Abstract
Viruses often encode viral silencing suppressors (VSSs) to counteract the hosts’ RNA silencing activity. The cricket paralysis virus 1A protein (CrPV-1A) is a unique VSS that binds to a specific Argonaute protein (Ago)—the core of the RNA-induced silencing complex (RISC)—in insects to suppress its target cleavage reaction. However, the precise molecular mechanism of CrPV-1A action remains unclear. Here we utilized biochemical and single-molecule imaging approaches to analyze the effect of CrPV-1A during target recognition and cleavage by Drosophila Ago2-RISC. Our results suggest that CrPV-1A obstructs the initial target searching by Ago2-RISC via base pairing in the seed region. The combination of biochemistry and single-molecule imaging may help to pave the way for mechanistic understanding of VSSs with diverse functions.
INTRODUCTION
RNA silencing is a highly conserved mechanism that regulates gene expression in various biological processes. RNA silencing also plays an important role in antiviral defense, especially in insects and plants (1–4). Upon infection, RNA silencing is triggered by double-stranded (ds) replicative intermediates of RNA viruses and/or highly structured regions of viral RNAs, which are processed into 20–25-nt small interfering RNAs (siRNAs) by the ribonuclease III enzyme Dicer (Dcr-2 in insects and DICER-LIKE2/4 in plants) (2,3,5–7). The siRNAs are then incorporated into a member of Argonaute family proteins (Ago) to form RNA-induced silencing complex (RISC), which cleaves complementary target RNAs and thereby restricts viral replication (2,3).
In order to counteract the hosts’ RNA silencing, viruses often encode viral silencing suppressors (VSSs) [also known as viral suppressors of RNA silencing (VSRs)]. Numerous VSSs have been discovered in insect and plant viruses, which block the RNA silencing pathway in diverse mechanisms (1,3). For example, many VSSs such as tombusvirus p19, Drosophila C virus 1A (DCV-1A) and flock house virus B2 (FHV-B2) bind to long dsRNAs and/or siRNAs, thereby sequestering their processing by Dicer or their assembly into Ago proteins (8–18). On the other hand, some VSSs including cricket paralysis virus 1A (CrPV-1A) and nora virus VP1 (NV-VP1) directly interact with a specific Ago protein (fly Ago2 for CrPV-1A and NV-VP1) to block the target cleavage activity of RISC, without affecting dsRNA processing by Dicer or siRNA loading into Ago (19,20).
It is thought that the target cleavage reaction of RISC can be separated into at least four steps: (i) initial searching of potential targets via base pairing in the ‘seed’ region of the guide strand (positions 2–7 or 8), (ii) propagation of base pairs toward the 3′ direction, (iii) extensive base pairing leading to a conformational change of Ago to its cleavage-competent form and (iv) catalysis of target cleavage by Ago (21–26). Although previous studies have demonstrated that CrPV-1A and NV-VP1 inhibit the target cleavage reaction of pre-assembled fly Ago2-RISC (19,20), it remains unknown at which fundamental step(s) of the above pathway is targeted for suppression. Here, by utilizing biochemical and single-molecule imaging techniques, we analyzed the effect of CrPV-1A during target recognition and cleavage. Our results together show that CrPV-1A obstructs the initial target searching by Ago2-RISC via the seed base pairing. The combination of biochemistry and single-molecule imaging may help to pave the way for precise understanding of the mode of action of VSSs with diverse functions.
MATERIALS AND METHODS
Plasmid constructions
pCold II-CrPV-1A, DCV-1A and FHV-B2
DNA fragments coding CrPV-1A (AF218039), DCV-1A (NP044945) and FHV-B2 (P68831) were amplified from synthetic DNAs (Genscript) and then cloned into pCold II (Takara) by In-Fusion HD cloning kit (Clontech).
Preparation of RNAs
The 182-nt RNAs for the target cleavage assay with let-7, 5GC, 3GC1 and 3GC2 siRNAs (23) were in vitro transcribed using T7-Scribe Standard RNA IVT Kit (CELLSCRIPT) from PCR products that were amplified with pGL3-basic (Promega) as a template using primer pairs (5′-GCGTAATACGACTCACTATAGTCACATCTCATCTACCTCC-3′) plus (5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTTGAGTGTAGAACGGTTGTATAAAAGGTTGAGGTAGTAGGTTGTATAGTGAAGAGAGGAGTTCATGA-3′), (5′-GCGTAATACGACTCACTATAGTCACATCTCATCTACCTCC-3′) plus (5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTTGAGTGTAGAACGGTTGTATAAAAGGTTCCGCCCGTAGGTTGTATAGTGAAGAGAGGAGTTCATGA-3′), (5′-GCGTAATACGACTCACTATAGTCACATCTCATCTACCTCC-3′) plus (5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTTGAGTGTAGAACGGTTGTATAAAAGGTTGAGGTAGTAGCCGCTATAGTGAAGAGAGGAGTTCATGA-3′), (5′-GCGTAATACGACTCACTATAGTCACATCTCATCTACCTCC-3′) plus (5′-CCCATTTAGGTGACACTATAGATTTACATCGCGTTGAGTGTAGAACGGTTGTATAAAAGGTTGAGGTAGTAGCCGCCCTAGTGAAGAGAGGAGTTCATGA-3′), respectively. The RNAs were gel purified and cap-radiolabeled using ScriptCap m7G Capping System (CELLSCRIPT) and [α-32P]-GTP. Target binding assay was performed using the following synthetic 31-nt 2′-O-methylaed RNA oligos: perfect target (5′-UCUUCACUAUACAACCUACUACCUCAACCUU-3′) and seed-only target (5′-UCUUCUGAUAUGUUGGAUCUACCUCAACCUU-3′) (GeneDesign). The target RNAs were 5′ 32P-radiolabeled using T4 Polynucleotide Kinase (Takara) and [α-32P]-ATP. Single-molecule experiments were performed with a 5′-biotinized 3′-Cy3-labeled synthetic 59-nt RNA oligo (5′-BiotinTEG-GGGCAACAACAACAACAACAACAACAACAACAACAAACUAUACAACCUACUACCUCACA-Cy3–3′) (IDT). 3′-Cy5-labeled guide RNA was previously described (23).
Expression and purification of recombinant VSSs
N-terminally His-tagged recombinant proteins of CrPV-1A, DCV-1A and FHV-B2 were expressed in Escherichia coli Rosetta2 strain (Merck Millipore). Typically, the cells in 1-litre cultures with 100 μg/ml ampicillin were cultivated to an OD600 of 0.4–0.5 at 37°C and then grown at 15°C overnight with 1 mM isopropyl-β-d-thiogalactoside (IPTG). The cell pellets were resuspended in His A buffer (30 mM HEPES–KOH pH 7.4, 200 mM KOAc, 2 mM Mg(OAc)2, 5% glycerol, 20 mM imidazole–HCl (pH 7.5), 0.2 mM TCEP) containing 1 × EDTA-free protease inhibitor cocktail (Roche), sonicated and centrifuged at 10 000 × g for 20 min. The supernatant was loaded onto HisTrap FF crude 5 ml (GE Healthcare Life Sciences) and eluted with a linear gradient from His A buffer to His B buffer (His A buffer containing 400 mM imidazole) using AKTA purifier (GE Healthcare Life Sciences). The peak fractions were collected and buffer-exchanged to 30 mM HEPES–KOH (pH 7.4), 100 mM KOAc, 2 mM Mg(OAc)2, 10% glycerol, 1 mM DTT with PD-10 (GE Healthcare Life Sciences). All purified proteins were flash-frozen in liquid nitrogen and stored at –80°C. The concentrations of the proteins were measured by SDS-PAGE with defined dilutions of BSA as concentration standards after CBB staining of the gel.
Expression of FLAG-Ago2
N-terminal 3 × FLAG-tagged proteins were expressed from pAFW (Drosophila Gateway vector collection) vectors in S2 cells. The cell lysate was prepared as previously reported (27).
Immunopurification of FLAG-Ago2-RISC
Typically for five reactions, 20 μl Dynabeads Protein G (Invitrogen) were equilibrated with lysis buffer (30 mM HEPES–KOH (pH 7.4), 100 mM KOAc, 2 mM Mg(OAc)2), incubated with 0.67 μl of 1 mg/ml anti-FLAG M2 antibody (F1804, Sigma) for 30 min at 4°C, and washed twice with lysis buffer. In parallel, lysate from S2 cells expressing FLAG-Ago2 was incubated with 10 nM let-7 siRNA duplex, 1 mM ATP, 25 mM creatine monophosphate (Fluka), 0.03 U/μl creatine kinase (Calbiochem), 0.1 U/μl RNasin Plus (Promega), and lysis buffer in 10 μl for 30 min at 25°C. After RISC assembly, 10 μl of the reaction mixture was mixed with the anti-FLAG beads and incubated for 2 h at 4°C. Beads were washed five times with wash buffer (lysis buffer containing 800 mM NaCl, 1% Triton X-100), rinsed twice with lysis buffer, and then split into tubes for further analysis.
Target RNA cleavage assay in S2 cell lysate
Lysate from S2 cells was incubated with 50 nM let-7, 5GC, 3GC1 or 3GC2 siRNA duplex (23), 1 mM ATP, 25 mM creatine monophosphate (Fluka), 0.03 U/μl creatine kinase (Calbiochem), 0.1 U/μl RNasin Plus (Promega), and lysis buffer in 10 μl for 30 min at 25°C. After RISC assembly, 0.35 μM recombinant VSSs were added and incubated for 10 min at 25°C. Then, 1 μl of 10 nM cap-radiolabeled target RNA (182-nt) was added to the reaction and incubated at 25°C for 10 min. Target RNAs were analyzed on 8% urea gel, as described previously (28). Gels were dried and imaged by Typhoon FLA 7000 (GE Healthcare Life Sciences). For Supplementary Figure S2, the reactions were diluted twice with lysis buffer after RISC assembly. Then, the indicated concentrations of CrPV-1A were added and incubated for 10 min at 25°C. Finally, 0.25 nM cap-radiolabeled target RNAs were added and incubated for 20 min at 25°C.
Target RNA binding and cleavage assay with immunopurified FLAG-Ago2
After immunopurification of let-7 programmed FLAG-Ago2-RISC, recombinant VSSs were added in lysis buffer at the indicated concentrations and incubated for 10 min at 25°C. For the cleavage assays, 1 μl of 10 nM cap-radiolabeled target RNA (182-nt) was added to the reaction and incubated at 25°C for 10 min. For the binding assays, 1 μl of 10 nM radiolabeled 2′-O-methylated target RNA (31-nt) was added to the reaction and incubated at 25°C for 30 min, followed by three times wash with lysis buffer containing 0.1% Triton X-100. Purification and detection of target RNAs were performed as described above.
Single-molecule imaging
Flow chambers in which Cy3-labeled 1–5 nM target RNAs were anchored on polyethylene glycol (PEG)-coated quartz slides were prepared and single-molecule images were visualized as described previously (23,29–31). Typically, lysate from S2 cells overexpressing FLAG-Ago2 was first programmed with 50 nM let-7 siRNA with Cy5-labeled guide RNA for 30 min. The mixture was then 5× diluted by naive S2 lysate and 20 μl of it was infused into the flow chamber under the O2 scavenger system (32). The Cy3 and Cy5 dyes were illuminated and images were acquired as described previously (23). Images were taken at a frame rate of 1 frame/s, and the observations were performed at 22 ± 1°C. Single-molecule images were visualized by a total internal reflection fluorescence (TIRF) microscope as previously described (23). The images were pseudo-colored in green for Cy3-labaled target and in red for Cy5-labaled Ago2-RISC in Figure 3C.
Figure 3.
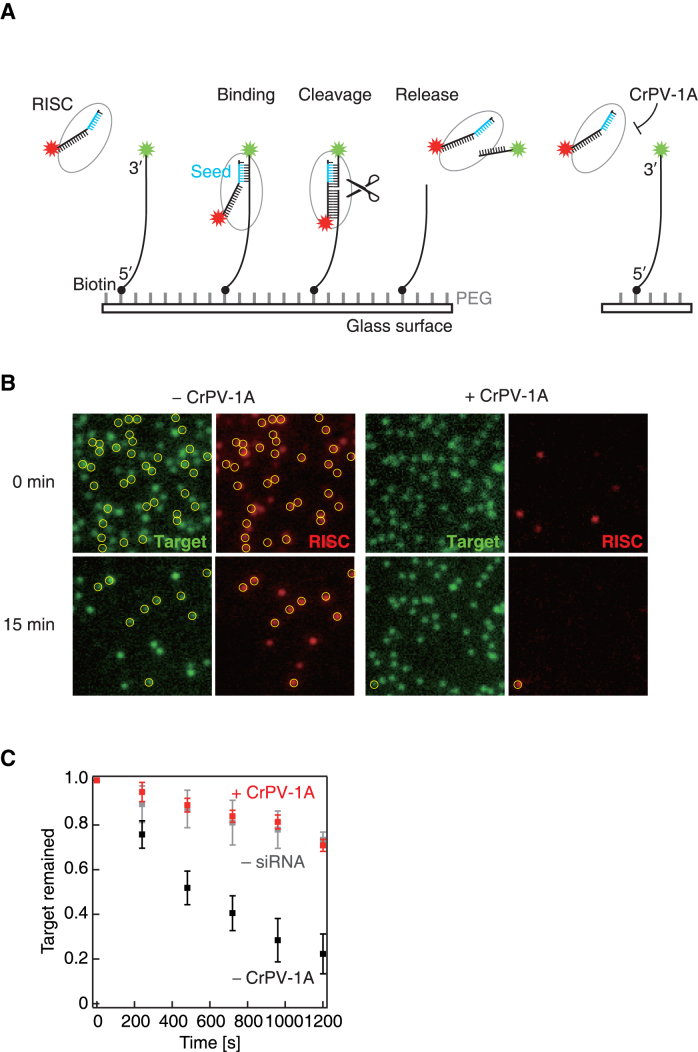
Single-molecule imaging of the target cleavage reaction. (A) Schematic representation of the single-molecule imaging. 3′-Cy3-labeled target RNA (green target) was tethered on the glass surface and incubated with S2 cell lysate containing Ago2-RISC with 3′-Cy5-labeled guide strand (red RISC) in the presence or absence of CrPV-1A. (B) Time-lapse single-molecule imaging of target cleavage with or without CrPV-1A. Yellow circles denote co-localization of the green target and the red RISC spots. (C) Quantification of the time-lapse single-molecule experiment. The number of green spots decreased remarkably in the absence of CrPV-1A (– CrPV-1A), but not in the presence of CrPV-1A (+ CrPV-1A) or without RISC assembly (– siRNA). Error bars indicate the SD from three independent experiments.
Data analysis
Images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/) as previously describe (29). The fluorescent intensity of the spots was measured using 6-pixel-diameter circular regions of interest (ROIs). For Figure 4, many green target spots were first picked up randomly, then the behavior of each spot was analyzed in an unbiased manner. After obtaining all the traces, those that do not match either of the three categories (e.g. disappearance of the green target spot without any co-localization of the red RISC spot—this presumably reflects either photobleaching of the green dye during observation or a contamination of the unlabeled guide strand in the chemical synthesis) were excluded from the analysis. Therefore, the three categories do not sum up to 100% in Figure 4. To determine the first binding time of Ago2-RISC in Figure 5, data were fitted by non-linear least-squares to the following equation: C1 × (1 – exp(–t/C2)), where C1 is a normalized parameter, C2 is the time for the first binding for Figure 5. All graphs and fitting curves were generated using KaleidaGraph (Synergy Software) or Igor Pro (Wavemetrics).
Figure 4.
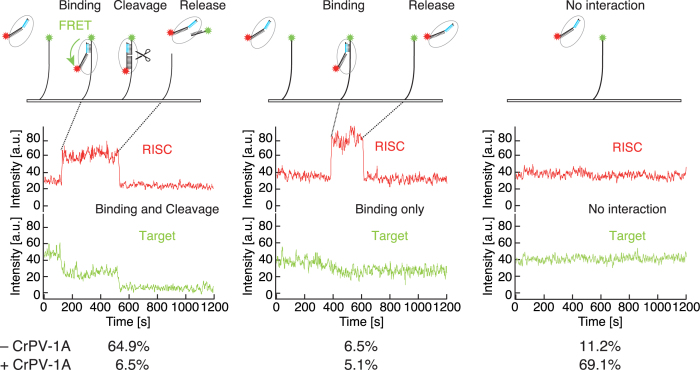
Continuous single-molecule imaging of the target cleavage reaction. Representative traces of red RISC and green target in the presence or absence of CrPV-1A. CrPV-1A strongly inhibited the interaction between Ago2-RISC and the target. a.u., arbitrary unit.
Figure 5.
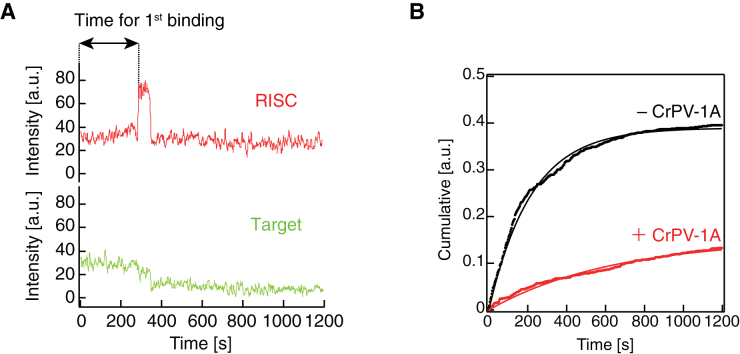
CrPV-1A inhibits initial target association by Ago2-RISC. (A) A schematic presentation of the time required for the first binding. (B) Cumulative plot for the time required for first binding. Ago2-RISC required ∼3-fold longer time to initiate target binding in the presence of CrPV-1A. a.u., arbitrary unit.
RESULTS AND DISCUSSION
CrPV-1A blocks an early step in target RNA recognition by Drosophila Ago2-RISC
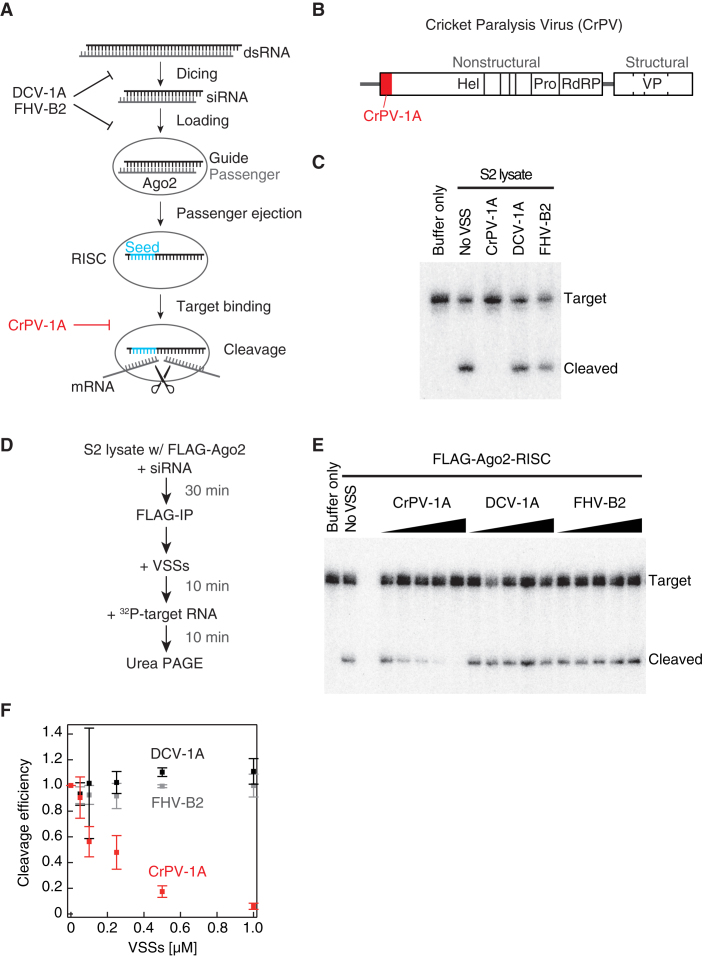
CrPV-1A is known as a VSS that specifically represses the target cleavage activity of Drosophila Ago2-RISC downstream of siRNA production (Figure 1A and B). Indeed, as previously reported, Ago2-RISC programmed with an artificial siRNA in crude lysate from Drosophila S2 cells failed to cleave a perfectly complementary target RNA in the presence of purified recombinant CrPV-1A, whereas two other insect VSSs, DCV-1A, FHV-B2, did not inhibit the target cleavage reaction (Figure 1C, Supplementary Figure S1) (15–17,19). The inhibitory effect of CrPV-1A on target cleavage was comparable when additional GC pairs were introduced in the seed region or the 3′ supplementary region (Supplementary Figure S2). To confirm that CrPV-1A acts directly on Ago2-RISC, we immunopurified pre-programmed Ago2-RISC onto paramagnetic beads, added increasing concentrations of recombinant VSSs, and then monitored the target cleavage reaction (Figure 1D). Again, CrPV-1A but not DCV-1A or FHV-B2 specifically inhibited target cleavage by immunopurified Ago2-RISC in a dose-dependent manner (Figure 1E and F).
Figure 1.
CrPV-1A inhibits the target cleavage reaction by Ago2-RISC. (A) A scheme for the mode of action of representative VSSs during the multi-step pathway of RNA silencing. (B) The genome structure of cricket paralysis virus. The silencing suppressor protein 1A is encoded at the N-terminal of ORF-1 (red rectangle). Hel: helicase, pro: protease, RdRP: RNA-dependent RNA polymerase, VP: viral structural ptotein. (C) Target cleavage assay in crude lysate from S2 cells. As previously reported, CrPV-1A, but not DCV-1A or FHV-B2, inhibited the cleavage reaction. The recombinant proteins were added at the concentration of 0.35 μM. (D) The experimental procedure of the target cleavage assay by immunopurified Ago2-RISC in E. (E) Target cleavage assay by immunopurified Ago2-RISC. The recombinant proteins were added at the concentrations of 0.05–1.0 μM. (F) Quantification of the cleavage assay in E. Error bars indicate the SD (standard deviation) from three independent experiments.
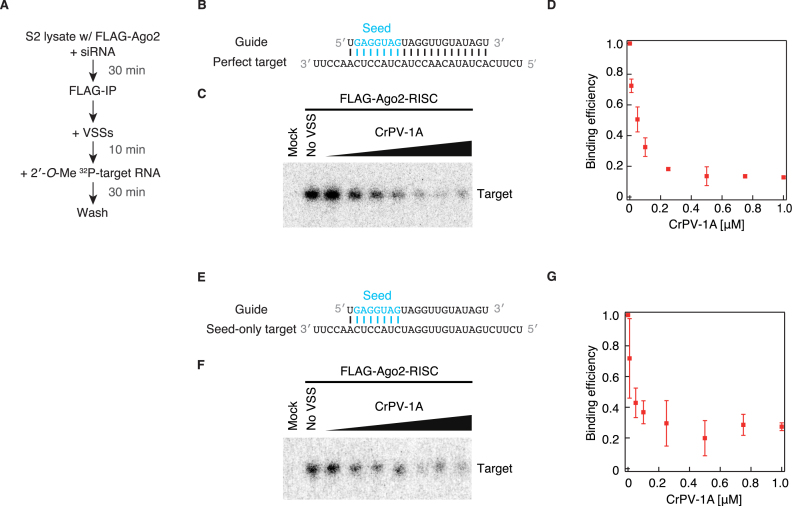
Might CrPV-1A block the base pairing between the guide strand of Ago2-RISC and its complementary target RNA, or interfere with the endonucleolytic ‘slicer’ activity of Ago2? To specifically monitor the target binding step independently of the subsequent slicing step, we utilized 2′-O-methylated (2′-O-Me) target RNA oligonucleotides that are refractory for RISC-mediated cleavage (Figure 2A and B). We first programmed and immunopurified Ago2-RISC, pre-incubated with increasing concentrations of CrPV-1A for 10 min, and then added a radiolabeled 2′-O-Me target RNA perfectly complementary to the guide strand. After 30-min incubation, we measured the amounts of the 2′-O-Me target bound to Ago2-RISC on beads. Importantly, CrPV-1A significantly inhibited the binding between Ago2-RISC and the target 2′-O-Me RNA (Figure 2C and D), whereas DCV-1A or BSA did not affect the binding (Supplementary Figure S3). Moreover, the inhibitory effect of CrPV-1A on target recognition was much stronger for Ago2-RISC than for Ago1-RISC (Supplementary Figure S4), in agreement with a previous notion that CrPV-1A preferentially interacts with Drosophila Ago2 but not Ago1 (19). These data together suggest that CrPV-1A blocks target RNA binding by Drosophila Ago2-RISC.
Figure 2.
CrPV-1A inhibits target binding by Ago2-RISC. (A) The experimental procedure of the target binding assay by immunopurified Ago2-RISC in C and F. (B) The guide-target configuration used in C. The base pairing was perfectly complementary between the guide strand and the target RNA. (C) CrPV-1A was added at the concentrations of 0.01–1.0 μM in the target binding assay. Mock indicates mock immunopurification from naive S2 lysate. (D) Quantification of the binding assay in C. Error bars indicate the SD from three independent experiments. (E) The guide-target configuration used in F. The base pairing was complementary only in the seed region (guide positions 2–8) between the guide strand and the target RNA. (F) CrPV-1A was added at the concentrations of 0.01–1.0 μM in the target binding assay. Mock indicates mock immunopurification from naive S2 lysate. (G) Quantification of the binding assay in E. Error bars indicate the SD from three independent experiments.
It is well established that Ago proteins divide their guide RNAs into individual functional domains. The seed region (guide positions 2–8) plays a central role in initial target searching, while subsequent propagation of base pairing through the central region toward the 3′ region is required for Ago to adopt a cleavage-competent conformation (21–26). This can be viewed as a sophisticated mechanism to limit ‘off-target’ cleavage of only partially complementary RNAs. To ask if CrPV-1A can block early target RNA recognition, we prepared a 2′-O-Me target RNA with seed complementarity only and repeated the target-binding assay by immunopurified Ago2-RISC (Figure 2E). The target-bound fraction of the ‘seed only’ 2′-O-Me target RNA was significantly decreased by increasing concentrations of CrPV-1A (Figure 2F and G), similarly to the case of the perfectly complementary 2′-O-Me target RNA (Figure 2C and D). Thus, CrPV-1A interferes target binding by Ago2-RISC as early as the seed recognition step.
Single-molecule observation of the effect of CrPV-1A in target recognition and cleavage
To better understand the mode of action of CrPV-1A during target recognition and cleavage by Ago2-RISC, we utilized single-molecule analysis by total internal reflection fluorescence (TIRF) microscopy (Figure 3A) (23,29–31). We first prepared Ago2-RISC programmed with 3′-Cy5-labeled guide strand (red RISC) in S2 cell lysate. We then tethered 3′-Cy3-labeled target RNA (green target) to a glass surface, incubated with S2 lysate containing red RISC in the presence or absence of CrPV-1A, and took snap-shot images at 0 and 15 min. Without CrPV-1A, we could observe many red RISC spots co-localizing with green target spots on the glass surface, indicative of target recognition by Ago2-RISC (Figure 3B, left). Moreover, the number of green target spots was markedly decreased with time (Figure 3C), reflecting target cleavage and release by Ago2-RISC. In contrast, in the presence of CrPV-1A, the number of the green target spots was only modestly decreased with time (Figure 3B, right and C). A similar decrease of green spots was observed with tethered targets alone and no S2 cell lysate, indicating that the modest decrease of the green spots in the presence of CrPV-1A was largely due to photobleaching and not target cleavage (Figure 3C). We concluded that the activity of CrPV-1A in blocking the function of Ago2-RISC can be faithfully monitored in our single-molecule observation setting.
We then continuously monitored target recognition and cleavage by Ago2-RISC for 20 min. In the absence of CrPV-1A, 64.9% of randomly chosen green target spots (N = 925) showed co-localization with red RISC spots, followed by the simultaneous disappearance of the red and green spots, indicating successful target cleavage and release (‘binding and cleavage’; Figure 4, left). 6.5% of analyzed green spots showed only temporary co-localization with red RISC spots, reflecting target binding without cleavage (‘binding only’; Figure 4, middle). Notably, a weak FRET (fluorescence resonance energy transfer) signal was observed between the co-localized green target and red RISC for ‘binding and cleavage’ but not for ‘binding only’. The apparent absence of the FRET signal in ‘binding only’ may reflect a transient binding of RISC to a non-target site (33,34) remote from the Cy3 dye at the 3′ end of the target RNA, but future studies are needed to reveal the actual situation. No red RISC binding was observed for 11.2% of the analyzed green target spots (‘no binding’; Figure 4, right) during 20 min.
In the presence of CrPV-1A, however, a remarkably different RISC population was observed. Among randomly chosen green target spots (N = 1123), 69.1% showed no co-localization with red RISC spots (Figure 4, right), supporting our biochemical finding that CrPV-1A blocks target recognition by Ago2-RISC (Figure 2). In contrast, only 6.5% showed successful target cleavage and release (Figure 4, left) and 5.1% showed target binding without cleavage. Interestingly, the time required for the first target binding by Ago2-RISC was much extended by CrPV-1A; the time constant (τ) for the first binding was ∼220 s in the absence of CrPV-1A but was ∼640 s in the presence of CrPV-1A (Figure 5A and B). On the other hand, when focused on the traces with successful target cleavage, only a single binding event was required before target cleavage in most cases, regardless of the presence (N = 64/65) or absence (N = 414/430) of CrPV-1A, indicating that essentially every Ago2-RISC cleaved the target before dissociating from it. Thus, CrPV-1A acts to slow the association rate (kon) in the initial interaction between Ago2-RISC and complementary target RNAs. This is reminiscent of the previously reported effect of a mismatch in the seed region, which showed ∼3-fold longer τ for the first target binding than the fully paired target binding and required virtually a single binding event before target cleavage (23); the situation was remarkably different when mismatches were present at the guide positions 14 and 15 in the 3′ supplementary region, where Ago2-RISC required multiple binding events before cleavage, presumably because Ago2-RISC dissociates from the target before being able to take the cleavage-competent conformation (23). Taken all together, we conclude that CrPV-1A interferes with the initial target recognition by Ago2-RISC via base pairing in the seed region. Accordingly, we envision that CrPV-1A directly binds and plug the target-binding cleft of Ago2-RISC. Future structural studies are warranted to understand the unique mechanism of how CrPV-1A suppresses the target recognition by Drosophila Ago2-RISC at the atomic level.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of our laboratories for fruitful discussions and critical comments on the manuscript.
H.I, H.T. and Y.T. designed and supervised the study. M.W. performed biochemical experiments, single-molecule imaging, and data analysis, with the help of H.I. and H.T. All authors discussed the results. M.W. H.I. and Y.T. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academic Research Grant for GSFS Doctor Course Students (to M.W.); Grants-in-Aid for Scientific Research on Innovative Areas (‘non-coding RNA neo-taxonomy’) [26113007 to H.T. and Y.T.]; Grant-in-Aid for Young Scientists (A) [16H06159 to H.T.]. Funding for open access charge: MEXT [26113007].
Conflict of interest statement. None declared.
REFERENCES
- 1. Marques J.T., Imler J.L.. The diversity of insect antiviral immunity: insights from viruses. Curr. Opin. Microbiol. 2016; 32:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ding S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010; 10:632–644. [DOI] [PubMed] [Google Scholar]
- 3. Csorba T., Kontra L., Burgyán J.. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015; 479–480:85–103. [DOI] [PubMed] [Google Scholar]
- 4. Merkling S.H., van Rij R.P.. Beyond RNAi: antiviral defense strategies in Drosophila and mosquito. J. Insect. Physiol. 2013; 59:159–170. [DOI] [PubMed] [Google Scholar]
- 5. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 6. Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W.. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004; 117:69–81. [DOI] [PubMed] [Google Scholar]
- 7. Bouché N., Lauressergues D., Gasciolli V., Vaucheret H.. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006; 25:3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iki T. Messages on small RNA duplexes in plants. J. Plant Res. 2017; 130:7–16. [DOI] [PubMed] [Google Scholar]
- 9. Lakatos L., Csorba T., Pantaleo V., Chapman E.J., Carrington J.C., Liu Y.P., Dolja V.V., Calvino L.F., López-Moya J.J., Burgyán J.. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006; 25:2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Omarov R., Sparks K., Smith L., Zindovic J., Scholthof H.B.. Biological relevance of a stable biochemical interaction between the tombusvirus-encoded P19 and short interfering RNAs. J. Virol. 2006; 80:3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silhavy D., Molnár A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyán J.. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002; 21:3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye K., Malinina L., Patel D.J.. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003; 426:874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vargason J.M., Szittya G., Burgyan J., Hall T.M.. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003; 115:799–811. [DOI] [PubMed] [Google Scholar]
- 14. Rawlings R.A., Krishnan V., Walter N.G.. Viral RNAi suppressor reversibly binds siRNA to outcompete Dicer and RISC via multiple turnover. J. Mol. Biol. 2011; 408:262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Rij R.P., Saleh M.C., Berry B., Foo C., Houk A., Antoniewski C., Andino R.. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006; 20:2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lingel A., Simon B., Izaurralde E., Sattler M.. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005; 6:1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chao J.A., Lee J.H., Chapados B.R., Debler E.W., Schneemann A., Williamson J.R.. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005; 12:952–957. [DOI] [PubMed] [Google Scholar]
- 18. Aliyari R., Wu Q., Li H.W., Wang X.H., Li F., Green L.D., Han C.S., Li W.X., Ding S.W.. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008; 4:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nayak A., Berry B., Tassetto M., Kunitomi M., Acevedo A., Deng C., Krutchinsky A., Gross J., Antoniewski C., Andino R.. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010; 17:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Mierlo J.T., Bronkhorst A.W., Overheul G.J., Sadanandan S.A., Ekström J.O., Heestermans M., Hultmark D., Antoniewski C., van Rij R.P.. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012; 8:e1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schirle N.T., Macrae I.J.. The crystal structure of human argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao C., Sasaki H.M., Ueda T., Tomari Y., Tadakuma H.. Single-molecule analysis of the target cleavage reaction by the Drosophila RNAi enzyme complex. Mol. Cell. 2015; 59:125–132. [DOI] [PubMed] [Google Scholar]
- 24. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fukaya T., Tomari Y.. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol. Cell. 2012; 48:825–836. [DOI] [PubMed] [Google Scholar]
- 28. Haley B., Tang G., Zamore P.D.. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003; 30:330–336. [DOI] [PubMed] [Google Scholar]
- 29. Iwasaki S., Sasaki H.M., Sakaguchi Y., Suzuki T., Tadakuma H., Tomari Y.. Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature. 2015; 521:533–536. [DOI] [PubMed] [Google Scholar]
- 30. Zhou Z.P., Shimizu Y., Tadakuma H., Taguchi H., Ito K., Ueda T.. Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. J. Biochem. 2011; 149:609–618. [DOI] [PubMed] [Google Scholar]
- 31. Miyazono Y., Hayashi M., Karagiannis P., Harada Y., Tadakuma H.. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010; 29:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harada Y., Sakurada K., Aoki T., Thomas D.D., Yanagida T.. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J. Mol. Biol. 1990; 216:49–68. [DOI] [PubMed] [Google Scholar]
- 33. Ameres S.L., Martinez J., Schroeder R.. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007; 130:101–112. [DOI] [PubMed] [Google Scholar]
- 34. Chandradoss S.D., Schirle N.T., Szczepaniak M., MacRae I.J., Joo C.. A dynamic search process underlies microRNA targeting. Cell. 2015; 162:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.