Abstract
The retargeting of protein–DNA specificity, outside of extremely modular DNA binding proteins such as TAL effectors, has generally proved to be quite challenging. Here, we describe structural analyses of five different extensively retargeted variants of a single homing endonuclease, that have been shown to function efficiently in ex vivo and in vivo applications. The redesigned proteins harbor mutations at up to 53 residues (18%) of their amino acid sequence, primarily distributed across the DNA binding surface, making them among the most significantly reengineered ligand-binding proteins to date. Specificity is derived from the combined contributions of DNA-contacting residues and of neighboring residues that influence local structural organization. Changes in specificity are facilitated by the ability of all those residues to readily exchange both form and function. The fidelity of recognition is not precisely correlated with the fraction or total number of residues in the protein–DNA interface that are actually involved in DNA contacts, including directional hydrogen bonds. The plasticity of the DNA-recognition surface of this protein, which allows substantial retargeting of recognition specificity without requiring significant alteration of the surrounding protein architecture, reflects the ability of the corresponding genetic elements to maintain mobility and persistence in the face of genetic drift within potential host target sites.
Understanding the molecular mechanisms that dictate the affinity and specificity of protein–DNA recognition is an area of investigation that remains critical for many fields of research, including protein engineering. This is particularly important for the purpose of creating novel target specificities for enzymes that act upon DNA targets, including those that are used for targeted genome modification (such as recombinases, integrases, transposases and especially endonucleases). Recent studies have greatly enhanced our understanding of the complex balance of contacts and forces that lead to protein–DNA recognition. In particular, examination of highly diverse DNA binding protein systems have demonstrated that recognition of the shape and structural features of a DNA target can greatly augment the specificity imparted by contacts to the chemically distinct sequence of individual nucleotide base pairs. This includes recognition of altered minor groove dimensions and corresponding changes in the surrounding surface electrostatic potential in response to DNA bending (1,2), recognition of altered DNA conformations as a result of cytosine methylation or other epigenetic modifications (again including altered minor groove dimensions) (3), recognition of altered DNA duplex shape and electrostatic potential corresponding to the presence of non-canonical base pairs in the target (4), and the contribution of surrounding DNA sequence on target shape and conformation (5).
A relatively recent review article (6), focused on how transcription factors limit their interactions with potential targets in various cell types and tissues, clearly outlines how DNA recognition involves the presence and exploitation of many layers of unique structural features beyond DNA sequence (including shape, flexibility, accessibility and cooperativity between multiple DNA binding proteins). Thus, simple codes or correlation between protein and DNA sequences that might be predictive of protein–DNA recognition are largely absent, except for rare examples of extremely modular DNA-binding proteins (e.g. TAL effectors).
DNA binding proteins and enzymes that are associated with mobile genetic elements face strong evolutionary pressure to rapidly and efficiently alter their DNA recognition specificity. By doing so, they maximize their ability to invade new DNA target sites, while also persisting in their existing target sites. Homing endonucleases (hereafter termed ‘meganucleases’) are microbial DNA cleavage enzymes that are encoded within mobile microbial intervening sequences (group I and group II introns and inteins). Meganucleases, and their associated introns and inteins, are encoded within phage, prokaryotes and single-cell eukaryotes. They recognize long DNA target sites (7): meganucleases from the LAGLIDADG protein family (reviewed in (8)) typically recognize and cleave DNA targets spanning 20–22 base pairs in length. The length of their targets provides sufficient specificity to avoid toxicity to their host organism, while their ability to tolerate polymorphisms within those targets allows them to remain active when encountering genetic drift within their host genes (9).
Even moderate evolutionary divergence from recent common ancestors allows wild-type meganucleases to establish entirely new DNA specificities, thus facilitating invasion of new genomic targets. A recent analysis has demonstrated that even when maintaining up to 50% amino acid sequence identity and very close structural similarity, homologous wild-type meganucleases can recognize and cleave highly diverged DNA targets (10). The ability of these endonucleases to readily adopt new DNA target specificities, with minimal resculpting of their overall folded topology and structure, may reflect their biological function as the enzymatic drivers of intron mobility, transfer and persistence (reviewed in (7)).
The mechanism of DNA recognition by meganucleases is typical of the structural and mechanistic features displayed by many DNA-binding proteins, involving a combination of (i) contacts between protein side chains and nucleotide bases throughout the major groove of the target site, (ii) significant DNA bending at the center of the target, causing a distortion of both major and minor groove dimensions and corresponding alteration to the shape and electrostatic surface of the target and (iii) additional contacts in and along the minor groove at the site of bending. At the same time, DNA recognition by a meganuclease is noteworthy with respect to the length of its target site (22 base pairs) and the corresponding expanse of its DNA-contacting surface (comprising ∼50 amino acids). Their recognition specificity is often enforced largely during DNA cleavage, rather than through modulation of binding affinity alone. Finally, they display highly variable specificity at individual positions in the protein–DNA complex (ranging from nearly exclusive recognition at some base pair positions, to considerable promiscuity at nearby positions) (9,11).
A large number of studies over the past 15 years have described a variety of engineering and selection experiments to alter meganuclease specificity. These experiments initially consisted of relatively simple experiments intended to alter meganuclease specificity at single base pairs (12,13). Subsequent experiments addressed the reprogramming of specificity across multiple base pairs and highlighted how context-dependent interactions between neighboring DNA base pairs and protein side chains could unexpectedly and significantly alter DNA conformation and shape. These studies illustrated that meganuclease engineering requires methods that treated DNA recognition as the product of more than the sum of individual contacts and interactions. A high-throughput selection method, in which the protein's cleavage activity was coupled to the homology-driven reconstitution of a reporter gene, successfully addressed this property (14). In that approach, multiple semi-randomized libraries of the meganuclease, where each library harbored collections of amino acid substitutions within ‘modules’ or ‘clusters’ of residues that collectively contacted several adjacent DNA base pairs, were screened. By doing so, investigators could isolate and combine a large number of individual protein variants, harboring multiple amino acid changes, that could accommodate multiple adjacent base pair substitutions at several distinct regions of the enzyme's target site (15).
Since then, multiple groups have described the creation of extensively altered variants of the I-CreI homing endonuclease and their successful application for nuclease-driven, targeted gene modification. Methods used for redirection of specificity include the phenotypic screens from semi-randomized protein libraries described above (14), as well as structure-based redesign of the wild-type protein (16). Using these approaches, these groups have created and employed redesigned variants of single-chain I-CreI endonuclease for a wide variety of purposes, such as modification and correction of the human XPC gene for the treatment of Xeroderma Pigmentosum (17–19), creation of cell lines harboring defined genetic insertions and alterations (18,20), generation of transgenic lines of maize containing heritable disruptions of the ligueleless-1 and MS26 loci (16,21), excision of defined genomic regions in Arabidopsis (22), insertion of multiple trait genes in cotton (23), generation of Rag1 gene knockouts in human cell lines (24,25) and in transgenic rodents (26), disruption of integrated viral genomic targets in human cell lines (27), and demonstration of the correction of exon deletions in the human DMD gene associated with duchenne muscular dystrophy (28). Crystallographic structures of two of these fully reengineered variants (against the human Rag1 and XPC targets have been solved and described (18,25).
Here we report crystallographic structure analyses of multiple fully reengineered variants of the I-OnuI meganuclease, and describe the manner in which this one individual DNA binding enzyme can be induced to bind and cleave several completely diverged DNA targets. The engineered variants of the starting enzyme were produced using an engineering pipeline that relies upon a combination of yeast surface display and high-throughput flow cytometry (Supplementary Figure S1) to screen semi-randomized endonuclease libraries for altered binding and cleavage specificity, followed by assembly of the final engineered nucleases (29). These redesigned enzymes cleave unrelated targets in human, viral or insect host genes, are highly active in transfected primary human cells or transgenic insects, and display specificity profiles that rival or exceed the parental meganuclease. The results of this study demonstrate the extent to which a single meganuclease protein can be substantially reprogrammed for recognition of multiple unique genomic target sites, without the need for significant alteration of the surrounding protein scaffold.
MATERIALS AND METHODS
Nomenclature
All sequences and/or structures of the engineered variants in this paper are deposited in in the RCSB structural database and named according to published nomenclature conventions (30). For brevity, these enzymes are referred to in the following text using a shorter convention (for example, ‘eOnuCCR5’ for the engineered ‘I-OnuI-e-hCCR5’ enzyme that targets the human CCR5 gene). Table 1 lists formal names, abbreviations, genomic targets, total number of mutations, PDB ID codes and in cellulo or in vivo cleavage activities of all constructs.
Table 1. Engineered enzymes in this study.
| Meganuclease (abbreviation) (PDB ID; # of mutations vs. WT) | Gene modification activity (method of analysis) | Genomic target | Reference |
|---|---|---|---|
| I-OnuI-e-hCCR5 (eOnuCCR5) (5THG; 48 mutations) | 80% disruption (MegaTAL); CCR5 cell staining & enzymatic T7 indel assay | Human HIV coreceptor (CCR5) | Sather et al. (7) |
| Ibarra et al. (8) | |||
| I-OnuI-e-hTCRα (eOnuTCRα) (5T2H; 43 mutations) | 38% disruption (Meganuclease); 70% disruption (MegaTAL); CD3 cell staining & MiSeq Target Analysis | Human T-cell receptor alpha chain (TCRα) | Boissel et al. (6) |
| I-OnuI-e-vHIVInt (eOnuHIVInt) (5T8D; 47 mutations) | 44% disruption (MegaTAL) Digital PCR target analysis | Viral integrase reading frame in HIV pol gene | Sedlak et al. (9) |
| I-OnuI-e-Ag7280 (eOnu7280) (5T2N; 38 mutations) | 63% disruption (Meganuclease) T7 indel assay & MiSeq target analysis of Anopheles gDNA | A. gambiae AGAP007280 (female fertility gene) | This study |
| I-OnuI-e-Ag11377 (eOnu11377) (5T2O; 53 mutations) | 22% disruption (Meganuclease) Disruption of integrated fluorescent reporter harboring target site | A. gambiae AGAP011377 (female fertility gene) | This study |
The PDB ID code and number of mutations relative to the wild-type enzyme is listed in the left-most column. The level of gene modification activity in cellular assays, the genomic target that is modified, and the citation for those experiments (for the three that have been published) are provided in the ensuing three columns.
Vectors
For yeast-based assays of endonuclease cleavage activity and specificity, the eOnu protein coding sequences were cloned into the pETCON yeast surface expression vector (Addgene #41522). The pETCON vector incorporates an N-terminal hemagglutinin (HA) epitope tag and a C-terminal Myc tag to allow for fluorescent antibody staining in flow cytometric assays and cell sorting. A modified version of this vector, containing the I-OnuI protein scaffold, was also used in all of the yeast libraries.
For protein production and crystallographic analyses, reading frames encoding engineered meganucleases were subcloned into commercially available bacterial pET expression vectors (Novagen, Inc.) for protein production. The commercially available expression vector pET21d was used to create T7-inducible constructs with no affinity purification tags. One enzyme (eOnu-CCR5) was also expressed using a GST fusion partner (which was subsequently removed proteolytically) to enhance expression levels and yield of the purified protein.
Protein engineering
The general methods used to reprogram the DNA binding specificity of the meganucleases in this study, while maintaining overall fidelity and cleavage activity, have been previously described in detail as outlined and cited here (31,32) and below. The basis for reprogramming the DNA cleavage specificity of a meganuclease is the use of yeast surface display coupled with flow cytometry (Supplementary Figure S1). This approach allows us to screen protein libraries for desired DNA cleavage activities and specificities, and to assay the activity of individual protein constructs at the end of each round of selections. Briefly, combinatorial libraries of meganuclease variants are expressed on the surface of transformed yeast, and individual constructs that display cleavage activity against a defined DNA target are identified and isolated. This process uses a flow-cytometric approach in which cleavage of a DNA substrate (harboring a fluorescent label on one end) in the presence of magnesium ions results in a reduction in cell staining intensity that allows for cell sorting and isolation of active constructs (31,32). The strategy for creating the combinatorial libraries of LAGLIDADG meganuclease variants, that harbor clusters of mutations at 6 to 9 residues (that surround the location of DNA base pair triplets that harbor one or more changes in the new target sequence) has been described in (33,34). Iterative, sequential steps of such selections spanning the entire protein–DNA interface is followed by assembly of fully retargeted enzymes that recognize completely novel genomic target sites.
The activity of individual clones against their genomic targets were determined by deep sequencing of the endogenous target locus and measuring of the frequency of mutated sites harboring indels or base pair substitutions as previously described in (35–37). These analyses were conducted either after transient expression in the appropriate primary cell line, or (in the case of gene-drive meganucleases) expressed in mosquito testes after isolation of appropriate tissue directly from a transgenic organism harboring the meganuclease gene, which was expressed under the control of a testes-specific promoter.
The specificity of the same engineered enzymes was further assessed both by measuring their ability to cleave target sites containing single base pair substitutions (a ‘one-off’ specificity profile) and by measuring their activity against potential genomic off-target sites (with the same assay used for analysis of the ‘on-target’ locus) using methods described in detail in (11,29,35,38).
The activity and specificity of a series of fully retargeted meganucleases and corresponding MegaTALs (fusions of TAL effectors and the engineered meganucleases) in primary human cells, including several described in the structural analyses in this study, have been described in (35) (eOnuTCRα) and (36,37) (eOnuCCR5). The cellular cleavage activities of constructs examined in this paper, including a cited recapitulation of published data, are summarized in Table 1.
Specificity assays
Wild-type I-OnuI, eOnuTCRα and eOnu7280 were expressed on the surface of yeast and tested for cleavage activity against 66 different targets, each harboring a single base pair substitution at one position in the enzyme's intended DNA target, using previously described flow cytometry methods (Supplementary Figure S1) (29). Cleavage activity was measured by quantifying the drop in mean A647 signal (corresponding to the fluorophore located on the end of the DNA substrate) between calcium (uncleaved) and magnesium (cleaved) samples, and the values are presented relative to the enzyme's activity against its wild type target sequence. (39)). For each construct, the experiment was repeated 3 times with separately transfected and induced yeast cultures; one representative set of results is displayed in the figure.
Mosquito transgenesis and characterization of in vivo gene modification activity
The method used for quantitative measurement of target gene modification by the eOnu7280 meganuclease (which was encoded under control of a testes-specific promoter in the germline of transgenic mosquitos and is expressed during male meiosis) has previously been described in (40). The eOnu11377 meganuclease, which is still under development as a gene drive meganuclease in mosquitos, was assayed in a cell line harboring an chromosomally integrated copy of its target site in a fluorescent reporter of cleavage activity (a method also described in (40)).
To create genetically modified insects (Supplementary Figure S2), Anopheles gambiae sensu stricto embryos (strain G3, referred to as wild-type) were injected with a mixture of 0.2 μg/μl of a transformation plasmid encoding the engineered meganuclease and 0.4 μg/μl of helper plasmid containing a vasa promoter driven piggyBac transposase, using a Femtojet Express injector and a Narishige 202ND micromanipulator mounted on an inverted microscope (Nikon TE-DH100W) Survivors were screened for transient expression of the DsRed marker at the larval stage. Adult transfectants were crossed to wild-type mosquitoes and their progeny was analyzed for DsRed fluorescence. Individual larvae showing expression of DsRed were then separated, and the adults that emerged were crossed individually to wild-type mosquitoes. The identity and independence of integration events was determined by inverse PCR.
Transgenic mosquitoes at different developmental stages were analyzed on a Nikon inverted microscope (Eclipse TE200) at a wavelength of 488 nm to detect eGFP expression (filter 535/20 nm emission, 505 nm dichroic) and 563 nm to detect DsRed expression (Filter 630/30 nm emission, 595 nm dichroic). The transgenic lines were maintained so that in each generation transgenic females were backcrossed back to G3 wild-type males. Genotyping was performed using inverse PCR. Briefly, 500 ng of genomic DNA was separately digested with 10 units of Sau3AI or HinP1I, 5 μl of each digestion was re-ligated with T4 DNA ligase (Takara) in a final volume of 20 μl, of which 5 μl were subjected to PCR. The piggyBac flanking regions were amplified with primers 5F1 (GACGCATGATTATCTTTTACGTGAC) and 5R1 (TGACACTTACCGCATTGACA) for 5′ junctions; or 3F1 (CAACATGACTGTTTTTAAAGTACAAA) and 3R1 (GTCAGAAACAACTTTGGCACATAT) for 3′ junctions, followed by a second inner PCR reaction using primers 5F2 (GCGATGACGAGCTTGTTGGTG) and 5R2 (TCCAAGCGGCGAATGAGATG) for 5′ junctions; or 3F2 (CCTCGATATACAGACCGATAAAAC) and 3R2 (TGCATTTGCCTTTCGCCTTAT) for 3′ junctions on 5 μl of the first reaction. Genomic insertion sites were sequenced using primers pB-5SEQ (CGCGCTATTTAGAAAGAGAGA) for 5′ junctions and pB-3SEQ (CGATAAAACACATGCGTCAATT) for 3′ junctions. Strains I1-H7280A1 carries the construct inserted on chromosome 2R (13C) at position 29 172 631. Primers 7280-F1 (GGGCTGTGGGATGGATCAG) and 7280-R1 (AGTCTCAGCTTCCGTTGTATCCAC) were used to amplify and sequence the target regions.
Protein overexpression and purification
Sequence verified plasmids were transformed into BL21(DE3) RIL Escherichia coli cells and plated on LB-Amp plates to grow at 37°C. Colonies were grown in 10 ml overnight cultures of LB + Amp (100 μg/ml) and diluted 1:100 the next day to a final volume of 1 l. Cell cultures were shaken at 37°C until the cells reached an OD600 between 0.6 and 0.8. Cells were then induced with 0.2 mM IPTG, and incubated overnight at 16°C. Induced cells were pelleted and stored at –20°C. Successful protein induction was verified by SDS-PAGE.
Cell pellets were resuspended in a buffer containing 25 mM Tris–HCl pH 7.5, 300 mM NaCl and 5% glycerol. PMSF and benzonase (Sigma-Aldrich) were added to 40 μM and 0.18 U/μl concentrations, respectively, prior to sonication. Cell debris was pelleted and the supernatant was filtered through a 0.45 μm filter. Untagged protein samples were loaded onto a 5 ml Heparin HP HiTrap column (GE Life Sciences) and eluted with a linear salt gradient (Buffer A: 25 mM Tris–HCl pH 7.5, 300 mM NaCl, 5% glycerol, Buffer B: 25 mM Tris–HCl pH 7.5, 1 M NaCl, 5% glycerol). The meganuclease constructs eluted in sharp individual peaks between 500 and 800 mM NaCl.
All constructs were then concentrated to 5–20 mg/ml and passed over a size exclusion column (15 ml Superdex 200 10/300 GL, GE Life Sciences) in the presence of 25 mM Tris–HCl pH 7.5, 200 mM NaCl and 5% glycerol. For eOnu7280, a higher salt concentration (500 mM NaCl) was necessary for optimal purification at this final step.
Meganuclease–DNA crystallization and data collection
Recombinant proteins were incubated with CaCl2 and a double-stranded DNA oligonucleotide (IDT) containing each meganuclease's 22 bp target site (underlined sequence in the text below), flanked by various lengths of random duplex sequences that terminated either with blunt ends, single base 3′ overhangs or single base 5′ overhangs. The initial mixtures were set up at 1:1.5 protein:DNA molar ratios. Crystallization of the protein/DNA complexes was initially screened in 96-well trays using a mosquito robot (TPP Labtech) with three pre-made crystallization grids: PEGs Suite (Qiagen), Index I & II (Hampton Research), and Wizard Classic (Rigaku/Emerald BioStructures). Crystal hits from initial screens were further optimized in larger scale 24-well hanging drop trays. Final DNA constructs and crystallization conditions for each structure are listed below.
Data was collected either on an in-house Rigaku Micromax 007HF rotating anode generator using an RaxisIV++ imaging plate detector or a Saturn 944+ CCD area detector or at the Advanced Light Source (ALS) synchrotron facility (Beamlines 5.0.1 or 5.0.2) at Lawrence Berkeley National Laboratory, using an ADSC Quantum 315R 3 × 3 CCD area detector or a Pilatus 6M silicon pixel detector. All data were processed using program HKL2000 (41). Phases were obtained by molecular replacement with PHASER (42) using the I-OnuI structure (PDB ID 3QQY) as a search model. Model building and refinement were performed using COOT (43) and PHENIX (44) or CCP4 (45,46), respectively. Structural analyses of superposition RMSD values and DNA bending parameters were done with PyMol (47), COOT (43), Visual Molecular Dynamics (48), and 3DNA (49).
eOnuCCR5 crystallized in 28% (w/v) PEG 8000, 100 mM HEPES pH 6.5 in the presence of a bound 28 base pair duplex DNA harboring single base 5′ C/G overhangs:
Top: 5′-CCACCTTCCAGGAATTCTTTGGCCTGCAC-3′
Bottom: 3′-GTGGAAGGTCCTTAAGAAACCGGAC GTGG-5′
The crystal was cryoprotected in artificial 80% mother liquor with 20% sucrose and flash frozen in liquid nitrogen. Data was collected with an in-house Rigaku Micromax 007HF rotating anode generator using a Saturn 944+ CCD area detector.
eOnuTCRα crystallized in 35% (v/v) pentaerythritol ethoxylate, 50 mM Bis–Tris pH 6.5 and 50 mM ammonium sulfate in the presence of a bound 25 base pair duplex DNA harboring single base 5′ G/C overhangs:
Top: 5′-GGGTGTCTGCCTATTCACCGATTTTG-3′
Bottom: 3′-CCACAGACGGATAAGTGGCTAAAA CC-5′
The crystal was cryoprotected in 80% mother liquor and 20% ethylene glycol and flash frozen in liquid nitrogen. Data was collected at ALS Beamline 5.0.1 using an ADSC Quantum 315R 3 × 3 CCD area detector.
eOnuHIVInt crystallized in 22.5% (w/v) PEG 3350, 100 mM HEPES pH 7.0, 200 mM ammonium sulfate, in the presence of a bound 25 base pair duplex DNA harboring single base 5′ T/A overhangs:
Top: 5′- GGGAATGGCAGTATTCATCCACAATG-3′
Bottom: 3′-CCTTACCGTCATAAGTAGGTGTTACC-5′
The crystal was cryoprotected in 80% mother liquor with 20% sucrose and flash frozen in liquid nitrogen. Data was collected at ALS Beamline 5.0.2 using a Pilatus 6M silicon pixel detector.
eOnu7280 crystallized in 35% pentaerythritol ethoxylate (15/4 EO/OH), 50 mM ammonium sulfate, 50 mM Bis–Tris pH 6.5, in the presence of a bound 25 base pair duplex DNA harboring single base 5′ G/C overhangs:
Top: 5′-GGGCCTCCTCACTTTCTTCCTCACCG-3′
Bottom: 3′-CCGGAGGAGTGAAAGAAGGAGTGG CC-5′
The crystal was cryoprotected in 80% mother liquor with 20% ethylene glycol and flash frozen in liquid nitrogen. Data was collected at ALS Beamline 5.0.2 using an ADSC Quantum 315R 3 × 3 CCD area detector.
eOnu11377 crystallized in 33% (v/v) pentaerythritol ethoxylate (15/4 EO/OH), 50 mM HEPES pH 7.5, 50 mM ammonium sulfate in the presence of a bound 25 base pair duplex DNA harboring single base 5′ G/C overhangs:
Top: 5′-GGGGCCGGAAAATTTCTACGTCTGCG-3′
Bottom: 3′-CCCGGCCTTTTAAAGATGCAGACG CC-5′
The crystal was cryoprotected in 80% mother liquor and 20% ethylene glycol and flash frozen in liquid nitrogen. Data was collected at ALS Beamline 5.0.2 using a Pilatus 6M silicon pixel detector.
RESULTS
Five separately reengineered meganucleases (Figure 1, Table 1 and Supplementary Figure S3) were generated that display highly efficient and specific cleavage of genomic target sites. Those sites are found in the human genome (in genes encoding the T-cell receptor alpha chain and the HIV coreceptor CCR5 (targeted by eOnuTCRα and eOnuCCR5), in a viral genome (in the integrase coding sequence within the HIV pol gene, targeted by eOnuHIVInt) and in the Anopheles gambiae genome (in two genes that encoded factors required for female reproductive development, targeted by eOnu7280 and eOnu11377). All five enzymes are highly active and specific in vitro, and when incorporated into a megaTAL gene editing architecture (35) they exhibit significant cleavage of their chromosomal target sites in transfected cells. The ability of two of these targeted nucleases (eOnuTCRα and eOnuCCR5) to efficiently disrupt and/or modify their endogenous genomic targets in primary human T-cells, as well as the ability of a third (eOnuHIVInt) to do so in an integrated proviral genome in a human cell line, has been previously described (36,37,50). A fourth enzyme (eOnu7280) drives highly efficient and specific gene disruption in vivo in transgenic mosquitos that transiently express the meganuclease during spermatogenesis. Measurements of the in cellulo and/or in vivo activities for these enzymes (including a recapitulation of the published activity data cited above) are summarized in Table 1.
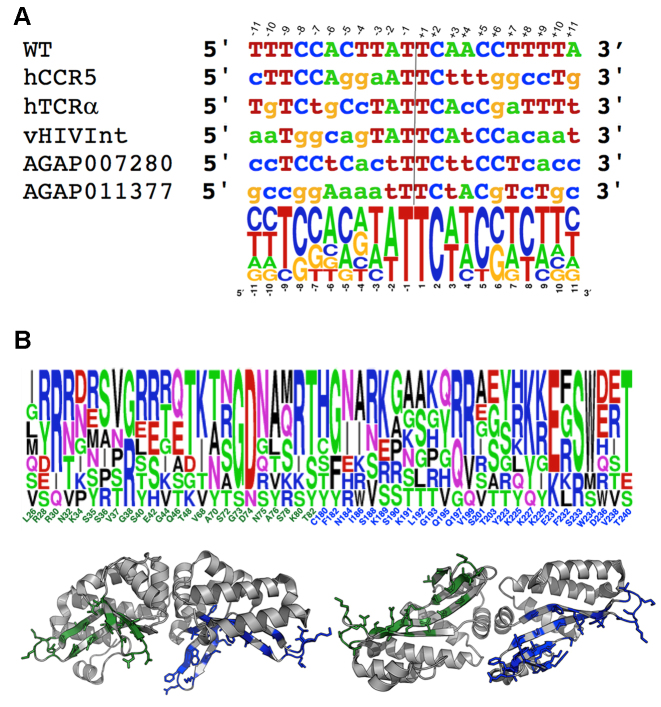
Figure 1.
Summary of engineered variants of I-OnuI. (A) DNA target sites and corresponding DNA logo plots of their alignment. The bases are numbered based on the exact center of the target site (indicated by the vertical line and flanked by positions –2, –1, +1 and +2, which lie inside the scissile phosphates that are cleaved by the enzyme to liberate 4 base, 3′ overhangs). The bases contacted by the enzymes' N-terminal domain are negatively numbered; the bases contacted by the C-terminal domain are positively numbered. (B) eOnu DNA binding surfaces and corresponding residue positions subjected to randomization and selection experiments to create individual eOnus. Positions and side chains shown in green correspond to the enzyme N-terminal domain (which contacts the 5′ half-site of the DNA target); those in blue correspond to the C-terminal domain (which contacts the 3′ half-site of the same DNA target). The logo plot shows the relative frequency of side chains at each position for the wild-type enzyme and the 5 variants described in this study. The wild-type residue is indicated below the logo plot. See Table 1 for a list of the enzymes and their functional properties and Supplementary Figure S3 for a multi-sequence alignment of the wild-type and engineered enzymes.
The target sites for the five reengineered meganucleases are shown in Figure 1A. Other than maintaining their original specificities across the central four base pairs of each target site (a constraint that is related to bending of the DNA upon protein binding (10)), the base pair identities are changed liberally throughout the remainder of the DNA target, and many base pairs are present at least once at each position.
The distribution of altered protein residues in the engineered nucleases are illustrated in Figure 1B and Supplementary Figure S3; the corresponding alteration of the topology and electrostatic charge distribution of the protein's DNA-binding surface is illustrated in Figure 2. Prior to individual engineering experiments, seven point mutations (indicated with black asterisks in the protein sequence alignment in Supplementary Figure S3) were incorporated on the solvent-exposed surface and inter-domain peptide linker of the wild-type enzyme to improve its solubility and solution behavior; those mutations are largely maintained in the redesigned meganucleases. A total of 50 additional residues in the protein–DNA interface (Figure 1B and Table 2) were then subjected to iterative rounds of randomization and selection for desired cleavage specificity. The final redesigned enzymes contain amino acid substitutions at anywhere from 38 (eOnu7280) to 53 (eOnu11377) positions, corresponding to alteration of up to 18% the wild-type I-OnuI enzyme's residues.
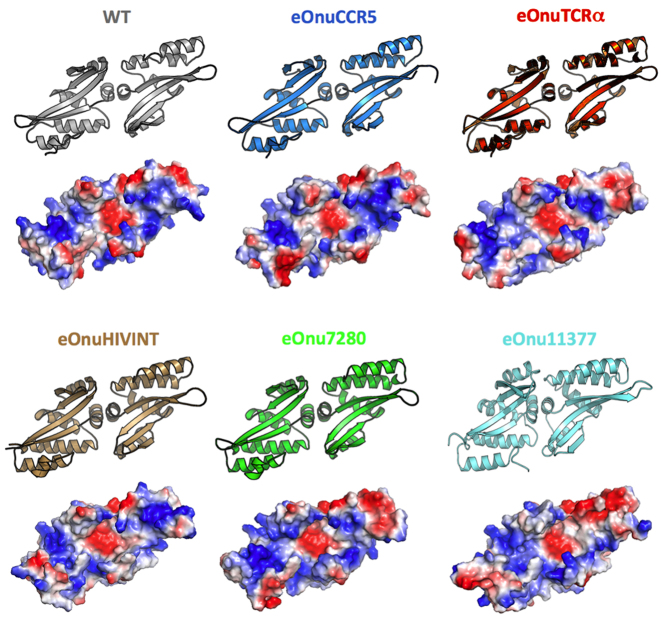
Figure 2.
DNA binding surfaces. Structures and electrostatic charge distribution of the DNA-binding surface of wild-type I-OnuI and its engineered variants. In the qualitative electrostatic charge distribution maps (calculated and visualized in PyMol (47)), blue corresponds to positively charged surface regions and red is negative. The structures are each viewed across the antiparallel DNA-contacting β-sheets presented by protein's N- and C-terminal domains. While the topology of the protein backbone across the β-sheets does not change dramatically (see also Figure 3), the amino acid changes create significant alterations in the details of the electrostatic potential across the surface.
Table 2. Structural and functional roles of residues in the protein–DNA interface.

|
All five engineered variants of the meganuclease were purified to homogeneity, and the crystallographic structures of each construct in complex with its DNA target site and calcium (i.e. in their pre-cleavage stage) was determined. Data collection and refinement statistics are provided in Supplementary Table S1. Superposition of the protein structures (Figure 3 and Supplementary Table S2) indicates that the alteration and diversity of DNA recognition specificity described above is accomplished with an overall shift in protein backbone positions across the entire protein scaffold well below 1 Å root mean square deviation (RMSD). The average RMSD values calculated across comparable superimposed DNA atom positions is 1.5 Å (Figure 4 and Supplementary Table S2).
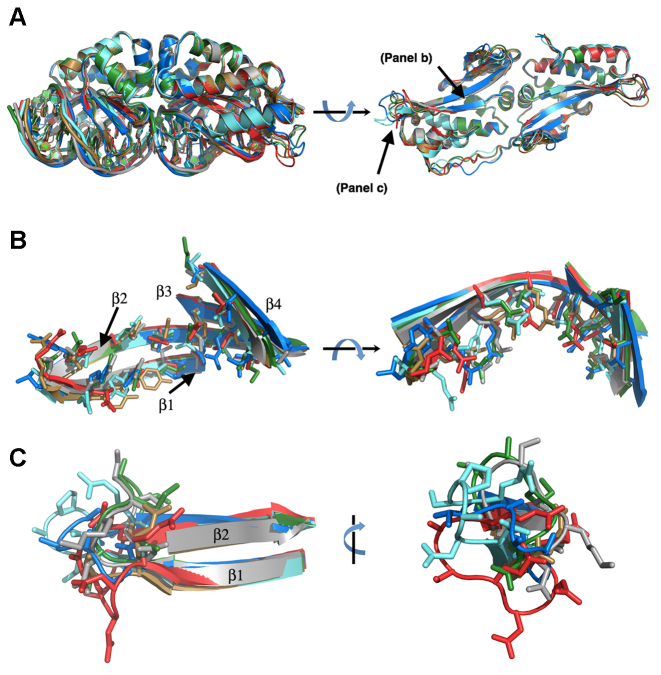
Figure 3.
Structural superposition of wild-type and eOnu-DNA complexes. (A) Full length enzyme scaffolds (left image viewed from the side bound to DNA; right image viewed into the DNA-binding surface of each enzyme in the absence of DNA). (B) Alignment of DNA-contacting β-sheets and corresponding side chains from the enzymes' N-terminal domains (DNA not shown). These residues form the surface that contacts base pair positions -8 to -3 in the 5′ half-site of the DNA target. (C) Alignment of DNA contacting loops connecting β-strands 1 and 2 in the N-terminal protein domain (DNA not shown). These residues contact the outer three bases (positions –11, –10 and –9) at the extreme 5′ end of the DNA target. See Supplementary Table S1 for crystallographic data and refinement statistics, Supplementary Table S2 for rmsd values of the superimposed coordinates, Figure 2 for additional views of the individual scaffolds and electrostatic surface potentials of the DNA-binding surfaces for the protein constructs, and Figure 4 for bending analyses of the DNA target sites. Colors: WT I-OnuI = gray, eOnuCCR5 = blue, eOnuTCRα = red, eOnuHIVInt = sand, eOnu7280 = green and eOnu11377 = aquamarine.
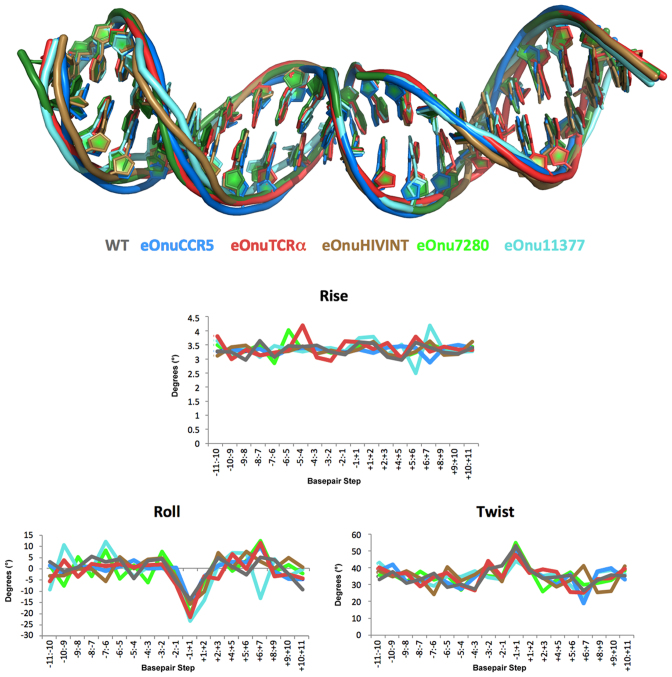
Figure 4.
DNA bending. Upper panel: superposition of the bound uncleaved DNA targets from each crystal structure. Lower panels: Output of 3DNA computational analyses (49) of three major DNA bending parameters (rise, roll and twist) for each crystal structure. Each engineered enzyme imparts a similar bend to the center of the DNA target, resulting in significant narrowing of the minor groove between the ‘central 4’ bases of the target (positions –2 to +2 in Figures 1 and 5).
The reorganization and structural changes in these proteins that facilitate recognition of alternate DNA targets can be described as the sum of: (i) small protein backbone motions involving DNA-contacting β-sheets (that contribute the largest share of contacts to nucleotide bases throughout the major groove) (Figure 3B); (ii) much larger reorganization of flanking protein loops at both ends of the β-sheets (Figure 3C), and (iii) extensive role-swapping throughout the entirety of the protein–DNA interface (explained further in the next paragraph).
As summarized in Table 2, ∼50 residues are localized to the protein DNA interface, and have the potential to form contacts to the target in a reengineered variant of the enzyme. These positions correspond to all residues in the DNA-bound structure of wild-type I-OnuI that are located on each of the eight separate DNA-contacting β-strands and that face towards the bound DNA, as well as all additional residues located at any position in the loop regions that connect those β-strands. In each structure (including the wild-type protein–DNA complex), a subset of those residues participates in DNA contacts, while the remainder are engaged in neighboring structural interactions that contribute to the overall structural organization of the protein's DNA-binding surface (but do not make obvious contacts to the DNA target). In the wild-type protein, 22 of these residues (44% of residues located in the protein–DNA interface) are clearly involved in nucleotide contacts. In the five reengineered variants of the enzyme, that same percentage ranges from a low of 17 DNA-contacting positions (34%) to 25 (50%). For each of the reengineered enzymes, approximately one-third of the side chains in the protein interface exchange roles in DNA recognition (converting between a DNA-contact and an indirect structural role or vice-versa) relative to the original wild-type enzyme.
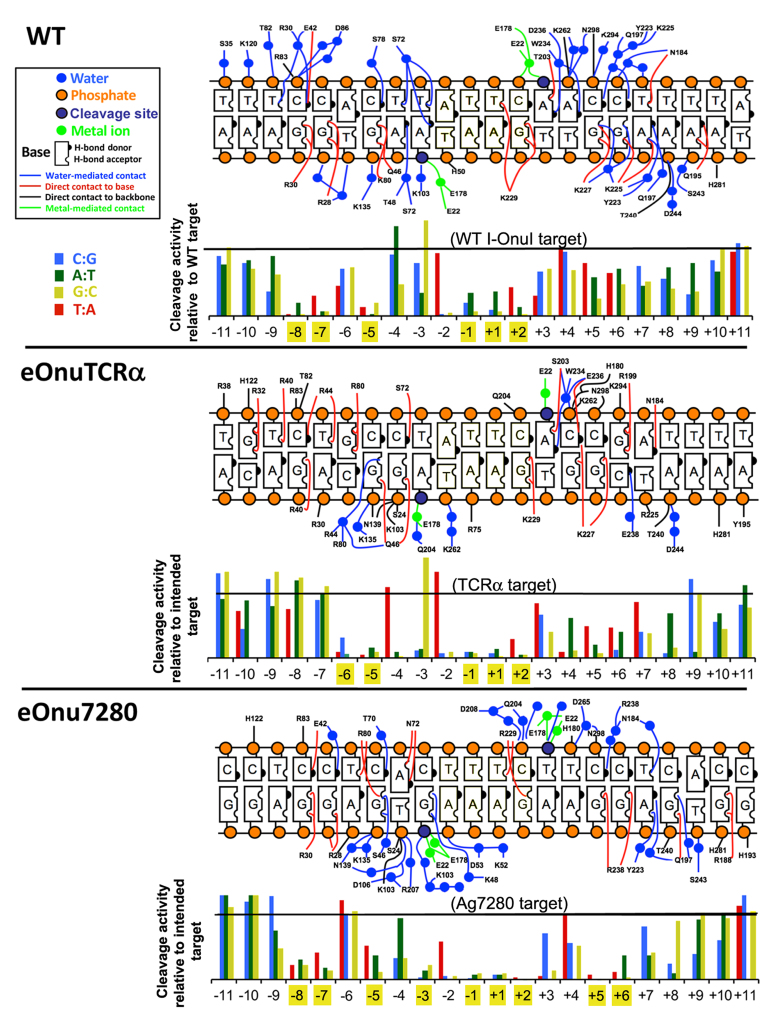
We next asked if the number and percentage of ‘DNA contacting’ residues observed in the interface of the various enzyme constructs (Table 2), as well as the overall composition of the protein surface (Figure 2), are closely correlated to the recognition specificity of the individual enzymes. To examine this question, we determined and compared the specificity profiles of the engineered eOnuTCRα enzyme (which exhibits 23 DNA contacting residues, corresponding to 46% of the surface) and the eOnu7280 enzyme (which only displays DNA contacts for 17 out of the same 50 residues, and contains significantly more non-polar and glycine residues) to one another, as well as to the specificity profile of the original wild-type enzyme (51). These three enzymes are all very active, driving high levels of gene modification in the original host (52), in primary human T-cells and in transgenic mosquitos (Table 1). This analysis (Figure 5 and Supplementary Figure S4) demonstrated that both redesigned enzyme variants are at least as specific as the original wild-type enzyme, and that their specificity profiles differ substantially from one another. The eOnu7280 enzyme displays significant discrimination at 9 individual positions in its target site (displaying greater than 50% reduction in cleavage activity as a result of any single base pair substitution at any of those positions), as compared to similar basepair discrimination at 5 or 6 positions by the eOnuTCRα and WT I-OnuI enzymes, respectively. All three enzymes display strong fidelity at positions –1, +1 and +2 (at the center of their DNA targets), which appears to indicate that they each read out, in a similar manner to one another, similar bent conformations of the DNA backbone across those positions (Figure 4).
Figure 5.
Contact maps and specificity plots for WT I-OnuI, eOnuTCRα and eOnu7280. Top of each panel: Residues that display direct (red or black), water-mediated (blue), or metal-mediated (green) contacts to the DNA bases and backbone by three variants of the enzyme. See also Table 2. Bottom of each panel: Specificity ‘information content’ plots of the same enzymes. At each position in the DNA target site, the effect of each of three separate possible single base pair substitutions on cleavage activity was measured. The cleavage activity of each enzyme against 66 separate substrates (three possible substitutions at 22 separate base pair positions; shown in the bar graphs) relative to the enzyme's intended ‘on-target’ substrate were measured (39). For each enzyme, positions in the DNA target that are limited to recognition and efficient cleavage in the presence of only one particular basepair (i.e. any basepair substitution at that position causes >50% reduction in cleavage) is highighted in yellow. For each construct, the experiment was repeated three times with biological replicates corresponding to separately transfected and induced yeast cultures; one representative set of results is displayed in the figure. Because the absolute magnitude of cell staining in each replicate depends upon overall enzyme expression in each replicate, presenting mean values and corresponding error bars representing standard deviation would require undesirable normalization between replicates; nonetheless the results shown here are reproducible and representative of each enzyme's behavior in those experiments. See Supplementary Figure S3 for example raw data for each enzyme.
The specificity profiles of the remaining three redesigned enzymes (eOnuCCR5, eOnuHIVInt and eOnu11377) have also been examined and agree with the analysis and conclusions above (reference (36) and unpublished data).
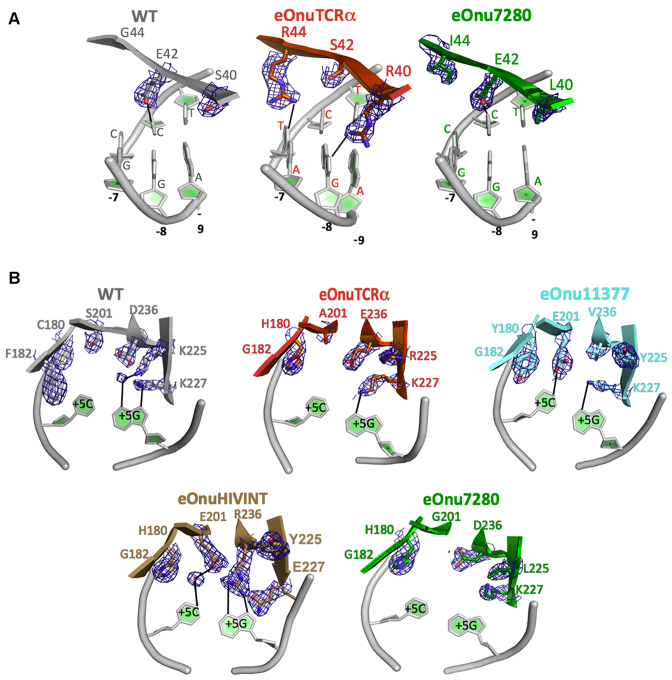
The comparison of structures of the wild-type and various redesigned meganucleases provides many explicit examples of role-swapping between immediately neighboring residues, from participating in direct or water-mediated contacts to DNA atoms, to instead playing an indirect structural role in the organization of the DNA binding surface (and vice-versa) (Figure 6A). Notable exchange of protein side chains in this process includes the introduction or removal of glycines, small aliphatic residues, and beta-branched residues at various positions, in order to accommodate and stabilize precise rotameric side conformations at neighboring residues that contact DNA atoms. In addition, long residues with high conformational freedom (particularly arginines) are able to either form DNA contacts, or to participate in purely structural contacts within the protein's binding surface. Even at a position where the identity of a particular DNA base pair is maintained across most of the protein–DNA complexes, the contacts and corresponding recognition mechanisms differ significantly from one another (Figure 6B).
Figure 6.
Examples of role-swapping between neighboring amino acid positions in the protein–DNA interface. (A) Residues 40, 42 and 44 are part of a cluster of amino acids that are located near DNA base pair positions –7, –8 and –9 near the 5′ end of the DNA target site. (B) Example of context-dependent diversity in recognition. Position +5 in the DNA target sites in this study corresponding to a C:G base pair for the wild-type enzyme and four of the five engineered variants of the meganuclease. A wide variety of amino acid identities and contacts, spanning residues 180, 182, 201, 225, 227 and 236, are required to dictate the same final specificity at that position, in a manner that is highly dependent on the surrounding sequence and structural context of each enzyme. Electron density corresponds to features observed in a simulated annealing composite omit map displayed at 1.5 sigma contour level.
DISCUSSION
The results of this study, along with prior crystallographic analyses of reengineered variants of the I-CreI meganuclease (18,25), demonstrate how a single LAGLIDADG meganuclease efficiently adopts multiple new DNA target specificities, even when changes to its sequence and composition is limited almost entirely to its DNA-binding surface (a fraction of the protein that corresponds to approximately one-sixth of the entire protein). Additional sequence changes in the surrounding protein scaffold does not appear to be an essential component of their ability to be extensively reprogrammed for new DNA targets. This property would seem to match the biological and genetic requirements placed upon a successful mobile element, that needs to be persistent in its recognition of an existing target site, as well as opportunistic when a new target site presents itself for ectopic transfer.
These results also reaffirm a broad body of published literature that collectively indicate that the balance of interactions that dictate protein–DNA recognition specificity comprises the formation of directional hydrogen bonds between protein residues and nucleotide bases, the overall steric complementarity of the protein–DNA interface, and upon recognition of the global structure and shape of the DNA target. Recognition specificity is derived from the combined contributions of both DNA-contacting and neighboring structural residues. In the case of the meganucleases studied here, the entire interface behaves as a highly fluid and readily malleable contact region during either evolutionary or man-made changes to DNA target specificity.
The combination of the highly conserved 3D structure and topology of LAGLIDADG meganucleases and the relatively facile manner by which their DNA recognition can be altered (largely via resculpting of only their protein–DNA interface) stands in contrast to restriction endonucleases, which are notable both for the significant divergence of their sequences and folded structures (while maintaining similar active site architectures) and their intransigence to protein engineering for the purpose of altering their DNA recognition and cleavage specificity (53–55). In the published examples of attempts to alter restriction endonuclease specificity, it has been noted that “even for very well characterized REases, the properties that determine specificity and selectivity are difficult to model with the available structural information…furthermore, the crystal structure of the recognition complex represents a form of the ‘ground state’, but catalysis involves the ‘transition state’, which may depend upon additional interactions not evident in the crystal structure" (55).
While this statement and conclusion is undoubtedly true, it could also be used to describe meganuclease activity and specificity, particularly the observation that a great deal of specificity is realized at the transition state of the cleavage reaction (11). It appears that very different selection pressures on these endonucleases have dictated the ‘reprogrammability’ of meganucleases (which, as the drivers of mobile genetic elements, must continuously adjust to new and shifting targets). In contrast, type II restriction endonucleases (which cannot dramatically alter their specificity without becoming toxic to their hosts) display quite different structural and energetic landscapes surrounding their cognate DNA complexes that are reflected in very different, more highly constrained routes for evolvability and engineerability.
In terms of protein engineering, particularly for the purpose of altering biomolecular recognition such as protein–DNA binding, a large body of historical literature has demonstrated an overall lack of consistent ‘codes’ or propensities for certain types of sidechains to consistently interact with certain base pairs (56). Considerable effort, involving extensive rounds of selections across wide regions of protein DNA-contacting surfaces, is often required when investigators attempt to retarget the specificity of the many types of nucleic acid-acting enzymes that are commonly used for molecular biology and biotechnology applications (recently reviewed in (57)). As illustrated in this study and many others, the contribution of ‘nearby neighbors’ in a protein–DNA interface should be accounted for in studies that are intended to either evaluate or actually redesign the specificity and function of a DNA-binding protein.
The eventual generation of a significant number of experimentally validated meganuclease–DNA cocrystal structures, featuring quite similar protein scaffolds that recognize considerably different target sequences, may eventually facilitate the development machine-learning algorithms as part of a redesign strategy that is considerably more automated and reliable. Such computational approaches would need to accurately model, with far greater precision than is currently possible, potential changes in the conformation of the DNA backbone and underlying base pairs, the conformation of protein loops at the distal ends of the complex, and the presence and participation of solvent molecules in the protein–DNA interface (Supplementary Figure S5). We envision that a dedicated effort in this direction may enable such an approach in the future.
Finally, these results illustrate the physical basis for the virtually limitless range of evolutionary retargeting of protein–DNA specificity for at least one type of mobile endonuclease. By empowering all residues through the molecular interface to potentially play a significant role in specificity determination, rather than relying only on a small subset of those that are observed to make DNA contacts in an individual complex. By sampling the considerable diversity of shapes and recognition mechanisms that can be generated during the modification of the extensive protein–DNA binding surface, and by doing so over an enormous scale of both time and protein variants, it is clear that even when constrained to a highly-conserved protein scaffold, nature can achieve virtually any recognition specificity required to satisfy the ongoing demands of selection pressures for fitness and survival.
DATA AVAILABILITY
All structures and corresponding processed X-ray data have been deposited in the RCSB protein data base (ID codes 5THG, 5T2H, 5T8D, 5T2N and 5T2O) for immediate release. All PDB deposition ID codes are also listed in Table 1 and Supplementary Table S1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Betty Shen, Ms Lindsey Doyle and Ms Ai Takeuchi for expert technical assistance. The Berkeley Center for Structural Biology, where the data for four of the protein–DNA complexes were collected, is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02–05CH11231.
Author contributions: J.J., K.H. and A.R.L. conducted the protein engineering of the I-OnuI DNA-binding protein; J.H. and A.R.L. contributed in vitro evaluations of enzyme specificity; R.W., J.H. and A.R.L. determined the crystal structures; T.N, R.G. N.W. and A.C. contributed mosquito transgenesis experiments and evaluation of in vivo enzyme activity. The manuscript was written by B.L.S. with continuous contributions and editing and final approval of all co-authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIGMS [R01 GM105691]; Bill and Melinda Gates Foundation; Fred Hutchinson Cancer Center; Bluebird Bio, Inc. Funding for open access charge: NIGMS [R01 R01 GM105691]; Fred Hutchinson Cancer Research Center.
Conflict of interest statement. J.J. and K.H. are employees of Bluebird Bio. Inc., which uses engineered meganucleases for genome engineering applications.
REFERENCES
- 1. Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S.. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007; 131:530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein–DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lazarovici A., Zhou T., Shafer A., Dantas Machado A.C., Riley T.R., Sandstrom R., Sabo P.J., Lu Y., Rohs R., Stamatoyannopoulos J.A. et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6376–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kitayner M., Rozenberg H., Rohs R., Suad O., Rabinovich D., Honig B., Shakked Z.. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2010; 17:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordan R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L.. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013; 3:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordan R., Rohs R.. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014; 39:381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stoddard B.L. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mobile DNA. 2014; 5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chevalier B., Monnat R.J.J., Stoddard B.L.. Belfort M, Wood DW, Stoddard BL, Derbyshire V. Homing Endonucleases and Inteins. 2005; 16, Berlin: Springer-Verlag; 33–47. [Google Scholar]
- 9. Scalley-Kim M., McConnell-Smith A., Stoddard B.L.. Coevolution of homing endonuclease specificity and its host target sequence. J. Mol. Biol. 2007; 372:1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lambert A.R., Hallinan J.P., Shen B.W., Chik J.K., Bolduc J.M., Kulshina N., Robins L.I., Kaiser B.K., Jarjour J., Havens K. et al. Indirect DNA sequence recognition and its impact on nuclease cleavage activity. Structure. 2016; 24:862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thyme S.B., Jarjour J., Takeuchi R., Havranek J.J., Ashworth J., Scharenberg A.M., Stoddard B.L., Baker D.. Exploitation of binding energy for catalysis and design. Nature. 2009; 461:1300–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ashworth J., Havranek J.J., Duarte C.M., Sussman D., Monnat R.J. Jr, Stoddard B.L., Baker D.. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006; 441:656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seligman L., Chisholm K.M., Chevalier B.S., Chadsey M.S., Edwards S.T., Savage J.H., Veillet A.L.. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002; 30:3870–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chames P., Epinat J.C., Guillier S., Patin A., Lacroix E., Paques F.. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005; 33:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith J., Grizot S., Arnould S., Duclert A., Epinat J.C., Chames P., Prieto J., Redondo P., Blanco F.J., Bravo J. et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006; 34:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao H., Smith J., Yang M., Jones S., Djukanovic V., Nicholson M.G., West A., Bidney D., Falco S.C., Jantz D. et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010; 61:176–187. [DOI] [PubMed] [Google Scholar]
- 17. Arnould S., Perez C., Cabaniols J.-P., Smith J., Gouble A., Grizot S., Epinat J.-C., Duclert A., Duchateau P., Paques F.. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007; 371:49–65. [DOI] [PubMed] [Google Scholar]
- 18. Redondo P., Prieto J., Munoz I.G., Alibes A., Stricher F., Serrano L., Cabaniols J.P., Daboussi F., Arnould S., Perez C. et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008; 456:107–111. [DOI] [PubMed] [Google Scholar]
- 19. Dupuy A., Valton J., Leduc S., Armier J., Galetto R., Gouble A., Lebuhotel C., Stary A., Paques F., Duchateau P. et al. Targeted gene therapy of Xeroderma pigmentosum cells using meganuclease and TALEN. PLoS One. 2013; 8:e78678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cabaniols J.P., Ouvry C., Lamamy V., Fery I., Craplet M.L., Moulharat N., Guenin S.P., Bedut S., Nosjean O., Ferry G. et al. Meganuclease-driven targeted integration in CHO-K1 cells for the fast generation of HTS-compatible cell-based assays. J. Biomol. Screen. 2010; 15:956–967. [DOI] [PubMed] [Google Scholar]
- 21. Djukanovic V., Smith J., Lowe K., Yang M., Gao H., Jones S., Nicholson M.G., West A., Lape J., Bidney D. et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013; 76:888–899. [DOI] [PubMed] [Google Scholar]
- 22. Antunes M.S., Smith J.J., Jantz D., Medford J.I.. Targeted DNA excision in Arabidopsis by a re-engineered homing endonuclease. BMC Biotechnol. 2012; 12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. D’Halluin K., Vanderstraeten C., Van Hulle J., Rosolowska J., Van Den Brande I., Pennewaert A., D’Hont K., Bossut M., Jantz D., Ruiter R. et al. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol. J. 2013; 11:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grizot S., Smith J., Daboussi F., Prieto J., Redondo P., Merino N., Villate M., Thomas S., Lemaire L., Montoya G. et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009; 37:5405–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munoz I.G., Prieto J., Subramanian S., Coloma J., Redondo P., Villate M., Merino N., Marenchino M., D’Abramo M., Gervasio F.L. et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011; 39:729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menoret S., Fontaniere S., Jantz D., Tesson L., Thinard R., Remy S., Usal C., Ouisse L.H., Fraichard A., Anegon I.. Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J. 2013; 27:703–711. [DOI] [PubMed] [Google Scholar]
- 27. Grosse S., Huot N., Mahiet C., Arnould S., Barradeau S., Clerre D.L., Chion-Sotinel I., Jacqmarcq C., Chapellier B., Ergani A. et al. Meganuclease-mediated Inhibition of HSV1 Infection in Cultured Cells. Mol. Ther. 2011; 19:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Popplewell L., Koo T., Leclerc X., Duclert A., Mamchaoui K., Gouble A., Mouly V., Voit T., Paques F., Cedrone F. et al. Gene correction of a duchenne muscular dystrophy mutation by meganuclease-enhanced exon knock-in. Hum. Gene Ther. 2013; 24:692–701. [DOI] [PubMed] [Google Scholar]
- 29. Jarjour J., West-Foyle H., Certo M.T., Hubert C.G., Doyle L., Getz M.M., Stoddard B.L., Scharenberg A.M.. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009; 37:6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S., Dryden D.T., Dybvig K. et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003; 31:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baxter S.K., Lambert A.R., Scharenberg A.M., Jarjour J.. Flow cytometric assays for interrogating LAGLIDADG homing endonuclease DNA-binding and cleavage properties. Methods Mol. Biol. 2013; 978:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baxter S.K., Scharenberg A.M., Lambert A.R.. Engineering and flow-cytometric analysis of chimeric LAGLIDADG homing endonucleases from homologous I-OnuI-family enzymes. Methods Mol. Biol. 2014; 1123:191–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takeuchi R., Choi M., Stoddard B.L.. Redesign of extensive protein–DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4061–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takeuchi R., Choi M., Stoddard B.L.. Engineering of customized meganucleases via in vitro compartmentalization and in cellulo optimization. Methods Mol. Biol. 2015; 1239:105–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boissel S.J., Astrakhan A., Jarjour J., Adey A., Shendure J., Stoddard B.L., Certo M., Baker D., Scharenberg A.M.. MegaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2013; 42:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ibarra G.S.R., Paul B., Sather B.D., Younan P.M., Sommer K., Kowalski J.P., Hale M., Stoddard B., Jarjour J., Astrakhan A. et al. Efficient modification of the CCR5 Locus in primary human T Cells with megaTAL nuclease establishes HIV-1 resistance. Mol. Ther.-Nucleic Acids. 2016; 5:e352–e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015; 7:307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thyme S.B., Boissel S.J., Arshiya Quadri S., Nolan T., Baker D.A., Park R.U., Kusak L., Ashworth J., Baker D.. Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res. 2014; 42:2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Workman C.T., Yin Y., Corcoran D.L., Ideker T., Stormo G.D., Benos P.V.. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005; 33:W389–W392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan Y.S., Takeuchi R., Jarjour J., Huen D.S., Stoddard B.L., Russell S.. The Design and In Vivo Evaluation of Engineered I-OnuI-Based Enzymes for HEG Gene Drive. PLoS One. 2013; 8:e74254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Otwinowski Z., Minor W.. Carter CWJ, Sweet RM. Methods in Enzymology. 1997; 276, Academic Press; 307–326. [DOI] [PubMed] [Google Scholar]
- 42. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystal. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Cryst. D. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 46. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. The PyMOL Molecular Graphics System, V. 2016; Schrödinger, LLC. [Google Scholar]
- 48. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 49. Zheng G., Lu X.-J., Olson W.K.. Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009; 37:W240–W246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sedlak R.H., Liang S., Niyonzima N., De Silva Feelixge H.S., Roychoudhury P., Greninger A.L., Weber N.D., Boissel S., Scharenberg A.M., Cheng A. et al. Digital detection of endonuclease mediated gene disruption in the HIV provirus. Sci. Rep. 2016; 6:20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeuchi R., Lambert A.R., Mak A.N.-S., Jacoby K., Dickson R.J., Gloor G.B., Scharenberg A.M., Edgell D.R., Stoddard B.L.. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13077–13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sethuraman J., Majer A., Friedrich N.C., Edgell D.R., Hausner G.. Genes within genes: multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol. Biol. Evol. 2009; 26:2299–2315. [DOI] [PubMed] [Google Scholar]
- 53. Lanio T., Jeltsch A., Pingoud A.. On the possibilities and limitation of rational protein design to expand the specificity of restriction enzymes: a scase study employing EcoRV as the target. Protein Eng. 2000; 13:275–281. [DOI] [PubMed] [Google Scholar]
- 54. Jeltsch A., Wenz C., Wende w., Selent U., Pingoud A.. Engineering novel restriction endonucleases: principles and applications. Trends Biotech. 1996; 14:235–238. [DOI] [PubMed] [Google Scholar]
- 55. Pingoud A., Wilson G.G., Wende W.. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res. 2014; 42:7489–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lavery R. Recognizing DNA. Q. Rev. Biophys. 2005; 38:339–344. [DOI] [PubMed] [Google Scholar]
- 57. Glasscock C.J., Lucks J.B., DeLisa M.P.. Engineered protein machines: emergent tools for synthetic biology. Cell Chem. Biol. 2016; 23:45–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All structures and corresponding processed X-ray data have been deposited in the RCSB protein data base (ID codes 5THG, 5T2H, 5T8D, 5T2N and 5T2O) for immediate release. All PDB deposition ID codes are also listed in Table 1 and Supplementary Table S1.