Abstract
Chloroplastic translation is mediated by a bacterial-type 70S chloroplast ribosome. During the evolution, chloroplast ribosomes have acquired five plastid-specific ribosomal proteins or PSRPs (cS22, cS23, bTHXc, cL37 and cL38) which have been suggested to play important regulatory roles in translation. However, their exact locations on the chloroplast ribosome remain elusive due to lack of a high-resolution structure, hindering our progress to understand their possible roles. Here we present a cryo-EM structure of the 70S chloroplast ribosome from spinach resolved to 3.4 Å and focus our discussion mainly on the architecture of the 30S small subunit (SSU) which is resolved to 3.7 Å. cS22 localizes at the SSU foot where it seems to compensate for the deletions in 16S rRNA. The mRNA exit site is highly remodeled due to the presence of cS23 suggesting an alternative mode of translation initiation. bTHXc is positioned at the SSU head and appears to stabilize the intersubunit bridge B1b during thermal fluctuations. The translation factor plastid pY binds to the SSU on the intersubunit side and interacts with the conserved nucleotide bases involved in decoding. Most of the intersubunit bridges are conserved compared to the bacteria, except for a new bridge involving uL2c and bS6c.
INTRODUCTION
Ribosomes are mega-Dalton ribonucleoprotein complexes which act as protein synthesis factories of the cell and are mostly conserved across all forms of life. However, ribosomes have evolved to include or exclude proteins and rRNA elements to serve specialized functions (1–3) in their unique environments. Historically, structural studies on ribosomes using X-ray crystallography and cryo-electron microscopy (cryo-EM) have been carried out with ribosomes purified from bacterial sources (Escherichia coli and Thermus thermophilus) providing a rich reservoir of knowledge on translation and its regulation inside bacterial cells. However, our current knowledge about structures of organellar ribosomes is still limited to mitochondrial ribosomes because of the lack of a high-resolution structure for the complete 70S chloroplast ribosome.
Chloroplasts were once free-living photosynthetic prokaryotic cells probably of cyanobacterial origin which according to the Endosymbiotic theory (4) were engulfed by other prokaryotic host cells. Evolution over a billion years have led the chloroplast to establish itself as an essential organelle in the plant and the algal cells, regulating myriad metabolic and signaling functions (5,6). Interestingly, over the time most of the chloroplastic genes have been transferred to the nucleus leaving behind a small genome in the plastid (plastome) (6). The plastome of higher plants contains a 120–220 kb circular DNA which harbors 120–130 genes arranged on a single chromosome (7–11). A vast majority (∼95%) of the proteins functioning inside chloroplasts are nuclear-encoded and post-translationally imported into the organelle by the help of transit peptides (6,10–12), necessitating coordination between the nuclear and the chloroplastic gene expressions (13,14). Gene expression in the chloroplast is mainly light-regulated (10,15) and mostly happens post-transcriptionally through mRNA processing, stabilization and during the initiation and elongation stages of translation (16). The fact that chloroplastic translation might follow routes resembling bacterial translation are reflected by the presence of bacterial-like 70S ribosomes in chloroplasts, Shine-Dalgarno (SD) sequences in many of the plastid-encoded mRNAs, the conservation of 3′-end of 16S rRNA (harboring anti-SD sequences) and the identification of conserved translation factors (7,17–20). However, chloroplastic translation system is unique (at least partly) as the putative SD sequences show poor conservation in its primary sequences and locations relative to the start codons demonstrating a diverse variance in the usage and function of ribosome binding site in chloroplasts (21,22). 5′ UTR elements in two-thirds of plastid genes are also reported to contain SD-like sequences (23–25) which might play significant roles in the translation initiation in chloroplasts. Apart from these cis-acting RNA elements, a set of trans-acting protein factors have been implicated in the translational regulation in the organelle (9,26). Interestingly, plastid-specific ribosomal proteins (PSRPs) and the N- and C-terminal extensions in the plastid ribosomal proteins (PRPs) have been implicated in playing a major role to regulate translation inside the chloroplast (27,28).
Chloroplast 70S ribosomes of spinach (Spinacia oleracea) harbor five PSRPs, namely cS22 (PSRP2), cS23 (PSRP3), bTHXc (PSRP4), cL37 (PSRP5) and cL38 (PSRP6) among which the first three resides in the 30S small subunit (SSU) and the latter resides in the 50S large subunit (LSU) (28–30). PSRP1 was initially considered as a bona fide protein of the chloroplast 70S ribosome. However, biochemical data showed that it is a translation factor (plastid pY) which co-purifies with spinach chloroplast 70S ribosome and also that it shares homology with the bacterial cold-shock protein pY (31). Three of the five PSRPs namely cS23, bTHXc and cL37 are proposed to be essential for the functioning of the chloroplast ribosome in Arabidopsis (32). PSRPs are suggested to play important roles in the light-dependent regulation of translation inside the chloroplast (27,28). In our previously published LSU structure of the spinach chloroplast ribosome (33), we localized and modeled cL37 and cL38 along with most of the PRPs and their plastid-specific protein extensions. The structure shed light on the possible functions of these two PSRPs in the context of translation regulation inside the chloroplast. However, the location and function of the three PSRPs residing in the SSU still remain elusive.
Here we present a cryo-EM reconstruction of the complete 70S chloroplast ribosome from spinach leaves resolved to an average 3.4 Å resolution where the LSU and the SSU are resolved to 3.3 and 3.7 Å, respectively. High-resolution map enabled us to build a near-complete atomic model of the chloroplast 70S ribosome. As model for the LSU was reported earlier (33), here we focus on the modeling of the SSU proteins and 16S rRNA. Localization and modeling of the three PSRPs (cS22, cS23, bTHXc) in the SSU provides important insights into the functions of the PSRPs in chloroplastic environment. In addition, we observe density for the N-terminal domain (NTD) of plastid pY bound to the SSU on the intersubunit side, in our 70S chloroplast ribosome. Moreover, we observe partial density for chloroplast ribosomal protein S1 (bS1c) which extensively interacts with protein bS2c on the solvent exposed side of the SSU. S1 has been recalcitrant to crystallographic and cryo-EM studies due to flexibility. Our finding of bS1c density on the SSU indicates that association of bS1c with the SSU is tighter than its bacterial counterpart. While our manuscript was under preparation, two cryo-EM structures of the chloroplast ribosome were reported by Graf et al. (34) and Bieri et al. (35). Overall, our findings regarding the LSU are in good agreement with both the structures however a comparison regarding the SSU could only be made with Bieri et al. (35) as SSU model of Graf et al. (34) was unavailable. Taken together, our study strives to elucidate the functional significance of the PSRPs residing in the SSU, which would be useful in designing further experiments for understanding of the translation regulation in the chloroplast.
MATERIALS AND METHODS
Purification of chloroplast ribosomes
Chloroplast ribosomes were purified from spinach leaves as described previously (33,36) with minor modifications in the protocol (Supplementary Figure S1a). In short, 6 kg of baby spinach leaves were deveined and washed with distilled water. The leaves were homogenized using 0.7 M Sorbitol in buffer A (10 mM Tris·HCl, pH 7.6, 50 mM KCl, 10 mM MgOAc and 7 mM β-mercaptoethanol) using a kitchen blender, throughout applying two shots of 10 s each. The homogenate was filtered through double-layered cheesecloth and one layer of Miracloth (Calbiochem) and centrifuged at 1200 × g for 15 min. The chloroplast pellet was resuspended in 0.4 M Sorbitol in buffer A and centrifuged again at 1200 × g for 15 min. The pellet was resuspended in buffer A supplemented with 2% (vol/vol) Triton X-100 and incubated on ice for 30 min. To achieve clarification, the lysed suspension was centrifuged at 26 000 × g for 30 min and thereafter centrifuged at 86 000 × g for 17 h through a sucrose cushion (1 M sucrose in buffer A) to pellet down crude ribosomes. The pellet was washed and gently resuspended in buffer B (buffer A supplemented with 10% glycerol). The crude ribosomes were clarified by centrifugation at 26 000 × g for 15 min and centrifuged at 111 000 × g for 4 h through 10–40% sucrose gradient in buffer A (with RNase inhibitor). 70S chloroplast ribosome fractions were collected and further centrifuged through a sucrose cushion (0.75 M sucrose in buffer A, with RNase inhibitor) at 84 000 × g for 2.5 h. The pelleted chloroplast ribosomes were suspended in a grid buffer containing 20 mM Tris·HCl, pH 7.6, 100 mM KCl, 10 mM MgOAc, 100 mM sucrose, 7 mM 2-mercaptoethanol, 1 unit/ml RNase inhibitor and 0.1% protease inhibitor. After taking out sample for grid preparation, the remaining chloroplast ribosomes were stored at −80°C.
Electron microscopy
Chloroplast ribosome particles were diluted to final concentration of 4 OD260/ml. A total of 4 μl of this sample was applied to glow-discharged 2-nm carbon coated holey grids (Quantifoil R2/2) and incubated for 30 s. FEI Vitrobot was used to blot the grids for 3 s in 100% humidity at 4°C and thereafter grids were flash frozen in liquid N2-cooled liquid ethane. Data were collected using a 300 kV FEI Titan Krios cryo-transmission electron microscope equipped with a back-thinned Falcon II direct electron detection device at an underfocus range of 0.4–3.7 μm. Electron micrographs were collected automatically by using Leginon software (37). A total of 3161 micrographs were recorded in a movie mode as a set of 25 frames (total dose of 37.5 electrons per Å2) at a calibrated magnification of 133 333 which rendered a pixel size of 1.05 Å on the object scale.
Image processing
A total of 3161 micrographs (Supplementary Figure S1b) were collected and were subsequently evaluated for drift. After the evaluation, 1285 good micrographs were subjected to whole-image drift correction using Motioncorr (38). Contrast transfer function (CTF) parameters were determined using CTFFIND3 (39) and semi-automated particle picking was carried out using e2boxer.py from EMAN 2.1 (40). Data were processed using RELION 1.4 (41). At the outset, 187 946 particles were subjected to reference-free 2D classification applying 2× decimation (Supplementary Figure S2a). After sorting the particles into good and bad classes, 132 904 good particles were further subjected to three-dimensional (3D) classification using 2× decimation guided by a 70S chloroplast ribosome map obtained previously (33). The dataset could be sorted into 22 737 (17%) poorly-aligned particles, 28 862 (22%) 50S and 81 305 (61%) 70S particles. 70S particles were used for high-resolution 3D refinement which yielded a 3.6 Å density map for the complete 70S chloroplast ribosome. Inclusion of movie frames for 3D refinement improved the resolution to 3.4 Å. Finally, Gaussian masks were used to perform 3D refinement on the ribosomal subunits which generated the final maps where the SSU and the LSU were resolved to 3.7 Å and 3.3 Å, respectively (Supplementary Figure S2a). Local resolution for the maps was calculated using ResMap (42) (Supplementary Figure S1d) and gold standard Fourier Shell Correlation (FSC) = 0.143 criterion (43) was used for determination of the resolution (Supplementary Figure S1e). The density maps were corrected for the modulation transfer functions and sharpened using automatically calculated B-factors (−72.6 Å2 for complete 70S ribosome, −97.9 Å2 for 30S subunit and −89.6 Å2 for 50S subunit) prior to visualization.
Model building and refinement
A combination of homology and de novo modeling approaches was used to build the atomic coordinates for the complete 70S ribosome. Models for the LSU components were extracted from our previously published structure of the LSU of spinach chloroplast ribosome (33) and were fitted initially as rigid bodies in our 3.3 Å LSU map and thereafter refined to improve model fitting using phenix.real_space_refine (44) with the application of RNA base-pairing restraints generated from ‘PDB to 3D Restraints’ server (http://rna.ucsc.edu/pdbrestraints/pdbtorestraints.html), RNA stacking restraints to maintain parallelity (default in phenix) and using secondary structure restraints for the proteins. For the structural modeling of the 16S rRNA, sequence information was obtained from the complete chloroplast genome sequence in NCBI (Accession no. AJ400848.1) and a homology model of 16S rRNA was generated using the ModeRNA server (45) and thereafter manually rebuilt in COOT (46) into our 3.7 Å SSU map using predicted secondary structure from the comparative RNA website (47). For SSU proteins, sequence information was obtained from UniProt (48) and GenBank (49) databases and homology models were generated using the ITASSER server (50) using E. coli ribosomal proteins from PDB ID: 4YBB (51) as templates. Because no suitable templates after extensive database search were found for cS22 and cS23, models for these two PSRPs were generated without specifying any templates using the ITASSER server (50) allowing the program to build the structures by performing multiple folding simulations from the identified templates in the Protein Data Bank (PDB) (52). Homology models for plastid pY and bTHXc were generated using YfiA and bTHX models (PDB ID: 4Y4O) (53) as templates using the ITASSER server (50). Guided by the positioning of the proteins in E. coli ribosome (51), the homology models for the chloroplast ribosome SSU proteins were thereafter docked in our cryo-EM reconstruction of the chloroplast ribosome SSU using ‘Fit in Map’ functionality in UCSF Chimera (54) and thereafter manually adjusted in COOT (46). Placement of the PSRPs and plastid pY were achieved by applying a combination of guidance from previous literature (28,53,55) and based on the size and the shape of the density. Extensions of the SSU proteins were built using ‘C-alpha baton mode’ and ‘add terminal residue’ in COOT (46). ‘Real Space Refine Zone’ and ‘Regularize Zone’ have been used throughout the manual model building to locally fit models inside density while keeping the geometry meaningful. Fittings for all the SSU proteins were initially improved using phenix.real_space_refine (44) on individual protein models using secondary structure restraints. The atomic coordinates for the SSU and the LSU were merged thereafter and clashes at the subunit interface were removed using COOT (46). To reduce overfitting, poorly resolved regions of the maps were identified by generating model-map cross-correlation values using phenix.map_model_cc (56) and models in these regions were refined locally using phenix.real_space_refine (44) applying restraints (mentioned above) and tighter RMSD values before adding them back to the final model. Finally, the complete 70S ribosome structure was subjected to global refinement and minimization using phenix.real_space_refine (44) applying default parameters. In spite of the high quality of our maps, register shifts could not be avoided at the poorly resolved flexible parts of the ribosome.
Model validation
The atomic coordinates of the spinach chloroplast ribosome were validated using the Molprobity server (57). The degree of model overfitting was evaluated by calculating FSC curves (Supplementary Figure S1e) between the models and the final cryo-EM maps. To generate the FSC curves, model derived maps (all frequencies until Nyquist included) were resampled onto the same grid as the final cryo-EM maps using UCSF Chimera (54) and FSC values were calculated using e2proc3d.py program in EMAN 2.1 (40). Maps were also colored to show local agreement between the models and the maps using ‘vop localCorrelation’ command in UCSF Chimera (54) (Supplementary Figure S3). The details about model validation and refinement statistics are presented in Supplementary Table S1.
Figure generation
UCSF Chimera (54) and PyMOL (58) were used to generate the figures in the manuscript. Plots showing local resolutions were generated using ResMap (42).
RESULTS AND DISCUSSION
Structure determination of the 70S chloroplast ribosome
70S ribosomes were purified (Supplementary Figure S1a) from spinach chloroplasts using sucrose density gradient centrifugation following previously published protocol (28,36). Cryo-EM and single particle analysis were employed to obtain the complete reconstruction of the 70S chloroplast ribosome (Figure 1A and Supplementary Figure S2a). A representative electron micrograph and some of the good 2D classes are displayed in Supplementary Figure S1b and c, respectively. Our previous attempts to obtain a high-resolution structure of the chloroplast ribosome were limited to only LSU (33) possibly due to the usage a 200 kV electron microscope (Arctica EM). In this study, we switched to a 300 kV Krios EM for imaging to improve the overall resolution of the 70S chloroplast ribosome to enable modeling of the SSU (details are in the ‘Materials and Methods’ section) using the same ribosome sample as before (33). 300 kV dataset yielded a high-resolution EM density map for the complete 70S chloroplast ribosome resolved to an average 3.4 Å (Supplementary Figure S1d and Table S1) as compared to a 4.5 Å map from the 200 kV dataset (33). The density maps were further improved using focused mask refinement procedures resulting in 3.3 and 3.7 Å resolved maps for the LSU and the SSU, respectively (Supplementary Figures S1d and 2a and Table S1). Representative images showing well resolved density from the LSU and the SSU maps are displayed in Supplementary Figure S4. The modeling of the LSU proteins and the rRNAs was carried out by fitting our previously published structure of the chloroplast ribosome 50S subunit (33) into the 3.3 Å LSU map (Supplementary Figure S2b and Table S2).
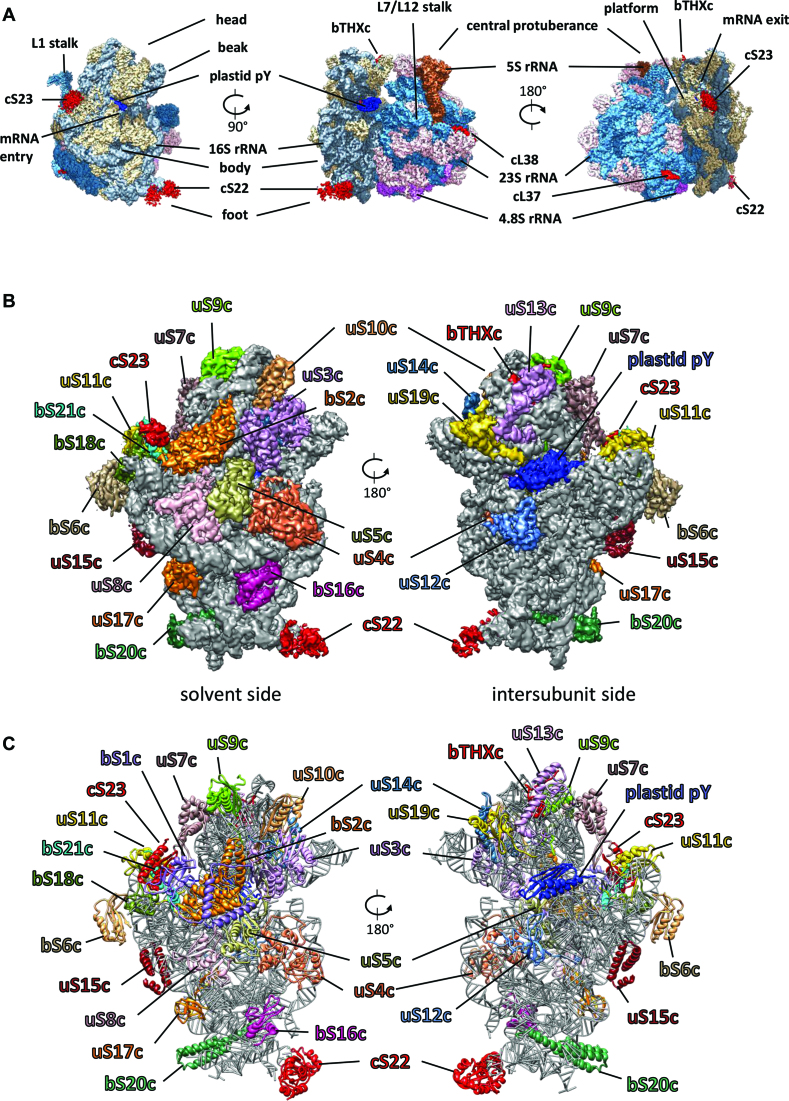
Figure 1.
Cryo-EM structure of the chloroplast 70S ribosome (A) Cryo-EM reconstruction (3.4 Å map) of the chloroplast 70S ribosome from spinach rendered at a density threshold value of 0.05 using UCSF Chimera (54). Density of 50S subunit proteins are shown in steel blue, 23S rRNA in pink, 5S rRNA in sienna, 4.8S rRNA in magenta, 30S subunit proteins in tan, 16S rRNA in slate gray, the translation factor pY in blue. PSRPs (cS22, cS23, bTHXc, cL37 and cL38) are shown in red. Structural landmarks of the ribosome are labeled. (B) Chloroplast ribosome SSU structure (3.7 Å map) from the solvent and the intersubunit side showing density for the PRPs and the PSRPs in differently colored surface at a density threshold value of 0.04 using UCSF Chimera (54). 16S rRNA is displayed in gray surface using the same threshold. Because of flexibility, density for bS1c does not appear at this threshold but can be observed in our 8-Å filtered map. (C) Models for the SSU proteins and 16S rRNA are displayed in the same colors as in (B).
Chloroplast ribosome SSU contains 24 proteins and 1491 nt of RNA (16S rRNA) (Figure 1B and C). All the 21 orthologs of bacterial ribosome SSU proteins are present in the chloroplast ribosome SSU while cS22, cS23 and bTHXc are plastid specific. Twelve SSU proteins (uS2c, uS3c, uS4c, uS7c, uS8c, uS11c, uS12c, uS14c, uS15c, bS16c, bS18c, uS19c) are encoded by the plastome while the remaining 12 proteins including the PSRPs (bS1c, uS5c, bS6c, uS9c, uS10c, uS13c, uS17c, bS20c, bS21c, cS22, cS23, bTHXc) are encoded by the nuclear genome and post-translationally imported into the chloroplast. The E. coli ribosome SSU model (PDB ID: 4YBB) (51) fitted quite well in the chloroplast SSU map and helped in identification and localization of all the PRPs and chloroplast 16S rRNA. The homology models of the PRPs and 16S rRNA were rigid body fitted in the bacterial ribosome and chloroplast-specific protein extensions were built de novo by tracing the density connected to the main bodies of the proteins (Supplementary Figure S5 and Table S3). After assigning densities for all the PRPs, clear mass of densities at four places remained unaccounted which we assigned to the three PSRPs and the translation factor plastid pY. Supplementary Figure S5 displays all the chloroplast SSU proteins fitted in their individual densities. In summary, we could localize all the PRPs and the PSRPs, and some PRP-extensions in our chloroplast ribosome SSU map. Our model of the chloroplast ribosome SSU comprises of 21 PRPs, 3 PSRPs, plastid pY and 16S rRNA which is consistent with the mass spectrometry studies reported earlier (30,59).
cS22 resides at the foot region of SSU
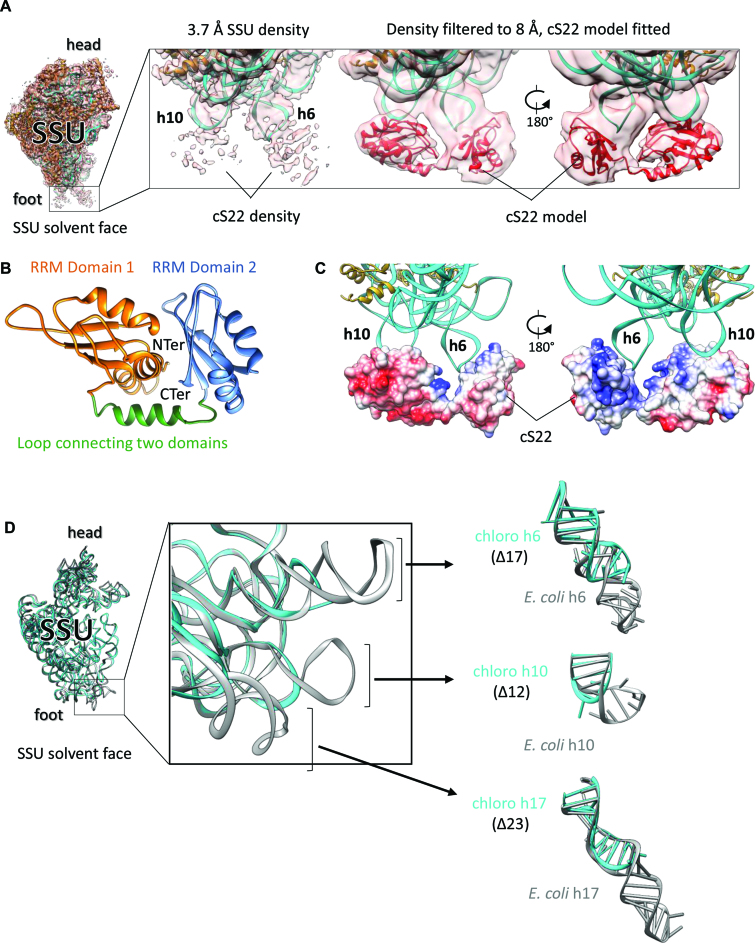
After localizing all the bacterial ribosome SSU homologous proteins, plastid-specific protein extensions and the 16S rRNA in the chloroplast ribosome SSU, densities at four places remained unaccounted which we assigned to cS22, cS23, bTHXc and plastid pY. One of these densities is present at the foot region of SSU and is highly fragmented in our 3.7 Å SSU map (Figure 2A, left). However, filtering the map to 8 Å reveals a bilobular shape of this density (Figure 2A, right) in which the homology model of cS22 (Figure 2B) could be accommodated. Our localization of cS22 at the chloroplast ribosome SSU foot is consistent with an earlier cryo-EM reconstruction by Sharma et al. (28). Homology model of cS22 revealed the presence of two globular RNA recognition motif (RRM) domains that were independently fitted in the two lobes of this density. The linker between the two RRM domains was subsequently modeled by tracing the density in the filtered map. Among the five PSRPs in the chloroplast 70S ribosome, cS22 is the only protein containing RRM domains (60). The RRMs are ubiquitous in eukaryotes and can bind a variety of RNA, DNA and protein sequences (61). The RRMs are characterized by the presence of a αβ-sandwich structure that folds in β1α1β2β3α2β4 fashion. Presence of high plasticity in RRM domains has been suggested to contribute toward high RNA-binding affinity and specificity in proteins (61). Analysis of our filtered map revealed that while bound to the chloroplast ribosome, cS22 adopts an open conformation where no interaction between the two RRM domains seems probable. It is possible that the flipped out bases in the hairpin loops of h6 and h10 stacks with the aromatic amino acids of cS22 to stabilize cS22 at this site (Figure 2A). The contribution of electrostatic forces between negatively charged rRNA and the basic patches of cS22 (Figure 2C) facing h6 and h10 can’t be ruled out either.
Figure 2.
Localization of cS22 at the chloroplast ribosome SSU foot (A) Fitting of cS22 at the SSU foot. SSU model fitted into high-resolution EM density showing extra density at the foot region near h6 and h10, which we assigned to cS22 (left). Proteins are in golden rod and 16S rRNA is in cyan. Zoomed-in view of the SSU foot showing high-resolution and 8 Å-filtered density map (right). Model of cS22 (in red) is fitted in the 8 Å-filtered map as indicated. (B) ITASSER (50) model of cS22 used for fitting the individual RRM domains in cS22 EM density. The two RRM domains of cS22 and connecting linker are labeled. (C) Coulombic surface coloring using UCSF Chimera (54) reveals that basic patches of cS22 interacts with negatively charged 16S rRNA (h6 and h10). (D) rRNA deletions in chloroplast ribosome 16S rRNA. Superimposition of 16S rRNA models from the chloroplast and the Escherichia coli ribosome (PDB ID: 4YBB) (51) (left). Zoomed view displays the three deletions in chloroplast ribosome 16S rRNA (center). Parts of h6, h10 and h17 are isolated to show comparison with E. coli counterparts (right). Chloroplast ribosome 16S rRNA is in cyan and E. coli 16S rRNA is in gray.
Structure of the chloroplast 16S rRNA is only altered at the foot region of SSU in the vicinity of cS22, compared to bacteria, due to deletions in h6, h10 and h17 (Figure 2D). cS22 seems to structurally compensate for the missing rRNA elements in h6 and h10 but the significance of such structural remodeling remains subtle considering that knockdown of cS22 does not affect ribosome assembly and translation in Arabidopsis under standard greenhouse conditions (32). Presence of cS22 is responsible for the increase in protein mass at the bottom of chloroplast ribosome SSU, compared to the bacterial ribosome (51). Given the diversity in the functioning of the RRM domains (61), it is possible that cS22 might act as an adaptor of protein factors important for ribosomal assembly and translation in chloroplast.
Localization of bTHXc at SSU head
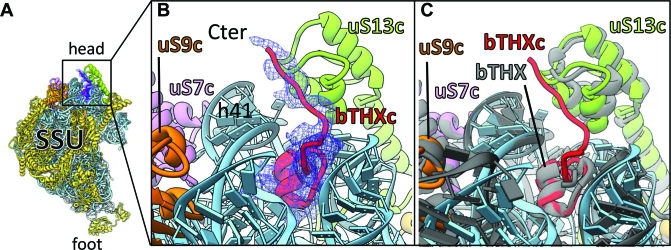
bTHXc (PSRP4) is a small, nuclear encoded and highly basic (pI 11.14) protein of the chloroplast ribosome SSU. Homologs of bTHXc exist in the ribosomes of T. thermophilus (bTHX) but are missing from the ribosomes of E. coli and Synechosystis (cyanobacteria). Consistent with the positioning of bTHX in T. thermophilus ribosome (62), we could localize the density for bTHXc at the chloroplast ribosome SSU head where it is buried in an rRNA environment (Figure 3). Our localization of bTHXc at the SSU head is consistent with Sharma et al. (28). Compared to bTHX of Thermus which is only 27 aas long, bTHXc is longer (47 aas) because of the chloroplast-specific C-terminal extension. Regardless of their lengths, superimposition of our bTHXc model to the Thermus bTHX (PDB ID: 4V4X) reveals that these two proteins share a common fold which interacts with the 16S rRNA helices h41 and h42 (h42 could not be seen in the displayed view in Figure 3) in the SSU. In bTHXc, the chloroplast-specific C-terminal extension further mediates hydrophobic interactions with uS13c (Figure 3B). uS13c is the only protein which is located in close vicinity of bTHXc. It is interesting to note that uS13c is an important constituent of two conserved intersubunit bridges (B1a and B1b) in chloroplast 70S ribosome. Since bTHX is only found in the ribosome of thermophilic bacteria, it seems plausible that chloroplasts have acquired bTHXc to stabilize the intersubunit bridge B1b under conditions of thermal fluctuations routinely experienced by plants. Importance of bTHXc in chloroplast ribosome is also evident from the knockdown studies in Arabidopsis which resulted in the reduced photosynthesis causing impaired growth (32).
Figure 3.
bTHXc localizes at the head of SSU (A) Position of bTHXc (red) at SSU head region. 16S rRNA is colored sky blue; ribosomal proteins except for those which surround bTHXc are colored golden rod. Isolated density of bTHXc is shown in blue mesh. (B) Zoomed-in view of (A) to show local environment of bTHXc. The core fold of bTHXc structure is buried in rRNA environment. (C) SSU structure of Thermus thermophilus ribosome (62) (gray) is overlaid with chloroplast ribosome SSU to show comparison between the structures of bTHXc and bTHX.
mRNA entry and exit sites
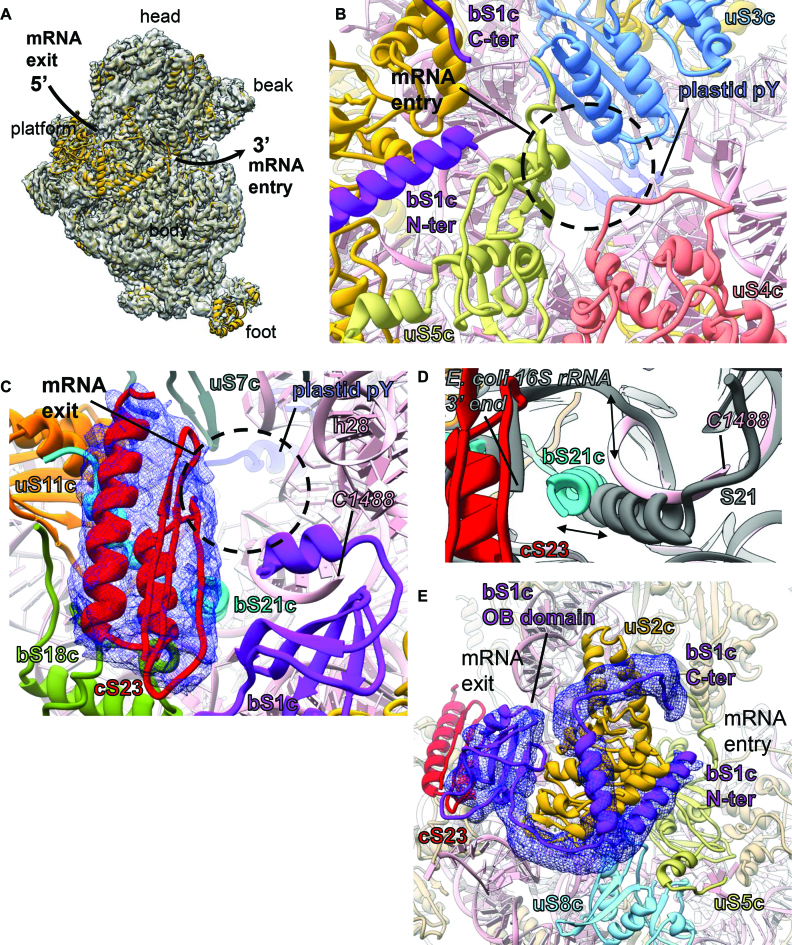
The mRNA channel is located on the intersubunit side of the neck region of the SSU. In the 70S ribosome, mRNA channel opens up to the solvent side of SSU via two openings: (i) the mRNA entry site which is located close to the beak and (ii) the mRNA exit site which is situated near to the platform (Figure 4A). The mRNA entry site in the chloroplast ribosome is surrounded by proteins uS3c, uS4c and uS5c (Figure 4B) in an orientation similar to the bacterial ribosome. Bacterial S3, S4 and S5 have been implicated in facilitating translation as S3 and S4 are reported to possess RNA-helicase activity and S5 being the protein involved in maintaining translation fidelity (63). It is interesting to note that, while uS3c and uS4c remains mostly conserved, uS5c has acquired an 86 aa-long extension (Supplementary Figure S6) at its N-terminal (NTE). Therefore, the mRNA entry site of the chloroplast ribosome is richer in protein content and appears to be little constricted compared to the bacterial ribosome (Supplementary Figure S6). However, most of the NTE of uS5c remain flexible and is not visible in our map.
Figure 4.
mRNA entry and exit sites, and localization of bS1c connecting both the sites (A) The mRNA entry and the exit sites are shown on the chloroplast ribosome SSU from the solvent exposed side. The arrow shows path of the mRNA. EM density for the SSU is shown in transparent surface (gray) with SSU model fitted. SSU proteins are colored golden rod and 16S rRNA is depicted in pink. (B) mRNA entry site. Three PRPs (uS3c, uS4c and uS5c) surround the mRNA entry site (black circle) in the chloroplast ribosome SSU. Fitted models of these PRPs are colored as: uS3c in sky blue, uS4c in salmon and uS5c in dark khaki. 16S rRNA is in pink. (C) mRNA exit site. Four PRPs (bS1c, uS7c, uS11c, bS21c) and one PSRP (cS23) along with h28 surround the mRNA exit site (black circle) in the chloroplast ribosome SSU and are colored as: bS1c in purple, uS7c in dim gray, uS11c in orange and bS21c in cyan. At the exit site, cS23 interacts with bS18c and bS21c. Isolated density of cS23 is displayed in blue mesh. 16S rRNA is colored pink. The nucleotide (C1488) at the 3′ end of our 16S rRNA model is indicated. (D) Shifted positions of bS21c (cyan) and the 3′ end of the chloroplast ribosome 16S rRNA (pink), compared to Escherichia coli (gray) ribosome SSU (PDB ID: 5AFI) (96). (E) Close association of bS1c (purple) with bS2c (golden rod). The N- and the C-terminals of bS1c extend toward the mRNA entry site while one of the three OB domains which is positioned closer to the C-terminal of bS1c reach out to the mRNA exit site. Proteins neighboring bS1c are colored as: cS23 in red, uS5c in dark khaki and uS8c in sky blue. 16S rRNA is in pink. Density for the N- and C-terminal of bS1c and the OB domain is displayed as blue mesh.
mRNA exit site (Figure 4C) harbors the 3′ end of the 16S rRNA and importance of this region is corroborated by the fact that it is the site of SD-anti SD interaction between the mRNA and the 16S rRNA in bacterial ribosomes (64–67) (Supplementary Figure S7a). SD-anti SD interaction plays a key role in translation initiation in the bacteria by ensuring correct positioning of the start codon at ribosomal P site (68). SD-like sequences are present among two-thirds of chloroplast mRNA but unlike the bacterial mRNA, their position is variable (19,25). Therefore, it remains elusive whether SD-anti SD interactions are necessary for translation of the chloroplastic mRNAs. After the assignment of all the PRPs and PRP-extensions, two PSRPs (bTHXc and cS22) and the 16S rRNA, we observed a mass of unaccounted density (Figure 4C) at the mRNA exit site in our SSU map. Considering that chloroplast SSU possesses only three PSRPs, we assigned this additional density to cS23 (Figure 4C). This density is poorly resolved compared to the densities of other proteins in close vicinity but the volume was big enough to accommodate the core fold of the cS23 homology model (Figure 4C). Although we do not have any further evidence for this density to be arising from cS23, we derived confidence in our assignment from mass spectrometry results which reported presence of only five PSRPs (29,30,59) (PSRP1 being the factor plastid pY) in the spinach chloroplast ribosome. Also, co-purification of any other proteins except for plastid pY (28,31) along with the 70S chloroplast ribosome has not been reported so far. Our localization of cS23 at mRNA exit site is at variance with the recently reported cryo-EM reconstruction of spinach chloroplast ribosome by Bieri et al. (35) where authors have localized cS23 at the SSU foot. In the light of accumulating evidences that translation initiation in chloroplast is far more complicated than anticipated (10,17,69), we observed major remodeling of the mRNA exit site in our structure (Figure 4C). The mRNA exit site of the chloroplast ribosome is more protein-rich (Supplementary Figure S7b and c) compared to the bacteria. The mRNA exit site is surrounded by proteins bS1c, uS7c, uS11c, bS21c, cS23 and 16S rRNA helix h28 in the chloroplast ribosome SSU. The most prominent difference compared to bacterial ribosomes is the presence of cS23 at this region (Supplementary Figure S7b and c). cS23 is positioned in a surface exposed fashion in the cleft formed between the neck (solvent side) and the platform of the SSU and mediates interactions with bS18c and bS21c (Figure 4C). bS21c (+66 aas) is significantly longer compared to its bacterial counterpart but most of the density for the extension is not visible in the map possibly due to flexibility. Additional differences at the mRNA exit site include repositioning of the C-terminal helix (residues 123–136) of bS21c which extensively interacts with the 3′ end of the 16S rRNA (Figure 4D). In this position, bS21c seems to lock and stabilize the 3′ end of 16S rRNA diminishing the possibility that 16S rRNA might contribute in SD-anti SD interaction.
Considering its location near to the platform, it seems that cS23 might form a scaffold to recruit trans-acting protein factors for efficient translation of certain mRNAs especially those lacking (or weak) SD sequences at the canonical position. Interestingly knockdown mutants of cS23 in Arabidopsis displayed severe defects in leaf anatomy affecting translation (32), which is suggestive of an important role played by cS23 in the chloroplastic translation. Apart from being a constituent protein of chloroplast ribosome, possibility of other roles (moon-lighting in gene expression or signaling) played by cS23 can’t be ruled out either, as speculated previously (32).
Localization of highly flexible protein bS1c which extends to both mRNA entry and exit sites
bS1c is the largest protein in the chloroplast ribosome that binds mRNA either in free form or while associated with the ribosome (70,71). Bacterial S1 is 557-amino acids in length comprising of six OB-fold (oligonucleotide/oligosaccharide-binding) domains. Chloroplastic S1 (Figure 4E) is truncated as it is 411-amino acids in length in spinach and contains only three OB-fold domains. Studies have shown that S1 is an essential player in translation initiation in bacteria (72–76) as it recruits mRNA to facilitate its binding to the mRNA channel, but the precise molecular mechanism has not been elucidated yet. Bacterial S1 interacts with the ribosome SSU in an on and off fashion (77) and is particularly essential in the translation of leaderless mRNAs or mRNAs containing weak SD sequences (74–76,78). The present consensus is that NTE of the bacterial S1 binds the SSU while its long C-terminal is flexible and due to its high affinity toward pyrimidine sequences, binds to the 5′ UTR of the mRNAs (79–83) to regulate the translation initiation in bacteria. It has been suggested in bacteria that a binary complex is formed between the protein S1 and the SSU (70,74,84) which is then recruited to 5′ UTR of mRNA to initiate translation. However, in contrast to bacterial S1, chloroplast bS1c (Figure 4E) by itself can’t direct the 30S complex for translation initiation and gene-specific nuclear-encoded translation factors have been proposed to play mediating roles in this process (83). In Chlamydomonas reinhardtii chloroplast, presence of three such mediator proteins had been reported which were suggested to act in a light-dependent manner (85). We observed partial density for bS1c (Figure 4E) on the chloroplast ribosome SSU after filtering the SSU map to 8 Å in which one of the OB domains and part of N- and C- terminal could be fitted. bS1c reaches to both the mRNA entry and the exit sites which is in agreement with an elongated S1 in bacterial SSU (86,87). Therefore, it seems that chloroplast bS1c plays an important role in the regulation of translation initiation in the chloroplast however the precise role remains unclear. Structurally, bacterial S1 is not well characterized due to its flexibility on the SSU. Compared to the bacterial counterpart we observe a stronger binding of bS1c with the chloroplast ribosome SSU. Previously, it has been shown in C. reinhardtii chloroplast that most of the S1 (CreS1) is SSU-associated during the whole translation process (70) supporting our notion that spinach bS1c is also associated with the spinach chloroplast ribosome SSU. Analysis of our structure reveals that, binding of bS1c on the SSU is predominantly mediated via uS2c although cS23, uS5c and uS8c also seem to contribute in stabilizing bS1c on the SSU (Figure 4E). Therefore, the N- and C-terminals and one of the three OB-fold domains of bS1c are involved in SSU binding. However, the other two OB-fold domains for which we did not observe any density possibly due to flexibility, might be involved in mRNA binding. Given the diversity in the functioning of the OB-fold domains (88) to bind a variety of substrates like DNA, RNA and proteins (89) it is also possible that OB-fold domains might even act as launching pads for translation factors mediating protein–protein or protein–mRNA interactions during translation initiation in the chloroplast. In this context, it seems probable that bS1c and cS23 might act in concert to regulate translation initiation in the chloroplast ribosome. In the light of accumulating evidences (73,90–93) that bacterial S1 contains RNA helicase activity, it is also possible that bS1c might also possess similar helicase activity to destabilize secondary structural elements in the 5′ UTR of chloroplastic mRNAs to facilitate translation initiation. However, further studies are required to elucidate any further details.
Interaction of plastid pY with the chloroplast ribosome SSU and conservation of inter-subunit bridges
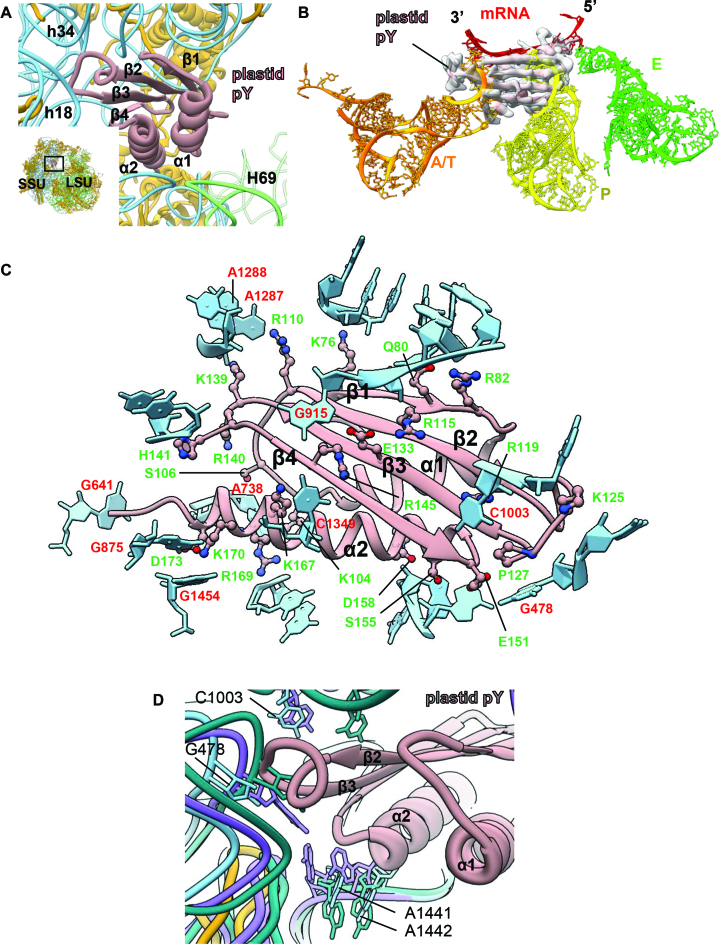
Consistent with earlier studies (28,31) translation factor plastid pY co-purified with our spinach chloroplast 70S ribosome (Figure 5A). Plastid pY was earlier considered as a PSRP1 due to its association with the chloroplast ribosome but later a biochemical study (31) revealed that it is rather a translation factor and homologous to bacterial cold shock protein pY (also known as YfiA, RaiA and SpotY). We observed density for plastid pY in our map at the intersubunit space, overlapping with the mRNA channel of the SSU (Figure 5B) which is in agreement with the cryo-EM reconstruction of the chloroplast ribosome by Sharma et al. (28). This is also consistent with the binding position of plastid pY-homolog, YfiA to the T. thermophilus 70S ribosome (53). Plastid pY is a 302-residue nuclear-encoded protein. Barring the first 66 residues which accounts for the signal peptide, the mature protein consists of 236 residues (67–302). Homology model of plastid pY reveals presence of two distinct domains in the protein where the NTD is well ordered and assumes a conserved (61,94) β1α1β2β3α2β4 fold while the C-terminal domain (CTD) is disordered. Homology model of the NTD of plastid pY fitted quite well into the plastid pY density in our SSU map. Well-resolved density of the NTD and the surrounding regions allowed us to identify the plastid pY residues which interacts with the SSU while the density for the CTD was not visible in our map possibly due to flexibility. Therefore, the role of the CTD of plastid pY still remains elusive although the interactions of the NTD with the chloroplast ribosome SSU (Figure 5C) provide important insights about its functioning as a hibernation factor in the context of translation regulation inside the chloroplast.
Figure 5.
Binding of the plastid translation factor pY on SSU (A) Position of plastid pY at the intersubunit space is shown in the snippet at the bottom left. Both the ribosomal subunits are indicated. SSU proteins are colored golden rod, 16S rRNA is colored sky blue and LSU rRNAs (4.8S, 5S and 23S) are in green. The association of plastid pY exclusively to the SSU is displayed in the enlarged view. Plastid pY is colored rosy brown; orientation of the α helices and the beta strands of plastid pY in close association with 16S rRNA is indicated. (B) Plastid pY precludes binding of mRNA as well as A- and P-site tRNAs on the chloroplast ribosome. Overlaid tRNAs and mRNA from the Escherichia coli ribosome (PDB ID: 5AFI) (96) are colored as: A/T-site in orange, P-site in yellow, E-site in green and mRNA in red. EM density for plastid pY is depicted in transparent surface (gray) and fitted model of plastid pY is colored rosy brown. (C) Interactions between plastid pY and 16S rRNA are demonstrated. Plastid pY residues which contribute toward its binding to SSU are displayed in ball-and-stick and labeled in green. 16S rRNA is colored sky blue; only the nucleotide bases which directly interact with plastid pY are labeled (red). Interaction with plastid pY is also mediated by sugar-phosphate backbone of 16S rRNA but labels are hidden for clarity. (D) The conserved nucleotide bases (G478, C1003, A1441, A1442) involved in decoding are indicated. 16S rRNA from the apo- (PDB ID: 4YBB, in green) (51) and the translating states (PDB ID: 5AFI, in purple) (96) of the E. coli ribosome are overlaid with the chloroplast ribosome 16S rRNA to show relative positions of these conserved bases in our translationally inactive chloroplast 70S ribosome.
Binding of the plastid pY at the intersubunit space precludes the binding of mRNA, A- and P-site tRNAs on the ribosome (Figure 5B). Helix α1 is situated facing the H69 of the LSU while helix α2 is positioned right above the mRNA channel while the four beta strands of plastid pY face the head of SSU (Figure 5A and B). Between the two helices of plastid pY, interaction of α2 with the ribosome seems to be more extensive than α1 (Figure 5A). Polar and charged amino acids in α2 (Ser89, Asp92, Lys101, Arg103, Lys104, Lys106 and Asp107) interacts with the negatively charged backbone of 16S rRNA while only two positively charged residues in α1 (Lys31 and Lys38) seems to stabilize the factor via interactions with the 16S rRNA backbone (Figure 5C). Encompassing the four beta strands, several residues like Lys10, Gln14, Arg49, Glu67 and Arg79 seems to mediate interactions mostly with the 16S rRNA backbone (Figure 5C). Interestingly, some of the bases of 16S rRNA (G641, A738, G875, C1003, A1287, A1288, G1454) interacts with both basic and acidic residues of plastid pY while stabilization of G478 (G530 in E. coli) is mediated via stacking interactions with Pro 127 (Figure 5C).
In bacteria, the universally conserved 16S rRNA bases A1492, A1493 and G530 have been implicated to play a major role during canonical decoding event (95). In the apo state of bacterial ribosome (PDB ID: 4YBB) (51), A1492 and A1493 maintain a flipped in conformation within the internal loop of h44 while in the translating bacterial ribosome (PDB ID: 5AFI) (96) these two bases bulge out (Figure 5D). In our translationally inactive chloroplast 70S ribosome, the equivalent bases A1441 (A1492 in E. coli) and A1442 (1493 in E. coli) assume an intermediate position while remaining stacked to each other and are stabilized by interacting with Glu 151, Ser 155 and Asp 158 (Figure 5C and D) of plastid pY. On the other hand, the orientation of G478 (G530 in E. coli) and C1003 (C1054 in E. coli) remains similar as compared to the bacterial ribosome (Figure 5D). The slight displacement in the positioning of G478 as noticed in our structure (Figure 5D) is caused by the complete head-swiveling of the chloroplast SSU. An important aspect of plastid pY structure is the presence of a longer loop between the beta strands β2 and β3 (Supplementary Figure S8), compared to its bacterial homolog, which is stabilized by interacting with h18 and h34 (Figure 5A). Taken together, interaction of plastid pY with the SSU serves two purpose: (i) it precludes the binding of mRNA and tRNA substrates by blocking the mRNA channel and the A- and P- sites of the SSU and (ii) while no substrate can enter ribosome, it locks the ribosome in a translationally inactive state (hibernation) during unfavorable conditions. The ribosome might go back to an active state probably with the help of elongation factor G (EF-G) and ribosome recycling factor when the conditions are conducive of protein synthesis but the mechanism remains unknown.
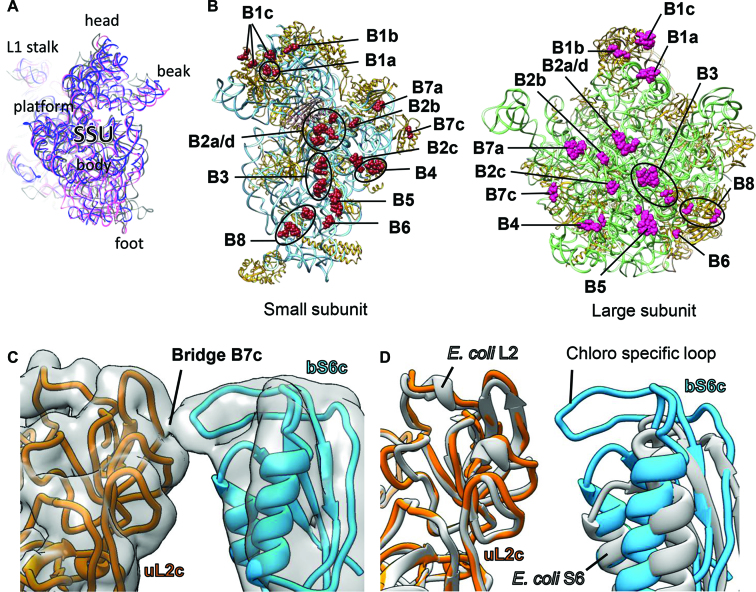
Interestingly, plastid pY stabilizes a non-rotated state of the chloroplast SSU which is similar to the release factor RF1-bound state (PDB ID: 4V63) (97) of the T. thermophilus 70S ribosome (Figure 6A) compared to the rotated state of the SSU (PDB ID: 4V6C) (98). Most of the intersubunit bridges observed for bacterial ribosomes (99) are also conserved in this state of the 70S chloroplast ribosome. Therefore, RNA–RNA interactions are the prevalent mode of interactions albeit some protein–protein and RNA–protein interactions contribute in bridge formation (Figure 6B and Supplemental Figure S9) as studied in the bacterial ribosome. All the intersubunit bridges observed in our structure are listed in Supplementary Table S4 and the corresponding densities are displayed in Supplementary Figure S9. Surprisingly, a conserved bacterial bridge, B7b is absent in our structure of 70S chloroplast ribosome. Mutagenesis studies on bacterial ribosome have reported B7b to contribute in maintaining the fidelity of the translation initiation in bacteria (100). However, the absence of B7b is compensated by a new bridge (B7c) near to the B7b site involving Lys161 and Ala162 of the chloroplast-specific loop of bS6c and Pro132, Leu133, Ala162 and Lys163 of uL2c (Figure 6B–D). This chloroplast-specific loop of bS6c (residues 160–168) is truncated (Figure 6D) in bacteria and therefore might explain why B7c is absent in bacterial ribosomes. Taken together, it seems that plastid pY prevents the fall-off of the subunits during translationally inactive state of the chloroplast ribosome by stabilizing a non-rotated state of the SSU.
Figure 6.
Intersubunit bridges of chloroplast 70S ribosome: (A) SSU models from PDB IDs: 4V6C (98) (gray) and 4V63 (97) (pink) overlaid with the chloroplast ribosome SSU displaying that plastid pY stabilizes a non-rotated state of the chloroplast ribosome. (B) The rRNA and protein residues of the SSU (left panel) and the LSU (right panel) which contribute in intersubunit bridge formation are represented as brown and pink spheres, respectively. 16S rRNA and the SSU proteins (left panel) are depicted in sky blue and golden rod, respectively. Plastid pY is displayed in rosy brown. 23S rRNA and LSU proteins (right panel) are depicted in green and golden rod, respectively. 5S rRNA and 4.8S rRNA (right panel) are colored in tan. In (B), both the ribosomal subunits are displayed from the subunit interface side. (C) Novel bridge in chloroplast 70S ribosome (B7c) is formed between proteins uL2c (golden rod) and bS6c (dodger blue). Density is 8 Å-filtered for clarity and shown in transparent surface (gray). Neighboring proteins and rRNAs are hidden for better visualization. (D) Models of the proteins S6 and L2 from Escherichia coli ribosome (PDB ID: 4YBB) (51), gray) overlaid with counterparts from chloroplast 70S ribosome showing absence of elongated loop in bacterial S6.
CONCLUSION
Here we report a complete near-atomic cryo-EM structure of the 70S chloroplast ribosome from spinach, predominantly focusing on the discussion of the SSU. The chloroplast ribosome LSU structure had earlier been discussed in details in our previously published article (33). While evolving from its bacterial ancestor, 16S rRNA of the chloroplast ribosome has acquired lesser changes compared to the rRNAs of the LSU. Among the three remodeled sites of the 16S rRNA which are caused exclusively by rRNA deletions, two are compensated by the presence of the PSRP cS22. Unlike the mRNA entry site which evolved to only procure a few PRP-extensions, we observe significant changes around the mRNA exit site primarily due to the presence of the PSRP cS23. Acquiring cS23 at this important regulatory site (platform) on the SSU might have been necessary for plants to cope with the varied positioning of the putative SD-like sequences in the chloroplastic mRNAs. The unique localization of the cS23 at the mRNA exit site also suggests an alternative mode of translation initiation in the chloroplast that might operate alongside the canonical bacterial pathway. Considering the role of SSU in the initiation of translation and the fact that translation initiation is far more complex in chloroplasts than previously anticipated (10,17,69), the unique localization of the PSRPs in our high-resolution structure of the 30S chloroplast ribosome thus reveals important insights about the functioning of the PSRPs in the context of translation regulation inside chloroplasts. While our manuscript was under preparation, two cryo-EM structures of the chloroplast 70S ribosome were reported by Graf et al. (34) and Bieri et al. (35). Graf et al. (34) reported the cryo-EM reconstitutions of the LSU and the SSU at 3.6 and 5.4 Å resolution, respectively. Although no meaningful comparison could be drawn for their SSU structure due to unavailability of the SSU model, our results concerning the chloroplast ribosome LSU are in good agreement with their LSU structure. In the report made by Bieri et al. (35), the SSU and the LSU structures are at 3.6 and 3.2 Å resolution, respectively. Superimposition of their map to ours reveals two main differences in the SSU: (i) the presence of an extra density in our map at the mRNA exit site, which we have assigned to cS23 and (ii) the presence of an additional mass of density at the SSU foot where we have fitted almost the complete model of cS22. Therefore, while Bieri et al. (35) mentioned that one of the two RRM domains of cS22 could not be observed in their SSU foot due to flexibility, we could accommodate both the RRM domains at this region in our low pass filtered map. Taken together, it can be said that the high-resolution structure of the chloroplast 70S ribosome would help in designing experiments to reveal further molecular details about the chloroplastic translation system.
ACCESSION NUMBERS
The 3.4 Å 70S, 3.7 Å SSU and 3.3 Å LSU cryo-EM density maps of the spinach chloroplast ribosome have been deposited in the Electron Microscopy Data Bank with accession codes EMD-6709, EMD-6710 and EMD-6711, respectively. The coordinates of the atomic structures of the 70S ribosome, SSU and LSU have been submitted to the Protein Data Bank as 5X8P, 5X8R and 5X8T, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andrew See Weng Wong for electron microscopy; Zhan Yin for help during data processing; Rikky Wenang Purbojati and Darshan Chandrakant Redij for help in computing and National Supercomputing Center (NSCC) Singapore for providing computing facility.
Author Contributions. S.B. conceived and supervised the project. T.A. and S.B. purified chloroplast ribosomes. J.S. collected the cryo-EM data. T.A. and S.B. carried out the cryo-EM data processing and interpreted the maps. T.A. built the models. T.A. and S.B. wrote the manuscript. All authors reviewed the final version of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nanyang Technological University Start Up Grant; Ministry of Education of Singapore [AcRF Tier 1, 2014-T1–001-019 (RG32/14) to S.B.]. Funding for open access charge: Research Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Greber B.J., Ban N.. Structure and Function of the Mitochondrial Ribosome. Annu. Rev. Biochem. 2016; 85:103–132. [DOI] [PubMed] [Google Scholar]
- 2. Hashem Y., des Georges A., Fu J., Buss S.N., Jossinet F., Jobe A., Zhang Q., Liao H.Y., Grassucci R.A., Bajaj C. et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013; 494:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gupta A., Shah P., Haider A., Gupta K., Siddiqi M.I., Ralph S.A., Habib S.. Reduced ribosomes of the apicoplast and mitochondrion of Plasmodium spp. and predicted interactions with antibiotics. Open Biol. 2014; 4:140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. 1970; New Have: Yale University Press. [Google Scholar]
- 5. Jensen P.E., Leister D.. Chloroplast evolution, structure and functions. F1000Prime Reports. 2014; 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin W., Herrmann R.G.. Gene transfer from organelles to the nucleus: how much, what happens, and why. Plant Physiol. 1998; 118:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris E.H., Boynton J.E., Gillham N.W.. Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 1994; 58:700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmitz-Linneweber C., Maier R.M., Alcaraz J.-P., Cottet A., Herrmann R.G., Mache R.. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001; 45:307–315. [DOI] [PubMed] [Google Scholar]
- 9. Manuell A., Beligni M.V., Yamaguchi K., Mayfield S.P.. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem. Soc. Trans. 2004; 32:601–605. [DOI] [PubMed] [Google Scholar]
- 10. Navarro M.J., Manuell A. L., Quispe J., Wu J., Mayfield S. P.. Chloroplast translation regulation. Photosynth. Res. 2007; 94:359–374. [DOI] [PubMed] [Google Scholar]
- 11. Sugita M., Sugiura M.. Filipowicz W, Hohn T. Post-Transcriptional Control of Gene Expression in Plants. 1996; Dordrecht: Springer; 315–326. [Google Scholar]
- 12. Zerges W. Translation in chloroplasts. Biochimie. 2000; 82:583–601. [DOI] [PubMed] [Google Scholar]
- 13. Barkan A., Goldschmidt-Clermont M.. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000; 82:559–572. [DOI] [PubMed] [Google Scholar]
- 14. Eberhard S., Drapier D., Wollman F.-A.. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002; 31:149–160. [DOI] [PubMed] [Google Scholar]
- 15. Surpin M., Larkin R.M., Chory J.. Signal transduction between the chloroplast and the nucleus. Plant Cell. 2002; 14:s327–s338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tiller N., Bock R.. The translational apparatus of plastids and its role in plant development. Mol. Plant. 2014; 7:1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sugiura M., Hirose T., Sugita M.. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998; 32:437–459. [DOI] [PubMed] [Google Scholar]
- 18. Liere K., Börner T.. Bock R. Cell and Molecular Biology of Plastids. 2007; Berlin: Springer; 121–174. [Google Scholar]
- 19. Peled-Zehavi H., Danon A.. Bock R. Cell and Molecular Biology of Plastids. 2007; Berlin: Springer; 249–281. [Google Scholar]
- 20. Beligni M.V., Yamaguchi K., Mayfield S.P.. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth. Res. 2004; 82:315–325. [DOI] [PubMed] [Google Scholar]
- 21. Drechsel O., Bock R.. Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res. 2011; 39:1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruf M., Kössel H.. Occurrence and spacing of ribosome recognition sites in mRNAs of chloroplasts from higher plants. FEBS Lett. 1988; 240:41–44. [Google Scholar]
- 23. Baecker J.J., Sneddon J.C., Hollingsworth M.J.. Efficient translation in chloroplasts requires element(s) upstream of the putative ribosome binding site from atpI. Am. J. Bot. 2009; 96:627–636. [DOI] [PubMed] [Google Scholar]
- 24. Bonham-Smith P.C., Bourque D.P.. Translation of chloroplast-encoded mRNA: potential initiation and termination signals. Nucleic Acids Res. 1989; 17:2057–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirose T., Sugiura M.. Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol. 2004; 45:114–117. [DOI] [PubMed] [Google Scholar]
- 26. Klaff P., Mundt S.M., Steger G.. Complex formation of the spinach chloroplast psbA mRNA 5′ untranslated region with proteins is dependent on the RNA structure. RNA. 1997; 3:1468–1479. [PMC free article] [PubMed] [Google Scholar]
- 27. Manuell A.L., Quispe J., Mayfield S.P.. Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 2007; 5:e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma M.R., Wilson D.N., Datta P.P., Barat C., Schluenzen F., Fucini P., Agrawal R.K.. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19315–19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamaguchi K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28466–28482. [DOI] [PubMed] [Google Scholar]
- 30. Yamaguchi K., von Knoblauch K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28455–28465. [DOI] [PubMed] [Google Scholar]
- 31. Sharma M.R., Donhofer A., Barat C., Marquez V., Datta P.P., Fucini P., Wilson D.N., Agrawal R.K.. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G). J. Biol. Chem. 2010; 285:4006–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tiller N., Weingartner M., Thiele W., Maximova E., Schöttler M.A., Bock R.. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 2012; 69:302–316. [DOI] [PubMed] [Google Scholar]
- 33. Ahmed T., Yin Z., Bhushan S.. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 2016; 6:35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Graf M., Arenz S., Huter P., Donhofer A., Novacek J., Wilson D.N.. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 2016; 45:2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N.. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2016; 36:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartsch M., Kimura M., Subramanian A.-R.. Purification, primary structure, and homology relationships of a chloroplast ribosomal protein. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:6871–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B.. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005; 151:41–60. [DOI] [PubMed] [Google Scholar]
- 38. Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y.. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Meth. 2013; 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mindell J.A., Grigorieff N.. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- 40. Bell J.M., Chen M., Baldwin P.R., Ludtke S.J.. High resolution single particle refinement in EMAN2.1. Methods. 2016; 100:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kucukelbir A., Sigworth F.J., Tagare H.D.. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenthal P.B., Henderson R.. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003; 333:721–745. [DOI] [PubMed] [Google Scholar]
- 44. Afonine P.V., Headd J.J., Terwilliger T.C., Adams P.D.. New tool: phenix.real_space_refine. Comput. Crystallogr. Newsl. 2013; 4:43–44. [Google Scholar]
- 45. Rother M., Milanowska K., Puton T., Jeleniewicz J., Rother K., Bujnicki J.M.. ModeRNA server: an online tool for modeling RNA 3D structures. Bioinformatics. 2011; 27:2441–2442. [DOI] [PubMed] [Google Scholar]
- 46. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:doi:10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004; 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2013; 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008; 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.D.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A.. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015; 22:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 55. Polikanov Y.S., Blaha G.M., Steitz T.A.. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012; 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schrödinger L.L.C. Py-MOL, The PyMOL Molecular Graphics System, V1.8. 2010. [Google Scholar]
- 59. Yamaguchi K., Subramanian A.R.. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur. J. Biochem. 2003; 270:190–205. [DOI] [PubMed] [Google Scholar]
- 60. Xu T., Lee K., Gu L., Kim J.I., Kang H.. Functional characterization of a plastid-specific ribosomal protein PSRP2 in Arabidopsis thaliana under abiotic stress conditions. Plant Physiol. Biochem. 2013; 73:405–411. [DOI] [PubMed] [Google Scholar]
- 61. Maris C., Dominguez C., Allain F.H.. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005; 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- 62. Yusupova G., Jenner L., Rees B., Moras D., Yusupov M.. Structural basis for messenger RNA movement on the ribosome. Nature. 2006; 444:391–394. [DOI] [PubMed] [Google Scholar]
- 63. Kurkcuoglu O., Doruker P., Sen T.Z., Kloczkowski A., Jernigan R.L.. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys. Biol. 2008; 5:046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Steitz J.A., Jakes K.. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jacob W.F., Santer M., Dahlberg A.E.. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hui A., de Boer H.A.. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shine J., Dalgarno L.. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Calogero R.A., Pon C.L., Canonaco M.A., Gualerzi C.O.. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997; 115:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Merendino L., Falciatore A., Rochaix J.-D.. Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii. Plant Mol. Biol. 2003; 53:371–382. [DOI] [PubMed] [Google Scholar]
- 71. Franzetti B., Carol P., Mache R.. Characterization and RNA-binding properties of a chloroplast S1-like ribosomal protein. J. Biol. Chem. 1992; 267:19075–19081. [PubMed] [Google Scholar]
- 72. S⊘rensen M.A., Fricke J., Pedersen S.. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo1. J. Mol. Biol. 1998; 280:561–569. [DOI] [PubMed] [Google Scholar]
- 73. Kolb A., Hermoso J.M., Thomas J.O., Szer W.. Nucleic acid helix-unwinding properties of ribosomal protein S1 and the role of S1 in mRNA binding to ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:2379–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boni I.V., Isaeva D.M., Musychenko M.L., Tzareva N.V.. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991; 19:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Farwell M.A., Roberts M.W., Rabinowitz J.C.. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 1992; 6:3375–3383. [DOI] [PubMed] [Google Scholar]
- 76. Tzareva N.V., Makhno V.I., Boni I.V.. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994; 337:189–194. [DOI] [PubMed] [Google Scholar]
- 77. Subramanian A.-R., van Duin J.. Exchange of individual ribosomal proteins between ribosomes as studied by heavy isotope-transfer experiments. Mol. Gen. Genet. 1977; 158:1–9. [DOI] [PubMed] [Google Scholar]
- 78. Komarova A.V., Tchufistova L.S., Dreyfus M., Boni I.V.. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005; 187:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Byrgazov K., Manoharadas S., Kaberdina A.C., Vesper O., Moll I.. Direct interaction of the N-terminal domain of ribosomal protein S1 with protein S2 in Escherichia coli. PLoS One. 2012; 7:e32702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sillers I.Y., Moore P.B.. Position of protein S1 in the 30 S ribosomal subunit of Escherichia coli. J. Mol. Biol. 1981; 153:761–780. [DOI] [PubMed] [Google Scholar]
- 81. Subramanian A.R., Rienhardt P., Kimura M., Suryanarayana T.. Fragments of ribosomal protein S1 and its mutant form m1-S1. Eur. J. Biochem. 1981; 119:245–249. [DOI] [PubMed] [Google Scholar]
- 82. McGinness K.E., Sauer R.T.. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shteiman-Kotler A., Schuster G.. RNA-binding characteristics of the chloroplast S1-like ribosomal protein CS1. Nucleic Acids Res. 2000; 28:3310–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Subramanian A.-R. Progress in Nucleic Acid Research and Molecular Biology. 1983; 28, Massachusetts: Academic Press; 101–142. [DOI] [PubMed] [Google Scholar]
- 85. Somanchi A., Mayfield S.P.. Nuclear–chloroplast signalling. Curr. Opin. Plant Biol. 1999; 2:404–409. [DOI] [PubMed] [Google Scholar]
- 86. Sengupta J., Agrawal R.K., Frank J.. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dunkle J.A., Cate J.H.D.. Ribosomes: Structure, Function, and Dynamics. 2011; Vienna: Springer; 65–73. [Google Scholar]
- 88. Flynn R.L., Zou L.. Oligonucleotide/oligosaccharide-binding (OB) fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010; 45:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002; 12:794–801. [DOI] [PubMed] [Google Scholar]
- 90. Rajkowitsch L., Schroeder R.. Dissecting RNA chaperone activity. RNA. 2007; 13:2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thomas J.O., Kolb A., Szer W.. Structure of single-stranded nucleic acids in the presence of ribosomal protein S1. J. Mol. Biol. 1978; 123:163–176. [DOI] [PubMed] [Google Scholar]
- 92. Bear D.G., Ng R., Van Derveer D., Johnson N.P., Thomas G., Schleich T., Noller H.F.. Alteration of polynucleotide secondary structure by ribosomal protein S1. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qu X., Lancaster L., Noller H.F., Bustamante C., Tinoco I.. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14458–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Birney E., Kumar S., Krainer A.R.. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993; 21:5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Laursen B.S., S⊘rensen H.P., Mortensen K.K., Sperling-Petersen H.U.. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005; 69:101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., Stark H.. Structure of the E. coli ribosome-EF-Tu complex at <3 a resolution by Cs-corrected cryo-EM. Nature. 2015; 520:567–570. [DOI] [PubMed] [Google Scholar]
- 97. Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H.F.. Structural basis for translation termination on the 70S ribosome. Nature. 2008; 454:852–857. [DOI] [PubMed] [Google Scholar]
- 98. Zhang W., Dunkle J.A., Cate J.H.. Structures of the ribosome in intermediate states of ratcheting. Science. 2009; 325:1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu Q., Fredrick K.. Intersubunit bridges of the bacterial ribosome. J. Mol. Biol. 2016; 428:2146–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sun Q., Vila-Sanjurjo A., O’Connor M.. Mutations in the intersubunit bridge regions of 16S rRNA affect decoding and subunit-subunit interactions on the 70S ribosome. Nucleic Acids Res. 2011; 39:3321–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.