Abstract
Background
RNAs within extracellular vesicles (EVs) have potential as diagnostic biomarkers for patients with cancer and are identified in a variety of biofluids. Glioblastomas (GBMs) release EVs containing RNA into cerebrospinal fluid (CSF). Here we describe a multi-institutional study of RNA extracted from CSF-derived EVs of GBM patients to detect the presence of tumor-associated amplifications and mutations in epidermal growth factor receptor (EGFR).
Methods
CSF and matching tumor tissue were obtained from patients undergoing resection of GBMs. We determined wild-type (wt)EGFR DNA copy number amplification, as well as wtEGFR and EGFR variant (v)III RNA expression in tumor samples. We also characterized wtEGFR and EGFRvIII RNA expression in CSF-derived EVs.
Results
EGFRvIII-positive tumors had significantly greater wtEGFR DNA amplification (P = 0.02) and RNA expression (P = 0.03), and EGFRvIII-positive CSF-derived EVs had significantly more wtEGFR RNA expression (P = 0.004). EGFRvIII was detected in CSF-derived EVs for 14 of the 23 EGFRvIII tissue-positive GBM patients. Conversely, only one of the 48 EGFRvIII tissue-negative patients had the EGFRvIII mutation detected in their CSF-derived EVs. These results yield a sensitivity of 61% and a specificity of 98% for the utility of CSF-derived EVs to detect an EGFRvIII-positive GBM.
Conclusion
Our results demonstrate CSF-derived EVs contain RNA signatures reflective of the underlying molecular genetic status of GBMs in terms of wtEGFR expression and EGFRvIII status. The high specificity of the CSF-derived EV diagnostic test gives us an accurate determination of positive EGFRvIII tumor status and is essentially a less invasive “liquid biopsy” that might direct mutation-specific therapies for GBMs.
Keywords: biomarker, CSF, EGFRvIII, glioma, vesicle
Importance of the study
In this article we investigate the utility of the EGFRvIII mutation as a biomarker in the CSF for patients with glioblastomas. This mutation is prevalent in the glioma population, and methods for DNA and RNA isolation from extracellular vesicles have improved our ability for detection of genetic alterations in human biofluids. The results from our translational investigation into a “liquid biopsy” would allow clinicians to not only diagnose EGFRvIII-positive glioblastomas, but also follow patients in the postoperative/chemotherapy/radiation period for declining levels of EGFRvIII in concordance with effective tumor treatment, as well as monitor for increasing EGFRvIII levels upon tumor recurrence. Furthermore, our methods may aid in the distinction of tumor necrosis versus tumor recurrence found on follow-up radiologic imaging and reduce the need for costly and unnecessary biopsy procedures. While EGFRvIII is glioma specific, we feel that this approach is applicable to a wide range of oncologic conditions.
Extracellular vesicles (EVs) obtained from biofluids, including microvesicles and exosomes, can contain tumor-derived RNAs which may be useful as diagnostic and prognostic biomarkers for patients with cancer. These EV-derived RNAs are present in many biofluids, including serum and cerebrospinal fluid (CSF). Glioblastomas (GBMs) are malignant infiltrative primary tumors of the central nervous system which release EVs into the serum and CSF.1–3 EVs containing RNAs can be readily isolated from these biofluids.4 While serum has promise as source material for these EV-derived RNAs, CSF analysis offers several advantages.3 The CSF biofluid can readily communicate with intrinsic brain tumors lying in proximity to the ventricular system of brain and its overlying subarachnoid spaces. Additionally, this fluid generally contains few inflammatory or brain-derived cells and is less likely than plasma or sera to be co-mixed with EVs of white cell or platelet origin, and less likely to contain EVs of non-brain tissue origin versus blood EVs. Thus, analyses of CSF-derived EV RNA offer the potential to detect genetic aberrations intrinsic to malignant gliomas.
Two common genetic alterations in glioblastomas are wild-type epidermal growth factor receptor (wtEGFR) amplification and EGFR variant (v)III mutation. Amplification and overexpression of wtEGFR is present in most GBMs and in virtually all of the classical subtype GBMs.5 The classical subtype is known to have high-level wtEGFR DNA copy number amplifications (97%), as well as significant increases (P < 0.01) in wtEGFR RNA expression. Furthermore, 23% of classical GBMs harbor the EGFRvIII mutation.5 EGFRvIII is the most common mutation of the wtEGFR gene and contains a deletion of exons 2 through 7, resulting in an in-frame deletion of 267 amino acids. This deletion leads to a truncated extracellular portion of the protein which cannot bind the endothelial growth factor ligand. However, the mutation also confers a constitutively active state, and is therefore associated with tumor progression through downstream effects separable from those of wtEGFR. Amplification of wtEGFR appears in about one-third of GBMs, and protein overexpression in about one-half of these tumors.5 EGFRvIII expression occurs in approximately one-third of GBM.5 Thus, both wtEGFR and EGFRvIII have been the subject of targeted drug and immunization trials in patients and are of interest as GBM-specific diagnostic and prognostic biomarkers.6
Here we assessed whether we could detect, isolate, and interrogate CSF-derived EV RNA from patients with presumptive GBMs prior to tumor resection. Our multi-institutional study included the analysis of CSF for EV RNA expression of both wtEGFR and EGFRvIII mutation. We then compared these CSF data with those of the tissue biopsy “gold standard” to evaluate the sensitivity and specificity of this CSF-derived EV RNA as a diagnostic biomarker for GBMs.
Materials and Methods
Patient Biospecimens
A consortium of institutions was developed and standardized institutional review board applications and consent forms and specimen handling protocols were created for the collection of CSF and brain tumor tissue from patients scheduled for surgical resection of presumed malignant glioma (GBM). All samples were collected with informed consent according to the appropriate protocols approved by the local institutional review board at each participating site. All patients were required to subsequently have confirmed pathologic diagnosis of glioblastoma to have tissue and CSF further analyzed in this study. Excluded from study were children, patients with presumed GBM who were not surgical candidates, patients with recurrent GBM, patients with presumed GBM and intracerebral infections, and those with hemorrhagic changes in CSF. CSF samples from patients diagnosed with de novo primary GBM were collected according to surgeon preference and usual clinical practice immediately prior to surgery (via lumbar puncture) or after opening the dura mater but prior to surgical tumor manipulation. Patients were provided “standard of care” at participant sites, including fasting prior to surgery, and most patients had received (as per practice) anti-edema corticosteroids and anticonvulsants, but not chemotherapy. A total of 71 patients were included in the study.
Pathologic Investigations and Tissue Handling
Two resected pieces of tumor were used from each patient as a “gold standard” for amplifications and mutations. Multiple tissue samples were used to minimize influence of tumor mutation heterogeneity. Tumor samples were collected intraoperatively at the time of tumor manipulation. Standard neuropathologic criteria were utilized for tumor grading. After frozen section confirmation of glioma and confirmation with the pathologist that sufficient material had been obtained for clinical diagnostic processing, 2 additional peanut-sized specimens of tumor were obtained. The tumor was accessioned into 1.8 mL cryotubes (Fisher Scientific) and stored at −80°C until use. For tumor samples, 100 ng RNA was analyzed from each resected tissue sample for EGFR and EGFRvIII expression. Genomic amplification of EGFR was also characterized in the tissues. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and 18S ribosomal RNA (rRNA) were analyzed as controls.
Standard Operating Procedures for CSF EV Specimen Handling and Processing
All site investigators received in-person or video training on tissue and CSF collection and handling procedures. Extracellular vesicles from 4 mL CSF were analyzed from each patient. CSF was filtered by slowly passing it through a 0.8 μm syringe filter into a 50 mL collection tube (Millipore) and aliquoted into each of the specified 1.8 mL cryotubes (Fisher Scientific), using a transfer pipette and stored at −80°C until use. For hemolyzed CSF, CSF was transferred to a Centrifuge BD PPT Tube in a balance, swing-out rotor type centrifuge at room temperature at 1500 relative centrifugal force for 10 minutes. Then the CSF was filtered through a 0.8 μm syringe filter, aliquoted into 1.8 mL cryotubes, and stored at −80°C.
Isolation of Extracellular Vesicle RNA
Isolation of RNA from extracellular vesicles was performed as previously described with a few modifications.7 Briefly, 1 mL of serum was transferred to an ultracentrifuge tube, diluted 1:3 with cold phosphate buffered saline (PBS) and centrifuged at 120000 g for 80 minutes at 8°C, and the supernatant was carefully aspirated without disturbing the extracellular vesicle pellet. The pellet was resuspended and treated for 15 minutes with 4 U of DNase I (Ambion) (in 25 μL of the accompanying buffer). Subsequently, 700 μL miRNeasy lysis buffer (Qiazol Reagent) was then added to the tube and the RNA was isolated following the manufacturer’s recommendations. After elution of the RNA from the column in 30 μL nuclease-free water (Ambion), the RNA was precipitated by adding 2.5 volumes of 100% ethyl alcohol and 1/10 volume of 3 M sodium acetate (pH 5.2), and incubated at −20°C for 1 hour. Samples were then centrifuged for 20 minutes at 16000 g and the supernatant was removed. The pellet was left to dry at room temperature and dissolved in 14 μL nuclease-free water and stored at −80°C until needed. RNA quality and concentration were assessed with the Agilent Bioanalyzer RNA Pico Chip and the Nanodrop 2000 (Thermo Scientific). The RNA was reverse transcribed and analyzed for EGFRvIII expression as well as EGFR, GAPDH, and 18S rRNA.
Tissue Preparation
The RNALater Ice (Ambion) was pre-chilled at −80°C and then added to the snap-frozen tumor biopsy (not greater than 0.5 cm in thickness) according to the manufacturer’s protocol. The biopsy sample (no more than 30 mg) was then placed in −20°C for at least 16 hours until ready to isolate.
DNA Extraction from Tissue
The tumor biopsy sample was minced using a motorized pestle in a total volume of 800 µL digestion buffer (60 mM Tris, pH 8.0, 100 mM EDTA, 0.5% sodium dodecyl sulfate). Proteinase K (Qiagen) was then added to a final concentration of 500 µg/mL, and the sample was mixed by inversion. The sample was then incubated at 55°C for approximately 4 hours with occasional mixing. The sample was subsequently transferred to a pre-spun Phase Lock Gel (PLG) Heavy tube and one volume of phenol:chloroform (pH 8) was added. The tube was rapidly inverted for 20 seconds and then centrifuged at 16000 × g for 5 minutes at room temperature. The aqueous phase was removed by aspiration and the phenol:chloroform step was performed 2 more times. The organic phase was then added to a new pre-spun PLG Heavy tube, with the addition of one volume of pure chloroform to wash off any residual phenol. The tube was then rapidly inverted for 20 seconds and subsequently centrifuged at 16000 × g for 5 minutes at room temperature. The aqueous phase was removed by aspiration and the volume of the residual organic phase was noted. One-tenth of the residual volume of 3M sodium acetate (pH 5.2) and one volume 100% ethanol was then added, and the tube was mixed by inversion at room temperature. The sample was then centrifuged at 16000 × g for 5 minutes at room temperature. The aqueous phase was removed by aspiration, and the residual pellet was washed once with 500 µL 70% ethanol and centrifuged again at 16000 × g for 5 minutes at room temperature. Residual ethanol was removed by aspiration and the pellet was allowed to air-dry at room temperature before resuspension in Tris/EDTA (TE) buffer. The sample was kept at 4°C overnight before DNA concentration measurement. Diluted DNA was then stored at 100 ng/µL in TE buffer at −80°C.
RNA Extraction from Tissue
The tumor sample was minced using a motorized pestle in a total volume of 700 µL Qiazol (Qiagen). The homogenate was allowed to sit at room temperature for 5 minutes to promote dissociation of nucleoprotein complexes. RNA was then purified from the sample using the miRNeasy extraction kit (Qiagen) according to manufacturer’s recommendation. Each RNA sample was measured using the NanoDrop spectrophotometer and diluted to 100 ng/µL prior to storage at −80°C.
Tissue Reverse Transcription
The tissue RNA was reverse transcribed using the VILO reverse transcription kit according to manufacturer’s recommendation. One hundred nanograms of RNA from each tissue was reverse transcribed.
CSF Extracellular Vesicle Isolation
Four milliliters of CSF was added to a 13 × 51 mm polyallomer tube containing 8 µL RNasin Plus RNase Inhibitor (Promega), then adjusted to 5 mL with PBS and incubated for 5 min at room temperature. The samples were spun at 120000 × g for 80 minutes at 8°C (MLS-50 rotor in an Optima Max-XP bench top ultracentrifuge; Beckman). Deceleration was set to 7. The extracellular vesicle pellet was pre-incubated with 8 μL RNase inhibitor in 42 μL PBS. Added to each sample was 700 μL Qiazol lysis buffer, and RNA was extracted using the miRNeasy micro isolation kit (Qiagen) according to manufacturer’s recommendation.
CSF Extracellular Vesicle RNA Reverse Transcription
The RNA was reverse transcribed using Sensiscript reverse transcriptase (Qiagen) according to manufacturer’s protocol and supplemented with 2 μg of T4 gene 32 protein (New England Biolabs).
Quantitative PCR for EGFR Amplification and EGFRvIII Mutation Detection
Twelve microliters of the RNA isolated from 1 mL of CSF were reverse transcribed using the Superscript VILO cDNA synthesis kit (Invitrogen), according to manufacturer’s recommendations. Samples were then preamplified using the TaqMan PreAmp Master Mix (Applied Biosystems). Briefly, 12.5 μL of the cDNA was added to the PreAmp Master Mix together with all the genes of interest and preamplified for 14 cycles, according to the manufacturer’s recommendations. The samples were then diluted 1:10 and TaqMan quantitative reverse transcription PCR was performed on all samples for all the selected genes. The amplification was performed using ABI PRISM 7500 with the following program: 50°C, 2 min; 95°C, 10 min; 40 cycles of 95°C, 15 s, 60°C, 1 min on standard mode. Logarithmic amplifications were interpreted as positive, and relative quantities versus GAPDH/18S were reported for each analyzed sample.
Wild-type EGFR primer sequences: 5ʹ-TATGTCCTCATTGC CCTCAACA
3ʹ-CTGATGATCTGCAGGTTTTCCA
EGFRvIII primer sequences: 5ʹ-CTGCTGGCTGCGCTCTG
3ʹ-GTGATCTGTCACCACATAATTACCTTTC
Statistical Analysis
Results were analyzed using GraphPad Prism 7.0a and R version 3.3.1. All tests were 2-tailed at the 5% significance level. For comparison of wtEGFR DNA amplification and RNA expression between EGFR-positive and -negative samples, we used the Wilcoxon signed rank test. For comparison of lumbar versus cisternal CSF sensitivity and specificity, we used Pearson’s chi-squared test.
Results
Patient Demographics
A total of 71 GBM patients—11 from the UCSD training set, 60 from the Extracellular RNA Collaborative Testing (ECT) validation set—entered the study for analysis, as described in Table 1. The median age of the 71 GBM patients was 61 years, with 51 (72%) being male. These metrics are in concordance with the published demographic data for glioblastomas.8 Tumor tissue and CSF from the UCSD training subjects were analyzed for EGFRvIII, while those from the ECT validation subjects were analyzed for both wtEGFR and EGFRvIII. CSF was collected from one of 2 sites: cisternal (40 patients) or lumbar (31 patients). EVs were subsequently isolated from the CSF by differential centrifugation and visualized by transmission electron microscopy.
Table 1.
Study demographics for the 71 high-grade glioma patients enrolled in the EGFRvIII CSF study
| N = 71 | |
|---|---|
| Age, median (range) | 61 y (21–82) |
| Gender | |
| Female | 20 (28.2%) |
| Male | 51 (71.8%) |
| CSF collection | |
| Cisternal | 40 (56.3%) |
| Lumbar | 31 (43.7%) |
| EGFRvIII tissue status | |
| Positive | 23 (32.4%) |
| Negative | 48 (67.6%) |
Detection of Wild-type EGFR and EGFRvIII
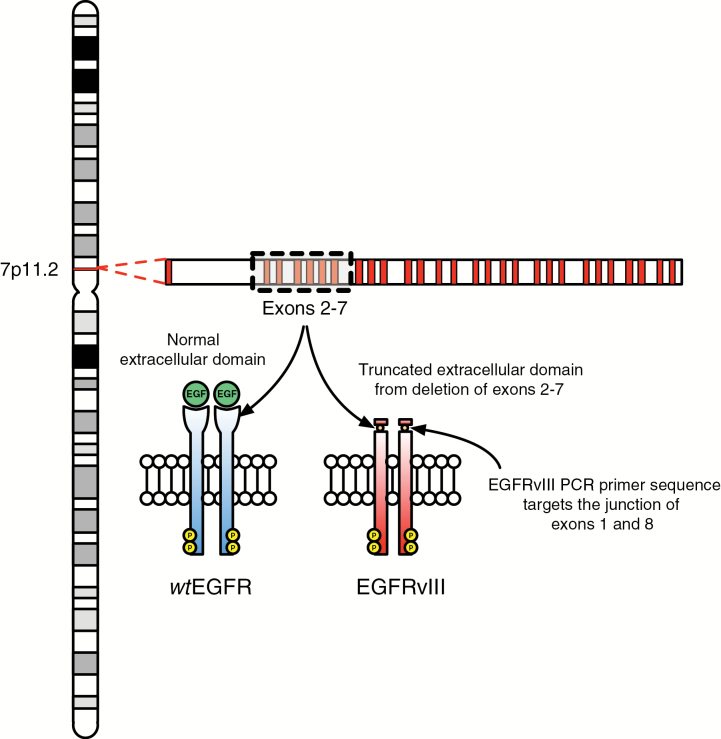
In order to distinguish between wtEGFR and EGFRvIII during quantitative (q)PCR analysis, we developed primer sequences to target different locations within the protein receptor. Fig. 1 shows that the primer sequence for EGFRvIII is at the junction of exons 1 and 8 as a result of an in-frame deletion of exons 2–7. This structural change in EGFRvIII allows for qPCR analysis distinct from wtEGFR. A subject was deemed to have an EGFRvIII-positive tumor if either of the 2 tumor samples exhibited logarithmic RNA amplification at higher cycle threshold values, as shown in Supplementary Figure S1. Of the 71 GBMs analyzed, 23 (32%) were positive for EGFRvIII in the tumor tissue.
Fig. 1.
EGFRvIII mutation. Comparison of wtEGFR vs EGFRvIII extracellular domains and depiction of target site for EGFRvIII PCR primer sequence.
Wild-type EGFR Amplification and Expression in Relation to EGFRvIII Tumor Status
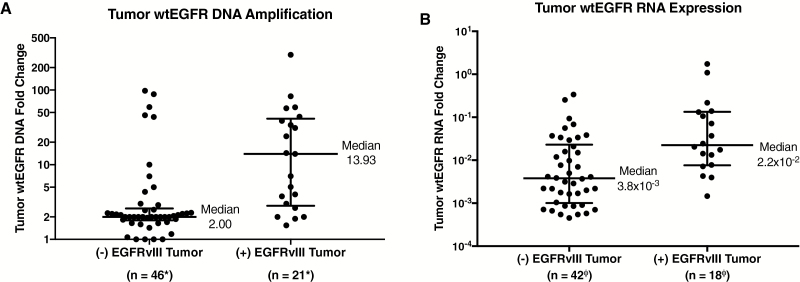
We assessed the relative DNA amplification of wtEGFR in tissue by qPCR, in relation to EGFRvIII expression in the 71 GBM patients (4 tissue samples excluded due to no DNA analysis). This interrogation revealed that the median tumor tissue wtEGFR copy number amplification was increased nearly 7.0-fold in EGFRvIII tissue–positive GBMs, as depicted in Fig. 2A. When normalized to genomic DNA RNaseP, EGFRvIII tissue–positive GBMs (median = 13.93) had significantly greater (P = 0.02, Wilcoxon test) amplification of wtEGFR DNA, compared with EGFRvIII tissue–negative GBMs (median = 2.00). This phenomenon also held true when evaluating the relative tissue RNA expression of wtEGFR in relation to the EGFRvIII tissue status of the 71 GBM patients (11 tissue samples excluded due to no wtEGFR RNA analysis). EGFRvIII tissue–positive GBMs demonstrated nearly a 6.0-fold greater median expression of wtEGFR mRNA, as depicted in Fig. 2B. When normalized to GAPDH mRNA and 18S rRNA, EGFRvIII tissue–positive GBMs (median = 2.2 × 10−2) had significantly increased (P = 0.03, Wilcoxon test) expression of wtEGFR mRNA, compared with EGFRvIII tissue–negative GBMs (median = 3.8 × 10−3). However, despite the large median shift, there is considerable overlap in the distributions of the wtEGFR expression values between EGFRvIII-positive and -negative subjects.
Fig. 2.
Comparison of EGFRvIII status and wtEGFR in tumor tissue. (A) DNA copy number amplification of wtEGFR in tumor tissue compared with EGFRvIII tumor status. EGFRvIII-positive tumors have significantly greater median wtEGFR DNA amplification. All DNA samples normalized to RNaseP (P = 0.02). (B) RNA expression of wtEGFR in tumor tissue compared with EGFRvIII tumor status. EGFRvIII-positive tumors have significantly greater median wtEGFR mRNA expression (P = 0.03). All RNA samples were normalized to GAPDH/18S. Medians and interquartile ranges depicted; P-values from 2-tailed Wilcoxon signed rank test.
*Two (−) EGFRvIII and 2 (+) EGFRvIII samples were not sent for wtEGFR analysis.
ΦSix (−) EGFRvIII and 5 (+) EGFRvIII samples did not have GAPDH and/or 18S for normalization.
Wild-type EGFR Expression in CSF-Derived EVs in Relation to EGFRvIII EV Status
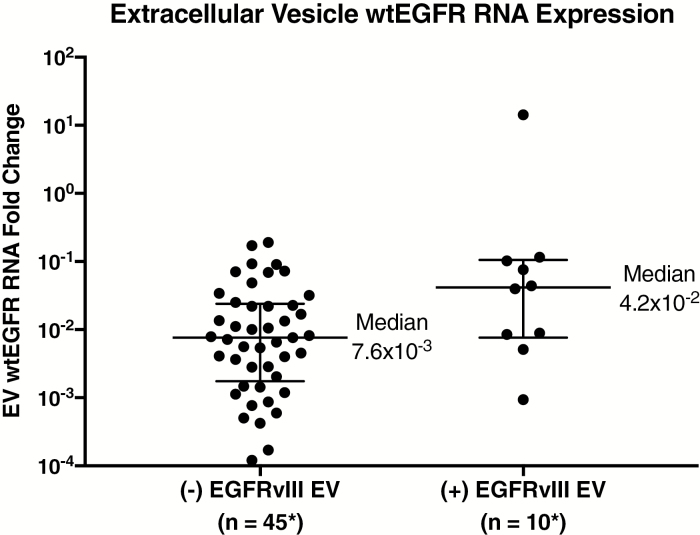
We assessed the relative expression of wtEGFR in CSF-derived EVs by qPCR, in relation to EGFRvIII RNA expression in CSF-derived EVs, for the 60 GBM CSF samples from the ECT validation cohort (11 patients from the UCSD cohort excluded due to no wtEGFR RNA analysis). This investigation revealed that EGFRvIII-positive CSF-derived EVs had a 5.5-fold greater median wtEGFR mRNA expression, as shown in Fig. 3. When normalized to GAPDH mRNA and 18S rRNA, EGFRvIII-positive CSF-derived EVs (median = 4.2 × 10−2) had significantly increased (P = 0.004, Wilcoxon test) expression of wtEGFR mRNA, compared with EGFRvIII-negative CSF-derived EVs (median = 7.6 × 10−3).
Fig. 3.
Comparison of EGFRvIII status and wtEGFR in extracellular vesicles. RNA expression of wtEGFR compared with EGFRvIII status in CSF-derived EVs. EGFRvIII-positive CSF-derived EVs have significantly greater median wtEGFR mRNA expression (P = 0.004). All RNA samples were normalized to GAPDH/18S. Medians and interquartile ranges depicted; P-value from 2-tailed Wilcoxon signed rank test.
*Six (−) EGFRvIII and 5 (+) EGFRvIII samples did not have GAPDH and/or 18S for normalization, and 5 samples had undetectable levels of wtEGFR.
Correlation Between EGFRvIII Expression in CSF-Derived EVs and Tumor Tissue
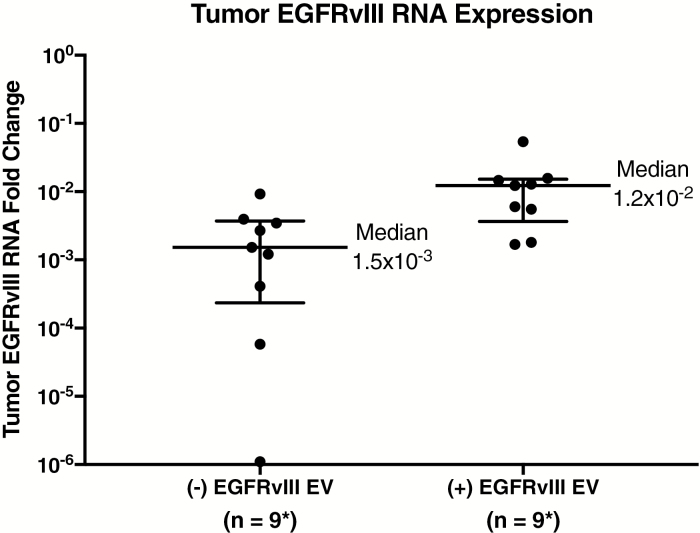
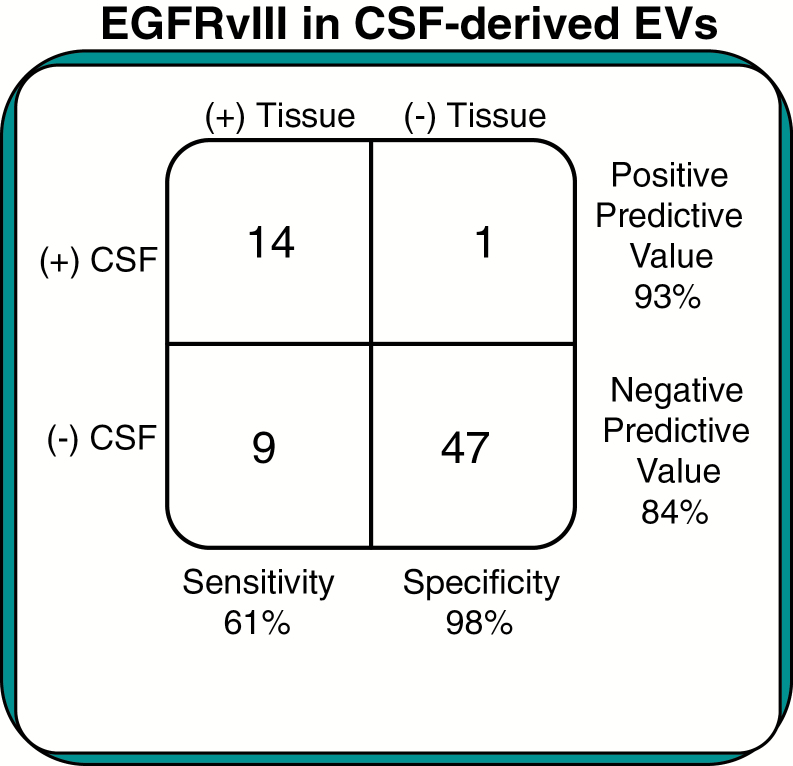
We evaluated the utility of EGFRvIII expression CSF-derived EVs as a biomarker for tumor EGFRvIII in the 71 GBM patients (11 tissue samples from the UCSD cohort excluded due to no GAPDH and/or 18S cycle threshold values for normalization). This analysis showed that EGFRvIII-positive CSF-derived EVs had higher median EGFRvIII mRNA expression in their corresponding tissue samples, as seen in Fig. 4. EGFRvIII-positive CSF-derived EVs (median = 1.2 × 10−2) had significantly greater (P = 0.04, Wilcoxon test) expression of EGFRvIII mRNA in their matching tumor samples, compared with EGFRvIII-negative CSF-derived EVs (median = 1.5 × 10−3). Of the 23 EGFRvIII tissue‒positive GBM samples, 14 demonstrated EGFRvIII expression in the EVs of CSF samples, resulting in a sensitivity of 61%, as shown in Fig. 5. Of the 48 EGFRvIII tissue‒negative GBM samples, only one demonstrated expression of EGFRvIII in CSF EVs but not in tissues, resulting in a specificity of 98%, as shown in Fig. 5.
Fig. 4.
Comparison of EGFRvIII in tumor and extracellular vesicles. RNA expression of EGFRvIII in tissue samples compared with EGFRvIII status in CSF EVs. EGFRvIII-positive CSF-derived EVs have significantly greater median EGFRvIII mRNA expression in corresponding tissue samples (P = 0.04). All RNA samples were normalized to GAPDH/18S. Medians and interquartile ranges depicted; P-value from 2-tailed Wilcoxon signed rank test.
*Five (+) EGFRvIII samples did not have GAPDH and/or 18S for normalization.
Fig. 5.
Classification error rates. Results for patients enrolled in the EGFRvIII study, depicting sensitivity and specificity of EGFRvIII CSF-derived EVs in diagnosing glioblastomas.
Discussion
Here we demonstrate that specific genetic alterations that are present in certain high-grade primary glial tumors can be detected by analysis of EV RNA. This represents a platform for future analytics using biofluid-derived EVs to provide a molecular diagnosis of GBMs.
In our study, CSF was obtained shortly before resection of the primary GBM. The availability of sensitive and specific analyses of EGFRvIII in EV-derived RNA from the CSF can direct tumor-specific therapies. In this case CSF might be obtained from lumbar puncture at the time of MRI detection of intracranial mass given that this procedure is relatively safe, with appropriate precautions, in patients with increased intracranial pressure.9 Importantly, no significant difference was identified between lumbar and cisternal CSF collections with regard to sensitivity (58% and 64%, respectively, P = 0.80, chi-squared) and specificity (100% and 97%, respectively, P = 0.45, chi-squared) of EGFRvIII in EV-derived RNA. This study indicates that EV CSF analysis can be readily performed as part of a standard of care approach to glial tumor CSF biofluid sampling. However, the relatively low sensitivity of 61% is concerning, given the high rate of falsely diagnosing a patient as negative for GBM. These results are likely due to the inability of the assay to detect such a small amount of EGFRvIII in a pool of wtEGFR, and is an area we are continuing to improve upon. Notably, the relatively high positive and negative predictive values of 93% and 84%, respectively, are more useful to clinicians when screening patients for GBM.
We have concentrated our efforts on the detection of the EGFRvIII mutation, which was detected in 21% of CSF samples and has not been shown to be expressed in normal tissue.10 The mutation is produced by an extracellular domain deletion of 267 amino acids in exons 2 through 7 of the EGFR gene. The mutation fails to bind the ligand and alters malignancy by downstream signal changes seemingly different from those of wtEGFR DNA amplification. EGFRvIII is present in 30%‒40% of GBMs and is seldom seen absent wtEGFR amplification.11 Amplification of wtEGFR is a unique feature of “primary” de novo GBMs but is found in less than 20% of less aggressive anaplastic astrocytoma and is rare in low-grade gliomas.11 Half of GBMs contain wtEGFR DNA amplification, and wtEGFR RNA overexpression is seen in over three quarters of tissues with amplification.11 This DNA amplification tends to be diffusely expressed within the tumor and is more prominent in small cell and oligodendroglial variants of GBM. EGFR targeting probes exist for imaging and at least 15 therapeutic agents are available to target this amplification. Although these overexpressions and amplifications have been reported, the literature lacks identification and quantification of wtEGFR in tissues or biofluids, for which the diagnosis of GBM can be based. This is of great importance, as wtEGFR amplification has been shown to be associated with a generally poor prognosis and yet amenable to “personalized” therapies.12,13 This report is the first to provide data upon which wtEGFR amplification findings may be provided in a fashion less invasive than craniotomy.
Our study suggests that amplification of wtEGFR can be detected in both tissue- and CSF-derived EVs when the EGFRvIII mutation is present, but we remain uncertain as to whether wtEGFR amplification is an independent or codependent diagnostic biomarker of GBM. Tumor wtEGFR RNA expression was almost 6.0-fold higher in the EGFRvIII-expressing tumors. This association between wtEGFR and EGFRvIII presence in tissue has been noted by others.14 A similar relation was noted between wtEGFR expression and EGFRvIII expression in CSF, with a 5.5-fold higher expression of wtEGFR when the EGFRvIII mutation was detected in this biofluid. However, a limitation in our investigation is the inability to determine the percentage of cells within the tumor that have amplified wtEGFR or EGFRvIII, which fails to address the degree of heterogeneity.
We note 2 patients who confirmed the known heterogeneity of EGFRvIII expression in tumor tissue sampling. Patients TUM-0021 and UMH-0021 each contained EGFRvIII in only one of 2 biopsy samples. However, patient TUM-0021 was EGFRvIII positive in CSF-derived EVs, whereas UMH-0021 was EGFRvIII negative in CSF-derived EVs. We postulate that this inconsistency of EGFRvIII detection in tissue reflects sampling error during operative biopsy. This issue raises the possibility that EVs in biofluid may provide access to a broader representation of tumor genetic alterations compared with point sampling within the tumor. This idea was supported by the results of the one patient who was deemed false positive for EGFRvIII. Patient UCS-0025 was noted to be EGFRvIII positive in CSF-derived EVs; however, both tissue biopsy samples were EGFRvIII negative. This patient was noted to have wtEGFR DNA amplification in tissue, which correlates with the presence of the EGFRvIII mutation, and we suspect there was a sampling error resulting in a false negative in tissue instead of a false positive in CSF-derived EVs.
Of particular interest was a patient who was categorized as a false negative for EGFRvIII but whose CSF-derived EVs contained an intriguing finding. Patient UCS-001 was positive for EGFRvIII in tumor tissue but was negative in CSF-derived EVs for the classical EGFRvIII mutation. Instead, this patient’s CSF-derived EVs were positive for a novel EGFR variant which has not yet been reported in the literature. Sequencing of this new EGFR mutation in both tissue and EV samples demonstrated a unique extracellular domain deletion compared with EGFRvIII, and is herein referred to as EGFRvVI (see Supplementary Figure S1). Interestingly, this patient also had wtEGFR DNA amplification in tumor tissue, suggesting that EGFRvVI is also associated with an overall amplification of EGFR signaling. This highlights the ability to discover de novo mutations in biofluid samples without the need for tissue sampling.
We chose to explore CSF as a source of EVs despite our previous experience of EGFRvIII detection in serum-derived EVs, as well as the relatively lower diversity of EVs in CSF compared with serum. CSF-derived EV studies reflect our careful evaluation of extracellular RNA in neurological diseases as part of the NIH Common Fund ExRNA project.15 Although it may be argued that CSF as a biofluid is not “minimally invasive,” both neurosurgeons and neuro-oncologists are comfortable with the safety of this approach, which offers many advantages over brain biopsy. Indeed, for malignant brain tumor, CSF-derived EV RNA studies inherently offer distinct advantages over serum-derived EV RNA studies, which have been described for other cancers such as non‒small cell lung cancer.16 Thus, we envision a pathway of translation for CSF-derived EV analysis in the clinical care of patients with intracranial tumors. A diagnostic biomarker from CSF can supplement a clinical exam and MRI images for the diagnosis of GBM while providing molecular subclassification of GBM, stratification of patients for targeted therapies, and metrics of response to therapy.
Further CSF studies of EV wtEGFR and EGFRvIII RNA expression have the potential to alter the current paradigm of GBM diagnosis and treatment. A CSF “liquid biopsy” can potentially direct non-operative therapies for specific mutations, such as rindopepimut (Celldex) for EGFRvIII-positive GBMs. Available imaging diagnostic studies are relatively limited at providing information necessary for a personalized genetic approach to cancer therapy. Although imaging studies are improving, MRI is sensitive but of low specificity for distinguishing GBM from other intracranial lesions, and does not speciate the molecular tumor subtype.17 In patients for whom a resection is not planned, a positive liquid biopsy may permit directed tumor-specific therapy. In some cases, a liquid biopsy may even obviate the need for a tissue biopsy by providing sufficient genetic information about a tumor for a molecular diagnosis. The liquid biopsy approach could also yield an improvement over the known morbidity of brain tumor stereotactic tissue biopsy.18 Additionally, future studies may permit better understanding of whether EV-based tumor molecular genetic sampling may improve upon the known sampling error of tissue-based approaches.19
While our methods of centrifuge EV isolation are relatively cumbersome, other approaches, such as precipitation and size-exclusion chromatography, are cheaper, faster, and nearly as efficient.20 Additionally, we have shown that EVs and isolated RNAs are stable enough at room temperature for transport prior to analysis.21 This approach suggests the potential of EV-based companion diagnostics for personalized medicine approaches to glioma therapy, yielding early disease monitoring and treatment assessment via the use of “liquid biopsies” to assess drug efficacy.
Our original cohort actually included 81 patients, 10 of whom were excluded when final pathology revealed grade III glioma. Interestingly, of the 10 grade III glioma patients, 2 harbored the EGFRvIII mutation in tumor tissue, and 1 of these also exhibited EGFRvIII in the CSF. Thus our CSF studies may ultimately address the issue of grade III gliomas with the potential to progress to GBM. Notably, if these 10 grade III gliomas had been included in the study, the results and statistical significance would be relatively unchanged.
In the future there is potential that EV-based RNA analysis of CSF could be expanded to develop a diagnostic tool for GBM more broadly beyond EGFRvIII positivity. By comparing CSF-derived EV profiles of miRs and mRNA species that are associated with glioma development and progression in the CSF of GBM patients and non-oncologic normal controls, we envision the possibility of a multiplexed extracellular RNA assessment that would improve the sensitivity of the EGFRvIII CSF test. Such data could significantly impact the field of neuro-oncology, as CSF-derived EVs could be utilized not only for diagnosing EGFRvIII-positive GBMs, but potentially for distinguishing patients with GBM from disease-free patients.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by TNIH; UH2—ExRNA Biomarkers in Glioma (grant# 5UH2TR000931), NIH; UH2 Supplement—ExRNA Biomarkers in Glioma (grant# UH2TR000931-S1), NIH; UH3—ExRNA Biomarkers in Glioma (grant# 4UH3TR000931-03), NIH; P01—Experimental Therapeutics and Biomonitoring of Glioma—Biorepository Core (grant# 2P01 CA069246-16), ABC2 Foundation; Consortium Study of EGFRvIII in Glioma Exosomes, ABC2 Foundation; miRNA Analysis of CSF Glioma Exosomes.
Funding sources had no role in study design, specimen collection, data analysis, manuscript preparation, or journal selection.
Conflict of interest statement. Drs Skog, LoGuidice, and Berghoff are employees of Exosome Diagnostics, Inc. Dr Breakefield is a consulting member of the Scientific Advisory Board of Exosome Diagnostics, Inc. All other authors have no conflicts of interests to disclose.
Supplementary Material
Acknowledgments
The authors acknowledge the Accelerate Brain Cancer Cure (ABC2) foundation and the National Institutes of Health (grant #5UH2/UH3TR000931) for their financial support.
References
- 1. Skog J, Würdinger T, van Rijn S et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Manterola L, Guruceaga E, Gállego Pérez-Larraya J et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akers JC, Ramakrishnan V, Kim R et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gonda DD, Akers JC, Kim R et al. Neuro-oncologic applications of exosomes, microvesicles, and other nano-sized extracellular particles. Neurosurgery. 2013;72(4):501–510. [DOI] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weller M, Kaulich K, Hentschel B et al. ; German Glioma Network. Assessment and prognostic significance of the epidermal growth factor receptor vIII mutation in glioblastoma patients treated with concurrent and adjuvant temozolomide radiochemotherapy. Int J Cancer. 2014;134(10):2437–2447. [DOI] [PubMed] [Google Scholar]
- 7. Chen WW, Balaj L, Liau LM et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ostrom QT, Gittleman H, Fulop J et al. CBTRUS statistical report: brain and central nervous system tumors diagnosed in the United States 2008–2012. Neuro Oncol. 2015;11(4): iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tuettenberg J, Czabanka M, Horn P et al. Clinical evaluation of the safety and efficacy of lumbar cerebrospinal fluid drainage for the treatment of refractory increased intracranial pressure. J Neurosurg. 2009;110(6):1200–1208. [DOI] [PubMed] [Google Scholar]
- 10. Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12(9):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benito R, Gil-Benso R, Quilis V et al. Primary glioblastomas with and without EGFR amplification: relationship to genetic alterations and clinicopathological features. Neuropathology. 2010;30(4):392–400. [DOI] [PubMed] [Google Scholar]
- 12. Shinojima N, Tada K, Shiraishi S et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 13. Ruano Y, Ribalta T, de Lope AR et al. Worse outcome in primary glioblastoma multiforme with concurrent epidermal growth factor receptor and p53 alteration. Am J Clin Pathol. 2009;131(2):257–263. [DOI] [PubMed] [Google Scholar]
- 14. Zadeh G, Bhat KP, Aldape K. EGFR and EGFRvIII in glioblastoma: partners in crime. Cancer Cell. 2013;24(4):403–404. [DOI] [PubMed] [Google Scholar]
- 15. Quinn JF, Patel T, Wong D et al. Extracellular RNAs: development as biomarkers of human disease. J Extracell Vesicles. 2015;4:27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol. 2011;84 Spec No 2:S107–S111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McGirt MJ, Woodworth GF, Coon AL et al. Independent predictors of morbidity after image-guided stereotactic brain biopsy: a risk assessment of 270 cases. J Neurosurg. 2005;102(5):897–901. [DOI] [PubMed] [Google Scholar]
- 19. Hall WA. The safety and efficacy of stereotactic biopsy for intracranial lesions. Cancer. 1998;82(9):1749–1755. [DOI] [PubMed] [Google Scholar]
- 20. Lobb RJ, Becker M, Wen SW et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akers JC, Ramakrishnan V, Yang I et al. Optimizing preservation of extracellular vesicular miRNAs derived from clinical cerebrospinal fluid. Cancer Biomark. 2016;17(2):125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.