisms of HP1-chromatin binding dynamics, we decided to model the binding process in an unbiased way. We thus developed a kinetic model accounting for all conceivable interactions between mono- (X) or dimeric HP1 (Y) and histone and DNA sites on the chromatin fibers and their respective transitions (Figure 5A, Supplementary Figure S10, Supplementary Methods and Tables S1 and 2). The model distinguishes between the possible combinations of DNA interactions (index 1) and specific histone PTM binding (index 2), including multivalent states combining both modes of interactions. As initial parameters, we used experimentally determined equilibrium constants for the elementary binding reactions and concentrations directly comparable to the experimental conditions in smTIRF. We then optimized the model output with respect to the measured single-molecule kinetics from HP1α and β, using a genetic algorithm (52). Based on the optimized parameter set (Supplementary Figure S9 and Table S3), we further performed stochastic simulations (53) of HP1-chromatin binding dynamics, allowing a direct comparison to the primary single-molecule data (Figure 5B–D). Importantly, our parameterized simulations revealed that the predominant binding pathways involved DNA bound states (Figure 5A). Further, we observed that individual observable binding events were composed of multiple rapid transitions between different H3K9me3 and intrinsically short-lived DNA bound states. Finally, the origin of the apparent bi-exponential kinetics observed for HP1 dissociation in the experimental data could be traced back to multivalent interactions: simulating HP1 interaction kinetics without multivalent binding did not reproduce the slow kinetic phase (τoff,2) whereas inclusion of multivalency gave rise a population HP1 molecules engaged in long-lived interactions (Supplementary Figure S10).
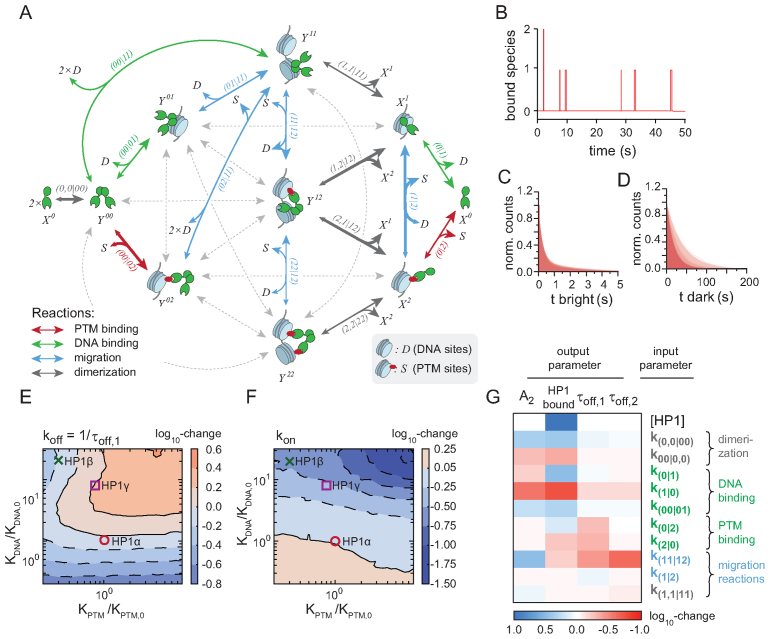
Figure 5.
Kinetic modeling of HP1-chromatin interaction network. (A) Full kinetic model of HP1—chromatin interactions, showing the pathways associated with high flux (arrow thickness). Gray arrows denote reactions associated with low but non-zero flux. Unbound states are labeled ‘0’, DNA bound states ‘1’ and H3K9me3 bound states ‘2’. Double-headed arrows indicate reversible reactions. Arrowhead size indicates the direction of net flux. For detailed information on all the parameters see Supplementary Tables S1–4 and Supplementary Methods). (B) Reaction trace from stochastic simulation based on the model shown in Figure 5 indicating binding and dissociation dynamics of HP1. (C) Overlay of cumulative histograms for HP1 dwell times obtained from stochastic simulations using the 185 best ranked parameter sets. (D) Overlay of cumulative histograms for HP1 unbound (dark) times obtained from stochastic simulations using the 185 best ranked parameter sets. (E) Dependence of simulated HP1 dissociation rate constants (koff = 1/τoff,1) on relative changes in KPTM and KDNA. Parameters for O: HP1α, X: HP1β.□: HP1γ. The color code and contour lines indicate changes in the dissociation on a logarithmic scale. (F) Dependence of simulated HP1 binding rate constants (kon) on relative changes in methyllysine (KPTM) and DNA (KDNA) interactions. Parameters for O: HP1α, X: HP1β.□: HP1γ. The color code and contour lines indicate changes in the binding rate on a logarithmic scale. (G) The heat map denotes the average order of magnitude change of the indicated model output with respect to the set of most sensitive model parameters, blue indicates correlation of parameter and output, red indicates anticorrelation.
We then applied our model to better understand how PTM- and DNA-dependent interactions modulate the chromatin recruitment kinetics of HP1α and HP1β. We thus probed the response of the apparent association (kon, Figure 5E) and dissociation rate constants (koff = 1/τoff,1, Figure 5F) to changes in either the equilibrium constant for H3K9me3 interactions (KPTM) or in the equilibrium constant for DNA binding (KDNA). Indeed, when systematically altering KPTM and KDNA over two orders of magnitude around the measured values for the HP1 isoforms (∼1–100 μM), kon was found to be more sensitive to changes in KDNA, whereas HP1 retention on chromatin (koff) depended more strongly on KPTM. Moreover, changes in KDNA could be partially compensated by respective changes in KPTM and vice versa. In accordance with results from current and earlier experiments (25), this analysis shows that charge-based DNA interactions are instrumental in guiding HP1 proteins to their binding sites.
Finally, our model allowed us to identify the key parameters governing chromatin interactions. We performed global sensitivity analysis (54), which revealed the set of microscopic rate constants that had the largest impact on a set of key observables: the amount of bound HP1, the two apparent residence times (τoff,2, τoff,2) as well as the fraction molecules engaged in long-lived interactions (i.e. the relative amplitude of the slow process, A2). Apart from the total HP1 concentration and the microscopic rate constants governing binding to DNA and to H3K9me3, HP1 dimerization and a number of internal migration pathways appeared as a key determinant for chromatin retention (Figure 5G). This allows us to make predictions about how the HP1 dynamics change in a cellular environment. Increasing the local HP1 concentration results in more chromatin bound protein. Cellular proteins that stabilize HP1 dimerization increase the chromatin bound population and prolong the residence times by inducing multivalent states. Contrariwise, an increase in migration reaction rates, potentially stimulated by local competition in the nucleus or active mechanisms such as chromatin remodeling, results in faster HP1 release times as they provide dissociation pathways. Finally, the combination of DNA and PTM interactions are responsible for chromatin targeting, as DNA binding has strong effects across all parameters, whereas histone PTM binding provides specificity for heterochromatic regions (Figure 5G).
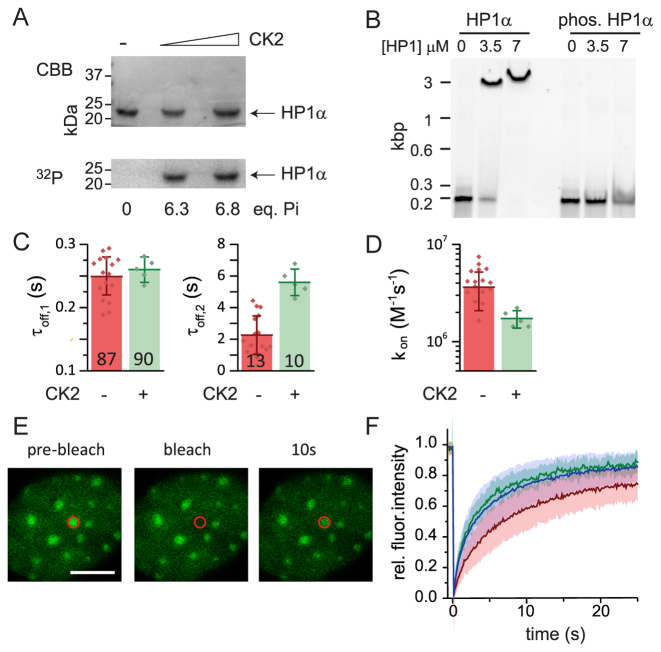
HP1α phosphorylation stabilizes bivalent binding
In the cell, PTMs on the HP1 proteins themselves provide a mechanism to regulate the strength of DNA binding. Indeed, mass-spectrometry studies allowed to map PTMs on each HP1 isoform revealing a complex modification landscape (55). In particular, a number of phosphorylation sites were identified on all HP1 proteins (Figure 4A). HP1α is unique in that it contains an N-terminal extension (NTE) containing for serine residues which have been found to be highly phosphorylated in vivo (29,50). Phosphorylation of these serines was shown to increase affinity for H3K9me3, while at the same time decreasing DNA binding (29,50). Intriguingly, these PTMs were also identified in stimulating HP1α multimerization, thereby increasing multivalency and inducing phase-separation behavior (23). We were thus interested in characterizing the effect of NTE phosphorylation on HP1α binding dynamics. We used casein kinase 2 (CK2) to phosphorylate fluorescently labeled HP1α, resulting in six phosphorylation marks as verified by quantification of the incorporation of 32P-labeled ATP (Figure 6A). Phosphorylated HP1α (pHP1α) exhibited markedly decreased equilibrium DNA binding in agreement with earlier experiments (29) (Figure 6B). We then measured its interaction dynamics with H3K9me3-containing chromatin fibers (Figure 6C and D). The reduced DNA interaction propensity of pHP1α resulted in two-fold slower chromatin binding (Figure 6D) compared to non-phosphorylated HP1α. In contrast, dissociation of pHP1α from chromatin was found to be slowed down as well, with a significant fraction of binding events (10 ± 1%) that decayed with a time constant of τoff,2 = 5.6 ± 0.37 s. Phosphorylation of HP1α thus not only decreases its binding rate by reducing DNA binding, but stabilizes multivalent interactions on chromatin, possibly through conformational changes within the protein (23) and additional interactions with the H3 N-terminal tail (29,50,56).
Figure 6.
Phosphorylation increases HP1α residence times. (A) CK2-mediated phosphorylation of HP1α using [γ-32P]-ATP for quantification. CBB: Coomassie Brilliant Blue staining, 32P: Autoradiography (B) EMSA for HP1α and phosphorylated HP1α at the indicated concentrations, analyzed by polyacrylamide gel electrophoresis and imaged by fluorescence detection of Cy5-labeled DNA. (C) Comparison of the chromatin residence times τoff,1 and τoff,2 for unmodified (CK2 −) and phosphorylated (CK2 +) HP1α. (D) Comparison of binding rate kon for unmodified (CK2 −) and phosphorylated (CK2 +) HP1α. (E) FRAP experiment with mEos3.2-HP1α showing dynamic recovery within 10 s. Scale bar: 5 μm. (F) FRAP analysis for HP1α (wild-type, red), HP1α (AAAA-mutant, blue) and HP1α(hinge-mutant, green) demonstrating decreased chromatin binding for the mutant proteins. Exponential fits to the FRAP data resulted in a recovery time constant of 8.1 ± 0.7 s for HP1α, a recovery time constant of 4.8 ± 0.6 s for HP1α(AAAA-mutant) and a recovery time of 5.1 ± 0.5 s for HP1α(hinge-mutant). Errors: s.d.
We then wondered how DNA interactions, modulated by phosphorylation, affect HP1α dynamics in cells. We thus performed FRAP experiments for wild type HP1α (fused to the monomeric fluorescent protein mEos3.2 via its N-terminus) and two mutant proteins: one construct corresponded to the previously used hinge-mutant, devoid of DNA binding affinity (Supplementary Figure S9). To probe the in vivo effect of HP1α phosphorylation, we produced a second HP1α construct where all four serines in the NTE were mutated to alanine (AAAA), thus preventing NTE phosphorylation. NIH 3T3 fibroblasts were transfected with the three mEos3.2-tagged HP1α variants. All constructs displayed heterochromatin localization (Figure 6E and Supplementary Figure S10). FRAP measurements targeting heterochromatin foci revealed an increased mobility for both the hinge-mutant HP1α (Figure 6F, green trace) as well as for the AAAA-mutant HP1α (Figure 6F, blue trace), as compared to wild-type HP1α (Figure 6F, red trace).Together, these findings correlate well with our in vitro findings that demonstrate the importance of DNA interactions, but also indicate the compensatory effect of NTE phosphorylation. The effects of hinge- or NTE-mutations on HP1α dynamics are however markedly smaller than mutations disrupting HP1α dimerization. The latter mutation drastically reduced chromatin retention (i.e. the while still retaining heterochromatin localization (25). This is in agreement with the importance of multivalent interactions for the organization of HP1α domains in the cell (23,24). Mutations targeting the CD finally result in complete lack of sub-nuclear targeting due to loss of any specific interactions with the chromatin landscape (20). Together, these experimental results strengthen the hierarchy of interactions observed in the kinetic model for HP1-chromatin interactions (Figure 5G), where HP1 dimerization is a key parameter governing chromatin recruitment, driven by DNA interactions, whereas histone PTM binding is responsible for targeting to heterochromatin sites.
DISCUSSION
Chromatin presents different chemical and structural features which are integrated by chromatin effectors and transcription factors to enact a biochemical response. Chromatin recognition by multivalent effectors not only depends on the presence of specific histone PTMs in a given chromatin state, but also on their spatial arrangement. Thus, the density of PTMs as well as spatial features, including positioning of nucleosomes, linker DNA lengths, higher order chromatin structure as well as asymmetrical modification patterns (57,58) are an important part of the recognition landscape and determine chromatin function. In addition to histone PTM-specific readers, effectors often contain additional interaction domains, enabling binding to DNA or RNA, to the nucleosome surface, to transcription factors or to other chromatin associated proteins (59–62). For a quantitative understanding of chromatin recognition, knowledge of these different factors is required.
Here, we probed the relative influences of spatial and chemical features in chromatin on the interaction dynamics on a class of multivalent effectors, the human HP1 family proteins. We employed single-molecule TIRF measurements to directly monitor chromatin binding and dissociation kinetics and, coupled with modeling the complete kinetic system, extracted mechanistic information on these transient interactions. We compared chromatin of different architecture (with linker lengths of 30–50 bp) and in either an open or a linker histone-induced compact conformation. We observed that HP1α prefers a compact fiber state, due to an increase in multivalent interactions. In agreement, linker histones are abundant in heterochromatic regions (63). HP1α has been reported to increase chromatin compaction upon binding (18). This results in positive feedback, which further stabilizes HP1α association to heterochromatin. Conversely, in embryonic stem cells, where chromatin adopts a globally open state, HP1 dynamics have been found to be markedly increased (64). Chromatin conformation is thus a direct modulator of multivalent effector interactions. The observed sensitivity to chromatin structure for HP1α was however smaller compared to the linker-DNA sensitivity of the Schizosaccharomyces pombe HP1 ortholog Swi6 (15), which shows a strong increase in H3K9me3-specific binding for short linker-DNA lengths. The preference of Swi6, which is quite divergent in sequence from the mammalian HP1 proteins, for short linker-DNA can be rationalized as S. pombe exhibits much shorter NRLs as compared to mammalian cells (65).
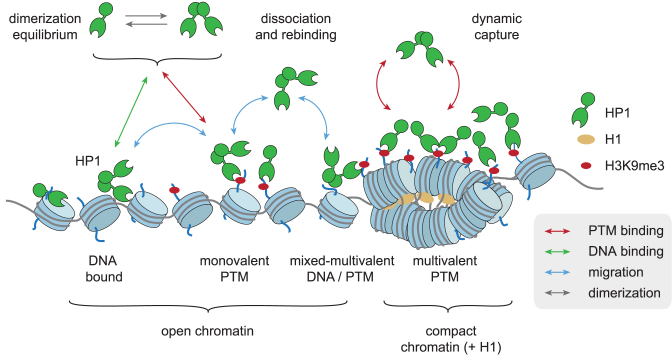
Combined single-molecule experiments comparing all three human HP1 subtypes, together with computational efforts further allowed us to generate a detailed picture of the dominant chromatin interaction pathways (Figure 7). In this dynamic capture model, the most common HP1 recruitment and retention pathways involve charge-dependent DNA interactions, which increase the capture radius for specific sites (i.e. H3K9me3 modified nucleosomes). This situation, where lower-affinity interactions guide an effector protein to its molecular target are reminiscent to the target search of transcription factors (66). From initial short-lived dynamic encounter complexes, HP1 molecules can transition into states involving more long-lived PTM-based interactions, either mono-, bivalent or mixed states where DNA and PTMs are simultaneously engaged. Moreover, DNA interactions enable rapid transitions between neighboring nucleosomes, facilitating rapid re-binding after dissociation from H3K9me3. Our previous results (25) indicated a critical importance of HP1 dimerization and the thus resulting multivalency. Here, we demonstrate that reducing the strength of charge-based interactions directly lowers the probability of H3K9me3 binding. Simultaneously the residence times of HP1 proteins is decreased as re-binding reactions are disfavoured. For this reason, the HP1β and HP1γ isoforms both exhibit slower binding and reduced chromatin retention in comparison to the HP1α isoform, in spite of their higher affinity for H3K9me3.
Figure 7.
Dynamic capture model of multivalent HP1 proteins. HP1 proteins are dynamically captured by histone PTMs (H3K9me3) and DNA interactions (for details see text). Compact chromatin, i.e. induced by H1, strengthens multivalent interactions. The color of the arrows denotes the reaction type (see also Figure 5A).
The molecular properties of HP1 proteins are modulated by various PTMs (55). Examples include HP1β phosphorylation at T51, which is important during initiation of the DNA damage response (22), or S83 phosphorylation of eukaryotic HP1γ (67). Moreover, both HP1α and HP1γ are phosphorylated throughout the cell cycle (68). The key phosphorylation sites in HP1α involve serine residues in the NTE, disrupting DNA binding but strengthening H3K9me3 interactions (29,50). In our kinetic assays, we demonstrate that phosphorylated HP1α can not only compensate the loss in DNA binding by its higher affinity for the H3 peptide (50), but even demonstrates increased chromatin retention, possibly involving further specific mechanisms. Importantly, NTE-phosphorylation has been implicated in phase-separation behavior (23,24), involving conformational changes of the HP1α dimer as well as interactions beyond the dimer. PTMs are thus clearly a critical mechanism for the cell to both regulate local dynamics as well as emerging behavior over larger spatial scales.
Taken together, we have gained insight into the mechanisms of chromatin interaction by multivalent effectors. Importantly, we have determined a key function of helper-interactions via the DNA in target search and chromatin retention for effector proteins. We propose that this is a general paradigm for chromatin associated proteins (Figure 7). Effectors are transiently trapped by multiple moderate-affinity interactions, and sequential dissociation and re-binding reactions result in effector accumulation in the high-avidity environment of chromatin. Such a dynamic capture mechanism enables a fast response to changes in the local environment or cell state (22,51). It further predicts that the interaction kinetics of effectors with chromatin depend on both the multivalency properties of effectors and the structural features of the chromatin fiber, modulating the local concentration of binding sites. Indeed, protein subdomains or regions conferring DNA or general nucleosome binding properties are commonly encountered among chromatin effectors, e.g. in SUV39H1 (59), PRC1 (60,62), PRC2 (61) or BRDT (69), in addition to specific PTM reader modules. These interactions are thus required for initial chromatin localization of effector complexes, whereas specific histone PTM recognition directs their accumulation in target chromatin domains.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Horst Pick for help with cell culture, Arne Seitz and Thierry Laroche (BIOP) for FRAP measurements, Maeva Tobler for help with the preparation of histones. We thank all LCBM members for discussions.
Footnotes
Present addresses:
Carolin C. Lechner, Bachem AG, Hauptstrasse 144, 4416 Bubendorf, Switzerland.
Pascal D. Odermatt, Department of Cell and Tissue Biology, HSW 601, 513 Parnassus Avenue, UCSF, San Francisco, CA 94143, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EPFL; Sandoz Family Foundation; Swiss National Foundation Grant [31003A_149789]; Systems X Grant [51PHP0_163580]. Funding for open access charge: Laboratory funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Dechassa M.L., Tremethick D.J.. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair?. Nat. Rev. Mol. Cell Biol. 2012; 13:436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith E., Shilatifard A.. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell. 2010; 40:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010; 471:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L.J., Lin M.F. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruthenburg A.J., Li H., Patel D.J., Allis C.D.. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007; 8:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi P., Allis C.D., Wang G.G.. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010; 10:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elgin S.C., Grewal S.I.. Heterochromatin: silence is golden. Curr. Biol. 2003; 13:R895–R898. [DOI] [PubMed] [Google Scholar]
- 8. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 9. Maison C., Almouzni G.. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004; 5:296–304. [DOI] [PubMed] [Google Scholar]
- 10. Dialynas G.K., Terjung S., Brown J.P., Aucott R.L., Baron-Luhr B., Singh P.B., Georgatos S.D.. Plasticity of HP1 proteins in mammalian cells. J. Cell Sci. 2007; 120:3415–3424. [DOI] [PubMed] [Google Scholar]
- 11. Singh P.B. HP1 proteins–what is the essential interaction. Genetika. 2010; 46:1424–1429. [PubMed] [Google Scholar]
- 12. Brown J.P., Bullwinkel J., Baron-Luhr B., Billur M., Schneider P., Winking H., Singh P.B.. HP1gamma function is required for male germ cell survival and spermatogenesis. Epigenet. Chromatin. 2010; 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 14. Thiru A., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D.. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004; 23:489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canzio D., Chang E.Y., Shankar S., Kuchenbecker K.M., Simon M.D., Madhani H.D., Narlikar G.J., Al-Sady B.. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell. 2011; 41:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiragami-Hamada K., Soeroes S., Nikolov M., Wilkins B., Kreuz S., Chen C., De La Rosa-Velazquez I.A., Zenn H.M., Kost N., Pohl W. et al. Dynamic and flexible H3K9me3 bridging via HP1beta dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 2016; 7:11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodcock C.L., Ghosh R.P.. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010; 2:a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Azzaz A.M., Vitalini M.W., Thomas A.S., Price J.P., Blacketer M.J., Cryderman D.E., Zirbel L.N., Woodcock C.L., Elcock A.H., Wallrath L.L. et al. Human heterochromatin protein 1alpha promotes nucleosome associations that drive chromatin condensation. J. Biol. Chem. 2014; 289:6850–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muller K.P., Erdel F., Caudron-Herger M., Marth C., Fodor B.D., Richter M., Scaranaro M., Beaudouin J., Wachsmuth M., Rippe K.. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys J. 2009; 97:2876–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheutin T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T.. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003; 299:721–725. [DOI] [PubMed] [Google Scholar]
- 21. Festenstein R., Pagakis S.N., Hiragami K., Lyon D., Verreault A., Sekkali B., Kioussis D.. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003; 299:719–721. [DOI] [PubMed] [Google Scholar]
- 22. Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R.. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008; 453:682–686. [DOI] [PubMed] [Google Scholar]
- 23. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domain formation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kilic S., Bachmann A.L., Bryan L.C., Fierz B.. Multivalency governs HP1alpha association dynamics with the silent chromatin state. Nat. Commun. 2015; 6:7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 27. Li F., Allahverdi A., Yang R., Lua G.B.J., Zhang X., Cao Y., Korolev N., Nordenskiöld L., Liu C.-F.. A direct method for site-specific protein acetylation. Angew. Chem. Int. Ed. Engl. 2011; 50:9611–9614. [DOI] [PubMed] [Google Scholar]
- 28. Vila-Perello M., Liu Z., Shah N.H., Willis J.A., Idoyaga J., Muir T.W.. Streamlined expressed protein ligation using split inteins. J. Am. Chem. Soc. 2013; 135:286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishibuchi G., Machida S., Osakabe A., Murakoshi H., Hiragami-Hamada K., Nakagawa R., Fischle W., Nishimura Y., Kurumizaka H., Tagami H. et al. N-terminal phosphorylation of HP1alpha increases its nucleosome-binding specificity. Nucleic Acids Res. 2014; 42:12498–12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aggarwal T., Materassi D., Davison R., Hays T., Salapaka M.. Detection of steps in single molecule data. Cell. Mol. Bioeng. 2012; 5:14–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J.. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007; 14:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002; 31:361–392. [DOI] [PubMed] [Google Scholar]
- 33. Zhu P., Li G.. Structural insights of nucleosome and the 30-nm chromatin fiber. Curr. Opin. Struct. Biol. 2016; 36:106–115. [DOI] [PubMed] [Google Scholar]
- 34. Mishima Y., Jayasinghe C.D., Lu K., Otani J., Shirakawa M., Kawakami T., Kimura H., Hojo H., Carlton P., Tajima S. et al. Nucleosome compaction facilitates HP1gamma binding to methylated H3K9. Nucleic Acids Res. 2015; 43:10200–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 36. Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L.. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006; 311:844–847. [DOI] [PubMed] [Google Scholar]
- 37. Fierz B., Chatterjee C., McGinty R.K., Bar-Dagan M., Raleigh D.P., Muir T.W.. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011; 7:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allahverdi A., Yang R., Korolev N., Fan Y., Davey C., Liu C.-F., Nordenskiöld L.. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011; 39:1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muir T.W., Sondhi D., Cole P.A.. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan L.Z., Dawson P.E.. Synthesis of peptides and proteins without cysteine residues by native chemical ligation combined with desulfurization. J. Am. Chem. Soc. 2001; 123:526–533. [DOI] [PubMed] [Google Scholar]
- 41. Wan Q., Danishefsky S.J.. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew. Chem. Int. Ed. Engl. 2007; 46:9248–9252. [DOI] [PubMed] [Google Scholar]
- 42. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.M., Zhu P., Li G.. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014; 344:376–380. [DOI] [PubMed] [Google Scholar]
- 43. Routh A., Sandin S., Rhodes D.. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bates D.L., Thomas J.O.. Histones H1 and H5: one or two molecules per nucleosome. Nucleic Acids Res. 1981; 9:5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woodcock C.L., Skoultchi A.I., Fan Y.. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006; 14:17–25. [DOI] [PubMed] [Google Scholar]
- 46. Yue H., Fang H., Wei S., Hayes J.J., Lee T.H.. Single-molecule studies of the linker histone H1 binding to DNA and the nucleosome. Biochemistry. 2016; 55:2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R.. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001; 7:729–739. [DOI] [PubMed] [Google Scholar]
- 48. Stutzer A., Liokatis S., Kiesel A., Schwarzer D., Sprangers R., Soding J., Selenko P., Fischle W.. Modulations of DNA contacts by linker histones and post-translational modifications determine the mobility and modifiability of nucleosomal H3 tails. Mol. Cell. 2016; 61:247–259. [DOI] [PubMed] [Google Scholar]
- 49. Bednar J., Garcia-Saez I., Boopathi R., Cutter A.R., Papai G., Reymer A., Syed S.H., Lone I.N., Tonchev O., Crucifix C. et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell. 2017; 66:384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hiragami-Hamada K., Shinmyozu K., Hamada D., Tatsu Y., Uegaki K., Fujiwara S., Nakayama J.. N-terminal phosphorylation of HP1alpha promotes its chromatin binding. Mol. Cell. Biol. 2011; 31:1186–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., Allis C.D.. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005; 438:1116–1122. [DOI] [PubMed] [Google Scholar]
- 52. Sun J.Y., Garibaldi J.M., Hodgman C.. Parameter estimation using metaheuristics in systems biology: a comprehensive review. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012; 9:185–202. [DOI] [PubMed] [Google Scholar]
- 53. Gillespie D.T. Exact stochastic simulation of coupled chemical-reactions. J. Phys. Chem. 1977; 81:2340–2361. [Google Scholar]
- 54. Pianosi F., Sarrazin F., Wagener T.. A Matlab toolbox for global sensitivity analysis. Environ. Modell. Softw. 2015; 70:80–85. [Google Scholar]
- 55. LeRoy G., Weston J.T., Zee B.M., Young N.L., Plazas-Mayorca M.D., Garcia B.A.. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell Proteomics. 2009; 8:2432–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shimojo H., Kawaguchi A., Oda T., Hashiguchi N., Omori S., Moritsugu K., Kidera A., Hiragami-Hamada K., Nakayama J., Sato M. et al. Extended string-like binding of the phosphorylated HP1alpha N-terminal tail to the lysine 9-methylated histone H3 tail. Sci. Rep. 2016; 6:22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rhee H.S., Bataille A.R., Zhang L., Pugh B.F.. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014; 159:1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voigt P., LeRoy G., Drury W.J. 3rd, Zee B.M., Son J., Beck D.B., Young N.L., Garcia B.A., Reinberg D.. Asymmetrically modified nucleosomes. Cell. 2012; 151:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Muller M.M., Fierz B., Bittova L., Liszczak G., Muir T.W.. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat. Chem. Biol. 2016; 12:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Francis N.J., Woodcock C.L., Kingston R.E.. Chromatin compaction by a polycomb group protein complex. Science. 2004; 306:1574–1577. [DOI] [PubMed] [Google Scholar]
- 61. Son J., Shen S.S., Margueron R., Reinberg D.. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013; 27:2663–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhen C.Y., Tatavosian R., Huynh T.N., Duc H.N., Das R., Kokotovic M., Grimm J.B., Lavis L.D., Lee J., Mejia F.J. et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife. 2016; 5:e17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hergeth S.P., Schneider R.. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006; 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Holde K. Chromatin. 1989; NY: Springer. [Google Scholar]
- 66. Wang Y., Guo L., Golding I., Cox E.C., Ong N.P.. Quantitative transcription factor binding kinetics at the single-molecule level. Biophys. J. 2009; 96:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grzenda A., Leonard P., Seo S., Mathison A.J., Urrutia G., Calvo E., Iovanna J., Urrutia R., Lomberk G.. Functional impact of Aurora A-mediated phosphorylation of HP1gamma at serine 83 during cell cycle progression. Epigenet. Chromatin. 2013; 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Minc E., Allory Y., Worman H.J., Courvalin J.C., Buendia B.. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999; 108:220–234. [DOI] [PubMed] [Google Scholar]
- 69. Miller T.C., Simon B., Rybin V., Grotsch H., Curtet S., Khochbin S., Carlomagno T., Muller C.W.. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat. Commun. 2016; 7:13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.