Abstract
While the use of RNA interference (RNAi) in molecular biology and functional genomics is a well-established technology, in vivo applications of synthetic short interfering RNAs (siRNAs) require chemical modifications. We recently found that amides as non-ionic replacements for phosphodiesters may be useful modifications for optimization of siRNAs. Herein, we report a comprehensive study of systematic replacement of a single phosphate with an amide linkage throughout the guide strand of siRNAs. The results show that amides are surprisingly well tolerated in the seed and central regions of the guide strand and increase the silencing activity when placed between nucleosides 10 and 12, at the catalytic site of Argonaute. A potential explanation is provided by the first crystal structure of an amide-modified RNA–DNA with Bacillus halodurans RNase H1. The structure reveals how small changes in both RNA and protein conformation allow the amide to establish hydrogen bonding interactions with the protein. Molecular dynamics simulations suggest that these alternative binding modes may compensate for interactions lost due to the absence of a phosphodiester moiety. Our results suggest that an amide can mimic important hydrogen bonding interactions with proteins required for RNAi activity and may be a promising modification for optimization of biological properties of siRNAs.
INTRODUCTION
Technologies for controlling gene expression in vivo require chemical modifications that optimize the properties of oligonucleotides without negatively affecting their biological activity. Recent developments in antisense (1,2), RNA interference (RNAi) (2–7) and, most recently, CRISPR-Cas9 technologies (8,9) continue driving the interest in synthetic chemistry of oligonucleotides. For short interfering RNAs (siRNAs), enzymatic stability, targeted delivery, biodistribution and pharmacokinetics, and cellular uptake are the main properties optimized using a variety of chemical modifications. Many modifications are available for improving enzymatic stability; however, insufficient delivery, biodistribution and cellular uptake remain the bottlenecks for in vivo application of siRNAs (2–7). Other major obstacles for RNAi success in vivo are limitations in potency, duration and specificity (off-target effects) of the gene knockdown by siRNAs. Compared to small molecule probes and drugs, the problems with intracellular in vivo delivery of oligonucleotides are related to their size, enzymatic lability, hydrophilicity and the negatively charged phosphodiester backbone (10). Despite substantial progress of the siRNA modification field, there is still a significant unmet need for new chemistries to improve performance of RNAi, especially for in vivo applications.
The traditional view is that the negatively charged phosphates are critical for siRNA recognition and binding by the Argonaute (Ago) proteins. Early structural studies showed that Ago proteins, which belong to the RNase H family of enzymes (11), bind siRNAs through electrostatic interactions with the phosphate backbone (12). The traditional view is also supported by recent crystal structures showing that almost all phosphates of the guide strand and most of the phosphates of the passenger strand are contacted by Ago primarily via electrostatic interactions (13–16). This notion has discouraged nucleic acid chemists from exploring siRNA modifications that neutralize the negative charge of the phosphodiester backbone, even though such modifications may be helpful in optimization of biodistribution and cellular uptake (17).
The most popular backbone modification to improve nuclease resistance and pharmacokinetic properties of antisense oligonucleotides and siRNAs has been the phosphorothioate (PS) linkage (1–4,18). Moderate PS modification is well tolerated at the ends of siRNA duplexes and improves duration of gene silencing (19–21), while extensive PS modification may lead to toxic side-effects (20,21). Apart from phosphorothioates, other backbone modifications, such as boranophosphates (22) and phosphorodithioates, alone (23) or in combination with 2′-OMe (24), have been successfully used in siRNAs to increase the nuclease resistance and silencing potency, but have not yet found broad applications. The phosphonoacetate and thiophosphonoacetate linkages have also been incorporated in the passenger strand of siRNA, but did not improve either silencing activity or cellular uptake of siRNAs (25). In a clever design that enables the use of a neutral backbone for delivery of siRNA, Dowdy and co-workers (17) recently demonstrated that replacement of phosphodiesters with neutral and bioreversible S-acyl-2-thioethyl phosphotriesters allowed conjugation of the neutralized siRNAs to cationic peptides and efficient cellular delivery of these prodrug conjugates. Once delivered in cells, the cytoplasmic thioesterases converted the phosphotriesters into native phosphodiesters that supported robust RNAi activity in vitro and in vivo. These intriguing results prompted us to explore if siRNAs could tolerate permanent non-ionic backbone modifications that may support the use of cationic peptide delivery agents.
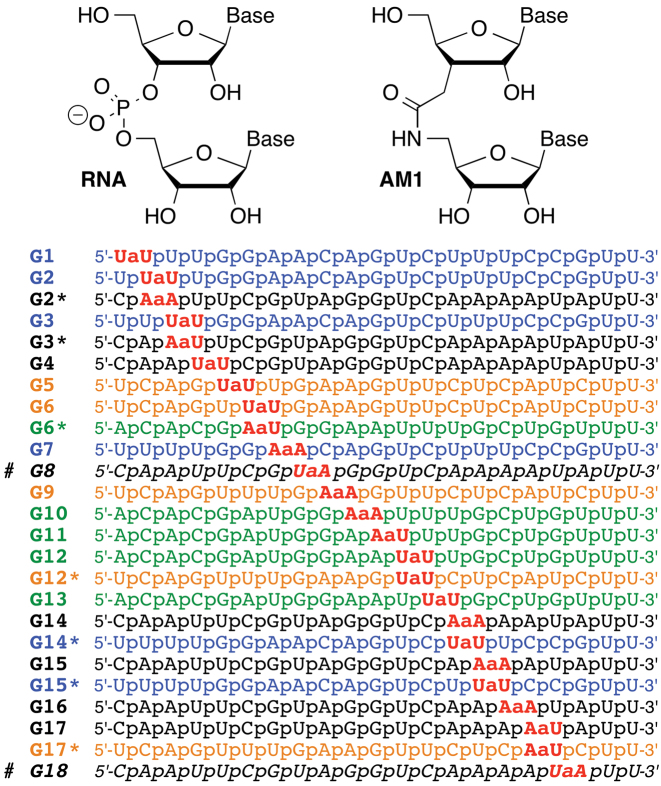
We hypothesized that an amide linkage (AM1, Figure 1) could serve as a non-ionic replacement for phosphate in siRNAs. Our hypothesis was based on earlier work (26–31) showing that isolated AM1 linkages were well accommodated in DNA–RNA heteroduplexes and all-RNA duplexes, with relatively small changes in thermal stability and overall structures of the modified duplexes. Later, we reported that isolated AM1 linkages were indeed surprisingly good structural mimics of phosphate and did not change the A-type conformation, thermal stability and hydration of short RNA duplexes (32,33). While three consecutive amide linkages in the middle of a short RNA duplex caused some loss of thermal stability, the overall RNA structure was only minimally altered (34). In all cases (32–34), the local conformational changes caused by the amide linkage were easily compensated by small adjustments in the RNA backbone, which suggested that amides may be promising modifications for optimization of siRNAs.
Figure 1.
Structure of amide (AM1) linkage and sequences of amide-modified siRNA guide strands. # G8 and G18 were not part of this study; the data for G8 and G18 are from our previous study (33).
Preliminary results on RNAi activity of amide-modified siRNAs were encouraging. Iwase et al. showed that modification of the 3′-overhangs of an siRNA with two AM1 linkages increased the enzymatic stability but did not significantly alter RNAi activity (35–37). Most recently, we reported initial results that AM1 linkages were tolerated at some internal positions of both guide and passenger strands of siRNAs (33). Most interestingly, AM1 modification increased the silencing activity when placed near the 5′-end of the passenger strand. As a result, an siRNA containing eight amide linkages was more active than the unmodified control (33). In another study (34), we showed that three consecutive amide linkages near the 5′-end of the passenger strand caused little change in siRNA activity. Herein we report a systematic study of how a single amide linkage affects silencing activity when placed across positions 1 through 18 of the guide strand of siRNAs. Our results show that amides are well tolerated in the seed and central regions of the guide strand and actually increase the silencing activity when placed in the middle of the guide strand, between nucleosides 10 and 11 and between nucleosides 11 and 12, at the catalytic site of Argonaute. We also report the first crystal structure of an amide modified RNA–DNA substrate with Bacillus halodurans RNase H1 (BhRNase H) that reveals how small changes in both RNA and protein conformation allow the amide moiety to establish new modes of hydrogen bonding interactions with the protein. Taken together with our earlier studies (31–34), the present data suggest that amides are not only excellent structural mimics of the phosphate backbone in RNA, but may also mimic functionally important hydrogen bonding interactions with proteins required for activation of the RNAi machinery. Our study shows that amides are promising backbone modifications for optimization of biological properties of siRNAs for in vivo applications.
MATERIALS AND METHODS
Synthesis of amide-modified RNA
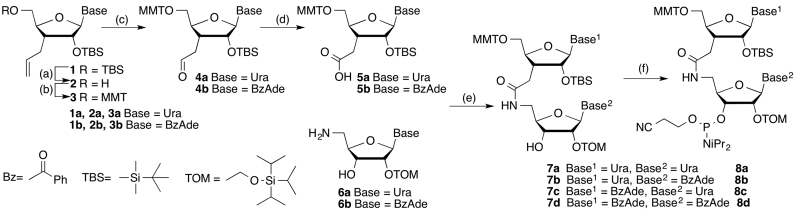
Amide-modified oligoribonucleotides were synthesized on a 1 μmol scale using the standard 2′-O-TOM RNA phosphoramidite (Glen Research) synthesis protocol on an Expedite 8909 Nucleic Acid Synthesis System and purified by reverse-phase HPLC as previously reported (33) (Supplementary Table S2 and Figures S4–S26). The UU (8a) and UA (8b) dimer phosphoramidates were synthesized as previously reported (32,33); the AU (8c) and AA (8d) were prepared using similar procedures (Scheme 1, for experimental details, see Supplementary Information). All modified guide strands were chemically 5′-phosphorylated to make sure that amide modifications do not interfere with enzymatic phosphorylation normally observed in cells.
Scheme 1.
Synthesis of dimeric r(UAM1U), r(UAM1A), r(AAM1A) and r(AAM1U) phosphoramidites.a. a Steps: (A) trifluoroacetic acid, THF, H2O, 0°C, 4 h, 92% (2a) and 92% (2b); (B) 4-methoxytrityl chloride, pyridine, 0°C to rt, 14 h, 85% (3a) and 90% (3b); (C) OsO4, 4-methylmorpholine N-oxide, dioxane, rt, 10 h, then NaIO4 in water, rt, 12 h, 95% (4a) and 98% (4b); (D) NaClO4, NaH2PO4, 2-methylbut-2-ene, t-BuOH, THF water, rt, 1 h, 82% (5a) and 92% (5b); (E) O-benzotriazole-N,N,N′,N′-tetramethyluroniumhexafluoro phosphate (HBTU), 1-hydroxybenzotriazole (HOBt), diisopropylethylamine, CH2Cl2, rt, 12 h, 89% (7c) and 97% (7d); (F) P(OCH2CH2CN)(N(iPr)2)2, tetrazole, CH2Cl2, rt, 7 h, 76%, (8c) and 85% (8d).
siRNA activity test
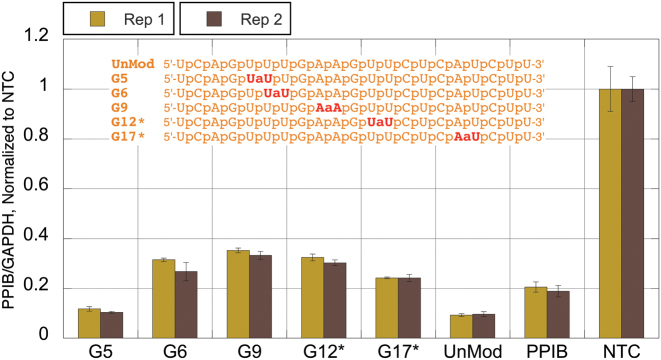
HeLa cells were plated in 96-well plates (1 × 104 cells/well) for 24 h before transfection. On the day of transfection, RNA–lipid complexes were introduced into each well of cells (1–100 nM siRNA duplexes formed by mixing the guide and passenger strands, 0.2 μl/well DharmaFECT 1 transfection reagent, GE Healthcare Dharmacon, catalogue# T-2001-01). siGENOME Non-Targeting siRNA #1 [guide strand 5′-r(UAGCGACUAAACACAUCAAUU)-3′ and passenger strand 5′-r(UUGAUGUGUUUAGUCGCUAUU)-3′, GE Healthcare Dharmacon, catalogue# D-001210-01] was used as a non-targeting control (NTC) and siGENOME Cyclophilin B [PPIB, guide strand 5′-r(GGAAAGACUGUUCCAAAAAUU)-3′ and passenger strand 5′-r(UUUUUGGAACAGUCUUUCCUU)-3′, GE Healthcare Dharmacon, catalogue# D-001136-01-05] was used as a positive control for transfection efficiency. Unmodified siRNA controls with sequences matching those of siRNAs with amide linkages were custom synthesized at GE Healthcare Dharmacon, and conclusions were based on their relative activities. NTC and siRNAs targeting the PPIB gene were both titrated separately at the same concentrations to show that they behave in the same way. Forty-eight hours post-transfection, the level of target knockdown was assessed using a branched DNA (bDNA) assay specific for the targets of interest according to the manufacturer's instructions (QuantiGene branched DNA signal amplification kit; Panomics). Cells were lysed and directly added to the bDNA assay plate without nucleic acid purification, cDNA synthesis steps nor amplification that can introduce bias. Experiments were performed 2–4 times; the results presented in Figures 2–5 are expressed as ratios of target PPIB mRNA expression and reference mRNA expression (GAPDH) and are averages of biological triplicates from three separate wells of cells and two replicate experimental plates.
Figure 2.
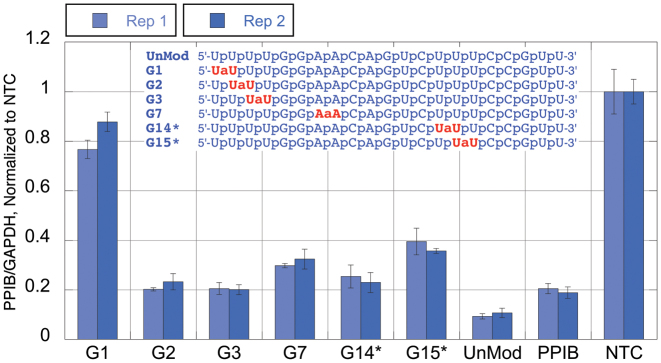
Silencing activity of amide-modified siRNAs. Duplexed siRNAs, formed from the respective guide strands 5′-UUUUGGAACAGUCUUUCCGUU-3′ (blue sequences in Figure 1) and the unmodified passenger strand 5′-CGGAAAGACUGUUCCAAAAUU-3′, at 1 nM final concentration were transfected in HeLa cells using DharmaFECT 1 transfection reagent for 48 h. The level of knockdown was assessed using a branched DNA (bDNA) assay (QuantiGene branched DNA signal amplification kit; Panomics). UnMod = unmodified, matched sequence siRNA, PPIB = transfection control siRNA targeting PPIB (different target sequence), NTC = non-targeting control siRNA.
Figure 5.
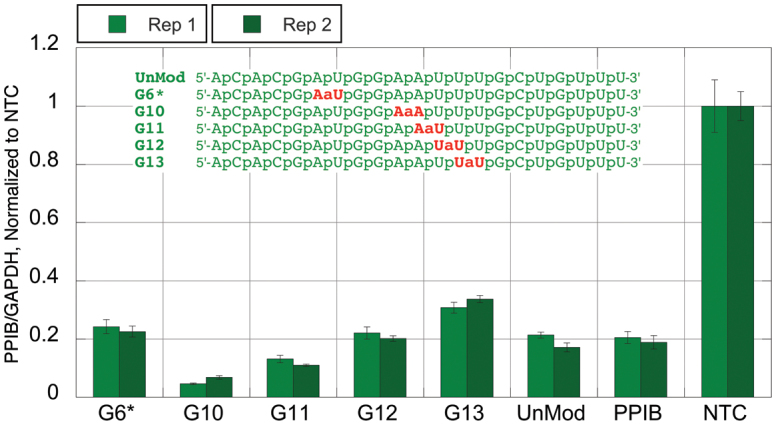
Silencing activity of amide-modified siRNAs. Duplexed siRNAs, formed from the respective guide strands 5′-ACACGAUGGAAUUUGCUGUUU-3′ (green sequences in Figure 1) and the unmodified passenger strand 5′-ACAGCAAAUUCCAUCGUGUUU-3′, at 1 nM final concentration were transfected in HeLa cells using DharmaFECT 1 transfection reagent for 48 h. The level of knockdown was assessed using a branched DNA (bDNA) assay (QuantiGene branched DNA signal amplification kit; Panomics). UnMod = unmodified, matched sequence siRNA, PPIB = transfection control siRNA targeting PPIB (different target sequence), NTC = non-targeting control siRNA.
Protein expression and purification
BhRNase H (D132N mutant; residues M58 to K196) was expressed in Escherichia coli and purified as previously reported (38).
Crystallization experiments
Solutions of BhRNase H and the duplex 5′-r(GACACCUGAUaUC)-3′ / 5′-d(GAATCAGGTGTC)-3′ were mixed in a 1:2 molar ratio in the presence of 5 mM MgCl2 and crystallization experiments were performed by the sitting drop vapor diffusion technique, using the Crystal Screen conditions (Hampton Research, Aliso Viejo, CA, USA) (39). Complex solution (200 nl) was mixed with an equal amount of reservoir solution and equilibrated against 60 mL reservoir well conditions using a Mosquito crystallization robot (TTP LabTech). Crystallization hits were then used to set up 2 ml sitting drops at 18°C. Crystals used in the diffraction experiments appeared in a crystallization condition containing 100 mM sodium cacodylate (pH 6.5) and 1.4 M sodium acetate. Crystals were mounted in nylon loops, cryo-protected in reservoir solution supplemented with 30% (w/v) glycerol and frozen in liquid nitrogen.
X-ray data collection, structure determination and refinement
Diffraction data were collected at a wavelength of 0.97856 Å with a MAR 300 CCD detector on the 21-ID-G beam line of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL, USA). Diffraction data were integrated and scaled with the program HKL2000 (40). The structure was phased with the Molecular Replacement approach using the program MOLREP (41,42) and the protein coordinates of the BhRNase H complex with PDB ID 1ZBI as the search model. Following initial refinement using the program REFMAC (43), and manual rebuilding of some protein residues in COOT (44), the amide linkage was inserted between uridine residues near the 3′- end of the RNA and the dictionary updated. Water molecules were then added gradually and isotropic refinement was continued with the program REFMAC. As the refinement proceeded, Mg2+ ions, acetate and glycerol molecules were added. A summary of crystal data, data collection parameters and crystallographic refinement statistics is provided in Supplementary Table S1. All crystallographic figures were generated with the program UCSF Chimera (45).
Data deposition
Atomic coordinates and structure factor data for the BhRNase H complex with amide-modified RNA:DNA have been deposited in the Protein Data Bank (http://www.rcsb.org, entry code 5VAJ).
NMR
For NMR studies, BhRNase H with uniform incorporation of 13C and 15N isotopic labels was expressed in minimal media containing 13C-glucose and 15N NH4Cl using the same plasmid as in preparation of protein for crystal growth. Purified 13C/15N-labeled protein was concentrated and exchanged into buffer containing 150 mM NaCl, 40 mM phosphate (pH 6.9) and 0.02% sodium azide for NMR studies at 25 °C. Backbone assignments of 118 of 126 residues in free BhRNase H (100 μM) were accomplished with HNCACB, CBCA(CO)NH and HNCO spectra acquired at 800 MHz with a 1H(13C,15N) cryo-probe. 1H–15N HSQC spectra of free BhRNase H (55 μM) and with added RNA/DNA were acquired at 600 MHz with a standard probe.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out in explicit solvent using AMBER 14 (ff14SB and parm10). Initial coordinates of the BhRNase H complex with modified RNA–DNA were taken from the X-ray structure described in this work. The initial coordinates of the full-length human Argonaute-2 (hAgo2) complexes with unmodified miRNA-20a 5′-(rUAAAGUGCUUCAGG)-3′ were taken from the structure described by Elkayam et al. (46) after removal of the PAZ domain and parts of the N domain. To obtain the starting structure of the complex with amide-modified RNA, the coordinates at each amide modification were determined by fitting the sugar rings of a dinucleotide extracted from the dominant conformation of a modified, unbound RNA duplex (32,33) to the sugar rings adjacent to the desired modification site in the Ago2 complex. The protein/nucleic acid complexes were surrounded with 6 Å of TIP3P water molecules in an octahedral box. Ions were added to neutralize the system, then Na+ and Cl− ions were added to a concentration of ∼0.1 M. All systems were subjected to two stages of minimization. In the first stage, all atoms in the complex were restrained while water, ions, and, in the case of Ago2 complexes with amide-modifications, the two nucleotide residues flanking the amide modification were minimized. In the second stage, protein residues that were distant from nucleic acid were weakly restrained (Supplementary Figure S1) while the rest of the system was minimized. During the first 20 ps of MD the system was heated to 298K. Then constant pressure dynamics was initiated at a pressure of 1 atm and a relaxation time of 2 ps. Temperature regulation was done with Langevin dynamics with a collision frequency of 1 ps−1. Bonds involving hydrogen atoms were constrained (SHAKE) and the time step was 2 fs. Distances, torsions, and RMSDs were calculated using the cpptraj module of the Amber software package. Partial charges for the amide-modification were derived using the restrained electrostatic potential (RESP) approach (47) based on an electrostatic potential grid calculated on a UaA dimer at the HF/6-31G* level using Gaussian 09, revision D.01.
RESULTS
Synthesis of amide-modified RNA
Synthesis of amide-linked UU, UA, AU and AA dimers and their phosphoramidates (Scheme 1) was done following our previously published procedures (33). Amide-linked dimeric phosphoramidites were used together with common 2′-O-TOM protected ribonucleoside phosphoramidites (Glen Research) to synthesize a series of amide-modified RNAs following the standard phosphoramidite chemistry protocol on an Expedite 8909 instrument. Using the Dharmacon bioinformatics database, we selected four siRNA sequences targeting PPIB gene that allowed a systematic replacement of a single phosphate with an AM1 linkage throughout positions 1 to 18 (G1–G18, Figure 1) of the guide strands using only the four amide-linked UU, UA, AA and AU dimers. The choice was determined by our experience that dimers built of U and A are easier to synthesize than dimers including C and G. PPIB is a common experimental reference because it is a highly expressed housekeeping gene. Knockdown of PPIB typically does not cause a negative phenotype and the cells maintain viability. The sequences allowed for double checking the effect of an amide linkage at several positions using different dimers, for example, UU in G2 and AA in G2*. The additional sequences (marked with *, e.g., G2*, G6*, etc.) allowed checking for potentially sequence-specific effects of amide modifications. We did not study G19 and G20 because Iwase and co-workers had previously shown that replacement of the siRNA overhang phosphates with AM1 linkages caused little change in the silencing activity of an siRNA (35,37). In our previous study (33) we showed that an amide at G8 decreased and at G18 did not change the silencing activity of the siRNA.
The effect of amide modifications in the guide strand on RNA interference activity
The relative activity of the siRNA duplexes formed by mixing the guide and passenger strands to knockdown the target sequence was initially performed in HeLa cells in a dose-dependent manner (1, 10 and 100 nM final concentration). To observe differences among the siRNAs we used a 1 nM final concentration as the other concentrations (10 and 100 nM) resulted in typically >95% knockdown (data not shown) and subtle changes in activity could not be distinguished. siRNA activity was compared to knockdown of the corresponding unmodified version of the same siRNA sequence containing the amide modification. Data were normalized to the siGenome non-targeting negative control, and the siGenome positive control siRNA targeting PPIB was included as a transfection and assay control. Further, experiments were performed on two separate plates (Rep1 and Rep 2) to show reproducibility of the data. The standard deviations are shown in the length of the error bars. Based on its interactions with Ago2 and functional roles, the guide strand of siRNAs can be categorized into five regions: anchor (nucleotide 1), seed (nucleotides 2–8), central (nucleotides 9–12), 3′-supplementary (nucleotides 13–17) and 3′-tail (nucleotides 18–21) (48). When discussing the results, we consider the amide modifications in the structural and functional context of these five regions of the guide strand.
Blue sequence
The unmodified blue siRNA (UnMod, Figure 2) was highly active producing ∼90% knockdown of PPIB. Amide modification in G1 (the anchor region) was not tolerated and almost completely abolished silencing. Surprisingly, amide modifications in the seed region were well tolerated in this siRNA. While G2, G3 and G7 showed notable loss of activity, these siRNAs still produced ∼70-80% knockdown of PPIB. G14* and G15*, modified in the supplementary region, showed similar trends.
Black sequence
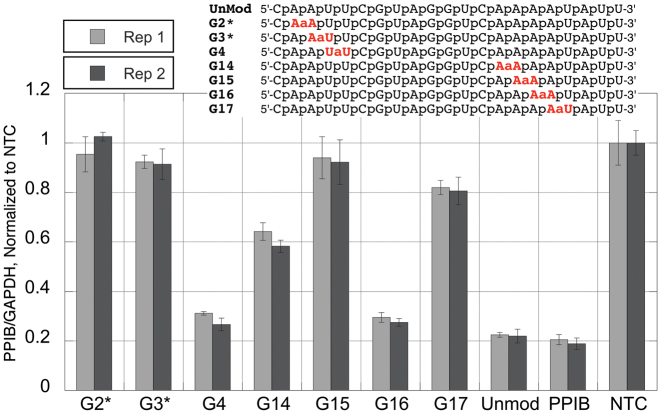
The unmodified black siRNA (UnMod, Figure 3) had good activity that was somewhat lower than that of the blue sequence, producing close to 80% knockdown of PPIB. In contrast to G2 and G3 in the blue sequence, G2* and G3* were not active suggesting that there is sequence dependence of amide tolerance. In a similar way, G14 and G15 in the black sequence were less active than G14* and G15* in the blue sequence (see Figure 2). Despite the low overall activity of amide modified black sequences, G4, having an amide linkage right in the middle of the seed region, and G16, having an amide linkage close to the 3′-end of the supplementary region, showed minimal loss of activity compared to the unmodified siRNA (Figure 3). Overall, the amide modifications in the black sequence were notably less tolerated compared to other siRNA sequences. This supports the notion that the effects of the amide linkages may depend not only on position, but also on the overall sequence and properties of the specific siRNA.
Figure 3.
Silencing activity of amide-modified siRNAs. Duplexed siRNAs, formed from the respective guide strands 5′-CAAUUCGUAGGUCAAAAUAUU-3′ (black sequences in Figure 1) and the unmodified passenger strand 5′-UAUUUUGACCUACGAAUUGUU-3′, at 1 nM final concentration were transfected in HeLa cells using DharmaFECT 1 transfection reagent for 48 h. The level of knockdown was assessed using a branched DNA (bDNA) assay (QuantiGene branched DNA signal amplification kit; Panomics). UnMod = unmodified, matched sequence siRNA, PPIB = transfection control siRNA targeting PPIB (different target sequence), NTC = non-targeting control siRNA.
Brown sequence
The unmodified brown siRNA (UnMod, Figure 4) was highly active producing >90% knockdown of PPIB. Consistent with the trend emerging in the blue and black sequences above, G5, with an amide modification in the seed region was well tolerated; however, G6 showed some loss of activity. G9 and G12*, the first and the last positions of the central region, were active albeit showing some loss in silencing compared to the unmodified siRNA. It is interesting to note that G12* having an amide linkage not far from the cleavage site was active. G17* that has an amide linkage close to the 3′-end of the guide showed good activity, although less than the unmodified siRNA, as expected from the general trend that the 3′-part of the guide strand is more tolerant to chemical modifications (5–7). Overall, the activity trends in the brown sequence were similar to those observed in the blue sequence.
Figure 4.
Silencing activity of amide-modified siRNAs. Duplexed siRNAs, formed from the respective guide strands 5′-UCAGUUUGAAGUUCUCAUCUU-3′ (brown sequences in Figure 1) and the unmodified passenger strand 5′-GAUGAGAACUUCAAACUGAUU-3′, at 1 nM final concentration were transfected in HeLa cells using DharmaFECT 1 transfection reagent for 48 h. The level of knockdown was assessed using a branched DNA (bDNA) assay (QuantiGene branched DNA signal amplification kit; Panomics). UnMod = unmodified, matched sequence siRNA, PPIB = transfection control siRNA targeting PPIB (different target sequence), NTC = non-targeting control siRNA.
Green sequence
The unmodified green siRNA (UnMod, Figure 5) had good activity and afforded close to 80% knockdown of PPIB, similar to the black sequence, but somewhat lower than those observed for the blue and brown sequences. In contrast to the brown sequence (Figure 4), G6* showed minimal loss of activity. Most surprisingly, G10, G11 and G12 with amide modifications in the central region close to the catalytic site of Argonaute were highly active. In fact, G10 and G11 were more active than the unmodified control. Moving the amide linkage further to the supplemental region, as in G13, resulted in some loss of activity.
X-ray crystal structure of amide-modified RNA–DNA in complex with RNase H
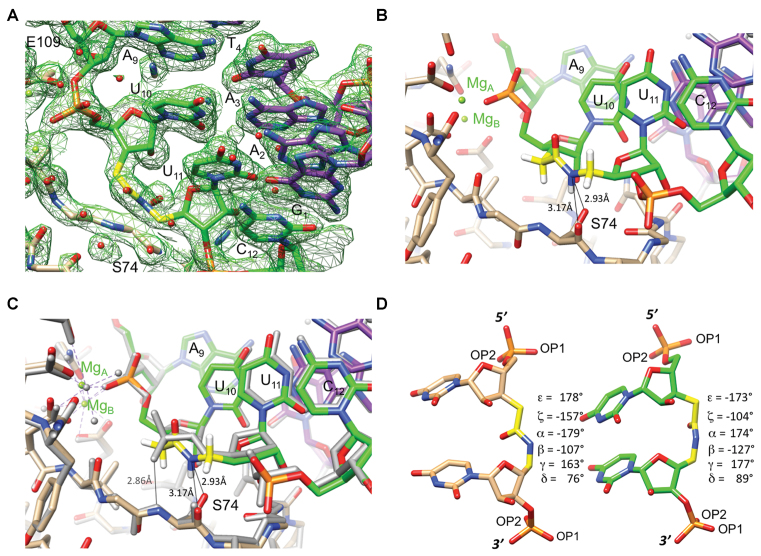
The silencing activity data in Figures 2–5 showed that amides were surprisingly well tolerated at some positions of the guide strand while causing almost complete loss of activity at others. Such large differences were somewhat surprising, given that almost all phosphates of the guide strand are involved in contacts to Ago2. It is possible that amides are able to maintain the interactions to the protein at some positions, whereas they are lost at others. To gain structural insight into how an amide linkage may mimic interactions of a phosphodiester linkage in protein-RNA complexes, we determined the 1.95 Å resolution crystal structure of the complex between BhRNase H and the r(GAC ACC UGA UaUC) - d(GAA TCA GGT GTC) hybrid duplex with a single AM1 linkage (UaU) in the RNA strand. BhRNase H was chosen as a model protein because it features a structural fold similar to the PIWI domain of Ago2 that harbors the endonuclease activity and crystal structures of its complexes with hybrid duplexes have been studied in detail (38). The BhRNase H enzyme employed contains the D132N mutation to prevent cleavage of the RNA strand in the crystallization drops in the presence of Mg2+. The structure is isomorphous to that of the complex between D132N BhRNase H and the native RNA–DNA hybrid [(38) PDB ID 1ZBI], with the asymmetric unit consisting of two protein molecules bound to a single duplex. An example of the quality of the final electron density is depicted in Figure 6A.
Figure 6.
Crystal structure of the complex between BhRNase H and the hybrid duplex r(GAC ACC UGA UaUC)–d(GAA TCA GGT GTC). (A) Quality of the final sum (2Fo – Fc) Fourier electron density (1 σ threshold) in the region around the amide-modified U10aU11 dimer step in the RNA strand. (B) A portion of the RNA strand viewed down the stack of bases from the 3′-end, with RNase H straddling the backbone and S74 engaging in two H-bonds to the amide nitrogen (thin lines; the position of the N(H) hydrogen is calculated). The active site is visible at the upper left and Mg2+ ions are green spheres with their coordination spheres indicated by dashed lines. Carbon atoms of RNA, AM1 linkage, DNA and protein are colored in green, yellow, purple and beige, respectively, H-bonds are indicated with thin lines, and selected residues are labeled. (C) Superimposition of the BhRNase H complexes with amide-modified RNA–DNA (identical to that seen in panel B) and the corresponding native RNA–DNA hybrid duplex (gray wire model). The view is identical to that in panel B. (D) Side-by-side comparison of the conformation of amide linkage in RNA/DNA bound to BhRNase H (left), and the conformation of amide linkage in protein-free RNA duplex from our previous study [right, (33) PDB ID 4O41]. The crystal structure of the latter RNA contains two independent duplexes per asymmetric unit, thus allowing observation of four independent UaU dimer steps. All four adopt a very similar conformation and only the dimer from strand A was used to produce the drawing shown here.
Both RNase H molecules contact the minor groove of the hybrid duplex and the amide modification sits adjacent to the scissile phosphate (Figure 6B). The amide nitrogen acts as a H-bond donor to the main chain oxygen and side chain Oγ of S74. The comparison with the structure of the native complex demonstrates that conformational changes between the two complexes are limited to the modification site. Thus, phosphate groups preceding (A9pU10) and succeeding (U11pC12) the amide that bridges residues U10 and U11 can be overlaid with minimal deviations and local base positions and stacking interactions are virtually unaffected by the replacement of the phosphate by amide (Figure 6C). As in the case of our recently reported (33) crystal structure of an RNA containing backbone amide modifications [PDB ID 4O41], the amide carbonyl bond in the modified RNA bound to RNase H assumes an orientation that mimics that of the P–OP2 bond (Figure 6C and D). Neither OP2 nor amide carbonyl oxygen participate in a direct interaction with a protein residue. In the structure of BhRNase H with the native substrate, the OP1 phosphate oxygen accepts a H-bond from the amide nitrogen of S74 (Figure 6C). The amide carbonyl, being aligned along the P–OP2 bond, is unable to mimic this interaction; instead, the amide N–H acts as a H-bond donor engaging in new stabilizing interactions with the main chain carbonyl oxygen and side chain Oγ of S74 (Figure 6B and C). As a result, both amide and phosphate enable stabilizing interactions between protein and RNA backbone. Importantly, the amide can serve as a H-bond donor creating new interactions between RNA and protein that are not available to native oligonucleotides.
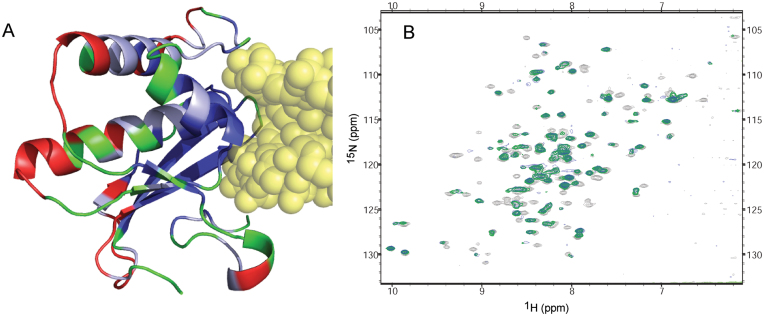
NMR data confirms that structure in crystal is maintained in solution
NMR spectroscopy provided further evidence that the structure of the amide-modified RNA/DNA/RNase H complex when in solution is similar to that observed in the crystal. BhRNase H residues that exhibit significant chemical shifts or loss of NMR signal when either amide-modified or unmodified RNA–DNA is added to the solution map closely with residues that are near the RNA–DNA in the X-ray structure (Figure 7A). Furthermore, a 2:1 complex of RNase H:RNA–DNA, as observed in the crystal, is indicated in solution by the fact that no further changes in the HSQC spectrum of RNase H are observed with RNA–DNA additions beyond half of one protein equivalent (Supplementary Figure S2). Finally, similarity of HSQC spectra of RNase H complexed with either unmodified or amide-modified RNA–DNA indicate that the amide-modification does not alter the binding mode in solution, in agreement with the crystal structures (Figure 7B).
Figure 7.
NMR studies of BhRNase H:RNA–DNA complex. (A) Crystal structure of BhRNase H in complex with amide-modified RNA–DNA (yellow), with protein residues colored according to chemical shift perturbation properties upon addition of RNA–DNA. Blue: residues whose NMR peaks disappear or are clearly shifted at least two line-widths due to added RNA–DNA. Light blue: peaks that shifted one to two line-widths. Red: peaks that shifted less than one line-width. Green: peaks where the shift is ambiguous (usually due to spectral crowding), or that were unassigned. (B) 15N-HSQC spectra of free BhRNase H (light grey), BhRNase H bound to unmodified RNA–DNA (green), and BhRNase H bound to amide-modified RNA–DNA (blue).
Molecular modeling of H-bonding interactions at amide-modified sites
The RNAi activity of amide-modified siRNAs may depend on the ability of amides to replace the interactions of Ago2 with phosphates of unmodified siRNA by similar or compensatory interactions, as illustrated above in the crystal structure of BhRNase H complex with amide-modified RNA–DNA (Figure 6). Molecular computation methods were used to examine the feasibility of compensatory interactions at modified sites. Since the complex of BhRNase H with amide-modified RNA–DNA provides the first detailed example of a protein-AM1 interaction, MD of this structure was carried out to see if the interactions present in the X-ray structure are maintained in the computational models. An MD trajectory in explicit solvent spanning 300 ns showed that the orientation of the AM1 linkage and interactions of the amide hydrogen with hydroxyl and carbonyl oxygen of a serine residue were maintained as observed in the crystal structure throughout the simulation (Supplementary Figure S3). This provides some confidence that simulations can correctly replicate, at least at short range, interactions of the amide-modification.
For the molecular modeling study of amide–Ago2 interactions, the crystal structure of miRNA-20a bound to human Ago2 by Elkayam et al. (46) was used for starting coordinates. MD simulations were carried out with amide modifications at various sites in the seed region on this physiologically relevant guide strand. Separate RNA:Ago-2 models were generated for each phosphate-to-amide modification. At each modified site, interactions formed between protein and the carbonyl oxygen of the amide linkage were compared with protein-phosphate oxygen interactions observed in the crystal structure. In some cases, flexibility of both protein and the amide-modified linkage allowed phosphate oxygen interactions to be replaced by carbonyl oxygens, but in other cases interactions were not recovered for most of the simulation.
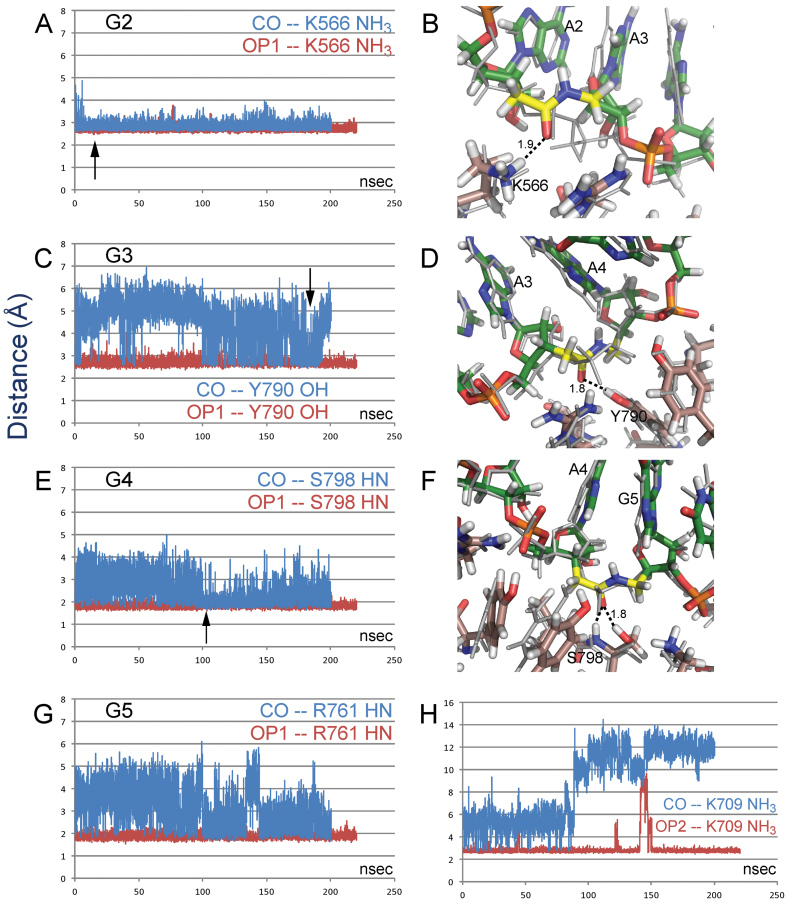
At the G2 site the initial orientation of the CO group was not suitable for an interaction with the side chain of lysine (K566) which interacts with OP1 in the unmodified complex. However, within 10 ns the amide reoriented to a conformation that allows the interaction to form and remain throughout the 200 ns simulation (Figure 8A). The recovery of the K566 interaction is consistent with biological results for this site in the blue sequence in Figure 2, but not the black sequence in Figure 3. One consequence of the reorientation of the G2 amide is an out-of-plane shift of the sugar moiety of the third residue (A3) which affects the position and orientation of the attached base (Figure 8B). This could affect the RNAi activity by changing the loading of the target strand.
Figure 8.
Distances between indicated interacting atoms in MD simulations of amide-modified (blue) and unmodified (red) RNA/hAgo-2 complexes. Interaction distances at: (A) site G2; (C) site G3; (E) site G4; (G, H) site G5. Representative frames show the region of the amide modification at (B) site G2; (D) site G3; (F) site G4. The crystal structure starting coordinates are shown as thin, gray lines. Carbon atoms of RNA, protein, and amide moiety are shown in green, copper and yellow, respectively. Arrows in the distance trajectories indicate time of the representative frames.
At the G3 site, as at the G2 site, the initial orientation of the amide CO group is not conducive to interactions with tyrosine (Y790) and arginine (R792) side chains that interact with OP1 in the unmodified complex. In contrast to the G2 site, the amide CO group of G3 is only transiently able to re-establish these interactions during the simulation (Figure 8C). More importantly, even for close interactions, the donor and acceptor are not well positioned for a hydrogen bond with the donor oriented at ca. 90° angle from the CO bond (Figure 8D). This result is consistent with the biological results for this site in the black sequence in Figure 3, but not the blue sequence in Figure 2. At both G2 and G3, no interactions with OP2 are present in the unmodified complex, and no interactions with the amide hydrogen are formed in the simulations.
At sites G4 and G5, both OP1 and OP2 interact with protein. Obviously, the amide modification, which offers just one carbonyl oxygen, is unlikely to replace all these interactions. In MD simulations of the G4 modification, the amide group reorients to place the carbonyl oxygen near the original OP1 position, and is eventually (∼100 ns) able to retain good geometry for the interaction with S798 HN and side chain HO, although there are substantial fluctuations and the amide linkage is slightly strained (Figure 8E and F). In addition, the amide hydrogen occasionally forms a weak interaction with the Y804 side chain, the original site for OP2. The recovery of these interactions is consistent with the high activity of the G4 modification in Figure 3. Displacement of adjacent sugar residues is less than at G2.
At G5 the interaction with a lysine side chain (K709) is unstable and the side chain moves far away. After losing the lysine side chain, the amide group reorients and recovers an interaction with R761 HN, formerly occupied by OP1. However, positions of residues in the 3′-direction appear to be affected by this switch (Figure 8G and H). Fluctuations in the OP2–K709 interaction were noted in the G4 simulations and even in simulations of the unmodified crystal structure.
DISCUSSION
RNAi has become a well-established and powerful tool for functional genomics (49). There is also the intriguing potential that RNAi may become a new therapeutic approach and currently more than 20 RNAi-based clinical trials are being pursued (50–52). While unmodified siRNAs perform well as research tools in cell cultures, in vivo applications require proper patterns of chemical modifications to insure potent and long lasting silencing activity (2–7). Chemical modifications of siRNAs must be compatible with the biochemistry of the RNA-degrading Ago2 enzyme that dictates precise requirements of the modification patterns. The purpose of this study was to systematically explore how Ago2 tolerates replacement of the phosphate with an amide linkage in the guide strand of siRNAs. We hypothesized that because of the similar orientation of the amide carbonyl and the OP2 non-bridging phosphate oxygen, the amide C=O will be able to mimic hydrogen bonding interactions between Ago2 and OP2 oxygens.
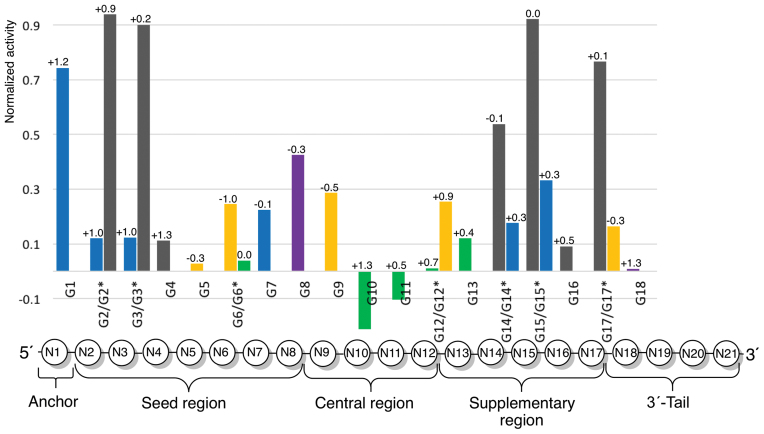
The conventional wisdom in the field is that the entire passenger strand and the 3′-end of the guide strand are relatively tolerant to chemical modifications, as long as the A-type RNA structure is not significantly distorted (5–7). In contrast, the 5′-end and central positions of the guide strand are sensitive, especially to multiple or bulky modifications (5–7). These notions are well supported by the structures determined for siRNA:Ago complexes (14,15,53–55). Of importance for replacement of phosphates with neutral amides is the observation that almost all phosphates of the guide strand are engaged in hydrogen bonding interactions to Ago residues in the crystal structures of complexes (13–16). Figure 9 presents a comparison of the effect on functionality of amide modification across the guide strand after normalization for the unmodified controls specific to each siRNA sequence (for sequences, see Figure 1). Contrary to the conventional wisdom, our data show that amide linkages are well tolerated in the middle of the seed region (G4 and G5) and that they can enhance the silencing activity when placed around the cleavage site (G10 and G11).
Figure 9.
Comparison of silencing activity across different siRNA sequences. The data present activity of the modified guide strand minus activity of the unmodified control divided by one minus activity of the unmodified control Y = (YGn – YUnMod)/(1 – YUnMod). After the normalization, a negative value indicates activity of amide-modified siRNA that is higher than that of unmodified siRNA, while ‘1’ indicates no activity (same as NTC). The data are color-coded as in Figure 1; violet data (G8 and G18) are from reference (33). N1, N2, …, N21 refer to nucleotides of the guide strand. The positions of amide linkages are numbered after the 5′-nucleotide, e.g. G1 has amide between N1 and N2. The number on top of the bar is the difference in melting temperature of the modified guide and unmodified control, Dtm = tm(modified guide) – tm(unmodified control).
The 5′-end phosphate and the first nucleotide (anchor in Figure 9) of the guide strand are docked in the MID domain and make essential interactions for recognition of the siRNA by Ago (15,53–55). The second phosphate that links the anchor with the seed region of the guide strand is kinked, thus assuming a non-canonical backbone conformation, and interacts with N551 and Q548 of the MID domain (15). Consequently, the 5′-end of the guide is sensitive to chemical modifications that may compromise siRNA binding by Ago2. Consistent with these observations, G1 showed reduced RNAi activity (Figure 2) and G2 and G3 were also sensitive, though only in the black sequence (Figure 3).
Nucleotides 2–8 constitute the seed region (Figure 9) that is responsible for initial recognition and guide-target duplex nucleation (48). The seed region, especially its central part, is highly sensitive to structural perturbation, such as mismatches and G–U wobble pairs (48). Surprisingly, amide modifications were very well tolerated in G4 and G5 and, while causing some reduction in activity, were not prohibitive in G6 and G7 (Figures 2–5). The high tolerance of amides in the seed region, especially, in the middle of that region, is contrary to the general perception that the seed region is highly sensitive to chemical modifications (5–7). MD simulations suggest that alternative conformations of the amide linkage are accessible and often allow recovery of some of the original interactions (e.g., G2 and G4). However, changes in the backbone which occur to accommodate the interaction may move the associated base (e.g. G2) and influence loading of the target strand. It is conceivable that such conformational changes around an amide linkage may be acceptable in some siRNA sequences, but cause decreased siRNA activity in others. Such an effect could help explain the different sensitivities of G2 and G3 positions in the blue and black sequences. While the MD calculations of amide-modifications in the seed region do not provide clear explanations of the siRNA activity results, the apparent availability of multiple conformations of amide linkages in RNA bound to Ago2 do not seem to be present in the complex with BhRNaseH where MD shows only a single, well-defined conformation in agreement with the crystal structure. Furthermore, as the simulations of the unmodified RNA:Ago2 complex reproduce the starting point crystal structure, it is possible that the simulations of the modified complex have simply not been carried out for sufficiently long periods to provide converged results of these as yet unknown structures.
The ability to introduce amide modifications in the seed region may be useful for reduction of miRNA-like off-target effects. Previous studies have shown that some seed modifications, for example, 2′-OMe at position 2 of the guide strand (56), replacement of the entire seed ribonucleotides with deoxynucleotides (57) and the strongly destabilizing unlocked nucleic acid at position 7 of the guide strand (58,59) reduced the off-target silencing due to miRNA-like activity. In our previous studies, amide modifications slightly reduced the thermal stability of RNA duplexes (32,34). Since the reduction of duplex thermal stability of the seed region also correlates with reduced miRNA-like off-target effects (57,60,61), it will be interesting to explore the effect of amide modifications in the seed region on siRNA sequence specificity.
The fact that amide modification decreased silencing activity in G6 through G9 may be related to kinks observed in crystal structures of Ago with RNA guide strand between nucleotides 6 and 7 (16,55,62,63) and nucleotides 9 and 10 (16). Upon binding of the target RNA, Ago undergoes conformational change, thus relaxing the kinks and avoiding steric clashes with the guide-target duplex. Gong and Desaulniers (64) reported that an amide linkage derived from insertion of a peptide nucleic acid monomer at position equivalent to G9 led to significant loss of silencing activity. We speculate that the amide modification may interfere with the tight RNA:Ago2 interactions or conformational movements of Ago2 in this region, leading to higher sensitivity to amide modification in G6 through G9. Further structural studies will be required to clarify this hypothesis.
The central region (nucleotides 9–12 in Figure 9) tolerates chemical modifications that maintain proper RNA A-form geometry around the cleavage site (65). The fact that amides are very well tolerated in the central region is consistent with our earlier findings that amide linkages cause negligible changes in the A-type conformation of duplex of RNA (32,33). In crystal structures of guide-bound Argonautes, phosphates at positions G11 and G12 have not been found to engage in interactions with protein (13,62,66), which correlates with excellent activity of amide-modified G11 in the green sequence (Figure 5). Another explanation for the increased activity may be local base-pairing destabilization by the amide modification. Previous studies have found that highly functional siRNAs have decreased internal base-pairing stability at positions 9–14 (67). In another study, placing thermally destabilizing modifications, such as noncanonical bases like 2,4-difluorotoluene or base pair mismatches at positions 9–12 significantly improved the potency of siRNAs (68). Overall, the crystal structures of Argonautes with guide RNAs (15,16,55,62) are missing most of the nucleotides in the central and supplemental regions, which prevents correlation of effects of amide modification with structural features of siRNA:Ago2 complexes. Nevertheless, the fact that amide modification at G10 and G11 increased the silencing activity is unexpected and encouraging for future studies.
Crystal structures of Ago2 bound to siRNA-like substrates suggest that only nucleotides 2–16 of the guide strand base pair with the mRNA target (62,66), which supports the observations that the 3′-end of the guide strand is tolerant to chemical modifications as long as they do not interfere with anchoring of the 3′-overhang in the PAZ domain during the assembly of RNA induced silencing complex (RISC). It is suggested that upon RISC assembly and activation the 3′-end of the guide strand is released from the PAZ domain after binding the target mRNA (62,66). Overall, the high activity of G16 and G18 was consistent with this notion, while low activity of G17 (black sequence) and good activity of G17* (brown sequence) was consistent with the sequence dependency observed at other positions (vide infra).
The unmodified siRNAs all resulted in high functionality, with the unmodified blue and brown sequences demonstrating ∼90% silencing and the unmodified black and green sequences were slightly less active with ∼80% silencing. Comparison of siRNA activity normalized to the corresponding unmodified siRNA across the four sequences (Figure 9) suggested that results depend on the siRNA sequence, rather than on the specific nucleosides surrounding the amide linkage. Examining the specific nucleotide sequences shows that at several positions (G2/G2*, G3/G3*, G14/G14*, G15/G15*) AA and AU modified siRNAs had lower activity compared to UU modified siRNAs. However, this was not a general trend and the opposite was observed for G6/G6*, and significant differences in functionality were observed for G12/G12* and G17/G17* both having the same modified dimer: UU at G12/G12* and AU at G17/G17*. Overall, the varying activity at these positions correlates better with different tolerance of amides in the four siRNA sequences—the black siRNA was the most sensitive to amide modifications, while the green siRNA was the most tolerant. The blue and brown siRNAs showed intermediate tolerance. Such differences may be caused by factors other than interactions of amide with Argonaute at the modified linkages. For example, the various siRNAs may have different silencing efficiencies and strand selectivity of RISC loading, which may be further modulated by the amide modifications. To test that the observed differences in RNAi activity are not a result of changes in thermal stability caused by the amide modifications, we measured the melting temperatures of all siRNA duplexes (Supplementary Table S3); the results are presented as Δtm values on the top of bars in Figure 9. Consistent with our previous observations (32,33), single amide modification caused little change in thermal stability (±1.3°C, Figure 9) that had no correlation with the differences in RNAi activity. While more studies will be needed to fully reveal the sequence dependency of amide modifications in siRNAs, our current results that active siRNAs may tolerate amide modifications in the middle of the seed and central regions are encouraging.
CONCLUSIONS
Our comprehensive study of systematic replacement of a single phosphate with an amide linkage shows that amides are unexpectedly well tolerated in the seed and central regions of the guide strand and even increase the silencing activity in G10 and G11. This is unexpected because Argonaute makes extensive hydrogen bonding contacts to the phosphate backbone of the guide strand. Moreover, G10 and G11 are in the middle of the guide strand at the catalytic site of Argonaute. The crystal structure of an amide-modified RNA–DNA substrate with BhRNase H provides intriguing insights into how small changes in both RNA and protein conformation allow the amide moiety to establish alternative modes of hydrogen bonding interactions with the protein. MD simulations suggest that these conformational changes and alternative binding modes may compensate for interactions that are lost due to the absence of a phosphodiester bond. Taken together with earlier studies (31–34), our results suggest that amides are not only excellent structural mimics of the phosphate backbone in RNA, but may also mimic functionally important hydrogen bonding interactions with proteins required for activation of the RNAi machinery and hence may be promising modifications for optimization of the biological properties of siRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Kaizhang He (GE Healthcare Dharmacon) for helpful discussions and Jianbo Zhao (Univeristy of Rochester) for calculation of amide-modification partial charges.
Footnotes
Present address: Daniel Mutisya, Department of Chemistry, Albany State University, Albany, GA 31707, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Institutes of Health [R01 GM71461 to E.R.]. The 600 MHz instrument at Binghamton University is supported by the National Science Foundation [CHE-0922815 to E.R.] and the 800 MHz instrument at Syracuse University is supported by the National Institutes of Health [1S10 OD012254]. Funding for open access charge: National Institutes of Health [R01 GM71461 to E.R.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wan W.B., Seth P.P.. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016; 59:9645–9667. [DOI] [PubMed] [Google Scholar]
- 2. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017; 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watts J.K., Corey D.R.. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012; 226:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deleavey G.F., Damha M.J.. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012; 19:937–954. [DOI] [PubMed] [Google Scholar]
- 5. Bramsen J.B., Kjems J.. Methods Mol. Biol. 2013; 942, NY: Springer; 87–109. [DOI] [PubMed] [Google Scholar]
- 6. Bramsen J.B., Grunweller A., Hartmann R.K., Kjems J.. Hartmann RK, Schon ABA., Westhof E. Handbook of RNA Biochemistry. 2014; 2nd edn, Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 1243–1277. [Google Scholar]
- 7. Bramsen J.B., Kjems J.. Development of therapeutic-grade small interfering RNAs by chemical engineering. Front. Genet.: Non-Coding RNA. 2012; 3:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Latorre A., Latorre A., Somoza A.. Modified RNAs in CRISPR/Cas9: An old trick works again. Angew. Chem., Int. Ed. 2016; 55:3548–3550. [DOI] [PubMed] [Google Scholar]
- 9. Kelley M.L., Strezoska Z., He K., Vermeulen A., Smith A.v.B.. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol. 2016; 233:74–83. [DOI] [PubMed] [Google Scholar]
- 10. Eguchi A., Dowdy S.F.. siRNA delivery using peptide transduction domains. Trends Pharmacol. Sci. 2009; 30:341–345. [DOI] [PubMed] [Google Scholar]
- 11. Joshua-Tor L., Hannon G.J.. Ancestral roles of small RNAs: an ago-centric perspective. Cold Spring Harbor Perspect. Biol. 2011; 3:a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carthew R.W., Sontheimer E.J.. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elkayam E., Kuhn Claus D., Tocilj A., Haase Astrid D., Greene Emily M., Hannon Gregory J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L.. The making of a slicer: activation of human argonaute-1. Cell Rep. 2013; 3:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meade B.R., Gogoi K., Hamil A.S., Palm-Apergi C., Berg A.v.d., Hagopian J.C., Springer A.D., Eguchi A., Kacsinta A.D., Dowdy C.F. et al. . Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014; 32:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eckstein F. Phosphorothioates, Essential Components of Therapeutic Oligonucleotides. Nucleic Acid Ther. 2014; 24:374–387. [DOI] [PubMed] [Google Scholar]
- 19. Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A., Corey D.R.. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003; 42:7967–7975. [DOI] [PubMed] [Google Scholar]
- 20. Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K., Tuschl T.. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucl. Acid Drug Dev. 2003; 13:83–105. [DOI] [PubMed] [Google Scholar]
- 21. Amarzguioui M., Holen T., Babaie E., Prydz H.. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003; 31:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall A.H.S., Wan J., Shaughnessy E.E., Ramsay Shaw B., Alexander K.A.. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 2004; 32:5991–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X., Sierant M., Janicka M., Peczek L., Martinez C., Hassell T., Li N., Li X., Wang T., Nawrot B.. Gene silencing activity of siRNA molecules containing phosphorodithioate substitutions. ACS Chem. Biol. 2012; 7:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu S.Y., Yang X., Gharpure K.M., Hatakeyama H., Egli M., McGuire M.H., Nagaraja A.S., Miyake T.M., Rupaimoole R., Pecot C.V. et al. . 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumor activity. Nat. Commun. 2014; 5:4459–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Threlfall R.N., Torres A.G., Krivenko A., Gait M.J., Caruthers M.H.. Synthesis and biological activity of phosphonoacetate- and thiophosphonoacetate-modified 2′-O-methyl oligoribonucleotides. Org. Biomol. Chem. 2012; 10:746–754. [DOI] [PubMed] [Google Scholar]
- 26. Idziak I., Just G., Damha M.J., Giannaris P.A.. Synthesis and hybridization properties of amide-linked thymidine dimers incorporated into oligodeoxynucleotides. Tetrahedron Lett. 1993; 34:5417–5420. [Google Scholar]
- 27. De Mesmaeker A., Waldner A., Lebreton J., Hoffmann P., Fritsch V., Wolf R.M., Freier S.M.. Amides as a new type of backbone modifications in oligonucleotides. Angew. Chem., Int. Ed. Engl. 1994; 33:226–229. [Google Scholar]
- 28. Lebreton J., Waldner A., Lesueur C., De Mesmaeker A.. Antisense oligonucleotides with alternating phosphodiester-“amide-3” linkages. Synlett. 1994; 137–140. [Google Scholar]
- 29. De Mesmaeker A., Lesueur C., Bevierre M.O., Waldner A., Fritsch V., Wolf R.M.. Amide backbones with conformationally restricted furanose rings: highly improved affinity of the modified oligonucleotides for their RNA complements. Angew. Chem., Int. Ed. 1996; 35:2790–2794. [Google Scholar]
- 30. De Mesmaeker A., Lebreton J., Jouanno C., Fritsch V., Wolf R.M., Wendeborn S.. Amide-modified oligonucleotides with preorganized backbone and furanose rings. Highly increased thermodynamic stability of the duplexes formed with their RNA and DNA complements. Synlett. 1997; 1287–1290. [Google Scholar]
- 31. Rozners E., Katkevica D., Bizdena E., Strömberg R.. Synthesis and properties of RNA analogs having amides as interuridyl linkages at selected positions. J. Am. Chem. Soc. 2003; 125:12125–12136. [DOI] [PubMed] [Google Scholar]
- 32. Selvam C., Thomas S., Abbott J., Kennedy S.D., Rozners E.. Amides as excellent mimics of phosphate linkages in RNA. Angew. Chem., Int. Ed. 2011; 50:2068–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mutisya D., Selvam C., Lunstad B.D., Pallan P.S., Haas A., Leake D., Egli M., Rozners E.. Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014; 42:6542–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanui P., Kennedy S.D., Lunstad B.D., Haas A., Leake D., Rozners E.. Synthesis, biophysical studies and RNA interference activity of RNA having three consecutive amide linkages. Org. Biomol. Chem. 2014; 12:1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwase R., Toyama T., Nishimori K.. Solid-phase synthesis of modified RNAs containing amide-linked oligoribonucleosides at their 3′-end and their application to siRNA. Nucleosides, Nucleotides Nucleic Acids. 2007; 26:1451–1454. [DOI] [PubMed] [Google Scholar]
- 36. Iwase R., Kurokawa R., Ueno J.. Synthesis of modified double stranded RNAs containing duplex regions between amide-linked RNA and RNA at both ends and enhanced nuclease resistance. Nucleic Acids Symp. Ser. 2009; 53:119–120. [DOI] [PubMed] [Google Scholar]
- 37. Iwase R., Miyao H., Toyama T., Nishimori K.. Synthesis and properties of modified siRNA having amide-linked oligoribonucleosides at their 3′ overhang regions. Nucleic Acids Symp. Ser. 2006; 175–176. [DOI] [PubMed] [Google Scholar]
- 38. Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W.. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005; 121:1005–1016. [DOI] [PubMed] [Google Scholar]
- 39. Jancarik J., Kim S.H.. Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 1991; 24:409–411. [Google Scholar]
- 40. Otwinowski Z., Minor W.. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 41. Vagin A., Teplyakov A.. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997; 30:1022–1025. [Google Scholar]
- 42. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011; D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997; D53:240–255. [DOI] [PubMed] [Google Scholar]
- 44. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004; D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 45. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 46. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bayly C.I., Cieplak P., Cornell W., Kollman P.A.. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993; 97:10269–10280. [Google Scholar]
- 48. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tripp R.A., Karpilow J.M.. Frontiers in RNAi. 2014; Bentham Science. [Google Scholar]
- 50. Bobbin M.L., Rossi J.J.. RNA interference (RNAi)-based therapeutics: delivering on the promise?. Annu. Rev. Pharmacol. Toxicol. 2016; 56:103–122. [DOI] [PubMed] [Google Scholar]
- 51. Zuckerman J.E., Davis M.E.. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discovery. 2015; 14:843–856. [DOI] [PubMed] [Google Scholar]
- 52. Wittrup A., Lieberman J.. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015; 16:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parker J.S., Roe S.M., Barford D.. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005; 434:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J.. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005; 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. et al. . Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006; 12:1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ui-Tei K., Naito Y., Zenno S., Nishi K., Yamato K., Takahashi F., Juni A., Saigo K.. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008; 36:2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bramsen J.B., Pakula M.M., Hansen T.B., Bus C., Langkjaer N., Odadzic D., Smicius R., Wengel S.L., Chattopadhyaya J., Engels J.W. et al. . A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010; 38:5761–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vaish N., Chen F., Seth S., Fosnaugh K., Liu Y., Adami R., Brown T., Chen Y., Harvie P., Johns R. et al. . Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011; 39:1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ui-Tei K. Optimal choice of functional and off-target effect-reduced siRNAs for RNAi therapeutics. Front. Genet. 2013; 4:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gu S., Zhang Y., Jin L., Huang Y., Zhang F., Bassik M.C., Kampmann M., Kay M.A.. Weak base pairing in both seed and 3′ regions reduces RNAi off-targets and enhances si/shRNA designs. Nucleic Acids Res. 2014; 42:12169–12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nakanishi K., Ascano M., Gogakos T., Ishibe-Murakami S., Serganov A.A., Briskin D., Morozov P., Tuschl T., Patel D.J.. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013; 3:1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gong W., Desaulniers J.-P.. Gene-silencing properties of siRNAs that contain internal amide-bond linkages. Bioorg. Med. Chem. Lett. 2012; 22:6934–6937. [DOI] [PubMed] [Google Scholar]
- 65. Alagia A., Eritja R.. siRNA and RNAi optimization. Wiley Interdiscipl. Rev. RNA. 2016; 7:316–329. [DOI] [PubMed] [Google Scholar]
- 66. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 68. Addepalli H., Meena, Peng C.G., Wang G., Fan Y., Charisse K., Jayaprakash K.N., Rajeev K.G., Pandey R.K., Lavine G. et al. . Modulation of thermal stability can enhance the potency of siRNA. Nucleic Acids Res. 2010; 38:7320–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.