Abstract
Ribosomal protein (RP) genes must be coordinately expressed for proper assembly of the ribosome yet the mechanisms that control expression of RP genes in metazoans are poorly understood. Recently, TATA-binding protein-related factor 2 (TRF2) rather than the TATA-binding protein (TBP) was found to function in transcription of RP genes in Drosophila. Unlike TBP, TRF2 lacks sequence-specific DNA binding activity, so the mechanism by which TRF2 is recruited to promoters is unclear. We show that the transcription factor M1BP, which associates with the core promoter region, activates transcription of RP genes. Moreover, M1BP directly interacts with TRF2 to recruit it to the RP gene promoter. High resolution ChIP-exo was used to analyze in vivo the association of M1BP, TRF2 and TFIID subunit, TAF1. Despite recent work suggesting that TFIID does not associate with RP genes in Drosophila, we find that TAF1 is present at RP gene promoters and that its interaction might also be directed by M1BP. Although M1BP associates with thousands of genes, its colocalization with TRF2 is largely restricted to RP genes, suggesting that this combination is key to coordinately regulating transcription of the majority of RP genes in Drosophila.
INTRODUCTION
The tight coordinate regulation of the ribosomal protein (RP) genes is common to all organisms (1). These regulatory mechanisms must ensure that each of the RPs are synthesized in the appropriate amounts to assemble ribosomes and at levels needed to meet the translational demands of each cell (1). In bacteria, coordinate regulation is achieved by organizing the RP genes into operons. In the yeast, Saccharomyces cerevisiae, transcription of the ribosomal genes is coordinated by a collection of well-characterized DNA binding proteins ((2) and references therein). Some of these proteins like Rap1, which is bound to essentially all of the RP promoters in S. cerevisiae, function at many genes in addition to the RP genes indicating that the regulatory network controlling the RP genes involves combinatorial control rather than a single master regulator. The evolution of these mechanisms is quite fluid because the Rap1-binding sites are absent from the RP genes in several strains of yeast that are evolutionarily distant from S. cerevisiae (3,4). Sequence comparisons of RP genes from other eukaryotes indicate there could be considerable diversity in the mechanisms that regulate RP genes (5,6), a result that is somewhat surprising given the functional conservation of the RPs.
Much less is known about the proteins that regulate the RP genes in higher eukaryotes. Conserved sequences shared among subsets of RP genes allude to several candidates but few of these have been tested directly (3,5,7,8). One conserved element whose function has been explored is the DNA replication-related element (DRE) (7). Available evidence indicates that the DRE-binding factor called DREF activates RP genes in human cells (9). Other candidate proteins implicated by the presence of conserved sequences include Sp1, NRF-2, Myc and YY1 (1,3,5,8,10).
Most of the sequences that have been implicated in regulating the RP genes are only present in a subset of RP genes. One exception is a pyrimidine-rich sequence called the TCT motif that encompasses the initiator of virtually all RP genes found in Drosophila and humans (11). This sequence might restrict RP genes from using the general transcription factor, TFIID, since TFIID binds poorly to RP gene promoters from Drosophila and swapping the TCT motif for the initiator sequence of the adenovirus major late promoter greatly reduces the affinity of TFIID for this mutated promoter even though a TATA box is still present (11,12).
Recently, the TATA-binding protein (TBP)-related factor called TATA-binding protein-related factor 2 (TRF2) was shown to be directly involved in transcription of RP genes (12). In addition to regulating RP genes, TRF2 is involved in regulating the histone H1 gene (but not the genes encoding core histones) and several genes involved in development (13,14). Like TBP, TRF2 associates with the general transcription factors, TFIIB and TFIIA; therefore, it is likely to provide a foundation much like TBP for assembling a pre-initiation complex (15). However, there is no evidence that TRF2 binds DNA and this lack of DNA-binding activity can be attributed to amino acid substitutions on the face of TRF2 that is homologous to the DNA-binding face of TBP (12,13,15,16). Thus, the mechanisms by which TRF2 associates with promoters are poorly understood. TRF2 has been detected in a complex that contains DREF, so the DRE found in a subset of RP genes could recruit TRF2 via DREF (17). An uncharacterized TRF2 complex has been shown to exhibit selective binding for the canonical initiator sequence and downstream promoter element (DPE) found in many Drosophila promoters (14). Since these elements are absent from most of the RP gene promoters, the recruitment mechanism for TRF2 to those RP promoters lacking the DRE is not known.
Here, we investigate the mechanism of transcriptional control of RP genes in Drosophila to gain insight into their coordinate regulation and how TRF2 associates with these promoters. This investigation was prompted by the observation that over half of the RP genes have a conserved core promoter motif known as Motif 1 (6). We recently identified a transcription factor called M1BP that associates with Motif 1 (18). M1BP is member of the ZAD-Znf family of zinc-finger proteins that has undergone a lineage-specific expansion in arthropods and could be the counterpart of KRAB-Znf family of proteins that are prevalent in mammals (19). M1BP associates with over 1500 promoters and most of these promoters drive constitutive expression of housekeeping genes (18). Here, we show that M1BP activates transcription of RP genes and that it could do so by directly interacting with TRF2 and recruiting TRF2 to the RP gene promoters. We also discover that although recent evidence indicates TBP and TFIID are not involved in RP gene transcription (12), TAF1, the largest subunit of TFIID, associates with all of the RP gene promoters in cells. The presence of TAF1 at RP gene promoters suggests the involvement of a TAF complex lacking TBP in transcribing the RP gene network. The specificity of this regulatory network appears to be defined in part by the combination of M1BP and TRF2 which is largely restricted to the RP genes. This work provides a mechanism for TRF2 recruitment to RP gene promoters and implicates a novel combination of both well conserved transcription factors (TAF1, TRF2 and DREF) and a lineage-specific transcription factor (M1BP), converging at core promoters to coordinately regulate this network of essential genes.
MATERIALS AND METHODS
RNAi knockdown in S2R+ cells followed by chromatin immunoprecipitation
dsRNA was generated byin vitro transcription with T7 polymerase on templates flanked by T7 promoter sequences. A total of 6 × 106 cells were plated in a 10 cm culture dish in 6 ml of serum-free M3+BPYE media (Drosophila Genome Resource Center) and were treated with 180 µg of the indicated dsRNA for 1 h after which the total media volume was brought up to 12 ml with a final fetal bovine serum concentration of 10%. Following this treatment, cells were allowed to incubate for the time specified in the figure legends. Following completion of the knockdown, cells were crosslinked with formaldehyde, chromatin was prepared and immunoprecipitations were performed as previously described (18). Percent recovery at designated genomic locations was determined by quantitative polymerase chain reaction (qPCR). Primers for dsRNA generation and qPCR are listed in the supplement. qPCR reactions were assembled using Bioline SensiMix SYBR Hi-ROX (QT605–20) master mix with reaction conditions matching the manufacturer’s recommendations. qPCR and analysis was performed using ABI StepOnePlus system. Reactions were heated to 95°C for 10 min, then underwent 40 cycles of 95°C for 15 s and 60°C for 60 s with data acquisition taking place during the 60°C step.
Western blots
Formaldehyde crosslinked chromatin lysates from RNAi-treated cells were heated to 75°C for 10 min in sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-Cl, pH 6.8, 2% SDS, 10% glycerol and 100 mM dithiothreitol (DTT)). Crosslinks were then reversed overnight at 65°C. Samples were electrophoresed on a 10% polyacrylamide gel, blotted to nitrocellulose and probed with antibodies against the indicated factors.
Nuclear extracts and immunodepletion of M1BP
Nuclear extracts were prepared from 0–12 h Oregon R embryos as previously described (20). Immunodepletion of nuclear extracts was performed as previously described (21).
In vitro transcription reactions and primer extension assay
In vitro transcription reactions and the primer extension assay were performed essentially as previously described (22). Twenty-five microliter reactions containing 125 µg nuclear extract, 32.5 mM HEPES (pH 7.6, K+), 20 mM KCl, 6.25 mM MgCl2, 0.05 mM ethylenediaminetetraacetic acid (EDTA), 5% glycerol, 1 mM DTT, 1% PEG (Sigma product number P2263, MW:15–20 kD), 10 µg/ml α-amanitin (where indicated), 2 units Promega Recombinant RNasin, 20 ng/µl plasmid template and 4.8 ng/µl recombinant M1BP (where indicated) were incubated at 24°C for 30 min. After pre-initiation complex formation, ribonucleoside triphosphates were added to a concentration of 0.5 mM and transcription occurred for 20 min at 24°C. Reactions were stopped by addition of 0.8% SDS, 16 mM EDTA, 160 mM NaCl, 0.2 mg/ml Torula yeast RNA and 0.08 mg/ml Proteinase K and incubated for at least 5 min at room temperature. Primer extension assays were then performed as previously described (23) and analyzed on a 10% sequencing gel containing 8 M urea.
Expression and purification of M1BP
Rosetta(DE3)pLysS competent cells (EMD millipore) were transformed with a previously reported M1BP expression vector (18). A total of 0.5 l of transformed cells were grown in LB media at 37°C to an OD600 of 0.4. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.25 mM IPTG and incubated overnight at 11°C. Cells were collected, resuspended and flash-frozen with liquid nitrogen in 25 ml Lysis buffer (25 mM HEPES (pH 7.6, K+), 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, 0.1% Triton X-100, 10 µM ZnCl2 with Protease inhibitors and 1 mg/ml lysozyme). Cells were thawed, incubated on ice for 15 min, sonicated and finally cleared by centrifugation at 20 000 × g. One-half milliliter of TALON(Clontech) resin previously equilibrated with lysis buffer was added to the cleared lysate and incubated with end-over-end mixing at 4°C for 1 h. Resin was collected and washed with lysis buffer for 15 min at 4°C, then poured into a column and washed with an additional 50 ml lysis buffer. Samples were eluted from the column in Elution buffer (25 mM HEPES (pH 7.6, K+), 150 mM NaCl, 250 mM imidazole, 10 mM β-mercaptoethanol, 0.1% Triton X-100, 10% glycerol and 10 µM ZnCl2 plus protease inhibitors). The samples were then dialyzed at 4°C overnight in Elution buffer lacking imidazole. Samples were centrifuged at 16 000 ×g for 10 mins at 4°C and the supernatant was analyzed by SDS-PAGE and used in the indicated experiments.
Synthesis of radiolabeled TRF2
A DNA fragment encoding TRF2 was amplified from S2R+ cDNA (See supplement for primer sequences). A total of 100 ng of the T7-flanked TRF2 coding region PCR product was added to the TnT Quick Coupled Transcription/Translation System (Promega) rabbit reticulocyte lysates and the reactions were carried out per the manufacturer’s protocol. A parallel reaction was done without adding the PCR template to produce a negative control for the immobilized template pulldown experiments.
Immobilized template pulldown experiments
Oligonucleotides corresponding to −52 to +8 of RpL30 or −36 to +14 of RpLP1 were annealed in 1 mM EDTA, 10 mM Tris–Cl pH 8.0 and 100 mM NaCl. The annealed oligonucleotides were purified from a polyacrylamide gel to ensure only hybridized oligos were used in the pulldowns. One oligonucleotide was biotinylated so that the template could be bound to Streptavidin Dynabeads as per the manufacturer’s protocol. Template bound beads were equilibrated in binding buffer consisting of 25 mM HEPES (pH 7.6, K+), 150 mM NaCl, 10 mM β-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride, 0.1% Triton X-100, 10% glycerol and 10 µM ZnCl2. A total of 3.5 µg of recombinant His-M1BP or control buffer and 10 µl 35S-TRF2 or control (No PCR template) TnT reactions were added as indicated. All components were incubated for 15 min at room temperature, then washed extensively with binding buffer. Beads were boiled in Laemmli sample buffer and the samples were analyzed by 10% SDS-PAGE. 35S-labeled TRF2 was detected with a phosphorimager.
Maltose-binding protein expression and pulldown experiments
BL21DE3 Escherichia coli cells expressing maltose-binding protein (Mal) alone or Mal fused to M1BP were grown to an OD600 of 0.4–0.5, IPTG was added and proteins were expressed in LB media overnight at 11°C. Cells were lysed cleared by spinning at 100 000 × g for 30 min. Mal was bound to amylose resin and washed with a buffer consisting of 50 mM HEPES, pH 7.6, 500 mM NaCl, 10% glycerol, 1% NP-40 and 1 mM DTT. Mal-M1BP was bound to amylose resin and washed with a buffer consisting of 25 mM HEPES, pH 7.6, 1 M NaCl, 200 mM KCl, 10% glycerol, 0.1% NP-40, 10 µM ZnCl2 and 1 mM DTT. A total of 10 µl of each type of protein-bound resin was equilibrated with several washes of pulldown buffer (25 mM HEPES, pH 7.6, 0.1 mM EDTA, 12.5 mM MgCl2, 10% Glycerol, 100 mM NaCl, 0.1% CHAPS and 0.1% NP-40). Following equilibration,100 µl of pulldown buffer, 2 µl of 35S-TRF2 and 20 µg bovine serum albumin was added to each sample. Mixtures were incubated for 15 min at room temperature while rotating. Samples were washed for 5 min with end-over-end rotation in 100 µl of pulldown buffer. Five washes were performed for each sample. Beads were boiled in Laemmli sample buffer and the samples were analyzed by 10% SDS-PAGE. 35S-labeled TRF2 was detected with a phosphorimager.
ChIP-exo
ChIP-exo was performed essentially as described in (24) with minor modifications. Libraries were quantified by qPCR and sequenced on an Illumina NextSeq 500. Basecalls were performed using Bcl2FastQ version 2.16.0. Sequenced reads were masked for low-quality sequence, then mapped to the Drosophila melanogaster dm3 whole genome using bwa mem (versions 0.7.9a, 0.7.12) with the default parameters. Heatmaps were generated with Homer bioinformatics software (25) and Java Treeview (26). Tables for composite plots were generated with HOMER and plots were visualized using R (27). Position weight matrices (PWM) for Motif 1 and DRE were obtained by performing a MEME-ChIP search of 200 bp regions centered around M1BP ChIP-exo or DREF ChIP-seq peak centers as determined by GEM using default settings (28). The PWM was fed into the FIMO tool (29) to identify motif locations genome-wide with a P-value cutoff <1E-04.
Bioinformatics
Our list of active genes was derived from the active gene list provided in (30). RP genes were selected and isolated from the list using their flybase annotation symbol. There are a total of 87 RP genes. Eight have duplicate isoforms and we eliminated one isoform of each duplicate if it lacked a TCT motif (Parry et al. (11)) or a TRF2 ChIP-seq peak (n = 79). RpL15 resides on Chr3LHet and, since our ChIP-exo data were not mapped to those regions, it was removed from the ChIP-exo heatmaps and composite plots thus bringing the final RP gene number to 78. PRO-seq bedgraph files were obtained from (31). Read pileups for heatmaps were performed with the HOMER bioinformatics tool using the annotatePeaks.pl script (25). Composite plots were generated in R. Venn Diagrams were generated with BioVenn (32). Heatmaps were generated with Java Treeview (26). STARR-seq heatmaps were generated using deepTools (33).
Peak calling and ChIP-seq analysis
DREF ChIP-seq (GSM1535985) (34) and input control experiment reads (34) were downloaded from http://www.ebi.ac.uk/ena in fastq format. DREF and Input reads were mapped to the dm3 genome in Galaxy with the bwa read aligner using default parameters. The GEM peak caller (28) was used to call peaks from the experiment and control bed files. Genes having transcription start sites (TSSs) within 200 bp of peak centers were designated M1BP-, TRF2- or DREF-associated.
Antibodies
The M1BP antibody was initially described and characterized in (18). The pre-immune sera comes from the same rabbit used to produce the M1BP antibody prior to injection with purified M1BP. The TRF2 antibody was described in (12). The TAF1 antibody was described in (35).
RESULTS
M1BP activates transcription of RP genes in cells
The conclusion that RP genes are highly expressed in coordinate fashion in metazoans is based largely on extrapolation of measurements of steady state mRNA levels in yeast (36). To more accurately assess RP gene transcription levels, we calculated PRO-seq read densities for each gene in Drosophila Schneider 2 (S2R+) cells (30). The region from the TSS to +100 bp was excluded to prevent bias arising from genes that are highly paused, but lowly transcribed (37). We ranked by PRO-seq reads per kilobase for all genes in the active gene list provided in (30) and assessed RP gene transcription activity relative to other actively transcribed genes. A total of 62 RP genes appear in the active gene list. The other 17 may have been filtered from the list either because they were not active or because their proximity to other genes could confound the bioinformatic analysis of PRO-seq signals (see (30)). Of the 62 genes on the list, 59 are transcribed in the top 10% of all active genes with the other 3 genes falling in the next decile (Supplementary Figure S1). Thus, most of the RP genes are transcribed at roughly equivalent high levels, suggesting that regulation at the level of transcription is important for coordinate RP expression. However, the factors involved in achieving this high level of coordinate transcription are largely unknown.
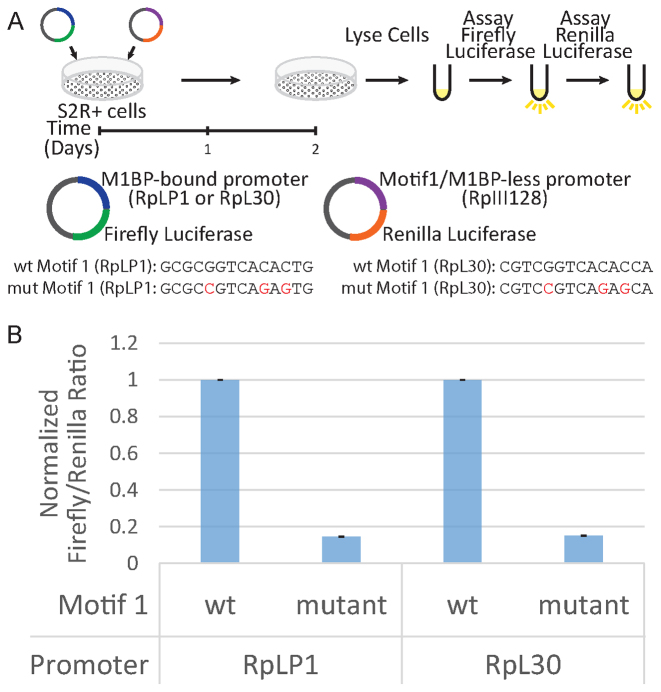
Previous analyses identified a conserved sequence called Motif 1 among the RP gene promoters (6) and we recently discovered a protein we named M1BP that associates with Motif 1 (18). To determine if Motif 1 and by extension M1BP, is involved in transcribing RP genes, we used a luciferase reporter assay (Illustrated in Figure 1A) with the promoter sequences (−500 to +50) from either RpLP1 or RpL30. These RP gene promoters had previously served as models for RP gene transcription (11,12). We also prepared mutant counterparts with three point mutations in Motif 1 that are known to abolish M1BP binding (18). Following transfection into Drosophila S2R+ cells, we saw >5-fold decline in luciferase levels when Motif 1 was mutated (Figure 1B). This demonstrates that Motif 1 contributes to transcription from RP gene promoters and implicates M1BP in transcriptional activation of RP gene promoters.
Figure 1.
Motif 1 is required for RP gene transcription in cells. (A) Schematic of the RP gene luciferase reporter assay. Mutant RpLP1 and RpL30 have three highly conserved nucleotides in Motif 1 mutated. The wt and mutant Motif 1 sequences for both promoters are shown below the illustration. The RpIII128 promoter lacks Motif 1 and serves an internal control. (B) Firefly/Renilla luciferase ratio of relative light unit measurements. Ratios are normalized to the wt Motif 1 sample for each promoter. Error bars represent standard deviation (n = 3).
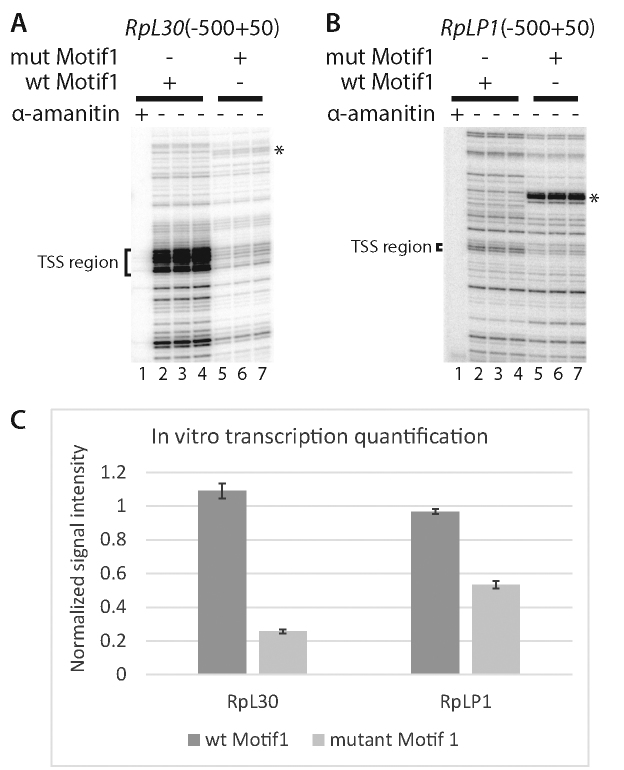
To directly test the role of M1BP in RP gene activation, we performed in vitro transcription in nuclear extracts which allowed us to determine the effects on RP gene transcription of both mutating Motif 1 or depleting M1BP. Mutating Motif 1 in the RpLP1 and RpL30 promoter caused about a 2- to 4-fold decrease in transcription which approximates the effect seen in cells (Figure 2A and B, cf. lanes 2–4 with 5–7 and Figure 2C). α-amanitin inhibited transcription of both promoters indicating that the transcription was mediated by Pol II (Figure 2A and B, cf. lane 1 with lanes 2–4).
Figure 2.
Motif 1 is required for transcription of RpL30 and RpLP1 in vitro. (A and B) Primer extension analysis of transcripts produced from the (A) RpL30 and (B) RpLP1 promoters (−500 to ∼+50) during transcription in Drosophila embryo nuclear extracts. Transcription reactions lacking α-amanitin were performed in triplicate. The bracketed region encompasses the M1BP-dependent TSS region and a portion of the TCT motif (Parry et al. (11). The M1BP-dependent transcription start sites (TSSs) observed in vitro correspond to the TSSs detected in vivo using PRO-cap (Kwak et al.(30)). (C) Quantification of bracketed TSS region transcripts from (A and B). Error bars represent standard deviation (n = 3). Samples have been normalized to the first wt replicate for each promoter. ‘*’ Denotes an artifact band arising in the Motif 1 region following mutation.
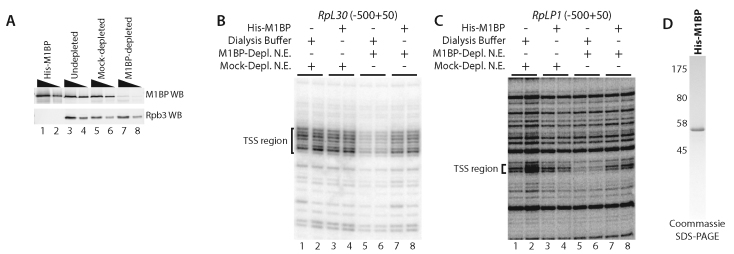
To determine if Motif 1 was functioning through M1BP, we immunodepleted M1BP from the nuclear extract (Figure 3A). Immunodepletion of M1BP caused a decrease in RP gene transcription (Figure 3B and C, lanes 1 and 2 versus 5 and 6). To establish that the immunodepletion of M1BP itself, rather than some associated protein was responsible for inhibiting transcription of the RP genesin vitro, we expressed and purified M1BP from E. coli (Figure 3D). Addition of recombinant M1BP to the M1BP-depleted nuclear extract restored RP gene transcription to its normal level (Figure 3B and C, cf. lanes 5 and 6 to lanes 7 and 8) indicating that M1BP activates transcription of the RP genein vitro. Addition of recombinant M1BP to the mock depleted extract had no effect on RP gene transcription suggesting the M1BP is not limiting in the mock-depleted extract (Figure 3B and C, cf. lanes 1 and 2 to lanes 3 and 4).
Figure 3.
M1BP is required for RP gene transcription in vitro. (A) M1BP-probed (top) and Rpb3-probed (bottom) western blot of purified His-M1BP and undepleted, mock-depleted or M1BP-depleted nuclear extracts from 0–12 h embryos. A total of 10 or 30 ng purified His-M1BP and 20 or 60 µg of each extract type was loaded for SDS-PAGE western blot analysis. (B and C) Primer extension analysis of transcripts produced from the (B) RpL30 or (C) RpLP1 promoter in embryo nuclear extracts. The bracketed region denotes the same TSS region described in Figure 2A. Each transcription reaction was performed in duplicate. Lanes 1 and 2: mock-depleted extract supplemented with dialysis buffer. Lanes 3 and 4: mock depleted extract supplemented with recombinant M1BP. Lanes 5 and 6: M1BP-depleted extract supplemented with dialysis buffer. Lanes 7 and 8: M1BP-depleted extract supplemented with enough recombinant M1BP to replace the amount that was immunodepleted. (D) Coomassie-stained SDS-PAGE analysis of purified, N-terminally His-tagged M1BP expressed in and purified from Escherichia coli.
M1BP recruits TRF2 to the RP gene promoter
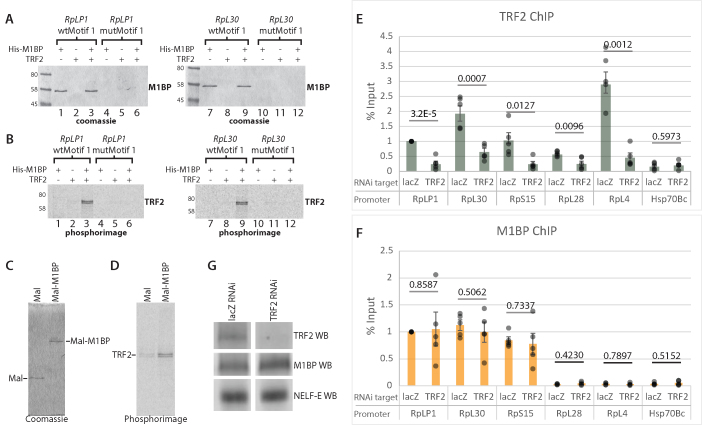
Recently, the TBP-related factor, TRF2, was found to be involved in transcription of RP genes (12). How TRF2 associates with promoters is enigmatic since, unlike its counterpart TBP, TRF2 has not been observed to bind directly to DNA. Motif 1, the binding site for M1BP, is typically located within 50 nts of the TSS of RP genes, making it a core promoter element (38). Hence, we investigated the possibility that M1BP might be recruiting TRF2 to RP gene promoters by performing immobilized template pulldown experiments with DNA template sequences corresponding to the core promoter regions of two RP genes. RpLP1 and RpL30 promoter sequences harboring normal or mutated Motif 1 elements were immobilized on beads. Following immobilization, purified recombinant M1BP andin vitro translated S35-labeled TRF2 were added either together or separately to the immobilized DNA and proteins that associated with the DNA templates were assessed by SDS-PAGE. M1BP binds in a Motif 1-dependent manner to both promoter sequences in the absence of TRF2 (Figure 4A, cf. lanes 1, 4, 7 and 10), while TRF2 alone does not bind to any of the promoter fragments (Figure 4B, lanes 2, 5, 8 and 11). In contrast, when M1BP is added together with TRF2, TRF2 associates with the Motif 1-containing M1BP-bound promoters (Figure 4A and B, lanes 3 and 9). Additionally, we expressed Mal fused to M1BP and were able to pulldown TRF2 indicating that TRF2 and M1BP interact in solution as well as on DNA templates (Figure 4C and D). These results show that M1BP can recruit TRF2 to the promoter but TRF2 is not required for M1BP to bind the promoter.
Figure 4.
M1BP recruits TRF2 to RP gene promoters. (A and B) Immobilized template pulldown assay. 35S-labeled TRF2 was synthesized in vitro using Promega’s TnT T7 Quick Coupled Transcription/Translation System. His-M1BP was expressed in and purified from Escherichia coli. His-M1BP, TRF2 or TRF2 and His-M1BP were added to either RpLP1 or RpL30 template-bound streptavidin dynabeads containing either a wild-type (wt) or mutant Motif 1 sequence (wtMotif 1 or mutMotif 1, respectively). Panel A shows coomassie-stained images from SDS-PAGE analysis of bound protein recovered from RpLP1 and RpL30 immobilized templates. Panel B shows phosphorimager scans of the same gels in panel A. M1BP binds only to the wt Motif 1 template regardless of whether TRF2 is present in the reaction. TRF2 only binds to the wt Motif 1 promoter template when M1BP is present. (C and D) Maltose-binding protein (Mal) and Mal-M1BP fusion pulldown assay. 35S-labeled TRF2 was synthesized as described above and added to either purified and amylose resin-bound Mal or Mal-M1BP. Panel C shows coomassie-stained images from SDS-PAGE analysis of bound protein recovered after binding and washing. Panel D shows the phosphorimage scans of the same gels in panel C. Recovery of TRF2 is increased with the Mal-M1BP fusion compared to the Mal alone. (E and F) RNAi-mediated depletion of TRF2. Following 3 days RNAi knockdown using dsRNA targeting either lacZ (negative control) or TRF2, cells were lysed and ChIP experiments were performed for TRF2 or M1BP. qPCR quantifications were normalized to lacZ RNAi signal at RpLP1. Hsp70Bc lacks both factors and thus serves as a negative control. RpL28 and RpL4 lack M1BP. Individual data points are displayed as gray dots. Each experiment was performed at least four-times. Error bars indicate standard deviation. P-values from two-tailed t-tests are provided for each promoter. (G) Western blots from S2R+ chromatin lysates used for ChIP following 3 days RNAi. The RNAi targets are indicated above the blot images. lacZ RNAi served as a negative control.
In order to determine if the immobilized template pulldowns reflect the binding properties in cells, we depleted TRF2 or M1BP using RNAi and monitored the association of each protein with representative RP genes using chromatin immunoprecipitation. We found that TRF2 depletion caused significant loss of TRF2 from RP promoters while M1BP levels remained unchanged (Figure 4E and F). A western blot confirmed that the RNAi worked as expected since we observed the total cellular TRF2 levels decreased, while the levels of M1BP and NELF-E remained unaffected (Figure 4G). Depletion of M1BP using RNAi caused a decrease of both M1BP and TRF2 from RP genes (Supplementary Figure S2). However, we were unable to conclude that TRF2’s association was directly linked to M1BP promoter binding in cells because depletion of M1BP also resulted in loss of TRF2 from RP genes that were bound and not bound by M1BP. Hence, the loss of TRF2 that occurred when M1BP was depleted could be a direct effect of the loss of M1BP or an indirect effect of the coordinate repression of RP genes that likely occurs as depletion of M1BP diminishes the rate of cell proliferation (18).
ChIP-exo analyses of M1BP and TRF2 provides evidence that M1BP recruits TRF2 to the majority of RP genesin vivo
Because of the pleiotropic effects that might accompany depleting M1BP from cells, we turned to ChIP-exo analysis to investigate the relationship between M1BP and TRF2 in cells. Recently, ChIP-exo analyses of factors associated with the RP genes in yeast provided insight into the protein–protein interactions that are involved in regulating these genes (2). ChIP-exo analysis maps at near single nucleotide resolution, the sites where a protein crosslinks to DNA by treating immunoprecipitated protein–DNA adducts with λ exonuclease and subjecting the digested DNA to high-throughput sequencing (24). Since λ exonuclease digests the DNA in a 5′ to 3′ direction and is blocked by protein–DNA crosslinks, protein-binding sites are demarcated by sequencing tags on opposite strands that manifest as peak pairs.
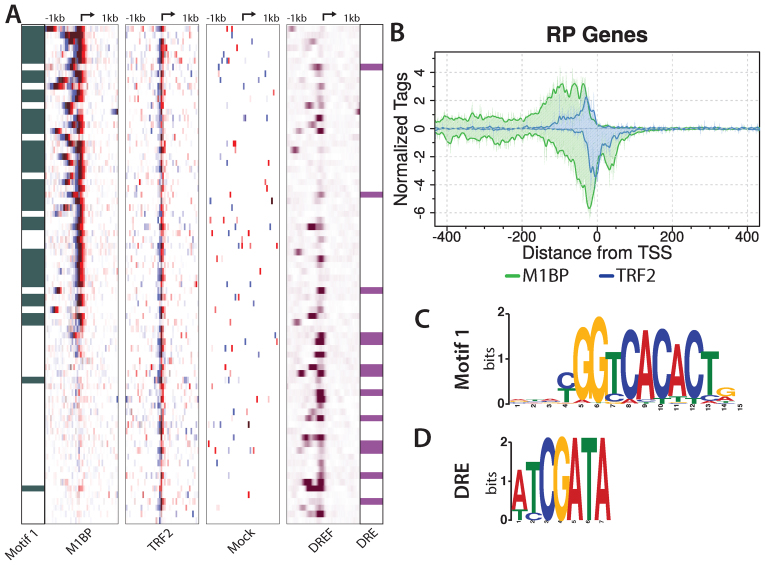
M1BP-binding sites on the DNA in cells were readily detected by ChIP-exo and are strikingly different from a mock ChIP-exo pattern (Figure 5A). We called peaks and the data confirmed that M1BP is highly enriched at RP gene promoters when compared to all active genes (P < 0.0001, Fisher’s exact test, two tailed). Composite plots using the TSS as a reference point reveal a complex pattern of crosslinking that extends from −150 on the top strand to +50 on the bottom strand (Figure 5B). The M1BP pattern is unlikely to be due solely to M1BP crosslinking directly to DNA since M1BP has five zinc fingers which are predicted to be just enough to bind the 15-nt long Motif 1 (Figure 5C). This ChIP-exo pattern resembles the broad pattern recently described for the yeast RP genes and was attributed to multi-protein complexes crosslinking to DNA (2). ChIP-exo analysis of TRF2 revealed a more compact pattern of crosslinks than M1BP. The majority of TRF2 crosslinks occurred immediately upstream from the TSS (Figure 5A and B). Comparison of the composite plots of TRF2 and M1BP revealed striking overlap of the TRF2 ChIP-exo pattern with the M1BP peak pair most proximal to the TSS. This type of overlap in ChIP-exo patterns has been interpreted to reflect the overlap between two factors binding in concert with one another (2), thus the data provide evidence consistent with our binding assays which indicates that TRF2 interacts with M1BP.
Figure 5.
M1BP and TRF2 co-occupy the majority of RP gene promoters. (A) M1BP, TRF2 and Mock ChIP-exo reads and DNA replication-related element-binding factor (DREF) ChIP-seq reads mapped relative to the TSS of 78 RP genes and sorted by M1BP ChIP-exo reads summed in a 2 kb window centered on the TSS. RP genes having Motif 1 or a DRE within 200 bp of the TSS are indicated in green in the far left panel or purple in the far right panel, respectively. The arrow at the top of each heatmap marks the TSS. Eight paralogs lacking a TCT motif or TRF2 peak have been removed. (B) ChIP-exo analysis of M1BP (green trace) and TRF2 (blue trace) for RP genes. Composite plots in single nucleotide bins were generated from the same RP gene list used for the heatmap in panel A. (C and D) Logo representation of the Motif 1 or DRE position weight matrices (PWM) used to identify genomic motif locations.
DREF is enriched among RP genes lacking M1BP and could provide an alternative mechanism for recruiting TRF2
TRF2 is present at almost all of the RP gene promoters yet M1BP is detected at approximately two-thirds of them (Figure 5A). TRF2 was previously found to be in a complex with DREF (17), so we wondered if the DRE (Figure 5D) and by extension, DREF might function in recruiting TRF2 to those RP genes that lack M1BP. To explore this possibility, we used previously published DREF ChIP-seq data (34) and determined that DREF is enriched at RP genes when compared to all active genes ((P = 0.0394, Fisher’s exact test, two tailed). Notably, DREF was further enriched among those RP gene promoters that lack M1BP (Figure 5A,P = 0.0009,Fisher’s exact test, two tailed). Thus, two mechanisms appear to function to recruit TRF2 to RP gene promoters.
ChIP-exo analysis detects TAF1 at RP gene promoters
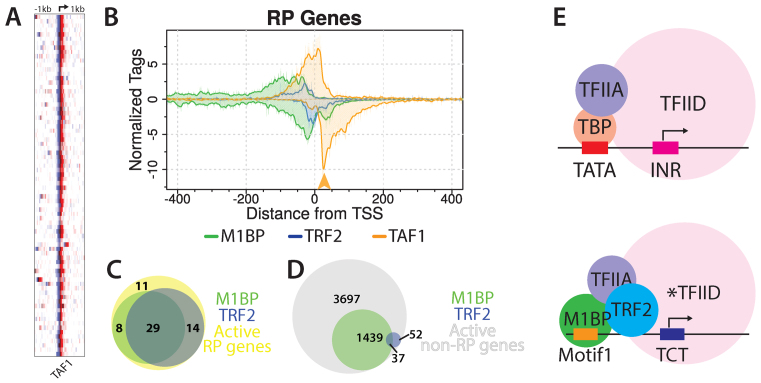
Our ChIP-exo pattern for M1BP revealed a peak of crosslinks on the bottom strand, 30–50 nts downstream from the TSS (Figure 5B). Since this region of the promoter was not required to bind M1BP to immobilized DNA, we suspected that these crosslinks might involve another protein that interacts with M1BP. Crosslinking and cryoelectron microscopy have shown that TAF1 contacts this region of the promoter (39–41). Our ChIP-exo analysis reveals that TAF1 is present at virtually all of the RP gene promoters (Figure 6A). TAF1 also appears to be present at virtually all genes with Pol II and most TRF2-associated promoters (Supplementary Figures S3 and 4). On RP genes, the downstream peak of TAF1 coincides well with the downstream peak of M1BP (Figure 6B, orange arrowhead) raising the possibility that this M1BP peak is the result of M1BP crosslinking to TAF1 which is in turn crosslinked to this downstream region.
Figure 6.
TAF1 is present at RP gene promoters. (A) Heatmaps display TAF1 ChIP-exo reads from S2R+ cells piled from −1 to +1 kb relative to RP gene TSS. Rows represent individual genes and are sorted by M1BP reads summed in a 2kb window as in Figure 5A. The TSS position is indicated by the arrow at the top of the panel. Duplicate genes were refined to a single isoform by removing eight paralogs lacking TRF2 or a TCT motif in the promoter. (B) Composite plots for M1BP, TRF2 and TAF1 were generated from the same RP gene list used for the heatmap. The orange arrow highlights a TAF1 peak that aligns with an M1BP peak. (C and D) Venn diagrams showing the overlap between M1BP and TRF2 peaks present at all active RP gene promoters (n = 62) or all other active gene promoters (n = 5225). (E) Model depicting M1BP’s recruitment of TRF2 at RP gene promoters. At TATA-containing promoters, TBP-bound TFIID engages with promoter sequences both up- and downstream of the TSS. At the majority of RP gene promoters, which lack both a TATA box and initiator sequence, M1BP and TRF2 bind the core promoter upstream of the TSS. The asterisk(*) denotes a noncanonical TFIID complex proposed to have TRF2 substituting for TBP.
Colocalization of M1BP and TRF2 is largely restricted to the RP genes
Genomic analysis indicates that M1BP and TRF2 associate with many genes (12,18). To determine if M1BP might function at other promoters by recruiting TRF2, we compared the distributions of M1BP and TRF2. M1BP and TRF2 show far less co-occupancy among nonRP genes (Figure 6C and D). Thus, these two factors appear to have converged on the RP genes to help drive their robust and coordinated expression. However, the association of TRF2 with the RP genes cannot be solely dependent on M1BP; otherwise, TRF2 would be present at other M1BP genes. A possible contributor to the specificity of TRF2 for M1BP-occupied RP genes is the TCT initiator element that is largely restricted to the RP gene promoters (11).
RP gene promoters act as enhancers of other RP genes
M1BP and TRF2, together with the TCT motif could play an essential role in coordinating expression of RP genes. A recent analysis of published STARR-seq (45) data concluded that housekeeping promoters themselves act as enhancers (46). We initially analyzed the STARR-seq data to see if we could detect enhancers that act upon the RP genes but found that the RP gene promoters themselves function as enhancers (Supplementary Figure S5). Thus, our analysis suggests that RP genes promoters could serve as enhancers of other RP genes, thus providing a way to coordinately regulate these promoters.
DISCUSSION
Here, we show that M1BP activates transcription of RP genes in Drosophila and that it can do so by recruiting TRF2 to RP gene promoters in cells. These conclusions are based on our demonstration that M1BP is detected in the core promoter region of the majority of RP genes in cells and that mutation of Motif 1 diminished the level of expression from RP reporter genes. Additionally, we have demonstrated that M1BP activates transcription of RP gene promoters in nuclear extracts. Also, we show that M1BP recruits TRF2 to promoter DNA in vitro and that M1BP and TRF2 colocalize on the RP gene promoters in cells. M1BP, therefore, is the first sequence-specific DNA-binding protein that has been directly shown to activate RP gene transcription in metazoans. DREF is possibly the only other protein, but it remains to be determined if it activates RP genes in vitro. Since these transcription factors associate with a broad spectrum of genes, loss of function assays in cells must be viewed with caution as it is difficult to distinguish between direct and indirect effects regardless of whether the protein can be detected at a particular gene.
Mechanisms by which TRF2 associate with promoters are not well understood. DREF was purified in a complex with TRF2 but no direct measurement of TRF2 recruitment to DNA by this complex was provided (17). An uncharacterized TRF2 complex associates with promoters bearing the DPE and canonical initiator (14), but RP genes lack both of these DNA elements. Here, we provide a direct mechanism that involves M1BP associating with its cognate-binding site and interacting directly with TRF2. Since there is little overlap between M1BP and TRF2 outside of RP gene promoters, it follows that additional cis-elements are required for TRF2’s association with M1BP. We suspect that the TCT motif, along with M1BP and DREF, may be an additional key contributor to TRF2’s association with gene promoters. Additionally, our ChIP-exo data reveals that TAF1 is present at virtually all promoters that are associated with Pol II including most promoters that associate with TRF2. Therefore, since TAF1 recognizes sequences at the initiator and the DPE (42,43), TAF1 could be part of the currently uncharacterized TRF2-containing complex that selectively binds the initiator-DPE-containing promoters (14).
The total number of TRF2 peaks that we observe is considerably lower than that reported previously (12). There could be a couple reasons for this discrepancy. First, the previous study used 2–4 h embryos, whereas we are using S2R+ cells. It is possible that TRF2 functions at a broader spectrum of developmentally regulated genes in the early embryo than in S2R+ cells. Additionally, the difference could be due to the increased signal to noise ratio afforded by ChIP-exo which results in more reliable peak detection.
Detection of TAF1 on RP gene promoters was unexpected because TAF1 is best known for being a subunit of the general transcription factor, TFIID and biochemical evidence argued against TFIID being involved in RP gene transcription (11). Moreover, previous analysis of the PCNA promoter showed that immunodepletion of TFIID with TAF1 antibody from a Drosophila transcription reaction did not inhibit transcription of a TRF2-dependent promoter (17). Our ChIP-exo data provide evidence for M1BP being in close proximity to and potentially interacting with, TRF2 and TAF1 on RP gene promoters. The ChIP-exo data showed a peak of M1BP contacts downstream from the TSS yet Motif 1 that binds M1BP typically resides upstream from the TSS. Since the ChIP-exo data for M1BP and TAF1 display overlapping peaks in the +30 to +50 region, we propose that M1BP is in contact with TAF1 and that the ChIP-exo signal for M1BP in this region is a consequence of M1BP crosslinking to TAF1 and TAF1 in turn crosslinking to the +30 to +50 region. In contrast, the ChIP exo signals for M1BP and TRF2 are shifted relative to each other by ∼10 nts suggesting that M1BP might position TRF2 on the DNA adjacent to M1BP.
A unique feature of the RP gene promoters in Drosophila and humans is the presence of the TCT motif located at the TSS (11). What recognizes this motif is currently not known. Since TAF1 is known to recognize the canonical initiator element (11,40,44), its presence at RP gene promoters raises the possibility that this TAF also recognizes the TCT motif. DNAse I footprinting analysis of TFIID binding to RP gene promoters indicated that binding was extremely weak. However, close inspection of the DNase I cutting patterns in the absence and presence of TFIID reveals the appearance of weak hypersensitive cut sites near the TCT initiator (11). One possibility is that M1BP together with TRF2 enhance the affinity of TAF1 for the RP gene initiator.
We propose that M1BP functions as a hub to recruit TRF2 and TAF1 (Figure 6E). Since the only known TAF1-containing complex in metazoans is TFIID, we propose that TFIID still binds to RP gene promoters along with TRF2. One possibility is that TRF2 displaces TBP at the RP gene promoter. A recent model of TFIID bound to promoter DNA indicates that TFIIA is involved in connecting TBP to TAF1 (41). Since TRF2 associates with TFIIA (15), displacement of TBP from TAF1 by TRF2 is tenable.
Our analysis of STARR-seq data indicates that RP gene promoters can act as enhancers and that they are selective in activating housekeeping gene core promoters and not core promoters of developmental and stress activated genes. The RP gene promoters more strongly activated the candidate RP gene promoter over all the other tested candidates. This selectivity could establish a network in which active RP genes and other housekeeping genes act reciprocally to activate each other. In addition, the selectivity of the enhancer activity of these RP promoters would prevent them from inadvertently activating nearby developmentally regulated genes.
ACCESSION NUMBER
Sequence data are in the Gene Expression Omnibus (GEO) database under accession number GSE97841.
Supplementary Material
ACKNOWLEDGEMENTS
We thank James T. Kadonaga and David A. Wassarman for providing the TRF2 and TAF1 antisera, respectively. We also thank members of the B. Frank Pugh lab for their assistance with ChIP-exo experiments. In particular, we thank Matthew J. Rossi, William K. Lai, Nina P. Farrell and Bongsoo Park. We thank members of the Gilmour lab for their suggestions.
Author contributions: D.G.B. and D.S.G. designed the project and planned experiments. D.G.B. performed the experiments. D.G.B. and D.S.G. interpreted the results and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM047477 to D.S.G.]. Funding for open access charge: NIH [GM047477].
Conflict of interest statement. None declared.
REFERENCES
- 1. Perry R.P. Balanced production of ribosomal proteins. Gene. 2007; 401:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reja R., Vinayachandran V., Ghosh S., Pugh B.F.. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015; 29:1942–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu H., Li X.. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics. 2007; 90:421–423. [DOI] [PubMed] [Google Scholar]
- 4. Tanay A., Regev A., Shamir R.. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:7203–7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X., Zhong S., Wong W.H.. Reliable prediction of transcription factor binding sites by phylogenetic verification. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16945–16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma X., Zhang K., Li X.. Evolution of Drosophila ribosomal protein gene core promoters. Gene. 2009; 432:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perry R.P. The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 2005; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roepcke S., Zhi D., Vingron M., Arndt P.F.. Identification of highly specific localized sequence motifs in human ribosomal protein gene promoters. Gene. 2006; 365:48–56. [DOI] [PubMed] [Google Scholar]
- 9. Yamashita D., Sano Y., Adachi Y., Okamoto Y., Osada H., Takahashi T., Yamaguchi T., Osumi T., Hirose F.. hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol. Cell. Biol. 2007; 27:2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown S.J., Cole M.D., Erives A.J.. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics. 2008; 9:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parry T.J., Theisen J.W.M., Hsu J.-Y., Wang Y.-L., Corcoran D.L., Eustice M., Ohler U., Kadonaga J.T.. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010; 24:2013–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y.-L., Duttke S.H.C., Chen K., Johnston J., Kassavetis G.A., Zeitlinger J., Kadonaga J.T.. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 2014; 28:1550–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R.. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007; 21:2936–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kedmi A., Zehavi Y., Glick Y., Orenstein Y., Ideses D., Wachtel C., Doniger T., Waldman Ben-Asher H., Muster N., Thompson J. et al. Drosophila TRF2 is a preferential core promoter regulator. Genes Dev. 2014; 28:2163–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rabenstein M.D., Zhou S., Lis J.T., Tjian R.. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4791–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moore P.A., Ozer J., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P.M.. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol. Cell. Biol. 1999; 19:7610–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hochheimer A., Zhou S., Zheng S., Holmes M.C., Tjian R.. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002; 420:439–445. [DOI] [PubMed] [Google Scholar]
- 18. Li J., Gilmour D.S.. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 2013; 32:1829–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stubbs L., Sun Y., Caetano-Anolles D.. Function and evolution of C2H2 zinc finger arrays. Subcell. Biochem. 2011; 52:75–94. [DOI] [PubMed] [Google Scholar]
- 20. Biggin M.D., Tjian R.. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988; 53:699–711. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y.-L., Duttke S.H.C., Chen K., Johnston J., Kassavetis G.A., Zeitlinger J., Kadonaga J.T.. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 2014; 28:1550–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wampler S.L., Kadonaga J.T.. Functional analysis of Drosophila transcription factor IIB. Genes Dev. 1992; 6:1542–1552. [DOI] [PubMed] [Google Scholar]
- 23. Carey M.F., Peterson C.L., Smale S.T.. The primer extension assay. Cold Spring Harb. Protoc. 2013; 2013:164–173. [DOI] [PubMed] [Google Scholar]
- 24. Rhee H.S., Pugh B.F.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. 2012; doi:10.1002/0471142727.mb2124s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 27. Venables W.N., Smith D.M. R Development Core Team . An Introduction to R. 2010; 2:The R Dev. Core Team; 1–90. [Google Scholar]
- 28. Guo Y., Mahony S., Gifford D.K.. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol. 2012; 8:e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwak H., Fuda N.J., Core L.J., Lis J.T.. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013; 339:950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duarte F.M., Fuda N.J., Mahat D.B., Core L.J., Guertin M.J., Lis J.T.. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 2016; 30:1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hulsen T., de Vlieg J., Alkema W.. BioVenn: a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008; 9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L., Lyu X., Hou C., Takenaka N., Nguyen H.Q., Ong C.-T., Cubeñas-Potts C., Hu M., Lei E.P., Bosco G. et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015; 58:216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maile T., Kwoczynski S., Katzenberger R.J., Wassarman D.A., Sauer F.. TAF1 activates transcription by phosphorylation of serine 33 in histone H2B. Science. 2004; 304:1010–1014. [DOI] [PubMed] [Google Scholar]
- 36. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 37. Adelman K., Lis J.T.. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012; 13:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohler U., Liao G.-C., Niemann H., Rubin G.M.. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002; 3, research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sypes M.A., Gilmour D.S.. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 1994; 22:807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu C.H., Madabusi L., Nishioka H., Emanuel P., Sypes M., Arkhipova I., Gilmour D.S.. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 2001; 21:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Louder R.K., He Y., López-Blanco J.R., Fang J., Chacón P., Nogales E.. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016; 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu C.H., Madabusi L., Nishioka H., Emanuel P., Sypes M., Arkhipova I., Gilmour D.S.. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 2001; 21:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Louder R.K., He Y., López-Blanco J.R., Fang J., Chacón P., Nogales E.. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016; 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Verrijzer C.P., Chen J.-L., Yokomori K., Tjian R.. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995; 81:1115–1125. [DOI] [PubMed] [Google Scholar]
- 45. Zabidi M.A., Arnold C.D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A.. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015; 518:556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cubeñas-Potts C., Rowley M.J., Lyu X., Li G., Lei E.P., Corces V.G.. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2016; 45:1714–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.