Abstract
G-quadruplex (G4) can be formed by G-rich DNA sequences that are widely distributed throughout the human genome. Although G-triplex and G-hairpin have been proposed as G4 folding intermediates, their formation still requires further investigation by experiments. Here, we employed single-molecule FRET to characterize the folding dynamics of G4 from human telomeric sequence. First, we observed four states during G4 folding initially assigned to be anti-parallel G4, G-triplex, G-hairpin and unfolded ssDNA. Then we constructed putative intra-strand G-triplex, G-hairpin structures and confirmed their existences in both NaCl and KCl. Further studies revealed those structures are going through dynamic transitions between different states and show relatively weak dependence on cations, unlike G4. Based on those results and molecular dynamics simulations, we proposed a multi-pathway folding mechanism for human telomeric G4. The present work may shed new light on our current understanding about the existence and stability of G4 intermediate states.
INTRODUCTION
G4s are four-stranded non-canonical nucleic acids structures held together by Hoogsteen H-bonds, and further stabilized by monovalent cations such as potassium or sodium (1,2). G4 forming sequences are prevalent throughout the human genome, and especially enriched in gene promoters and telomere regions (3). Over 375 000 putative G4 forming sequences have been revealed in human genome by initial computational analyses (4,5). Recent high-resolution sequencing based method has identified 716 310 G4 structures in the human genome which is two times higher than the previous prediction (6), highlighting the importance of G4 in genome integrity. Indeed, the existence of G4 in living cell has been confirmed using an antibody which recognizes G4 with high affinity and specificity (7,8). Due to their essential biological functions in critical cellular processes, including initiation of DNA replication at the origin, regulation of DNA transcription, restart of collapsed replication fork, telomere maintenance and also anti-cancer drug targeting (9), G4s have attracted increased attentions in recent years.
The 3′ single-stranded overhang of human telomere consists of ∼30 tandem TTAGGG repeats which can fold into G4 with a variety of topologies (10). Nuclear magnetic resonance (NMR) and X-ray crystallographic studies have shown that, in the presence of Na+ human telomeric sequence d[AGGG(TTAGGG)3] folds dominantly into anti-parallel basket conformation (11); while in K+ buffer, parallel, hybrid and 2-tetrad anti-parallel conformations have been resolved (12–15). In addition to the dependence on ion type and concentration, the folding conformation and stability of G4 are also sensitive to other factors such as number of G-tetrad layers (16), loop length and sequence (17,18), molecular crowding reagent (19), 5′ and/or 3′ flanking DNA sequences which may interact by base stacking with the main quadruplex body (20).
The folding mechanism of human telomeric G4 in vitro is a long-standing question, and one debated issue is to identify the structure of different folding states. Although the dynamic conversion of G4 between different conformations have been suggested by several studies (21–25), evidences for a simple sequential folding with intermediates G-triplex and G-hairpin started to emerge by molecular dynamics (MD) (26), circular dichroism (CD), differential scanning calorimetry, isothermal titration calorimetry analyses (27–29), stopped-flow mixing (30), fluorescence emission and single-molecule methods (31–34). In particular, our earlier work by magnetic tweezers has identified G-triplex as an in-pathway intermediate in G4 (32). The step-wise G4 unfolding mediated by helicases Pif1 (35,36), BLM (37,38), FANCJ (39) and telomere binding protein POT1 (40) also implies there should be stable G4 folding intermediate states.
In addition to above circumstantial evidences, more direct evidences for the existence of those intermediate states emerged recently: (i) the atomic structure of G-triplex from thrombin binding aptamer has been resolved by NMR (41,42); (ii) inter-strand G-triplex/G-hairpin have been visualized in solution state by Atomic Force Microscope (31,34); (iii) using ultrafast microfluidic mixer, the hairpin formation was observed in the early stage of G4 structure (43); (iv) G-hairpin was predicted to participate in folding of human telomeric sequence quadruplexes by standard and T-REMD simulations (44,45) and real-time NMR spectroscopy (46). However, to the best of our knowledge, the conformational dynamics and stability of those intermediate states have never been investigated experimentally. In this regard, there are at least three fundamental questions. First, can intra-strand G-triplex/G-hairpin be detected at single-molecule level? Second, will those structures keep static or go through dynamic transitions? Third, what factors may affect the folding conformation and stability of those structures?
In this report, using single-molecule florescence resonance energy transfer (smFRET) (47), we first characterized the conformational dynamics of human telomeric G4 in NaCl and observed four folding states initially assigned to be anti-parallel G4, G-triplex, G-hairpin and ssDNA. Afterward, putative G-triplex and G-hairpin structures were constructed. We showed intra-strand G-triplex/G-hairpin can indeed be formed and undergo dynamic transitions between different states. Interestingly, the dominant population in G-triplex folding is G-hairpin instead of full G-triplex in NaCl, highlighting the stability of G-hairpin. Besides, the effects of cation type and concentration on G4, G-triplex and G-hairpin folding were carefully characterized. MD simulations further corroborate our assignments of the middle FRET states. In addition, MD simulations suggest hybrid G4 and 2-tetrad anti-parallel G4 may also participate in the folding of human telomeric G4. Accordingly, we proposed a multi-pathway folding mechanism for human telomeric G4. We also discovered the relative position of those structures to adjacent duplex DNA can disturb their folding conformation and stability significantly. Our results may deepen our understanding about G4 folding mechanism and its interaction with proteins in cell.
MATERIALS AND METHODS
DNA constructs
All oligonucleotides required to make the DNA substrates were purchased from Sangon Biotech (Shanghai, China). Sequences and labeling positions of all the oligonucleotides were listed in Supplementary Table S1. For DNA constructs used in smFRET measurements, DNA was annealed with 1:3 mixture of the stem and G4 or ssDNA strands by incubating the mixture at 95°C for 5 min, then slowly cooling down to room temperature in about 7 h. The strand without biotin was used in excess to reduce the possibility of having non-annealed strand anchored at the coverslip surface. The concentration of stem strand was 2.5 μM and all annealing were carried out in annealing buffer containing 20 mM Tris–HCl, pH 8.0 with different concentrations of NaCl or KCl.
Buffers
Buffers containing 20 mM Tris–HCl, pH 8.0 with different concentrations of NaCl or KCl were used. For single-molecule FRET measurements, 0.8% D-glucose, 1 mg/ml glucose oxidase (266600 units/g, Sigma), 0.4 mg/ml catalase (2000–5000 units/mg, Sigma) and 4 mM Trolox were added to prevent the photobleaching and photoblinking.
Single-molecule fluorescence data acquisition
Single-molecule FRET study was carried out with a home-built objective-type total-internal-reflection microscopy. Cy3 was excited by 532 nm Sapphire laser (Coherent Inc., USA). An oil immersion objective (100 ×, N.A.1.49) was used to generate an evanescent field of illumination. Fluorescence signal from Cy3 and Cy5 were split by a dichroic mirror, and finally collected by an electron-multiplying charge-coupled device camera (iXON, Andor Technology, South Windsor, CT, USA). The coverslips (Fisher Scientific, USA) and slides were cleaned thoroughly by a mixture of sulfuric acid and hydrogen peroxide, acetone and sodium ethoxide, then the surfaces of coverslip were coated with a mixture of 99% mPEG (m-PEG-5000, Laysan Bio, Inc.) and 1% of biotin-PEG (biotin-PEG-5000, Laysan Bio, Inc.). Streptavidin (10 μg/ml) were added to the microfluidic chamber made of the PEG coated coverslip, and incubated for 10 min. After washing, 50 pM DNA were added to the chamber and allowed to be immobilized for 10 min. Then free DNA was removed by washing with the imaging buffer (20 mM Tris–HCl, pH 8.0, different concentrations of NaCl or KCl, 0.8% D-glucose, 1 mg/ml glucose oxidase, 0.4 mg/ml catalase and 4 mM Trolox). We used an exposure time of 100 ms for all single-molecule measurements at a constant temperature of 22°C.
FRET data analyses
The FRET efficiency was calculated using IA/(ID+IA), where ID and IA represent the intensity of donor and acceptor respectively. The FRET value above 1 is due to background subtraction from very low intensity in the donor channel, giving rise to negative donor intensity. Basic data analysis was carried out by scripts written in Matlab, and all data fitting were generated by Origin 8.0. Single-molecule FRET histograms were generated by picking the initial 50–100 frames of each trace from ∼300 molecules, and fitted by multi-peak Guassian distributions, with the peak position unconstrained. Transition density plots (TDP) (48) were constructed from smFRET traces showing dynamic transitions using free available Hidden Markov Model (HMM) and TDP software from Professor Taekjip Ha’s website (http://bio.physics.illinois.edu/HaMMy.asp).
Circular dichroism spectropolarimetry
CD experiments were performed with a Bio-Logic MOS450/AF-CD optical system (BioLogic Science Instruments, France) equipped with a temperature controlled cell holder, using a quartz cell with 1-mm path length. A 2 μM solution of DNA was prepared in 20 mM Tris–HCl, pH 8.0 with different cation. The solution was incubated at 95°C for 5 min, then slowly cooled down to room temperature in about 7 h. CD spectra were recorded in the UV (220–320 nm) regions in 0.75 nm increments with an averaging time of 2 s at 25°C.
Gel electrophoresis assay
A total of 12 nt poly-T sequence was labeled by FAM fluorophore at 3′ end for visualization (T12-FAM, Supplementary Table S1). DNA for gel assay was annealed with 1:1 mixture of T12-FAM and 3G2 or 4G2 sequences by incubating the mixture at 95°C for 5 min in 100 mM NaCl or KCl, then slowly cooling down to room temperature in about 7 h. Afterward both T12-FAM and annealing products were subjected to 15% native PAGE (polyacrylamide gel electrophoresis), run at room temperature and visualized by Gel imaging analysis system (Gel Doc-It310, UVP, USA).
MD simulations
MD simulations and available volume (AV) simulations were performed to calculate the FRET efficiencies for several DNA constructs (25,49). To get the initial conformations for AV simulations, we carried out all-atom MD simulations for anti-parallel G4, hybrid-2 G4, 2-tetrad anti-parallel G4, G-triplex and G-hairpin using GROMACS5.0.6 with AMBER99SB-ILDN force field (50). The solvent (water model: TIP3P) and 100 mM KCl were added to build a charge-neutral system. We chose KCl as G4 was known to be more stable when the central ion is K+ compared with Na+ (17). All systems were energy-minimized and equilibrated successfully. MD simulations were performed at 1 bar and 298 K with Parrinello-Rahman pressure coupling (51) and Velocity-rescaling temperature coupling schemes (52). For each system, the simulations were run for 100 ns according to previous studies (25,44).
The constructs of anti-parallel G4 (2MCO.pdb), hybrid-2 G4 (2JSL.pdb) and 2-tetrad anti-parallel G4 (2KF8.pdb) were taken from RCSB Protein Data Bank. The anti-parallel G4 structure in 2MCO is highly consistent with 143D which has been chosen by several earlier MD simulation studies (44,53). G-triplex and G-hairpin were constructed from the mutation of anti-parallel G4 according to the sequences in Supplementary Table S1. Relevant nucleotides of anti-parallel G4 were replaced with dTs, and energy-minimization was run for 100 ps before further MD simulations. The double-stranded DNA was built in Nucleic Acid Builder.
Available volume simulations
The AV simulations were performed to calculate the FRET efficiency for each system (49). Frames for AV simulations were extracted from the MD simulation trajectories by every 10 ps, implying that there are 10 000 frames for each of the five systems. The parameters for Cy3/Cy5 dye pairs are listed in Supplementary Table S2. During AV simulations, the atom O3′ of the last nucleotide in the G4 constructs was selected as the attachment point for Cy3 and the methyl group carbon atom C7 of T6 in the complementary stand was selected as the attachment point for Cy5. Hydrogen atoms which were attached to the O3′ and C7 atoms were deleted before AV simulations. The Förster radius was set as 60 Å (47).
RESULTS
Human telomeric G4 displays four folding states in NaCl
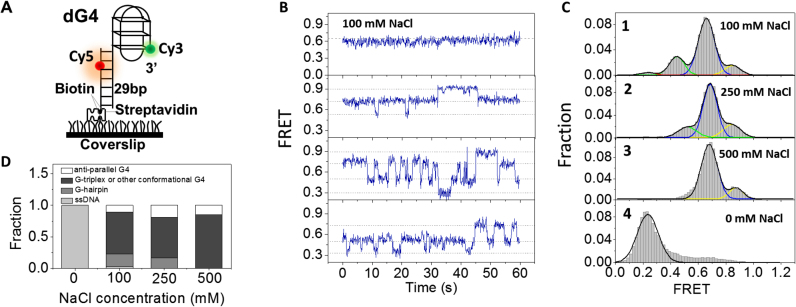
To understand how G4 structures are folded at single-molecule level, we first characterized the conformational dynamics of G4 with human telomere repeats in NaCl by smFRET. The substrate was constructed with an ssDNA sequence containing donor fluorophore (Cy3) attached to the 3′ end of G4 motif, and its 5′ tail was hybridized with a complementary stem strand modified by biotin at the 3′ end for immobilization and attached by an acceptor fluorophore (Cy5) at the sixth nucleotide from the 5′ end (Figure 1A, referred as dG4). The fluorophores were so spaced that FRET signal can sensitively report the conformational change of G4. DNA was surface-immobilized using a biotin-streptavidin bridge onto a polyethylene glycol passivated microscope coverslip, then buffer solution containing both 20 mM Tris–HCl, pH 8.0 with different concentrations of NaCl and oxygen scavenger system was flowed into the chamber.
Figure 1.
Conformational dynamics of G4 at the 3′ end of duplex DNA in NaCl. (A) Schematic representation of experimental set-up. (B) Representative smFRET traces of dG4 in 100 mM NaCl. (C) FRET histograms of dG4 in 20 mM Tris–HCl, pH 8.0 buffer containing different concentrations of NaCl. Multi-peak and single-peak Gaussian distributions were used to fit those histograms. (D) Fractions of different folding structures at increasing concentrations of NaCl, obtained from the area of individual Gaussian peaks in (C).
First, we examined G4 structures in 20 mM Tris–HCl, pH 8.0 with 100 mM NaCl. CD spectrum in Supplementary Figure S1A confirms G4 formation with dGGG[TTAGGG]3 sequence according to previous report (11). In Figure 1B and Supplementary Figure S2, smFRET traces show that individual dG4 is either being static during 1 min (∼30%) or going through dynamic transitions between 3 and 4 states (EFRET ∼ 0.9, 0.7, 0.5 and 0.3), consistent with Noer et al. (24). The two dimensional TDP from dynamic smFRET traces provides information about the transition frequency between different FRET states (48). Supplementary Figure S3A shows that, the most frequent transitions are between E0.7 and E0.5, and transitions between E0.9 and E0.5 are less frequent. There are also a small amount of transitions between E0.5 and E0.3. However, due to the low probability, direct transitions between E0.9 and E0.7 in Figure 1B and Supplementary Figure S2 were not able to show up in TDP figure.
The constructed FRET histogram from above 300 molecules can be well fitted by multi-peak Gaussian distribution with three major peaks at E0.85, E0.66, E0.44 and one tiny peak at E0.24 (Figure 1C, panel 1). The FRET distribution of dG4 in 20 mM Tris–HCl without any salt (Figure 1C, panel 4) suggests the minor peak at E0.24 should be completely unfolded G4, as G4 cannot be formed in the absence of monovalent cation. According to previous smFRET study (18), the peak at E0.85 here is attributed to anti-parallel G4 due to the maximum proximity of two fluorophores. Regarding to the two middle peaks, here we temporarily designated them to be folding intermediates G-hairpin and G-triplex (or other G4 folding conformation such as hybrid structure) and will be discussed in detail later.
One may query that, in several early smFRET studies, only three states have been identified in G4 folding (21,22,54). We think this discrepancy might be due to the different position of fluorescence labeling: in those studies, Cy5 was labeled at longer distance away from Cy3, therefore unfolded state and G-hairpin may be mixed together due to the insensitivity of FRET efficiency at low FRET range. Indeed, four FRET states have been observed in NaCl with similar DNA substrate and fluorophore labeling positions by Noer et al. (24), in agreement with our observations.
We noticed even in 100 mM NaCl, only ∼10% dG4 molecules can be folded into anti-parallel G4 (E0.85 in Figure 1C, panel 1). To examine whether high concentration of cation can stabilize G4, we further increased NaCl concentration to 250 mM. Multi-peak Gaussian fitting of FRET histograms generates three peaks at E0.84, E0.68 and E0.52 respectively (Figure 1C, panel 2). Even in extreme high concentration of NaCl (500 mM), the FRET histograms still consist of two peaks at E0.87 and E0.68 (Figure 1C, panel 3). Above findings suggest: (i) with the increase of NaCl concentration, unfolded ssDNA and G-hairpin disappear gradually, however the fraction of anti-parallel G4 does not have much variation, and G-triplex (or other G4 folding conformation) is still the dominant population (Figure 1D); (ii) according to the fraction of dG4 keeping static during 1 min (64% in 500 mM NaCl, compared to 30% in 100 mM NaCl), we think high concentration of NaCl can indeed stabilize those related structures.
Formation of G-triplex with human telomere repeats in NaCl
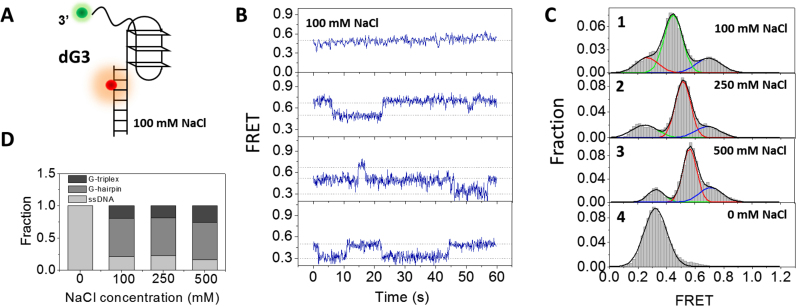
We were wondering whether the two middle peaks in panel 1 of Figure 1C indeed include G-hairpin and G-triplex. For this purpose, the formation of G-triplex was testified first. As CD might provide useful diagnostic evidence for DNA conformation (55), we measured the CD spectrum of GGGTTAGGGTTAGGG sequence in buffer containing 100 mM NaCl. There are two major positive peaks around 255 and 290 nm (Supplementary Figure S1D), different from CD spectrum of both 3G4 (Supplementary Figure S1A) and non-structured ssDNA (Supplementary Figure S1C). The positions of peaks are consistent with the result from Koirala et al. with the same DNA sequence and similar buffer condition (56). Therefore, results in Supplementary Figure S1D may suggest the formation of higher order structures for 3G3 in NaCl. However, triplexes DNA do not have a characteristic CD spectrum like the A-form, Z-form or tetraplexes (55), additional methods are still required to identify those structures. Then mutations were introduced to G4 forming sequence GGGTTAGGGTTAGGGTTAGGG with one G-column replaced by TTT which was further characterized by smFRET (Figure 2A, referred as dG3). In Figure 2B and Supplementary Figure S3B, smFRET traces and TDP map show that in 100 mM NaCl, individual dG3 is either being static during 1 min (∼42%) or going through dynamic transitions between 2 and 3 states (EFRET ∼ 0.7, 0.5 and 0.3). FRET distribution consists of three Gaussian peaks at E0.26, E0.45 and E0.70 respectively (Figure 2C, panel 1), very close to the FRET values of the three lower EFRET peaks in G4 folding (E0.24, E0.44 and E0.66). However, the peak at E0.85 in dG4 folding was missing here, further confirming the peak at E0.85 in Figure 1C should be well-folded G4.
Figure 2.
Conformational dynamics of G-triplex at the 3′ end of duplex DNA in NaCl. (A) Design of dG3 substrate. (B) Representative traces of dG3 in 100 mM NaCl. (C) FRET histograms of dG3 in 20 mM Tris–HCl, pH 8.0 buffer containing different concentrations of NaCl. Multi-peak and single-peak Gaussian distributions were used to fit those histograms. (D) The fractions of different folding structures at increasing concentrations of NaCl.
FRET distribution of dG3 in 20 mM Tris–HCl without any salt contains only one Gaussian peak at E0.3 (Figure 2C, panel 4), suggesting the formation of G-triplex is also disfavored in the absence of monovalent cation. Besides, this result further confirms the peak at E0.26 in 100 mM NaCl is the completely unfolded G-triplex. To examine whether high concentration NaCl can stabilize G-triplex, we increased NaCl concentration to 250 and 500 mM (Figure 2C, panels 2 and 3). However, FRET distribution has no significant change: the well-folded G-triplex only occupies a small fraction (Figure 2D). In addition, we noticed the peaks slightly shift to higher FRET values with the increasing of NaCl concentration in Figure 2C. The possible reason might be high concentration of cation reduces the electrostatic interaction in the 6-nt ssDNA and increases its flexibility, according to previous study (57).
Altogether, results from dG3 folding suggest: (i) G-triplex structure can be formed in 100 mM NaCl and the FRET value indicates the peak at E0.7 in dG4 folding may include G-triplex; (ii) dG3 is unstable, going through transitions between G-triplex, G-hairpin and ssDNA; (iii) the dominant population of dG3 in 100–500 mM NaCl might be G-hairpin at E0.5 rather than G-triplex, indicating the folding of G-hairpin is more favored in the buffer condition we used.
In addition to dG3, formation of G-triplex may also take place at 3′ end of G4 sequence. Then we constructed a substrate named dG3* with TTTTTTGGGTTAGGGTTAGGG (Supplementary Figure S4A). In 100 mM NaCl, individual dG3* is either being static or going through dynamic transitions similarly as dG3 (Supplementary Figure S4B). FRET distribution consists of 3 Gaussian peaks at E0.27 (28.9%), E0.43 (46.9%) and E0.67 (24.2%) respectively (Supplementary Figure S4C), slightly different from that of dG3 at E0.26 (20.9%), E0.45 (60.0%) and E0.70 (19.1%). Therefore, G-triplex can be formed at either 5′ or 3′ end of G4 sequence, but they may not be easily distinguished just by FRET efficiency.
Ion-dependent G4 and G-triplex folding
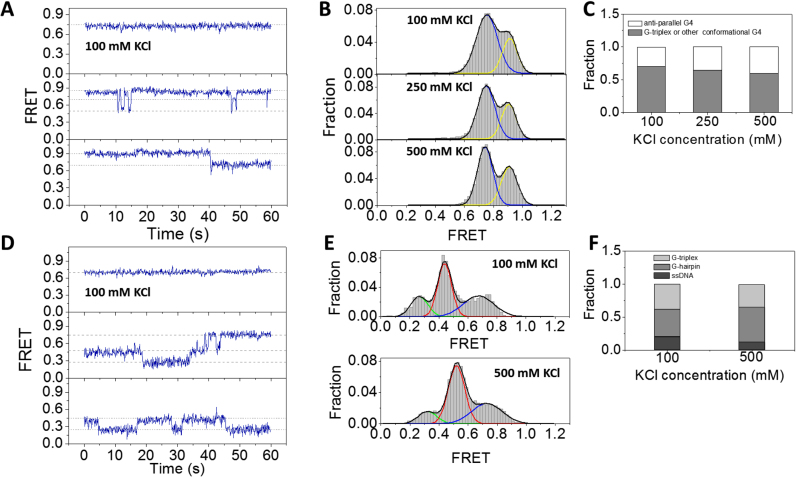
As G4 was known to be more stable when the central ion is K+ compared with Na+ (17), we next employed similar methods to characterize the folding dynamics of G4 in KCl. CD spectrum in Supplementary Figure S1B confirms the formation of G4 structure with dGGG[TTAGGG]3 sequence according to previous report (14,27). In 100 mM KCl, majority of dG4 are keeping static in 1 min, although transitions between two, three and even four states can also be occasionally observed (Figure 3A and Supplementary Figure S5). The FRET histogram displays two peaks at E0.75 and E0.91 (Figure 3B, upper panel). According to the analysis in 100 mM NaCl, we assume peak at E0.91 to be anti-parallel G4, and peak at E0.75 to be folding intermediate state or other G4 conformation. Although in previous structural studies, parallel, hybrid and 2-tetrad anti-parallel G4 structures have been reported for telomeric repeat sequence in KCl (12–15), the presence of proximal duplex DNA may alter the folding conformations significantly. Compared with the results in NaCl (Figure 1B and C), dG4 is more stable in 100 mM KCl as most molecules are being static and no populations of unfolded G4 and G-hairpin can be observed (Figure 3B). With the increasing of KCl concentration from 100 to 500 mM, there is no significant change in the FRET distribution (Figure 3B and C), indicating 100 mM KCl should be enough to stabilize the G4 structure.
Figure 3.
Conformational dynamics of G4 and G-triplex at 3′ end of duplex DNA in KCl. (A) Representative traces of dG4 in 100 mM KCl. (B) In 100–500 mM KCl, the FRET histograms of dG4 display two peaks at E0.9 and E0.75. (C) The fractions of different folding structures at increasing concentrations of KCl. (D) Representative traces of dG3 in 100 mM KCl. (E) FRET histograms of dG3 in 20 mM Tris–HCl, pH 8.0 buffer containing different concentrations of KCl. Multi-peak Gaussian distributions were used to fit those histograms. (F) The fractions of different folding structures at increasing concentrations of KCl.
We also examined the folding of G-triplex in KCl. CD spectrum of GGGTTAGGGTTAGGG sequence in KCl buffer shows two major positive peaks at ∼265 and 285 nm (Supplementary Figure S1E), therefore also suggesting the formation of higher order DNA structures. Individual smFRET traces of dG3 in 100 mM KCl show both static state (∼52%) and transitions between different states (Figure 3D and Supplementary Figure S3C). FRET histogram has three peaks at E0.27, E0.45 and E0.67 which should be completely unfolded ssDNA, G-hairpin and G-triplex (Figure 3E, upper panel). When KCl concentration was further increased to 500 mM, the folding of dG3 has no much variation (Figure 3E, lower panel). Comparative analysis between Figures 2D and 3F imply in the presence of KCl, the fractions of G-triplex are slightly higher than that in NaCl. The fractions of dG3 keeping in static states in 1 min (52% in KCl and 42% in NaCl) also indicate K+ is able to stabilize G-triplex structure, however the contrast is not as much as that for dG4.
Formation of G-hairpin with human telomere repeats
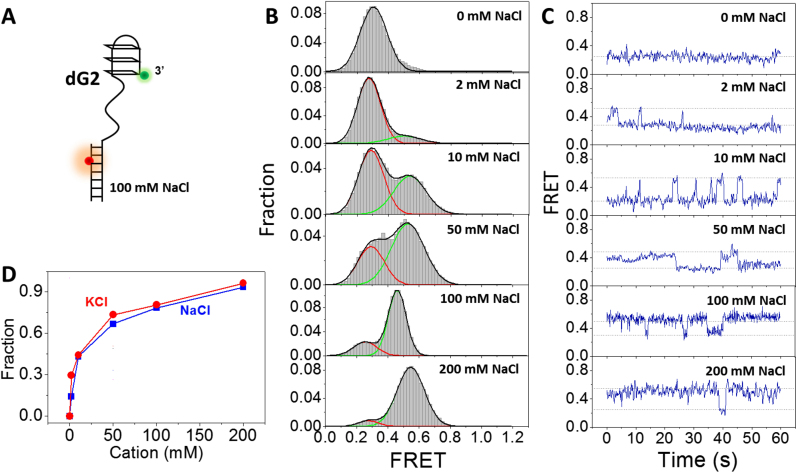
Above results suggest G-hairpin might be formed in G4 and G-triplex folding in both NaCl and KCl. CD spectrum of GGGTTAGGG sequence also suggest the formation of higher order DNA structures but different from G4 and G-triplex (Supplementary Figure S1F and G). Therefore, we would like to know whether the formation of G-hairpin can indeed be captured at single-molecule level. For this purpose, two adjacent G-columns in GGGTTAGGGTTAGGGTTAGGG were both replaced by TTT (Figure 4A, referred as dG2). Afterward, the formation of G-hairpin was examined by increasing NaCl concentration from 0 to 200 mM (Figure 4B). Without NaCl, dG2 exists in unfolded ssDNA form at E0.3. Surprisingly, we found with the increasing of NaCl concentration, a second peak at E0.5 shows up which is supposed to be G-hairpin. In 200 mM NaCl, dG2 predominantly folds into G-hairpin. The FRET value of G-hairpin (∼0.5) is consistent with that in Figures 1C, 2C and Supplementary Figure S4C, further supporting G-hairpin participates in the folding of both G4 and G-triplex. The selected FRET traces in Figure 4C and TDP in Supplementary Figure S3D demonstrate transitions between two distinct states, and with the increasing of NaCl concentration, dG2 prefers to stay at the higher FRET state. Those evidences together strongly support the formation of G-hairpin in NaCl.
Figure 4.
Formation of G-hairpin at the 3′ end of ss/dsDNA. (A) Design of dG2 substrate. (B) FRET distributions of dG2 in 0–200 mM NaCl. Single-peak and two-peak Gaussian distributions were used to fit those histograms. (C) Selected FRET traces of dG2 in 0–200 mM NaCl. (D) Fractions of G-hairpin in different concentrations of NaCl and KCl.
The dependence of G-hairpin formation on cation type has also been carefully characterized. The FRET distributions of dG2 were measured at a series of KCl concentrations. Supplementary Figure S3E shows the transitions between ssDNA and hairpin states in 100 mM KCl. Similar as G-hairpin folding in NaCl, with the increasing of KCl concentration, the fraction of G-hairpin increases (Figure 4D). However, it is only slightly higher than that in NaCl. Therefore, G-hairpin can be formed in KCl and NaCl, but the dependence on cation type is less significant compared to G4, consistent with the simulation results that no major difference can be found in KCl and NaCl for G-hairpin (44). In addition to Na+ and K+, the effect of Mg2+ on G-hairpin stability has also been examined. In 20 mM Tris–HCl, pH 8.0 containing no NaCl or KCl, with the increasing of MgCl2 from 0 to 10 mM, the fractions of folded G-hairpins increase significantly (Supplementary Figure S6A and B), indicating Mg2+ is able to stabilize G-hairpins, in agreement with the results from Rajendran et al. (31).
To make sure the increase in FRET value in dG2 indeed reflects the formation of G-hairpin, two control experiments have been performed. (i) the FRET distributions of a partial duplex DNA dT20 with poly-T overhang which cannot fold into higher order structures were measured at a series of KCl concentrations (Supplementary Figure S7). It can be clearly seen that with the increasing of KCl concentration there is no appearance of additional FRET band, suggesting no formation of higher order structures. The slight shift of FRET distribution to higher value is due to the increased electronic screening with increase of ion concentrations (57). (ii) hairpins formed by G-T base pairs between poly-T linker and G-rich part has also been ruled out by gel electrophoresis assay. A 12-nt poly-T sequence was labeled by FAM fluorophore at 3′ end for visualization (Supplementary Figure S8A). If poly-T and G-rich sequences can form stable base pairs, the annealed products between T12-FAM and 3G2 will migrate slower in native PAGE. Based on Supplementary Figure S8B and C, there are unlikely stable base pairs between poly-T and G-rich sequences in either 100 mM NaCl or KCl.
We further investigated whether the formation of G-hairpin can be more stabilized with GGGGTTAGGGG sequence (d4G2, Supplementary Figure S9A). In both 100 mM NaCl and 100 mM KCl, the behaviors of individual d4G2 molecules show similar pattern as dG2 (Supplementary Figure S9B and C). The FRET distributions in both 100 mM NaCl and KCl include one major Gaussian peak at ∼E0.5 and one minor peak at ∼E0.3, which should be G-hairpin and ssDNA respectively (Supplementary Figure S9D and E). Based on the FRET distributions (G-hairpin occupies ∼87% for d4G2 and ∼80% for dG2), we think the stability of d4G2 is slightly higher than dG2.
MD simulations
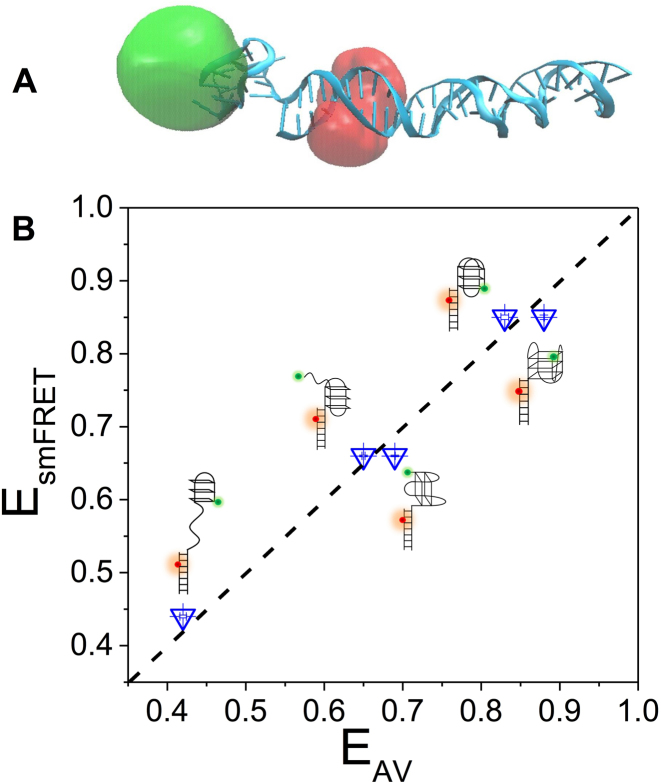
To further identify the observed FRET states in dG4, we performed MD simulations with structures of different G4 conformations and intermediate states respectively (Figure 5A and Supplementary Figure S10). Afterwards, we employed AV approach to assess the FRET efficiency of those simulated DNA structures. Figure 5B shows the simulated transfer efficiencies (EAV) for anti-parallel G4, G-triplex and G-hairpin match well with smFRET experiments (EsmFRET), further supporting our previous assignments of different FRET states for dG4.
Figure 5.
Molecular dynamics simulations. (A) Selective structure of anti-parallel G4 adjacent to duplex DNA. Donor and acceptor positions were obtained using AV approach and shown as clouds. (B) FRET efficiencies obtained from smFRET experiments (EsmFRET) and AV calculations based on the MD simulations (EAV). Standard errors from Gaussian fitting to obtain the peak values of EsmFRET and EAV are <0.01. The diagonal dashed line indicates the identity line.
All three types of G4 keep being folded during the simulation process. Anti-parallel and hybrid G4 are predominantly positioned at one side of duplex rather than right on top (Supplementary Figure S10). The direction of G-tetrad plane is almost perpendicular to the long axis of duplex. It has been proposed anti-parallel G4 may have higher FRET efficiency than other G4 (18). However due to the steric hindrance between G4 and duplex, as well as the structural asymmetry of G4, FRET efficiency of hybrid G4 is almost consistent with anti-parallel G4 as shown by AV simulations (Figure 5B), suggesting both anti-parallel and hybrid G4 may contribute to the E0.9 state. As for 2-tetrad anti-parallel G4, the structure is predominantly tilted relative to duplex, and G-tetrad plane is almost parallel to long axis of duplex. The FRET efficiency is lower than anti-parallel and hybrid G4, but quite close to G-triplex (Figure 5B), therefore may also contribute to the E0.7 state. It is worth to note in the work by Aznauryan et al. (25), there is an additional nucleotide between G4 motif and duplex which can make more spatial freedom for G4 relative to duplex. Therefore, in that study G4 structure is predominantly positioned on top of the duplex displayed by MD simulations, while in current study G4 prefers to be at one side of duplex instead of on the top possibly due to the steric hindrance as G4 is right adjacent to duplex. Supplementary Figure S11 shows once hybrid G4 is on the top of duplex occasionally by free motion, the first base pair in duplex can even be disrupted. Therefore, the additional nucleotide may result in very different G4-duplex structures, affect the Cy3-Cy5 distance and then change the FRET value of each folding state.
As for G-triplex, we observed some subtle structural differences during the simulation. For instance, GGGTTAGGGTTAGGG sequence forms three layered triplex by well-organized G:G:G plane at some time; while the G-triad plane seems deformed to some extent at another time (Supplementary Figure S12). Nevertheless, G-triplex structure keeps being folded without disruption and the orientation of G-triad plane tends to be almost perpendicular to duplex (Supplementary Figure S10).
In the simulation process, we also discovered the polymorphism for G-hairpin structures (Supplementary Figure S13). At 10 and 94 ns, G-hairpin has three G-G Hoogsteen base pairs. However, it appears to only have two G-G base pairs at 45 and 75 ns, and there is even one transient T-G base pair at 45 ns. However, no hairpins of pure G-T base pairs have been observed. Therefore, G-hairpin may have different forms based on different base paring.
Taken together, MD simulations not only corroborate our previous assignments of FRET states, but also imply hybrid and 2-tetrad anti-parallel G4 may participate in the folding of human telomeric G4. MD simulations also provide more detailed structural information about G-triplex and G-hairpin than smFRET experiment.
Adjacent DNA disturbs G-quadruplex folding
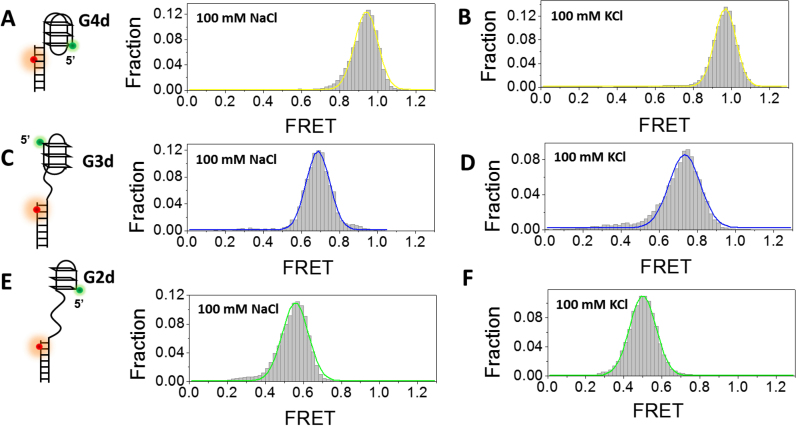
In addition to dG4, G4 structure may also locate at the 5′ end of duplex DNA resembling an ongoing synthesis of lagging strand stalled by G4 (35). To examine the effects of duplex DNA position on G4 folding, a 17-bp partial duplex DNA with Cy3 at the 5′ end of the ssDNA overhang harboring G4 forming sequence and Cy5 at the fifth nucleotide from the 3′ end of the complementary strand was designed (referred as G4d, Figure 6A, left panel). In either 100 mM NaCl or KCl, smFRET distribution of G4d only has one peak at E0.95 (Figure 6A and B), significantly different from dG4 in same buffer condition. As Cy5 was labeled one base closer to G4/duplex junction in G4d, the FRET value is slightly higher than dG4. Based on the high FRET value, we speculate G4 sequence may possibly fold into stable anti-parallel and/or hybrid form when linked directly to the 5′ end of duplex DNA.
Figure 6.
Formation of G4, G-triplex and G-hairpin at the 5′ end of a duplex DNA in 100 mM NaCl or KCl. (A) smFRET histograms of G4d in 100 mM NaCl has 1 peak at E0.94. (B) smFRET histograms of G4d structures in 100 mM KCl has 1 peak at E0.96. (C) smFRET histograms of G3d structures in 100 mM NaCl has 1 peak at E0.69. (D) smFRET histograms of G3d structures in 100 mM KCl has 1 peak at E0.73. (E) smFRET histograms of G2d structures in 100 mM NaCl has 1 peak at E0.56. (F) smFRET histograms of G-hairpin structures in 100 mM KCl has 1 peak at E0.50.
To eliminate the influences of adjacent duplex DNA on G4 folding, we have designed another kind of substrates, in which the G4 motif and duplex DNA was separated by a 12-nt ssDNA linker. Such a G4 structure may be formed in the telomerase-mediated reverse transcription. Supplementary Figure S14 displays the FRET distributions of two DNA substrates dsG4 (G4 at the 3′ end of the ss/ds DNA) and G4sd (G4 at the 5′ end of the ss/ds DNA). In either 100 mM NaCl or KCl, the FRET histogram of dsG4 differs significantly from G4sd. Once again, those results suggest the folding of G4 is dependent on the polarity, i.e. which terminal of G4 (3′ or 5′) is linked to the partial duplex DNA. Therefore, we think detailed DNA structures should be taken into account when assessing the stabilities of those structures.
Finally, we placed G-triplex and G-hairpin at the 5′ end of a partial duplex DNA (referred to as G3d and G2d). In 100 mM NaCl or KCl, single populations at ∼E0.7 (G3d) and ∼E0.5 (G2d) have been observed (Figure 6C–F) which should be completely folded G-triplex and G-hairpin. Therefore, the stability of G-triplex/G-hairpin may also be dependent on adjacent DNA environment.
DISCUSSION
In this report, using smFRET method, we have shown that human telomeric G4 structure undergoes repetitive transitions between four different states in Na+ buffer. To testify whether the two intermediate states include G-triplex and G-hairpin hypothesized by a lot of recent studies, we constructed DNA substrates derived from G4 sequences which may fold into higher order structures. Investigation of the conformational dynamics and stability of those substrates show G-triplex and G-hairpin can indeed be formed in both Na+ and K+ buffer.
G4 structure folds and unfolds in a multi-pathway
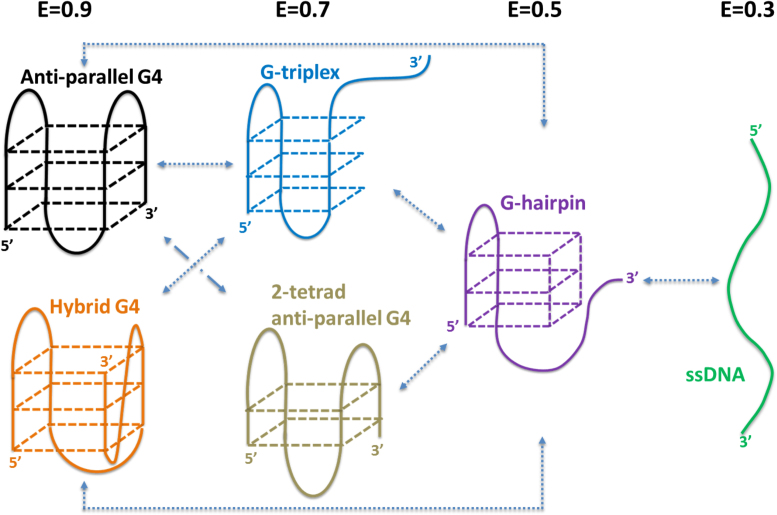
Based on smFRET experiments and MD simulations, here we propose a mechanism for the folding of human telomeric G4 at the 3′ end of duplex DNA resembling the telomere structure (Figure 7). First, in either Na+ or K+ buffer, anti-parallel G4 unfolds into ssDNA with G-triplex and G-hairpin as intermediate states. That is why direct conversions of E0.9-E0.7-E0.5 can be observed (Supplementary Figures S2 and 5). 2-tetrad anti-parallel G4 may also be converted directly from anti-parallel G4 to hairpin as hypothesized by previous study (15). Second, since hybrid G4 has been suggested to form with hairpin and triplex intermediates (26,29), we think conversion from hybrid G4 to G-triplex and G-hairpin may also be included in E0.9-E0.7-E0.5 transition. However, direct conversion from hybrid G4 to 2-tetrad anti-parallel G4 should be inhibited due to the spatial orientation of G-column. Third, there are a large amount of indirect transitions between E0.9-E0.7 via E0.5 (∼80% in all the transitions between E0.9 and E0.7). This kind of transition could happen between anti-parallel G4 (or hybrid G4), G-hairpin and 2-tetrad anti-parallel G4 (or G-triplex), suggesting G-hairpin may play an important role in the interconversion of different G4 conformations. In addition, we speculate 2-tetrad anti-parallel G4 might be the main conformation adopted by the GGG(TTAGGG)3 sequence in E0.7 state according to the moderate stability of G-triplex (Figures 2C, 3E and Supplementary Figure S4C). Therefore, the folding dynamics of human telomeric G4 might be rather complicated. It includes not only the step-wise folding of ssDNA, G-hairpin and G-triplex, but also the interconversion of G4 between different conformations.
Figure 7.
Proposed folding pathway of human telomeric G4 at 3′ end of duplex DNA.
It should be noted that intra-molecular G4 folds into a variety of topologies depending on cation, G-tract length, loop length and sequence, the presence of 5′ and 3′ flanking sequences, therefore we think our model might only be applied to human telomeric G4 at 3′ terminal of duplex DNA, but is less likely to be a uniform mechanism valid for all sequences and conditions.
Four G4 folding states have also been detected by Aznauryan et al. in a similar smFRET study which are attributed to be anti-parallel G4, parallel/anti-parallel hybrid, 2-tetrad basket and ssDNA (25). However, stable G-triplex has not been detected in their work possibly due to following reasons: (i) low concentration of KCl was used. As even in 100 mM KCl, only ∼37% dG3 molecules may fold into G-triplex (Figure 3E), the population might be much lower in 25 mM KCl therefore difficult to detect; (ii) different experimental strategy was employed. In the work by Aznauryan et al., single molecules were initially imaged in 25 mM LiCl for 100 s, then buffer with 25 mM KCl was manually injected to induce folding. As no change in transfer efficiency was observed upon buffer exchange, the authors were not able to detect folded structures. In present study, dG3 molecules were prepared by denaturing at 95°C for 5 min in 100 mM NaCl or KCl to make sure all DNA transform to unfolded form, then slowly cooling down to room temperature in about 7 h which was commonly used in G4 studies (14,30). We think this procedure may assist DNA to adopt high order structures.
Dynamic folding/unfolding of G-triplex and G-hairpin
Using DNA origami method and AFM imaging, Rajendran et al. have captured images of solution-state structures of a tetra-molecular anti-parallel and (3+1)-type G4 intermediates, such as G-hairpin and G-triplex with nanometer precision (31). In this report, we have further confirmed the formation of G-hairpin and G-triplex by smFRET in both NaCl and KCl. Importantly, those G-hairpin and G-triplex are intra-molecular folding structures which should be more biological relevant. We have observed the dynamic transitions of both G-triplex and G-hairpin for the first time. The majority transition rates between G4, triplex, hairpin and unfolded states are in the order of 10−2 s (Supplementary Table S3). G-hairpins were observed to occupy the majority population in triplex folding in NaCl, highlighting the stability of G-hairpin structure. In our previous study of BLM-catalyzed G4 unfolding with similar experimental design, at low concentration of BLM and ATP, G4 structure has been shown to unfold partially into hypothesized G-hairpin structure at E0.5 (37), consistent with the FRET value of G-hairpin here. Werner and RecQ helicases also displayed similar properties to unfold G4 structure to a stable state at E0.5 (unpublished results). In addition, RHAU was reported to only partially disrupt G4 structure by Tippana et al. (58). Therefore, we think G-hairpin can be a stable obstacle for those specialized helicases in G4 unfolding process.
Effects of proximal DNA environments on G4 folding conformation and stability
Previous biophysical stability studies indicate that formation of G4 and i-motif conformations do destabilize proximal duplex DNA structures (59). On the other side, addition of 3′ or 5′ nucleotides to G4 sequences also strongly affects the folding conformation and stability of G4s as assessed by NMR, UV-spectroscopy and CD (15,16,60,61). The influences of duplex DNA on G4 structure have been characterized by several studies. For instance, in quadruplex-duplex hybrids quadruplex stability generally increases with the length of the incorporated stem loop (60), and in quadruplex-duplex junctions, a pocket can be formed providing a potential molecular target (61). Here, we further discovered the proximal duplex DNA position as well as the nature of the sequences linking G4 and duplex motifs have impact on the folding conformation and stability of G4. When G4 is linked directly to the 5′ end of duplex DNA (G4d), it may fold into stable anti-parallel and/or hybrid form, while linked to the 3′ end of duplex DNA (dG4), it may fold into a variety of states, and go through dynamic transitions. However, when G4 is linked by ssDNA to duplex, G4 at 3′ end of ss/dsDNA (dsG4) prefers higher FRET state compared with G4sd, suggesting different folding conformations. As for G-triplex and G-hairpin, they both show higher stability when linked to the 5′ end of ss/dsDNA.
It is worth mentioning that, usually in smFRET studies G4 sequence of interest was tethered to duplex DNA for immobilization. Formation of a G4-duplex conjugate may change the stability and folding conformation of G4 significantly as shown in current study. Furthermore, we also discovered incorporation of additional ssDNA tail to G4 motif for enzyme loading in G4 unwinding assays will lead to further alteration of G4 structure (37). For instance, dG4 folds into two species at ∼E0.9 and E0.7 in 100 mM KCl as shown in Figure 3B, however dG4 with 12 nt ssDNA tail at 3′ end displays mainly one peak at ∼E0.9 in our previous study on BLM-catalyzed G4 unfolding (37), indicating the E0.7 state is disfavored with the existence of 3′ ssDNA tail. Therefore, the folding of G4 is not only sensitive to the relative position of G4 to duplex but also the detailed adjacent DNA context. In the future, we will place G4 structures in different DNA environments to systematically investigate the effects of duplex DNA, ssDNA, adjacent G4 and fork structure on G4 folding conformation and stability in more complex condition.
Based on above analysis, we think the kinetic and thermodynamic properties obtained from G4-duplex in smFRET experiments might be different from the simple untethered G4 in ensemble studies. In addition, although G4 structures have been well characterized by NMR in recent years, it needs to be cautious to extrapolate G4 structures from simple sequences based on NMR to complex G4 derivatives when it is linked to duplex or ss/dsDNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wei Li in the Institute of Physics, Chinese Academy of Sciences and Dr Xing-Hua Zhang in Wuhan University for critical reading of the manuscript. We also thank Xi’s laboratory members for insightful discussions. The Research was conducted within the context of the International Associated Laboratory ‘Helicase-mediated G-quadruplex DNA unwinding and Genome Stability’. The MD simulations were carried out at National Supercomputer Center in Tianjin, and the calculations were performed on TianHe-1 (A).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31370798, 11574252]; Chinese Universities Scientific Fund [Z109021504, Z109021704, Z109021718]; National Key R&D Program of China [2016YFA0301500]. Funding for open access charge: College of Life Sciences, Northwest A&F University; National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENECS
- 1. Lane A.N., Chaires J.B., Gray R.D., Trent J.O.. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008; 36:5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balasubramanian S., Neidle S.. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009; 13:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neidle S., Parkinson G.N.. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003; 13:275–283. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d AG(3)(T(2)AG(3))3 G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 12. Dai J.X., Carver M., Yang D.Z.. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008; 90:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T.. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009; 131:4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Dai J., Veliath E., Jones R.A., Yang D.. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010; 38:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 17. Guedin A., Gros J., Alberti P., Mergny J.L.. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010; 38:7858–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tippana R., Xiao W.K., Myong S.. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014; 42:8106–8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue Y., Kan Z.Y., Wang Q., Yao Y., Liu J., Hao Y.H., Tan Z.. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007; 129:11185–11191. [DOI] [PubMed] [Google Scholar]
- 20. Arora A., Nair D.R., Maiti S.. Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 2009; 276:3628–3640. [DOI] [PubMed] [Google Scholar]
- 21. Lee J.Y., Okumus B., Kim D.S., Ha T.J.. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:18938–18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Long X., Parks J.W., Bagshaw C.R., Stone M.D.. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013; 41:2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. You H.J., Zeng X.J., Xu Y., Lim C.J., Efremov A.K., Phan A.T., Yan J.. Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 2014; 42:8789–8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noer S.L., Preus S., Gudnason D., Aznauryan M., Mergny J.L., Birkedal V.. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016; 44:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aznauryan M., Sondergaard S., Noer S.L., Schiott B., Birkedal V.. A direct view of the complex multi-pathway folding of telomeric G-quadruplexes. Nucleic Acids Res. 2016; 44:11024–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H.. Folding pathways of human telomeric Type-1 and Type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010; 132:14910–14918. [DOI] [PubMed] [Google Scholar]
- 27. Boncina M., Lah J., Prislan I., Vesnaver G.. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012; 134:9657–9663. [DOI] [PubMed] [Google Scholar]
- 28. Gray R.D., Buscaglia R., Chaires J.B.. Populated Intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 2012; 134:16834–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gray R.D., Trent J.O., Chaires J.B.. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014; 426:1629–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang A.Y.Q., Balasubramanian S.. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012; 134:19297–19308. [DOI] [PubMed] [Google Scholar]
- 31. Rajendran A., Endo M., Hidaka K., Sugiyama H.. Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew. Chem. 2014; 53:4107–4112. [DOI] [PubMed] [Google Scholar]
- 32. Li W., Hou X.M., Wang P.Y., Xi X.G., Li M.. Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures. J. Am. Chem. Soc. 2013; 135:6423–6426. [DOI] [PubMed] [Google Scholar]
- 33. Koirala D., Ghimire C., Bohrer C., Sannohe Y., Sugiyama H., Mao H.B.. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013; 135:2235–2241. [DOI] [PubMed] [Google Scholar]
- 34. Rajendran A., Endo M., Hidaka K., Teulade-Fichou M.P., Mergny J.L., Sugiyama H.. Small molecule binding to a G-hairpin and a G-triplex: a new insight into anticancer drug design targeting G-rich regions. Chem. Commun. 2015; 51:9181–9184. [DOI] [PubMed] [Google Scholar]
- 35. Hou X.M., Wu W.Q., Duan X.L., Liu N.N., Li H.H., Fu J., Dou S.X., Li M., Xi X.G.. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015; 466:189–199. [DOI] [PubMed] [Google Scholar]
- 36. Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T.. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014; 3:e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu W.Q., Hou X.M., Li M., Dou S.X., Xi X.G.. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015; 43:4614–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chatterjee S., Zagelbaum J., Savitsky P., Sturzenegger A., Huttner D., Janscak P., Hickson I.D., Gileadi O., Rothenberg E.. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 2014; 5:doi:10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C.G., Spies M.. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016; 44:8742–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hwang H., Buncher N., Opresko P.L., Myong S.. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012; 20:1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cerofolini L., Amato J., Giachetti A., Limongelli V., Novellino E., Parrinello M., Fragai M., Randazzo A., Luchinat C.. G-triplex structure and formation propensity. Nucleic Acids Res. 2014; 42:13393–13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Limongelli V., De Tito S., Cerofolini L., Fragai M., Pagano B., Trotta R., Cosconati S., Marinelli L., Novellino E., Bertini I. et al. The G-triplex DNA. Angew. Chem. Int. Ed. Engl. 2013; 52:2269–2273. [DOI] [PubMed] [Google Scholar]
- 43. Li Y., Liu C., Feng X.J., Xu Y.Z., Liu B.F.. Ultrafast microfluidic mixer for tracking the early folding kinetics of human telomere G-quadruplex. Anal. Chem. 2014; 86:4333–4339. [DOI] [PubMed] [Google Scholar]
- 44. Stadlbauer P., Kuhrova P., Banas P., Koca J., Bussi G., Trantirek L., Otyepka M., Sponer J.. Hairpins participating in folding of human telomeric sequence quadruplexes studied by standard and T-REMD simulations. Nucleic Acids Res. 2015; 43:9626–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luo D., Mu Y.G.. Computational insights into the stability and folding pathways of human telomeric DNA G-quadruplexes. J. Phys. Chem. B. 2016; 120:4912–4926. [DOI] [PubMed] [Google Scholar]
- 46. Bessi I., Jonker H.R.A., Richter C., Schwalbe H.. Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem. Int. Ed. Engl. 2015; 54:8444–8448. [DOI] [PubMed] [Google Scholar]
- 47. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat. Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McKinney S.A., Joo C., Ha T.. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006; 91:1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sindbert S., Kalinin S., Nguyen H., Kienzler A., Clima L., Bannwarth W., Appel B., Muller S., Seidel C.A.. Accurate distance determination of nucleic acids via Forster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011; 133:2463–2480. [DOI] [PubMed] [Google Scholar]
- 50. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E.. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010; 78:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parrinello M., Rahman A.. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981; 52:7182–7190. [Google Scholar]
- 52. Bussi G., Donadio D., Parrinello M.. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007; 126:014101. [DOI] [PubMed] [Google Scholar]
- 53. Stadlbauer P., Trantirek L., Cheatham T.E. 3rd, Koca J., Sponer J.. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie. 2014; 105:22–35. [DOI] [PubMed] [Google Scholar]
- 54. Ying L.M., Green J.J., Li H.T., Klenerman D., Balasubramanian S.. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14629–14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kypr J., Kejnovska I., Renciuk D., Vorlickova M.. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009; 37:1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koirala D., Mashimo T., Sannohe Y., Yu Z.B., Mao H.B., Sugiyama H.. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. 2012; 48:2006–2008. [DOI] [PubMed] [Google Scholar]
- 57. Murphy M.C., Rasnik I., Cheng W., Lohman T.M., Ha T.J.. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004; 86:2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tippana R., Hwang H., Opresko P.L., Bohr V.A., Myong S.. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konig S.L.B., Huppert J.L., Sigel R.K.O., Evans A.C.. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013; 41:7453–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lim K.W., Khong Z.J., Phan A.T.. Thermal stability of DNA quadruplex-duplex hybrids. Biochemistry. 2014; 53:247–257. [DOI] [PubMed] [Google Scholar]
- 61. Krauss I.R., Ramaswamy S., Neidle S., Haider S., Parkinson G.N.. Structural insights into the quadruplex-duplex 3΄ interface formed from a telomeric repeat: a potential molecular target. J. Am. Chem. Soc. 2016; 138:1226–1233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.