Abstract
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) plays a key role in mediating non-homologous end joining (NHEJ), a major repair pathway for DNA double-strand breaks (DSBs). The activation, function and dynamics of DNA-PKcs is regulated largely by its reversible phosphorylation at numerous residues, many of which are targeted by DNA-PKcs itself. Interestingly, these DNA-PKcs phosphorylation sites function in a distinct, and sometimes opposing manner, suggesting that they are differentially regulated via complex actions of both kinases and phosphatases. In this study we identified several phosphatase subunits as potential DSB-associated proteins. In particular, protein phosphatase 1 (PP1) is recruited to a DSB-mimicking substrate in Xenopus egg extracts and sites of laser microirradiation in human cells. Depletion of PP1 impairs NHEJ in both Xenopus egg extracts and human cells. PP1 binds multiple motifs of DNA-PKcs, regulates DNA-PKcs phosphorylation, and is required for DNA-PKcs activation after DNA damage. Interestingly, phosphatase 1 nuclear targeting subunit (PNUTS), an inhibitory regulator of PP1, is also recruited to DNA damage sites to promote NHEJ. PNUTS associates with the DNA-PK complex and is required for DNA-PKcs phosphorylation at Ser-2056 and Thr-2609. Thus, PNUTS and PP1 together fine-tune the dynamic phosphorylation of DNA-PKcs after DNA damage to mediate NHEJ.
INTRODUCTION
DNA double strand breaks (DSBs) are frequently induced in eukaryotic cells by various exogenous and endogenous DNA damaging agents. Non-homologous end joining (NHEJ) is a major DSB repair pathway that directly re-ligates the DSB ends after they are processed to become compatible for ligation. Unlike homologous recombination (HR), NHEJ is executed without the need for a template, and is active in almost all stages of the cell cycle. Malfunction of NHEJ has been linked to premature aging, neurological diseases, immunodeficiency and cancer (1–3).
The DNA-dependent protein kinase (DNA-PK) complex composed of Ku70, Ku80 and DNA-PK catalytic subunit (DNA-PKcs) is the core component of NHEJ. The assembly of DNA-PK at DSB ends serves as a platform to recruit Artemis, DNA ligase IV and other NHEJ factors that are involved in end-processing and ligation (4,5). Within the DNA-PK complex, Ku proteins confer high affinity to DSB ends, and function as early sensors. The subsequent recruitment of DNA-PKcs to DSBs via the Ku proteins triggers the activation of DNA-PKcs, a member of the phosphatidylinositol 3-kinase-related kinase (PIKK) family. Upon activation, DNA-PKcs phosphorylates a number of substrates, including H2AX, XRCC4, Artemis and most importantly, DNA-PKcs itself. Autophosphorylation of DNA-PKcs occurs at numerous Ser/Thr residues throughout the kinase, and has been shown to mediate NHEJ (4–7). Intriguingly, mounting evidence revealed that these phosphorylation events control DNA-PKcs activity and configuration, and the subsequent end-processing, in a complex and sometimes opposing, manner. For example, DNA-PKcs phosphorylation at Thr-2609 and other sites of the ABCDE cluster by both ATM and DNA-PKcs facilitates Artemis recruitment and end resection, whereas DNA-PKcs autophosphorylation at Ser-2056 and surrounding sites within the PQR cluster limits end-processing, but may enable end-ligation by promoting the removal of DNA-PK (8–10). Moreover, three DNA-PKcs autophosphorylation sites, including Ser-56 and Ser-72 at the N-terminus and Thr-3950 at the C-terminal kinase domain, can lead to kinase inactivation (8,11–13). These findings suggest a dynamic and, possibly, site-specific regulation of DNA-PKcs autophosphorylation after DNA damage. In principle, the complex and differential pattern of DNA-PKcs phosphorylation cannot result solely from autophosphorylation. Instead, regulated actions of protein phosphatases toward distinct sites/clusters of DNA-PKcs may be crucially involved. In vitro enzymatic analysis indicated that dephosphorylation of DNA-PK by PP1 or PP2A-like phosphatases is necessary to activate the kinase activity of DNA-PKcs (5,14). Although a number of protein phosphatases have been implicated in NHEJ, molecular mechanisms by which specific phosphatases regulate DNA-PKcs remain to be uncovered (15–17).
Protein phosphatase 1 (PP1) is a major Ser/Thr phosphatase that accounts for ∼50% of all Ser/Thr phosphatase activities (18). Vertebrate cells encode several isoforms of PP1, including PP1α, β and γ. These isoforms of PP1 are highly similar in sequences and functions, but may differ in tissue-specific expression and subcellular localization. Existing studies indicate that PP1 relies on additional regulatory/targeting subunits to achieve their specific functions. It has been estimated that over 200 PP1 binding proteins may exist to regulate PP1 activities, as well as the subcellular localization and substrate recognition of PP1 (18–21). One of these PP1 regulators is phosphatase 1 nuclear targeting subunit (PNUTS, also known as PPP1R10), which was originally described as a nuclear regulator of PP1 that retains a portion of PP1 in the nucleus (22). PNUTS is predominantly localized to the interphase nucleus where it is partially chromatin-bound (22,23). Previous studies have implicated PNUTS in transcriptional regulation, cell cycle control, apoptosis, and telomere maintenance (23–30). A recent study also linked PNUTS to DNA damage response (31), but the underlying mechanism is unknown. In this study, we show that PP1 and PNUTS act in NHEJ by modulating the phosphorylation and activation of DNA-PKcs.
MATERIALS AND METHODS
Cell culture and treatment
The Human cervix carcinoma (HeLa) cell line, authenticated by ATCC, was maintained in Dulbecco's modified Eagle medium (DMEM, Hyclone) with 10% fetal bovine serum (FBS, Hyclone). Cell proliferation and death assays were performed as in our previous study (32). Ionized radiation was performed using an X-ray cabinet (RS-2000 Biological irradiator). Multiple siRNAs targeting PP1γ (target sequence CUAUCAACAUUAGGAGUA or AACUGUUGAUGGCAUAUU), PNUTS (target sequence GGCGGCUACAAACUUCUU or UCUGACAAGUACAACCUU), and Ku70 (target sequence UUAGUGAUGUCCAAUUCA) were purchased from Integrated DNA Technologies (IDT), and transfected into cells using a protocol recommended by the manufacturer. A non-targeting control siRNA was used as control.
Immunoblotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were carried out as previously described (33), using the following antibodies: Artemis (A300-234A), Chk1 phospho-S317 (A304-673A), Ku80 (A302-627A), γ-H2AX (A300-081A), PP1β (A300-905A), PP1γ (A300-906A), PP5 (A300-909A), PP6 (A300-844A), PNUTS (A300-439A) and Smc1 phospho-S957 (A304-147A) antibodies from Bethyl Laboratories (Montgomery, TX, USA); GFP (sc-9996) and Ku70 (sc-56129) antibodies from Santa Cruz Biotechnology (Dallas, TX, USA); ATM phospho-S1981 (ab36810), Chk2 phospho-T387 (ab195783), DNA-PKcs (ab70250), DNA-PKcs phospho-S2056 (ab18192), DNA-PKcs phospho-T2609 (ab18356) and H2B (ab1790-100) antibodies from Abcam (Cambridge, MA, USA); β-actin (#4970), BRCA1 phospho-S1524 (#9009P), Chk1 (#2345), Chk1 phospho-S296 (#2349), Chk2 phospho-T68 (#2661) and Rad52 (#3425P) antibodies from Cell Signaling Technology (Beverly, MA, USA); DNAL IV (GTX100100), and XRCC4 (GTX100094) antibodies from GeneTex (Irvine, CA, USA).
Immunofluorescence
Immunofluorescence was performed as previous described (34). Briefly, cells were fixed in 3% formaldehyde with 0.1% Triton X-100, washed, and blocked in 10% goat serum in PBS. Primary antibodies to γ-H2AX, PNUTS, PP1β, NBS1 phospho-S343 and ATM phospho-S1981 were diluted in blocking buffer, and incubated with the cells for 2 h. The cells were then incubated with the Alexa Fluor 488/555 conjugated secondary antibody (Invitrogen, 1: 2000) for 1 h. Imaging was performed using a Zeiss Axiovert 200M inverted fluorescence microscope at the UNMC Advanced Microscopy Core Facility. Laser microirradiation was performed using 405 nm laser under the Zeiss Axiovert 200M Microscope with Marianas Software (Intelligent Imaging Innovations, Inc. Denver, CO, USA).
NHEJ assays in Xenopus egg extracts and human cells
Repair assays in Xenopus egg extracts were performed as previously described (35). Briefly, plasmid pMBP-parallel2 (pMBPII) was linearized with Kpn1 and Xho1, or Xho1 alone, and incubated in Xenopus egg extracts for DNA repair. The reaction was terminated and resuspended in 500 μl of Proteinase K Buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 50 mM EDTA, 0.5% Sarkosyl, 10 μg/ml RNase A and 1 mg/ml Proteinase K) at 42°C for 1 h. The DNA template was re-isolated by phenol: chloroform extraction and isopropanol precipitation, and transformed into DH5-alpha cells (New England Biolabs, MA, USA) to measure the repair efficiency. The colonies were subjected to sequencing analysis. A plasmid-based NHEJ assay was also performed in HeLa cells. Briefly, pEGFP-C1 vector was linearized with Nhe1 and transfected into HeLa cells. Twenty four hours after transfection, cells were collected and analyzed by immunoblotting for GFP expression. The level of GFP was quantified using NIH ImageJ. Finally, the chromosomal NHEJ assay was performed in a U2OS-derived cell line stably integrated with a single GFP-reporter cassette (generous gift from Dr Jeremy Stark at the Beckman Research Institute of the City of Hope) (36). Briefly, cells were seeded at 3 × 105 cells per well in a six-well plate one day before siRNA treatment. The cells were treated with 100 nM siRNA for 5 h. After removing the siRNA, the cells were grown for 48 h in fresh medium, and transfected with an expression vector of I-SceI endonuclease (generous gift from Dr Maria Jasin at Memorial Sloan Kettering Cancer Center). In this assay, GFP is expressed only after DSBs introduced by I-SceI endonuclease are repaired, and the level of GFP expression was quantified by immunoblotting.
PP1 and PNUTS depletion
For PP1 depletion in Xenopus egg extracts, the middle segment (aa 320–539) of PNUTS was purified on magnetic glutathione beads (Pierce), and incubated in extracts for 30 min. The beads were removed with a magnet, and remaining extracts were collected. For immunodepletion of PP1 or PNUTS from Xenopus egg extract, antibodies were conjugated on anti-rabbit magnetic beads (Pierce), and incubated in extracts for 30 min. The beads were removed with a magnet, and remaining extracts were collected. Recombinant PP1γ or PNUTS with MBP-tag was expressed in bacteria and purified. These purified proteins were added back to PP1 or PNUTS depleted extracts for the ‘rescue’ experiments.
Protein expression, pull-down, kinase assay and mass spectrometry analysis
Five segments of DNA-PKcs, including N (aa 1–174), JK (aa 873–1100), PQR (aa 1800–2200), ABCDE (aa 2300–2800), and C (aa 2984–4146), were cloned from a Xenopus oocyte cDNA library, and then inserted into pMBP-parallel vector with an N-terminal MBP-tag. The recombinant proteins were expressed in BL21 bacterial cells and purified on amylose beads. Xenopus PNUTS expression vectors were described in our previous study (33). For pull-down assays, MBP- or GST-tagged proteins were conjugated on amylose or glutathione beads, and incubated in Xenopus egg extracts or human cell lysate. The beads were re-isolated using low speed centrifugation, washed five times, and then resolved by SDS-PAGE. For DNA-PKcs kinase assay using XRCC4 as substrate, XRCC4 cDNA from the pBMM42 vector (a gift from Dr Martin Gellert, Addgene plasmid #13331) was inserted into the pGEX4T vector, expressed in bacteria cells, purified on beads, and incubated in Xenopus extracts. The beads were then incubated in a kinase buffer (50 mM HEPES, 100 mM KCl, 10 mM MgCl, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT with [γ-32P]-ATP) at 37°C for 30 min. The reaction was stopped by the addition of Laemmli buffer. To re-isolate biotinylated DNA oligonucleotides, biotinylated (dA-dT)70 was bound to streptavidin-coated beads, added to Xenopus egg extracts, and re-isolated with a magnet. Samples were resolved by SDS-PAGE and analyzed by immunoblotting or mass spectrometry. For the mass spectrometric analysis, (dA-dT)70 or PNUTS pull-down samples were first resolved by SDS-PAGE. The gel sections were dissected and subjected to mass spectrometric analysis (Taplin mass spectrometry facility, Harvard).
Statistical analysis
A minimum of three experiments were carried out and the results are shown as the mean values and standard deviations. Statistical significance was analyzed using an unpaired two-tailed Student's t-test. A P-value<0.05 was considered statistically significant.
Xenopus egg extracts
Xenopus egg extracts were prepared as previously described (33). Briefly, eggs were rinsed in distilled water and then incubated in 2% cysteine in 1× XB (1 M KCl,10 mM MgCl2, 100 mM HEPES pH 7.7 and 500 mM sucrose) to remove the outer layers of jelly. Eggs were washed in 0.2× MMR buffer (100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 0.1 mM EDTA, 10 mM HEPES), and activated with Ca2+ ionophore. Eggs were then washed and crushed by centrifugation at 10 000 g. The cytoplasmic layer was transferred to new tubes, supplemented with an energy mix (7.5 mM creatine phosphate, 1 mM ATP, 1 MgCl2), and then further separated by centrifugation at 10 000 g for 15 min.
RESULTS
PP1 and PNUTS are recruited to a DSB-mimicking substrate in Xenopus egg extracts and sites of laser microirradiation in human cells
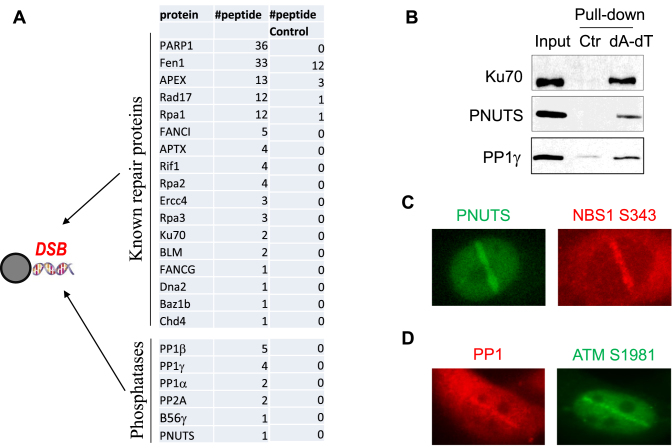
It is well established that supplementation of double stranded oligonucleotides that mimic free DSBs in Xenopus egg extracts activates the DNA damage response, in a manner similar to that in intact mammalian cells (37–39). Taking advantage of this cell-free system, we incubated biotin-labeled poly-(dA-dT)70 oligonucleotides in Xenopus egg extracts, and then recovered the oligonucleotides while they were in complex with DSB response and repair proteins in the extracts. As expected, the subsequent proteomic analysis identified a large number of proteins that were known to function in DNA repair, including PARP1, Fen1, APEX, Rad17, RPA and others (Figure 1A). Interestingly, several catalytic and regulatory subunits of protein phosphatases, including PP1 and PNUTS, were recovered with the DSB template (Figure 1A). The association of PP1 and PNUTS to poly-(dA-dT)70 oligonucleotides was confirmed by immunoblotting (Figure 1B). Importantly, both PNUTS and PP1 were recruited to the sites of laser micro-irradiation in human cells (Figure 1C and D), suggesting that these phosphatase subunits may play a role in the cellular responses to DNA damage, especially DNA DSBs.
Figure 1.
PP1 and PNUTS are recruited to DNA damage sites in vitro and in vivo. (A) As described in Materials and Methods, beads conjugated with biotin-(dA-dT)70 were incubated in Xenopus egg extracts for 30 min, re-isolated, resolved by SDS-PAGE and analyzed by mass spectrometry. The identified repair proteins and phosphatase subunits are shown, along with the numbers of peptides. A control pull-down was performed in Xenopus egg extracts using beads without oligonucleotides. (B) The biotin-(dA-dT)70 pull-down was performed as in panel A. The extract input, control pull-down and biotin-(dA-dT)70 pull-down were analyzed by immunoblotting for Ku70, PNUTS and PP1γ. (C) HeLa cells transfected with GFP-PNUTS were subjected to laser microirradiation as described in Materials and Methods. The fluorescent signal of GFP and immunofluorescent signal of NBS1 phospho-S343 are shown. (D) HeLa cells were subjected to laser microirradiation. The immunofluorescent signals of PP1β and ATM phospho-S1981 are shown.
PP1 is required for efficient DSB repair via NHEJ
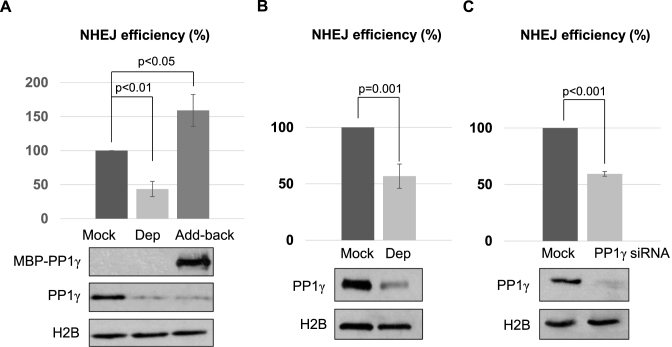
With the identification of PP1 as a protein recruited to DNA damage sites, we sought to determine its function in DNA repair. We previously characterized a plasmid-based DSB repair assay in Xenopus egg extracts to recapitulate the canonical, Ku-dependent NHEJ pathway (35). Interestingly, we depleted PP1 from Xenopus egg extracts, and observed a significant reduction of NHEJ (Figure 2A). The repair deficiency was rescued with the add-back of purified PP1γ (Figure 2A). In our previous study we showed that the repair of a DSB template with non-compatible ends required both DNA-PKcs and ATM, whereas the repair of a ligation-compatible template was dependent on DNA-PKcs but not ATM (35). Here, we noted that PP1 was required for the repair of both types of NHEJ templates (Figure 2A and B). PP1 depletion, however, did not significantly alter the mode of end processing, as judged by the sequence of repair products (Supplementary Figure S1). Moreover, suppression of PP1 expression in human cells disrupted the repair of a linearized GFP plasmid, confirming the role of PP1 in NHEJ in mammalian cells (Supplementary Figure S2). Similarly, downregulation of PP1 reduced the repair of a chromosomal, I-SceI-induced DSB via NHEJ (Figure 2C). This NHEJ assay was characterized in a previous study (36), and confirmed in our experiments using Ku70 suppression (Supplementary Figure S3). PP1 depletion caused a stronger deficiency in the plasmid-based assay than in the chromosomal-based assay, potentially due to the different complexity of chromosomal environment in these systems.
Figure 2.
PP1 is required for NHEJ. (A) PP1 depletion was performed in Xenopus egg extracts using beads conjugated with a PP1-binding motif derived from PNUTS, as described in Materials and Methods. Extracts were mock treated with beads alone, depleted of PP1γ (Dep), or PP1γ-depleted and then reconstituted with purified recombinant MBP-PP1γ (Add-back). As in our previous study (35), a non-compatible NHEJ template, linearized with Xho1 and Kpn1, was incubated and the repair efficiency, measured by colony formation, is shown. The level of PP1γ was monitored by immunoblotting, and histone H2B served as a loading control. (B) As in our previous study (35), a compatible NHEJ template linearized with Xho1 was incubated in Xenopus egg extracts with or without PP1 depletion. The repair efficiency, measured by colony formation, is shown. (C) The chromosome-integrated, I-SceI-induced NHEJ reporter cell line was characterized in a previous study (36), and described in Materials and Methods. The cells were transfected with or without PP1γ siRNA as indicated. The repair efficiency, measured by GFP expression, is shown. The cell lysates were analyzed by immunoblotting for PP1γ and H2B. The above experiments were performed at least three times, and the results are shown as the mean values and standard deviations. Statistical significance was analyzed using an unpaired 2-tailed Student's t-test.
PP1 regulates the phosphorylation and activation of DNA-PKcs
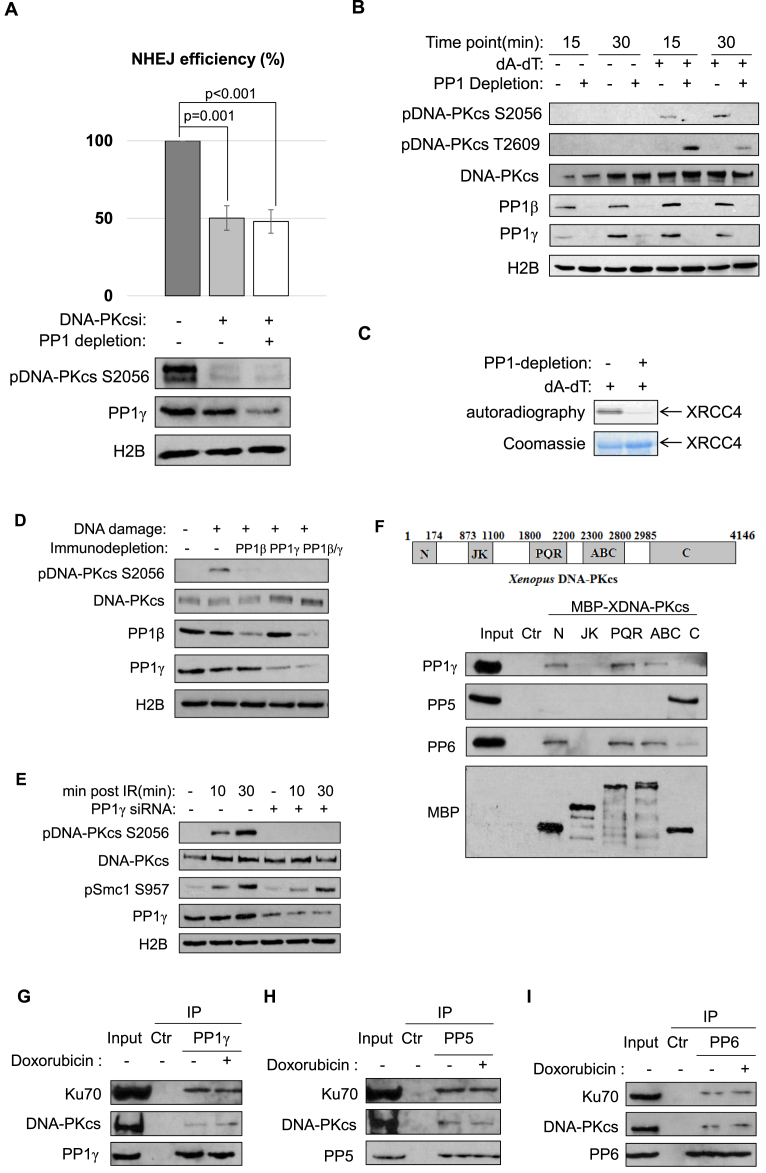
A number of NHEJ proteins are phosphorylated after DNA damage. Among them, phosphorylation of DNA-PKcs plays a crucial role in NHEJ (4–7). We speculated that PP1 may function by modulating DNA-PKcs, in part, because we showed that PP1 was required for the repair of both compatible and non-compatible NHEJ templates. In support of this notion, PP1 depletion did not impact NHEJ in the presence of a DNA-PKcs inhibitor (Figure 3A). In dissecting the molecular details by which PP1 modulates DNA-PKcs, we discovered the loss of DNA-PKcs autophosphorylation at Ser-2056 with the depletion of PP1 (Figure 3B). By comparison, DNA-PKcs phosphorylation at Thr-2609, mediated primarily by ATM was rather increased (Figure 3B). We further confirmed that PP1 depletion blocked DNA-PKcs kinase activation after DNA damage, as measured by an in vitro kinase assay using XRCC4 as substrate (Figure 3C) (40). The reduced DNA-PKcs phosphorylation at Ser-2056 was also seen with immunodepletion of PP1γ, or, to a lesser extent, PP1β (Figure 3D). Importantly, a conserved role of PP1 was observed in human cells: knockdown of PP1γ using two distinct siRNAs significantly reduced DNA-PKcs autophosphorylation at Ser-2056, but not Smc1 phosphorylation mediated by ATM/ATR (Figure 3E and Supplementary Figure S4).
Figure 3.
PP1 functions in NHEJ via regulation of DNA-PKcs. (A) The linearized NHEJ template was repaired in Xenopus egg extracts, as in Figure 2B. The extracts were depleted of PP1 (using the PP-binding motif of PNUTS, as in Figure 2A), or treated with a DNA-PKcs inhibitor (NU7441, 20 uM), as indicated. The repair efficiency, measured by colony formation, is shown. Extract samples were analyzed by immunoblotting for DNA-PKcs phospho-S2056, PP1γ and H2B. The experiment was performed at least three times, and the results are shown as the mean values and standard deviations. Statistical significance was analyzed using an unpaired two-tailed Student's t-test. (B) Xenopus egg extracts with or without PP1 depletion (using the PP-binding motif of PNUTS) were treated with 20 ng/μl of (dA-dT)70 as indicated. Immunoblots of DNA-PKcs phospho-S2056, DNA-PKcs phospho-T2609, DNA-PKcs, PP1β, PP1γ and H2B are shown. (C) In vitro DNA-PKcs kinase assay was performed using XRCC4 as substrate, as described in Materials and Methods. The autoradiography and Coomassie stain of XRCC4 are shown. (D) Xenopus egg extracts were immunodepleted of PP1β, PP1γ or both. The extracts were treated with (dA-dT)70 for 30 min, and analyzed by immunoblotting for DNA-PKcs phospho-S2056, DNA-PKcs, PP1β, PP1γ and H2B. (E) HeLa cells were transfected with control or PP1γ siRNA, treated with 10 Gy IR, and incubated as indicated. Immunoblots of DNA-PKcs phospho-S2056, DNA-PKcs, Smc1 phospho-S957, PP1γ and H2B are shown. (F) Five segments of DNA-PKcs (N, JK, PQR, ABCDE (ABC) and C) were expressed with MBP-tag, and purified on amylose beads, as described in Materials and Methods. The beads were then incubated in HeLa cell lysates, re-isolated, and analyzed by immunoblotting. The ‘Ctr’ sample was a mock pull-down using empty amylose beads. (G–I) Immunoprecipitation of PP1γ (G), PP5 (H) and PP6 (I) was performed in the lysates of HeLa cell with or without doxorubicin treatment. The lysate input, control IP with blank beads, and PP1γ IP products were analyzed by immunoblotting for DNA-PKcs, Ku70 and PP1γ, PP5 and PP6, as indicated.
To shed more light on PP1-dependent regulation of DNA-PKcs, and evaluate the hypothesis that various clusters of DNA-PKcs phosphorylation sites are differentially regulated by distinct phosphatases, we expressed several motifs of DNA-PKcs and probed for their associations with phosphatases in human cell lysates. Interestingly, PP1 bound the far N-terminal region, PQR cluster, and ABCDE cluster of DNA-PKcs. PP6 exhibited the same pattern of DNA-PKcs association as PP1, whereas PP5 associated exclusively with the C-terminal region that contains the kinase domain (Figure 3F). A similar fashion of protein associations was also observed in Xenopus egg extracts (Supplementary Figure S5). Furthermore, co-immunoprecipitation experiments confirmed the association of DNA-PKcs and Ku70 with PP1, PP5 and PP6 at the endogenous level (Figure 3G–I).
PNUTS controls the DNA damage response and repair
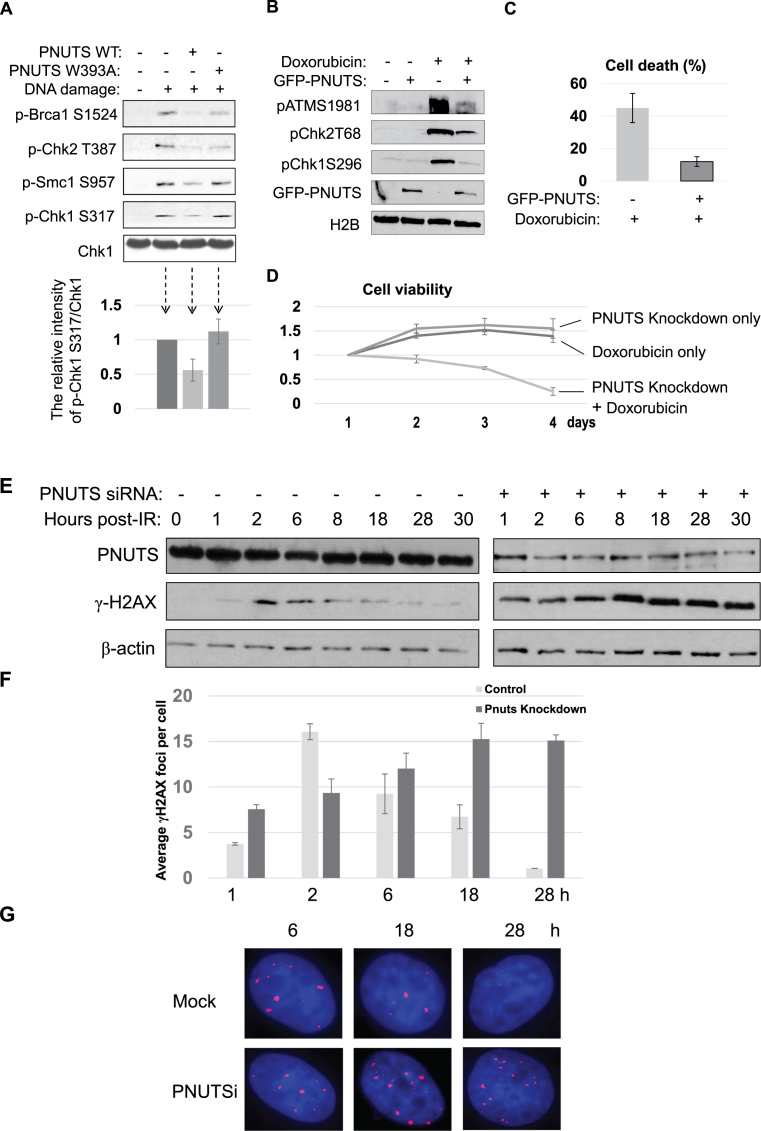
In addition to PP1, we identified PNUTS as a protein that associated with a DSB-mimicking substrate in Xenopus egg extracts, and was recruited to sites of laser microirradiation in human cells. Because PNUTS is known as an inhibitory regulator of PP1, we hypothesized that PNUTS plays a role in the DNA damage response via modulating PP1, and sought to investigate the functional impact of PNUTS upregulation and downregulation on DNA damage-induced signaling, repair and survival. Interestingly, addition of PNUTS in Xenopus egg extracts reduced checkpoint signaling, as judged by the phosphorylation of BRCA1, Chk2, Smc1 and Chk1 (Figure 4A). By comparison, a PP1-binding deficient mutant of PNUTS exhibited either no suppression or less-efficient suppression of Chk1 phosphorylation or other phosphorylation events, suggesting that PNUTS functions in the DNA damage response largely via PP1 (Figure 4A). Similarly, expression of exogenous PNUTS rendered human cells resistant to doxorubicin-induced DNA damage, as shown by the diminished DNA damage signaling and attenuated induction of cell death (Figure 4B and C). Conversely, PNUTS knockdown sensitized the cell response to doxorubicin (Figure 4D). Moreover, we showed that PNUTS knockdown impaired DNA repair, as indicated by the persistence of γ-H2AX after IR (Figure 4E–G). The repair deficiency caused by PNUTS knockdown was rescued with the expression of exogenous PNUTS (Supplementary Figure S6). Thus, these findings underscore an essential role of PNUTS in the DNA damage response, and the potential of targeting PNUTS as a new approach to sensitize cancer chemotherapy.
Figure 4.
PNUTS mediates the DNA damage response and repair. (A) Xenopus egg extracts were supplemented with (dA-dT)70 (DNA damage), WT or W393A PNUTS, as indicated, and incubated for 30 min. The extract samples were analyzed by immunoblotting for Brca1 phospho-S1524, Chk2 phospho-T387, Smc1 phospho-S957, Chk1 phospho-S317 and Chk1. In the lower panel, the relative intensity of Chk1 phospho-S317/Chk1 was measured using NIH ImageJ, and shown by the mean values and standard deviations. (B) HeLa cells were transfected with GFP-PNUTS, and treated with 5 μM doxorubicin for 4 h, as indicated. The cell lysates were analyzed by immunoblotting for ATM phospho-S1981, Chk2 phospho-T68, Chk1 phoshpo-S296, GFP and H2B. (C) HeLa cells with or without GFP-PNUTS expression were treated with doxorubicin for 24 h. Cell death was measured by trypan blue excursion. (D) HeLa cells were treated with PNUTS siRNA and doxorubicin, as indicated. The cell number of each day was normalized to that of the first day for the relative cell viability. (E) HeLa cells with control or PNUTS siRNA were irradiated with 4 Gy IR and incubated as indicated. The cell lysates were analyzed by immunoblotting for γ-H2AX, PNUTS and β-actin. (F, G) HeLa cells treated as in panel E were examined by immunofluorescence for γ-H2AX. The average numbers of γ-H2AX foci were counted (F) and representative images of cells stained for γ-H2AX (in red) and DAPI (in blue) are shown (G). At least 100 cells were analyzed for γ-H2AX foci in panel F.
PNUTS is required for NHEJ in Xenopus egg extract and human cells
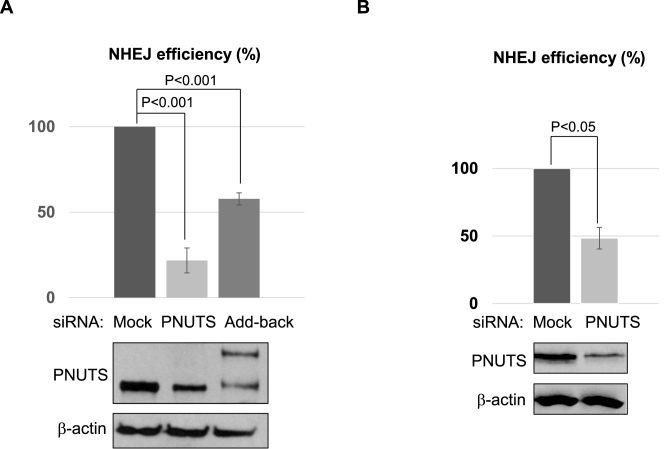
As we characterized PNUTS as an important regulator of DNA repair after IR or doxorubicin, we further reasoned that PNUTS is involved in NHEJ, given that we revealed the role of PP1 in regulation of NHEJ via DNA-PKcs. Interestingly, PNUTS knockdown significantly reduced the repair efficiency of the plasmid-based NHEJ template in HeLa cells, and expression of exogenous PNUTS partially restored the repair deficiency (Figure 5A). A consistent role of PNUTS in NHEJ was also demonstrated using the chromosomal, I-SceI-induced NHEJ template (Figure 5B). Furthermore, in Xenopus egg extracts, the depletion of PNUTS reduced NHEJ, which was partially rescued with the add-back of purified PNUTS protein (Supplementary Figure S7).
Figure 5.
PNUTS is required for NHEJ in Xenopus egg extract and human cells. (A) As described in Materials and Methods, HeLa cells were treated with control (Ctr) or PNUTS siRNA, or with PNUTS siRNA and then reconstituted with siRNA resistant GFP-PNUTS (Add-back). NHEJ assay was performed using a GFP-expressing template linearized with Nhe1, as described in Material and Methods. The repair efficiency, measured by GFP expression, is shown. The cell lysates were analyzed by immunoblotting for PNUTS and β-actin. (B) The chromosomal NHEJ reporter cells were transfected with control or PNUTS siRNA. The cell lysates were analyzed by immunoblotting for PNUTS and β-actin. The chromosomal NHEJ assay was performed as in Figure 2C. The repair efficiency, measured by GFP expression, is shown. A minimum of three experiments were carried out in the above panels, and the results are shown as the mean values and standard deviations. Statistical significance was analyzed using an unpaired two-tailed Student's t-test.
PNUTS associates with Ku proteins and regulates DNA-PKcs phosphorylation
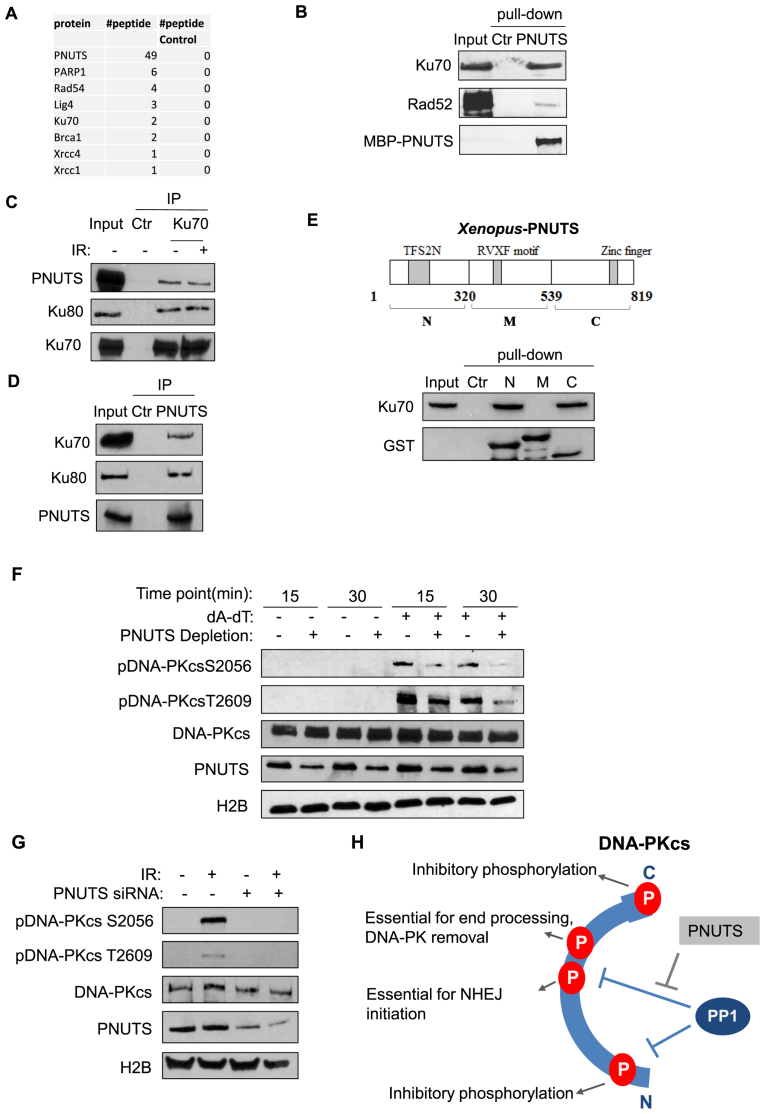
To reveal unbiased insights into the role of PNUTS, we biochemically isolated PNUTS from Xenopus egg extracts, and identified the associated proteins using mass spectrometry (Figure 6A). Consistent with the role of PNUTS in DNA repair, this proteomic analysis recovered a number of DSB repair proteins (Figure 6A). Among these proteins, we consistently observed a strong association between PNUTS and Ku70 (Figure 6B). The PNUTS and Ku70/80 association in human cells was confirmed at the endogenous level by the reciprocal IP of Ku70 or PNUTS (Figure 6C and D). The PNUTS and Ku association appeared constitutive, and was not dependent on DNA damage (Figure 6C). In Xenopus egg extracts, both the N- and C-termini of PNUTS bound Ku70, suggesting the existence of multiple Ku-binding motifs (Figure 6E). To mitigate the concern that PNUTS and Ku associate indirectly via DNA, we showed that treatment with ethidium bromide (EtBr) or DNase I did not affect the PNUTS/Ku70 association (Supplementary Figure S8A and B). Furthermore, reciprocal pull-down assays using purified Ku70 and PNUTS suggested a direct protein interaction between Ku70 and PNUTS (Supplementary Figure S8C and D).
Figure 6.
PNUTS associates with the Ku protein complex and regulates DNA-PKcs phosphorylation. (A) MBP-PNUTS pull-down was performed in Xenopus egg extracts. The pull-down product was resolved by SDS-PAGE, and analyzed by mass spectrometry. A number of DNA repair proteins were identified as binding patterns of PNUTS, as shown here with the numbers of peptides. A control pull-down was performed in Xenopus egg extracts using beads without MBP-PNUTS. (B) PNUTS pull-down was performed as in panel A. The extract input, control pull-down, and PNUTS pull-down samples were analyzed by immunoblotting for Ku70, Rad 52 and PNUTS. The ‘Ctr’ sample was a mock pull-down using empty amylose beads. (C) Ku70 IP was performed in the lysates of HeLa cell with or without IR-treatment. The lysate input, control IP, and Ku70 IP products were analyzed by immunoblotting for PNUTS, Ku80 and Ku70. (D) PNUTS IP was performed in HeLa cell lysates. The lysate input, control IP and PNUTS IP products were analyzed by immunoblotting for PNUTS, Ku70 and Ku80. (E) Three segments of PNUTS (N, M and C) were expressed with an N-terminal GST tag, and purified on glutathione beads, as described in Materials and Methods. The beads were incubated in Xenopus egg extracts, re-isolated, and analyzed by immunoblotting for Ku70 and GST. A mock pull-down using empty glutathione beads was performed as control (Ctr). (F) Xenopus egg extracts with or without PNUTS depletion were treated with (dA-dT)70, as indicated. The extract samples were analyzed by immunoblotting for DNA-PKcs phospho-S2056, DNA-PKcs phospho-T2609, DNA-PKcs, PNUTS and H2B. (G) HeLa cells were treated with PNUTS siRNA and 10 Gy IR, as indicated. The cell lysates were analyzed by immunoblotting for DNA-PKcs phospho-S2056, DNA-PKcs phospho-T2609, DNA-PKcs, PNUTS and H2B. (H) PP1 and PNUTS fine-tune the dynamic, and complex pattern of DNA-PKcs phosphorylation. PP1 is required for DNA-PKcs activation after DNA damage, presumably via dephosphorylation of multiple sites within the N, PQR and ABCDE regions. PNUTS modulates the action of PP1 toward S2056 and T2609, as the proper phosphorylation of these sites is required for end processing and ligation.
Our earlier finding that PP1 regulates DNA-PKcs activation and phosphorylation prompted us to examine the role of PNUTS in this process. Interestingly, immunodepletion of PNUTS in Xenopus egg extracts reduced DNA-PKcs phosphorylation at both Ser-2056 and Thr-2609 (Figure 6F). Consistently, PNUTS knockdown in human cells also decreased the phosphorylation of DNA-PKcs at Ser-2056 and Thr-2609 (Figure 6G).
DISCUSSION
Using an unbiased proteomic approach, our study identified PP1 and PNUTS as proteins associated with a DNA DSB-mimicking substrate in Xenopus egg extracts. We subsequently confirmed their recruitment to laser-induced DNA damage sites in human cells. To our knowledge, this study provides the first evidence that PP1, a major Ser/Thr phosphatase, accumulates at sites of DNA damage. On the other hand, the identification of PNUTS as a DNA damage response factor is consistent with several existing studies. PNUTS was found as one of the proteins that bound DNA adducts generated by platinum-based DNA damaging drugs (25). Moreover, a recent study reported that PNUTS is recruited to DNA damage sites and that PNUTS depletion led to prolonged DNA damage foci (31). Expression of PNUTS suppressed the apoptosis of cardiomyocytes treated with DNA damaging agents (27). These existing findings suggested that PNUTS may function to suppress DNA damage signaling, and/or promote DNA repair. Here we characterized the roles of PP1 and PNUTS in NHEJ, a major DSB repair mechanism. Our study further discovered that PP1 and PNUTS fine-tune the DNA damage-induced phosphorylation and activation of DNA-PKcs (Figure 6H).
DNA-PK complex recognizes DSB ends, and plays a central role in NHEJ by orchestrating end processing and ligation. The function and dynamics of DNA-PK are regulated by its phosphorylation, catalyzed largely by DNA-PKcs itself. It has been shown that DNA damage induces phosphorylation of DNA-PKcs at several dozen sites throughout the protein. Mutational studies revealed at least 13 sites that are functionally important. Many of these phosphorylation sites are localized within several clusters, and phosphorylation of each cluster may influence the function of DNA-PK in a distinct manner (8,10). As the better-studied examples, DNA-PKcs phosphorylation at the ABCDE cluster, including Thr-2609, by both ATM and DNA-PKcs facilitates Artemis recruitment and end resection, whereas autophosphorylation at Ser-2056 in the PQR cluster limits end-processing. A number of additional DNA-PKcs autophosphorylation sites were identified within the N-terminal and C-terminal motifs, and studies revealed that these modifications suppress the kinase activation of DNA-PKcs (8–13). Presumably, a phosphatase is required to dephosphorylate these inhibitory phosphorylation sites to permit DNA-PKcs activation after DNA damage. The complex and sometimes opposing role of DNA-PKcs phosphorylation suggests dynamic and site-specific regulation of these modifications. Because most of these sites are commonly phosphorylated by DNA-PKcs and ATM/ATR, a very interesting idea to account for the regulatory complexity is that these phosphorylation clusters are differentially dephosphorylated by various phosphatases.
We showed in this study that PP1 binds the ABCDE and PQR clusters, and the far N-terminal region, but not the JK cluster or the C-terminal kinase domain of DNA-PKcs. We confirmed that PP1 is required for the kinase activation of DNA-PKcs after DNA damage, consistent with the role of PP1 in NHEJ. As autophosphorylation of residues within the N-terminal region inactivates DNA-PKcs, we reason that PP1 is responsible for the dephosphorylation of these residues to facilitate DNA-PKcs activation. This notion shall be further investigated in the future when phospho-specific antibodies against these sites become available. Furthermore, depletion of PP1 led to accumulation of Thr-2609 phosphorylation, supporting a role of PP1 in dephosphorylation of Thr-2609 and other sites of the ABCDE cluster. In contrast, PP1 depletion reduced the level of Ser-2056 autophosphorylation, which was seemingly counterintuitive, as phosphatases are known to mediate protein dephosphorylation. However, the loss of Ser-2056 phosphorylation after PP1 depletion is in line with our finding that PP1 was required for DNA-PKcs activation, given that Ser-2056 is an autophosphorylation site. Thus, we speculate that, under the condition of PP1 depletion, phosphorylation of Ser-2056 is inefficient due to the lack of DNA-PKcs activation, whereas dephosphorylation of Ser-2056 can still be mediated by other phosphatases, such as PP6 which binds this region of DNA-PKcs.
PNUTS has been characterized as an inhibitory regulator of PP1 in the nucleus. Our study indicated that PNUTS is a strong binding partner of the DNA-PK complex. We speculated that PNUTS participates in the DNA damage response via modulation of PP1, because the expression of WT PNUTS, but not a PP1-binding deficient mutant form of PNUTS significantly influenced DNA damage signaling. Consistent with the role of PP1 in regulation of DNA-PKcs phosphorylation, we showed that PNUTS is indispensable for the phosphorylation of DNA-PKcs at both Ser-2056 and Thr-2609, the essential sites for the subsequent end processing and ligation. We hypothesize that, regardless if PP1 targets Ser-2056 and Thr-2609 under normal circumstance, in the absence of PNUTS, these sites are prematurely dephosphorylated by PP1, resulting in deficient NHEJ (Figure 6H). Finally, we ruled out the possibility that PP1 or PNUTS indirectly influences NHEJ by modulating the expression of core NHEJ factors. The levels of DNAL IV, XRCC4, Ku70, Ku80, DNA-PKcs and Artemis were unchanged with PP1 or PNUTS depletion (Supplementary Figure S9).
PP1 and other phosphatases have been previously shown to deactivate the DNA damage response after the completion of DNA repair, and thereby, returning the cell to the unperturbed state. Importantly, our study revealed a new role of PP1 as an early and upstream regulator of DSB repair by controlling DNA-PKcs dephosphorylation and activation. Interestingly, RIF1, a repair factor that promotes NHEJ and suppresses HR, is itself a targeting subunit of PP1 (41–44). BRCA1, another important DSB repair factor, is also a PP1-binding protein. Previous studies demonstrated that mutations within the PP1-binding motif of BRCA1 impaired DNA repair, and that BRCA1 modulates DNA-PKcs autophosphorylation in S phase (45,46). Therefore, the function and regulation of PP1 in DSB repair, and its relationship with other repair proteins, remain to be clarified. Previous studies also suggested the involvement of other phosphatases in regulation of DNA-PKcs, including PP5 and PP6 (15–17). We showed that PP6 binds the ABCDE and PQR clusters and the N-terminal region, suggesting that PP6 may play a redundant role with PP1. On the other hand, PP5 binds exclusively the C-terminal kinase domain of DNA-PKcs. Because it has been shown that PP5 positively modulates the kinase activation of DNA-PKcs, ATM and ATR (16,47,48), a general role of PP5 in regulation of the kinase domains of these PIKK family members is worthy of future investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jay Reddy (University of Nebraska-Lincoln) for technical support with X-ray irradiation; Dr Li Lan (University of Pittsburgh) for assistance with imaging; Drs Jeremy Stark (City of Hope) and Maria Jasin (Memorial Sloan Kettering Cancer Center) for reagents; Drs Gregory Oakley and Ali Nawshad for stimulating discussions; Dr James Wahl and Nicholas Eurek for assisting with the microscopic studies. The UNMC Advanced Microscopy Core Facility was supported by the Nebraska Research Initiative, the Fred and Pamela Buffett Cancer Center Support Grant (P30CA036727), and an Institutional Development Award (IDeA) from the NIGMS of the NIH (P30GM106397).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [1R01CA172574 to A.P.]. Funding for open access charge: National Institutes of Health [1R01CA172574 to A.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davis A.J., Chen D.J.. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013; 2:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Radhakrishnan S.K., Jette N., Lees-Miller S.P.. Non-homologous end joining: emerging themes and unanswered questions. DNA Repair. 2014; 17:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis A.J., Chen B.P., Chen D.J.. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair. 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jette N., Lees-Miller S.P.. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015; 117:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobbs T.A., Tainer J.A., Lees-Miller S.P.. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair. 2010; 9:1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pawelczak K.S., Bennett S.M., Turchi J.J.. Coordination of DNA-PK activation and nuclease processing of DNA termini in NHEJ. Antioxid. Redox Signal. 2011; 14:2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neal J.A., Sugiman-Marangos S., VanderVere-Carozza P., Wagner M., Turchi J., Lees-Miller S.P., Junop M.S., Meek K.. Unraveling the complexities of DNA-dependent protein kinase autophosphorylation. Mol. Cell. Biol. 2014; 34:2162–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang W., Crowe J.L., Liu X., Nakajima S., Wang Y., Li C., Lee B.J., Dubois R.L., Liu C., Yu X. et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell. 2015; 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J.. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007; 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meek K., Lees-Miller S.P., Modesti M.. N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res. 2012; 40:2964–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Douglas P., Cui X., Block W.D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S.P., Meek K.. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 2007; 27:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Summers K.C., Shen F., Sierra Potchanant E.A., Phipps E.A., Hickey R.J., Malkas L.H.. Phosphorylation: the molecular switch of double-strand break repair. Int. J. Proteomics. 2011; 2011:373816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douglas P., Moorhead G.B., Ye R., Lees-Miller S.P.. Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 2001; 276:18992–18998. [DOI] [PubMed] [Google Scholar]
- 15. Mi J., Dziegielewski J., Bolesta E., Brautigan D.L., Larner J.M.. Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS One. 2009; 4:e4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wechsler T., Chen B.P.C., Harper R., Morotomi-Yano K., Huang B.C.B., Meek K., Cleaver J.E., Chen D.J., Wabl M.. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Douglas P., Zhong J.N., Ye R.Q., Moorhead G.B.G., Xu X.Z., Lees-Miller S.P.. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol. Cell. Biol. 2010; 30:1368–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moorhead G.B.G., Trinkle-Mulcahy L., Ulke-Lemee A.. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell. Biol. 2007; 8:234–244. [DOI] [PubMed] [Google Scholar]
- 19. Bollen M., Gerlich D.W., Lesage B.. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 2009; 19:531–541. [DOI] [PubMed] [Google Scholar]
- 20. Virshup D.M., Shenolikar S.. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009; 33:537–545. [DOI] [PubMed] [Google Scholar]
- 21. Peng A., Maller J.L.. Serine/threonine phosphatases in the DNA damage response and cancer. Oncogene. 2010; 29:5977–5988. [DOI] [PubMed] [Google Scholar]
- 22. Allen P.B., Kwon Y.G., Nairn A.C., Greengard P.. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J. Biol. Chem. 1998; 273:4089–4095. [DOI] [PubMed] [Google Scholar]
- 23. Kim Y.M., Watanabe T., Allen P.B., Lee S.J., Greengard P., Nairn A.C., Kwon Y.G.. PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J. Biol. Chem. 2003; 278:13819–13828. [DOI] [PubMed] [Google Scholar]
- 24. Lee J.H., You J., Dobrota E., Skalnik D.G.. Identification and characterization of a novel human PP1 phosphatase complex. J. Biol. Chem. 2010; 285:24466–24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bounaix Morand du Puch C, Barbier E., Kraut A., Coute Y., Fuchs J., Buhot A., Livache T., Seve M., Favier A., Douki T. et al. TOX4 and its binding partners recognize DNA adducts generated by platinum anticancer drugs. Arch. Biochem. Biophys. 2011; 507:296–303. [DOI] [PubMed] [Google Scholar]
- 26. Kim H., Lee O.H., Xin H., Chen L.Y., Qin J., Chae H.K., Lin S.Y., Safari A., Liu D., Songyang Z.. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009; 16:372–379. [DOI] [PubMed] [Google Scholar]
- 27. Boon R.A., Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S., Kaluza D., Treguer K., Carmona G., Bonauer A. et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013; 495:107–110. [DOI] [PubMed] [Google Scholar]
- 28. Udho E., Tedesco V.C., Zygmunt A., Krucher N.A.. PNUTS (phosphatase nuclear targeting subunit) inhibits retinoblastoma-directed PP1 activity. Biochem. Biophys. Res. Commun. 2002; 297:463–467. [DOI] [PubMed] [Google Scholar]
- 29. Krucher N.A., Rubin E., Tedesco V.C., Roberts M.H., Sherry T.C., De Leon G.. Dephosphorylation of Rb (Thr-821) in response to cell stress. Exp. Cell Res. 2006; 312:2757–2763. [DOI] [PubMed] [Google Scholar]
- 30. Kavela S., Shinde S.R., Ratheesh R., Viswakalyan K., Bashyam M.D., Gowrishankar S., Vamsy M., Pattnaik S., Rao S., Sastry R.A. et al. PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res. 2013; 73:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Landsverk H.B., Mora-Bermudez F., Landsverk O.J., Hasvold G., Naderi S., Bakke O., Ellenberg J., Collas P., Syljuasen R.G., Kuntziger T.. The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO Rep. 2010; 11:868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang L., Luong V.Q., Giannini P.J., Peng A.. Mastl kinase, a promising therapeutic target, promotes cancer recurrence. Oncotarget. 2014; 5:11479–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fisher L.A., Wang L., Wu L., Peng A.. Phosphatase 1 nuclear targeting subunit is an essential regulator of M-phase entry, maintenance, and exit. J. Biol. Chem. 2014; 289:23745–23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng A.M., Lewellyn A.L., Schiemann W.P., Maller J.L.. Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Curr. Biol. 2010; 20:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu S., Peng A.. Non-homologous end joining repair in Xenopus egg extract. Sci. Rep. 2016; 6:27797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gunn A., Stark J.M.. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012; 920:379–391. [DOI] [PubMed] [Google Scholar]
- 37. Guo Z.J., Dunphy W.G.. Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends. Mol. Biol. Cell. 2000; 11:1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lupardus P.J., Van C., Cimprich K.A.. Analyzing the ATR-mediated checkpoint using Xenopus egg extracts. Methods. 2007; 41:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garner E., Costanzo V.. Studying the DNA damage response using in vitro model systems. DNA Repair. 2009; 8:1025–1037. [DOI] [PubMed] [Google Scholar]
- 40. Leber R., Wise T.W., Mizuta R., Meek K.. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 1998; 273:1794–1801. [DOI] [PubMed] [Google Scholar]
- 41. Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J.. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013; 49:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 43. Feng L., Fong K.W., Wang J., Wang W., Chen J.. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 2013; 288:11135–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T.. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013; 339:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen B.Y., Huang C.H., Lin Y.H., Huang C.C., Deng C.X., Hsu L.C.. The K898E germline variant in the PP1-binding motif of BRCA1 causes defects in DNA Repair. Sci. Rep. 2014; 4:5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davis A.J., Chi L., So S., Lee K.J., Mori E., Fattah K., Yang J., Chen D.J.. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 2014; 42:11487–11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ali A., Zhang J., Bao S.D., Liu I., Otterness D., Dean N.M., Abraham R.T., Wang X.F.. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004; 18:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang J., Bao S.D., Furumai R., Kueera K.S., Ali A., Dean N.A., Wang X.F.. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol. Cell. Biol. 2005; 25:9910–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.