Abstract
The Clustered Regularly Interspaced Palindromic Repeats (CRISPR)-CRISPR-associated proteins (Cas) systems of bacterial and archaeal adaptive immunity show multifaceted evolutionary relationships with at least five classes of mobile genetic elements (MGE). First, the adaptation module of CRISPR-Cas that is responsible for the formation of the immune memory apparently evolved from a Casposon, a self-synthesizing transposon that employs the Cas1 protein as the integrase and might have brought additional cas genes to the emerging immunity loci. Second, a large subset of type III CRISPR-Cas systems recruited a reverse transcriptase from a Group II intron, providing for spacer acquisition from RNA. Third, effector nucleases of Class 2 CRISPR-Cas systems that are responsible for the recognition and cleavage of the target DNA were derived from transposon-encoded TnpB nucleases, most likely, on several independent occasions. Fourth, accessory nucleases in some variants of types I and III toxin and type VI effectors RNases appear to be ultimately derived from toxin nucleases of microbial toxin–antitoxin modules. Fifth, the opposite direction of evolution is manifested in the recruitment of CRISPR-Cas systems by a distinct family of Tn7-like transposons that probably exploit the capacity of CRISPR-Cas to recognize unique DNA sites to facilitate transposition as well as by bacteriophages that employ them to cope with host defense. Additionally, individual Cas proteins, such as the Cas4 nuclease, were recruited by bacteriophages and transposons. The two-sided evolutionary connection between CRISPR-Cas and MGE fits the “guns for hire” paradigm whereby homologous enzymatic machineries, in particular nucleases, are shuttled between MGE and defense systems and are used alternately as means of offense or defense.
Keywords: CRISPR-Cas systems, mobile genetic elements, casposons, CRISPR adaptation, CRISPR effector modules
Introduction
Thanks to the striking success of the Cas9 endonucleases as new generation of genome editing tools, in recent years, comparative genomics, structures, biochemical activities and biological functions of Clustered Regularly Interspaced Palindromic Repeats (CRISPR)-CRISPR-associated proteins (Cas) systems and individual Cas proteins have been explored with an unprecedented intensity (Sorek etal. 2013; Barrangou and Marraffini 2014; Doudna and Charpentier 2014; Hsu etal. 2014; Mohanraju etal. 2016; Wright etal. 2016; Barrangou and Horvath 2017; Komor etal. 2017; Koonin etal. 2017). The CRISPR-Cas are adaptive (acquired) immune systems with memory of past encounters with foreign DNA that is stored in unique spacer sequences derived from viral and plasmid genomes and inserted into CRISPR arrays. Transcripts of the spacers, along with portions of the surrounding repeats, are utilized as guide CRISPR (cr)RNAs to recognize the cognate sequences in foreign genomes and thus direct Cas nucleases to their unique cleavage sites.
Because CRISPR-Cas are programmable immune systems that can adapt to target any sequence, they are not subject to extreme diversifying selection that led to the evolution of the immense variety of restriction-modification enzymes, the most abundant form of innate immunity in prokaryotes (Pingoud etal. 2016). Nevertheless, CRISPR-Cas systems evolve in a regime that is common to all defense system, namely continuous arms race with genetic parasites, primarily viruses, resulting in rapid evolution of at least some cas gene sequences (Takeuchi etal. 2012). Furthermore, the notable diversity of the gene compositions and genomic architectures of the CRISPR-cas loci translates into diversification of the molecular mechanisms of defense (Makarova etal. 2011, 2015).
The CRISPR-Cas belong to the class of nucleic acid-guided defense systems, along with eukaryotic RNAi and prokaryotic Argonaute-based machinery (Hutvagner and Simard 2008; Shabalina and Koonin 2008; Hur etal. 2014; Swarts etal. 2014; Koonin 2017). Unlike the Argonaute systems and most of the forms of the eukaryotic RNAi but similarly to the piRNA branch of RNAi, CRISPR-Cas mediates bona fide adaptive immunity (Makarova etal. 2006; Koonin and Makarova 2009; van der Oost etal. 2009; Marraffini and Sontheimer 2010). The CRISPR-cas genomic loci are modified to target the genome of a unique pathogen or its closest relatives with exceptional specificity and efficiency. These loci typically consist of a CRISPR array, that is, from several to several hundred direct, often partially palindromic, exact repeats (25–35 bp each), separated by unique spacers (typically, 30–40 bp each) and the adjacent cluster of multiple cas genes that are organized in one or more operons. The CRISPR-Cas immune response consists of three stages: 1) adaptation, 2) expression, and 3) interference. At the adaptation stage, a distinct complex of Cas proteins binds to a target DNA molecule, migrates along that molecule and, typically after encountering a distinct, short (2–4 bp) motif known as Protospacer-Adjacent Motif (PAM), cleaves out a portion of the target DNA, the protospacer, and inserts it into the CRISPR array between two repeats (most often, at the beginning of the array) so that it becomes a spacer (Amitai and Sorek 2016; Jackson etal. 2017). Some CRISPR-Cas systems possess an alternative mechanism of adaptation, namely spacer acquisition from RNA via reverse transcription by a reverse transcriptase (RT) encoded in the CRISPR-cas locus (Silas etal. 2016; Silas etal. 2017). At the expression stage, the CRISPR array is transcribed into a single, long transcript, the pre-crRNA, that is processed into mature crRNAs, each consisting of a spacer and a portion of an adjacent repeat, by a distinct complex of Cas proteins or a single, large Cas protein (Charpentier etal. 2015; Hochstrasser and Doudna 2015) (and see below). At the final, interference stage, the crRNA that typically remains bound to the processing complex is employed as the guide to recognize the protospacer or a closely similar sequence in an invading genome of a virus or plasmid that is then cleaved and inactivated by a Cas nuclease (Plagens etal. 2015; Nishimasu and Nureki 2017). The CRISPR-Cas systems modify the genome content in response to an environmental cue (an invader genome) and store the memory of such encounters, allowing them to efficiently and specifically protect the host from the same or related parasites. Accordingly, these systems are often regarded as an “evolvability device” implementing Lamarckian-type inheritance (Koonin and Wolf 2009, 2016). This brief description is an oversimplified schematic that inevitably omits many important details of CRISPR-Cas functioning. Such details can be found in many recent reviews on different aspects of CRISPR-Cas biology (Sorek etal. 2013; Barrangou and Marraffini 2014; Doudna and Charpentier 2014; Hsu etal. 2014; Charpentier etal. 2015; Hochstrasser and Doudna 2015; Plagens etal. 2015; Amitai and Sorek 2016; Mohanraju etal. 2016; Wright etal. 2016; Barrangou and Horvath 2017; Jackson etal. 2017; Komor etal. 2017; Koonin etal. 2017; Nishimasu and Nureki 2017).
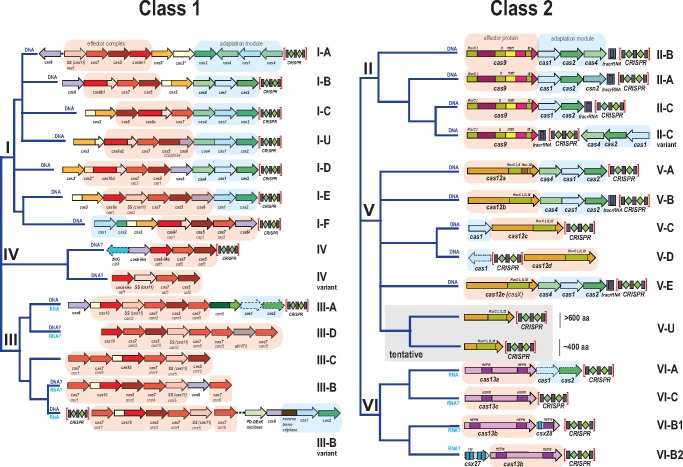
At the molecular level, the CRISPR-Cas systems possess a readily definable modular organization (Makarova etal. 2013a; Makarova etal. 2015). The two principal parts of the CRISPR-Cas systems are the adaptation and effector modules that consist, respectively, of the suites of genes encoding proteins involved in spacer acquisition (adaptation) and genes encoding Cas proteins involved in pre-crRNA processing that is followed by the target recognition and cleavage (interference). In most of the CRISPR-Cas systems, the adaptation module consists of the Cas1 and Cas2 proteins that form a complex, in which Cas1 is the endonuclease (integrase) involved in the cleavage of both the source, protospacer-containing DNA and the CRISPR array, whereas Cas2 forms the structural scaffold (Nunez etal. 2014, 2015; Wang etal. 2015; Amitai and Sorek 2016). In many variants, additional Cas proteins, such as Cas4 or Cas3 also contribute to the adaptation stage, in some cases forming fusions with Cas1 or Cas2 (Li etal. 2014; Kunne etal. 2016; Fagerlund etal. 2017).
In a sharp contrast to the relatively simple and uniform organization of the adaptation module, the effector modules are highly diverse, and their variation forms the basis of the current classification of CRISPR-Cas systems (Makarova etal. 2015; Koonin etal. 2017). On the basis of the organizational principles of the effector modules, all CRISPR-Cas systems are divided into Class 1, with multisubunit effector complexes comprised of several Cas proteins, and Class 2, in which the effector is a single, large, multidomain protein. Among other distinctions, Class 1 and Class 2 CRISPR-Cas systems substantially differ in the mechanisms of pre-crRNA processing. In Class 1 systems, the crRNAs are generated by a dedicated complex of multiple Cas proteins (Brouns etal. 2008; Wiedenheft etal. 2011; Rouillon etal. 2013; Spilman etal. 2013; Staals etal. 2013). In Class 2 systems, processing is catalyzed either by an external bacterial enzyme, RNAse III, with the help of an additional RNA species, the transacting CRISPR (tracr) RNA (Deltcheva etal. 2011; Jinek etal. 2012, 2014; Shmakov etal. 2015; Jiang and Doudna 2017; Liu etal. 2017a), or by the same effector protein that is involved in target cleavage (East-Seletsky etal. 2016; Fonfara etal. 2016; Liu etal. 2017b; Zetsche etal. 2017).
The differences between Class 1 and Class 2 CRISPR-Cas systems extend to the interference stage. In Class 1, the processing complex containing the guide crRNA recognizes the target site and recruits an additional Cas protein (Cas3 in type I and Cas10 in type III) that contains the nuclease domain directly responsible for the target cleavage (Sinkunas etal. 2011; Gong etal. 2014; Redding etal. 2015). In Class 2, cleavage is performed by the nuclease domain(s) of the large effector protein (Deltcheva etal. 2011; Gasiunas etal. 2012; Jinek etal. 2012; Zetsche etal. 2015; Abudayyeh etal. 2016; Dong etal. 2016; Jiang etal. 2016a; Yamano etal. 2016; Jiang and Doudna 2017; Liu etal. 2017a, 2017b; Smargon etal. 2017) (see more below). The composition and organization of the genes encoding effector module components have been comprehensively compared with delineate 6 types and 24 subtypes within the two CRISPR-Cas classes (Makarova etal. 2015; Koonin etal. 2017) (fig. 1). Various proteins involved in ancillary roles, such as regulation of the CRISPR response and other, still poorly characterized functions, can be assigned to a third, accessory module (Makarova etal. 2013a, 2014, 2015; Mohanraju etal. 2016). The modules of the CRISPR-Cas systems are partially autonomous as demonstrated by their frequent recombination as well as by the existence of isolated adaptation and effector modules in many bacterial and archaeal genomes (Makarova etal. 2015; Silas etal. 2017). However, it is important to note that the functional separation between the modules is only a rough approximation because some Cas proteins, in particular, Class 2 effectors, appear to be involved in all stages of the CRISPR response (Heler etal. 2015, 2017).
Fig. 1.
—Current classification of CRISPR-Cas systems. The organization of the CRISPR-cas loci and domain architectures of the effector proteins as well as the (predicted) target (DNA or RNA, or both) are shown for each subtype. The trees reflect the latest classifications for the Class 1 and Class 2 CRISPR-Cas systems (Makarova etal. 2015; Koonin etal. 2017) and are not traditional phylogenetic trees. The block arrows represent cas genes (not to scale); homologous genes are shown by the same color. For each cas gene, the systematic name and the legacy name (if any) are indicated below the respective arrow. The adaptation and effector modules are shaded in blue and light brown, respectively. A shaded outline for an arrow depicting a gene indicates that the gene in question is present only in a subset of CRISPR-cas loci of the respective subtype. For subtype III-D, a locus with a reverse transcriptase fused to cas1 is included; other reverse transcriptase-containing variants, from subtypes III-A and III-D, are not shown. SS, small subunit; TM, predicted transmembrane segment.
The currently characterized diversity and classification of CRISPR-Cas systems are covered in several recent reviews. Comparative genomics analyses aiming at the reconstruction of the origins and evolution of the CRISPR-Cas systems have revealed a pervasive trend, namely multiple contributions of several classes of mobile genetic elements (MGE) to the emergence and diversification of the CRISPR-Cas immunity (Makarova etal. 2013a, 2015; Chylinski etal. 2014; Koonin and Krupovic 2015a; Mohanraju etal. 2016; Shmakov etal. 2017). In this article, we summarize the evidence on the roles of MGE in CRISPR-Cas evolution and combine it with the indications of “reverse flow,” that is, hijacking of CRISPR-Cas systems and their components by MGE. We conclude that in prokaryotes, MGE and defense systems, in particular, CRISPR-Cas, form a dynamic network of genetic elements that continuously exchange genes.
Origin of the Adaptation Module and Adaptive Immunity in Prokaryotes: The Casposons
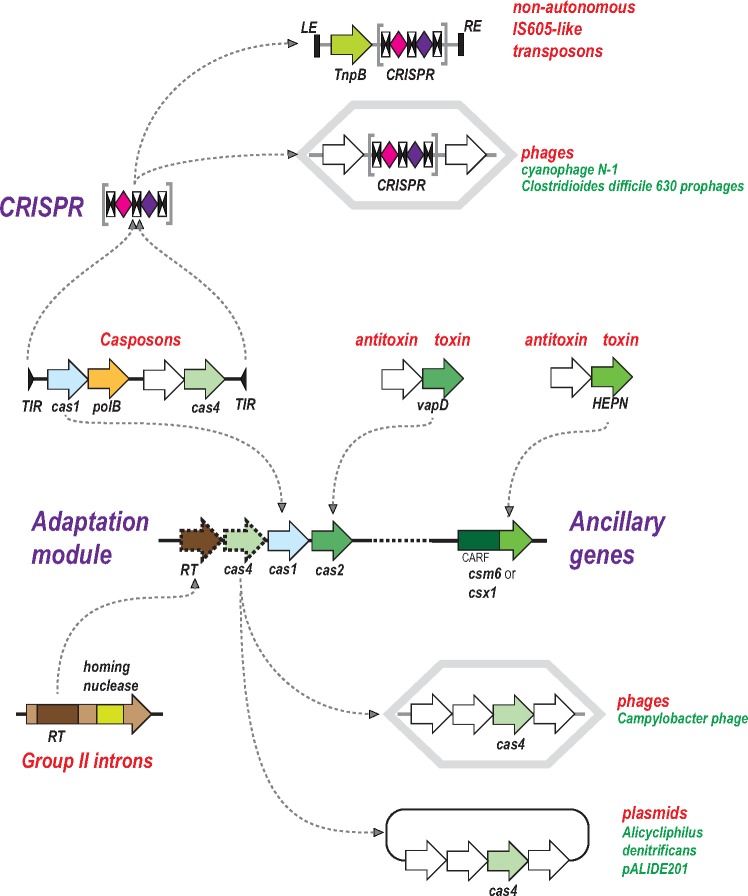
Cas1, the endonuclease responsible for spacer integration into CRISPR arrays, is not always encoded within CRISPR-cas loci. Analysis of the genomic surroundings of the “solo” cas1 homologs led to an unexpected discovery: the genomic context of these genes closely resembled transposable elements, with terminal inverted repeats (TIR) flanking DNA regions of 12–18 kb (Krupovic etal. 2014). These putative transposons all shared two genes encoding Cas1 and a DNA polymerase of family B, and additionally, encompassed variable sets of genes, mostly various nucleases and predicted DNA-binding containing helix-turn-helix (HTH) domains (fig. 2). Given the close similarity between the mechanisms of the reactions catalyzed by Cas1 during spacer integration into CRISPR arrays and by integrases during transposon integration, a natural prediction was that Cas1 functions as a bona fide integrase, hence the name Casposons for this class of MGE and casposase for the enzyme itself (Krupovic etal. 2014; Krupovic and Koonin 2016). Indeed, the integrase activity of the casposase has been promptly validated experimentally (Hickman and Dyda 2015), and moreover, similar target site specificities of Casposon integration and CRISPR spacer incorporation have been demonstrated (Beguin etal. 2016). The invariable presence of a DNA polymerase gene indicates that Casposons are self-synthesizing transposons that catalyze their own replication during transposition via a copy-and-paste mechanism. Before the discovery of the Casposons, self-synthesizing transposons, those of the Polinton class (also known as Mavericks), have been identified only in eukaryotes (Kapitonov and Jurka 2006; Pritham etal. 2007; Krupovic and Koonin 2015). The Casposons are not one of the particularly abundant prokaryotic MGE classes but nevertheless, show considerable diversity of genome organization, with four families characterized by distinct gene compositions. Transposition of Casposons has not been demonstrated directly but comparative genomic analysis of numerous strains of the archaeon Methanosarcina mazei has revealed clear signs of recent mobility, leaving little doubt that at least some of the Casposons are active MGE (Krupovic etal. 2016).
Fig. 2.
—Contributions of mobile genetic elements to the origin and evolution of CRISPR-Cas systems: adaptation module and CRISPR. The block arrows show genes (not to scale); the gene organizations of the depicted genetic elements are shown schematically. The curved arrows show inferred ancestor–descendent relationships. The double black arrowheads represent CRISPR repeats (to emphasize their palindromic organization in many although not all CRISPR arrays). The diamonds represent spacers that are colored differently, to emphasize that these sequences are unique. Each CRISPR array is schematically shown as three repeats and two spacers although the actual size differs from such minimal units to hundreds of repeats and spacers. Abbreviations: CARF, CRISPR-Associated Rossmann Fold (domain); HEPN, Higher Eukaryote and Prokaryote Nucleotide-binding (domain). LE, left end; RE, right end; RT, reverse transcriptase; TIR, terminal inverted repeat. VapD is a toxin with the activity of an interferase, that is, an RNase that cleaves ribosome-associated mRNA.
It appears likely that the entire adaptation module and perhaps even additional Cas proteins were contributed to the emerging CRISPR-Cas system by the ancestral Casposon (Koonin and Krupovic 2015a). The phylogenetic tree of the Cas1 family splits into two major branches, one of which consists of the casposases and the other one of CRISPR-associated Cas1 proteins, a topology that is compatible with a founding role of the Casposase in the evolution of CRISPR-Cas. Although the currently identified Casposons do not encode Cas2, some encode PD-DExK family nucleases homologous to Cas4, which is a component of the adaptation module in several CRISPR-Cas variants, and additional nucleases (Krupovic etal. 2014; Krupovic and Koonin 2016). Conceivably, the ancestral Casposon architecture including a gene for a Cas2 homolog remains to be discovered. Furthermore, the prototype CRISPR repeats and the leader sequence could have originated from either the TIRs or a duplicated target site of the ancestral Casposon (Krupovic etal. 2017).
The key role of Casposons in the origin of CRISPR-Cas is strongly supported by the close mechanistic similarity between the reactions catalyzed by the casposase during integration and by Cas1 during spacer incorporation into CRISPR arrays (Beguin etal. 2016). The founding event of CRISPR-Cas evolution could have been a random insertion of a Casposon in a vicinity of a putative ancestral innate immunity locus followed by immobilization of the inserted transposon and loss of some of its genes including the DNA polymerase (but see an alternative scenario below). The nature of the putative ancestral innate immunity system is perhaps the hardest puzzle of CRISPR-Cas evolution (Makarova etal. 2013a; Shmakov etal. 2015). Potentially, this locus could have encoded an ancestral Class 1 effector module that functioned without an adaptation module and CRISPR arrays, by directly employing guide RNA derived from transcripts of foreign genomes, analogously to Argonaute-based defense systems (Hur etal. 2014; Swarts etal. 2014). The organization of such potential ancestral innate immunity loci could resemble type IV CRISPR-Cas (Makarova etal. 2015) and the “minimal” variants of type I encoded by a distinct family of Tn7-like transposons (fig. 2; and see below).
CRISPR-Cas Adaptation Modules Containing RTs
A distinct variety of the CRISPR-Cas adaptation module that is widely represented in type III includes a RT that, in many cases, forms a fusion protein with Cas1 or alternatively, is encoded by a separate gene adjacent to cas1 (fig. 2). It has been shown that the RT-containing CRISPR-Cas systems are capable of acquiring spacers from RNA via reverse transcription, in addition to the spacer acquisition from DNA that is universal in CRISPR-Cas (Silas etal. 2016). Phylogenetic analysis of the RT superfamily indicates that most of the CRISPR-associated RTs form a monophyletic group that is affiliated with the RTs of Group II introns (Toro etal. 2014; Silas etal. 2017). The RT-Cas1 fusion represented in diverse type III loci appears to have emerged at a single point in evolution, conceivably, subsequent to a random insertion of a Group II intron into a type III CRISPR-cas locus. There is little if any correlation between the RT phylogeny and the CRISPR-Cas subtypes that are defined primarily from the organization of the effector modules (fig. 1). Furthermore, many RT-cas fusions are not associated with an effector module subunits but often are located adjacent to a CRISPR array. Taken together, these observations indicate that the RT-containing adaptation modules can combine or function in trans with most Type III systems. Thus, the Cas1-RT combination itself can be viewed as a MGE with an organization resembling that of Group II introns that combine RT with a homing endonuclease and eukaryotic retroelements encoding RT and integrases (Eickbush and Jamburuthugoda 2008; Zimmerly and Wu 2015; McNeil etal. 2016) although this element lacks the typical RNA structures of Group II introns and is not a Group intron per se (Silas etal. 2017).
In addition, two cases of apparent independent acquisition of RT by CRISPR-cas loci have been detected (Silas etal. 2017). One of these includes a small group of type III-B loci that are represented primarily in mesophilic archaea of the genus Methanosarcina. Phylogenetic analysis of the RT indicates that these loci have independently acquired the RT gene from a Group II intron. The second case includes type I-E CRISPR-Cas systems in bacteria of the genus Streptomyces to which the RT gene was apparently transferred from a type III CRISPR-cas locus. In these minor groups of the RT-containing CRISPR-Cas systems, RT is not fused to Cas1 or any other Cas protein, and the type III-B loci of Methanosarcina lack the cas1 gene altogether suggesting that RT interacts with Cas1 in trans. The independent fixation of the captured RT in these groups of archaea and bacteria seems to be indicative of the utility of the RT-mediated acquisition of spacers from RNA for increasing efficiency of the CRISPR response.
Type II and Type V Effectors: Parallel Capture of Transposon-Encoded Nucleases
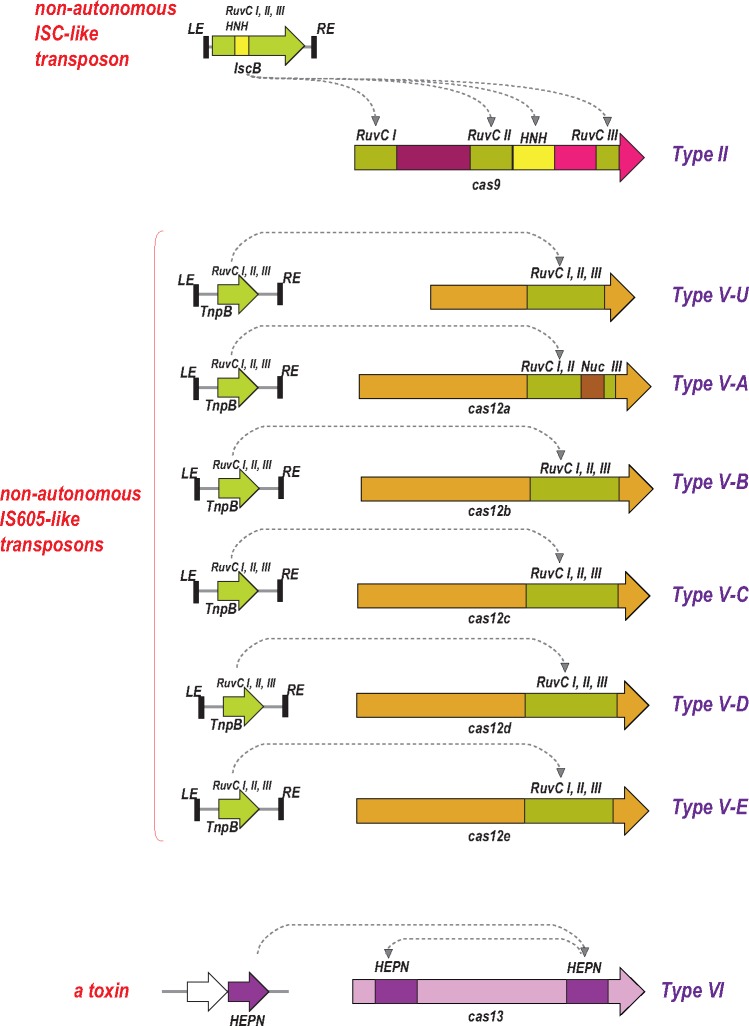
The common feature of all type II and type V effectors (Cas9 and Cas12 proteins, respectively) is the presence of a RuvC-like endonuclease domain (fig. 3). However, the sequence similarity between the RuvC-like domains of Cas9 and Cas12, and even between different subtypes within each type is very low such that these proteins can be recognized as homologs only by highly sensitive profile searches or structural comparisons. Moreover, other portions of Cas9 and Cas12 show no similarity to each other and appear not to be homologous (Shmakov etal. 2015, 2017). The structures of several Cas9 variants (Jinek etal. 2014; Nishimasu etal. 2014, 2015; Hirano etal. 2016), Cas12a (Cpf1) (Dong etal. 2016; Yamano etal. 2016), and Cas12b (C2c1) (Liu etal. 2017a; Yang etal. 2016) complexed with the guide RNA, target DNA and, in the cases of Cas9 and Cas12b, tracrRNA have been reported. All these effectors proteins are similar in size and general shapes, that is, a bilobed, “jaw-like” structure that accommodates the target DNA and the guide RNA between the lobes. However, the structures cannot be superimposed apart from the RuvC-like domains (Lewis and Ke 2017). Furthermore, the RuvC-like domains of Cas9 and Cas12a contain inserts, in similar but not identical positions, that represent nonhomologous domains, the HNH family nuclease and a novel NUC domain, respectively. In type II systems, the HNH domain of Cas9 cleaves the target DNA strand (i.e., the strand complementary to the crRNA) whereas the RuvC-like nuclease cleaves the nontarget strand (Gasiunas etal. 2012; Jinek etal. 2012). It has been initially concluded that in Cas12a, the NUC domain is the nuclease responsible for the cleavage of the target strand (Dong etal. 2016; Yamano etal. 2016). However, a subsequent, detailed biochemical study has suggested that the RuvC-like nuclease of Cas12a was responsible for the cleavage of both strands whereas the NUC domain facilitated the cleavage of the target strand in a nonenzymatic fashion (Swarts etal. 2017). Similarly, although the RuvC-like nuclease of Cas12b also contains a unique inserted domain, both DNA strands apparently are cleaved by the RuvC-like nuclease, which undergoes a major conformational change triggered by the initial, nontarget strand cleavage (Liu etal. 2017a; Yang etal. 2016). Thus, comparative analysis of the sequences and structures of type II and type V effectors reveals a remarkable interplay between homology, diversification and convergence which reflects their complex evolutionary history.
Fig. 3.
—Contributions of mobile genetic elements to the origin and evolution of CRISPR-Cas systems: Class 2 effector proteins. The genes for Class 2 effector proteins and the homologous proteins encoded by transposons or toxin–antitoxin modules are shown by block arrows (roughly to scale). Different colors denote distinct domains or uncharacterized regions in the effector proteins. The curved arrows show putative ancestor–descendent relationships. RuvC I, II, III are distinct amino acid motifs that jointly comprise the catalytic site of the RuvC-like nuclease.
The key insight into the evolution of the type II and type V effectors has been the observation that, among other members of the RuvC-like nuclease family, these effectors, particularly Cas12, show the highest sequence similarity to TnpB proteins of the IS605 and other related families of transposons (Chylinski etal. 2014; Shmakov etal. 2015). The tnpB genes are extremely abundant in bacterial and archaeal genomes and belong to autonomous transposons, which additionally encode a transposase (TnpA) but more frequently nonautonomous transposons, in which TnpB is the only protein product including the eukaryotic Fanzor transposons (Bao and Jurka 2013). The role of TnpB in the transposon life cycle remains unclear given that this protein is not required for transposition (Pasternak etal. 2013), but the conservation of the RuvC-like endonuclease catalytic sites in most TnpB sequences indicates that these proteins are active nucleases.
Remarkably, the effectors of type II and different subtypes of type V (Cas9 and Cas12a, 12b, 12c, respectively) showed the highest similarity to different groups of TnpB proteins, suggesting independent origins for the effectors of different types and subtypes of Class 2. In these cases, informative phylogenetic trees could not be constructed due to the low sequence conservation. Nevertheless, the specific ancestry of Cas9 could be readily traced to a distinct family of transposons (denoted ISC, after Insertion Sequences Cas9-related) thanks to the shared domain architectures of the IscB and Cas9 proteins (fig. 3), in which an HNH endonuclease domain is inserted into the RuvC-like domain (Chylinski etal. 2014; Kapitonov etal. 2015).
The likely scenario for the origin of type V effectors from TnpB has become clearer with the identification of a distinct variety of putative CRISPR-Cas systems, denoted subtype V-U (after Uncharacterized), that lack adaptation modules and consist of TnpB homologs encoded next to CRISPR arrays (Shmakov etal. 2017). The putative V-U effector proteins are much smaller than Cas9 or Cas12 and similar in size or only slightly larger than typical, transposon-encoded TnpB. Moreover, unlike Cas9 and Cas12, the TnpB homologs from the V-U loci show highly significant similarity to the transposon proteins, allowing construction of robust phylogenetic trees, which strongly support independent origin of Class 2 effectors from different TnpB subfamilies (Shmakov etal. 2017). The functionality of the type V-U systems so far has not been demonstrated in direct experiments. Nevertheless, multiple lines of evidence, namely evolutionary conservation in diverse groups of bacteria, the difference between the spacer complements of even closely related bacteria, and the presence of spacers homologous to phage genome sequences, indicate that at least some V-U variants are active immune systems. The close relationship between the putative V-U effectors and bona fide TnpB implies that V-U are recently evolved CRISPR-Cas variants. The current scenario for type II and type V evolution includes multiple, random insertions of nonautonomous TnpB-encoding transposons next to CRISPR arrays, with subsequent convergent “maturation” of the effectors that involved acquisition of additional domains (Shmakov etal. 2017). The newly gained domains are unrelated between different subtypes, but in each case, the outcome is the emergence of a full-fledged effector protein capable of accommodating the complex of the crRNA and the target DNA (fig. 3).
Contributions of Toxin–Antitoxin Modules to CRISPR-Cas Evolution
In addition to two distinct classes of transposons and group II introns, multiple contributions to the evolution of CRISPR-Cas systems apparently come from a very different type of MGE, the toxin–antitoxin (TA) modules. These modules consist of a toxin which, in most common variants, is an RNase (known as interferase) that specifically cleaves ribosome-associated mRNAs and an antitoxin that forms a complex with the toxin and reversibly inhibits its activity (Gerdes etal. 2005; Makarova etal. 2009; Van Melderen and Saavedra De Bast 2009; Van Melderen 2010; Gerdes 2012). The TA modules lack mechanisms of active mobility but are typically transferred on plasmids and are “addictive” to the host cells, which die if they do not receive the TA-carrying plasmid upon segregation. This postsegregational cell killing occurs because antitoxins are unstable proteins, compared with toxins, with a much shorter half-life, and therefore, unless the toxin and antitoxin are continuously produced in stoichiometric amounts, the unleashed toxin exerts its deleterious effect on the cell lacking the TA genetic locus. Apart from plasmids, numerous TA loci are carried by bacterial chromosomes, and in addition to their selfish properties, appear to perform defense functions, namely inducing dormancy or programmed cell death (PCD) as an “altruistic” defense strategy.
Notably, at least two unrelated classes of TA modules contributed to the evolution of CRISPR-Cas (fig. 3). The structural subunit of the adaptation complex, Cas2, is derived from the VapD family of interferases, which adopt a distinct version of the RNA Recognition Motif fold (Makarova etal. 2006). The interferase catalytic site is conserved in Cas2 proteins from some CRISPR-Cas systems but disrupted in others, and the function of the demonstrated nuclease activity of Cas2 (Beloglazova etal. 2008; Ka etal. 2014; Dixit etal. 2016) in the CRISPR-mediated defense remains obscure given that this activity is not required for adaptation. One possibility appears to be that Cas2 actually functions as a toxin that could induce dormancy or PCD in cases when the immune function of CRISPR-Cas fails (Makarova etal. 2012; Koonin and Zhang 2017). A different, unrelated toxin RNase, the HEPN domain, is present in several Cas proteins. The HEPN domain-containing Cas proteins include the effectors of the RNA-cleaving type VI CRISPR-Cas systems effectors (Cas13), which contain two diverged HEPN domain copies (Shmakov etal. 2015, 2017; Abudayyeh etal. 2016; Liu etal. 2017b; Smargon etal. 2017), and the Csm6 and Csx1 proteins that are responsible for the ancillary RNA cleavage in type III-A systems and type III-B systems, respectively (Elmore etal. 2016; Jiang etal. 2016b; Niewoehner and Jinek 2016; Sheppard etal. 2016). A highly diverged HEPN domain is contained also in a small protein that has been shown to regulate the activity of the effector protein Cas13b in one of the variants of the type VI-B CRISPR-Cas systems (Smargon etal. 2017). In the Csm6 and Csx1 proteins, the HEPN RNase is fused to a CRISPR-Associated Rossmann Fold (CARF) domain, which implies regulation by a nucleotide ligand (Makarova etal. 2014). In a striking recent development, the ligands of the CARF domain of Csm6 have been identified as cyclic oligoadenylates that are synthesized by the Cas10 proteins, the polymerase-cyclase large subunit of type III effectors complexes, in response to the target RNA recognition (Kazlauskiene etal. 2017; Niewoehner etal. 2017).
The HEPN superfamily, which includes primarily RNases involved in various defense-related functions in both prokaryotes and eukaryotes, in general, and the CRISPR-associated HEPN domains in particular, show extreme sequence divergence such that their identification often requires careful manual examination of protein alignments (Anantharaman etal. 2013; Shmakov etal. 2015; Smargon etal. 2017). Due to this low sequence conservation, reliable phylogenetic analysis and confident identification of the specific ancestral relationships of the HEPN domains are impractical. Nevertheless, with the exception of the CRISPR-associated HEPN RNases, all other prokaryotic HEPN domains appear to belong to TA or Abortive Infection modules, which are extremely abundant in many bacteria and archaea, especially thermophiles (Makarova etal. 2009, 2013b). These TA modules are, in general, poorly characterized, but the toxin activity of one of the HEPN domains from such a module has been demonstrated experimentally (Yao etal. 2015). Thus, there is little doubt that the CRISPR-Cas systems have originally coopted a toxin RNase on at least one but possibly multiple occasions (fig. 3).
There is an intriguing possibility that the toxicity of the HEPN domain RNases remains relevant in the context of the CRISPR-mediated defense as indicated by the recent characterization of the type VI interference activity. Once activated by the recognition of the cognate target RNA, the Cas13a and Cas13b proteins become promiscuous RNases that cleave RNA nonspecifically (Abudayyeh etal. 2016; Smargon etal. 2017). Expression of the Cas13 proteins together with the guide and target RNAs is toxic for bacteria, suggesting induction of dormancy or PCD in bacteria (Abudayyeh etal. 2016). This function of the type VI effectors remains to be studied in detail but appears compatible with the hypothesis on coupling between immunity and dormancy induction and/or PCD in prokaryotes (Makarova etal. 2012; Koonin and Zhang 2017).
Capture of CRISPR-Cas Systems and Individual Cas Genes by MGEs
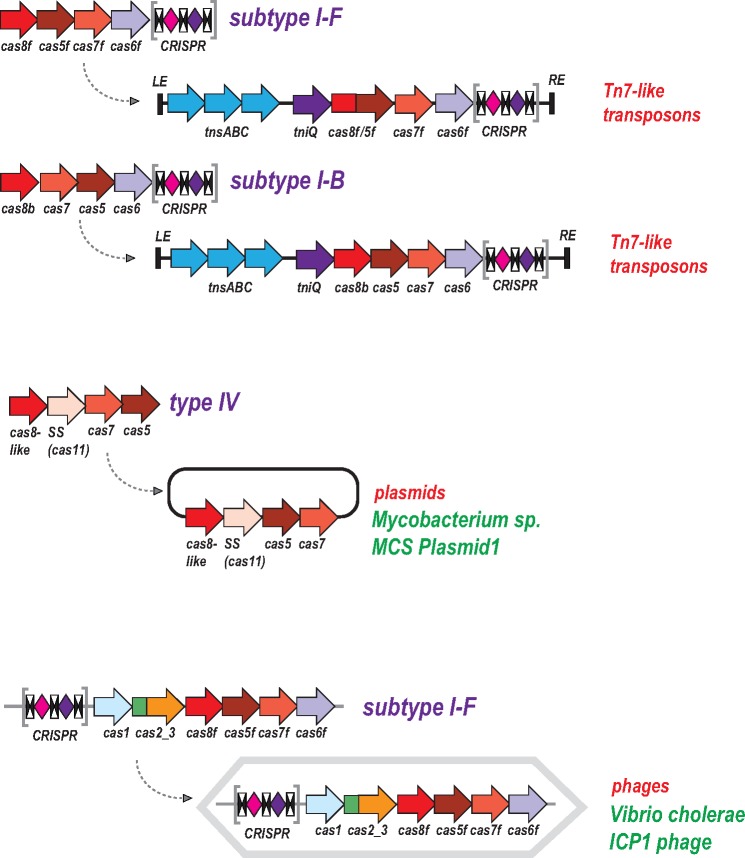
In the preceding section, we described the recruitment of proteins, primarily nucleases, from widely different MGE for functions in the CRISPR-mediated adaptive immunity. Here we address the reverse gene flow, from CRISPR-Cas systems to MGE. The most wide spread case of such recruitment includes a distinct group of Tn7-like transposons, which contain “minimal” type I-F CRISPR-Cas systems that consist of a reduced effector module and a short CRISPR array (fig. 4) (Peters etal. 2017). Several smaller groups of Tn7-like transposons encompass similarly truncated type I-B CRISPR-Cas systems. Notably, these type I variants lack the Cas3 helicase and the associated HD domain that are required for target cleavage (Beloglazova etal. 2011; Mulepati and Bailey 2011; Huo etal. 2014). Accordingly, they are predicted to be active in pre-crRNA processing yielding mature crRNAs as well as target binding but not interference that requires target cleavage. Phylogenetic analysis indicates a single, ancestral capture of a type I-F CRISPR-cas locus by the founder of a specific subfamily of Tn7-like transposons and two independent instances of type I-B loci capture (Peters etal. 2017). The transposon-associated CRISPR arrays contain multiple spacers homologous to plasmid and bacteriophage sequences, and in several cases, also bacterial chromosomal sequences adjacent to the transposon. Taken together, these observations prompt the hypothesis that the transposon-encoded CRISPR-Cas systems generate displacement (R-loops) in the cognate DNA sites, targeting the transposon to these sites and thus facilitating their spread, particularly, via plasmids and phages (Peters etal. 2017). Experimental study of the transposon-encoded CRISPR-Cas systems and their involvement in the transposon life cycle can be expected to shed new light on the functional and evolutionary interplay between CRISPR-Cas and MGE. More specifically, we do not currently understand what features of the Tn7 biology make this particular transposon family prone to capture and maintain CRISPR-Cas systems. Elucidation of the role of CRISPR-Cas in the life cycle of Tn7 is likely to uncover such features.
Fig. 4.
—Capture of CRISPR-Cas systems and cas genes by mobile genetic elements. The genes are shown by block arrows (not to scale). The CRISPR arrays are shown as in figure 2. The curved arrows show recruitment of CRISPR-Cas systems or individual cas genes by mobile genetic elements.
Similarly to the above cases, many type IV CRISPR-Cas systems are encoded on plasmids and might contribute to the plasmid–host interaction, in particular inhibiting host defense including resident CRISPR-Cas. The complete type IV systems encompass genes for the effector complex, Cas6 and CRISPR arrays but many lack an array, suggesting that they rely on arrays in the host genome (fig. 1) (Makarova etal. 2015).
In addition to transposons and plasmids, CRISPR-Cas systems are also scattered among bacteriophage genomes. In particular, a type I-F system, complete with an adaptation module and two CRISPR arrays, is inserted into the genomes of a group of ICP1-related phages that infect Vibrio cholerae (Seed etal. 2013). Strikingly, the majority of the spacers in the phage arrays are homologous to sequences from a host antiphage defense island, and targeting of this island by the phage-encoded CRISPR-Cas is essential for productive infection (Seed etal. 2013). Additional evidence of the presence of type I CRISPR-Cas or stand-alone CRISPR arrays in genomes of bacteriophages and archaeal viruses comes from metagenomics sequencing, and in at least some cases, the viral arrays include spacers homologous to other viral genomes (Minot etal. 2011; Garcia-Heredia etal. 2012; Bellas etal. 2015). Taken together, these findings clearly indicate that, at least sporadically, prokaryotic viruses acquire CRISPR-cas loci or CRISPR arrays alone from the hosts and deploy them as counter-defense in the virus-host arms race and/or as protection device from other viruses. This counter-defense strategy complements the dedicated anti-CRISPR proteins that are encoded by at least some and perhaps many prokaryotic viruses (Bondy-Denomy etal. 2015; Pawluk etal. 2016a, 2016b).
Finally, on multiple occasions, MGE recruit individual cas genes that either interact with the host CRISPR-Cas or are exapted for unrelated functions. The most prominent case in point is the Cas4 endonuclease that, in addition to Casposons, is encoded by numerous bacterial and archaeal viruses. In two families of Campylobacter jejunii phages, the phage-encoded Cas4-like protein affects the spacer acquisition by the host II-C CRISPR-Cas system, apparently promoting spacer capture from the host DNA (Hooton and Connerton 2015). The biological role of this effect of the phage Cas4 on adaptation is not entirely clear but the result is a stable persistence of the phage in the C. jejunii culture, so the host spacers might act as “decoys” to prevent acquisition of spacers from the phage genomes (Hooton etal. 2016). In an example of Cas4 exaptation, a virus of the archaeon Thermoproteus tenax has been shown to employ an inactivated derivative of Cas4 as the capsid protein (Krupovic etal. 2015).
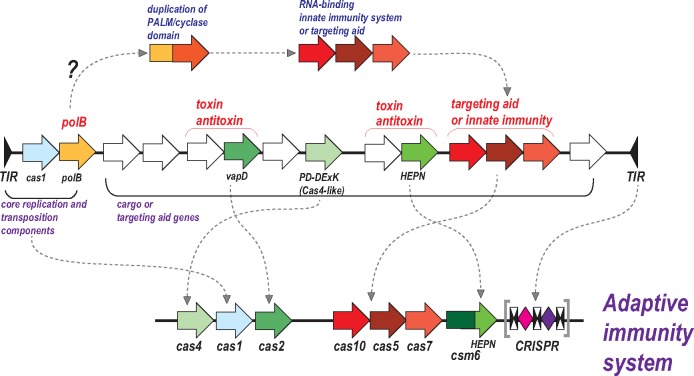
An “All from the Casposon” Scenario for the Origin of CRISPR-Cas
As described in the preceding section, a large subfamily of Tn7-like transposons encode a “minimal” CRISPR-Cas system that is predicted to mediate crRNA processing and target recognition but not interference, and thus might facilitate the transposon targeting for integration into a new site. The incorporation of type I-F CRISPR-Cas into a Tn7-like transposon definitely was a secondary, relatively late event in evolution because it involves a specific transposon subfamily incorporating a distinct variant of CRISPR-Cas, which could occur only after the diversification of both CRISPR-Cas systems and the transposons (Peters etal. 2017). Nevertheless, the predicted involvement of CRISPR-Cas in transposition potentially might recapitulate some of the earliest stages in CRISPR-Cas history, suggesting an alternative scenario. In this version, the entire CRISPR-Cas system evolved within a Casposon such that the predecessor of the effector module initially functioned as an ancillary, RNA-guided integration mechanism or, alternatively or additionally, as a distinct defense mechanisms preventing replication of other MGE in the same host (fig. 5). Further study of the Casposon diversity can shed light on the key aspects of CRISPR-Cas origin and evolution.
Fig. 5.
—An alternative hypothetical scenario of the CRISPR-Cas origin: both modules from the same Casposon? The putative ancestral Casposon is a hypothetical construct that does not precisely correspond to any Casposon so far identified. The curved arrows show putative ancestor–descendent relationships. The CRISPR array (depicted as in fig. 2) is tentatively derived from the casposon TIR. The evolution of the effector complex is speculated to have involved an initial duplication of the PALM domain of the Casposon DNA polymerase, in a development of the previously proposed evolutionary scenario (Makarova etal. 2013a). The abbreviations are as in the other figures.
Conclusions
The findings outlined in this article reveal multiple contributions of widely different classes of MGE to the origin and evolution of the CRISPR-Cas adaptive immune systems. These contributions come from two unrelated families of DNA transposons (Casposons and TnpB-encoding transposons, IS605 and ISC), retrotransposons (Group II introns) and at least two classes of TA systems. There is also substantial reverse flow of genetic information, thats is, recruitment of CRISPR-Cas systems and individual cas genes by various MGE, followed by their repurposing for counter-defense or, in some cases, other functions. This multiplicity of exchanges between the immune system and MGE clearly indicates that the connection is not random but rather reflects a deep evolutionary unity that is not limited to CRISPR-Cas but involves the entirety of defense mechanisms. Indeed, simple defense systems in prokaryotes, such as TA and restriction-modification modules themselves possess properties of MGE (Kobayashi 2001; Van Melderen and Saavedra De Bast 2009; Van Melderen 2010; Furuta and Kobayashi 2011). A more complex interplay between parasitism and defense can be captured in the “guns for hire” paradigm whereby homologous proteins, such as endonucleases, are utilized as offensive and defensive “weapons,” by MGE and defense systems, respectively (Koonin and Krupovic 2015b). Recruitment of transposons or their components apparently was central not only to the evolution of CRISPR-Cas but also to the origin of adaptive immunity in vertebrates (Kapitonov and Jurka 2005; Kapitonov and Koonin 2015; Koonin and Krupovic 2015a), the system of DNA elimination and rearrangement in ciliates (Nowacki etal. 2011; Dubois etal. 2012; Betermier and Duharcourt 2014; Allen and Nowacki 2017), and the piRNA machinery of germ line defense in animals (Aravin etal. 2007). Perhaps, there is ground for a sweeping generalization: all defense systems that are involved in some form of genome manipulation are evolutionarily linked to MGE. Elucidation of the diversity and the intricacies of the interactions between MGE and defense machineries, and development of a general theory of their coevolution are research directions for decades to come.
Acknowledgments
E.V.K. and K.S.M. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Literature Cited
- Abudayyeh OO, et al. 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299):aaf5573.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SE, Nowacki M.. 2017. Necessity is the mother of invention: ciliates, transposons, and transgenerational inheritance. Trends Genet. 33(3):197–207. [DOI] [PubMed] [Google Scholar]
- Amitai G, Sorek R.. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 14(2):67–76. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L.. 2013. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8(1):15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J.. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318(5851):761–764. [DOI] [PubMed] [Google Scholar]
- Bao W, Jurka J.. 2013. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob DNA 4(1):12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Horvath P.. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2:17092.. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA.. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54(2):234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguin P, Charpin N, Koonin EV, Forterre P, Krupovic M.. 2016. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 44(21):10367–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellas CM, Anesio AM, Barker G.. 2015. Analysis of virus genomes from glacial environments reveals novel virus groups with unusual host interactions. Front Microbiol. 6:656.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, et al. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 283(29):20361–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, et al. 2011. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. Embo J. 30(22):4616–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M, Duharcourt S.. 2014. Programmed rearrangement in ciliates: paramecium. Microbiol Spectr. 2(6). doi: 10.1128/microbiolspec.MDNA3-0035-2014. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, et al. 2015. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526(7571):136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321(5891):960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J, White MF.. 2015. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 39(3):428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV.. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42(10):6091–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, et al. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340):602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit B, et al. 2016. Dual nuclease activity of a Cas2 protein in CRISPR-Cas subtype I-B of Leptospira interrogans. FEBS Lett. 590(7):1002–1016. [DOI] [PubMed] [Google Scholar]
- Dong D, et al. 2016. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532(7600):522–526. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E.. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096.. [DOI] [PubMed] [Google Scholar]
- Dubois E, et al. 2012. Transposon invasion of the paramecium germline genome countered by a domesticated PiggyBac transposase and the NHEJ pathway. Int J Evol Biol. 2012:436196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, et al. 2016. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538(7624):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK.. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134(1–2):221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, et al. 2016. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 30(4):447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund RD, et al. 2017. Spacer capture and integration by a type I-F Cas1-Cas2-3 CRISPR adaptation complex. Proc Natl Acad Sci U S A. 114(26):E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, et al. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532(7600):517–521. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Kobayashi I.. 2011. Restriction-modification systems as mobile epigenetic elements In: Roberts AP, Mullany P, editors. Bacterial integrative mobile genetic elements. Austin (TX: ): Landes Bioscience. [Google Scholar]
- Garcia-Heredia I, et al. 2012. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One 7(3):e33802.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V.. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 109(39):E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. 2012. Prokaryotic toxin-antitoxins Berlin: Springer. [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A.. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 3(5):371–382. [DOI] [PubMed] [Google Scholar]
- Gong B, et al. 2014. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. Proc Natl Acad Sci U S A. 111(46):16359–16364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, et al. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519(7542):199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, et al. 2017. Mutations in Cas9 enhance the rate of acquisition of viral spacer sequences during the CRISPR-Cas immune response. Mol Cell 65(1):168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Dyda F.. 2015. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 43:10576–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, et al. 2016. Structure and engineering of Francisella novicida Cas9. Cell 164(5):950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA.. 2015. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 40(1):58–66. [DOI] [PubMed] [Google Scholar]
- Hooton SP, Brathwaite KJ, Connerton IF.. 2016. The bacteriophage carrier state of Campylobacter jejuni features changes in host non-coding RNAs and the acquisition of new host-derived CRISPR spacer sequences. Front Microbiol. 7:355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton SP, Connerton IF.. 2015. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol. 5:744.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F.. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, et al. 2014. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 21(9):771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Olovnikov I, Aravin AA.. 2014. Prokaryotic Argonautes defend genomes against invasive DNA. Trends Biochem Sci. 39(6):257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ.. 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 9(1):22–32. [DOI] [PubMed] [Google Scholar]
- Jackson SA, et al. 2017. CRISPR-Cas: adapting to change. Science 356(6333):eaal5056.. [DOI] [PubMed] [Google Scholar]
- Jiang F, Doudna JA.. 2017. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 46:505–529. [DOI] [PubMed] [Google Scholar]
- Jiang F, et al. 2016a. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351(6275):867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Samai P, Marraffini LA.. 2016b. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164(4):710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, et al. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, et al. 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka D, Kim D, Baek G, Bae E.. 2014. Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem Biophys Res Commun. 451(1):152–157. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J.. 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3(6):e181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J.. 2006. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. 103(12):4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Koonin EV.. 2015. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct 10:20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Makarova KS, Koonin EV.. 2015. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J Bacteriol. 198(5):797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V.. 2017. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357:605–609. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29(18):3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR.. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168(1–2):20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2017. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct 12(1):5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M.. 2015a. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 16(3):184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M.. 2015b. A Movable Defense. The Scientist, January 1.
- Koonin EV, Makarova KS.. 2009. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep. 1:95.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F.. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2009. Is evolution Darwinian or/and Lamarckian? Biol Direct 4:42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2016. Just how Lamarckian is CRISPR-Cas immunity: the continuum of evolvability mechanisms. Biol Direct 11(1):9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Zhang F.. 2017. Coupling immunity and programmed cell suicide in prokaryotes: life-or-death choices. Bioessays 39(1):1–9. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Beguin P, Koonin EV.. 2017. Casposons: the mobile elements that gave rise to the adaptation module of CRISPR-Cas systems. Curr Opin Microbiol. 38:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Cvirkaite-Krupovic V, Prangishvili D, Koonin EV.. 2015. Evolution of an archaeal virus nucleocapsid protein from the CRISPR-associated Cas4 nuclease. Biol Direct 10:65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV.. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. 13(2):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV.. 2016. Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems. Curr Opin Microbiol. 31:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV.. 2014. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 12:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV.. 2016. Recent mobility of casposons, self-synthesizing transposons at the origin of the CRISPR-Cas immunity. Genome Biol Evol. 8(2):375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne T, et al. 2016. Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell 63(5):852–864. [DOI] [PubMed] [Google Scholar]
- Lewis KM, Ke A.. 2017. Building the class 2 CRISPR-Cas Arsenal. Mol Cell 65(3):377–379. [DOI] [PubMed] [Google Scholar]
- Li M, Wang R, Zhao D, Xiang H.. 2014. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42(4):2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, et al. 2017a. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol Cell 65(2):310–322. [DOI] [PubMed] [Google Scholar]
- Liu L, et al. 2017b. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168(1–2):121–134. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Aravind L, Koonin EV.. 2012. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct 7:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L.. 2014. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet. 5:102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV.. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 13(11):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV.. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4(1):19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV.. 2013a. The basic building blocks and evolution of CRISPR-cas systems. Biochem Soc Trans. 41(6):1392–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV.. 2013b. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41(8):4360–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ.. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 11(3):181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BA, Semper C, Zimmerly S.. 2016. Group II introns: versatile ribozymes and retroelements. Wiley Interdiscip Rev RNA 7(3):341–355. [DOI] [PubMed] [Google Scholar]
- Minot S, et al. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21(10):1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P, et al. 2016. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353(6299):aad5147.. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S.. 2011. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J Biol Chem. 286(36):31896–31903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, et al. 2017. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. [DOI] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M.. 2016. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22(3):318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, et al. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156(5):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, et al. 2015. Crystal structure of Staphylococcus aureus Cas9. Cell 162(5):1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Nureki O.. 2017. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr Opin Struct Biol. 43:68–78. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF.. 2011. RNA-mediated epigenetic programming of genome rearrangements. Annu Rev Genomics Hum Genet. 12:367–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, et al. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 21:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, Doudna JA.. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak C, et al. 2013. ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol Microbiol. 88(2):443–455. [DOI] [PubMed] [Google Scholar]
- Pawluk A, et al. 2016a. Naturally occurring Off-switches for CRISPR-Cas9. Cell 167(7):1829–1838, e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, et al. 2016b. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol. 1(8):16085.. [DOI] [PubMed] [Google Scholar]
- Peters JE, Makarova KS, Shmakov S, Koonin EV.. 2017. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A. pii:201709035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A, Wilson GG, Wende W.. 2016. Type II restriction endonucleases – a historical perspective and more. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Richter H, Charpentier E, Randau L.. 2015. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 39(3):442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ, Putliwala T, Feschotte C.. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390(1–2):3–17. [DOI] [PubMed] [Google Scholar]
- Redding S, et al. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163(4):854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, et al. 2013. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell 52(1):124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Lazinski DW, Calderwood SB, Camilli A.. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494(7438):489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV.. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 23(10):578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard NF, Glover CV, Terns RM, Terns MP.. 2016. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22(2):216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, et al. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60(3):385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, et al. 2017. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silas S, et al. 2016. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351(6276):aad4234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silas S, et al. 2017. On the origin of reverse transcriptase-using CRISPR-Cas systems and their hyper-diverse, enigmatic spacer repertoires. MBio 8(4):e00897-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, et al. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, et al. 2017. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B.. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 82:237–266. [DOI] [PubMed] [Google Scholar]
- Spilman M, et al. 2013. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 52(1):146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, et al. 2013. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52(1):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, et al. 2014. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 21(9):743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, van der Oost J, Jinek M.. 2017. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell 66(2):221–233, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Wolf YI, Makarova KS, Koonin EV.. 2012. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 194(5):1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Nisa-Martínez R, Cordaux R.. 2014. Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One 9(11):e114083.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ.. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 34(8):401–407. [DOI] [PubMed] [Google Scholar]
- Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol. 13(6):781–785. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M.. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5(3):e1000437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2015. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163(4):840–853. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, et al. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477(7365):486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA.. 2016. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164(1–2):29–44. [DOI] [PubMed] [Google Scholar]
- Yamano T, et al. 2016. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165(4):949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, Patel DJ.. 2016. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167(7):1814–1828, e1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, et al. 2015. Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis. Microb Biotechnol. 8(6):961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, et al. 2017. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 35(2):31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Wu L.. 2015. An unexplored diversity of reverse transcriptases in bacteria. Microbiol Spectr. 3(2):MDNA3–M0058. 2014. [DOI] [PubMed] [Google Scholar]