Abstract
Red fluorescent proteins (RFPs) have emerged as valuable biological markers for biomolecule imaging in living systems. Developing artificial fluorogenic systems that mimic RFPs remains an unmet challenge. Here, we describe the design and synthesis of six new chromophores analogous to the chromophores in RFPs. We demonstrate, for the first time, that encapsulating RFP chromophore analogues in canonical DNA G-quadruplexes (G4) can activate bright fluorescence spanning red and far-red spectral regions (Em = 583−668 nm) that nearly match the entire RFP palette. Theoretical calculations and molecular dynamics simulations reveal that DNA G4 greatly restricts radiationless deactivation of chromophores induced by a twisted intramolecular charge transfer (TICT). These DNA mimics of RFP exhibit attractive photophysical properties comparable or superior to natural RFPs, including high quantum yield, large Stokes shifts, excellent anti-photobleaching properties, and two-photon fluorescence. Moreover, these RFP chromophore analogues are a novel and distinctive type of topology-selective G4 probe specific to parallel G4 conformation. The DNA mimics of RFP have been further exploited for imaging of target proteins. Using cancer-specific cell membrane biomarkers as targets, long-term real-time monitoring in single live cell and two-photon fluorescence imaging in tissue sections have been achieved without the need for genetic coding.
INTRODUCTION
Green fluorescent proteins (GFP) are essential tools in biomedical and pharmacologic studies due to their genetic encoding properties and intrinsic high fluorescence quantum yields (1–3). When fused with proteins of interest, they can be utilized as a fluorescent marker suitable for visualizing protein localization and biochemical trafficking in live cells (4). Red fluorescent proteins (RFP) have, to date, received more attention than GFPs as they are indispensable complements to the FP spectral palette and have a number of advantages in bio-imaging. RFPs exhibit fluorescence emission maxima beyond 570 nm, thus they have less autofluorescence interference of cells, lower light-scattering, and lower photo-toxicity (5). These make RFPs especially valuable for deep tissue and whole body imaging. RFPs have a chromophore similar to the p-hydroxybenzylidine-imidazolinone (HBI) core of GFP, except that it possesses an additional N-acylimine substituent on the HBI motif. The acylimine extends the π-electron delocalized network of HBI and results in a red-shift in absorption and emission relative to GFP (6). The RFP protective β-barrel structure provides a rigid and hydrophobic environment which confines a coplanar chromophore structure, and ensures the emission pathway dominant in excited state energy release.
Developing synthetic materials mimicking natural fluorescent proteins is crucial for elucidating photo-physical mechanism of FPs and creating new emission-responsive materials and biosensors. Unfortunately, most of synthetic chromophores of RFP are non-fluorescent similar to denatured RFP, owing to ultrafast intramolecular vibration (7,8). Developing highly emissive molecular mimics of RFP is still challenging. Currently, only two cases of emissive RFP analogues with decent quantum yield (>0.02) are available based on confinement of RFP chromophore analogues via covalent structure locking or the assembly of chromophores with proteins (9,10). Given the significance of RFP in in vivo imaging, the scarcity of fluorescent RFP mimics greatly restricts the available spectral region of synthetic FP-mimicking materials and their use in multi-color or deep-tissue imaging.
Unlike the largely unexplored fluorescent RFP mimics, emissive GFP mimics have been extensively studied. A number of approaches have achieved emissive GFP analogues via mimicking the natural confined effect of GFP β-barrel to restrict the intramolecular motions of GFP chromophores. They include: structure locking by chemical linkage (11–16), both covalent and non-covalent; and, GFP chromophore encapsulation into a rigid environment, such as supra-molecular hosts (17,18), metal-organic frameworks (19), the framework of aggregation-induced emission (AIE) (20,21), protein assembly (22,23), and ribonucleic acid (RNA) aptamers. A fluorogenic RNA aptamer ‘Spinach’ developed by Jaffrey et al. is an intriguing GFP mimic with strong emission comparable to native GFPs (24–26), which was selected by the systematic evolution of ligands by exponential enrichment (SELEX) process. Its crystal structures demonstrated that ‘Spinach’ contain a unique G-quadruplex (G4)-containing binding pocket, which is composed of a non-canonical G4 RNA module, an unpaired G, and an adjacent base-triplet, to accommodate GFP chromophore analogues (27,28). This significant finding implied the potential of G-quadruplex as a promising structural motif for construction of fluorogenic GFP mimics. However, several important issues still need to be addressed: (i) except the selected G4-containing RNA aptamers, whether canonical G4 structures enable fluorescent activation of FP chromophores have not been investigated; (ii) given the polymorphism of G4, the impact of G4 topology on its ability to activate FP mimics is also unexplored; (iii) most relevantly, except GFP chromophores analogues, whether RFP chromophores could be fluorescently activated by G4 or other nucleic acid structures is totally unknown.
Herein, we report the design and synthesis of a set of new multi-color chromophores resembling the RFP chromophores (Scheme 1). We prove, for the first time, that RFP chromophore analogues are a new type of G-quadruplexes ligand and encapsulating RFP chromophore analogues in canonical DNA G4s can elicit bright fluorescence spanning red and far-red spectral regions (Em = 583−668 nm), which perfectly match existing RFP palettes. G-quadruplex is an alternative secondary nucleic acid structure formed by guanine-rich (G-rich) DNA sequences. The G4 G-tetrad, which is formed by a four guanine via Hoogsteen hydrogen-bonding, provides an ideal docking site for RFP chromophore analogue stacking and restricts them to a planar conformation for efficient emission, which resemble the confinement of natural RFP β-barrel structure (Scheme 1A). These unique DNA RFP mimics have several photo-physical merits, such as high quantum yields (maximum = 0.39), large Stokes shifts (maximum = 112 nm), and superior anti-photobleaching properties. It is worth to note that the G4-confined RFP chromophores exhibit unique two-photon fluorescence and holds promise for live systems imaging. Moreover, the proposed RFP chromophore analogues perform as ‘smart’ G-quadruplex ligands having the dual-functionalities of being potent structural stabilizers and topology-selective fluorescent sensors capable of recognizing parallel G4 against other G4 or DNA conformations. These DNA RFP mimics can be further exploited to ‘role-play’ as natural RFPs for imaging of target proteins. Both one-photon and two-photon fluorescent imaging of cancer-specific cell membrane biomarkers, such as protein tyrosine kinase-7 (PTK7), has been achieved in a live cell and tissue setting without genetic coding being required.
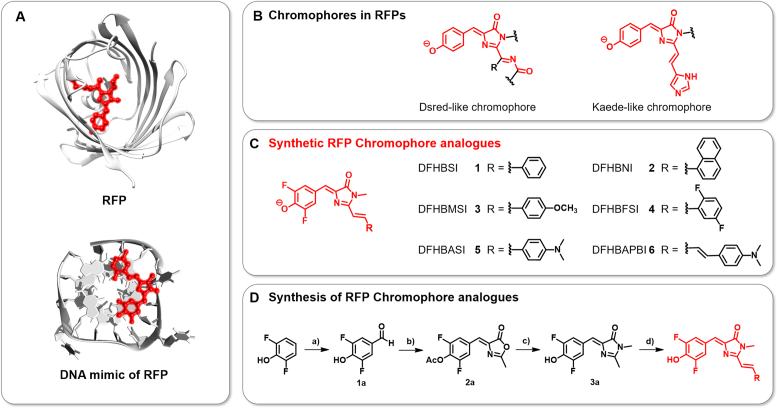
Scheme 1.
(A) Structures of a RFP (upper) and a DNA mimic of RFP composed of a DNA G-quadruplex and a synthetic RFP chromophore analogue (bottom). Chromophores are emphasized in red. (B) Chemical structures of the chromophores in RFPs. (C) Structures of the RFP chromophore analogues synthesized in this study. (D) Synthetic route of the RFP chromophore analogues using the following reagents and conditions: a) hexamethylenetetramine, TFA, 120°C, 12 h; b) N-Acetylglycine, anhydrous sodium acetate, acetic anhydride, 120°C, 4 h; c) 40% aq. CH3NH2, K2CO3, EtOH, 80°C, 4 h; and d) ZnCl2, THF, RCHO, 80°C.
MATERIALS AND METHODS
Unless otherwise stated, all the organic reagents were purchased from commercial suppliers and used without further purification. THF were dried and purified by distillation from sodium/benzophenone and CaH2. Tris, KCl, EDTA and MgCl2 were purchased from Sangon Biotech (Shanghai, China). DPBS, penicillin, streptomycin, RPMI 1640, McCoy's 5a Medium Modified and fetal bovine serum were obtained from Thermo Scientific (USA). Ultrapure water (18.2 MΩ·cm) from the Millipore Milli-Q system was used in the experiments.
NMR spectra were recorded on a Bruker-400 spectrometer, using TMS as an internal standard. Chemical shifts (δ) are quoted in parts per million (ppm) and coupling constant (J) are recorded in Hertz (Hz). Mass spectra were performed using an LCQ Advantage ion trap mass spectrometer from Thermo Finnigan. All reactions were monitored by TLC with Macherey-Nagel pre-coated Glassic sheets (0.20 mm with fluorescent indicator UV254). Compounds were visualized with UV light at 254 nm and 365 nm. Flash column chromatography was carried out using silica gel from Merck (200–300 mesh). The pH measurements were carried out on a Mettler-Toledo Delta 320 pH meter. Fluorescence lifetime measurements were performed on a FLS980 fluorescence spectrofluorometer (Edinburgh Instruments, USA).
Oligonucleotides (HPLC-purified, sequences were shown in Supplementary Tables S1 and S6) were synthesized and purified by Sangon Biotech (Shanghai, China). Oligonucleotides were dissolved in buffer solution (25 mM Tris and pH 8.0) at 100 μM and annealed (heated to 95°C for 10 min and then cooled slowly to 4°C) prior to be used in each experiment. The dsDNA was prepared by annealing equal amounts of complementary strands in buffer solution (25 mM Tris, 10 mM MgCl2 and pH 8.0).
Circular dichroism (CD) spectroscopy
The CD spectra were recorded on MOS-500 spectrophotomer (BioLogic, France) at room temperature (RT, 25°C). Before the CD measurement, the oligonucleotides were annealed following the above-mentioned conditions prior to measurements. For CD measurements, samples were transferred into 2 mm path length quartz cuvettes, scanning from 220 nm to 360 nm at a rate of 50 nm/min for three times with the slit of 5 nm. CD melting curve was performed in the temperature range of 24–96°C at an interval of 2.5°C with a heating rate of 1°C/min.
Ultraviolet–visible (UV–Vis) spectroscopy
UV–Vis spectra were recorded on a Varian Cary 100-Bio at RT using a 10-mm path length quartz cuvette. For titration experiment, small aliquots of a stock solution of oligonucleotide were added into the solution containing DFHBFSI (10 μM) in Tris-HCl buffer (25 mM Tris, 100 mM KCl, 50 mM MgCl2 and pH 8.0). The final concentrations of oligonucleotide were varied from 0 to 15 μM. The absorption spectra from 400 nm to 700 nm were measured after incubation at RT for 20 min.
Fluorescence Spectroscopy and Real-time Fluorescent Monitoring the Topological Switch of G4. Fluorescence experiments were carried out using a QuantaMaster™ fluorescence spectrophotometer (PTI, Canada). A quartz cuvette with 2 mm × 10 mm path length was used for spectrum measurements, and both excitation and emission slit widths were 10 nm unless otherwise specified. For fluorescence titration experiments, DFHBFSI (3 μM) and pre-annealed oligonucleotides (0–15 μM) were incubated for 20 min in Tris–HCl buffer (25 mM Tris, 100 mM KCl, 50 mM MgCl2 and pH 8.0). For real-time fluorescent monitoring, the topological switch of G4, DFHBFSI (1 μM) and PW17 (G-quadruplex, 1 μM) were incubated in 100 μl of Tris–HCl buffer (25 mM Tris, 2 mM KCl, 50 mM MgCl2 and pH 8.0) for 5 min. Then, 3 μl of 5 mM Pb2+ was added into the solution. Ten minutes later, 3 μl of 5 mM EDTA was added into the solution. During these process, fluorescence intensity at 606 nm was monitoring with an excitation wavelength of 513 nm on the fluorescence spectrometer.
Gel electrophoresis studies
Twenty microliters of NG16 at various concentrations or different G4 oligonucleotides were loaded and separated by native electrophoresis using a 20% acrylamide gel, stained with DFHBFSI (2 μM) in Tris–HCl buffer (25 mM Tris, 100 mM KCl, 50 mM MgCl2 and pH 8.0) and visualized via fluorescence detection using a ChemiDoc MP system (Bio-Rad, USA).
Please see supporting information (SI) for the experimental details of measurement of two-photon cross section, pKa measurements, quantum yield calculations, dissociation constant measurements, photo-bleaching analysis, molecular modelling, DFT calculations, cell culture, cytotoxicity measurements, construction of plasmids and real-time quantitative reverse transcription PCR, flow cytometry and confocal laser scanning microscopy assay, tissue imaging.
RESULTS AND DISCUSSION
G-quadruplex-chromophore complexes as DNA RFP mimics
Inspired by that RNA aptamer ‘Spinach’ contains a composite binding pocket with a non-canonical G4 module to accommodate the GFP chromophore analogue DFHBI, an optimized derivative of HBI (27,28), we thus tested whether a canonical DNA G4 structure could elicit the fluorescence of synthetic FP chromophores. Given that both GFP and RFP share the common HBI core, we first examined the influence of G4 structures on the fluorescence of HBI and its analogue DFHBI prior to investigating RFP chromophores. HBI and DFHBI were prepared via an Erlenmeyer azlactone synthesis (15,24) and treated with a known G4-forming DNA sequence NG16 in the presence of 100 mM K+ ion. The sequence of NG16 is shown in Supplementary Table S1, and the CD characterization of its G4 conformation is described below. As shown in Figure 1A, NG16 shows negligible influence on the absorption properties of HBI/DFHBI and can’t activate their fluorescence even at considerably high stoichiometric ratio (50 eq., Supplementary Table S2). Similar results were also observed for other G4-forming sequences (Supplementary Table S3). Hence, conventional G4 structures alone, without any other additional specific contacts as found in spinach, are not efficient for GFP chromophores binding and fluorescent activation. This might result from that the p-aromatic surface of HBI analogues is relative small thus unable to interact with a terminal G-quartet strongly via π–π stacking.
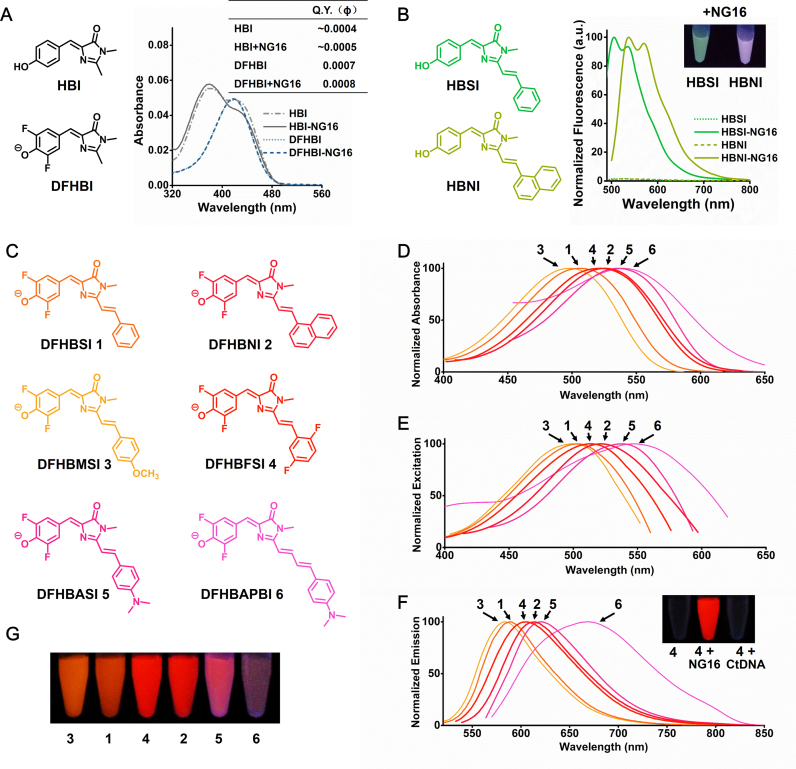
Figure 1.
(A) Absorption spectra and Q.Y. of HBI (3 μM) and DFHBI (3 μM) in the absence and presence of NG16 (30 μM). (B) NG16 (15 μM) enhances the fluorescence of HBSI (3 μM) and HBNI (3 μM). Inset is the photograph of HBSI and HBNI with NG16 under illumination of ultraviolet (UV, 365 nm) light. (C) Structures of the RFP chromophore analogues. (D) Absorption, (E) excitation and (F) emission spectra of chromophore–NG16 complexes (3 μM). Inset of F is the solutions containing DFHBFSI, DFHBFSI with NG16, and DFHBFSI with calf thymus DNA (CtDNA) photographed under illumination of UV light. (G) Chromophore-NG16 complexes were illuminated with UV light and photographed. The image is a montage obtained under identical image-acquisition conditions.
We envisaged that the RFP chromophores, especially Kaede-like chromophores (Scheme 1B), bearing additional aromatic ring conjugated with HBI core would lead to two desirable features: one is the spectroscopic red-shift compared with GFP chromophores due to extended conjugation; the other is the strengthened interaction with G-quartet of G4 structure by the chromophores’ enlarged π-ring system. To test this assumption, we have synthesized two RFP chromophores with aromatic substituents linking to HBI core via a vinyl group according to a literature procedure (29), termed HBSI and HBNI (Figure 1B). The theoretical prediction was performed by the online molecular docking system of the G-quadruplex Ligands Database (G4LDB) (30), and the results suggested that HBSI and HBNI are preferable ligands of G4 compared to HBI and DFHBI due to their additional π-ring (Supplementary Figure S1). Inspiringly, the experimental data (Figure 1B) showed that HBSI and HBNI fluorescence markedly increased by 66.8 and 68.5 folds, respectively, while bound with NG16, which is closely consistent with the theoretical predictions. This indicates that these RFP chromophores are fluorescently activatable through their binding with a DNA G4 structure. However, a limitation is that they fluoresce green and purplish green with emission maxima at 505 and 537 nm, respectively, which are significantly short than those of RFPs (>570 nm). An analysis of absorption properties of HBSI and HBNI indicated that their phenol form (protonated) was predominant at neutral pH (Supplementary Figure S2). This is consistent with their relative high pKa (the dissociation constant of phenolic OH group) which are 8.8 and 9.3 for HBSI and HBNI, respectively (Supplementary Figure S3). Supplementary Figure S2 shows that the chromophores in 0.1 M NaOH exhibit remarkable absorption bathochromic shifts (∼49 nm and ∼52 nm for HBSI and HBNI). This indicates their phenolate forms are more relevant for mimicking RFPs, which is in accordance with natural chromophores in RFPs and other synthetic ones (6,29).
To obtain G4-chromophore complexes with red emission comparable with RFPs, we sought to further adjust spectroscopic red-shift of chromophores via introducing electron withdrawing substituents to the phenolic ring to facilitate its deprotonation at neutral pH. Two new RFP chromophores with fluorine substituents, DFHBSI and DFHBNI (Figure 1C), were synthesized from the condensation of DFHBI with corresponding arylaldehydes catalyzed by ZnCl2 (Scheme 1D). As expected, their phenolic acidities were obviously enhanced (pKa = 6.4 and 7.3 for DFHBSI and DFHBNI, Supplementary Figure S3) compared to their unsubstituent counterparts, HBSI and HBNI. Correspondingly, the chromophore-NG16 complexes of DFHBSI and DFHBNI exhibited a bright red fluorescence with a single emission at 590 and 612 nm (Figure 1D–G), respectively, which is identical to conventional RFPs (Supplementary Table S4). Furthermore, to determine whether the DNA RFP mimics could be spectrally modulated by the diverse chromophores, another four novel RFP chromophores were generated by introducing either electron-donating (methoxyl or dimethylamino) or electron-withdrawing groups (fluorines) to the additional aromatic ring or elongating olefinic linkage (Figure 1C). When bound with NG16, these compounds exhibited significantly increased fluorescence with emission maxima ranging from 583 to 668 nm (Figure 1D–G), which spans the spectrum from red to far-red, nearly overlapping the entire existing RFP palette.
Unique photo-physical properties of DNA RFP mimics
A panel of DNA mimics of RFP with different chromophores allows direct comparison of spectral properties and corresponding substituent's effect (Table 1). All synthesized chromophores weakly fluoresce in aqueous solution at pH 8 with quantum yields below 0.007, as do denatured RFP and other reported RFP chromophores (6,29,31,32), which results from the nonradioactive decay of their photo-excited state via the intramolecular motion about the ethylenic bridge (33). The quantum yields of these RFP chromophores are relatively higher than those of GFP chromophores partly due to the bulky aromatic group may hinder the exocyclic C–C bond rotation, and a similar effect was also observed for the synthetic Kaede chromophore (32). The observed fluorescence activation by G4 indicated that G-quadruplex might provide a rigid environment like the β-barrel structure of fluorescent proteins to lock the planar conformation of the RFP chromophores and prevent intramolecular motions, which makes fluorescence predominant in energy dissipation of the excited state. This is confirmed by the significantly increased excited-state lifetimes of all G4-chromophores complexes compared to individual RFP chromophores (Supplementary Figure S4). Under the optimized condition with 100 mM K+ and 50 mM Mg2+ (Supplementary Figure S5) in the Tris buffer (pH 8.0), most of the RFP chromophores exhibited at least 50-fold increase of fluorescence quantum yield (ϕ) in the presence of NG16, and the maximum up to 75-fold was observed for DFHBFSI. The quantum yield of DFHBFSI-NG16 (λem = 606 nm) is above 0.39, which is pronouncedly higher than that of all reported synthetic RFP mimics, including unlocked (<0.007) (29,31,32), protein-confined (0.052) (10), and intramolecularly locked RFP chromophores (0.19) (9). It is even better than that of most of RFPs with similar emission maxima (600–620 nm), such as mRuby (0.35) (5), mCherry (0.22), FusionRed (0.19) (34), and also its structure parent Kaede (0.33) (35). Substituent comparisons would provide useful information for directing the spectral turning of DNA RFP mimics (Table 1). Replacing the benzene ring (DFHBSI) with a naphthalene ring (DFHBNI) leads to an 18 nm red shift in excitation and a 22 nm red shift in emission due to the expanded conjugated system. Intriguingly, introducing dimethylamino substituent (DFHBASI), a strong electron-donating group, on the benzene ring causes a substantial red-shift in both excitation (35 nm) and emission (30 nm) which is even superior to the naphthalene-containing chromophore (DFHBNI), changing the fluorescence from red to far-red. The most remarkable bathochromic effect derived from olefinic linkage elongation by an additional double bond, contributing to an emission maxima change from 620 nm of DFHBASI to 668 nm of DFHBAPBI. However, DFHBAPBI–NG16 complex have a relatively low quantum yield (0.032) compared to other chromophore-NG16 complexes. Two reasons may account for this phenomenon: (i) the relatively low quantum yield is conventionally observed for far-red or near infrared fluorophores, such as the far-red FP eqFP670 (φ = 0.06) (36) and near-infrared RFP AQ143 (φ = 0.04) (37); (ii) its molecular length is too long to perfectly match the size of G-quartet. As a result, unlike the fixed spectral properties of each RFPs, the emission of DNA RFP mimics can be facilely modulated in a controllable manner by changing various synthetic chromophores that hold great potential to be rationally designed.
Table 1. Photophysical and binding properties of chromophores and chromophore-NG16 complexes.
| Chromophore | Exmax (nm) | Emmax (nm) | ϵ (M−1 cm−1) | Stokes shift (nm) | Q.Y.b | K d (μM) |
|---|---|---|---|---|---|---|
| DFHBMSI-NG16 | 499 | 583 | 16 950 | 84 | 0.37 | 1.56 ± 0.31 |
| DFHBMSI | 500 | 570 | 18 200 | 70 | 0.0072 | N/Aa |
| DFHBSI-NG16 | 503 | 590 | 17 350 | 87 | 0.31 | 1.76 ± 025 |
| DFHBSI | 501 | 576 | 19 210 | 75 | 0.0066 | N/A |
| DFHBFSI-NG16 | 513 | 606 | 28 200 | 93 | 0.39 | 1.27 ± 0.11 |
| DFHBFSI | 511 | 594 | 32 000 | 83 | 0.0052 | N/A |
| DFHBNI-NG16 | 521 | 612 | 37 500 | 91 | 0.30 | 1.37 ± 0.19 |
| DFHBNI | 517 | 600 | 38 840 | 83 | 0.0048 | N/A |
| DFHBASI-NG16 | 538 | 620 | 22 800 | 82 | 0.19 | 2.77 ± 0.58 |
| DFHBASI | 540 | 614 | 23 890 | 74 | 0.0028 | N/A |
| DFHBAPBI-NG16 | 547 | 668 | 15 500 | 121 | 0.032 | 24.88 ± 0.82 |
| DFHBAPBI | 548 | 662 | 16 120 | 114 | 0.0011 | N/A |
| mCherry | 587 | 610 | 72 000 | 23 | 0.22 | N/A |
aNot applicable.
bQuantum yields were calculated using rhodamine 6G (Φf = 0.95 in water) as standard for DFHBMSI, DFHBSI, DFHBFSI, DFHBNI and DFHBASI, and sulfoindo cyanine dye Cy5 (Φf = 0.20 in PBS) as standard for DFHBAPBI.
Another spectral feature of the new RFP mimics is their large Stokes shifts. In contrast to small Stokes shifts of conventional RFPs (summarized in Supplementary Table S4), such as 23 nm for mCherry (shown in Figure 2A), the proposed DNA RFP mimics have large Stokes shifts exceeding 80 nm (shown in Table 1 and Supplementary Figure S6), for instances 93 nm for DFHBFSI (shown in Figure 2A) and 112 nm for DFHBAPBI. A larger Stokes shift can reduce the self-quenching and fluorescence detection errors caused by excitation back scattering effects, which is beneficial for high contrast fluorescence imaging (38). The large Stokes shift is probably due to the excited-state relaxation of the RFP chromophore itself (39). Moreover, the photo-stability of G4-based RFP mimics was also determined. As depicted in Figure 2B, the fluorescence signal of DFHBFSI-NG16 complex shows negligible decrease upon continuous illumination over 30 min, while those of mCherry, a typical RFP, and Lyso-Tracker Red, a model of small molecular red fluorophore, quickly declined 93.5% in 10 min and 80% in 30 min, respectively. This result suggested that the DNA RFP mimics possess much better photo-stability against photo-bleaching than RFPs, probably resulting from that partial exchange of bound chromophore with free chromophore in solution prevents the accumulation of photo-bleached complexes (24,40,41).
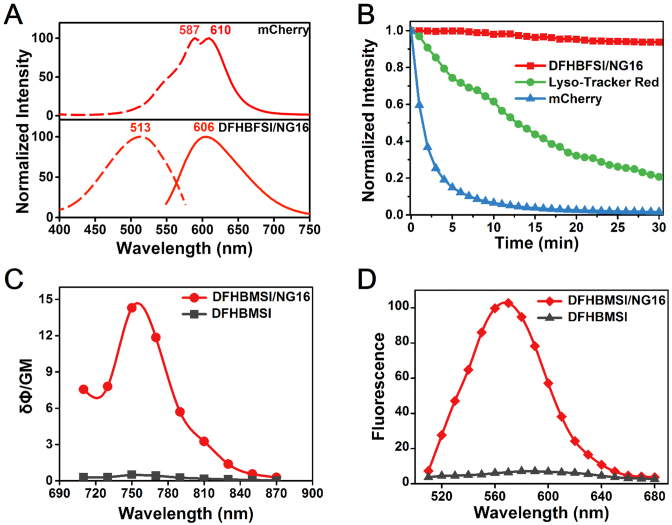
Figure 2.
(A) Comparison between mCherry and DFHBFSI–NG16 in excitation and emission spectra. (B) Photobleaching curves of DFHBFSI-NG16 (1 μM), Lyso-Tracker Red (1 μM) and mCherry (1 μM). (C) Two-photon excitation action cross-section of DFHBMSI (3 μM) and DFHBMSI with NG16 (15 μM). (D) Two-photon excited emission spectra of DFHBMSI (3 μM) and DFHBMSI with NG16 (15 μM, Ex = 750 nm).
Next, we examined the two-photon fluorescence property of the proposed DNA RFP mimics. Two photon fluorophores, cooperating with two-photon excitation fluorescence (TPEF) microscopy, possesses several advantages over conventional one-photon imaging, including increased tissue penetration, reduced out-of-focus photodamage, negligible background fluorescence, and high three-dimensional resolution (42). To the best of our knowledge, the two-photon properties of synthetic RFP chromophore analogues are totally unknown. As shown in Figure 2C, the DFHBMSI–NG16 complex exhibited an intensive two-photon absorption with a maximum peak at around 750 nm (Figure 2D), which resulted from unique S0 to Sn electronic transitions (43). Its TPEF spectrum possesses a maximal emission peak at 579 nm, which is slightly hypsochromically shifted relative to its one-photon emission peak (583 nm). Unexpectedly, although DFHBMSI is a RFP chromophore analogue with an unlocked structure, the confining effect by G4 successfully endow DFHBMSI with decent two-photon properties with a two-photon cross-section (σ) of 38.6 GM and a two-photon excitation action cross-section (brightness, σ′) of 14.3 GM (Supplementary Table S5). They are comparable with those of its protein counterpart, mStrawberry (σ = 35 GM, σ′ = 12 GM) (43). Compounds DFHBSI and DFHBFSI shows similar two-photon properties upon G4 binding (Supplementary Table S5). Significantly, the S0 to Sn transition of these RFP mimics at around 750 nm perfectly matches the tuning range of femtosecond Ti:sapphire lasers, which make them well applicable to most of commercial TPEF microscopic systems.
The synthetic RFP chromophore is a selective fluorescent dye for parallel G4 structures
To our knowledge, the proposed RFP chromophores are structurally dissimilar to any structural scaffolds of known G4 ligands or fluorogens, presenting a new category of G4 fluorescent probes. Given that G4s have gained substantial interest due to their increasing biological significance and applications in biosensing, supramolecular chemistry, and bionanotechnology (44–46), the development of small-molecule ligands with fluorogenic ability is highly important for G4 structure investigation and G4-targeting drug discovery (47–51). Thus, the recognition characteristics of these compounds with G-quadruplexes warrant further examination. Taking DFHBFSI as a model, the gradual addition of NG16 to DFHBFSI solution in the presence of 100 mM K+ and 50 mM Mg2+ lead to a distinct red shift (Δλ = 25 nm) in absorption with a clear isosbestic point at 534 nm (Figure 3A), indicating that the spectroscopic properties of DFHBFSI can be altered via a strong interaction with G4. The fluorescence titration at the same condition exhibited a 12 nm bathochromic shift of DFHBFSI emission maxima concomitant with significant fluorescence enhancement upon binding with NG16 (Figure 3B). The spectral bathochromic shift of DFHBFSI caused by G4 binding is probably due to the π−π stacking interactions between the phenolic ring of DFHBFSI and the G-tetrad plane (6). The fluorescent signal of DFHBFSI shows a good linear response in the range of 0.02–0.5 μM of NG 16 with the lowest detectable concentration of 0.02 μM (Supplementary Figure S7). A 1:1 binding model for DFHBFSI with NG16 was confirmed by a Job's plot analysis (Supplementary Figure S8), and the fitting results of fluorescence titration curves provided a Kd value of 1.27 μM, indicating the high affinity of DFHBFSI with G4 structure. All the other RFP analogues have similarly low Kd value ranging from 1.37 to 2.77 μM except for DFHBAPBI (Kd = 24.9 μM) which is consistent with its relative low quantum yield.
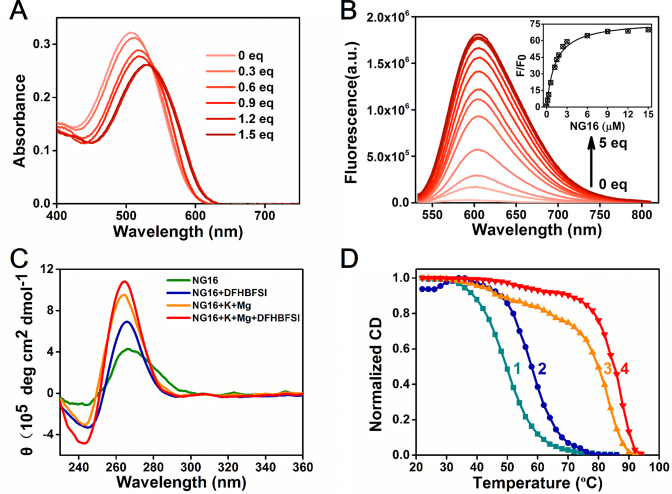
Figure 3.
(A) Absorption and (B) emission spectra of DFHBFSI (10 and 3 μM) upon stepwise addition of NG16. Inset of B is plot of fluorescence enhancement ratio (F/F0) versus the concentration of NG16. (C) Changes in CD spectra of NG16 (10 μM) upon the addition of DFHBFSI (50 μM) in different buffer conditions. K+, 100 mM; Mg2+, 50 mM. (D) CD melting curves of NG16 (5 μM) at 265 nm under different conditions. NG16 (curve 1) and NG16 with DFHBFSI (25 μM, curve 2) in buffer without metal ions; NG16 (curve 3) and NG16 with DFHBFSI (25 μM, curve 4) in buffer containing K+ (100 mM) and Mg2+ (50 mM).
Next, we investigated the topology and thermal stability of G-quadruplexes upon the addition of DFHBFSI by using circular dichroism (CD) spectroscopy. G4s are structurally polymorphic and can be classified as parallel, antiparallel, or mixed parallel/antiparallel structures based on the orientation of the G-tracts in the quartet core (52). The G4s folding topologies depend on sequences, loop geometry, and the local environment, especially the central metal ions in G4s (53). Under the optimum emission condition (25 mM Tris-HCl buffer, 100 mM KCl, 50 mM MgCl2 and pH 8.0), the CD spectra of NG16 exhibited a typical characteristic of a parallel structure with a positive peak at 265 nm and a negative peak at 240 nm (Figure 3C). It marginally changed upon the addition of DFHBFSI, indicating a slight stabilizing effect of DFHBFSI on the parallel G4 structure of NG16 in the presence of K+/Mg2+ salts. It is noteworthy that under the condition without metal ions, the parallel G4 structure of NG16 is poorly formed, whereas addition of DFHBFSI significantly improve the parallel G4 formation reflected by a marked enhancement of their CD characteristic peaks (Figure 3C), indicating its role as the parallel G4 inducer. Subsequent CD melting experiments (Figure 3D) showed that DFHBFSI can raise G4 thermal stabilization as the melting temperature (Tm) of DFHBFSI-NG16 was 5.4°C higher than that of NG16 alone in the cases involving K+/Mg2+ salts, whereas the ΔTm between DFHBFSI–NG16 and NG16 further increased to 8.1°C without salts. These CD results also evidence the crucial influence of metal ions on G4 topology and G4-chromophore interaction (Supplementary Figure S9). This is consistent with the fluorescence optimization data presented above (Supplementary Figure S5). K+ could account for 62% of the fluorescence because of its known ability and preference to support parallel structures of short loop-sized DNA G4s. The divalent ion, Mg2+, also enhances fluorescence activation in the presence of K+, indicating a possible role for Mg2+ in improving DFHBFSI binding with negatively-charged G4 structures via neutralizing its phenolate anion and screening their electrostatic repulsion (27, 28).
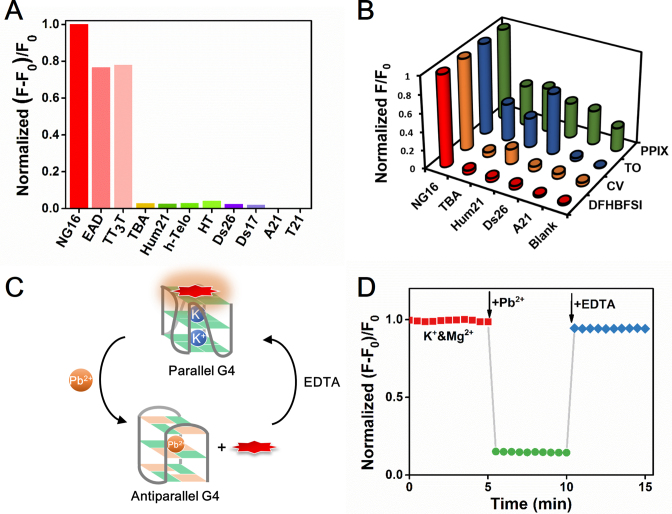
Among the known G4 probes, only a few molecules have selectivity for G4s over duplex DNA and even fewer are selective for a particular G4 structure (47–51,54,55). To test its structural specificity, DFHBFSI was challenged by various DNA conformations (their CD characterizations are shown in Supplementary Figure S10) to measure its fluorescence response. As depicted in Figure 4A, marked fluorescence enhancement was exclusively observed for parallel G4s, such as NG16, EAD, and TT3T. The fluorescence increment of DFHBFSI induced by parallel NG16 was ∼35-fold higher than that by the antiparallel G4 (TBA), 25–41-fold higher than those by hybrid-type G4s (Hum21, h-Telo, HT), 42–51-fold higher than those by double-stranded DNA (Ds26, Ds17), and over 650-fold higher than those by single stranded DNA (A21 and T21), indicating the RFP chromophore is a topology-specific turn-on probe for parallel G4s. In a sharp contrast, several widely used G4-binding dyes, includes thiazole orange (TO) (56), crystal violet (CV) (57), and N-methyl mesoporphyrin IX (NMM) (58), show poor discriminating abilities for parallel G4s from other G4 or DNA conformations (Figure 4B). Meanwhile, native gel electrophoresis experiments also confirmed that DFHBFSI has a good specificity for parallel G4 structures (Supplementary Figure S11), demonstrating the potential of DFHBFSI as a gel-staining reagent for visualizing parallel G4s. A dose–response experiment showed that G-quadruplex (NG16) is still detectable at 0.25 μM, indicating an approximate detection limit of 25.6 ng for gel staining.
Figure 4.
(A) Fluorescence changes of DFHBFSI (3 μM) in response to G-quadruplex (15 μM), double-strand (15 μM) and single-strand oligonucleotides (15 μM). F and F0 are the fluorescence intensity of DFHBFSI in the presence and absence of the oligonucleotides, respectively. (B) Fluorescence responses of G-quadruplex-specific dyes (3 μM) toward different kinds of oligonucleotides (15 μM). (C) Reversible topological switch between parallel and anti-parallel G-quadruplexes. (D) Real-time monitoring the topological switch of G4 based on DFHBFSI-PW17 (1 μM). K+, 2 mM; Mg2+, 50 mM; Pb2+, 150 μM; EDTA, 150 μM.
The unique specificity of DFHBFSI for parallel G4 structure enables dynamic monitoring of topological switching of G4s in real-time, which is seldom explored in previous research on G4-targeting dyes. Due to the higher efficiency of Pb2+ in stabilizing G4 structures, substituting K+ by Pb2+ as the G4 chelating cation causes a topological transformation from parallel to anti-parallel G4 (59), presenting an ideal model of G4 structural switch. As depicted in Figure 4C and D, in the presence of DFHBFSI, Pb2+-driven parallel-to-antiparallel conformation transition can be directly monitored by sharp quenching of fluorescence at 606 nm to 14% as a result of negligible fluorogenic response of DFHBFSI to antiparallel G4s. Introducing a Pb2+ chelator, EDTA, quickly restored fluorescence to 95%, indicating that the extraction of Pb2+ by EDTA from Pb2+-G4 complex leads to re-formation of parallel G4s via re-binding with K+. Further CD measurements confirmed this K+/Pb2+-driven G4 conformational switch (Supplementary Figure S12). This clearly demonstrated that the RFP chromophore is a smart G4 dye competent to visualize G4 structural switches in real-time, which is highly promising for functional investigation of G4 and G4-based DNA nano-machines.
Theoretic calculations and molecular dynamics simulations for elucidating fluorogenic G4 RFP mimics
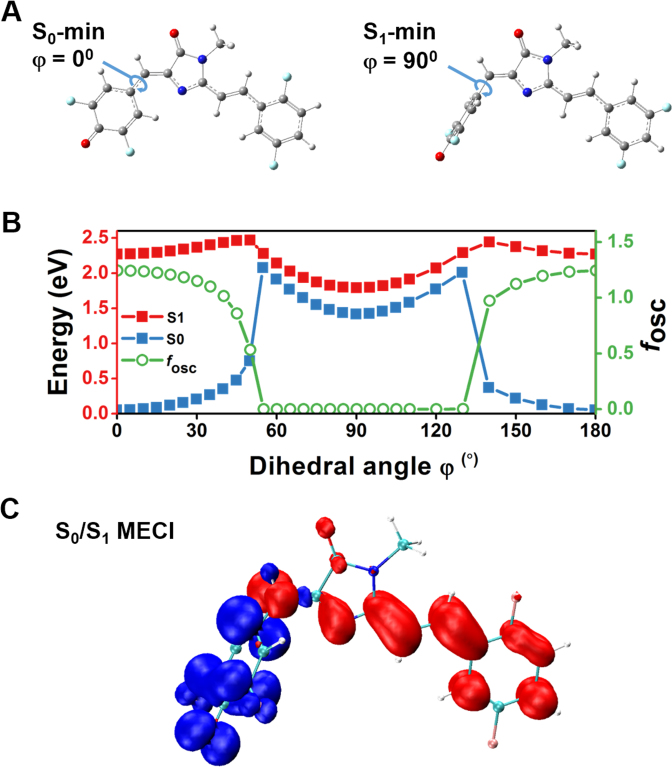
To gain more insight into the mechanism of the on/off optical switch of RFP chromophores upon binding with G4, long-range-corrected time-dependent density functional theory (LRC-TD-DFT) calculations and molecular dynamics (MD) simulations were performed using DFHBFSI as a model. In the case of anionic form of DFHBFSI which is dominant in neutral pH, LRC-TD-DFT calculation results shows that the minimum energy conformation in the first excited state (S1-min) is substantially non-planar with an almost perpendicular plane of the phenolic ring, whereas its counterpart in the ground state (S0-min) is nearly coplanar (Figure 5A). Further coordinate-driving potential surface scans indicate that the potential energy surfaces of S0 and S1 are approaching upon increasing the twist angle (φ) at the bridging bond of the phenolic moiety. When DFHBFSI in its S0-min state is excited to S1 by photoirradiation, the relaxation process starts with the barrierless rotation of the phenolic moiety to φ = 55°, leading to the minimum energy conical intersection (MECI) between S1 and S0 (Figure 5B). Electron density analysis suggest that in this highly twisted S1 state, the electronic charge obviously transfers from phenolic moiety to imidazolinone, vinyl linkage, and aromatic ring moieties (Figure 5C). This intersection causes a twisted intramolecular charge transfer (TICT) to facilitate radiationless relaxation, and results in an extremely low oscillator strength (f1) of 0.0005 (Figure 5B), implying a low fluorescence. This deactivation pathway is similar to that reported previously for the green and red fluorescent protein chromophores (33,60). Hence, the LRC-TD-DFT results manifested that the proposed RFP chromophores without confinement by G4 are non-fluorescent due to intramolecular rotation-caused TICT.
Figure 5.
Radiationless decay of RFP chromophore (DFHBFSI) via twisted intramolecular charge-transfer state. (A) Equilibrium geometry configurations at the minimum energy point of ground state S0 (S0-min) and the minimum energy point of excited state S1 (S1-min). (B) Potential energy surfaces of S1 (red) and S0 (blue) as well as oscillator strength (fosc) along the phenoxy-twisted photoisomerization pathway. (C) Charge density difference isosurfaces (isovalue = ±0.0075) at the phenoxy-twisted minimum energy conical intersection (MECI) between S1 and S0 (S0/S1 MECI, φ = 55o). Positive isosurfaces are red, and negative isosurfaces are blue.
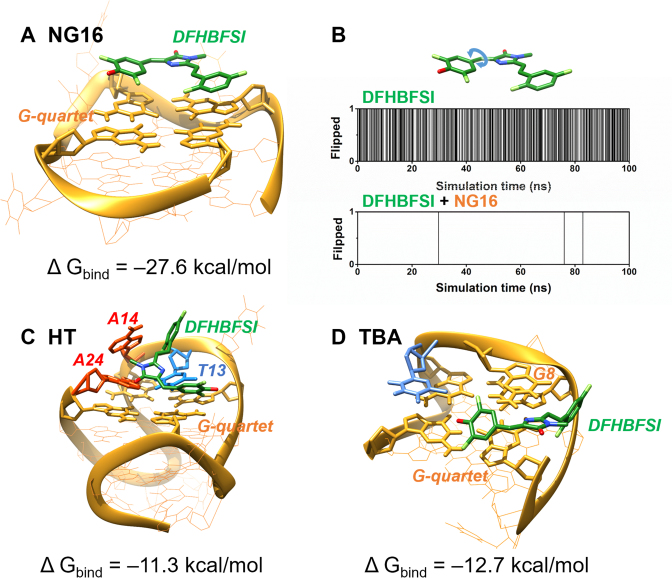
In order to further elucidate G4-activable DFHBFSI fluorescence, MD simulations were carried out to investigate the binding mode and interaction of G4 and DFHBFSI. A parallel NMR G-quadruplex structure for NG16 (PDB ID: 2LXV) (61) was used as a template. The starting conformations of the DFHBFSI-NG16 complex were obtained by molecular docking using Autodock, and then the MD simulations were performed using GROMACS for 100 ns molecular dynamic runs. Three representative complexes as starting conformations were simulated: (i) DFHBFSI with end stacking mode; (ii) DFHBFSI with groove binding; (iii) free DFHBFSI as a control. The simulation results showed that the groove-binding complexes were considerably unstable, in which the DFHBFSI quickly moved away from the G4 after roughly 10 ns (data not shown), whereas DFHBFSI perfectly end-stacks on the NG16 structure throughout simulation (Figure 6A). The end-stacking mode of DFHBFSI was further experimentally supported by the competitive inhibition of DFHBFSI-NG16 fluorescence by 5, 10, 15, 20-tetra (N-methyl-4-pyridyl) porphyrin (TMPyP4) (Supplementary Figure S13), a known end-stacking ligand of G4 (62). With respect to intramolecular motion of DFHBFSI (Figure 6B), in its free form, the phenyl moiety was constantly rotating (50% of the simulation), which broke the conjugated system essential for high quantum yield therefore it does not fluoresce. In contrast, the phenyl moiety of DFHBFSI complexed with NG16 rarely rotated (∼0.01% of the simulation), evidencing that the strong π-π stacking interaction between DFHBFSI and G-quartet significantly restricts the chromophore's range of motion and improve its rigidity. NG16-confined DFHBFSI exhibited a stable conformer with 14° and 12° for the dihedral angles of phenoxy-bridge and imidazolinone-bridge bond torsion, respectively (Figure 6A). Such torsion angles are very close to planarity and comparable to those of the mCherry chromophore (13.7° and 11.3°) (3), which leads to increased fluorescence.
Figure 6.
Averaged structures of DFHBFSI and G-quadruplex-DFHBFSI obtained through MD simulations. (A) Complex model of DFHBFSI with NG16. (B) The flipping of phenoxy ring in DFHBFSI in 100 ns MD simulation. In the y axis, 0 stands for no flipping, and 1 stands for flipping. Flipping is defined as the phenoxy ring rotates across the plane perpendicular to the imidazole ring. Complex models of DFHBFSI with (C) HT and (D) TBA. DFHBFSI, G-quartet, adenine and thymine are shown using green, yellow, red and blue sticks, respectively.
Furthermore, MD simulations are also helpful for elucidating the G4 topological preference of DFHBFSI. A hybrid-type NMR G-quadruplex structure for HT (PDB ID: 2GKU) (63) and an antiparallel NMR G-quadruplex structure for TBA (PDB ID: 5MJX) (64) were also used. The examined models and the estimated free energies of the binding of DFHBFSI with NG16, HT, and TBA are shown in Figure 6. Unlike the perfect stacking of DFHBFSI on the ending G-quartet of NG16 which confined DFHBFSI to the co-planar conformation, DFHBFSI bound to both HT and TBA twisted dramatically to form favorable interactions with both G-quartet and several bases in the flanking and loop sequences, for instances T13, A14, A24 in HT and G8 in TBA, respectively (Figure 6C and D). As a consequence, both the conformations of DFHBFSI in their complexes with HT and TBA are considerably non-planar and thus adverse to its fluorescence. In addition, DFHBFSI exhibited stronger binding with NG16 (–27.6 kcal/mol) than with HT (–11.3 kcal/mol) and TBA (–12.7 kcal/mol), which show good agreement with the observed G4 selectivity of DFHBFSI. Overall, the quantum chemical calculation and molecular modeling reasoned the fluorogenic property of DFHBFSI upon G4 binding and its unique specificity to parallel G4.
One- and two-photon bio-imaging in live cells and tissues
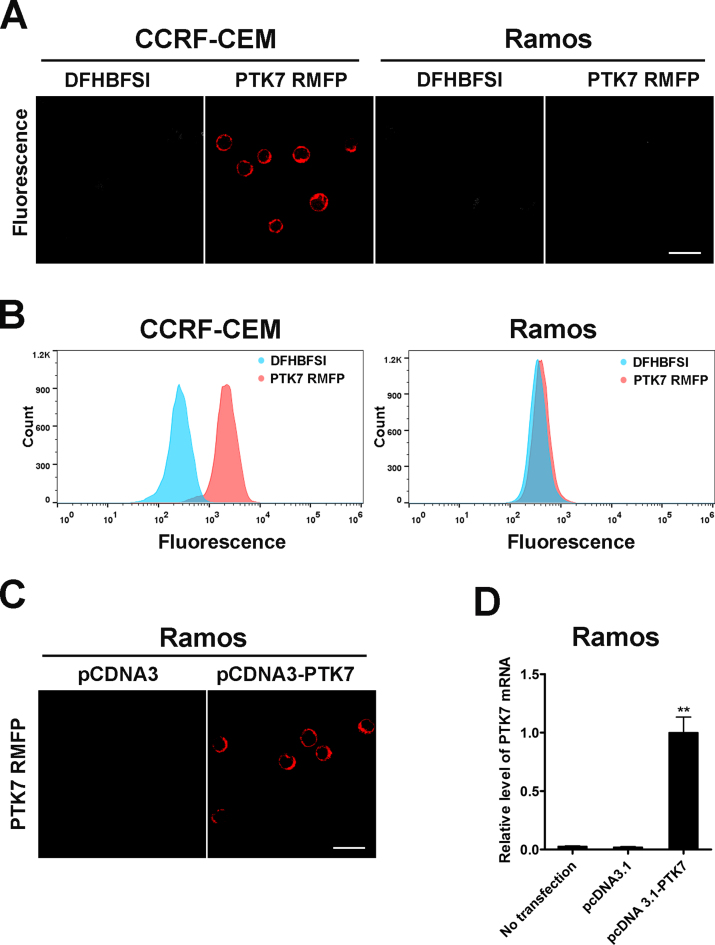
Encouraged by their prominent spectral merits, including bright red luminescence, large Stokes shifts, anti-photobleaching and two-photon fluorescent properties, we further exploited DNA mimics of RFP as fluorescent probes for living cell and tissue bio-imaging. DFHBFSI was chosen as the model because it had the highest quantum yield of all the proposed RFP chromophores. Prior to imaging experiments, we confirmed that the G4-activated fluorescence of DFHBFSI was barely influenced by the presence of cell lysates and cell culture medium with or without live cells (Supplementary Figure S14). The cell counting kit-8 (CCK-8) assay revealed that DFHBFSI has negligible influence on cell viability over the 24-h observation period (Supplementary Figure S15). These results imply the reliability and biocompatibility of DFHBFSI-G4 complexes for bio-imaging. To selectively target protein biomarkers displaying on cancer cell surface, DFHBFSI-G4 complexes was employed as RFP mimics to specifically ‘tag-and-lighten’ cancer-associated membrane proteins through conjugation with their targeting-aptamers. Aptamers are single-stranded DNA or RNA oligonucleotides with unique molecular recognition and binding capabilities specific to various targets, including small molecules, proteins, and live cells (65–67). The proposed membrane protein-specific RFP-mimicking fluorescent probe (RMFP) is composed of two segments: a reporter strand bearing two repeat NG16 sequences binding with DFHBFSI and a recognition strand with the aptamer sgc8 targeting to protein tyrosine kinase-7 (PTK7), a transmembrane receptor protein closely relative to a number of cancers (68–71), which was readily prepared by simple hybridization (Supplementary Table S6). As shown in Figure 7A, based on highly specific aptamer recognition, PTK7 RMFP are selectively targeted to CCRF-CEM cells, a T cell acute lymphoblastic leukemia (ALL) cell line with high expression of PTK7, reflected by bright red fluorescence on the cellular periphery, whereas the control Ramos cells shows weak fluorescence. It is noteworthy that DFHBFSI, by itself, is not detectably fluorescent upon incubation with either CCRF-CEM cells or Ramos cells, indicating negligible background. Flow cytometry results (Figure 7C) also revealed that the PTK7-specific RMFP selectively binds to CCRF-CEM cells. To further elucidate the specificity of PTK7-targeting RMFP, we engineered Ramos cells by electroporation with plasmids expressing PTK7. Fluorescence was readily detectable on the cytoplasmic membrane of individual engineered Ramos cells (Figure 7D), which accords with the quantification data of PTK7 mRNA by real-time quantitative reverse transcription PCR (Figure 7E). These data suggested that the proposed RMFP is a feasible tool for probing expression profiles of PTK7 in different cells.
Figure 7.
(A) Confocal microscope image of PTK RMFP treated CCRF-CEM cells and Ramos cells at 570–620 nm emission channel using 488 nm excitation, Scale bar: 20 μm. (B) Flow cytometry of PTK RMFP treated CCRF-CEM cells and Ramos cells. (C) Confocal microscope image of PTK RMFP treated Ramos cells with transfection of pcDNA3-PTK7 at 570–620 nm channel, Scale bar: 20 μm. (D) Relative mRNA level of PTK7 in Ramos cells with transfection of pcDNA3-PTK7 using real-time quantitative reverse transcription PCR. Values are means with SD (n = 3), **P < 0.001.
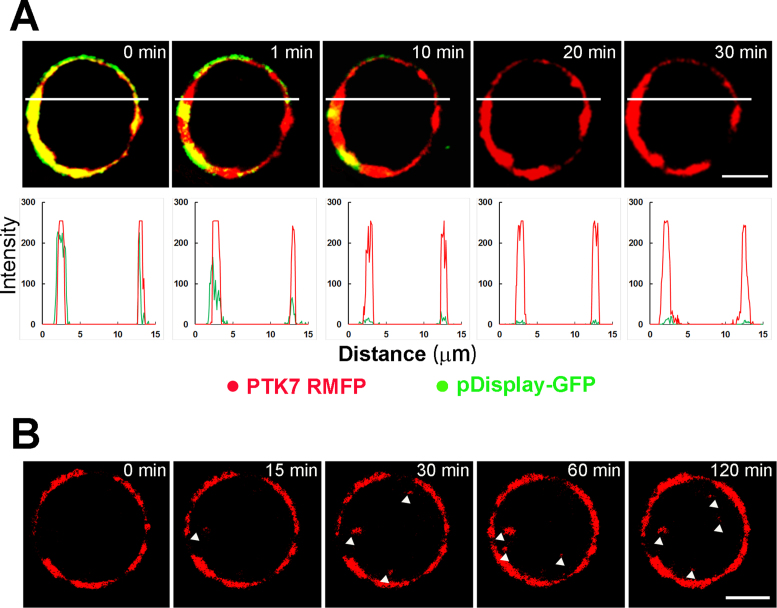
To explore the plasma membrane localization of PTK7 RMFP, we transfected CCRF-CEM cells with pDisplay-EGFP for a co-localization analysis. The pDisplay system is a mammalian expression vector that allows display of enhanced GFPs (EGFPs) on the extracellular side of cell plasma membrane. Given that the large Stokes shifts of DFHBFSI-G4 complexes (Ex = 513 nm, Em = 606 nm) greatly facilitates simultaneous imaging of multi-color fluorophores, RMFP and EGFP were simultaneously excited by the same 488 nm channel and monitored separately by emission channels of 500–550 nm and 570–620 nm, respectively, without mutual interference. A perfect co-localization of RMFP and EGFP was observed (Figure 8A) and further quantified using Manders’ overlap coefficient (R). A high overlap coefficient (0.90) was obtained and confirmed that PTK7-bound RMFP was predominately located on the membrane extracellular surface. Moreover, this RMFP/EGFP system is desirable for a simultaneously comparison of the anti-photo-bleaching properties of fluorescent protein and its DNA mimics. As shown in Figure 8A, DFHBFSI–G4 complexes retain distinctive red fluorescence throughout 30 min irradiation. In contrast, EGFP quickly lost its fluorescence in 10 min irradiation due to severe photo-bleaching. Furthermore, the real-time imaging of a single live CCRF-CEM cell confirmed strong membrane staining could be still measured even after 2 h continuous observation, which allows monitoring the dynamic processes of membrane proteins, such as cellular internalization of PTK7 (Figure 8B). These results proved that DNA mimics of RFPs are suitable for long-term and real-time cellular imaging due to their superior anti-photo-bleaching properties.
Figure 8.
(A) Time-lapse confocal microscope images of PTK RMFP treated CCRF-CEM cells with transfection of pDispaly-GFP for different time spans. The images were acquired using 488 nm excitation and presented as overlay images (top) from 500 to 550 nm emission channel (green) and 570–620 nm emission channel (red), respectively, Scale bar: 5 μm. Co-localization analysis of red and green fluorescence channels of upper images (bottom). (B) Long duration confocal microscope image at different time span of PTK RMFP treated CCRF-CEM cells at 570–620 nm emission channel using 488 nm excitation, Scale bar: 5 μm.
The generality of DNA RFP mimics as potent probes for tagging cell membrane biomarkers was also tested. We further prepared RMFPs targeting to hepatocyte growth factor receptor (HGFR) or epithelial cell adhesion molecule (EpCAM), two representatively overexpressed tumor antigens, through integrating with their specific aptamers, SL1 (72) and SYL3C (73), respectively. Both RMFPs exhibit great potency for cell-specific imaging based upon target-expression differences (Supplementary Figures S16 and S17), which can efficiently discriminate targeting antigen-positive cells from negative cells.
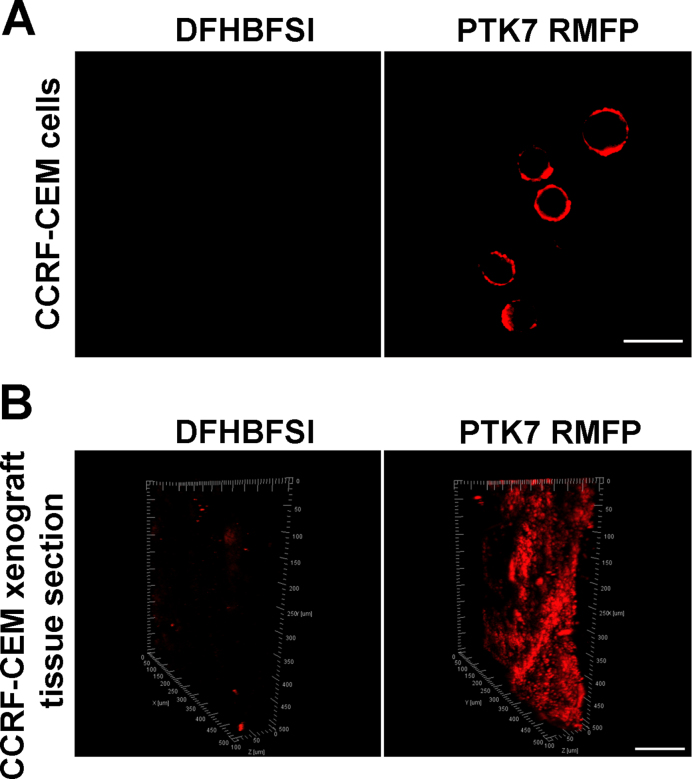
Taking advantage of the two-photon properties of DFHBFSI/G4 complexes, we further investigated the possibility of conducting two-photon imaging based on the specificity of aptamer-derived RMFP probes in living cells or tissues. As a proof-of-concept, PTK7 RMFP-treated CCRF-CEM cells were employed for two-photon imaging upon excitation of the sample at λ = 750 nm. As shown in Figure 9A, strong red fluorescence was detected on the surface of CCRF-CEM cells. Similar two-photon imaging was further undertaken by incubating the PTK7 RMFP probe with tissue sections from CCRF-CEM cell xenograft mice. The images were collected at different depths and reconstructed in a three-dimensional box (3D) to present the spatial distribution of PTK7-overexpressed cells (Figure 9B), and the fluorescence of RMFPs could be evidently detected at depths of ∼80 μm. The bright fluorescence detected clearly indicated that the developed probe could efficiently monitor PTK7 expression in real tissue samples.
Figure 9.
(A) Two-photon image of PTK RMFP treated CCRF-CEM cells at using 750 nm excitation, Scale bar: 20 μm. (B) 3D reconstruction from two-photon images of PTK RMFP treated tissue section from CCRF-CEM cells xenograft using 750 nm excitation, Scale bar: 100 μm.
CONCLUSIONS
In summary, we have successfully developed a palette of G4-chromophore complexes spanning most of spectrum region of existing red fluorescent proteins, providing an unprecedented complement to conventional synthetic mimics of GFPs. These novel RFP chromophore analogues, which possess an extended π-ring system, not only cause a significant emission shift to red and far-red but also strengthen the interactions with DNA G4 structures via π-π stacking, presenting a simple, potent, and versatile strategy to develop highly emissive RFP mimics via G4 binding and activation. Detailed theoretical calculation and molecular dynamics simulations suggest that the confinement effect of G4 is analogous to RFP, in which the end-stacking mode of RFP chromophores bound to G4 greatly restricts their intramolecular rotation and TICT-induced radiationless relaxation, ensuring their bright fluorescence. These DNA RFP mimics offer attractive photo-physical advantages over RFPs in that: spectral properties are readily tunable by changing specific synthetic chromophores; large Stokes shifts enable high-contrast multicolor imaging using only a single excitation; and excellent resistance to photobleaching allows long-term real-time monitoring of live cells. Furthermore, despite the unlocked structure of RFP chromophores, G4 confinement endows the RFP mimics with a unique S0 to Sn two-photon excitation with red emission, which is highly desirable for in vivo imaging. On the other hand, the proposed RFP chromophores affords a novel and distinctive category of G-quadruplex probes, possessing a selective light-up function specific for parallel conformation, which is greatly useful for discrimination of different G4 topologies and real-time detection of G4 structural switches. This work will shed light on multifaceted researches: (i) the discovery of a new generation of FP mimicking system with valuable features, including red, near-infrared, and two-photon fluorescence; (ii) the development of novel topology-selective G4 probes for G4-relative detection and nano-system; (iii) potential toolkits for biomedical imaging and clinical diagnosis based on the integration of DNA RFP mimics with functional nucleic acids.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Tianyou Zhai and Prof. Zhihong Liu for assistance with fluorescence lifetime and two-photon fluorescence measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSF of China [21235002, 21575038]; Foundation for Innovative Research Groups [21521063]; Young Top-notch Talent for Ten Thousand Talent Program; NSF of Hunan Province [2015JJ1005]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zimmer M. ChemInform Abstract: Green Fluorescent Protein (GFP): Applications, Structure, and Related Photophysical Behavior. Chem. Rev. 2002; 102:759–781. [DOI] [PubMed] [Google Scholar]
- 2. Lei C., Huang Y., Nie Z., Hu J., Li L., Lu G., Han Y., Yao S.. A supercharged fluorescent protein as a versatile probe for homogeneous DNA detection and methylation analysis. Angew. Chem. Int. Ed. 2014; 53:8358–8362. [DOI] [PubMed] [Google Scholar]
- 3. Acharya A., Bogdanov A.M., Grigorenko B.L., Bravaya K.B., Nemukhin A.V., Lukyanov K.A., Krylov A.I.. Photoinduced chemistry in fluorescent proteins: curse or blessing. Chem. Rev. 2016; 117:758–795. [DOI] [PubMed] [Google Scholar]
- 4. Tsien R.Y. Constructing and exploiting the fluorescent protein paintbox. Angew. Chem. Int. Ed. 2009; 48:5612–5626. [DOI] [PubMed] [Google Scholar]
- 5. Shcherbakova D.M., Subach O.M., Verkhusha V.V.. Red fluorescent proteins: advanced imaging applications and future design. Angew. Chem. Int. Ed. 2012; 51:10724–10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Subach F.V., Verkhusha V.V.. Chromophore transformations in red fluorescent proteins. Chem. Rev. 2012; 112:4308–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker C.L., Lukyanov K.A., Yampolsky I.V., Mishin A.S., Bommarius A.S., Durajthatte A.M., Azizi B., Tolbert L.M., Solntsev K.M.. Fluorescence imaging using synthetic GFP chromophores. Curr. Opin. Chem. Biol. 2015; 27:64–74. [DOI] [PubMed] [Google Scholar]
- 8. Deng H., Zhu X.. Emission enhancement and application of synthetic green fluorescent protein chromophore analogs. Mater. Chem. Front. 2017; 1:619–629. [Google Scholar]
- 9. Baleeva N.S., Myannik K.A., Yampolsky I.V., Baranov M.S.. Bioinspired fluorescent dyes based on a conformationally locked chromophore of the fluorescent protein Kaede. Eur. J. Org. Chem. 2015; 26:5716–5721. [Google Scholar]
- 10. Povarova N.V., Bozhanova N.G., Sarkisyan K.S., Gritcenko R., Baranov M.S., Yampolsky I.V., Lukyanov K.A., Mishin A.S.. Docking-guided identification of protein hosts for GFP chromophore-like ligands. J. Mater. Chem. C. 2016; 4:3036–3040. [Google Scholar]
- 11. Baranov M.S., Lukyanov K.A., Borissova A.O., Shamir J., Kosenkov D., Slipchenko L.V., Tolbert L.M., Yampolsky I.V., Solntsev K.M.. Conformationally locked chromophores as models of excited-state proton transfer in fluorescent proteins. J. Am. Chem. Soc. 2012; 134:6025–6032. [DOI] [PubMed] [Google Scholar]
- 12. Hsu Y.H., Chen Y.A., Tseng H.W., Zhang Z.Y., Shen J.Y., Chuang W.T., Lin T.C., Lee C.S., Hung W.Y., Hong B.C. et al. . Locked ortho-and para-Core chromophores of green fluorescent protein; dramatic emission enhancement via structural constraint. J. Am. Chem. Soc. 2014; 136:11805–11812. [DOI] [PubMed] [Google Scholar]
- 13. Yuan L., Lin W., Chen H., Zhu S., He L.. A unique family of rigid analogues of the GFP chromophore with tunable two‐photon action cross‐sections for biological imaging. Angew. Chem. Int. Ed. 2013; 52:10018–10022. [DOI] [PubMed] [Google Scholar]
- 14. Baldridge A., Solntsev K.M., Song C., Tanioka T., Kowalik J., Hardcastle K., Tolbert L.M.. Inhibition of twisting of a green fluorescent protein-like chromophore by metal complexation. Chem. Commun. 2010; 46:5686–5688. [DOI] [PubMed] [Google Scholar]
- 15. Wu L., Burgess K.. Syntheses of highly fluorescent GFP-chromophore analogues. J. Am. Chem. Soc. 2008; 130:4089–4096. [DOI] [PubMed] [Google Scholar]
- 16. Paolino M., Gueye M., Pieri E., Manathunga M., Fusi S., Cappelli A., Latterini L., Pannacci D., Filatov M., Léonard J.r.m.. Design, synthesis, and dynamics of a green fluorescent protein fluorophore mimic with an ultrafast switching function. J. Am. Chem. Soc. 2016; 138:9807–9825. [DOI] [PubMed] [Google Scholar]
- 17. Baldridge A., Samanta S.R., Jayaraj N., Ramamurthy V., Tolbert L.M.. Steric and electronic effects in capsule-confined green fluorescent protein chromophores. J. Am. Chem. Soc. 2010; 133:712–715. [DOI] [PubMed] [Google Scholar]
- 18. Zheng Y.J., Li G.L., Deng H.P., Su Y., Liu J.H., Zhu X.Y.. Temperature-induced fluorescence enhancement of GFP chromophore containing copolymers for detection of Bacillus thermophilus. Polym. Chem. 2014; 5:2521–2529. [Google Scholar]
- 19. Williams D.E., Dolgopolova E.A., Pellechia P.J., Palukoshka A., Wilson T.J., Tan R., Maier J.M., Greytak A.B., Smith M.D., Krause J.A. et al. . Mimic of the green fluorescent protein β-barrel: photophysics and dynamics of confined chromophores defined by a rigid porous scaffold. J. Am. Chem. Soc. 2015; 137:2223–2226. [DOI] [PubMed] [Google Scholar]
- 20. Ghodbane A., Fellows W.B., Bright J.R., Ghosh D., Saffon N., Tolbert L.M., Feryforgues S., Solntsev K.M.. Effects of the benzoxazole group on green fluorescent protein chromophore crystal structure and solid state photophysics. J. Mater. Chem. C. 2016; 4:2793–2801. [Google Scholar]
- 21. Carayon C., Ghodbane A., Gibot L., Dumur R., Wang J., Saffon N., Rols M.P., Solntsev K.M., Feryforgues S.. Conjugates of benzoxazole and GFP chromophore with aggregation-induced enhanced emission: influence of the chain length on the formation of particles and on the dye uptake by living cells. Small. 2016; 12:6602–6612. [DOI] [PubMed] [Google Scholar]
- 22. Zhou Q., Wu F., Wu M., Tian Y., Niu Z.. Confined chromophores in tobacco mosaic virus to mimic green fluorescent protein. Chem. Commun. 2015; 51:15122–15124. [DOI] [PubMed] [Google Scholar]
- 23. Baldridge A., Feng S., Chang Y.T., Tolbert L.M.. Recapture of GFP chromophore fluorescence in a protein host. ACS Comb. Sci. 2011; 13:214–217. [DOI] [PubMed] [Google Scholar]
- 24. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song W.J., Strack R.L., Svensen N., Jaffrey S.R.. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 2014; 136:1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. You M., Jaffrey S.R.. Structure and mechanism of RNA mimics of green fluorescent protein. Annu. Rev. Biophys. 2015; 44:187–206. [DOI] [PubMed] [Google Scholar]
- 27. Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Na.t Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warner K.D., Chen M.C., Song W.J., Strack R.L., Thorn A., Jaffrey S.R., Ferré-D’Amaré A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chuang W.T., Chen B.S., Chen K.Y., Hsieh C.C., Chou P.T.. ChemInform abstract: fluorescent protein red Kaede chromophore; one-step, high-yield synthesis and potential application for solar cells. Chem. Commun. 2009; 41:6982–6984. [DOI] [PubMed] [Google Scholar]
- 30. Li Q., Xiang J.F., Yang Q.F., Sun H.X., Guan A.J., Tang Y.L.. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013; 41:D1115–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He X., Bell A.F., Tonge P.J.. Synthesis and spectroscopic studies of model red fluorescent protein chromophores. Org. Lett. 2002; 4:1523–1526. [DOI] [PubMed] [Google Scholar]
- 32. Yampolsky I.V., Kislukhin A.A., Amatov T.T., Shcherbo D., Potapov V.K., Lukyanov S., Lukyanov K.A.. Synthesis and properties of the red chromophore of the green-to-red photoconvertible fluorescent protein Kaede and its analogs. Bioorg. Chem. 2008; 36:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olsen S., Smith S.C.. Radiationless decay of red fluorescent protein chromophore models via twisted intramolecular charge-transfer states. J. Am. Chem. Soc. 2007; 129:2054–2065. [DOI] [PubMed] [Google Scholar]
- 34. Shemiakina I.I., Ermakova G.V., Cranfill P.J., Baird M.A., Evans R.A., Souslova E.A., Staroverov D.B., Gorokhovatsky A.Y., Putintseva E.V., Gorodnicheva T.V. et al. . A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012; 3:1204–1210. [DOI] [PubMed] [Google Scholar]
- 35. Ando R., Hama H., Yamamoto-Hino M., Mizuno H., Miyawaki A.. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12651–12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shkrob M.A., Yanushevich Y.G., Chudakov D.M., Gurskaya N.G., Labas Y.A., Poponov S.Y., Mudrik N.N., Lukyanov S., Lukyanov K.A.. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem. J. 2005; 392:649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shcherbo D., Shemiakina I.I., Ryabova A.V., Luker K.E., Schmidt B.T., Souslova E.A., Gorodnicheva T.V., Strukova L., Shidlovskiy K.M., Britanova O.V. et al. . Near-infrared fluorescent proteins. Nat. Methods. 2010; 7:827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin W.Y., Yuan L., Cao Z.M., Feng Y.M., Song J.Z.. Through‐bond energy transfer cassettes with minimal spectral overlap between the donor emission and acceptor absorption: coumarinth minimal spectral overlap between‐Stokes shifts and emission shifts. Angew. Chem. Int. Ed. 2010; 49:375–379. [DOI] [PubMed] [Google Scholar]
- 39. Faraji S., Krylov A.I.. On the nature of an extended Stokes shift in the mPlum fluorescent protein. J. Phys. Chem. B. 2015; 119:13052–13062. [DOI] [PubMed] [Google Scholar]
- 40. Han K.Y., Leslie B.J., Fei J., Zhang J., Ha T.. Understanding the photophysics of the spinach–DFHBI RNA aptamer–fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 2013; 135:19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang P., Querard J., Maurin S., Nath S.S., Le Saux T., Gautier A., Jullien L.. Photochemical properties of Spinach and its use in selective imaging. Chem. Sci. 2013; 4:2865–2873. [Google Scholar]
- 42. Kim H.M., Cho B.R.. Two-photon probes for intracellular free metal ions, acidic vesicles, and lipid rafts in live tissues. Acc. Chem. Res. 2009; 42:863–872. [DOI] [PubMed] [Google Scholar]
- 43. Drobizhev M., Makarov N.S., Tillo S.E., Hughes T.E., Rebane A.. Two-photon absorption properties of fluorescent proteins. Nat. Methods. 2011; 8:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug. Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 46. Wang S.-R., Min Y.-Q., Wang J.-Q., Liu C.-X., Fu B.-S., Wu F., Wu L.-Y., Qiao Z.-X., Song Y.-Y., Xu G.-H.. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti–hepatitis C target. Sci. Adv. 2016; 2:e1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin B., Zhang X., Zheng W., Liu X., Qi C., Wang F., Shangguan D.. Fluorescence light-up probe for parallel G-quadruplexes. Anal. Chem. 2013; 86:943–952. [DOI] [PubMed] [Google Scholar]
- 48. Alexander H., Wu Y., Huang Y.C., Chavez E.A., Jesse P., Brad J.F., Jr B.R.M., Dipankar S., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhasikuttan A.C., Mohanty J.. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: promising fluorescence sensors. Chem. Commun. 2015; 51:7581–7597. [DOI] [PubMed] [Google Scholar]
- 50. Xu S.J., Li Q., Xiang J.F., Yang Q.F., Sun H.X., Guan A.J., Wang L.X., Liu Y., Yu L.J., Shi Y.H.. Directly lighting up RNA G-quadruplexes from test tubes to living human cells. Nucleic Acids Res. 2015; 43:9575–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen S.-B., Hu M.-H., Liu G.-C., Wang J., Ou T.-M., Gu L.-Q., Huang Z.-S., Tan J.-H.. Visualization of NRAS RNA G-quadruplex structures in cells with an engineered fluorogenic hybridization probe. J. Am. Chem. Soc. 2016; 138:10382–10385. [DOI] [PubMed] [Google Scholar]
- 52. Collie G.W., Parkinson G.N.. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011; 40:5867–5892. [DOI] [PubMed] [Google Scholar]
- 53. Zhou W.H., Saran R., Liu J.W.. Metal Sensing by DNA. Chem. Rev. 2017; 117:8272–8325. [DOI] [PubMed] [Google Scholar]
- 54. Zhu L.N., Wu B., Kong D.M.. Specific recognition and stabilization of monomeric and multimeric G-quadruplexes by cationic porphyrin TMPipEOPP under molecular crowding conditions. Nucleic Acids Res. 2013; 41:4324–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang X.X., Zhu L.N., Wu B., Huo Y.F., Duan N.N., Kong D.M.. Two cationic porphyrin isomers showing different multimeric G-quadruplex recognition specificity against monomeric G-quadruplexes. Nucleic Acids Res. 2014; 42:8719–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang P., De Cian A., Teulade- Fichou M.P., Mergny J.L., Monchaud D.. Engineering bisquinolinium/thiazole orange conjugates for fluorescent sensing of G‐quadruplex DNA. Angew. Chem. Int. Ed. 2009; 48:2188–2191. [DOI] [PubMed] [Google Scholar]
- 57. Kong D.M., Ma Y.E., Wu J., Shen H.X.. Discrimination of G‐quadruplexes from duplex and single‐stranded DNAs with fluorescence and energy‐transfer fluorescence spectra of crystal violet. Chem. Eur. J. 2009; 15:901–909. [DOI] [PubMed] [Google Scholar]
- 58. Kreig A., Calvert J., Sanoica J., Cullum E., Tipanna R., Myong S.. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015; 43:7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li T., Wang E.K., Dong S.J.. Potassium-lead-switched G-quadruplexes: a new class of DNA logic gates. J. Am. Chem. Soc. 2009; 131:15082–15083. [DOI] [PubMed] [Google Scholar]
- 60. Martin M.E., Negri F., Olivucci M.. Origin, nature, and fate of the fluorescent state of the green fluorescent protein chromophore at the CASPT2//CASSCF resolution. J. Am. Chem. Soc. 2004; 126:5452–5464. [DOI] [PubMed] [Google Scholar]
- 61. Kuryavyi V., Cahoon L.A., Seifert H.S., Patel D.J.. The RecA-binding pilE G4 sequence essential for Pilin antigenic variation forms parallel-stranded monomeric and 5′-end stacked dimeric G-quadruplexes. Structure. 2012; 20:2090–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ali A., Bansal M., Bhattacharya S.. Ligand 5, 10, 15, 20-tetra (N-methyl-4-pyridyl) porphine (TMPyP4) prefers the parallel propeller-type human telomeric G-Quadruplex DNA over its other polymorphs. J. Phys. Chem. B. 2014; 119:5–14. [DOI] [PubMed] [Google Scholar]
- 63. Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J.. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006; 128:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lietard J., Assi H.A., Gómezpinto I., González C., Somoza M.M., Damha M.J.. Mapping the affinity landscape of Thrombin-binding aptamers on 2′F-ANA/DNA chimeric G-Quadruplex microarrays. Nucleic Acids Res. 2017; 45:1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu J.W., Cao Z.H., Lu Y.. Functional nucleic acid sensors. Chem. Rev. 2009; 109:1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tan W.H., Donovan M.J., Jiang J.H.. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013; 113:2842–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang N., Bing T., Shen L., Song R., Wang L., Liu X., Liu M., Li J., Tan W., Shangguan D.. Intercellular connections related to cell–cell crosstalk specifically recognized by an aptamer. Angew. Chem. Int. Ed. 2016; 55:3914–3918. [DOI] [PubMed] [Google Scholar]
- 68. Shangguan D.H., Li Y., Tang Z.W., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W.H.. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi H., He X.X., Wang K.M., Wu X., Ye X.S., Guo Q.P., Tan W.H., Qing Z.H., Yang X.H., Zhou B.. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. You M., Peng L., Shao N., Zhang L., Qiu L., Cui C., Tan W.. DNA “nano-claw”: logic-based autonomous cancer targeting and therapy. J. Am. Chem. Soc. 2014; 136:1256–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. You M., Zhu G., Chen T., Donovan M.J., Tan W.. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc. 2015; 137:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ueki R., Sando S.. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun. 2014; 50:13131–13134. [DOI] [PubMed] [Google Scholar]
- 73. Pu Y., Liu Z.X., Lu Y., Yuan P., Liu J., Yu B., Wang G.D., Yang C.J., Liu H., Tan W.H.. Using DNA aptamer probe for immunostaining of cancer frozen tissues. Anal. Chem. 2015; 87:1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.