Abstract
Cytoplasmic maturation of precursors to the small ribosomal subunit in yeast requires the intervention of a dozen assembly factors (AFs), the precise roles of which remain elusive. One of these is Rio1p that seems to intervene at a late step of pre-40S particle maturation. We have investigated the role played by Rio1p in the dynamic association and dissociation of AFs with and from pre-40S particles. Our results indicate that Rio1p depletion leads to the stalling of at least 4 AFs (Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p) in 80S-like particles. We conclude that Rio1p is important for the timely release of these factors from 80S-like particles. In addition, we present immunoprecipitation and electron microscopy evidence suggesting that when Rio1p is depleted, a subset of Nob1p-containing pre-40S particles associate with translating polysomes. Using Nob1p as bait, we purified pre-40S particles from cells lacking Rio1p and performed ribosome profiling experiments which suggest that immature 40S subunits can carry out translation elongation. We conclude that lack of Rio1p allows premature entry of pre-40S particles in the translation process and that the presence of Nob1p and of the 18S rRNA 3′ extension in the 20S pre-rRNA is not incompatible with translation elongation.
INTRODUCTION
Production of the small ribosomal subunit in yeast starts in the nucleolus with the formation on the nascent RNA pol I transcript of a ribonucleoprotein particle termed the small subunit processome (SSU processome) (1) that can be visualized on chromatin spreads as terminal balls (2) (for reviews, see (3,4)). The SSU processome contains small subunit ribosomal proteins, small nucleolar ribonucleoprotein particles (snoRNPs) and scores of proteins not present in the mature ribosomal subunits, diversely called non ribosomal proteins or maturation/assembly factors (AFs) (5–7). SnoRNPs and AFs assemble in a stepwise fashion as transcription of the nascent pre-rRNA proceeds (8–10). The first pre-40S pre-ribosomal particles are released in the nucleus by Utp24p-catalyzed endonucleolytic cleavage (11) of the RNA pol I transcript at site A2 within the internal transcribed spacer 1 that separates the sequences of the 18S and 5.8S rRNAs. These early nuclear pre-40S particles contain the 20S pre-rRNA and a subset of AFs already present in the SSU processome, including Enp1p, the endonuclease Nob1p and its partner Pno1p/Dim2p, the methyltransferase Dim1p, the GTPase-like Tsr1p and the kinase Hrr25p. Prior to their export to the cytoplasm, nuclear pre-40S particles acquire, in addition to the previously mentioned AFs, Ltv1p and the ATPase/kinase Rio2p. Once in the cytoplasm, pre-40S particles undergo final maturation steps leading to the production of the mature small ribosomal subunits. These maturation steps include RNA restructuring events coupled with the stepwise dissociation of most AFs and the proper positioning of several ribosomal proteins (12). Prior to cleavage of the 20S pre-rRNA by Nob1p at the D site to yield mature 18S rRNA (13), late cytoplasmic pre-40S particles undergo a quality control step involving transient interaction with a mature 60S ribosomal subunit (14,15) in the absence of mRNA. This interaction is promoted by Fun12p/eIF5B and the resulting so-called 80S-like particles are then disrupted by the intervention of Rli1p (15).
The timing of intervention and dissociation of AFs and their precise molecular roles remain uncertain and the subject of intense research. The first AF to dissociate following export to the cytoplasm may be Ltv1p. Ltv1p is phosphorylated by the Hrr25p kinase in yeast (16–18) or the casein kinase 1 isoforms δ and ε in humans (19). Ltv1p phopshorylation by Hrr25p promotes its dissociation from pre-40S particles prior to 80S-like particle formation (17). In addition, correct Ltv1p release seems to require Tsr1p, since Ltv1p shifts to 80S-containing fractions when extracted from Tsr1p-depleted cells (20). Ltv1p forms a complex with Enp1p and ribosomal protein Rps3 (21), both of which are also phosphorylated, probably by Hrr25p (16,21) or CK1 δ and ε in the case of human ENP1 (19), although this remains disputed (17). Whether Enp1p is released together with Ltv1p is debated, some authors proposing that Enp1p remains present until 80S-like particle formation (15,17). Rio2p release could occur following that of Ltv1p. Rio2p may function as an ATPase rather than a kinase and while its catalytic activity is not required for its association with pre-40S particles, it promotes Rio2p dissociation (16). Tsr1p release then occurs following 80S-like ribosome formation and requires the intervention of the adenylate kinase/ATPase Fap7p (15). The last AFs to remain in 80S-like particles are likely Rio1p, Pno1p/Dim2p and Nob1p (22). D site cleavage requires nucleotide binding by Rio1p which may induce removal of Dim2p from the D site to allow access of the Nob1p endonuclease (7,22).
Several roles have been attributed to AFs during cytoplasmic maturation of pre-40S particles. High-throughput probing of pre-rRNA structure suggests that AFs in early and intermediate cytoplasmic pre-40S particles maintain a flexible pre-rRNA structure, in particular in the 3′ end domain, and delay access of some ribosomal proteins to their final binding site (12). Some AFs seem to participate in the control of the stepwise incorporation of a subset of ribosomal proteins by direct protein-protein interactions, as in the case of Fap7p and Rps14 (23,24) or Ltv1p and Rps3 (17,18). The CRAC analyses of the binding sites of AFs (25) and the resolution of the cryo-EM structures of early cytoplasmic yeast pre-40S particles purified via Ltv1p or Rio2p as bait (20) show that Rio2p, Tsr1p and Dim1p binding sites overlap those of translation initiation factors eIF1 and eIF1A on the mature 40S subunits. Nob1p and Pno1p/Dim2p are positioned over the binding site of translation initiation factor eIF3 and the binding of Enp1p and Ltv1p is incompatible with mRNA channel opening. Likewise, TSR1 in early cytoplasmic human pre-40S particles is expected to block binding of translation initiation factors eIF1A, DHX29 and eIF5b, while RIO2 likely inhibits tRNA loading (26). Thus, early cytoplasmic pre-40S particles are prevented from engaging in translation by all the AFs they contain.
Strikingly, Lacombe and colleagues showed that removal of the ubiquitin-coding sequence from the UBI3 gene encoding the Ubi3-Rps31 fusion protein (ubi3Δub mutant) led to the appearance of 20S pre-rRNA in all polysome fractions of sucrose gradients (27). Moreover, the sedimentation of 20S pre-rRNA in heavy gradient fractions was lost under polysome run-off conditions. This led the authors to the conclusion that in the ubi3Δub mutant, aberrant pre-40S particles could engage in translation initiation and possibly elongation (27). Likewise, sedimentation of 20S pre-rRNA in polysome fractions was detected when using extracts from certain mutant strains impaired in 20S pre-rRNA processing, including a strain depleted for the assembly factors Ltv1p and Pfa1p (13) or the rpl3[W255C] strain expressing a mutant Rpl3 ribosomal protein (28). Moreover, we showed that a small proportion of 20S pre-rRNA sediments on sucrose gradients in fractions containing 80S ribosomes or polysomes in wild-type yeast cells and that it almost entirely shifts to these fractions when Rio1p or the Nob1p endonuclease is depleted (29). We also detected an association between 20S pre-rRNA and the poly-A binding protein or several translation initiation factors and reported that several abundant mRNAs can be precipitated together with Nob1p. Collectively, these data lead us to propose that a minority of late pre-40S particles can engage in translation in wild-type cells and that this phenomenon is exacerbated in certain mutant conditions that impair pre-40S particle maturation. It remained unclear whether these particles could engage in productive translation elongation, although Garcia-Gomez et al. argue that pre-40S particles that accumulate in rpl3[W255C] cells can support translation elongation since 20S/18S and 20S/25S ratios are similar for light and heavy polysome fractions (28).
In the present work, we set out to characterize further the pre-40S particle maturation defects caused by Rio1p depletion. We show that Rio1p depletion affects the correct release of AFs Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p and we provide strong evidence that a subset of Nob1p-containing pre-40S particles carry out productive translation elongation under these conditions.
MATERIALS AND METHODS
Yeast strains and plasmids
A TetO7-RIO1, NOB1-FPZ Saccharomyces cerevisiae strain was constructed as follows. A plasmid encoding the FPZ-tag (Flag—PreScission protease cleavage site—Z sequence derived from S. aureus Protein A) was constructed by hybridixing two kinased complementary primers (Primers 2XFlag FW / 2XFlag Rev, see Table 1) to create two tandem repeats of the Flag DNA sequence flanked at the 5′ end by the overhang of a cut NcoI restriction site and at the 3′ end by the overhang of a cut NheI restriction site, allowing their insertion into the dephosphorylated pBS1539 plasmid (30) at NcoI/NheI restriction sites, leading to the substitution of the CBP sequence by the 2 x Flag sequence. The new plasmid was called pBS1539Flag. The PreScission protease cleavage site was then constructed by hybridizing two complementary primers (Primers PPX_FW / PPX_REV, see Table 1) to create the PreScission protease cleavage site DNA sequence flanked at the 5′ side by the overhang of a cut NheI restriction site. This fragment was then inserted into pBS1539Flag cut at the NheI/Ecl136II restriction sites, to substitute the sequence encoding the TEV cleavage site by the PreScission protease cleavage site. The plasmid obtained was named pBS1539-FPZ.
Table 1. Oligonucleotides used.
| 2XFlag FW | 5′CATGGACTACAAGGACGACGATGACAAAGGTACCGATTACAAAGATGATGACGACAAAG3′ |
| 2XFlag Rev | 5′CTAGCTTTGTCGTCATCATCTTTGTAATCGGTACCTTTGTCATCGTCGTCCTTGTAGTC3′ |
| PPX_FW | 5′CTAGCCTGGAAGTTCTGTTCCAGGGGCCCGAG3′ |
| PPX_REV | 5′CTCGGGCCCCTGGAACAGAACTTCCAGG3′ |
| Nob1 MFM Tag-Nat-Fd | 5′ACGTCCGCATTGGTAAGGGAAGGTACGTCAACAGTTCCAAAAGGAGAAGTTCCATGGACTACAAGGACGAC3′ |
| Nob1 MFM Tag-Nat-Rev | 5′TCTTCAAAGTGCTGTAACACCATTCATCTCATAAGGGGAGGGCATACTGAGCAATTACGACTCACTATAGG3′ |
| Nob1–1081 FD | 5′AACAGATATTCTGTAGCCAG3′ |
| tRNAiMet probe | 5′TCGGTTTCGATCCGAGGACATCAGGGTTATGA3′ |
A derivative of TAP-tag encoding plasmid pBS1479 (30) containing the nourseothricin resistance gene NAT instead of the TRP1 gene was produced by inserting a NotI/EcoRV fragment containing the NAT gene extracted from plasmid pAG25 (Euroscarf) into pBS1479 cut at EcoRV/Bsp120I sites, creating pBS1479-NAT. In order to replace the TAP-tag sequence of pBS1479-NAT by the FPZ-tag sequence, the PciI/EcoRV fragment of pBS1539-FPZ containing the FPZ sequence was inserted into pBS1479-NAT digested by the same enzymes, creating plasmid pBS1479- FPZ-NAT.
Finally, a TetO7-RIO1, NOB1-FPZ strain was constructed by the substitution of the TAP tag sequence by the FPZ tag sequence in the TetO7-RIO1, NOB1-TAP strain (29). To this end, a fragment containing the FPZ tag sequence and the NAT gene was PCR amplified using oligonucleotides Nob1 MFM Tag-Nat-Fd and Nob1 MFM Tag-Nat-Rev (see Table 1) and plasmid pBS1479-FPZ-NAT as template and transformed into the TetO7-RIO1, NOB1-TAP strain. Transformants were selected for resistance to 80 μg/ml nourseothricin and screened by PCR (oligonucleotides Nob1–1081 FD and Nob1 MFM Tag-Nat-Rev, see Table 1) and western blot.
Saccharomyces cerevisiae strains were grown in YP medium (1% yeast extract, 1% peptone) supplemented with 2% glucose as the carbon source (YPD). When required, doxycycline was added at 200 μg/ml final concentration.
Sucrose gradient fractionation of cells extracts
Yeast cells grown in 400 ml YPD medium to an optical density (O.D.) of 0.6–0.9 (600 nm) and treated with 100 μg/ml cycloheximide (CH) for 10 min were harvested by centrifugation and washed in 4 ml ice-cold H2O (MilliQ) containing 100 μg/ml CH. After centrifugation, the pellet was washed in 2 ml buffer A (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.1% Igepal) containing 100 μg/ml CH. The cell pellet was resuspended in an equivalent volume of buffer A containing the same concentration of CH plus 0.1 unit/μl RNasin (Promega), protease inhibitors (Complete EDTA free, Roche) and 1 mM DTT. Whole cell extracts (WCE) were prepared by breaking the cells by vortexing with glass beads (425–600 μm, Sigma). Lysates were cleared briefly at 9300 × g for 5 min followed by a 10 min 9300 × g centrifugation (Eppendorf 5415D) to obtain the final WCEs. WCEs weighing 1 mg of proteins were layered on a 10–50% sucrose gradient prepared in buffer K (20 mM Tris–HCl pH 7.5, 50 mM KCl, 10 mM MgCl2) and centrifuged for 2 h 40 min at 36 000 rpm in a Beckman SW41 rotor. Positions of 40S, 60S ribosomal subunits, 80S ribosomes and polysomes in the gradient were determined by A254 scanning with the ISCO UA-6 gradient fraction collector. Fractions of 500 μl were collected. 100 μl of each fraction were precipitated with 400 μl of 25% trichloroacetic acid (TCA) in the presence of 0.6 μl of glycogen (20 μg/μl). After 20 min on ice, samples were centrifuged 15 min at 4°C and 16 000 × g in a microcentrifuge (Eppendorf 5415D). The supernatants were removed and the pellets were washed once with 1 ml acetone and centrifuged 5 min at 4°C. Pellets were dryed and resuspended in 30 μl of loading buffer (19.5 μl H2O, 7.5 μl NuPAGE LDS sample buffer 4×, 3 μl NuPAGE sample reducing agent 10×, Life Technology). Samples were heated at 72°C for 10 min before proceeding to polyacrylamide gel electrophoresis (PAGE).
Western analysis
Proteins obtained from gradient fractions after TCA precipitation were subjected to PAGE in 4–20% or 8% pre-cast NuPAGE Bis Tris gels (Life technology) and transferred to nitrocellulose membranes (ThermoFisher Scientific, iBlot Gel Transfer Stacks). Nob1p-FPZ protein was detected either by using monoclonal anti-FLAG® M2-Peroxidase (HRP) antibody produced in mouse (Sigma-Aldrich) and diluted 5000-fold, or anti-Nob1p serum diluted 1000-fold. Ltv1p, Tsr1p, Dim1p, Pno1p/Dim2p and Fap7p were detected by use of sera diluted 1000-fold. Rio2p and Enp1p were detected by use of anti-Rio2p and anti-Enp1p sera diluted 5000- and 20 000-fold, respectively. The anti-Ltv1p, anti-Tsr1p, anti-Dim1p, anti-Pno1p/Dim2p and anti-Rio2p antibodies were raised in rabbits and a kind gift of Katrin Karbstein (15). The anti-Nob1p, anti-Enp1p and anti-Fap7p antibodies were raised in rabbits against purified proteins by Eurogentec (Speedy program). The primary antibodies were detected using anti-rabbit IgG-HRP conjugate (Promega) diluted 10 000-fold.
Total RNA extractions and Northern hybridizations
RNA extractions from sucrose gradient fractions were performed using the phenol–chloroform method. 200 μl of 4 M guanidinium isothiocyanate solution, 2 μl glycogen (Roche, 20 mg/ml), 150 μl of a 100 mM NaAc (pH 5), 10 mM Tris–HCl (pH 8.0), 1 mM EDTA solution, 225 μl water-saturated phenol, 225 μl chloroform were added to 150 μl of each gradient fraction, vortexed 30 s, incubated at 65°C during 5 min and placed on ice 5 min. After 5 min centrifugation at 14 000 rpm, 4°C, 400 μl of aqueous phase were recovered and precipitated with 1 ml absolute ethanol overnight at –20°C. After 45 min centrifugation at 14 000 rpm, 4°C, the pellet was washed with 1 ml 70% ethanol. After 5 min centrifugation at 14 000 rpm, 4°C, the supernatant was removed and the pellet was dried at room temperature during 5–10 min and then dissolved in 15 μl H2O.
Electrophoresis of glyoxal-treated RNAs through agarose gels was performed as reported (29). Northern analyses of pre-rRNAs, mature rRNAs and tRNAiMet were carried out by use of 32P-labelled oligodeoxynucleotide probes. Sequences of antisense oligonucleotides used to detect pre- and mature rRNAs have been reported earlier (31,32). The sequence of the probe used to detect tRNAiMet is indicated in Table 1. Blots were hybridized with 5′ end-labelled oligonucleotide probes using Rapid-Hyb Buffer (GE Healthcare).
Tandem-affinity purification of Nob1p-containing pre-40S particles
Particles containing Nob1p-FPZ expressed in TetO7-RIO1, NOB1-FPZ strain grown without or with 200 μg/ml doxycycline (Dox) during 6 and 16 h, respectively, were affinity purified from 70 g (without Dox) or 30 g (with Dox) of cells grown to mid log phase. Cells were quickly cooled on ice and harvested by centrifugation after a 10 min treatment with CH (100 μg/ml). Cell pellets rinsed with one cell volume of lysis buffer A (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.1% Igepal, 100 μg/ml CH) were directly frozen as noodles in liquid nitrogen and were crushed in liquid nitrogen for nine cycles, each of 3 min/1 min reverse rotation at 15 Hz, on a Retsch PM100 planetary ball mill. Sample chamber and grinding stainless steel balls were pre-chilled in liquid nitrogen and re-chilled between each pulverization cycle. Broken cell powder was stored at –80°C or immediately thawed in 10 ml (per 10 g of powder) buffer A plus 100 units/ml RNasin (Promega), protease inhibitors (Complete EDTA free, Roche) and 1 mM DTT. Cell debris were removed by spinning at 4°C, 25 000 × g for 25 min. The supernatant was recovered and pH adjusted to 7.5. Aliquots of this extract were flash-frozen in liquid nitrogen. Small amounts (50 μl) of each extract were phenol-chloroform extracted for RNA analysis or precipitated with TCA for further protein analysis as described below. Typical yields were 20–120 ml of extract with an estimated protein concentration at 15–20 mg/ml. 20 ml extract aliquots were incubated with 600 μl of IgG sepharose beads (GE Healthcare) in buffer A (plus RNasin, protease inhibitors and DTT) under agitation for 1 h 15 min at 4°C. Beads were washed three times in 20 ml cold buffer B (20 mM Hepes pH 7.4, 100 mM KoAc, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.02% Tween 20, 0.1% Triton), resuspended in 800 μl of buffer B containing 50 μl PreScission protease (100 units, Sigma) plus 100 units/ml RNasin. Protease digestion was carried out under agitation during 2 h at 4°C. The bead eluate was recovered and pooled with three bead washing (200 μl of buffer B each). Pooled eluate was carefully added to the surface of an anti-Flag M2 affinity gel (500 μl, Sigma) packed in buffer B in a 2 ml column. Flow-through was recovered and beads washed four times with 1 ml cold buffer C (10 mM Tris–HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT). Elution of the particles was obtained by adding five times 300 μl FLAG elution buffer (buffer C containing 2× Flag peptide (IGBMC synthesis) at 200 μg/ml). At each step of the purification, small amounts (50 μl) of each sample was phenol-chloroform extracted for RNA analysis or precipitated with TCA for further protein analysis.
Electron microscopy
3.5 μl of purified particles were deposited onto glow-discharged, 300 Mesh Copper carbon-coated grids (Quantifoil, Germany). After a 1 min incubation at room temperature, excess liquid was blotted and the grids were stained with two successive drops of 1% uranyl acetate (3.5 μl each). The first drop was immediately blotted, the second was left for ∼20 s on the grids before blotting. Finally, the grids were air-dried before observation on a Jeol 2100 microscope of the METI platform (Centre de Biologie Intégrative, Toulouse, France), equipped with a LaB6 cathode and operating at 200 kV. Images were recorded with defocus ranging from 0.6 to 2.0 μm, on a Gatan US1000 CCD digital camera and DigitalMicrograph (Gatan, Inc.), at various magnifications corresponding to calibrated pixel sizes of 3.4, 1.7 and 1.1 Å.
Ribosome protected fragments (RPF) purification
Total RPFs from strain TetO7-RIO1, NOB1-FPZ expressing or depleted of Rio1p were obtained as follows. Six liters cultures of this strain were grown to an O.D. 600 of 0.8 in YPD medium without or with 200 μg/ml doxycycline during 8 or 20 h, respectively. CH was added to a final concentration of 100 μg/ml and culturing was continued for 10 min. Cells were collected by centrifugation at 6000 rpm, 10 min at 4°C and rapidly frozen as ‘spaghettis’ in liquid nitrogen. Cells were crushed in liquid nitrogen for nine cycles, each of 3 min/1 min reverse rotation at 15 Hz, on a Retsch PM100 planetary ball mill. Total cellular extracts were then prepared as described in the ‘sucrose gradient fractionation of cells extracts’ paragraph above. RPFs were obtained by incubation of a volume of extract corresponding to 500–1000 μg of total proteins with 750 units of RNase I (Ambion) during 1 h, 20°C. Digestion was stopped by addition of 100 units of SUPERase•In RNase inhibitor (Ambion). Monosomes thus obtained were centrifuged in a 4.5–45% sucrose gradient for 2.5 h at 39 000 rpm in a Beckman SW41 rotor. RNAs present in the 80S gradient fractions were then extracted using hot phenol as described in the ‘total RNA extractions’ paragraph above. 15–20 μg of RNAs obtained were treated with the ‘RiboZero Magnetic Gold’ solution and beads (Illumina) following the manufacturer's protocol to remove rRNA fragments. The remaining RNA fragments were then ethanol precipitated, RNA pellets were washed, dried and dissolved in 10 μl 10 mM Tris–HCl pH 7.4. RNAs were denatured by incubation at 80°C for 90 s and cooled for 2 min at 37°C. 33 μl RNase-free H2O, 5 μl 10× T4 polynucleotide kinase (PNK, Thermo Scientific) buffer, 1 μl SUPERase•In RNase inhibitor and 1 μl (10 units) PNK were then added. The resulting solution was incubated 30 min at 37°C. 4 μl of 10 mM ATP were then added and the 37°C incubation was continued for an additional 30 min. The reaction was stopped by a 5 min incubation step at 70°C and the 3′ dephosphorylated, 5′ phosphorylated RNA fragments were precipitated at –20°C overnight after addition of 2 μl glycogen (Roche, 20 mg/ml), 5 μl of a 100 mM NaAc (pH 5), 10 mM Tris–HCl (pH 8.0), 1 mM EDTA solution and 500 μl ethanol. The RNA pellets were washed with 70% ethanol, dried and dissolved in 5 μl RNase-free H2O. RNA fragments were submitted to PAGE in a 15% acrylamide/8M urea denaturing gel. A gel slice containing RNA fragments of 28–32 nucleotides was cut out and RNA fragments present in the slice were eluted using the ‘crushed acryl’ solution from the ‘ARTseq Ribosome Profiling (yeast)’ kit (Illumina) following the manufacturer's recommendations. RNAs were extracted with phenol/chloroform and precipitated with ethanol as described in (33). 60 ng of purified total RPFs from Rio1p-expressing or Rio1p-depleted cells were used for deep-sequencing library construction (called ‘+ Rio1’ and ‘– Rio1’, respectively) by the IMAGIF platform using the TruSeq Small RNA kit (Illumina). Banks were sequenced on a HiSeq (SNL176) line in 51 cycles 51 bases single read, producing ∼240 million sequences per bank.
RPFs due to Nob1p-containing pre-40S particles were purified and identified as follows. Two 12 L YPD/200 μg/ml doxycycline cultures of TetO7-RIO1, NOB1-FPZ cells were grown during 20 h to an O.D. 600 of 0.8. Samples derived from these two cultures were processed in parallel throughout. Cells were harvested and total cellular extracts were prepared as described in the ‘tandem-affinity purification’ paragraph above. The extracts were treated with RNase I as described for total RPFs above. Affinity purification on IgG-sepharose beads followed by a PreScission protease digestion step were then carried out as described in the ‘tandem-affinity purification’ paragraph above. Purified RNA fragments were then extracted using the hot phenol procedure. The rest of the procedure is identical to that described for total RPFs above.
Analysis of deep sequencing data
Reads from Next-Generation Sequencing (NGS) were trimmed by Trim Galore! (Galaxy v0.4.2) in order to remove Illumina small RNA adapters and were aligned to the S. cerevisiae sacCer3 reference genome by Hisat (Galaxy v2.0.3, k = 5). The aligned reads were filtered for MapQ >= 30 by BamFilter (Galaxy v0.0.2) and sorted with Samtools (Galaxy v2.0). The genomic regions corresponding to the aligned reads were intersected with the regions of known yeast mRNA coding sequences (Bedtools v2.25.0; CDS intervals, positions of stop and start codons of SGD genes were downloaded from UCSC Genome Browser as a gtf file). For each aligned read corresponding to an mRNA CDS, the middle position of the corresponding genomic interval was calculated using R (RStudio v0.99.896 with R v3.3.2). The distance between the middle positions and either the translation start site or the position of the stop codon of the corresponding mRNA was calculated. The distribution of obtained distances from the translation start site or the stop codon were visualized on a density plot using ggplot2 (v2.2.0). Code for distance calculation and plots is published on https://github.com/jadybea/Rio1p.
RESULTS
A subset of pre-40S AFs accumulate in gradient fractions containing 80S ribosomes when Rio1p is depleted
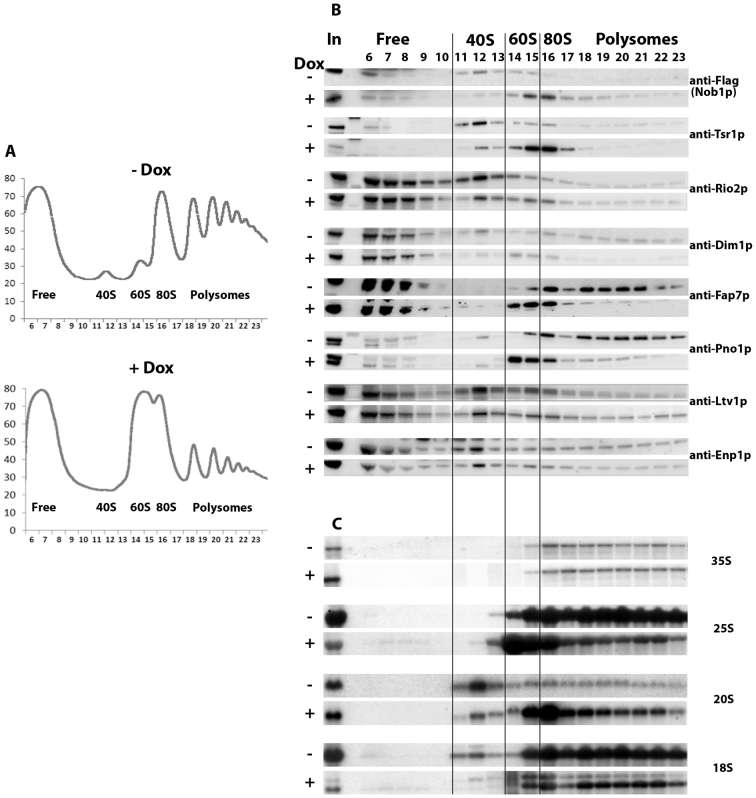
To investigate the effects of Rio1p depletion on the dynamics of association and dissociation of AFs with and from pre-40S particles, we fractionated extracts from TetO7-RIO1 cells grown in the absence (Rio1p expressed) or presence of doxycycline (Rio1p depleted) on sucrose gradients and assessed the sedimentation pattern of AFs by western blot using specific antibodies. Cycloheximide was added shortly before cell harvest and during extract preparation to stabilize the polysome complexes. Depletion of Rio1p leads to a striking decrease and increase in the 40S and 60S peaks, respectively and a strong reduction of polysomes (Figure 1A). Whereas Nob1p and Tsr1p sediment predominantly in the 40S fractions when Rio1p is present, they accumulate in 80S fractions when Rio1p is depleted (Figure 1B). A similar phenomenon is observed for Rio2p, although to a far lesser extent. Dim1p from non depleted extracts displays a complex sedimentation pattern, being present in light fractions, 40S, 60S, 80S and polysome fractions. The presence of Dim1p in the latter fractions may reflect its early incorporation in 90S pre-ribosomal particles. Depletion of Rio1p does not dramatically alter Dim1p sedimentation pattern, although the proportion of the protein in the 60S fractions increases while that in the 40S fractions decreases. Fap7p and Pno1p/Dim2p sediment in 80S and polysome fractions when extracted from Rio1p-expressing cells and become concentrated in 60S fractions and 80S fractions, when extracted from Rio1p-depleted cells. In contrast, the sedimentation pattern of Ltv1p and Enp1p is little affected by Rio1p depletion, suggesting that Rio1p is not essential for their association with and dissociation from pre-40S particles. Northern analysis shows that 20S pre-rRNA, the RNA component of pre-40S particles strongly increases in the 80S and polysome fractions when Rio1p is absent (Figure 1C). These results suggest that depletion of Rio1p leads to the stalling of a subset of AFs in pre-40S particles that co-sediment with 80S ribosomes and likely correspond to the previously described 80S-like particles (14,15). These particles contain at least Nob1p, Tsr1p, Dim1p, Pno1p/Dim2p, Rio2p and Fap7p. Such retention is most evident for Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p. Surprisingly, we also noted that Nob1p, Tsr1p, Fap7p and Pno1p accumulate in 60S fractions upon Rio1p depletion. This may reflect the presence of 80S-like particles in the 60S fractions due to insufficient separation of the 60S and 80S peaks. To determine whether this is the case, we repeated the fractionation experiment using slightly altered conditions to improve resolution (lower amounts of total extracts and higher centrifugation speed). This led to an improved resolution of the 60S and 80S peaks in the Rio1p-depleted conditions (Supplementary Figure S1A). Strikingly, in these conditions, the 20S pre-rRNA is present in the 60S fractions (12 and 13), as well as the AFs tested (Nob1p, Tsr1p and Fap7p), while little 18S rRNA is found in these fractions, showing that they contain few 80S ribosomes, if at all (Supplementary Figure S1B and C). These results lend support to the notion that Rio1p depletion leads to the accumulation of not only 80S-like particles but also to pre-40S particles that co-sediment with 60S particles, although we cannot totally rule out the possibility that the presence of 20S pre-rRNA in 60S fractions results from a spill over of the 40S and 80S fractions.
Figure 1.
Sucrose gradient fractionation of total extracts from a TetO7-RIO1, NOB1-FPZ strain grown in YPD or in YPD plus doxycycline for 16 h to repress RIO1 expression. (A) Recording of A254 nm profiles. Top: Rio1p-expressing cells (- Dox); bottom: Rio1p-depleted cells (+ Dox). The peaks of free proteins, free 40S and 60S subunits, 80S ribosomes and polysomes are indicated as well as fraction numbers. (B) Sedimentation profile of pre-40S particle AFs analyzed by western blot. Proteins were purified from the indicated fractions (top) of gradients loaded with extracts from Rio1p-expressing cells (–Dox) or Rio1p-depleted cells (+Dox). The antibodies used for the detection of specific factors are indicated. The anti-FLAG antibody allows the detection of tagged Nob1p. (C) Sedimentation profile of (pre)-rRNAs analyzed by northern. Same as in (B) except that RNAs were extracted from gradient fractions, separated by agarose gel electrophoresis in denaturing conditions, transferred to a nylon membrane and detected using various specific oligonucleotide probes.
Rio1p depletion leads to a significant increase of 20S pre-rRNA in polysome-containing fractions. Strikingly, none of the tested AFs show a clear increase in polysome fractions following Rio1p depletion, with the exception of Nob1p, suggesting that such depletion leads to the formation of a third type of pre-40S particles that can engage in translation and contain few AFs.
Ltv1p is underphosphorylated in Rio1p-depleted cells
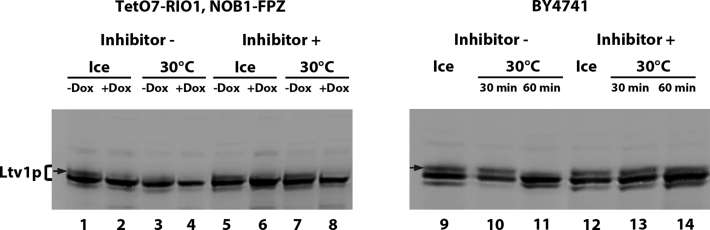
Ltv1p extracted from Rio1p-expressing cells, but not Rio1p-depleted cells, appears as a doublet in the 40S, 80S and polysome-containing gradient fractions (Figure 1B). As Ltv1p has been described as a phosphorylated protein, we reasoned that the upper band of the doublet may correspond to a (pluri)-phosphorylated form of Ltv1p, that is not formed when Rio1p is absent. To confirm this hypothesis, we treated Rio1p-expressing cell extracts with λ phosphatase and tested by western blot whether the upper Ltv1p band could still be detected. However, the incubation of the extract at 30°C led to the disappearance of the upper band, whether the phosphatase was added or not, probably due to the action of endogenous phosphatases present in the extract (data not shown). We thus decided to use phosphatase inhibitors to counteract the action of such endogenous phosphatases. Extracts from TetO7-RIO1 cells grown with or without doxycycline were incubated at 30°C in the presence or absence of phosphatase inhibitors. As seen previously, the 30°C incubation step without inhibitor led to the disappearance of the upper Ltv1p band (Figure 2, lane 3). Strikingly, the addition of phosphate inhibitors prevented the disappearance of the upper Ltv1p band (Figure 2, lane 7), strongly suggesting that it corresponds to (a) phosphorylated form(s) of Ltv1p. Since the strain used for these experiments (TetO7-RIO1) might over-express Rio1p when grown without doxycycline, leading to abnormal levels and/or forms of phosphorylated Ltv1p, we tested whether equivalent levels of differentially migrating forms of Ltv1p could also be detected in a standard laboratory strain (BY4741). Faster and slower migrating forms of Ltv1p could also be detected using a total BY4741 extract (Figure 2, lane 9), of similar relative intensities to those observed in the case of the TetO7-RIO1 strain. Again, the disappearance of the upper band after incubation at 30°C was prevented by addition of phosphatase inhibitors (Figure 2, lane 14). Collectively, these results strongly suggest that Rio1p is involved in the production of at least some phosphorylated form(s) of Ltv1p.
Figure 2.
Rio1p-dependent formation of phosphorylated form(s) of Ltv1p. Total protein extracts from TetO7-RIO1, NOB1-FPZ strain grown with or without doxycycline as indicated (lanes 1–8) or BY4741 (lanes 9–14) were incubated at 30°C for 30 min (lanes 3, 4, 7, 8, 10 and 13) or 1 h (lanes 11 and 14) or stored on ice as control (lanes 1, 2, 5, 6, 9 and 12), in the presence (lanes 5–8, 12–14) or absence (lanes 1–4, 9–11) of phosphatase inhibitors (50 mM β-glycerophosphate, 10 mM sodium fluoride, 2 mM sodium orthovanadate). The extracts were analyzed by western blot using anti-Ltv1p antibodies. The putative phosphorylated form(s) of Ltv1p are indicated by an arrow.
A subset of Nob1p-containing pre-40S particles are incorporated into polysomes when Rio1p is depleted
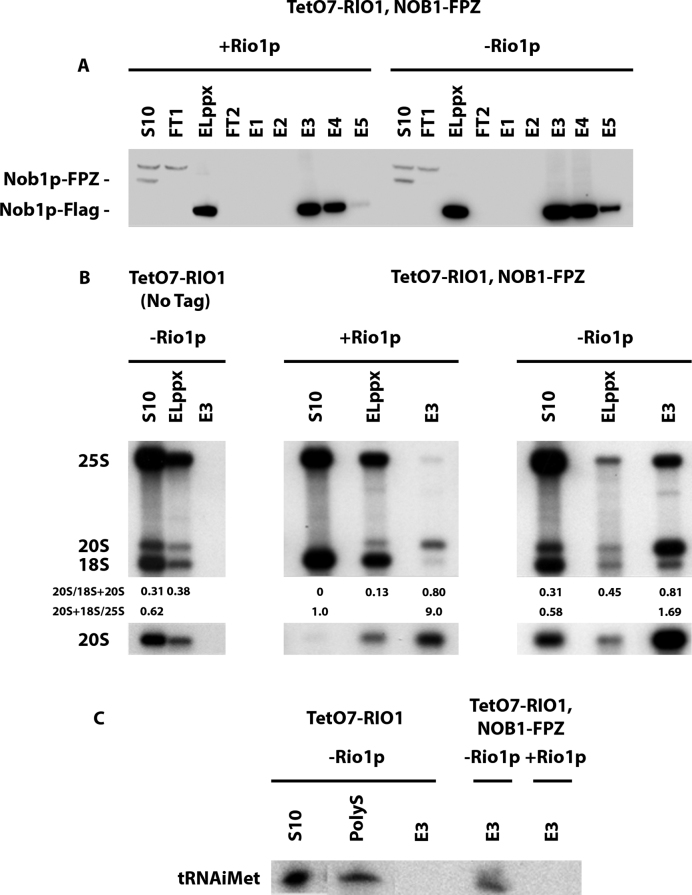
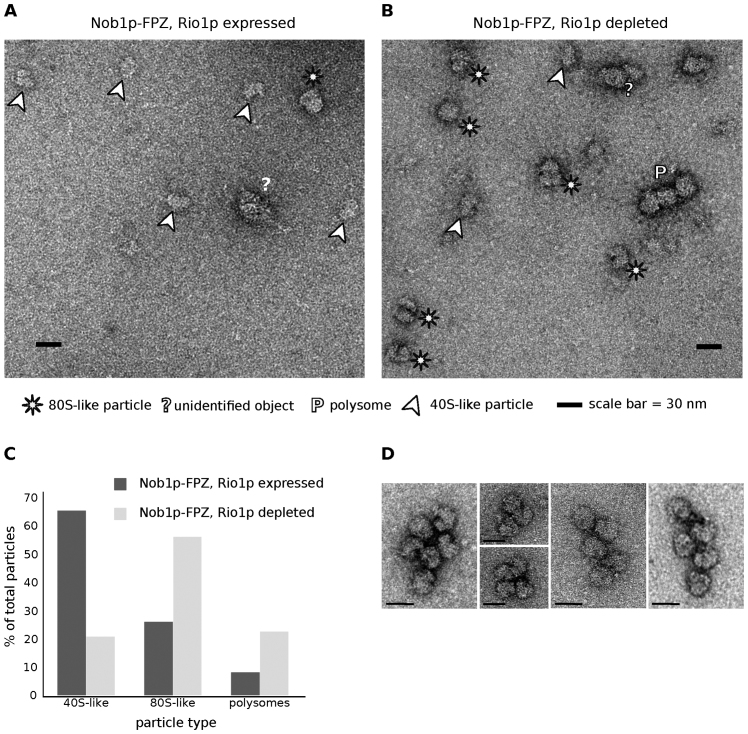
To further characterize the nature of pre-40S particles containing Nob1p that accumulate in Rio1p-depleted cells, we purified these particles by tandem affinity purification via Nob1p fused to the FPZ tag (Flag-PreScission cleavage site-ZZ) from cells containing or lacking Rio1p, under conditions of polysome stabilization (cycloheximide addition). Western blot analysis shows an equivalent recovery of Nob1p at the end of the purification procedure from extracts containing or lacking Rio1p (Figure 3A). 20S pre-rRNA is strongly enriched under both conditions (Figure 3B). Strikingly, 25S rRNA is recovered with Nob1p to a far greater extent from cells lacking Rio1p (Figure 3B). This is compatible with Nob1p being present in the previously described 80S-like particles (14,15), as well as in pre-40S particle-containing ribosomes formed on mRNAs undergoing translation. To investigate the second possibility, the presence of initiator tRNA (tRNAiMet) in the purified samples was tested by northern blot, since tRNAiMet is not present in the 80S-like particles in which pre-40S particles undergo quality control. Indeed, initiator tRNA was detected in the purified sample obtained from Rio1p-depleted cells, but not from cells expressing the protein (Figure 3C). To obtain more direct evidence for the presence of Nob1p-containing pre-40S particles in polysomes when Rio1p is depleted, the Nob1p-FPZ tandem affinity purification samples were imaged following negative staining by transmission electron microscopy. Images of purified samples obtained from Rio1p-expressing cells show mostly isolated particles of the size and shape of 40S subunits that most likely correspond to pre-40S particles (Figure 4A). In contrast, images of samples obtained from Rio1p-depleted extracts show a mixture of isolated particles the size and shape of 40S subunits (20%) or 80S ribosomes (60%), as well as concatenated 80S ribosomes (20%) (Figure 4B and C). These particles likely correspond to pre-40S particles, 80S-like ribosomes (pre-40S + 60S subunits) and polysomes, respectively. Indeed, the concatenated 80S ribosome structures are very similar to the polysomes observed by negative staining by Kopeina et al. (34) as well as Bonderoff et al. (35). The interpretation that the concatenated 80S ribosomes (Figure 4D) correspond to polysomes is reinforced by the finding that mRNAs can be detected by RT-PCR in the purified samples obtained from Rio1p-depleted extracts (Supplementary Figure S2). Since they were purified via Nob1p-FPZ, the detected polysomes must comprise at least one Nob1p-containing pre-40S particle. We conclude that Rio1p depletion leads to both the accumulation 80S-like particles as well as premature entry of Nob1p-containing pre-40S particles into translation.
Figure 3.
Tandem-affinity purification of Nob1p-containing pre-40S particles. Purification was performed using extracts from the TetO7-RIO1, NOB1-FPZ strain, grown in YPD (+ Rio1p) or in YPD plus doxycycline for 16 h (– Rio1p) to repress RIO1 expression. (A) Western analysis of tagged Nob1p purification. Aliquots from the initial extract (S10), flow-through of the first IgG-sepharose column (FT1), eluted sample from the IgG-sepharose affinity column (ELppx), flow-through of the second anti-Flag column (FT2), eluted samples from the anti-Flag column (E1–E5) were subjected to western analysis using anti-Flag antibodies. The positions of Nob1p-FPZ and Nob1p-Flag are indicated. (B) Northern analysis of 20S pre-rRNA, 18S and 25S rRNA levels in aliquots of the S10, ELppx and E3 samples from purification experiments performed with the TetO7-RIO1 strain grown in the presence of doxycycline as control (left), the TetO7-RIO1, NOB1-FPZ strain grown in the absence (middle) or presence (right) of doxycycline. Quantifications of the 20S/(18S+20S) and the (20S+18S)/25S ratios were performed by phosphorimager analysis. (C) Northern blot analysis of initiator tRNA levels in aliquots of the E3 anti-Flag affinity column elutions from the three purifications described in (B). Aliquots of S10 and polysomal gradient fraction (PolyS) are used as control.
Figure 4.
Negative staining electron microscopy images of Nob1p-containing particles obtained by tandem affinity purification from (A) TetO7-RIO1, NOB1-FPZ cells grown in YPD without doxycycline (Rio1p expressed), or (B) TetO7-RIO1, NOB1-FPZ cells grown in YPD plus doxycycline for 16 h (Rio1p depleted). Putative pre-40S particles (40S-like particles), 80S-like particles and polysomes are highlighted as indicated below the images. (C) Percentage of 40S-like, 80S-like particles and putative polysomes in Rio1p-expressing and Rio1p-depleted cells. (D) Examples of polysomes seen in images obtained from TetO7-RIO1, NOB1-FPZ cells grown in YPD plus doxycycline.
Nob1p-containing pre-40S particles engage in translation elongation
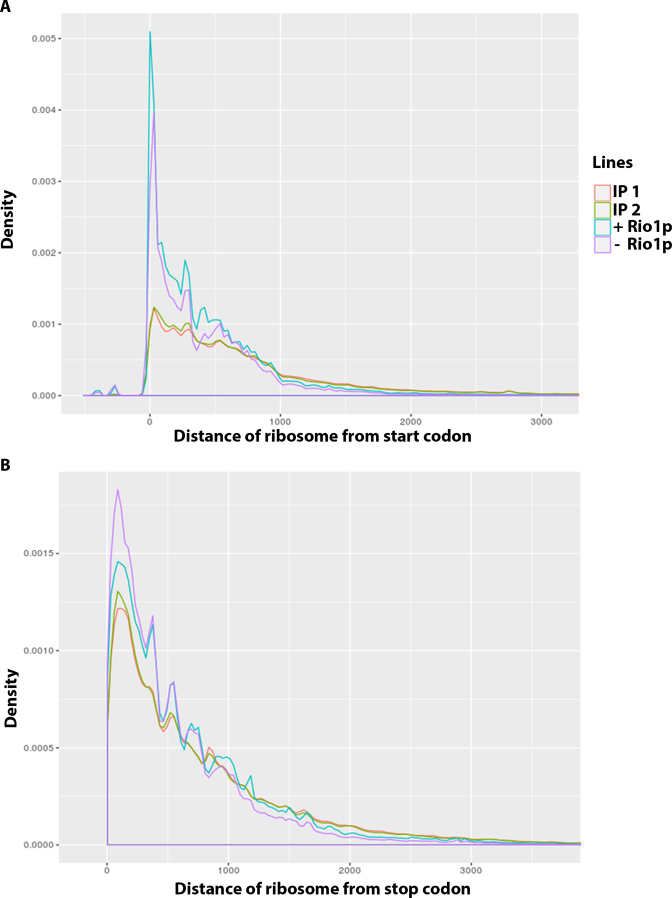
To determine whether Nob1p-containing pre-40S particles can carry out translation elongation, we adapted the ribosome profiling experiment protocol to map the mRNA fragments protected by ribosomes containing immature 40S particles (see Materials and Methods for details) (33,36). Briefly, the total extract obtained from TetO7-RIO1, NOB1-FPZ cells grown with doxycycline to repress Rio1p expression and harvested in the presence of cycloheximide to stabilize polysome complexes was treated with RNase I to digest mRNA regions not protected by translating ribosomes. Nob1p-containing ribosomes were then purified using IgG-sepharose beads. The purified RNAs were extracted and rRNA fragments present in the sample were eliminated using the ‘RiboZero’ procedure. The remaining 28–32 nt long RNA fragments (Ribosome Protected Fragments, RPFs) were then identified by NGS. As controls, total RPFs obtained using a standard ribosome profiling protocol from Rio1p-expressing or Rio1p-depeleted cells were also identified. Analysis of the reads corresponding to the immuno-purified RPFs does not point to a preferential association of Nob1p-containing ribosomes with a specific subset of mRNAs (data not shown). The distribution of total and purified RPFs with respect to the translation initiation codon was analyzed genome wide (Figure 5A). This analysis shows that purified RPFs globally display a density profile somewhat similar to that of total RPFs. Likewise, mapping of purified RPFs on the corresponding yeast coding DNA sequences shows that pre-40S particle-containing ribosomes are present throughout the body of mRNAs (Supplementary Figure S3). In addition, for several mRNAs, the density profile of purified RPFs follows that of total RPFs. These data strongly suggest that pre-40S particle-containing ribosomes can carry out translation elongation. Nevertheless, the genome-wide analysis (Figure 5A) also reveals that the ratio of purified RPFs positioned inside open reading frames relative to those placed close to the initiation codon is higher than in the case of total RPFs. For total RPFs of Rio1p-expressing and Rio1p-depleted cells, 16.17% and 13.77% of mapped reads were positioned in close vicinity of ±15 nucleotides to start codons, respectively. In contrast, only 4.61% and 4.21% of mapped reads of purified RPFs were localised in close vicinity to start codons. Moreover, we note that the density of Nob1p-containing ribosomes at the initiation codon of the open reading frames specifically analyzed is systematically lower than that of non-selected ribosomes (Supplementary Figure S3). Altogether, these findings may indicate that Nob1p-containing pre-40S particles carry out translation initiation at a reduced pace and/or that the kinetics of translation elongation of pre-40S particle-containing ribosomes are slower than those of wild-type ribosomes. In contrast to the distribution relative to the start codon, the RPF distribution relative to the stop codon is not significantly different between the total RPFs and purified RPFs (Figure 5B). In addition, no significant amount of purified RPFs were mapped to genomic regions following coding DNA sequences (data not shown). These data suggest that ribosomes containing pre-40S particles can recognise and dissociate at canonical stop codons.
Figure 5.
Genome-wide distribution relative to the start codon (A) or the stop codon (B) of total ribosome protected fragments (RPFs) from Rio1p-expressing cells (+ Rio1) or Rio1p-depleted cells (– Rio1) or immuno-purified RPFs via Nob1p-FPZ from Rio1p-depleted cells (two separate experiments termed IP1 and IP2).
DISCUSSION
The timing of Rio1p association with pre-40S particles and its mode(s) of action still remain ill defined. Analysis of pre-40S particles purified using Rio1p as bait suggests that in wild-type conditions, Rio1p is mostly associated with 80S-like particles containing Nob1p and Dim2p (12,22). Studies on human cells also demonstrate that hRio1 is present in late pre-40S particles containing hDim2/PNO1 and hNob1 (37). Our results indicate that lack of Rio1p leads to the retention of Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p within 80S-like particles. This strongly suggests that Rio1p joins 80S-like particles prior to or concomitant with Tsr1p dissociation and moreover, that Rio1p is required not only for Pno1p/Dim2p and Nob1p release as previously proposed (22,37), but also for Fap7p and Tsr1p dissociation. Rio1p and Tsr1p binding sites determined by CRAC are partially overlapping or very close in the structure of mature 18S rRNA (20,22,25) and a recent modelling study led to the conclusion that Tsr1p occludes Rio1p binding site (38). Hence our results are best explained by the proposal that Rio1p integration in pre-40S particles drives Tsr1p eviction. While our work was in progress, the distribution on sucrose gradients of AFs extracted from cells over-expressing a catalytically inactive version of Rio1p was determined (39). This analysis showed that the catalytic activity of Rio1p is required for efficient release of Nob1p, Tsr1p and Pno1p/Dim2p from 80S-like ribosomes (Fap7p was not analyzed). Unexpectedly, we observed that a sizable proportion of 20S pre-rRNA extracted from Rio1p-depleted cells sediments on sucrose gradients in fractions containing 60S subunits. Moreover, these fractions are also enriched for AFs Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p. It seems as if Rio1p depletion leads not only to the accumulation of 80S-like particles containing Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p, but also to that of particles co-sedimenting with 60S ribosomal subunits containing the very same AFs and the 20S pre-rRNA. The latter particles have not been described before and it remains a mystery, how they can acquire such sedimentation behavior. However, we note that a substantial amount of 20S pre-rRNA sediments in 60S-containing fractions when extracted from Tsr1p depleted cells (20), Fap7p depleted cells (15), from cells expressing the Rpl3 W255C mutant (28) or a catalytically inactive version of Rio1p (39). One possibility is that these unknown particles correspond to pre-40S particles stalled with translation initiation factors at the 48S pre-initiation complex stage on large ribosome-free mRNAs.
Our finding that the accumulation of (a) phosphatase-sensitive form(s) of Ltv1p is dependent upon the presence of Rio1p was unexpected since it suggested that Rio1p could be involved in Ltv1p phosphorylation events. Indeed, it has been proposed that Rio1p, like Rio2p, functions primarily as an ATPase rather than a kinase (16,39). Moreover, as discussed previously, it is currently thought that Rio1p associates with pre-40S particles after the dissociation of Ltv1p promoted by its phosphorylation by Hrr25p (17). Nevertheless, the possibility that Rio1p can function as a kinase in vivo cannot be ruled out, since Rio1p from C. thermophilum displays weak auto-phosphorylation activity in vitro (39). Of course, whether Rio1p is directly involved in Ltv1p phopshorylation remains to be determined. If that is the case, Rio1p must phosphorylate Ltv1p outside pre-40S particles, unless contrary to current thinking, Rio1p in fact associates with pre-40S particles prior to Ltv1p departure. Interestingly, our data suggest that such phosphorylated form(s) of Ltv1p can be maintained in pre-ribosomal particles since they are found in 40S gradient fractions (Figure 1).
In addition to 80S-like particles and pre-40S particles co-sedimenting with 60S ribosomal subunits that contain AFs Nob1p, Tsr1p, Pno1p/Dim2p and Fap7p, Rio1p-depleted cells also accumulate pre-40S particles that sediment in polysome-containing fractions. Our previous studies (29) and the present work clearly establish that the majority of these latter particles are engaged in translation. At least a subset of these translating pre-40S particles contain Nob1p, since Nob1p associates with mRNAs as well as the initiator tRNA (Supplementary Figure S2 and Figure 3C) and polysomes can be directly visualised by electron microscopy in tandem affinity purification samples obtained from Rio1p-depleted cells using Nob1p as bait (Figure 4). The presence of additional AFs in pre-40S particles engaged in translation remains unknown but is unlikely. Fap7p, Tsr1p and Pno1p/Dim2p are found in heavy gradient fractions when extracted from Rio1p-expressing cells, likely reflecting, at least for Tsr1p and Pno1p/Dim2p, their integration within early nuclear pre-ribosomal particles since these fractions contain 35S pre-rRNA. The sedimentation of these AFs in polysome-containing fractions is strongly reduced when they are extracted from Rio1p-depeleted cells, suggesting they are not part of pre-40S particles engaged in translation. This is fully in accordance with the hypothesis that the simultaneous binding of Tsr1p and Pno1p/Dim2p would prevent the interaction with translation initiation factors (20). Our data therefore suggest that while Rio1p depletion affects Tsr1p, Pno1p/Dim2p and Fap7p dissociation from 80S-like particles, the block is not complete and that some 80S-like particles manage to shed these assembly factors. The Nob1p-containing pre-40S particles that are released can enter translation. While initial structural work suggested that Nob1p position within early pre-40S particles blocks the association of translation initiation factor eIF3 (20), more recent data point to an inherent flexibility of Nob1p and the platform region on which it is bound (40), leading us to propose that some conformations adopted by Nob1p may allow eIF3 binding. The Nob1p-containing pre-40S particles that enter translation are unable to carry out D site cleavage, suggesting that Rio1p action is not restricted to late AF removal but also induces conformational changes within pre-40S particles that are required for cleavage to occur. In addition, or alternatively, translation may promote Nob1p dissociation as proposed by Garcia-Gomez et al., rendering D site cleavage impossible (28). They found that cells expressing the mutant ribosomal protein gene allele rpl3[W255C] display a very similar phenotype to the one of Rio1p-depleted cells, with accumulation of 20S pre-rRNA in 60S, 80S and polysome-containing fractions. Strikingly, they noticed that reducing translation by various means lowers 20S accumulation in rpl3[W255C] cells, hence the proposal that translation induces Nob1p loss. However, our data clearly indicate that translation does not lead to the dissociation of all Nob1p molecules from pre-40S particles.
Whether immature pre-40S particles could support translation elongation remained unclear until now. Garcia-Gomez and colleagues argued that pre-40S particles from rpl3[W255C] cells could support translation elongation since the 20S/18S and 20S/25S ratios remained constant for most polysomal fractions. This is also what we find for polysomal fractions obtained from Rio1p-depleted extracts, supporting the view that pre-40S particles from cells lacking Rio1p can also perform translation elongation. This conclusion is strongly reinforced by our mapping of mRNA fragments protected by pre-40S particle-containing ribosomes. Thus it appears that the presence of Nob1p and of 20S pre-rRNA instead of 18S rRNA does not block translation elongation, although our data suggest that the pace of translation initiation and/or elongation may be slowed. Our data argue that even if pre-40S particle-containing ribosomes move on mRNAs with altered kinetics, they are able to recognise and dissociate at canonical stop codons. Remarkably, it was recently proposed that immature large ribosomal subunits containing various 5.8S rRNA 3′extended precursors can also support translation elongation (41). Thus, the phenomenon of premature entry into translation is not restricted to small ribosomal subunit precursors.
Our previous results suggest that pre-40S particles containing Nob1p can enter translation even in wild-type cells. This conclusion stems from the findings that in wild-type conditions, a small amount of 20S pre-rRNA is found in polysome-containing fractions, associates with polyA-binding protein and translation initiation factors and that certain abundant mRNAs can be co-precipitated with Nob1p (29). Whether pre-40S particles can support translation elongation under wild-type conditions remains to be formally established but is likely in light of the present study. Pre-40S particles entering translation in wild-type cells may serve a regulatory purpose or correspond to aberrant pre-40S particles that have improperly shed most of their AFs and retain an incorrectly positioned Nob1p protein unable to process 20S pre-rRNA. Cytoplasmic steps of pre-40S particle maturation may be intrinsically error prone, in spite of quality control steps, such as 80S-like particle formation.
AVAILABILITY
Analysis of ribosome profile data: code for distance calculation and plots is published on https://github.com/jadybea/Rio1p. NGS analysis files of raw and processed data were deposited in Gene Expression Omnibus database under the accession number GEO: GSE98333.
Supplementary Material
ACKNOWLEDGEMENTS
With thank Katrin Karbstein for the gift of antibodies, Marlène Faubladier for plasmid construction, Natacha Pérébaskine and Sébastien Fribourg for the gift of purified proteins, Stéphanie Balor and Vanessa Soldan (METI platform, Toulouse) for technical support in electron microscopy, Yves Romeo and Jean-Paul Gélugne for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS); The University of Toulouse; Agence Nationale de la Recherche (ANR) [ANR-2010-BLAN-1224 to Y.H.]. Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L.. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004; 16:943–954. [DOI] [PubMed] [Google Scholar]
- 3. Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y.. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008; 65:2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woolford J.L. Jr, Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2016; aal1880. [DOI] [PubMed] [Google Scholar]
- 6. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R. et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 7. Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., Chen R., Wang B., Zheng S., Zhang C. et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017; 6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaker-Margot M., Hunziker M., Barandun J., Dill B.D., Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015; 22:920–923. [DOI] [PubMed] [Google Scholar]
- 9. Perez-Fernandez J., Roman A., De Las Rivas J., Bustelo X.R., Dosil M.. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007; 27:5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L., Wu C., Cai G., Chen S., Ye K.. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016; 30:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C.. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016; 44:5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hector R.D., Burlacu E., Aitken S., Le Bihan T., Tuijtel M., Zaplatina A., Cook A.G., Granneman S.. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014; 42:12138–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pertschy B., Schneider C., Gnadig M., Schafer T., Tollervey D., Hurt E.. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009; 284:35079–35091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Bottcher B., Granneman S., Watkins N.J., Tollervey D.. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012; 19:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strunk B.S., Novak M.N., Young C.L., Karbstein K.. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012; 150:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferreira-Cerca S., Sagar V., Schafer T., Diop M., Wesseling A.M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N.. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 2012; 19:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., Karbstein K.. Hrr25/CK1delta-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 2015; 208:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitterer V., Murat G., Réty S., Blaud M., Delbos L., Stanborough T., Bergler H., Leulliot N., Kressler D., Pertschy B.. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun. 2016; 7:10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zemp I., Wandrey F., Rao S., Ashiono C., Wyler E., Montellese C., Kutay U.. CK1delta and CK1epsilon are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 2014; 127:1242–1253. [DOI] [PubMed] [Google Scholar]
- 20. Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L. 3rd, Karbstein K., Skiniotis G.. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011; 333:1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schafer T., Maco B., Petfalski E., Tollervey D., Bottcher B., Aebi U., Hurt E.. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006; 441:651–655. [DOI] [PubMed] [Google Scholar]
- 22. Turowski T.W., Lebaron S., Zhang E., Peil L., Dudnakova T., Petfalski E., Granneman S., Rappsilber J., Tollervey D.. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 2014; 42:12189–12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellmich U.A., Weis B.L., Lioutikov A., Wurm J.P., Kaiser M., Christ N.A., Hantke K., Kotter P., Entian K.D., Schleiff E. et al. Essential ribosome assembly factor Fap7 regulates a hierarchy of RNA-protein interactions during small ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loc’h J., Blaud M., Réty S., Lebaron S., Deschamps P., Bareille J., Jombart J., Robert-Paganin J., Delbos L., Chardon F. et al. RNA mimicry by the fap7 adenylate kinase in ribosome biogenesis. PLoS Biol. 2014; 12:e1001860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Granneman S., Petfalski E., Swiatkowska A., Tollervey D.. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010; 29:2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larburu N., Montellese C., O’Donohue M.F., Kutay U., Gleizes P.E., Plisson-Chastang C.. Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res. 2016; 44:8465–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lacombe T., Garcia-Gomez J.J., de la Cruz J., Roser D., Hurt E., Linder P., Kressler D.. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol. Microbiol. 2009; 72:69–84. [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Gomez J.J., Fernandez-Pevida A., Lebaron S., Rosado I.V., Tollervey D., Kressler D., de la Cruz J.. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet. 2014; 10:e1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soudet J., Gélugne J.P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A.. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010; 29:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B.. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001; 24:218–229. [DOI] [PubMed] [Google Scholar]
- 31. Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gélugne J.P., Caizergues-Ferrer M.. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998; 17:7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lebaron S., Froment C., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y.. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005; 25:9269–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kopeina G.S., Afonina Z.A., Gromova K.V., Shirokov V.A., Vasiliev V.D., Spirin A.S.. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008; 36:2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonderoff J.M., Lloyd R.E.. Time-dependent increase in ribosome processivity. Nucleic Acids Res. 2010; 38:7054–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G. et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011; 147:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U.. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell. 2012; 23:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCaughan U.M., Jayachandran U., Shchepachev V., Chen Z.A., Rappsilber J., Tollervey D., Cook A.G.. Pre-40S ribosome biogenesis factor Tsr1 is an inactive structural mimic of translational GTPases. Nat. Commun. 2016; 7:11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferreira-Cerca S., Kiburu I., Thomson E., LaRonde N., Hurt E.. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 2014; 42:8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnson M.C., Ghalei H., Doxtader K.A., Karbstein K., Stroupe M.E.. Structural heterogeneity in pre-40S ribosomes. Structure. 2017; 25:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodriguez-Galan O., Garcia-Gomez J.J., Kressler D., de la Cruz J.. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol. 2015; 12:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.