Abstract
CHD3 and CHD4 (Chromodomain Helicase DNA binding protein), two highly similar representatives of the Mi-2 subfamily of SF2 helicases, are coexpressed in many cell lines and tissues and have been reported to act as the motor subunit of the NuRD complex (nucleosome remodeling and deacetylase activities). Besides CHD proteins, NuRD contains several repressors like HDAC1/2, MTA2/3 and MBD2/3, arguing for a role as a transcriptional repressor. However, the subunit composition varies among cell- and tissue types and physiological conditions. In particular, it is unclear if CHD3 and CHD4 coexist in the same NuRD complex or whether they form distinct NuRD complexes with specific functions. We mapped the CHD composition of NuRD complexes in mammalian cells and discovered that they are isoform-specific, containing either the monomeric CHD3 or CHD4 ATPase. Both types of complexes exhibit similar intranuclear mobility, interact with HP1 and rapidly accumulate at UV-induced DNA repair sites. But, CHD3 and CHD4 exhibit distinct nuclear localization patterns in unperturbed cells, revealing a subset of specific target genes. Furthermore, CHD3 and CHD4 differ in their nucleosome remodeling and positioning behaviour in vitro. The proteins form distinct CHD3- and CHD4-NuRD complexes that do not only repress, but can just as well activate gene transcription of overlapping and specific target genes.
INTRODUCTION
In eukaryotes the nuclear DNA is organized in form of chromatin. A nucleosome, in which 147 bp of double-stranded DNA are wrapped in a left-handed helix around a histone octamer, represents the smallest structural unit of chromatin. On the one hand this organization compacts the DNA and on the other hand it causes accessibility problems for DNA-dependent processes like transcription, replication or DNA repair. To circumvent such chromatin mediated repression, eukaryotic cells are able to modify chromatin structure by post translational modifications (PTMs) of histones (1), DNA modifications (2) or the association of RNA with chromatin (3,4). In addition, cells possess chromatin remodeling enzymes, which are able to move, evict or rearrange nucleosomes upon hydrolysis of ATP (5). In vivo, many remodeling enzymes act in the context of protein complexes, comprising up to 10 subunits or more, which can modify and control the activity and behaviour of those enzymes (6,5).
Nucleosome remodeling enzymes possess a helicase-like region of similar primary sequence to Saccharomyces cerevisiae Snf2p, placing them among the Snf2 family of the so called helicase-like superfamily 2 (SF2) (7). Beside the common ATPase domain, remodelers possess diverse unique domains (5). In combination with biochemical assays and sequence analyses, the individual domain composition of a remodeler forms the basis to classify the diverse enzymes into 24 Snf2 subfamilies (7,8). Up to now, there are 53 Snf2 proteins listed in total (9). Among those proteins are diverse isoforms and highly identical proteins: hBrm and hBrg1 [Snf2 subfamily], hSnf2h, hSnf2l, hSnf2l+13, yeast Isw1p and Isw2p [Iswi subfamily] or hCHD3 and hCHD4 [Mi-2 subfamily] (10–13). This raises the question about the functional differences between those enzymes. Why do cells invest so much effort in the expression of so many enzymes which are very similar regarding their sequence?
Notably, human Brg1 and Brm, two SWI-SNF2 homologs with 75% sequence identity (13) were found to reside in separate complexes, named Numac (Brg1), PBaf (Brg1) or Baf (Brg1 or Brm) (14–17), suggesting specific roles for each of the two enzymes. Indeed, experiments in mouse showed that Brg1-null mice die already at periimplantation stage (18,19), whereas Brm-null mice only exhibit minor defects like increased body weight (20,19). Similar to Brg1 and Brm, two representatives of the Iswi subfamily, named Snf2h and Snf2l, were also found in distinct complexes (17). Human and mouse Snf2h were found in most of the Iswi complexes, i.e. in WCRF, a complex comprising hSnf2h with subunits of Cohesin and NuRD complex, hACF, hRSF, hCHRAC (predominantly Snf2h but as well Snf2l), mNoRC and mCERF (Snf2h or Snf2l, cell type dependent) (21–29). Up to now, hNURF is the only complex reported to contain exclusively hSnf2l as the motor subunit (30). In correlation with the presence of human Snf2h/l (80% sequence identity) (31) in distinct chromatin remodeling complexes the murine homologs, mSnf2h and mSnf2l (83% overall identity) (10) were reported to exert distinct roles at specific time points during development (10,32). Furthermore mSnf2h is expressed ubiquitously, whereas the expression of mSnf2l seems to be restricted to the brain and gonadal tissues (10).
In contrast to the examples mentioned above, the highly similar Mi-2 subfamily members hCHD3 and hCHD4 (71.6% amino acid identity) (Figure 1A and Supplementary Figure S1 and (11)) were both described to act solely in the context of NuRD, a protein complex that comprises both nucleosome remodeling and deacetylase activities (33–37). Beside the CHD proteins, which represent the motor subunit of the complex (38), proteins such as MTA1/2/3, MBD2/3, HDAC1/2, RBBP4/7 (RbAp48/46), p66α or p66β have been reported to be core subunits of the NuRD complex (33–38). Since several of those core subunits are associated with transcriptional repression, NuRD is thought to act mainly as a transcriptional repressor (37–39). Interestingly, different physiological conditions or different cell/tissue types seem to influence the subunit composition of NuRD complexes (38).
Figure 1.
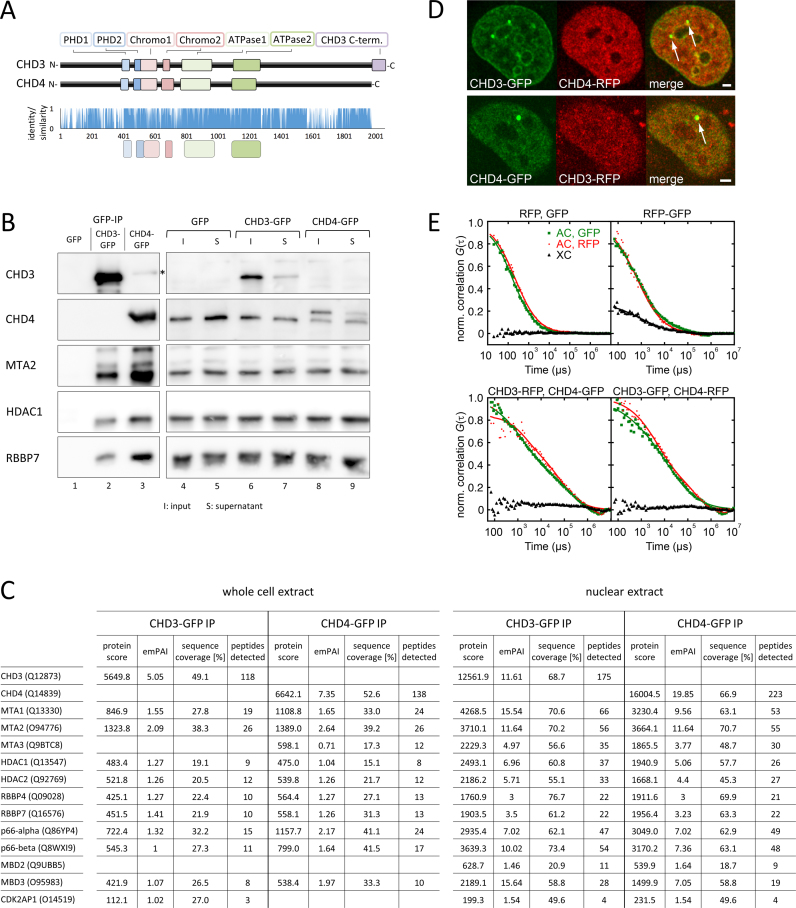
The highly similar CHD3 and CHD4 proteins form isoform specific NuRD complexes in living cells. (A) Upper panel: Schematic representation for human CHD3 (UniProt Q12873) and CHD4 (UniProt Q14839) with the annotated functional domains like the paired PHD domains 1 and 2 (light and dark blue), the paired Chromodomains 1 and 2 (light and dark rose) and the Helicase 1 (ATP binding) and the Helicase 2 domain (C-terminal) (light and dark green). The unique C-terminal domain of CHD3 is presented in violet (see also (11)). Lower panel: Sequence alignement for CHD3 and 4 by Clustal Ω (120). Identical amino acids were assigned a value of 1, groups of strongly similar amino acids were assigned a value of 0.66 and weakly similar amino acids a value of 0.33. Mismatches were scored with a value of 0. The ‘shared cores’ of the annotated domains are highlighted in the same colors like in the upper panel, below the diagram. CHD3 and CHD4 share an overall identity of 71.6%. (B) The expression of GFP (control), CHD3-GFP or CHD4-GFP in stably transfected Flp-In™ T-Rex™ 293 cells was induced by doxycycline treatment for 24 hours. Subsequently, whole cell extract was prepared and subjected to IP reactions, using GFP-Trap®_A beads. The IP reactions were analysed by western blot with the indicated antibodies (black asterisk indicates signal, derived from antibody cross reaction). To emphasise a cross reaction signal of the CHD4 antibody, the CHD4 blot of the GFP-IP reactions (lane 1–3) is presented as a longer exposure (shown in Supplementary Figure S6D). (C) The expression of GFP (control), CHD3-GFP or CHD4-GFP in stably transfected Flp-In™ T-Rex™ 293 cells was induced by doxycycline treatment for 24 h. Subsequently, whole cell extracts or nuclear extracts (NE) were prepared and subjected to IP reactions, using GFP Trap®_A beads. The reactions were finally analysed by mass spectrometry. A quantitative difference in protein amount levels is reflected by the emPAI-values (exponentially modified protein abundance index) (121). By reason of clear arrangement we only present the Mi-2 IP mass spectrometry data, since the control reactions (GFP) either did not show any signal or for the listed proteins or only insufficient signals according to the quality criteria mentioned in Material and Methods. (D) U2OS cells with stably integrated lacO repeats were transiently transfected with GBP-LacI and the indicated GFP and RFP constructs. 24 hours after transfection, the cotransfected cells were analyzed for colocalization of the lacO-bound GFP protein and the respective RFP protein via fluorescence microscopy. Scale bar, 2 μm (white arrows in the ‘merge picture’ indicate lacO spots). (E) FCCS measurements performed in U2OS cells transiently transfected with the indicated GFP and RFP constructs. Auto-(AC) and cross-(XC) correlation functions of the two colour channels are presented as green/red and black curves. Control experiments with GFP and RFP alone or a GFP-RFP fusion protein were performed to illustrate XC functions in the absence or presence of interactions between GFP and RFP proteins. Number of replicates: (B) ≥3; (C) 3; (D) (upper panel with CHD3-GFP and CHD4-RFP: 8; lower panel with CHD4-GFP and CHD3-RFP: 5); (E) (RFP,GFP: 5; RFP-GFP: 8; CHD3-RFP, CHD4-GFP: 24; CHD3-GFP, CHD4-RFP: 15).
CHD3 (also named Mi-2 alpha) and CHD4 (also named Mi-2 beta) are dermatomyositis specific autoantigens (40,41). Their common association with NuRD core subunits and the fact that both enzymes are coexpressed in many cell lines and tissues (Supplementary Figure S2) suggests that they might exert similar functions. Up to now, there are only functional studies available characterizing one of the two Mi-2 isoforms. Furthermore, it is also still controversial, whether CHD3 and CHD4 coexist in the same NuRD complex or if they form isoform-specific NuRD complexes (37,42,43).
In the present study, we therefore focused on two aspects: mapping the CHD composition in NuRD complexes in living cells and comparing both remodeling enzymes in several in vitro and live cell assays. Combining biochemical and microscopy based techniques, we report here a functional comparison of the CHD3 and CHD4 isoforms with the following findings: (i) NuRD complexes in mammalian cells are isoform-specific: They contain only one of the two types of motor proteins, which seem to be associated with the chromatin remodeling complex as monomers. (ii) These isoform-specific NuRD complexes exhibit similar mobility within the nucleoplasm, interact with HP1 and are recruited to laser-induced DNA repair sites within two minutes. Nevertheless, CHD3 and CHD4 exhibit differential nuclear localization patterns in unperturbed cells. (iii) Recombinant CHD3 and CHD4 show differences in their remodeling behaviour on reconstituted chromatin. (iv) Both CHD proteins seem to regulate the transcription of distinct genes. Taken together our data therefore suggest that CHD3 and CHD4 form separate NuRD complexes with common and distinct biological functions.
MATERIALS AND METHODS
Plasmids and cloning
The cDNA sequences for hCHD3 (UniProt: Q12873) and hCHD4 (UniProt Q14839) were subcloned into pDONR™221 (Life Technologies) according to the BP recombination protocol from the Gateway® Technology manual (Life Technologies). The cDNA encoding hCHD3 was a kind gift from Prof. O.S. Gabrielsen, Centre d’Etudes Nucléaires de Saclay, Gif-sur-Yvette, cedex, France. The cDNA encoding hCHD4 was obtained by supplementing/modifying a hCHD4 cDNA sequence (kind gift of Prof. M. Takahashi, Nagoya University, Japan) with a DNA fragment from pCMV-SPORT6 hMi2b (cat. no.: IRAT p970A0270D; ImaGenes Berlin, Germany). The mammalian expression vectors encoding C-terminal GFP- and RFP-tagged hCHD3/4 and the insect cell expression vectors encoding C-terminally Flag-tagged hCHD3/4 were created by recombining the respective entry clones with pEGFP-N1_GWc (kind gift from Prof. G. Schotta; Biomedical Center Munich-Molecular Biology; Ludwig-Maximilians-University, Planegg-Martinsried, Germany), pTagRFP-N DEST (in-house production) and pDFB6 C-Flag (in-house production) according to the LR recombination reaction protocol (see above). The cDNA for GFP and hCHD3/4-GFP were subcloned into pcDNA™5/FR/TO which was purchased like pOG44 from Life Technologies. The plasmid named pPCRScript_slo1-gla75 encodes for a dimeric nucleosome positioning sequence with rDNA and hsp 70 DNA flanking sites (see below) and was purchased from Sloning BioTechnology GmbH Puchheim, Germany. The plasmid pUC18 12x-601 contains 12 copies of a nucleosome positioning sequence (in house production). The plasmids ‘pUC19 10 x -190/+90’ and ‘pUC19 7 x HSP70’ contain 10 copies of the mouse rDNA promotor from bp –190 to + 90 in regard to the transcription start site and 7 copies of the drosophila melongaster HSP70 promotor, respectively (in-house production). The construct pBS 601-10N-601 encoding a paired nucleosome positioning sequence (NPS) sequence separated by 10 nucleotides was obtained from Joseph C. Reese (44). The cDNAs for human H2A, H2B, H3 and H4 were subcloned into pET21a (Novagen). The constructs encoding RFP tagged HP1 alpha were described in (45). All constructs were verified by sequencing.
Reagents and antibodies
Gateway® BP Clonase® Enzyme Mix and Gateway® LR Clonase® Enzyme mix (Life Technologies); anti HDAC1 ab (Santa Cruz: SC-7872); anti MTA2 ab (Abcam: ab8106); antiRbAp46 ab (Abcam: ab3535); anti Tubulin ab [DM1A] (Abcam: ab7291); anti CHD3 ab and anti CHD4N ab (Prof. Weidong Wang, National Institute on aging, Baltimore, Maryland, USA or (35)); anti Mi-2 antibody H-242 (Santa-Cruz: sc-11378 X); anti HP-1 α ab (Merck Millipore: MAB3446); anti HP-1 β ab (Abcam: ab10478); anti HP-1 γ ab (Merck Millipore: MAB3450); HRP anti rabbit (Jackson Immuno Research Laboratories Inc.: 111-035-144) and HRP anti mouse (Jackson Immuno Research Laboratories Inc.: 115-035-146); mouse monoclonal anti rabbit IgG light chain HRP conjugated ab (Abcam ab 99697) and mouse monoclonal anti rabbit IgG heavy chain HRP conjugated ab (Abcam ab 99072); Alexa 594-anti rabbit (Molecular Probes: A-11012); GFP-Trap®_A Beads and Binding control-agarose beads for preclearing (from Chromotek); ANTI-FLAG® M2 Affinity Gel and FLAG® Peptide (Sigma Aldrich); HiTrap SP FF and HiLoad 16/60 Superdex 200 (Amersham); Protein A-Sepharose GE CL-4B 17-0780-01 (Sigma Aldrich); Amicon Ultra Centrifugal Filter Unit [MWCO 10 kDa] (Millipore); (protease free, fatty acid free, essentially globulin free) BSA (Sigma Aldrich); gelatin from cold water fish skin (Sigma Aldrich); NuPAGE® Novex® 4–12% Bis-Tris Protein Gels, 1.0 mm, 10 well (Life Technologies); NucleoSpin RNA Kit (Macherey-Nagel); TruSeq RNA Sample Preparation Kit v2 (Illumina); iScript™ cDNA Synthesis Kit (Bio-Rad); FCS [‘Tetracycline free’] (Biochrom); FCS (Gibco); DMEM (1x) + GlutaMAX™-I [1g/l glucose] (Life Technologies); Gibco Sf-900™ II SFM (1×) Serum free Medium (Life Technologies); Doxycycline (Clontech Laboratories, Inc.); Zeozin (Life Technologies), Hygromycin (Life Technologies), Blasticidin (Life Technologies); FuGENE® HD transfection reagent (Promega); Effectene transfection reagent (Qiagen); HotStarTaq®Plus DNA Polymerase (Qiagen); SuperSignal West Dura Extended Duration Substrate and SuperSignal West Femto Trial Kit (Thermo Scientific); Triton X-100 (Sigma Aldrich); Igepal CA-630 (Sigma Aldrich); Tween-20 (Roth); Hydroxylapatite, Fast Flow (Calbiochem); 2-Mercaptoethanol (Merck); PEG4000 (Roth); Trypsin Gold, mass spectrometry grade (Promega); 32P-γ-ATP (Hartmann Analytic); Benzonase ≥ 250 U/μl (E1014 Sigma-Aldrich); Ethidium bromide (Roth).
Cell lines and strains
Cells of the human U2OS osteosarcoma cell line (HTB-96) and U2OS cells, containing stably integrated lacO-repeats (46), were cultured in DMEM/10% FCS (47). Flp-In™ T-REx™ 293 cells (Life technologies) were cultured at in DMEM/10% tetracyclin free FCS/100 μg/ml Zeozin/10 μg/ml Blasticidin (before transfection). BL21(DE3) strain was obtained from Novagen.
Preparation of chicken histones
All steps were carried out at 4°C in the presence of proteinase inhibitors. Red blood cells from 130 ml (adult) chicken blood were pelleted by centrifugation at 3893g for 5 min. Afterward, the pellet (in total: 30 ml red blood cells) was washed two times in the following buffer: 10 mM Tris pH 7.5/140 mM NaCl/15 mM NaCitrate. Finally the cells were lysed by resuspending them in 90 ml of buffer A (15 mM Tris pH 7.5/15 mM NaCl/60 mM KCl/340 mM Sucrose/1.5 mM 2-mercaptoethanol/0.5 mM spermidine/0.15 mM spermin), supplemented with 0.5% Igepal CA-630. The cell lysate was immediately filtered through 8 layers of cheese cloth (Stricker). The whole flow-through was diluted 1:2 in buffer A and centrifuged for 10 min at and 3893g. To remove the remaining haemoglobin from the nuclei-containing pellet, the latter was subsequently washed several times at in buffer A until the supernatant became transparent. The pellet was finally resuspended in 10 ml buffer A and stored in 1–1.5 ml aliquots at –80°C. 1.5 ml of white coloured nuclei were finally lysed for 10 min in 25 ml of 50 mM sodiumphosphate pH 6.8, supplemented with 480 mM NaCl and 1 mM 2-mercaptoethanol under continuous stirring. The nuclear lysate was sonicated with a Branson sonifier (Emerson Electric Co) until there were no (DNA) particles visible any longer. Afterwards 10 g hydroxylapatite and 50 ml 50 mM sodiumphosphate pH 6.8, supplemented with 480 mM NaCl and 1 mM 2-mercaptoethanol were added under continuous stirring. After pelleting the hydroxylapatite for 5 min at 3893g, the latter was washed 3× in 50 mM sodiumphosphate pH 6.8, supplemented with 480 mM NaCl and 1 mM 2-mercaptoethanol and subsequently 5× in 50 mM sodiumphosphate pH 6.8, supplemented with 700 mM NaCl and 1 mM 2-mercaptoethanol. The histones were eluted by adding 50 mM sodiumphosphate pH 6.8, supplemented with 2.5 M NaCl and 1 mM 2-mercaptoethanol (5 × 15 ml steps). Fractions were pooled, dialyzed (MWCO 6–8 kDa) against 20 mM Tris–HCl pH 7.6/2 M KCl/1.5 mM MgCl2/0.5 mM EGTA/25% glycerol and concentrated via PEG4000. The histone solution was analyzed via SDS gel electrophoresis for stoichiometrically correct histone distribution and quantified via absorption measurement at A 230 nm (Abs of 4.3 corresponds to 1 mg/ml histones).
Preparation of human histones
Recombinant human histones were purified using an adapted protocol from (48) and detailed protocol can be obtained upon request. In brief, the expression of human H2A, H2B, H3 and H4 in BL21(DE3) strains was induced at OD 0.6 with 2 mM IPTG for 2–3 h at 37°C. The proteins were purified from inclusion bodies in 20 mM Tris–HCl pH 7.5/10 mM DTT/7 M guanidinium hydrochloride (buffer A) and dialyzed (MWCO 6–8 kDa) against 20 mM NaAC pH 5.2/200 mM NaCl/1 mM Na-EDTA/5 mM 2-mercaptoethanol/7 M urea. The denatured, single histones were loaded on a HiTrap SP FF column and eluted by a salt gradient (0.2–1 M). The eluted histones were dialyzed against water, supplemented with 2 mM 2-mercaptoethanol (MWCO 6–8 kDa) and lyophilized with a Speed Vac Concentrator (Savant). For reconstituting histone octamers, single histones in buffer A were mixed at equimolar amounts, followed by a renaturing step (dialysis) in 10 mM Tris–HCl pH 7.5/1 mM Na-EDTA/5 mM 2-mercaptoethanol/2 M NaCl (buffer B). The reconstituted octamers were separated from dimers and high molecular weight aggregates by a gel filtration step with a HiLoad 16/60 Superdex 200 column. The protein concentration was adjusted to 1 mg/ml in buffer B with Amicon Ultra Centrifugal Filter Units. Afterwards, octamers were analyzed via SDS gel electrophoresis for stoichiometrically correct histone distribution and supplemented with an equal volume of 100% glycerol.
Nucleosome assembly
For the assembly of symmetric and asymmetric mononucleosomes, the plasmids pPCRScript_slo1-gla75 (i) and pUC18 12x-601 (ii) were digested with Bgl II (i) and NotI (ii). The released fragments were purified via gel extraction and served as a PCR template to generate the diverse assembly templates (Table 1).
Table 1. Primers for generating nucleosomal DNA-templates.
| Forward primer (5′-3′): | Reverse Primer (5′-3′): | Symmetrically positioned mononucleosomes |
| atc ttt tga ggt ccg gtt ctt t | gta cag aga ggg aga gtc aca aaa c | 77-NPS-77 |
| atg ttt ggg cc acct ccc c | taa cgg cct taa gag aaa ttt ct | 40-NPS-40 |
| gat cca gaa tcc tgg tgc tga g | tag ctg tat ata tct gac aca tg | 0-NPS-0 |
| Forward primer (5′-3′): | Reverse Primer (5′-3′): | Asymmetrically positioned mononucleosomes |
| gat cca gaa tcc tgg tgc tga g | gta cag aga ggg aga gtc aca aaa c | 0-NPS-77 |
| ggccgccctggagaatc | Cy3-gcgtatagggtccatcacataacc | 6-NPS-47 (pUC18 12x-601) assembly with human octamers |
| Forward primer (5′-3′): | Reverse Primer (5′-3′): | Dinucleosome |
| gcgaattggagctccaccgc | ggaattcctgcagaccgggg | 601-10N-601 |
agatcttttgaggtccggttcttttcgttatggggtcatatgtttgggccacctcc ccatggtatgacttccaggtatggatccagaatcctggtgctgaggctgctcaatt ggttgtagcaagctctagcactgcttaaatgcatgtacgcgcggtcccctgtgttt taactgccaaggggattactccctagtctccaggcatgtgtcagatatatacag ctagctacaaagaaaactcgagaaatttctcttaaggccgttattctctagattcg ttttgtgactctccctctctgtactaagatct (pPCRScript_slo1-gla75; sequence between two BglII sites: 310 bp)
gcggccgccctggagaatcccggtgccgaggccgctcaattggtcgtagcaa gctctagcaccgcttaaacgcacgtacgcgctgtcccccgcgttttaaccgcca aggggattactccctagtctccaggcacgtgtcagatatatacatcctgtgcatgt attgaacagcgactcgggttatgtgatggaccctatacgcggccgc (pUC18 12x-601; sequence between two NotI sites: 208 bp)
For the assembly of the mouse rDNA promotor and the drosophila HSP70 promotor mononucleosomes the plasmids ‘pUC19 10x -190/+90’ (i) and ‘pUC19 7x HSP70’ (ii) were digested with AvaI (i) and EcoRI (ii). The restriction digest reactions with the released fragments were used as a template for the assembly procedure (see below).
cccgggtatcagttctccgggttgtcaggtcgaccagttgttcctttgaggtcc ggttcttttcgttatggggtcatttttgggccacctccccaggtatgacttccagg tattctctgtggcctgtcactttcctccctgtctcttttatgcttgtgatcttttctatc tgttcctattggacctggagataggtactgacacgctgtcctttccctattaacac taaaggacactataaagagaccctttcgatttaaggctgttttgcttgtccagcct attcccggg (pUC19 10x -190/+90 mouse rDNA promotor; sequence between two AvaI-sites: 291 bp)
gaattcggatcccacgataagcataaccaagctctgcgattatctctaccataa ttaatttaagcagccgtatttataaagaaatttccaaaataaagcgaatattctag aatcccaaaacaaactggttgttgcggtaggtcatttgtttggcagaaagaaaa ctcgagaaatttctctggccgttattctctattcgttttgtgactctccctctctgta ctattgctctctcactctgtcgcacagtaaacggcacactgttctcgttgcttcga gagagcgcgcctcgaatgttcgcgaaaagagcgccggagtataaatagaggc gcttcgtcgacggaacgtcaattcagatctgaattc (pUC19 7x HSP70 drosophila HSP70 promotor; sequence between two EcoRI sites: 366 bp)
For the assembly of the dinucleosomes, the sequence below was amplified via PCR (Table 1) from the plasmid pBS 601-10N-601:
gcgaattggagctccaccgcggtggcggccggccgccctggagaatcccggt gccgaggccgctcaattggtcgtagacagctctagcaccgcttaaacgcacgta cgcgctgtcccccgcgttttaaccgccaaggggattactccctagtctccaggcac gtgtcagatatatacatcctgtgccgctctagaagccctggagaatcccggtgccg aggccgctcaattggtcgtagacagctctagcaccgcttaaacgcacgtacgcg ctgtcccccgcgttttaaccgccaaggggattactccctagtctccaggcacgtgt cagatatatacatcctgtgccgctctagaagtattgtattccccggtctgcaggaa ttcc (pBS 601-10N-601; 388 bp)
The nucleosome assembly was performed via salt gradient dialysis. Briefly, an assembly reaction (50–200 μl) contained chicken/human octamers and DNA (both in the μg range) in ratios of 0.6–1.6: 1 (histones: DNA) in high salt buffer (10 mM Tris pH 7.6/2M NaCl/1 mM EDTA/1 mM 2-mercaptoethanol/0.04–0.05% Igepal CA-630), supplemented with 200 ng/μl BSA. The reaction was pipetted into a 1.5 ml Protein LoBind tube (Eppendorf) which was placed head first in a foam-floater in 300 ml high salt buffer. The tubes were manipulated in advance by introducing a hole of 6 mm diameter into the lid and by removing the bottom. Subsequently, a 1 cm2 piece of a buffer equilibrated dialysis membrane (MWCO 6–8 kDa) was mounted between lid and tube. After removing air bubbles between dialysis membrane and buffer, 3 L of low salt buffer (10 mM Tris pH 7.6/50 mM NaCl/1 mM EDTA/1 mM 2-mercaptoethanol/0.05% Igepal CA-630) were pumped with a flow rate of 200 ml/h into the beaker containing the dialysis reaction. The success of an assembly reaction was checked by loading 200–500 ng of the nucleosomes (concentration determined via the applied DNA amount per reaction) on a PAA-gel, using non-assembled DNA as a control.
Nucleosome remodeling assay
A nucleosome remodeling assay was performed in 20 mM Tris pH 7.6/80–120 mM KCl/1.5 mM MgCl2/0.5 mM EGTA/10% glycerol/1–1.8 mM ATP (in case of the dinucleosome reaction extra BSA was added to reach a final concentration of 90 ng/μl) at 26°C/30°C for 30–80 min (10–11 μl reaction volume in total). Nucleosomes were added in concentrations ranging from 130 to 300 nM (77-NPS-77; 40-NPS-40; 0-NPS-0; 0-NPS-77 and 6-NPS-47) or 13.6–44 ng/μl (601-10N-601; drosophila HSP70 promotor and –190/+90 mouse rDNA promotor) respectively. The final concentration of recombinant nucleosome remodeling enzymes varied from 25 to 400 nM. The enzymatic reactions were stopped by adding 300–1000 ng competitor (plasmid) DNA for 5 min (at 30°C or on ice). The nucleosome movements were visualized by supplementing the reactions with glycerol (4–5%) and loading them on 5–6% PAA gels, which were subsequently scanned on a Fluorescence Image Reader FLA-5000 (Fujifilm) or stained with ethidium bromide.
ATPase assay
Recombinantly purified CHD3-FLAG or CHD4-FLAG (both 40 nM) were incubated with 130 nM mononucleosomes (77-NPS-77) in 20 mM Tris–HCl pH 7.6/120 mM KCl/1.5 mM Mg2+/0.5 mM EGTA/10% glycerol in the presence of 500 μM ATP and 0.2 μCi 32P-γ-ATP for 40 min at 30°C. Released 32P-γ phosphate was separated from non-hydrolyzed 32P-γ-ATP by TLC on PEI-Cellulose F plates (Merck) [mobile phase: 50% acetic acid, 0.5 mM LiCl]. 32P-γ and 32P-γ-ATP ratios were calculated after phosphoimaging (Fuji Film FLA-3000), using Fuji Multi Gauge Software and Excel. Concentration of hydrolyzed ATP was calculated from these ratios and the (total) initial ATP concentration in the assay. Averages and standard deviations for the ATPase rates represent three experiments, done with two to three independent protein preparations.
Purifying recombinant hCHD3/4 from insect cells
V1 virus encoding for C-Flag hCHD3/4 were created according to (49). Briefly, pDFB6 C-Flag with hCHD3/4 was transformed into chemically competent DH10Bac EM YFP (kind gift from Dr. Imre Berger, EMBL Grenoble, Grenoble Cedex 9, France) (49,50), whereby positive transformants were screened via blue-white screening. Several clones were used to isolate the bacmid DNA (created via Tn7 transposition) by alkaline lysis. The isolated DNA was transfected afterwards into Spodoptera frugiperda (Sf21) cells using Fugene HD. The initial virus (V0) was collected from YFP positive cells and used for producing the generation 1 virus (V1), which was in turn used for performing large scale infections. Forty-eight hours after V1-infected Sf21 cells stopped dividing (day of proliferation arrest), cells were harvested by centrifugation (10 min/RT/800g). For purifying recombinant C-Flag hCHD3 or CHD4, 80–120 × 106 cells were lysed in 20 mM Tris–HCl pH 7.6/500 mM KCl/1.5 mM MgCl2/0.5 mM EGTA/10% glycerol/0.1% Igepal CA-630 (30 ml), accompanied by three cycles of freezing and thawing in liquid nitrogen. The thawed cell extract was sonicated using a Branson Sonifier 250 (Emerson Electric Co.), followed by a 20 min centrifugation step at 4°C (18 000g). The supernatant, containing the Flag-tagged proteins, was incubated with 250 μl ANTI-FLAG® M2 Affinity Gel (gravity flow method). After washing the beads with lysis buffer and 20 mM Tris–HCl pH 7.6/300 mM KCl/1.5 mM MgCl2/0.5 mM EGTA/10% glycerol/0.1% Igepal CA-630, the protein was eluted in 20 mM Tris–HCl pH 7.6/300 mM KCl/1.5 mM MgCl2/0.5 mM EGTA/10% glycerol/0.1% Igepal CA-630 containing 500 ng/μl and FLAG® Peptide (5 × 250 μl elution steps). The protein concentration was estimated via Bradford assay and the purity was checked with Coomassie-stained SDS gels. The activity of the respective enzyme fractions was tested by nucleosome remodeling assays (see above).
Stable cell lines
Before transfection, Flp-In™ T-REx™ 293 cells (Life technologies) were cultured in DMEM/10% tetracyclin free FCS/100 μg/ml Zeozin/10 μg/ml Blasticidin. The cells were transfected with the pOG44 plasmid and the respective pcDNA 5/FRT/TO constructs encoding CHD3-GFP, CHD4-GFP or GFP (see above), using Fugene HD according to the Fugene and TRex Flp-In system manuals. Twenty-four hours after transfection, the selection process was started by incubating the cells with DMEM/10% tetracycline free FCS/100 μg/ml Hygromycin/10 μg/ml Blasticidin. After 10–14 days several individually visible colonies were selected and screened for reliable induction after adding 1 ng/μl doxycycline via western blot and fluorescence microscopy.
Preparation of whole cell extract (WCE)
Flp-In™ T-REx™ 293 cells with GFP, CHD3-GFP and CHD4-GFP were induced with 1 ng/μl doxycycline at ∼70% confluency for 24 h. Cells from two 150 mm (⊘) culture dishes were lysed in 20 mM Tris–HCl pH 7.6/1.5 mM MgCl2/0.5 mM EGTA/150 mM KCl/10% glycerol/1 mM DTT/1.0% IGEPAL CA-630. Protein concentrations were determined by Bradford assay.
Preparation of nuclear extract (NE)
For preparation of nuclear extract, Flp-In™ T-REx™ 293 cells or stably transfected Flp-In™ T-REx™ 293 cells with GFP, CHD3-GFP and CHD4-GFP were used. The stably transfected cells were induced with 1 ng/μl Doxycycline at ∼70% confluency for 24 h. Cells from four 150 mm (⊘) culture dishes were lysed in 4 ml buffer A (10 mM Tris–HCl pH 7.6/3 mM MgCl2/10 mM NaCl/0.5% IGEPAL CA-630) by vortexing. The cell extract was centrifuged (10 min, 4°C, 500g) and the nuclei-pellet was washed twice with buffer A. The nuclei were finally lysed on a vortex by adding 800 μl buffer B (15 mM Tris–HCl pH 7.6/60 mM KCl/500 mM NaCl). The released DNA was pelleted by ultra-centrifugation (120 000g, 10 min, 4°C, rotor: Beckman Coulter TLA100.4, Beckman Coulter Optima TL Ultracentrifuge 100 000 rpm). The supernatant (nuclear extract) was quantified via Bradford assay.
Co-IP experiments
Co-IP experiments were performed with either 5 mg whole cell extract (WCE) or 1 mg nuclear extract (NE) in IP buffer (20 mM Tris–HCl pH 7.6/1.5 mM MgCl2/150 mM KCl for WCE and 150 mM NaCl for NE/0.5 mM EGTA/10% glycerol/1 mM DTT/0.25% IGEPAL CA-630) in a total volume of 800 μl. For IP with GFP-Trap A beads the lysates were precleared in advance in IP buffer for 1 h at 4°C using 20 μl Chromotek BAB-20 agarose beads. Lysates for IP with Protein A beads (+ anti Mi-2 antibody and no antibody for negative control) were not precleared. Both, GFP-Trap A beads and Protein A beads were preincubated for 1 hour at 4°C in buffer A (20 mM Tris–HCl pH 7.6/1.5 mM MgCl2/150 mM KCl for WCE and 150 mM NaCl for NE/0.5 mM EGTA/10% glycerol/1 mM DTT) supplemented with 1% gelatin and 200 ng/μl BSA. In case of mass spectrometry experiments the GFP-Trap_A beads were not blocked. For GFP-IP experiments, the precleared lysate was incubated for 3–4 h with GFP-Trap A beads on a rotating wheel at 4°C. For Mi-2-IP reactions, the lysate was incubated in the presence or absence of ≥ 125 U Benzonase or 200 ng/μl Ethidium bromide with 5 μg Mi-2 antibody for 2 h on a rotating wheel at 4°C, whereby after two hours 20 μl blocked protein A beads were added for a further incubation time of 2 h. After the Co-IP reaction, all supernatants were collected by centrifugation (2700g) and the beads were washed four times in buffer A. For mass spectrometry, beads from Co-IP experiments were washed once in buffer A (see above), once in buffer A with 500 mM KCl (see above), followed by two further washing steps in buffer A (see above). For Mi-2-IP reactions, beads were washed once in buffer A, 3× in buffer A with 500 mM KCl and once more with buffer A. All beads were finally resuspended in 2× Laemmli buffer and boiled for 5 min at 80°C. 5–25% of one IP reaction and 15 μg of the input and supernatant fraction were used for the western blot or mass spectrometry analyses.
Western blot
After semi dry western blotting, the PVDF membrane (Millipore) was blocked over night at 4°C in 5% milk powder in PBS with 0.2% Tween. Primary antibodies were diluted 1:1000 or 1:2000 in blocking solution (see above) and incubated for 1 h at RT. Afterwards, the membranes were washed five times for 5 min in PBS with 0.2% Tween. HRP-coupled secondary antibodies were diluted 1:5000 in blocking solution (see above) and incubated for 1 h at RT. The membranes were washed again five times (see above) and incubated with the SuperSignal West Dura Extended Duration Substrate or SuperSignal West Femto Trial Kit according to the manufacturers protocol. Images were recorded on a FujiFilm LAS-3000 image reader (Fujifilm Holdings Corporation).
Mass spectrometry
25% of one Co-IP reaction were loaded on a NuPAGE® Novex® 4–12% Bis-Tris Protein Gels, 1.0 mm, 10 well. The gels were run in 1 x MOPS buffer according to the manufacturer's protocol. For mass spectrometric analysis of proteins, a gel lane was cut into 12 consecutive slices. The gel slices were then transferred into 2 ml micro tubes (Eppendorf) and washed with 50 mM NH4HCO3, 50 mM NH4HCO3/acetonitrile (3/1) and 50 mM NH4HCO3/acetonitrile (1/1) while shaking gently in an orbital shaker (VXR basic Vibrax, IKA). Gel pieces were lyophilized after shrinking by 100% acetonitrile. To block cysteines, reduction with DTT was carried out for 30 min at 57°C followed by an alkylation step with iodoacetamide for 30 min at room temperature in the dark. Subsequently, gel slices were washed and lyophilized again as described above. Proteins were subjected to in gel tryptic digest overnight at 37°C with approximately 2 μg trypsin per 100 μl gel volume. Peptides were eluted twice with 100 mM NH4HCO3 followed by an additional extraction with 50 mM NH4HCO3 in 50% acetonitrile. Prior to LC–MS/MS analysis, combined eluates were lyophilized and reconstituted in 20 μl of 1% formic acid. Separation of peptides by reversed-phase chromatography was carried out on an UltiMate 3000 RSLCnano System (Thermo Scientific, Dreieich), which was equipped with a C18 Acclaim Pepmap100 preconcentration column (100μm i.D.x20mm, Thermo Fisher) in front of an Acclaim Pepmap100 C18 nano column (75 μm i.d. × 150 mm, Thermo Fisher). A linear gradient of 4% to 40% acetonitrile in 0.1% formic acid over 90 min was used to separate peptides at a flow rate of 300 nl/min. The LC-system was coupled on-line to a maXis plus UHR-QTOF System (Bruker Daltonics, Bremen) via a CaptiveSpray nanoflow electrospray source (Bruker Daltonics). Data-dependent acquisition of MS/MS spectra by CID fragmentation was performed at a resolution of minimum 60 000 for MS and MS/MS scans. The MS spectra rate of the precursor scan was 2 Hz processing a mass range between m/z 175 and m/z 2000. Via the Compass 1.7 acquisition and processing software (Bruker Daltonics) a dynamic method with a fixed cycle time of 3 s and a m/z dependent collision energy adjustment between 34 and 55 eV was applied. Raw data processing was performed in Data Analysis 4.2 (Bruker Daltonics), and Protein Scape 3.1.3 (Bruker Daltonics) in connection with Mascot 2.5.1 (Matrix Science) facilitated database searching of the Swiss-Prot Homo sapiens database (release November 2015, 20 194 entries). Search parameters were as follows: enzyme specificity trypsin with two missed cleavages allowed, precursor tolerance 0.02 Da, MS/MS tolerance 0.04 Da, carbamidomethylation or propionamide modification of cysteine, oxidation of methionine, deamidation of asparagine and glutamine were set as variable modifications. Mascot peptide ion-score cut-off was set 25. Protein list compilation was done using the Protein Extractor function of Protein Scape. Furthermore, we considered protein identification as confident, if the following criteria were met: at least two unique peptides found and a minimum protein score of 100. If necessary, fragment spectra were validated manually.
Fluorescence microscopy and colocalization analysis
Images of induced and non-induced Flp-In™ T-REx™ 293 cells with CHD3-GFP, CHD4-GFP and GFP were recorded using an Axiovert 200M (Carl Zeiss AG) microscope and the Axiovision Rel. 4.7 software (Carl Zeiss AG). Briefly, cells were fixed, washed 3× with PBS, stained with DAPI (see above), washed again 3× with PBS and directly mounted in 50% glycerol/PBS (see above).
Live cell imaging of U2OS cells expressing different combinations of GFP/RFP-tagged proteins was conducted at a Leica SP5 confocal microscope as previously described (45).
Colocalization of CHD3-GFP and CHD4-RFP was assessed using the Pearson correlation coefficient (51) and the intensity correlation coefficient (ICQ) (52) as described. First, the background intensity IBG in a region outside of the cell nucleus and the average intensity IAVG within the cell nucleus were determined for each colour channel. Next, normalized intensity values were calculated according to Inorm = (I - IBG)/(IAVG - IBG). Based on these normalized intensities, the Pearson correlation coefficient was calculated according to r = Cov(Inorm,red, Inorm,green)/SD(Inorm,red)/SD(Inorm,green), where Cov and SD denote the covariance and the standard deviation, respectively. The slope m and the y-intercept c of the regression line were obtained according to m = Cov(Inorm,red, Inorm,green)/SD(Inorm,green)2 and c = 1 – m. Covariation plots, ICQ values and psign test values were calculated as previously described (52).
Fluorescent three-hybrid (F3H) assay
F3H assays were conducted as described previously (53). In brief, U2OS cells containing stably integrated lacO arrays were plated on coverslips in DMEM supplemented with 10% FCS. On the next day, cells were transfected with plasmids encoding a bacterial LacI repressor fused to GFP-binding protein (LacI-GBP), a GFP-tagged bait protein (GFP-, CHD3-GFP or CHD4-GFP), and optionally an RFP-tagged prey protein (CHD3-RFP or CHD4-RFP) using Effectene. After 24 h, live cells were either imaged by confocal microscopy, or cells were fixed for 10 min in 4% paraformaldehyde/PBS and washed 3× with PBS. Next, the cells were permeabilised with 0.25% Triton X-100/PBS for 25 min at RT and washed 3× in PBS. Afterward, the cells were blocked in 2% BSA/PBS for 1 h. Primary antibodies were diluted 1:200 or 1:1000 in blocking solution (see above) and incubated for 1 h at RT. The cells were washed again 3× with PBS and incubated for 1 h at RT with an Alexa-594 labeled secondary antibody which was diluted 1:500 in blocking solution (see above). Finally the cells were washed 3× with PBS and stained for 2 min with DAPI (50 ng/μl)/PBS. After this step, the cells were washed 3× in PBS and mounted on microscopy slides with 50% glycerol/PBS.
Images for the PFA-fixed cells were acquired using a Leica SP8 confocal microscope with the Leica LAS AF V4.0 software. The following excitation and emission wavelengths were used: DAPI (405 nm/415–480 nm), GFP (488 nm/500–550 nm) and Alexa-594 (561 nm/570–680 nm). Images were evaluated using the Leica LAS AF V4.0 software and ImageJ 1.49s. The lacO sites in the nucleus recruiting GFP, CHD3-GFP and CHD4-GFP were evaluated as colocalized with the protein of interest, if the Alexa-594 signal at the lacO site was at least 1.7-fold higher than in the rest of the nucleus.
Fluorescence cross-correlation spectroscopy (FCCS)
For FCCS experiments, U2OS cells were transfected with different combinations of GFP/RFP-tagged proteins as indicated using Effectene. On the next day, measurements were conducted for 60 seconds using a Zeiss LSM 710 microscope equipped with a 63×/NA 1.2 water immersion objective and a Confocor3 unit. GFP and RFP were excited with a 488 nm Argon laser line and a 561 nm DPSS laser line, respectively. Emitted signals were recorded through a BP 505–540 IR filter and a LP 580 filter. Data were analysed as described previously (45). In brief, auto- and cross-correlation functions were computed using the self-written STC- or software. After removal of spectral cross talk, functions were fitted to an anomalous diffusion model to quantify the mobility of GFP/RFP-tagged proteins and the abundance of double-labeled complexes containing at least one GFP-tagged and one RFP-tagged protein.
Laser microirradiation
Laser microirradiation experiments were conducted as previously described (54). In brief, U2OS cells expressing HP1α-RFP and CHD3-GFP or CHD4-GFP were mounted on a Leica SP5 confocal microscope and imaged using a 63× oil immersion objective. Subsequently, a circular region of interest with a diameter of 1 μm was selected and irradiated with a 405 nm diode laser at a power of 1.5 mW in the back aperture of the objective. For irradiation, the region was scanned 16-times, corresponding to a net exposure time of ∼0.1 ms for each pixel or ∼1.5 s for the entire region. After irradiation, image series in the GFP and RFP channel were acquired and the enrichment of the different proteins over time was quantified.
RNA isolation, transcription to cDNA, library preparation and sequencing
Total RNA was prepared from 24 h induced (1 ng/μl Dox) and non-induced Flp-In™ T-REx™ 293 cells (2 million cells per sample were seeded one day before induction). For harvesting the cells, they were scraped from the culture dishes and pelleted at 1600 g and 4°C for 10 min. The RNA isolation procedure was performed with the NucleoSpin RNA Kit according to the manufacturer's instructions. Five independent induction series served as RNA sources for the RNA-seq and qPCR experiments, presented in this study. The (i) sequencing and (ii) qPCR experiments were both performed with (i) two and (ii) three complete sets of total RNA preparations, respectively.
Reverse transcription of 1 μg total RNA (quantified with a NanoDrop 1000 Spectrophotometer [Thermo Scientific] Nucleic Acids-RNA-40 measurement mode) into cDNA (for qPCR), using oligo(dT) and random hexamer primers, was performed with the iScript™ cDNA Synthesis Kit according to the manufacturer's protocol.
Library preparation, comprising rRNA depletion, polyA enrichment, fragmentation to ∼270 nucleotide length and reverse transcription to cDNA, using random hexamer primers and adapter ligation, was accomplished with the TruSeq RNA Sample Preparation Kit v2 according to the manufacturer's protocol by EMBL GeneCore facility in Heidelberg (Dr. Vladimir Benes). Libraries were sequenced on Illumina HiSeq2000 platform resulting in 37–71 Mio 50 bp paired-end reads per sample.
RNA-seq analysis
Raw reads were mapped to the UCSC human genome version 37 (hg19) gene annotation using TopHat2 (55) with following parameters: -g 1 -N 1 - -no-novel-juncs - -realign-edit-dist 2. Mapped reads were further analyzed in the R environment (56) using Rsamtools, GenomicFeatures and GenomicAlignments Bioconductor packages (57,58). Mapped reads were counted over the transcript model using the union mode. Statistical analysis of differentially expressed genes was performed with the DESeq2 Bioconductor package (59). Samples without doxycycline induction (uninduced) were considered as ground state and differential expression was determined as a statistical significant (FDR < 0.1) change in expression over the ground state.
Coding DNA sequences of CHD3 (RefSeq-ID: NM_001005271) and CHD4 (RefSeq-ID: NM_001273) were obtained from the Ensembl data base (December 2013 release) using the biomaRt package (60,61). The GFP-Sequence corresponds to the GFP-sequence in pEGFP-N1 (Clontech) and is available at: https://www.addgene.org/vector-database/2491 or upon request. RNA-seq reads were mapped to the cDNA sequences using bowtie2 (62) with following parameters: -a - -very-sensitive - -no-unal - -no-mixed - -no-discordant. Properly aligned reads of each construct were counted and normalized by the size factors obtained from DESeq analysis (see above) and the length of the constructs.
Quantitative PCR (qPCR) and data analysis
Primers for quantitative PCR were designed with Primer3web version 4.0.0 (free software). The quality of the designed primer pairs was checked with PerlPrimer (free) software v1.1.21 [http://perlprimer.sourceforge.net] and by aligning them to the human genome and transcriptome [Human genomic plus transcript (Human G + T)] using nucleotide blast (blastn) feature from NCBI. The primers were ordered from Sigma Aldrich (desalted) and are listed in Table 2.
Table 2. Primers for quantitative PCR (qPCR).
| Gene-/_Primer-name | Sequence in 5′-3′- orientation |
|---|---|
| hZNF467_1fw | agctccctgggattctctgt |
| hZNF467_1re | catctgctcctgggcattat |
| hRGS19_5fw | cgaggctcatctacgaggac |
| hRGS19_5re | ggctcctgcatcttcttgtt |
| hUSE1_2fw | ggacaaccagaccctgtcac |
| hUSE1_2re | gacgctctgactccgtcttc |
| hSumo3_6fw | ggcagccaatcaatgaaact |
| hSumo3_6re | aacacgtcgatggtgtcctc |
| hLaminA/C_fw | tgcgtacggctctcatcaact |
| hLaminA/C_re | ctcgtcgtcctcaaccacagt |
A qPCR reaction (∑ 20 μl) contained 0.4 μM of the respective forward and reverse primers and 0.2 μl of cDNA (10 ng of total RNA). The PCR buffer was a 1:10 dilution of 10× PCR buffer (Qiagen) supplemented with 0.2 mM dNTPs (each) and 1 mM MgCl2 (both from Qiagen). The final reaction comprised furthermore a 1:32.000.000 dilution of a 10.000× Sybr green I stock (Roche) and 0.4 U of HotStarTaq®Plus DNA Polymerase.
Data were collected with a Rotor-Gene 3000 system (Corbett Research/Qiagen) and analyzed using the comparative analysis software module. All samples were run in triplicate and Lamin A/C (Table 2) was used for normalization. The mean values and error bars (calculated in Excel) are derived from three biological replicates (with at least two technical replicates for each biological replicate). Relative expression values for induced (+dox) and non-induced (–dox) conditions were tested for significance using a two tailed, homoscedastic t-test in Microsoft Excel. P-values are given for sets with significant changes (*P < 0.05, **P < 0.01, ***P < 0.001).
RESULTS
CHD3 and CHD4 form isoform-specific NuRD complexes
The two Mi-2 subfamily members CHD3 and CHD4 are coexpressed in many cell lines and tissues and show an overall identity of 71.6% (Figure 1A and Supplementary Figures S1 and S2). The high similarity between both proteins impedes the development of molecular biological and biochemical tools for the detection and functional characterization of the respective CHD-isoform. In order to examine whether CHD3 and CHD4 coexist in the same NuRD complex or if they form isoform-specific NuRD complexes, we generated a doxycycline inducible Hek293-based cell line (Flp-In™ T-REx™ 293) expressing either human CHD3 or CHD4 with a C-terminal GFP tag, or GFP alone (Supplementary Figure S3). The exogenously expressed proteins were immunoprecipitated, using GFP-Trap®_A beads and the precipitation reactions were consequently analysed via western blot and mass spectrometry (Figure 1B, C and Supplementary Figures S4 and S5D). As expected, we observed several core factors like HDAC1, RBBP7 (RbAp46) and MTA2 specifically coprecipitating with both CHD proteins by western blot and mass spectrometry analyses (Figure 1B,C and Supplementary Figure S4). We further detected reported core factors like MTA1/3, p66 (alpha and beta), RBBP4 (RbAp48) and MBD2/3 via mass spectrometry (Figure 1C and Supplementary Figure S4), suggesting that the GFP epitope did not interfere with proper formation of the NuRD complex. To analyse the precipitation reactions with respect to endogenous CHD proteins, we used antibodies directed against human CHD3 and CHD4 (Supplementary Figure S5B). Both antibodies exhibit weak cross reactivity towards the other Mi-2 isoform, most probably due to the above-mentioned high degree of similarity between both isoforms (Figure 1A and Supplementary Figures S1 and S5B). We therefore propose that the band obtained with the CHD3 antibody in lane 3 of Figure 1B ( = CHD4-GFP IP reaction) corresponds to the precipitated CHD4-GFP protein (Figure 1B, CHD3 blot, lane 3 with black asterisk and Supplementary Figure S5). Similarly, the antibody signal in lane 2 of Supplementary Figure S5D ( = CHD3-GFP IP reaction) of the CHD4 western blot likely represents the bait protein CHD3-GFP, which is recognized by the antibody against CHD4 (Figure 1B and Supplementary Figure S5A-C and CHD4 blot, lane 2 with black cross in Supplementary Figure S5D). Mass spectrometry analyses from whole cell extract and nuclear extract IP experiments did not reveal any significant enrichment of unique CHD3 or CHD4 peptides in the reaction of the respective other isoform (Figure 1C and Supplementary Figure S4). Our IP experiments therefore suggest that the two CHD isoforms do not coexist in the same NuRD complex.
To validate this finding in the context of living cells we used two microscopy-based approaches, namely the fluorescent three-hybrid (F3H) assay and fluorescence cross correlation spectroscopy (FCCS). For F3H assays (53), U2OS cells containing stably integrated lacO repeats (46) were transfected with LacI-GBP (GFP-binding protein), CHD3-GFP and CHD4-RFP (or CHD3-RFP and CHD4-GFP), which resulted in recruitment of the GFP-tagged protein to the lacO sites (Figure 1D). Even though immobilized CHD3/4-GFP proteins were able to associate with endogenous NuRD factors like HDAC1, RbAp46 and MTA2 (Supplementary Figure S6A,C), suggesting the assembly/recruitment of NuRD complexes at the lacO arrays, we did not observe any colocalization of RFP-tagged CHD3 or CHD4 with the opposite or identical GFP-tagged CHD isoform (Figure 1D and Supplementary Figure S6B,C). Next, we used FCCS to test if there are mobile NuRD complexes containing both CHD isoforms (Figure 1E). Due to its high time resolution, FCCS can also detect short-lived complexes that diffuse through the observation volume within a few milliseconds. Autocorrelation functions show that a considerable fraction of CHD3-GFP/RFP and CHD4-GFP/RFP is mobile (red/green curves in Figure 1E, bottom and Supplementary Figure S6D). Based on the small amplitudes of the cross-correlation curves obtained for CHD3-GFP/CHD4-RFP and CHD3-RFP/CHD4-GFP (black curves in Figure 1E, bottom), we conclude that these mobile CHD3/4 molecules are not in complex with a labelled member of the other isoform. Furthermore, these molecules are neither in complex with another labelled member of the same isoform (black curves in Supplementary Figure S6D).
These data strongly suggest that cells contain isoform-specific NuRD complexes, which use either CHD3 or CHD4 as the motor subunit. In contrast to remodeling enzymes such as human Snf2h/Acf (47), the human CHD proteins do not tend to form homomers. Since many cell lines and tissues coexpress CHD3 and CHD4 (Supplementary Figure S2), which share moreover many core subunits like HDAC1/2, MTA2/3 or RBBP4/7 (and others) (Figure 1B,C and Supplementary Figure S4), this observation raises the question about the physiological relevance of the coexistence of such structurally similar NuRD complexes: Do CHD3 and CHD4-NuRD complexes exhibit redundant and/or distinct functions?
Recombinant CHD3 and CHD4 exhibit distinct, sequence-specific nucleosome positioning behaviour
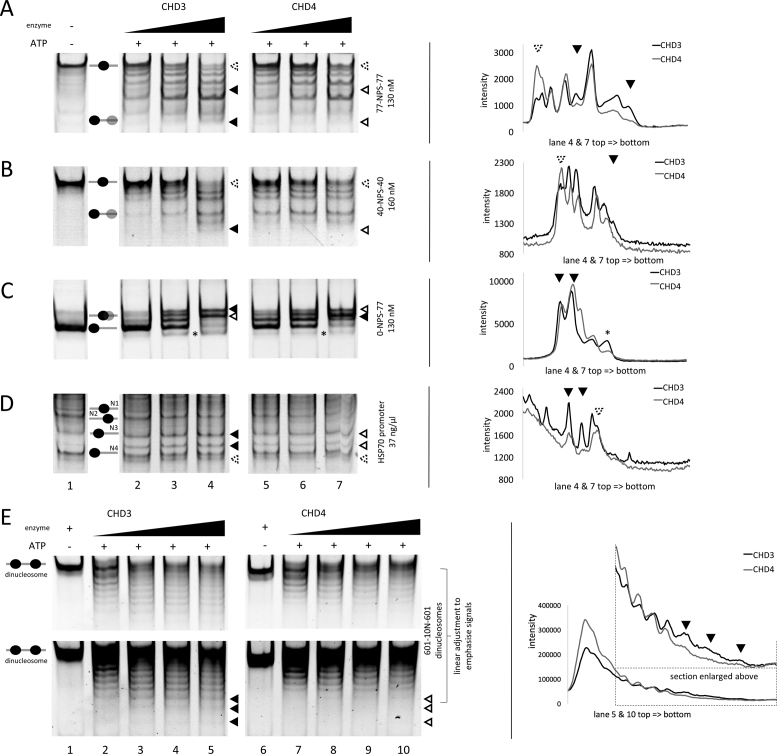
In order to analyse whether the respective CHD remodeling enzymes alone already harbour distinct enzymatic properties, which in turn might lend certain functional specificity to NuRD complexes in vivo, we compared the chromatin remodeling behaviour of the recombinant CHD3 and CHD4 proteins on various nucleosomal templates (Figure 2 and Supplementary Figures S5A and S7A-D). We found that in contrast to yeast Ino80, Isw1a or CHD1 (63–65), CHD3 and as well CHD4 seem to remodel nucleosomes independent of linker length and the initial position (Figure 2 and Supplementary Figure S7A–C). In line with data from other remodeling enzymes like dCHD3 and plant PICKLE (66,67), we observe for both CHD reactions remodeling end products that migrate below endpositioned (asymmetric) nucleosomes, arguing for nucleosome species in which less than 147 base pairs are in contact with the histone octamer (Figure 2C, lanes 4/7 and Supplementary Figure S7C, lanes 5/9 black asterisks). Comparing the remodeling patterns between CHD3 and CHD4, we can see that the majority of final nucleosome positions are the same between CHD3 and CHD4, but the relative nucleosome distributions do vary and distinct positions do appear (Figure 2A–D lanes 4/7, Figure 2E lanes 5/10 and Supplementary Figure S7A lanes 5/9, filled and empty triangles and as well intensity profiles). Nucleosome species, which according to Hamiche and coworkers (68) correspond to nucleosomes positioned close to the DNA ends were enriched in the CHD3 remodeling reactions (Figure 2A–D lanes 4/7, Figure 2E lanes 5/10 and Supplementary Figure S7A lanes 5/9, filled and empty triangles and as well intensity profiles). We therefore suggest that CHD3 reveals a stronger preference for moving nucleosomes to the end of the DNA strand than CHD4.
Figure 2.
Recombinant CHD3 and CHD4 exhibit distinct, sequence-specific nucleosome positioning behaviour. Recombinantly purified human CHD3/4 were titrated in increasing concentrations (A/C/E: 25, 50, 75 and 100 nM; B/D: 50, 100 and 200 nM) to the indicated mono- and dinucleosomal templates containing different configurations of linker DNA (see also Materials and Methods). The reactions were started by adding ATP. Nucleosomes in the absence or presence of enzyme (without ATP) served as reference. To visualize the nucleosome movements, the reactions were loaded on PAA gels (ethidium bromide stain). Positions of mono- and dinucleosomes are indicated by ovals, according to (68). Filled triangles represent more intense bands, empty triangles less intense bands or no signal, comparing CHD3 and CHD4 remodeling patterns. Triangles with a dashed rim were added for better orientation in the intensity profiles (see below). Black asterisks indicate nucleosomes, which were probably pushed over the edge of the DNA strand. On the right side of each remodeling gel are intensity profiles of the indicated gel lanes, based on Multi Gauge software analysis. The triangles indicate the positions of the bands highlighted by the triangles in the respective lanes of the corresponding gel picture. All lanes shown for one remodeling template are from one gel. Number or replicates: (A) ≥3; (B) 3; (C) ≥3; (D) 3; (E) ≥3.
Since both remodeling enzymes show overall comparable ATP hydrolysis rates and remodeling kinetics (Figure 2 and Supplementary Figure S7A—C, E), we consider the differences in the remodeling patterns significant, pinpointing to distinct remodeler-DNA interactions, resulting in turn in nucleosome positioning differences.
In living cells, the functional correlation between nucleosome repositioning and/or displacement at gene promotors and transcription is well documented in the literature (69–71). That applies as well for nucleosome remodeling at DNA ends in regard to DNA repair (72,73). The fact that CHD3 and CHD4 exhibit distinct remodeling properties already in the absence of accessory core subunits suggests that isoform-specific NuRD complexes also possess distinct remodeling features and activities regarding the regulation of DNA dependent processes like repair or transcription.
CHD3 and CHD4 exhibit differential localization patterns and similar mobility in living cells
To examine whether CHD3 and CHD4 exhibit similar or different functions in living cells, we decided to use transient transfection approaches and/or the inducible cell line system, described above. To compare the localization pattern of both proteins we cotransfected human U2OS cells with CHD3-GFP and CHD4-RFP (Figure 3).
Figure 3.
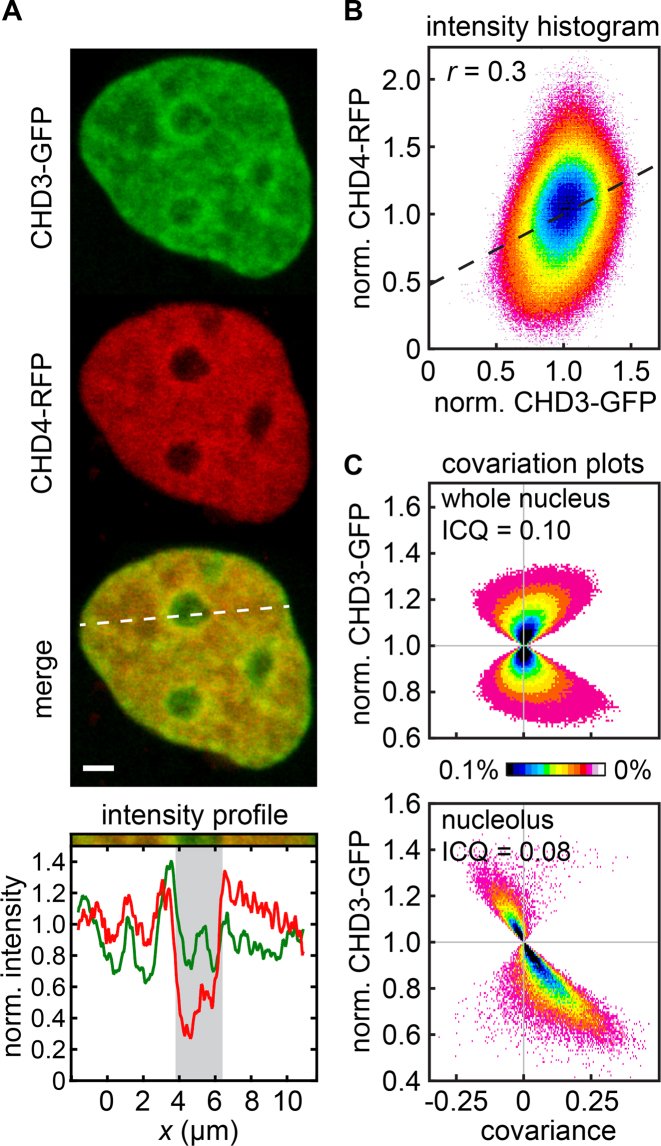
CHD3 and CHD4 exhibit differential nuclear localization patterns. (A) Representative image of a living U2OS cell transiently transfected with CHD3-GFP and CHD4-RFP. Both proteins were enriched in perinuclear heterochromatin. CHD4-RFP was more strongly depleted in nucleoli than CHD3-GFP. Scale bar, 2 μm. (B) Scatterplot for GFP- and RFP signals in the same cells. A Pearson correlation coefficient of r = 0.3 was obtained, indicating moderate co-localization. Fifteen images were analyzed. (C) Covariation analysis for the data in panel B. The intensity correlation quotient (ICQ) value of 0.1 within the whole nucleus suggests partial but significant co-localization (psign test < 0.001) of both proteins (top). A similar result was obtained if only pixels within nucleoli were considered (bottom). In the latter case, we observed a subpopulation of pixels with anti-correlated signals, which reflects nucleolar regions in which CHD3 was enriched and CHD4 was depleted. An anti-correlated subpopulation of pixels was present, which reflects CHD3 enrichment and CHD4 depletion. Fifteen images were analysed.
Both isoforms were enriched in perinucleolar heterochromatin and depleted in nucleoli, whereas exclusion from nucleoli was particularly pronounced for CHD4-RFP (see representative image in Figure 3A). To quantitatively compare the localization of both proteins we conducted a colocalization analysis using the pixels within the nucleus. CHD3-GFP and CHD4-GFP exhibited moderate co-localization throughout the entire nucleus, which is reflected by the intensity scatter plot in Figure 3B that was created based on images from several cells. The Pearson correlation coefficient for the plotted pixels amounted to 0.3 (Figure 3B) and the intensity correlation coefficient (ICQ) (52) to 0.1 (Figure 3C, top). Pixels in the left quadrants of the covariance plots in Figure 3C represent positions where one of the CHD proteins is enriched and the other one is depleted, whereas pixels in the right quadrants represent positions where both CHD proteins are co-depleted or co-enriched. The correlation analyses suggest that both proteins colocalize in some but not all regions of the nucleus, consistent with the images shown in Figure 3A. Notably, the probability to obtain this ICQ value for two completely independent distributions is very low (P < 0.001, see Materials and Methods), indicating that the localization patterns of CHD3-GFP and CHD4-RFP share significant, but only limited similarity. When considering only pixels within nucleoli, a similar but slightly lower ICQ value was obtained (Figure 3C, bottom). Notably, we observed a distinct subpopulation of pixels coinciding with CHD3-GFP enrichment and CHD4-RFP depletion in nucleoli (top left quadrant of the covariance plot), corroborating the finding that CHD4 is stronger depleted in nucleoli than CHD3.
Next, we compared the nuclear mobility parameters of CHD3 and CHD4 based on the FCCS measurements shown in Figure 1E and Supplementary Figure S6E. Both CHD3- and CHD4-containing NuRD complexes exhibited similar effective diffusion coefficients (Supplementary Figure S6E), which are comparable to those found for other remodeling enzymes in U2OS cells (47). This result suggests that a large fraction of NuRD complexes samples chromatin in transient binding interactions rather than being stably tethered to their target sites.
CHD3 and CHD4 are recruited to laser-induced DNA damage sites
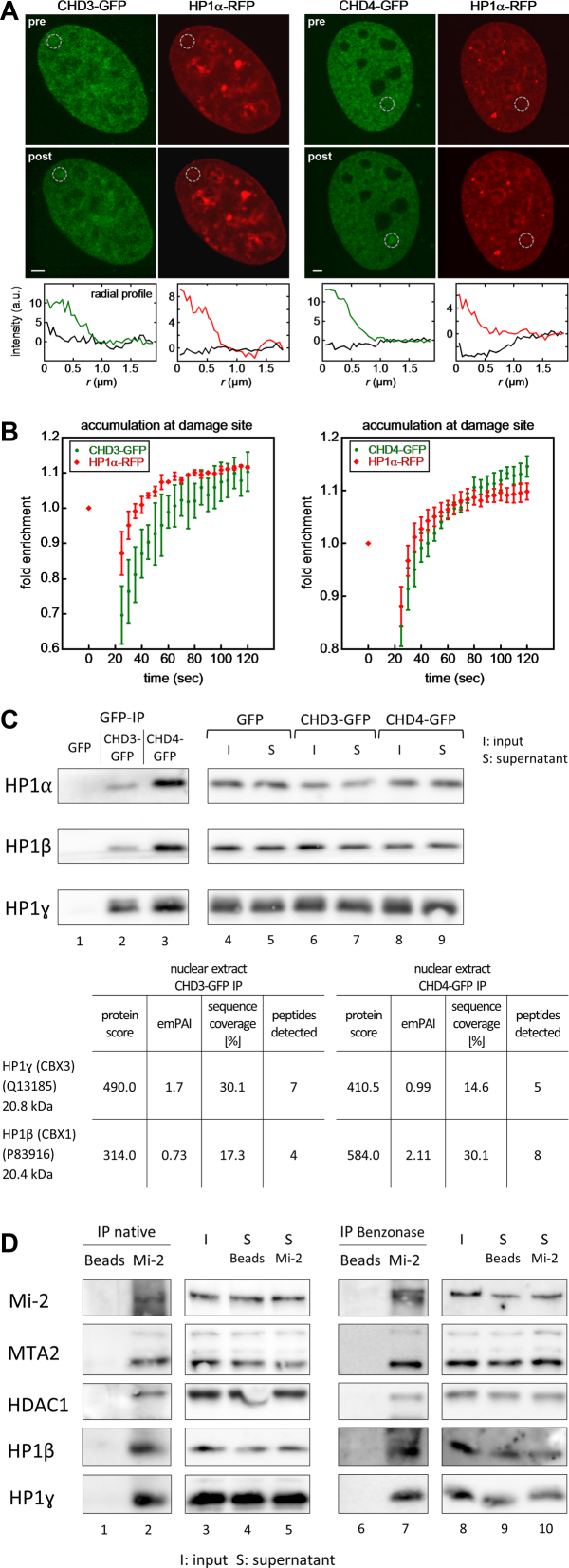
Several chromatin remodelers are recruited to laser-induced DNA damage sites, which contain pyrimidine dimers and DNA intrastrand cross-links (47,74–77). Recent studies in mammalian cells reported a deviance of CHD3/4-NuRD complexes in the repair of DNA damage (78–81). Here, we compared the behaviour of CHD3- and CHD4-GFP upon laser-treatment of U2OS cells. In these experiments, cotransfected RFP-tagged HP1-alpha (CBX5), which has been shown to be recruited to laser-induced damage sites (76,82) served as a positive control (Figure 4A and B). Both CHD3 and CHD4 accumulated at laser-induced lesions (Figure 4A), with association kinetics comparable to each other and to HP1-alpha (Figure 4B). HP1-alpha and NuRD are localized to pericentric heterochromatin (83–86) and HP1-IP reactions revealed that HP1-alpha coprecipitates with CHD4 (87), whereas HP1-beta (CBX1) and gamma (CBX3) coprecipitate with CHD3 and CHD4 (85). This prompted us to examine immunoprecipitation reactions of CHD3- and CHD4-GFP for the presence of the different HP1 isoforms (Figure 4C). In western blot experiments we found that all three HP1 isoforms were associated with both CHD3- and CHD4-GFP in the context of NuRD in the absence of DNA damage (Figure 4C, lane 2 and 3). Mass spectrometry analyses of CHD3- and CHD4-GFP-IP samples showed a clear coprecipitation of HP1-gamma with both Mi2-isoforms and also suggest a coprecipitation of HP1-beta (Figure 4C, table). We even successfully verified the interaction of Mi-2 proteins with HP1 proteins on endogenous protein level, by detecting HP1-gamma and HP1-beta in western blots of Mi-2-IP reactions (Figure 4D, lane 2). Since we still see a coprecipitation of endogenous HP1-beta and gamma with endogenous Mi-2 in the presence of Benzonase or Ethidium bromide (Figure 4D, lane 7 and Supplementary Figure S5E), we propose that the interaction is not DNA mediated (88,89). Taken together our IP data and the colocalization of HP1 and NuRD in pericentric heterochromatin and at laser-induced DNA lesions suggest a functional connection between both CHD proteins and HP1.
Figure 4.
CHD3 and CHD4 are rapidly recruited to laser-induced DNA repair sites and interact with HP1. (A) U2OS cells transiently expressing HP1α-RFP and CHD3/4-GFP were subjected to localized microirradiation with a 405 nm laser. CHD3-GFP, CHD4-GFP and HP1α-RFP accumulated at the damage site (gray circles). Radial intensity profiles around the damage site are shown below the images for clarity. Scale bar, 2 μm. (B) Accumulation kinetics of the indicated constructs at the damage site. (C) The expression of GFP (control), CHD3-GFP or CHD4-GFP in stably transfected Flp-In™ T-Rex™ 293 cells was induced by doxycycline treatment for 24 h. Subsequently, (i) whole cell extracts (WCE) and (ii) nuclear extracts (NE) were prepared and subjected to IP reactions, using GFP-Trap®_A beads. The IP reactions were finally analysed by (i) western blot with the indicated HP1 antibodies or (ii) mass spectrometry. The reactions were finally analyzed by mass spectrometry. A quantitative difference in protein amount levels is reflected by the emPAI-values (exponentially modified protein abundance index) (121). (*) Only one unique peptide was found in the mass spectrometry analysis (see also Materials and Methods). (D) Nuclear extract (NE) from non-transfected Flp-In™ T-Rex™ 293 cells was prepared and subjected to IP reactions in the absence or presence of Benzonase (see also Materials and Methods), using Protein A beads (+ Mi-2 antibody or no antibody as control). The IP reactions were finally analysed by western blot with the indicated HP1 and Mi-2-NuRD antibodies. Number of replicates: (A/B) (left) CHD3-GFP and HP1-alpha-RFP (12); (right) CHD4-GFP and HP1-alpha-RFP (12); (C) 3; (D) 3.
CHD3 and CHD4 exert common and distinct gene regulatory functions
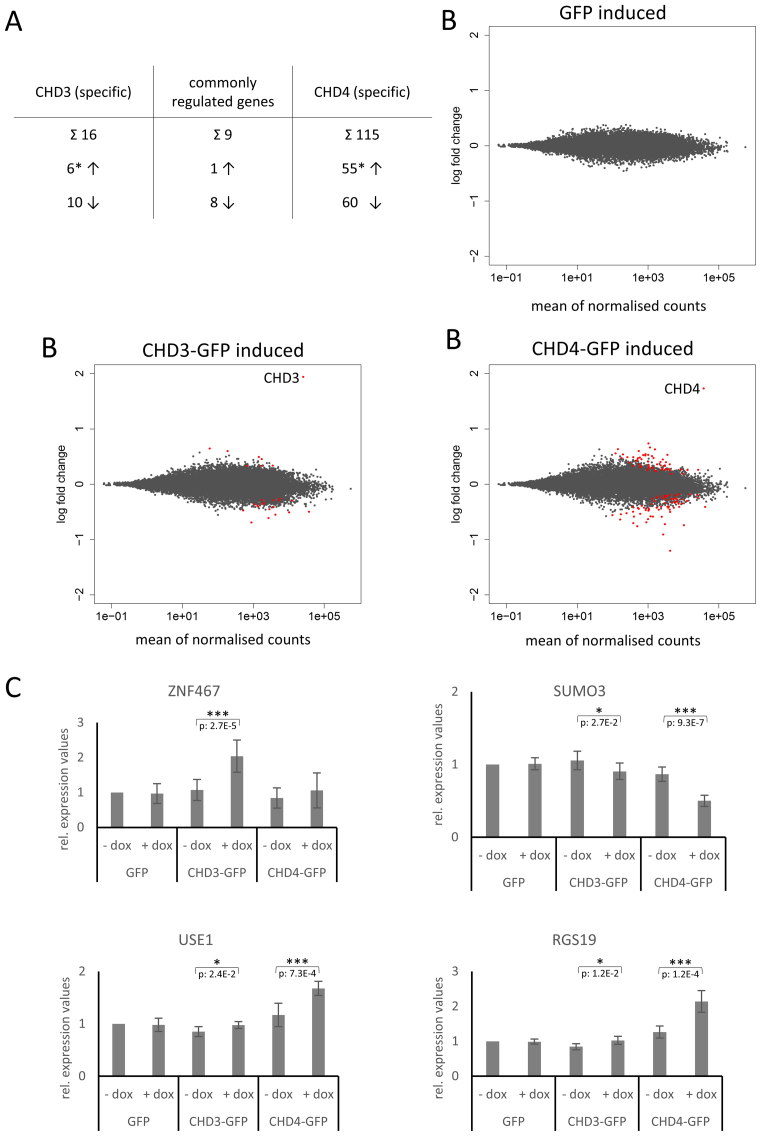
The differences in intranuclear localization and the differences in the remodeling behaviour found above for CHD3 and CHD4 might reflect distinct activities in different genomic regions with a putative effect on gene activity. We were therefore interested to examine if CHD3 and CHD4 are responsible for the regulation of different genes, since chromatin remodeling enzymes are involved in the control of various DNA-dependent processes like transcription (5). Analysing RNA-seq data of two independent (total) RNA preparations from doxycycline-induced (24 hours) and non-induced Flp-In™ T-REx™ 293 cells (Supplementary Figure S3) via DESeq revealed 16 (i) and 115 (ii) distinctly regulated genes when CHD3-GFP (i) and CHD4-GFP (ii) were overexpressed, respectively (Figure 5A and B and Supplementary Figure S8: black/blue labelled gene names). Nine genes are equally influenced by both types of NuRD complexes (Figure 5A and Supplementary Figure S8: red labeled gene names). The RNA-seq analysis furthermore suggests that almost the same number of genes is up- and downregulated in the presence of exogenous CHD3 and CHD4 (Figure 5A, B and Supplementary Figure S8). These findings are somewhat surprising, since most of the publications describe NuRD in the context of transcriptional repression (37,39).
Figure 5.
CHD3/4 containing NuRD complexes regulate distinct genes in living cells. Stably transfected Flp-In™ T-Rex™ 293 cells were incubated in the absence (non-induced) or presence (induced) of doxycycline for 24 h. Five independent sets of total RNA were prepared. Two sets of total RNA were subjected to Illumina Sequencing (A and B): differential gene expression was analysed using the DESeq2 R package (59). Samples without doxycycline induction were considered as ground state and differential expression was determined by a false discovery rate below 0.1. (A) The table presents the total number of specifically and commonly up- (↑) and downregulated (↓) genes upon CHD3/4 expression (* the number of specifically upregulated genes does not include the upregulated CHD3/4 genes due to doxycycline treatment). (B) MA-plots illustrate log2 fold changes (y-axis) for each gene in comparison to the average count (x-axis). The red points indicate differentially expressed genes determined by DESeq2 analysis (FDR < 0.1), including the doxycycline induced CHD3/4 proteins. (C) The other three sets of total RNA were reversely transcribed into cDNA for qPCR experiments: Data were collected by the Rotor-Gene 3000 system, analyzed using the comparative analysis software module and presented as relative expression values, using Lamin A/C for normalization. The mean values and standard deviations are derived from three biological replicates (with at least two technical replicates for each biological replicate). Relative expression values for induced (+dox) and non-induced (–dox) conditions were tested for significance using a two tailed, homoscedastic t-test in Microsoft Excel. P-values are given for sets with significant changes (*P < 0.05, **P < 0.01, ***P < 0.001). Number of replicates: (A/B) 2 (see above); (C) 3 (see above) → in total: five independent total RNA preparations (see above). The raw data for the RNA-seq analysis are deposited in the GEO database (accession number: GSE93543).
We validated the RNA-seq results via quantitative PCR (qPCR) with cDNA from three new (independent) doxycycline inductions. The alterations in gene expression, determined by our qPCR experiments, were comparable to those from the RNA-seq analysis (Figure 5B, C and Supplementary Figure S8). We found four genes that are predominantly regulated by either CHD3 or CHD4: ZNF467 was upregulated by CHD3-GFP (Figure 5C), RGS19 and USE1 were upregulated by CHD4-GFP and SUMO3 was downregulated by CHD4-GFP (Figure 5C).
Taken together our data therefore suggest that NuRD complexes either contain CHD3 or CHD4 as a motor subunit. We furthermore postulate that the two remodeling enzymes are associated with the NuRD complex as monomers. Our data in vitro and in living cells reveal that CHD3 or CHD4 comprising NuRD complexes exert similar and as well distinct functions (Figure 6).
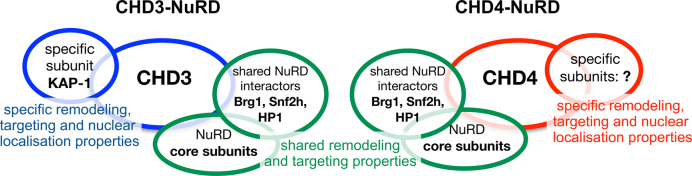
Figure 6.
CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Our experiments propose that CHD3 and CHD4 form isoform-specific NuRD complexes in living cells, harboring one single copy of the respective remodeling enzyme. Beside well known ‘shared’ core subunits like HDAC1/2 or MTA2/3 proteins, we found Brg1, Snf2h and HP1 to be associated with both CHD3- and CHD4-containing NuRD complexes. DNA damage experiments, performed in this study, suggest that both CHD3- and CHD4-containing NuRD complexes exert a role in the DNA damage response. Nevertheless, an individual role of mouse CHD3 in DSB repair was proposed by the CHD3 specific binding protein KAP-1 (11,81). In addition to interaction partners that might act as functional NuRD regulators, we propose intrinsic remodeling properties as another possibility to influence the physiological role of NuRD complexes in vivo. Nucleosome remodeling assays with recombinant, single CHD3 and CHD4 proteins revealed that both enzymes move nucleosomes to distinct, sequence-specific positions, supporting the idea of CHD remodeling enzymes acting as chromatin specific organizers. In line with this, we observe mainly distinct nuclear localization patterns for CHD3- and CHD4-NuRD complexes in living cells, arguing for the existence of structurally and spatially separated complexes acting independently from each other in different genomic regions with a putative effect on gene activity. Indeed, our RNA-seq and qPCR experiments showed that CHD3 and CHD4 mainly regulate distinct genes. Taken together our data suggest that CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality (see also Results and Discussion).
DISCUSSION
Approximately twenty years ago, the purification and biochemical characterization of a large macromolecular complex, coupling chromatin remodeling properties with histone deacetylase function was reported simultaneously by several laboratories (33–36). This complex, named Mi-2 NuRD, contains several repressors like HDAC1/2, MTA2/3 or MBD2/3 and CHD3/4 [Mi-2 subfamily] as the motor subunit (38). The composition of this complex seems to vary upon distinct physiological circumstances and it is still matter of debate whether CHD3 and CHD4 coexist in the same complex or not (37,38). CHD3 and CHD4 are highly identical in their amino acid sequence (Figure 1A and Supplementary Figure S1). Therefore the functional characterization of this complex in vivo is difficult, since many molecular biological and biochemical detection tools only allow a statement about the two Mi-2 representatives in general. Since many studies in the past focused only on one of the two Mi-2 isoforms, the question about potential structure-function relationships remained enigmatic. Here, we shed light on this question by comparing both Mi-2 isoforms in four different types of experiments: (i) We showed in living cells that the presence of CHD3 and CHD4 in NuRD is mutually exclusive. (ii) Somewhat surprisingly, not only CHD4 but also CHD3 accumulated at sites of DNA damage. (iii) CHD3 and CHD4 have distinct regulatory functions for gene expression as inferred from a comparison of their target genes. (iv) The detailed analysis of CHD3 vs. CHD4 showed an overall similar activity with small but significant differences in their enzymatic translocation properties.
CHD3 and CHD4 exist in distinct NuRD complexes
Several mammalian remodelers like Snf2h [Iswi subfamily] (47) and yeast Fun30 [Etl1 subfamily] (90) tend to form homodimers (or higher-order complexes), which raises the questions if the closely related CHD3 and CHD4 proteins act as homo- and/or heterodimers in NuRD complexes of living cells. Using different biochemical and microscopy-based techniques we clearly demonstrate here that CHD3 and CHD4 do not tend to self-associate in living cells (Figure 6 and Supplementary Figure S6B–D). Rather, our data suggest that cells contain isoform-specific NuRD complexes, supporting previous findings of (36) and (11) (Figures 1B–E and 6 and Supplementary Figure S4). Interestingly, a NuRD-like complex with CHD5 [Mi-2 subfamily] as the motor subunit did not show a coexistence of the closely related enzymes CHD5 and CHD3/4 in the same complex (91,92). Consistently, our mass spectrometry analyses also argue for the presence of only one CHD type per NuRD complex (Figures 1C and 6 and Supplementary Figure S4). In line with this, Bergs and colleagues (93) showed that CHD3/4 and CHD5 have different expression patterns in mouse testis and brain, whereupon CHD3/4 and CHD5 are expressed in a mutually exclusive manner in adult mouse testis.
By contrast, remodelers from other subfamilies seem to interact with CHD3/4 NuRD complexes. The mass spectrometry and western blot data showed a noticeable enrichment of Brg1 and Snf2h signals in the CHD-IP reactions in comparison to the control reaction (Figure 6 and Supplementary Figures S4 and S9). These findings are in line with data from (26,94,95). Thus, the presence of two remodeling enzymes from different subfamilies within one NuRD complex might provide sufficient functional complexity without requiring a coexistence of CHD3 and CHD4. In this regard it is interesting to note that Morris and coworkers (96) performed ChIP-seq and DNase I-seq experiments arguing for a co-occupancy of mouse Brg1, CHD4 and Snf2h at certain genomic loci, where they act to some extent synergistically.
Beside Brg1 and Snf2h, we confirmed that HP1 is a subunit that is also shared among CHD3- and CHD4 containing NuRD complexes (Figures 4C, D and 6 and Supplementary Figure S4). To our knowledge, we are the first to show a DNA-independent interaction of Mi-2 and HP1 proteins on endogenous protein level (Figure 4D and Supplementary Figure S5E), suggesting a functional connection between the two proteins (Figure 4). Interestingly, Sun et al. (97) propose a coregulation of SOX9 gene transcription by HP1-beta (CBX1), CHD4 and KAP-1 (TRIM28) on the basis of consecutive ChIP experiments.
Finally, our mass spectrometry and western blot analyses predominantly detected shared core subunits and shared interacting partners (Figures 1B,C and 4C,D and Supplementary Figures S4 and S9), but no (new) isoform-specific binding protein. In line with this, there are primarily ‘shared binding partners’ documented in the literature. Regarding the high degree of similarity between CHD3 and CHD4 and the small unique amino acid region in CHD3, representing a putative binding platform for specific interaction partners (Figure 1A and Supplementary Figure S1), these findings don’t appear too surprising. To our knowledge, KAP-1 is so far the only verified binding protein, that exclusively interacts with hCHD3 in its (mostly unique) C-terminal amino acids 1909–2000 (Figures 1A and 6 and Supplementary Figure S1 and (11)). Interestingly we could detect KAP-1 in CHD3 and as well CHD4-IP-reactions (Supplementary Figure S4), which might be explained by the use of different IP protocols and cell lines. For CHD4, no specific interaction partner has been published until now.
CHD4 and as well CHD3 accumulate at sites of DNA damage
The complex interplay between common and specific binding partners could explain why CHD3 and CHD4 are involved in the same, similar or distinct intracellular processes (Figure 6). Even though both Mi-2 isoforms seem to exhibit opposite interaction behaviour regarding heterochromatic DSBs (induced by ionizing radiation), which is mediated by KAP-1 (78–81), we do not observe the same for laser-induced DNA damage in human U2OS cells (Figure 4A, B). Surprisingly, this kind of DNA damage resulted in the recruitment of both, CHD3- and as well CHD4-NuRD complexes to the damage sites (Figure 4A, B), an observation which might be explained by the fact that we induced DNA damage in a different way and within bona fide euchromatin. Our Co-IP data moreover suggest that HP1 and CHD proteins might be recruited to the damage sites in the context of a common (NuRD) complex (Figures 1B, C and 4C, D and Supplementary Figure S4). Interestingly data from (98) and (99) suggest that the recruitment of CHD4/NuRD and as well HP1 to DNA damage sites depends on PARP (Poly (ADP-ribose) polymerase) activity. In contrast to HP1, both CHD proteins and their Drosophila homologue Mi-2 seem to possess a conserved PAR binding motif (99–101). In case of CHD4 this motif seems to span the first N-terminal 360 aa and seems to form a HMG box like structure which is important for directing the remodeling enzyme to DSB sites (101). Therefore, association of CHD3/4 with PARylated chromatin might promote binding of the NuRD-HP1 complex to DNA lesions. Interestingly, HP1 also interacts with other proteins that accumulate at DNA lesions, e.g. Brg1 and Acf (75,76,102–104). Taking into account our Co-IP data, showing an interaction of Brg1 and Snf2h (the catalytic subunit of the ACF complex) with CHD containing NuRD complexes (Figures 1B, C and 6 and Supplementary Figures S4 and S9), one could speculate whether the CHD3/4 mediated recognition of PARylated chromatin might also play a role for binding of these factors to DNA damage sites.
CHD3 and CHD4 showed small but significant differences in their enzymatic translocation properties
Beside common or unique binding partners, our data suggest that intrinsic remodeling properties have also to be taken into account as an important factor influencing the functional properties of CHD-containing chromatin remodeling complexes (Figures 2 and 6 and Supplementary Figure S7A). Several in vitro studies, comparing remodeling enzymes representing distinct subfamilies, revealed obvious disparities in the qualitative remodeling behaviour of those proteins (6,105). Furthermore those enzymes exhibited distinct stimulation properties by features like DNA linker length or histone tails concerning their ATPase activity (6,105). In the light of these findings it is therefore interesting to note that we even observed differences in the nucleosome positioning between CHD3 and CHD4 (Figure 2 and Supplementary Figure S7A), which belong to the same subfamily [Mi-2] (7) and exhibit moreover a high degree of identity in the known functionally relevant motifs (Figure 1A and Supplementary Figure S1 and (11)). The fact that the observed differences in nucleosome positioning already occur in the absence of potentially regulatory NuRD subunits (Figures 2 and 6 and Supplementary Figure S7A) shows that they are intrinsic to the remodeling enzymes themselves. Assuming that CHD3- and CHD4-NuRD complexes move nucleosomes in vivo to distinct positions would provide an attractive mechanism for how the balance of both proteins affects the accessibility or inaccessibility of DNA regions for regulatory factors, like for example transcription factors. This emphasizes the idea of CHD nucleosome remodelers (and nucleosome remodelers in general) acting as functionally specific chromatin organizers, as favoured by (70,106–108). Indeed, chromatin remodeling enzymes from three different subfamilies, namely Snf2h (NoRC) [Iswi], CSB [ERCC6] and CHD4 (NuRD) [Mi-2] seem to regulate the transcription status of mammalian rDNA genes via distinct nucleosome repositioning at the rDNA promotor (7,70,109–111). Interestingly, we also observe differences in the remodeling pattern between CHD3 and CHD4 on the drosophila HSP70 and the mouse rDNA promotor sequence (Figure 2D and Supplementary Figure S7A).
CHD3 and CHD4 have distinct regulatory functions for gene expression
The aspect of CHD chromatin remodelers acting as functional specific chromatin organizers is furthermore supported by our fluorescence microscopy data (Figure 3). Transient transfections in U2OS cells show that CHD3 and CHD4, being coexpressed in one cell, share significant but only limited similarities in their nuclear localization patterns. Both were enriched in perinucleolar heterochromatin, but CHD4 seemed to be stronger depleted in nucleoli than CHD3 (Figure 3). Consistently, immunocytochemical experiments (human protein atlas) performed with antibodies directed against the endogenous proteins in U2OS, A-431, U-251 MG and HeLa cells, solely ascribe a nucleolar localization to CHD3, not to CHD4 (112–115). These differential localization patterns for CHD3 and CHD4 argue for the existence of structurally and spatially separated complexes acting independently from each other in different genomic regions with a putative effect on gene activity. Indeed, our RNA-seq and qPCR data reveal that the majority of expression changes concerns genes which seem to be under the control of one particular Mi-2 isoform (Figure 5A, C and Supplementary Figure S8): The opposite isoform either shows insignificant effects compared to the GFP control or, in some cases, subtle synergistic effects (Figure 5C). In this context it is interesting to note that recent studies, performed in G1E-ER-GATA-1 cells and 0308 TICs, fortify our RNA-seq and qPCR experiments by attributing a regulatory function on RGS19 and SUMO3 gene transcription to CHD4 as well (116,117). However, while we found that RGS19 was up- and SUMO3 was downregulated by CHD4 overexpression, (116) and (117) report the opposite regulation of these genes by CHD4. This discrepancy might be explained by the different cellular environment. Furthermore, ChIP-seq experiments in TICs showed that CHD4 is associated with the USE1 gene, a gene which we found upregulated by CHD4 (117).
Our RNA-seq data also give reason to reconsider our view on NuRD acting predominantly as a transcriptional repressor (37–39). The definition of NuRD as transcriptional silencer mainly derives from the fact that many core subunits are associated with transcriptional repression and that many studies only focused on certain sets of genes or even single gene analysis (37–39,97). To our knowledge we are the first to present a comparative (general) screen of genes, regulated by WT Mi-2 proteins (Figure 5 and Supplementary Figure S8). Since we observe nearly the same number of genes up- and downregulated in the presence of CHD3 and CHD4 (Figure 5A,B and Supplementary Figure S8), we suggest that NuRD rather functions equally as a transcriptional activator and silencer.
Furthermore our RNA-seq analysis shows that the amount of CHD4 regulated genes seems to be nearly seven times higher than the number of CHD3 regulated genes (Figure 5A,B and Supplementary Figure S8). Looking at the same time at the endogenous expression levels of (human and mouse) Mi-2 proteins in vivo, an imbalance of the expression levels in favour of CHD4 becomes obvious: Despite the coexpression of both Mi-2 proteins in many tissues, CHD4 very often reaches higher expression levels than CHD3 (Supplementary Figure S2) and (118,119). Accordingly, we also observe more endogenous CHD4 than CHD3 transcripts in our Flp-In™ T-REx™ 293 cells (Supplementary Figure S10A). Taken together, the expression level disequilibrium - even though we modulate it by overexpressing CHD3 or CHD4 (Supplementary Figure S10A,B) - and the higher amount of CHD4 regulated genes (Figure 5A,B and Supplementary Figure S8) might argue for a dominant role of CHD4 over CHD3 regarding transcription regulation.
Concluding remarks
In mouse thymus CHD3 levels increased upon CHD4 knockdown, whereas the rest of the NuRD core subunits remained unaffected (119). Interestingly, we see a decrease of endogenous CHD4 mRNA levels upon induction of CHD3-GFP in our RNA-seq data (Supplementary Figure S8). Similar observations were made for the highly identical proteins Snf2h and Snf2l. Data from (embryonic) mouse experiments showed that a time-specific loss of Snf2h seems to be compensated by rising amounts of Snf2l (reconstituting amongst other parameters En1 mRNA levels) (7,32). One could therefore speculate that the coexpression of both Mi-2 proteins might serve the following purpose: CHD3, due to its high similarity and the partially redundant functions, might be able to replace CHD4 to some extent (and vice versa). On the other hand, our data argue that the coexistence of CHD3- and CHD4 proteins in many cells might originate from the partly functional difference between both remodelers (Figure 6).
ACCESSION NUMBERS
The raw data for the RNA-seq analysis are deposited in the GEO database under accession number: GSE93543.
Supplementary Material
ACKNOWLEDGEMENTS
We thank PD Dr. A. Rascle and PD Dr. J. Griesenbeck (University of Regensburg, Germany) for advice in RNA preparation and qPCR. We acknowledge Prof. Dr. R. Wagner (University of Regensburg, Germany) for providing us with Flp-In™ T-REx™ 293 cell line and the respective plasmids. We thank Dr. K. Müller-Ott (University of Heidelberg, Germany) for providing us with HP-1 antibodies. We thank Dr. Max Felle, Elisabeth Silberhorn, Dr. Ancilla Neu and Dr. J. Exler and Julia Roth for providing us with several in house produced plasmids and histones. We also thank Maria Engelhart for her contribution by doing a practical training.
Footnotes
Present address: Claudia Huber, Department of Chemistry, University of Konstanz, 78457 Konstanz, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG; SFB 960 (Ribosome formation: principles of RNP biogenesis and control of their function). Funding for open access charge: University of Regensburg.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breiling A., Lyko F.. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015; 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang R.C., Bonner J.. Histone-bound RNA, a component of native nucleohistone. Proc. Natl. Acad. Sci. U.S.A. 1965; 54:960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodríguez-Campos A., Azorín F.. RNA Is an Integral Component of Chromatin that Contributes to Its Structural Organization. PLoS ONE. 2007; 2:e1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 6. Rippe K., Schrader A., Riede P., Strohner R., Lehmann E., Langst G.. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:15635–15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flaus A., Martin D.M.A., Barton G.J., Owen-Hughes T.. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006; 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eisen J.A., Sweder K.S., Hanawalt P.C.. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995; 23:2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prasad P., Lennartsson A., Ekwall K.. The Roles of SNF2/SWI2 Nucleosome Remodeling Enzymes in Blood Cell Differentiation and Leukemia. BioMed Res. Int. 2015; 2015:e347571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lazzaro M.A., Picketts D.J.. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 2001; 77:1145–1156. [DOI] [PubMed] [Google Scholar]
- 11. Schultz D.C., Friedman J.R., Rauscher F.J.. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2? subunit of NuRD. Genes Dev. 2001; 15:428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barak O., Lazzaro M.A., Cooch N.S., Picketts D.J., Shiekhattar R.. A tissue-specific, naturally occurring human SNF2L variant inactivates chromatin remodeling. J. Biol. Chem. 2004; 279:45130–45138. [DOI] [PubMed] [Google Scholar]
- 13. Watanabe T., Semba S., Yokozaki H.. Regulation of PTEN expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. Br. J. Cancer. 2011; 104:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W., Côté J., Xue Y., Zhou S., Khavari P.A., Biggar S.R., Muchardt C., Kalpana G.V., Goff S.P., Yaniv M. et al. . Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996; 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 15. Xue Y., Canman J.C., Lee C.S., Nie Z., Yang D., Moreno G.T., Young M.K., Salmon E.D., Wang W.. The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W., Cho H., Kadam S., Banayo E.M., Anderson S., Yates J.R., Emerson B.M., Evans R.M.. A methylation-mediator complex in hormone signaling. Genes Dev. 2004; 18:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eberharter A., Becker P.B.. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 2004; 117:3707–3711. [DOI] [PubMed] [Google Scholar]
- 18. Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G. et al. . A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000; 6:1287–1295. [DOI] [PubMed] [Google Scholar]
- 19. Flowers S., Nagl N.G., Beck G.R., Moran E.. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J. Biol. Chem. 2009; 284:10067–10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reyes J.C., Barra J., Muchardt C., Camus A., Babinet C., Yaniv M.. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 1998; 17:6979–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. LeRoy G., Orphanides G., Lane W.S., Reinberg D.. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998; 282:1900–1904. [DOI] [PubMed] [Google Scholar]
- 22. Bochar D.A., Savard J., Wang W., Lafleur D.W., Moore P., Côté J., Shiekhattar R.. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poot R.A., Dellaire G., Hülsmann B.B., Grimaldi M.A., Corona D.F., Becker P.B., Bickmore W.A., Varga-Weisz P.D.. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000; 19:3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeRoy G., Loyola A., Lane W.S., Reinberg D.. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 2000; 275:14787–14790. [DOI] [PubMed] [Google Scholar]
- 25. Strohner R., Nemeth A., Jansa P., Hofmann-Rohrer U., Santoro R., Längst G., Grummt I.. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001; 20:4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hakimi M.-A., Bochar D.A., Schmiesing J.A., Dong Y., Barak O.G., Speicher D.W., Yokomori K., Shiekhattar R.. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002; 418:994–998. [DOI] [PubMed] [Google Scholar]
- 27. Loyola A., Huang J.-Y., LeRoy G., Hu S., Wang Y.-H., Donnelly R.J., Lane W.S., Lee S.-C., Reinberg D.. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol. Cell. Biol. 2003; 23:6759–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banting G.S., Barak O., Ames T.M., Burnham A.C., Kardel M.D., Cooch N.S., Davidson C.E., Godbout R., McDermid H.E., Shiekhattar R.. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum. Mol. Genet. 2005; 14:513–524. [DOI] [PubMed] [Google Scholar]
- 29. Thompson P.J., Norton K.A., Niri F.H., Dawe C.E., McDermid H.E.. CECR2 is involved in spermatogenesis and forms a complex with SNF2H in the testis. J. Mol. Biol. 2012; 415:793–806. [DOI] [PubMed] [Google Scholar]
- 30. Barak O., Lazzaro M.A., Lane W.S., Speicher D.W., Picketts D.J., Shiekhattar R.. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003; 22:6089–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rippe K. Genome Organization And Function In The Cell Nucleus. 2012; John Wiley & Sons. [Google Scholar]
- 32. Alvarez-Saavedra M., De Repentigny Y., Lagali P.S., Raghu Ram E.V.S., Yan K., Hashem E., Ivanochko D., Huh M.S., Yang D., Mears A.J. et al. . Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun. 2014; 5:4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tong J.K., Hassig C.A., Schnitzler G.R., Kingston R.E., Schreiber S.L.. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998; 395:917–921. [DOI] [PubMed] [Google Scholar]
- 34. Wade P.A., Jones P.L., Vermaak D., Wolffe A.P.. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. CB. 1998; 8:843–846. [DOI] [PubMed] [Google Scholar]
- 35. Xue Y., Wong J., Moreno G.T., Young M.K., Côté J., Wang W.. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998; 2:851–861. [DOI] [PubMed] [Google Scholar]
- 36. Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D.. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998; 95:279–289. [DOI] [PubMed] [Google Scholar]
- 37. Marfella C.G.A., Imbalzano A.N.. The Chd family of chromatin remodelers. Mutat. Res. 2007; 618:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Denslow S.A., Wade P.A.. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007; 26:5433–5438. [DOI] [PubMed] [Google Scholar]
- 39. Cai Y., Geutjes E.-J., de Lint K., Roepman P., Bruurs L., Yu L.-R., Wang W., van Blijswijk J., Mohammad H., de Rink I. et al. . The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2014; 33:2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seelig H.P., Moosbrugger I., Ehrfeld H., Fink T., Renz M., Genth E.. The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum. 1995; 38:1389–1399. [DOI] [PubMed] [Google Scholar]
- 41. Seelig H.P., Renz M., Targoff I.N., Ge Q., Frank M.B.. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 1996; 39:1769–1771. [DOI] [PubMed] [Google Scholar]
- 42. Smits A.H., Jansen P.W.T.C., Poser I., Hyman A.A., Vermeulen M.. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013; 41:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kloet S.L., Baymaz H.I., Makowski M., Groenewold V., Jansen P.W.T.C., Berendsen M., Niazi H., Kops G.J., Vermeulen M.. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 2015; 282:1774–1785. [DOI] [PubMed] [Google Scholar]
- 44. Krajewski W.A., Reese J.C.. SET domains of histone methyltransferases recognize ISWI-remodeled nucleosomal species. Mol. Cell. Biol. 2010; 30:552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Müller‐Ott K., Erdel F., Matveeva A., Mallm J.-P., Rademacher A., Hahn M., Bauer C., Zhang Q., Kaltofen S., Schotta G. et al. . Specificity, propagation, and memory of pericentric heterochromatin. Mol. Syst. Biol. 2014; 10:746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jegou T., Chung I., Heuvelman G., Wachsmuth M., Görisch S.M., Greulich-Bode K.M., Boukamp P., Lichter P., Rippe K.. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol. Biol. Cell. 2009; 20:2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Erdel F., Schubert T., Marth C., Längst G., Rippe K.. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19873–19878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luger K., Rechsteiner T.J., Richmond T.J.. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. Clifton NJ. 1999; 119:1–16. [DOI] [PubMed] [Google Scholar]
- 49. Fitzgerald D.J., Berger P., Schaffitzel C., Yamada K., Richmond T.J., Berger I.. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006; 3:1021–1032. [DOI] [PubMed] [Google Scholar]
- 50. Berger I., Fitzgerald D.J., Richmond T.J.. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004; 22:1583–1587. [DOI] [PubMed] [Google Scholar]
- 51. Manders E.M., Stap J., Brakenhoff G.J., van Driel R., Aten J.A.. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992; 103:857–862. [DOI] [PubMed] [Google Scholar]
- 52. Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E.F.. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. Off. J. Soc. Neurosci. 2004; 24:4070–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chung I., Leonhardt H., Rippe K.. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 2011; 124:3603–3618. [DOI] [PubMed] [Google Scholar]
- 54. Erdel F., Rippe K.. Binding kinetics of human ISWI chromatin-remodelers to DNA repair sites elucidate their target location mechanism. Nucl. Austin Tex. 2011; 2:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. R Core Team R: A Language and Environment for Statistical Computing. 2015; Vienna: R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- 57. Morgan M., Pagès H., Obenchain V., Hayden N.. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 1.20.4. http://bioconductor.org/packages/release/bioc/html/Rsamtools.html. [Google Scholar]
- 58. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W.. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005; 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 61. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stockdale C., Flaus A., Ferreira H., Owen-Hughes T.. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 2006; 281:16279–16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McKnight J.N., Jenkins K.R., Nodelman I.M., Escobar T., Bowman G.D.. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol. Cell. Biol. 2011; 31:4746–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Udugama M., Sabri A., Bartholomew B.. The INO80 ATP-dependent chromatin remodeling complex Is a nucleosome spacing factor. Mol. Cell. Biol. 2011; 31:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murawska M., Kunert N., van Vugt J., Längst G., Kremmer E., Logie C., Brehm A.. dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol. Cell. Biol. 2008; 28:2745–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ho K.K., Zhang H., Golden B.L., Ogas J.. PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta. 2013; 1829:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hamiche A., Sandaltzopoulos R., Gdula D.A., Wu C.. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999; 97:833–842. [DOI] [PubMed] [Google Scholar]
- 69. Brown C.R., Mao C., Falkovskaia E., Law J.K., Boeger H.. In vivo role for the chromatin-remodeling enzyme SWI/SNF in the removal of promoter nucleosomes by disassembly rather than sliding. J. Biol. Chem. 2011; 286:40556–40565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H.G., Grummt I., Tao W.. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8161–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu S., Ge Y., Huang L., Liu H., Xue Y., Zhao Y.. BRG1, the ATPase subunit of SWI/SNF chromatin remodeling complex, interacts with HDAC2 to modulate telomerase expression in human cancer cells. Cell Cycle. 2014; 13:2869–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shim E.Y., Hong S.J., Oum J.-H., Yanez Y., Zhang Y., Lee S.E.. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007; 27:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lake R.J., Geyko A., Hemashettar G., Zhao Y., Fan H.-Y.. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell. 2010; 37:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y., Rohde L.H., Wu H.. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr. Genomics. 2009; 10:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang L., Zhang Q., Jones K., Patel M., Gong F.. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle. 2009; 8:3953–3959. [DOI] [PubMed] [Google Scholar]
- 76. Luijsterburg M.S., Dinant C., Lans H., Stap J., Wiernasz E., Lagerwerf S., Warmerdam D.O., Lindh M., Brink M.C., Dobrucki J.W. et al. . Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009; 185:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Larsen D.H., Poinsignon C., Gudjonsson T., Dinant C., Payne M.R., Hari F.J., Danielsen J.M.R., Menard P., Sand J.C., Stucki M. et al. . The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 2010; 190:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P.. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010; 29:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H.. The NuRD chromatin–remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010; 190:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goodarzi A.A., Kurka T., Jeggo P.A.. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011; 18:831–839. [DOI] [PubMed] [Google Scholar]
- 82. Baldeyron C., Soria G., Roche D., Cook A.J.L., Almouzni G.. HP1α recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011; 193:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hayakawa T., Haraguchi T., Masumoto H., Hiraoka Y.. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003; 116:3327–3338. [DOI] [PubMed] [Google Scholar]
- 84. Helbling Chadwick L., Chadwick B.P., Jaye D.L., Wade P.A.. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009; 118:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rosnoblet C., Vandamme J., Völkel P., Angrand P.-O.. Analysis of the human HP1 interactome reveals novel binding partners. Biochem. Biophys. Res. Commun. 2011; 413:206–211. [DOI] [PubMed] [Google Scholar]
- 86. Sims J.K., Wade P.A.. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol. Biol. Cell. 2011; 22:3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lechner M.S., Schultz D.C., Negorev D., Maul G.G., Rauscher F.J.. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005; 331:929–937. [DOI] [PubMed] [Google Scholar]
- 88. Lai J.S., Herr W.. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. van den Berg D.L.C., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R.A.. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010; 6:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Awad S., Ryan D., Prochasson P., Owen-Hughes T., Hassan A.H.. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J. Biol. Chem. 2010; 285:9477–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M.. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013; 49:368–378. [DOI] [PubMed] [Google Scholar]
- 92. Quan J., Adelmant G., Marto J.A., Look A.T., Yusufzai T.. The chromatin remodeling factor CHD5 Is a transcriptional repressor of WEE1. PLoS ONE. 2014; 9:e108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bergs J.W., Neuendorff N., van der Heijden G., Wassenaar E., Rexin P., Elsässer H.-P., Moll R., Baarends W.M., Brehm A.. Differential expression and sex chromosome association of CHD3/4 and CHD5 during spermatogenesis. PLOS ONE. 2014; 9:e98203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. O’Neill D.W., Schoetz S.S., Lopez R.A., Castle M., Rabinowitz L., Shor E., Krawchuk D., Goll M.G., Renz M., Seelig H.P. et al. . An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 2000; 20:7572–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shimono Y., Murakami H., Kawai K., Wade P.A., Shimokata K., Takahashi M.. Mi-2β associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J. Biol. Chem. 2003; 278:51638–51645. [DOI] [PubMed] [Google Scholar]
- 96. Morris S.A., Baek S., Sung M.-H., John S., Wiench M., Johnson T.A., Schiltz R.L., Hager G.L.. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 2014; 21:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sun F., Yang Q., Weng W., Zhang Y., Yu Y., Hong A., Ji Y., Pan Q.. Chd4 and associated proteins function as corepressors of Sox9 expression during BMP-2–induced chondrogenesis. J. Bone Miner. Res. 2013; 28:1950–1961. [DOI] [PubMed] [Google Scholar]
- 98. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ayrapetov M.K., Gursoy-Yuzugullu O., Xu C., Xu Y., Price B.D.. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U. S. A. 2014; 111:9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A.. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011; 7:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Silva A.P.G., Ryan D.P., Galanty Y., Low J.K.K., Vandevenne M., Jackson S.P., Mackay J.P.. The N-terminal region of chromodomain helicase DNA-binding protein 4 (CHD4) is essential for activity and contains a high mobility group (HMG) box-like-domain that can bind poly(ADP-ribose). J. Biol. Chem. 2016; 291:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nielsen A.L., Sanchez C., Ichinose H., Cerviño M., Lerouge T., Chambon P., Losson R.. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. EMBO J. 2002; 21:5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Eskeland R., Eberharter A., Imhof A.. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 2007; 27:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nozawa R.-S., Nagao K., Masuda H.-T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C.. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 2010; 12:719–727. [DOI] [PubMed] [Google Scholar]
- 105. Brehm A., Längst G., Kehle J., Clapier C.R., Imhof A., Eberharter A., Müller J., Becker P.B.. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 2000; 19:4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gao H., Lukin K., Ramírez J., Fields S., Lopez D., Hagman J.. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:11258–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Moshkin Y.M., Chalkley G.E., Kan T.W., Reddy B.A., Ozgur Z., van Ijcken W.F.J., Dekkers D.H.W., Demmers J.A., Travers A.A., Verrijzer C.P.. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell. Biol. 2012; 32:675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F.. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012; 149:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H.J., Vermeulen W.. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000; 20:7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li J., Längst G., Grummt I.. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006; 25:5735–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhao Z., Dammert M.A., Grummt I., Bierhoff H.. lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 2016; 14:1876–1882. [DOI] [PubMed] [Google Scholar]
- 112. Uhlén M., Björling E., Agaton C., Szigyarto C.A.-K., Amini B., Andersen E., Andersson A.-C., Angelidou P., Asplund A., Asplund C. et al. . A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics MCP. 2005; 4:1920–1932. [DOI] [PubMed] [Google Scholar]
- 113. Pontén F., Jirström K., Uhlen M.. The Human Protein Atlas–a tool for pathology. J. Pathol. 2008; 216:387–393. [DOI] [PubMed] [Google Scholar]
- 114. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. . Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
- 115. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. . Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 116. DeVilbiss A.W., Boyer M.E., Bresnick E.H.. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc. Natl. Acad. Sci.. U.S.A. 2013; 110:E3398–E3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chudnovsky Y., Kim D., Zheng S., Whyte W.A., Bansal M., Bray M.-A., Gopal S., Theisen M.A., Bilodeau S., Thiru P. et al. . ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor initiating cell state. Cell Rep. 2014; 6:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T. et al. . Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999; 10:345–355. [DOI] [PubMed] [Google Scholar]
- 119. Williams C.J., Naito T., Arco P.G.-D., Seavitt J.R., Cashman S.M., De Souza B., Qi X., Keables P., Von Andrian U.H., Georgopoulos K.. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004; 20:719–733. [DOI] [PubMed] [Google Scholar]
- 120. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al. . Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M.. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol. Cell. Proteomics. 2005; 4:1265–1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.