Abstract
The FASTK family proteins have recently emerged as key post-transcriptional regulators of mitochondrial gene expression. FASTK, the founding member and its homologs FASTKD1–5 are architecturally related RNA-binding proteins, each having a different function in the regulation of mitochondrial RNA biology, from mRNA processing and maturation to ribosome assembly and translation. In this review, we outline the structure, evolution and function of these FASTK proteins and discuss the individual role that each has in mitochondrial RNA biology. In addition, we highlight the aspects of FASTK research that still require more attention.
INTRODUCTION
The founding member of the FASTK family, FASTK (Fas-activated serine/threonine kinase) itself, was discovered two decades ago as an interacting partner of TIA-1 (T-cell intracellular antigen-1) in Jurkat leukemia T cells. TIA-1 is an RRM-type RNA-binding protein that acts as a regulator of alternative splicing of nuclear pre-mRNA and translation of cytoplasmic mRNAs (1). FASTK was initially proposed to act as a protein kinase activated upon Fas ligation (1), but critical active site residues are not conserved within the family and this activity has been questioned since (2). Later studies reported that FASTK counteracts TIA-1-mediated inhibition of mRNA translation and localizes to stress granules and P-bodies during stress (3,4). Similar to TIA-1, FASTK also regulates the alternative splicing of exons flanked by weak splice site-recognition sequences. Accordingly, FASTK promotes the inclusion of exon IIIb of the fibroblast growth factor receptor 2 pre-mRNA and exon 6 of Fas pre-mRNA (5,6).
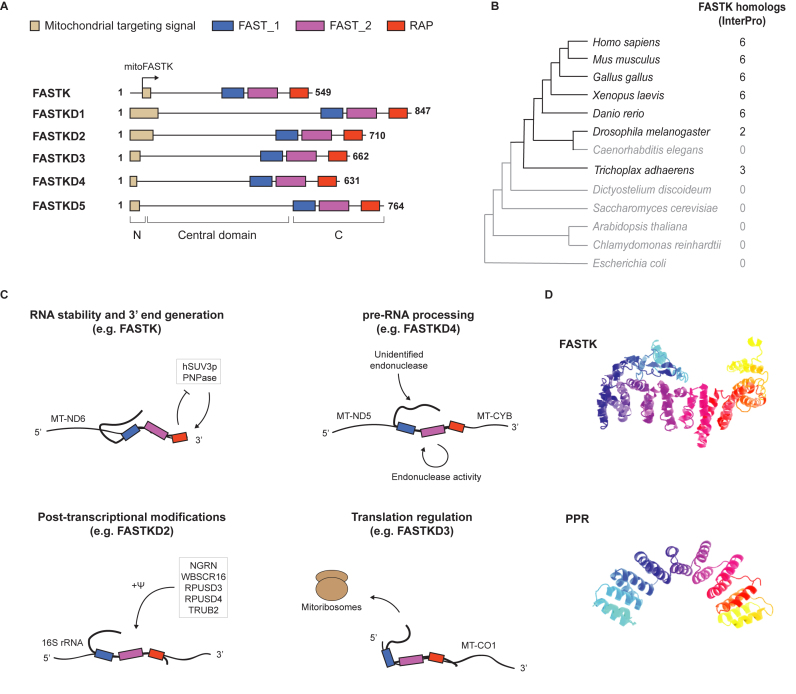
The identification of a cryptic mitochondrial targeting signal (MTS) in the FASTK sequence and its mitochondrial isoform, as well as the discovery of FASTK homologs, human FASTKD1–5, were reported only much later (2,7). The mitochondrial isoform of FASTK is synthesized from an alternative downstream translation initiation site and lacks the first 34 amino acids present at the N-terminus of the previously described FASTK protein, exposing an MTS (Figure 1A). The FASTKD1–5 proteins also contain an amino-terminal MTS and all localize to mitochondria. In addition to the MTS, members of the FASTK family of proteins share a C-terminal region made of three conserved domains called FAST_1, FAST_2 and RAP (Figure 1A). According to homology predictions, the ∼60-amino acid RAP domain (for RNA binding domain abundant in Apicomplexans) is likely to bind RNA (8), and indeed all members of the FASTK family have recently been found to bind to mRNAs in vivo in crosslinking-mass spectrometry studies (9,10). The RAP domain is an unusually small RNA binding domain and its boundaries should be re-explored. As we discuss below, structural modeling of the RAP domain predicts that it may also adopt an endonuclease-like structure (11). To date, the precise function of the leucine rich domains FAST_1 (∼70 amino acids) and FAST_2 (∼90 amino acids) remains unknown. The region of the FASTK family members between the MTS and the conserved C-terminal domains (central domain) shows little conservation in sequence or length among the family members and do not contain any recognizable domains (Figure 1A). Importantly, all six members of the FASTK family are ubiquitously expressed, although some FASTK proteins are particularly overexpressed in tissues with marked abundance of mitochondria (2).
Figure 1.
(A) Schematic representation of the FASTK family proteins. N: Amino-terminal domain. C: carboxy-terminal domain. The arrow indicates the internal translation start site in FASTK mRNA that generates mitoFASTK (B) Conservation of the FASTK family across evolution according to InterPro (12). Only proteins containing a FAST_1, FAST_2 and a RAP domain on the same polypeptide are reported. Redundant proteins from the database have been filtered out. (C) Proposed mechanism of action of the members of the FASTK family, as detailed in the main text. Ψ: pseudouridine. (D) (top) I-TASSER prediction of FASTK structure (40) and (bottom) crystal structure of a designed PPR protein (PDB ID: 4WSL) (41).
EVOLUTIONARY CONSERVATION OF FASTK FAMILY PROTEINS
The particular architecture of the C-terminal domain of FASTK proteins, the signature of this family, is conserved across evolution. Database searches for canonical FASTK family proteins (defined by the presence of a FAST_1, FAST_2 and a RAP domain on the same polypeptide) suggests that the FASTK family has appeared early in the history of multicellular animals (Metazoa). Accordingly, the ancestral placozoan Trichoplax adhaerens, at the root of metazoan evolution, presents three FASTK homologs, whereas the canonical FASTK proteins are absent in yeasts or plants (Figure 1B) (12). Genes have undergone duplications and losses over evolutionary time and most vertebrate genomes encode all the six members of the family whereas Drosophila melanogaster encodes only two of them. Interestingly, in Drosophila, both FASTK proteins (CG31643, CG13850) are predicted to be targeted to the mitochondria (13,14), suggesting that they are true homologs of the human FASTK proteins.
Proteins containing the three signature domains together (FAST_1, FAST_2 and RAP) are not found in other kingdoms of life, but we have noted the presence of a considerable number of genes containing a combination of two of the three domains. For example, sponges present a combination of a FAST_1 and a RAP domain in certain polypeptides and a similar domain organization is observed in certain green algae (e.g. Chlorella variabilis) or in Apicomplexa, which not only express proteins that have only a RAP domain, but also present proteins with a FAST_1 and a RAP domain together. Finally, the three domains can also be found separately: RAP is the most abundant domain found and it is present in some eubacteria, archaea, viruses or in plants. For example, in the land plant Arabidopsis thaliana the RAP domain-containing protein ‘RAP’ is required for the 16S rRNA maturation in the chloroplast (15). Similarly, the FAST_1 domain exists in plants and animals, but does not seem to be present in prokaryotes, whereas the FAST_2 domain seems to be unique to animals. This suggests that the RAP domain is the most ancestral and has been combined with the FAST_1 and FAST_2 domains over the course of evolution and evolved into the current FASTK family of proteins. Importantly, it is possible that proteins with a single domain may act in concert to control a specific cellular process. This is for example the case of Raa1 and Raa3, which contain a FAST_1 and a RAP domain, respectively and are required for photosynthesis in Chlamydomonas reinhardtii. Both genes encode proteins that are targeted to the chloroplast and together participate in the trans-splicing reaction of PsaA mRNA (16,17). Thus, whereas the canonical FASTK family of proteins is specific to metazoans, other FASTK-related proteins exist and regulate RNA-related processes not only in mitochondria, but also in chloroplasts.
REGULATION OF MITOCHONDRIAL RNA BIOLOGY BY THE FASTK FAMILY
In 2008, the team led by Massimo Zeviani showed for the first time the presence of a FASTK family member, namely FASTKD2, in the mitochondrial matrix, and found that a loss-of-function mutation in FASTKD2 was the cause of a human mitochondrial disease (18). In subsequent years, it became clear that all FASTK proteins can localize to the mitochondrial matrix and that they regulate mitochondrial gene expression. Human mitochondrial DNA (mtDNA) encodes 37 genes that are required for oxidative phosphorylation (OXPHOS), altered expression of which is associated with human diseases and aging (19). Thirteen of these genes encode protein subunits of the OXPHOS machinery, which closely interact with nuclear DNA-encoded subunits within four of the five OXPHOS complexes (complexes I, III, IV and V). In addition, mtDNA encodes 2 rRNAs and 22 tRNAs necessary for translation of the mitochondrial mRNAs. Each strand of mtDNA (historically termed ‘heavy’ and ‘light’) is transcribed as a long polycistronic transcript that is processed to yield individual RNAs. In most cases this is achieved according to the tRNA punctuation model, which implies that the folding of the tRNAs and their processing by the action of RNAse P and RNAse Z results in the concomitant release of flanking mRNAs and rRNAs (20). Because of the polycistronic nature of mitochondrial transcription, post-transcriptional events are thought to be the main regulators of mitochondrial gene expression (21,22). Accumulating evidences indicate that the early steps of transcript processing take place in punctate structures named mitochondrial RNA granules (MRGs) (23,24). MRGs concentrate key RNA processing and maturation factors including RNase P subunits, RNase Z and GRSF1, as well as part of the hSuv3p–PNPase complex or degradosome (25). Some members of the FASTK family, including FASTK, FASTKD1, FASTKD2 and FASTKD5 are intimately linked to MRGs.
It is important to note that the many factors required for mitochondrial gene expression are products of nuclear genes that are imported into the mitochondrial matrix. Noteworthy with six members, the FASTK family represents one of the largest family of mitochondrial gene expression regulators described so far. One of the unique characteristics of this family is that its members have distinct and sometimes even opposing effects on mitochondrial RNA biology as we will review here (summarized in Table 1).
Table 1. The names, localization and functions of the FASTK family proteins.
| Subcellular localization | RNAs regulated | Mitochondrial function | |
|---|---|---|---|
| FASTK | Nucleus, cytosol and mitochondria (MRGs) | MT-ND6 | MT-ND6 3′ end generation and stability |
| FASTKD1 | Mitochondria (nucleoids and MRGs) | MT-ND3 | mRNA stability |
| FASTKD2 | Mitochondria (MRGs) | MT-RNR2 (16S rRNA), MT-ND6 | rRNA/mRNA stability |
| FASTKD3 | Mitochondria | MT-ND2, MT-ND3, MT-ATP8/6, MT-CYB, MT-CO2 | mRNA stability, MT-CO1 translation |
| FASTKD4 | Mitochondria | MT-ND3, MT-ND5, MT-CYB, MT-CO1, MT-CO2, MT-CO3, MT-ATP8/6 | mRNA stability and processing |
| FASTKD5 | Mitochondria (MRGs) | All MT-mRNAs | mRNA processing |
FASTK
FASTK localizes to different cellular compartments due to the presence of an alternative translational initiation site. In cultured mouse embryonic fibroblasts, about 50% of the endogenous FASTK protein resides in mitochondria (mitoFASTK, ∼50 kDa species), whereas the rest distributes between the nucleus and the cytosol (cytoFASTK, ∼60 kDa species) (Figure 1A) (7). In mitochondria, mitoFASTK concentrates in MRGs, where it interacts with the G-rich sequence factor 1 (GRSF1) and MT-ND6 mRNA. MT-ND6 mRNA is unique in being the only protein-coding transcript encoded on the light strand and it is also an exception to the tRNA punctuation model, because it lacks a tRNA gene at the 3′ end of its coding sequence. Hence, 3′-end formation of the MT-ND6 transcript must occur independently of the classical mitochondrial tRNA processing machinery. FASTK in fact interacts with the 3′-ends of the MT-ND6 mRNA and precursor RNAs in a RAP domain-dependent manner. Through this interaction, FASTK protects MT-ND6 from degradation by the hSuv3p–PNPase complex (Figure 1C) (7). Depletion of FASTK results in the loss of MT-ND6 mRNA, whereas FASTK over-expression leads to its stabilization. Accordingly, FASTK knock-out mice present a 50–60% decrease in complex I activity. The defect in complex I activity is not sufficient to compromise the viability of FASTK knock-out neonates, and the complete description of the mitochondrial phenotype associated with FASTK deficiency in mice still needs to be reported. Further studies are necessary to determine whether the two isoforms of FASTK, with different cellular locations and functions, cooperate in the regulation of mitochondrial metabolism. It will also be interesting to unravel the mechanisms that control differential isoform expression via alternative translation initiation.
FASTKD1
FASTKD1 has recently been found to localize to MRGs (11). An interactome study has identified FASTKD1 as an interaction partner of Twinkle, a known nucleoid protein (26), and as such, FASTKD1 partially colocalizes with mtDNA. This indicates that FASTKD1 may interact with mitochondrial RNAs shortly after transcription. Interestingly, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/nuclease-mediated depletion of FASTKD1 causes the accumulation of MT-ND3 mRNA and leads to elevated complex I activity (11,26). Thus, FASTKD1 has the opposite effect of FASTK as it negatively regulates activity of complex I. FASTKD1 requires its RAP domain to regulate MT-ND3 mRNA levels (11). However, the detailed molecular mechanisms underlying its function still remain elusive.
FASTKD2
Genome-wide linkage analysis led to the identification of a homozygous nonsense mutation in FASTKD2 gene in two siblings affected with mitochondrial encephalomyopathy associated with developmental delay, hemiplegia, convulsions and low cytochrome C oxidase activity in skeletal muscle (18). A second report recently identified FASTKD2 compound heterozygous mutations in a patient with adult-onset MELAS (Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-like episodes)-like syndrome (27). These are to date the only pathogenic mutations described in a member of the FASTK family. The molecular understanding of FASTKD2 action remained unexplored for several years but was addressed later by the independent work of four research teams. FASTKD2 was found to interact with GRSF1 and fluorescence studies have confirmed its presence in MRGs (7,28,29). Moreover, two immunoprecipitation studies showed that FASTKD2 has very strong affinity for the 16S mitochondrial rRNA (MT-RNR2) (28,29) and its RNAi- and CRISPR/nuclease-mediated depletion causes a decrease in the 16S rRNA which results in impaired ribosome assembly and translation (28–30). High-throughput sequencing of the cDNA libraries from the FASTKD2-bound RNAs suggests that FASTKD2, in addition to 16S rRNA, also associates with MT-ND6. In this respect, depletion of FASTKD2 leads to aberrant processing and expression of MT-ND6 mRNA (29). More recently, FASTKD2 was identified as part of a functional module that contains the RNA-binding proteins NGRN and WBSCR16 as well as three mitochondrial pseudouridine synthases: RPUSD3, RPUSD4 and TRUB2 (31). This module plays an important role in the pseudouridylation and stability of 16S rRNA, mitochondrial ribosome assembly and translation (30–32). It is tempting to speculate that FASTKD2 plays a role as a determinant of RNA binding specificity within the pseudouridylation module (Figure 1C). Intriguingly, depletion of FASTKD2 leads to different effects on the OXPHOS system depending on the cell type: FASTKD2 deficiency compromises biogenesis and activity of the OXPHOS complexes in 143B osteosarcoma cells and in HEK 293 epithelial cells (29,30). On the other hand, depletion of FASTKD2 in K562 myelogenous leukemia cells does not lead to an appreciable decrease of mitochondrial protein synthesis nor does it apparently alter OXPHOS (31). It is important to note that the activities of all respiratory complexes were within normal ranges in fibroblasts isolated from the two patients with FASTKD2 mutations mentioned above, which contrasts with the deficiency in cytochrome C oxidase activity found in the muscle. The mechanisms underlying the apparently differing effects of FASTKD2 depletion demands further investigation. One explanation may be varying expression levels of other FASTK family proteins that might counter-balance the effects of the loss of FASTKD2. On the other hand, impaired mitochondrial ribosome assembly in the absence of FASTKD2 might affect individual activities of OXPHOS components in a tissue-specific manner. These possibilities are supported by the finding of tissue- and age-dependent effects of a mutation in the mitochondrial ribosomal protein S34 on individual OXPHOS complex activities in mice (33).
FASTKD3
FASTKD3 was the first of the FASTK family proteins to be described as a mitochondrial protein essential for OXPHOS since its RNAi-mediated depletion causes a drastic decrease in basal and maximal oxygen consumption rates in living cells (2). In the same report, the preliminary analysis of the FASTKD3 interactome identified proteins involved in mitochondrial RNA processing and translation, suggesting a function of FASTKD3 in post-transcriptional control of mitochondrial gene expression. The actual mechanism by which FASTKD3 controls respiration has been characterized very recently (34). Targeted disruption of the FASTKD3 gene increases the half-lives and steady-state levels of the mature mitochondrial transcripts MT-ND2, MT-ND3, MT-CYB, MT-CO2 and MT-ATP8/6. In addition, FASTKD3 is required for efficient COX1 protein synthesis, importantly without altering MT-CO1 mRNA levels (Figure 1C). Accordingly, cells depleted of FASTKD3 exhibit decreased levels of COX1 protein and reduced mitochondrial complex IV assembly and activity (34). An intact RAP domain is required for FASTKD3 to function, suggesting a direct interaction of FASTKD3 with its mRNA targets. Yet, the identity of the FASTKD3 interacting transcripts is presently unknown and demands further investigation. Taken together, these observations suggest that the function of FASTKD3 extends beyond RNA processing, ribosome assembly and RNA stability, and includes the regulation of translation within mitochondria. However, as a reconstituted mitochondrial translation system still needs to be established (35), it remains challenging to experimentally address aspects of regulation of mitochondrial translation.
FASTKD4
Similar to FASTKD3, FASTKD4 (TBRG4) is not enriched in MRGs but rather appears to be present throughout the mitochondrial matrix, where it loosely interacts with the inner mitochondrial membrane (11,36). Within the organelle, FASTKD4 binds to the majority of heavy strand encoded transcripts and its depletion leads to decreased levels of MT-ATP8/6, MT-CO1, MT-CO2, MT-CO3, MT-ND3, MT-CYB and MT-ND5 mRNAs (11,36). RNA decay analysis showed that the decrease in MT-CO1, MT-ND3 and MT-CO2 is associated with an increased degradation rate, thus implicating FASTKD4 and its RAP domain in the regulation of the stability of these transcripts (36). Importantly, the decrease of the mature forms of MT-ND5 and MT-CYB mRNAs is accompanied by a substantial accumulation of non-canonical MT-ND5+MT-CYB precursor RNA (Figure 1C) (11). The processing of this precursor does not obey the tRNA punctuation model since the two coding sequences are not separated by an intervening tRNA. As mentioned before, structural modeling predicts that the RAP domain of FASTKD4 may adopt a PD-(D/E)-XK nuclease superfamily fold. Mutational studies of this domain revealed that a conserved aspartate in a putative catalytic nuclease motif is required for MT-ND5+MT-CYB precursor processing and it is tempting to speculate that FASTKD4 may carry a specific nucleolytic activity for the cleavage of non-canonical junctions (11). Further studies will be required to determine whether FASTKD4 displays a nuclease activity. Whereas there is currently no evidence regarding a potential nuclease activity in other members of the FASTK family, this important and novel aspect of the function of the RAP domain of FASTKD4 raises new questions for further investigation that may involve the RAP domains of other FASTK proteins. Surprisingly, despite the critical role of FASTKD4 in mitochondrial mRNA metabolism, its depletion neither leads to a significant reduction in mitochondrial translation nor does it affect mitochondrial respiration in cultured cells as judged by oxygen consumption measurements (11).
FASTKD5
The remaining member of the family, FASTKD5, is present in MRGs and, like GRSF1 and FASTKD4, is required for maturation of mRNAs that cannot be processed by the activities of RNase P and RNase Z (28). Depletion of FASTKD5 results in a massive accumulation of the MT-ATP8/6+MT-CO3, MT-ND5+MT-CYB and ncRNA+MT-CO1 precursor transcripts. FASTKD5 regulates the abundance of virtually all mitochondrial-encoded mRNAs and its depletion leads to an overall 50% decrease in protein synthesis, which is particularly pronounced for COX1 protein. Accordingly, FASTKD5 siRNA-treated cells present a complex IV disassembly. The global depletion of mitochondrial translation might be due to the accumulation of mitochondrial mRNA precursors, which cannot be efficiently translated and therefore lead to stalling and subsequent disassembly of the mitochondrial ribosome (28). RNA immunoprecipitation experiments showed that FASTKD5 binds with high affinity to mitochondrial mRNAs encoding complex IV subunits, consistent with its role in processing of MT-CO1 and MT-CO3 mRNAs. As for the other FASTK proteins, the molecular mechanisms underlying FASTKD5 function remain to be addressed in greater depth.
PROPOSED MECHANISM OF ACTION
Despite the fact that proteins of the FASTK family share the same domains, they exhibit various—sometimes opposing—functions in almost all steps of mitochondrial RNA metabolism. FASTK proteins have been implicated in processing of tRNA-containing and tRNA-less precursor transcript junctions, RNA stability, pseudouridylation of 16S rRNA, ribosome assembly and eventually mitochondrial translation. The enormous functional flexibility of FASTK proteins is reminiscent of the pentatricopeptide-repeat (PPR) protein family, another family of RNA-binding proteins that act on organellar RNAs to modulate various post-transcriptional events. PPR proteins constitute one of the largest protein families in land plants, where they are involved in RNA editing, stability, processing and translation in chloroplasts and mitochondria (37). In contrast, the human genome only encodes a handful of PPR proteins that all localize to mitochondria: the RNA polymerase POLRMT, the catalytic subunit of the RNAse P MRPP3 (KIAA0391), the ribosomal protein MRPS27, the leucine-rich PPR protein LRPPRC as well as PTCD1–3 (38). Structurally, the PPR domains contain a tandem of PPR repeats, and each PPR repeat forms a pair of anti-parallel α-helices made of a repetition of ∼35 amino acids. The series of pairs of anti-parallel α-helices throughout the protein produces a superhelix with a central groove capable of sequence-specific recognition of RNA strands (39). Interestingly, structural modeling of FASTK proteins indicates that FAST_1, FAST_2 and RAP domains share a similar global architecture to PPR proteins, including the repeated α-helix motifs (7,40,41) (Figure 1D). Indeed, previous studies already suggested that the domains in FASTK proteins may be structurally related to the plant octotricopeptide repeat (OPR) domains, which have been proposed to structurally and functionally resemble PPR domains in plants (15,42,43), and accordingly out of 44 OPR proteins present in C. reinhardtii, 16 also contain a FAST-like domain (42). The insoluble nature of bacterially expressed FASTK proteins has hitherto impeded studies attempting to confirm the superhelical structure of these proteins. We predict that the X-ray structures of FASTK proteins will soon reveal their exact domain architecture and boundaries as well as the molecular aspects of sequence-specific RNA recognition by these proteins.
Based on the above mentioned structural predictions, we envision a model by which proteins of the FASTK family act as adaptors, serving as guides to recognize and bind specific RNA sequences encoded by the mitochondrial genome. Hence, FASTK family members would recognize specific RNA sequences through their conserved carboxy-terminal domains, and their central domain would facilitate, or prevent, the recruitment of trans-acting factors to their target RNAs (Figure 1C). We hypothesize that the members of the family involved in RNA stability (e.g. FASTKD1, FASTKD3 and FASTKD4) may regulate the activity and accessibility of specific substrate cleavage sites for mitochondrial endo- or exonucleases, such as the degradosome. Similarly, the members of the family involved in atypical RNA processing (FASTKD4, FASTKD5) may recruit a yet unidentified endonuclease to perform processing of the tRNA-less RNA junctions such as MT-CYB+MT-ND5 or MT-ATP8/6+MT-CO3. FASTK family members may also contribute to the generation of mRNA 3′-ends as has been shown for the FASTK and MT-ND6 mRNAs. Furthermore, we hypothesize that members of the FASTK family might regulate the recruitment of RNA-modifying enzymes to specific sites of the mitochondrial RNAs. These modifications may involve for example methylation, polyadenylation or pseudouridylation as in the case of FASTKD2, which interacts with mitochondrial pseudouridine synthases (30–32). Finally, in a recent publication Boehm et al. reported the intriguing observation that the RAP domain may be homologous to a PD-(D/E)-XK nuclease fold, which, in the case of FASTKD4, is similar to the bacterial very short patch repair (VSR) endonucleases (11). Although this activity has not yet been reconstituted in vitro, these observations raise the intriguing possibility that certain members of the family not only act as adaptors but may also carry a catalytic domain with endonuclease activity.
CONCLUDING REMARKS
In conclusion, the six members of the FASTK family represent an emerging but already key group of proteins that regulate mitochondrial gene expression and function. Whereas previous work has resulted in a clearer picture of the spectrum of mitochondrial transcripts affected by the depletion of each individual family member, the precise mechanism by which FASTK proteins exert their functions is still mysterious. Further experiments will certainly help to refine the models described in this review, and the precise identification of both protein and RNA interaction partners of all six members should contribute to our understanding of how the FASTK proteins exert their functions. In particular, we predict that the use of chimeric proteins that combine the putative protein-interacting central domains with the RNA-interacting carboxy-terminal domains from the different FASTK proteins may unveil fundamental aspects of their function. Moreover, the degree of redundancy of FASTK proteins is only beginning to emerge and may hamper the analysis of phenotypes of the deletion of individual family members. One way of addressing this issue may be combinatorial genomic deletion studies. However, so far only partially overlapping functions of individual FASTK family proteins have been described and therefore the success of such reverse genetics approaches remains uncertain. FASTK family proteins have so far been refractory to the preparation in sufficient amounts using recombinant expression systems. Nevertheless, we hope that the in vitro reconstitution of the FASTK family members bound to their RNA targets will soon drastically transform our understanding of this particular aspect of mitochondrial biology, and may potentially unveil an amino acid code by which members of the FASTK family recognize their mRNA substrates.
Footnotes
Present address: Alexis A. Jourdain, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
FUNDING
Funding for open access charge: Sundry funds. Swiss National Science Foundation 310030B_160257/1 to J.-C.M. Consejeria de Sanidad JCyL Grants BIO/VA20/15 to M.S. and BIO/VA21/15 to M.A.d.l.F.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tian Q., Taupin J., Elledge S., Robertson M., Anderson P.. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med. 1995; 182:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simarro M., Gimenez-Cassina A., Kedersha N., Lazaro J.B., Adelmant G.O., Marto J.A., Rhee K., Tisdale S., Danial N., Benarafa C. et al. . Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 2010; 401:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P.. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005; 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li W., Simarro M., Kedersha N., Anderson P.. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 2004; 24:10718–10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Izquierdo J.M., Valcarcel J.. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007; 282:1539–1543. [DOI] [PubMed] [Google Scholar]
- 6. Simarro M., Mauger D., Rhee K., Pujana M.A., Kedersha N.L., Yamasaki S., Cusick M.E., Vidal M., Garcia-Blanco M.A., Anderson P.. Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:11370–11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jourdain A.A., Koppen M., Rodley C.D., Maundrell K., Gueguen N., Reynier P., Guaras A.M., Enriquez J.A., Anderson P., Simarro M. et al. . A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 2015; 10:1110–1121. [DOI] [PubMed] [Google Scholar]
- 8. Lee I., Hong W.. RAP–a putative RNA-binding domain. Trends Biochem. Sci. 2004; 29:567–570. [DOI] [PubMed] [Google Scholar]
- 9. Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. . The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 10. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. . Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 11. Boehm E., Zaganelli S., Maundrell K., Jourdain A.A., Thore S., Martinou J.C.. FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain. Nucleic Acids Res. 2017; 45:6134–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztanyi Z., El-Gebali S., Fraser M. et al. . InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Claros M.G., Vincens P.. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996; 241:779–786. [DOI] [PubMed] [Google Scholar]
- 14. Emanuelsson O., Nielsen H., Brunak S., von Heijne G.. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000; 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- 15. Kleinknecht L., Wang F., Stube R., Philippar K., Nickelsen J., Bohne A.V.. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell. 2014; 26:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merendino L., Perron K., Rahire M., Howald I., Rochaix J.D., Goldschmidt-Clermont M.. A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 2006; 34:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivier C., Goldschmidt-Clermont M., Rochaix J.D.. Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 2001; 20:1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghezzi D., Saada A., D’Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V., Elpeleg O., Zeviani M.. FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 2008; 83:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallace D.C. Mitochondrial genetics: a paradigm for aging and degenerative diseases. Science. 1992; 256:628–632. [DOI] [PubMed] [Google Scholar]
- 20. Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. [DOI] [PubMed] [Google Scholar]
- 21. Nagao A., Hino-Shigi N., Suzuki T.. Measuring mRNA decay in human mitochondria. Methods Enzymol. 2008; 447:489–499. [DOI] [PubMed] [Google Scholar]
- 22. Piechota J., Tomecki R., Gewartowski K., Szczesny R., Dmochowska A., Kudla M., Dybczynska L., Stepien P.P., Bartnik E.. Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim. Pol. 2006; 53:157–168. [PubMed] [Google Scholar]
- 23. Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E.A.. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013; 17:386–398. [DOI] [PubMed] [Google Scholar]
- 24. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jourdain A.A., Boehm E., Maundrell K., Martinou J.C.. Mitochondrial RNA granules: compartmentalizing mitochondrial gene expression. J. Cell Biol. 2016; 212:611–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han S., Udeshi N.D., Deerinck T.J., Svinkina T., Ellisman M.H., Carr S.A., Ting A.Y.. Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells. Cell Chem. Biol. 2017; 24:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoo D.H., Choi Y.C., Nam D.E., Choi S.S., Kim J.W., Choi B.O., Chung K.W.. Identification of FASTKD2 compound heterozygous mutations as the underlying cause of autosomal recessive MELAS-like syndrome. Mitochondrion. 2017; 35:6135–6146. [DOI] [PubMed] [Google Scholar]
- 28. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 29. Popow J., Alleaume A.M., Curk T., Schwarzl T., Sauer S., Hentze M.W.. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015; 21:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Antonicka H., Choquet K., Lin Z.Y., Gingras A.C., Kleinman C.L., Shoubridge E.A.. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 2017; 18:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arroyo J.D., Jourdain A.A., Calvo S.E., Ballarano C.A., Doench J.G., Root D.E., Mootha V.K.. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 2016; 24:875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zaganelli S., Rebelo-Guiomar P., Maundrell K., Rozanska A., Pierredon S., Powell C.A., Jourdain A.A., Hulo N., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. et al. . The pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules. J. Biol. Chem. 2017; 292:4519–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richman T.R., Ermer J.A., Davies S.M., Perks K.L., Viola H.M., Shearwood A.M., Hool L.C., Rackham O., Filipovska A.. Mutation in MRPS34 compromises protein synthesis and causes mitochondrial dysfunction. PLoS Genet. 2015; 11:e1005089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boehm E., Zornoza M., Jourdain A.A., Delmiro Magdalena A., Garcia-Consuegra I., Torres Merino R., Orduna A., Martin M.A., Martinou J.C., De la Fuente M.A. et al. . Role of FAST kinase domains 3 (FASTKD3) in post-transcriptional regulation of mitochondrial gene expression. J. Biol. Chem. 2016; 291:25877–25887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mai N., Chrzanowska-Lightowlers Z.M., Lightowlers R.N.. The process of mammalian mitochondrial protein synthesis. Cell Tissue Res. 2017; 367:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolf A.R., Mootha V.K.. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 2014; 7:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 38. Lightowlers R.N., Chrzanowska-Lightowlers Z.M.. Human pentatricopeptide proteins: only a few and what do they do. RNA Biol. 2013; 10:1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hall T.M. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016; 36:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coquille S., Filipovska A., Chia T., Rajappa L., Lingford J.P., Razif M.F., Thore S., Rackham O.. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014; 5:5729. [DOI] [PubMed] [Google Scholar]
- 42. Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., Wollman F.A.. Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. Cell Mol. Biol. 2011; 67:1055–1066. [DOI] [PubMed] [Google Scholar]
- 43. Hammani K., Bonnard G., Bouchoucha A., Gobert A., Pinker F., Salinas T., Giege P.. Helical repeats modular proteins are major players for organelle gene expression. Biochimie. 2014; 100:141–150. [DOI] [PubMed] [Google Scholar]