Abstract
Bacterial degraders of chlorophenoxy herbicides have been isolated from various ecosystems, including pristine environments. Among these degraders, the sphingomonads constitute a prominent group that displays versatile xenobiotic-degradation capabilities. Four separate sequencing strategies were required to provide the complete sequence of the complex and plastic genome of the canonical chlorophenoxy herbicide-degrading Sphingobium herbicidovorans MH. The genome has an intricate organization of the chlorophenoxy-herbicide catabolic genes sdpA, rdpA, and cadABCD that encode the (R)- and (S)-enantiomer-specific 2,4-dichlorophenoxypropionate dioxygenases and four subunits of a Rieske non-heme iron oxygenase involved in 2-methyl-chlorophenoxyacetic acid degradation, respectively. Several major genomic rearrangements are proposed to help understand the evolution and mobility of these important genes and their genetic context. Single-strain mobilomic sequence analysis uncovered plasmids and insertion sequence-associated circular intermediates in this environmentally important bacterium and enabled the description of evolutionary models for pesticide degradation in strain MH and related organisms. The mobilome presented a complex mosaic of mobile genetic elements including four plasmids and several circular intermediate DNA molecules of insertion-sequence elements and transposons that are central to the evolution of xenobiotics degradation. Furthermore, two individual chromosomally integrated prophages were shown to excise and form free circular DNA molecules. This approach holds great potential for improving the understanding of genome plasticity, evolution, and microbial ecology.
Keywords: xenobiotics degradation, mobilome, circular DNA molecules, Nanopore, PacBio, Sphingomonadaceae
Introduction
The chlorophenoxy herbicides, including 2-methyl-4-chlorophenoxyacetic acid (MCPA), 2,4-dichlorophenoxyacetic acid (2, 4-D), methylchlorophenoxypropionic acid (MCPP), and dichlorprop (DCPP), are widely applied herbicides that frequently contaminate the environment, including groundwater (McManus etal. 2014). As recently reviewed (Huang etal. 2016), many microorganisms that degrade these pesticides have been isolated, but only two isolates capable of degrading the phenoxypropionic herbicides MCPP and DCPP have been described, namely Sphingobium herbicidovoransMH (Horvath etal. 1990) and Delftia acidovorans MC1 (Müller etal. 1999). Both of these strains have been studied with regards to the enantiomer-specific enzymes 2,4-dichlorophenoxypropionate dioxygenases RdpA and SdpA and their corresponding genes (Nickel etal. 1997; Westendorf etal. 2002; Müller etal. 2004, 2006; Schleinitz etal. 2004). Furthermore, enantioselective degradation and transport of MCPP and DCPP have been investigated (Zipper etal. 1996, 1998; Kohler 1999) along with other features (Benndorf etal. 2004; Hoffmann and Müller 2006; Müller and Hoffmann 2006; Westendorf etal. 2006; Leibeling etal. 2010). Although RdpA and SdpA seem to be specific for MCPP and DCPP degradation, the canonical 2-oxoglutarate dependent dioxygenase TfdA and the alternative Rieske non-heme iron oxygenase CadA are involved in the first step of degradation of 2,4-D and MCPA (Fukumori and Hausinger 1993; Kitagawa etal. 2002). The complete pathway for degradation of DCPP to 3-oxoadipate in Sphingobium herbicidovorans MH was previously proposed with all enzymes identified (Müller etal. 2004).
Despite its relevance in microbial ecology, only two short genetic fragments from Sphingobium herbicidovorans MH were previously isolated. They both contain a transposase gene and an sdpA or rdpA gene similar to those observed in Delftia acidovorans MC1 (Müller etal. 2004). These enantiomer-specific genes were later cloned and expressed in E. coli, showing that these genes were linked to the first step in degradation of phenoxypropionic acids (Müller etal. 2006). Likewise, only two chlorocatechol gene clusters were identified in strain Sphingomonas sp. TFD44 (Thiel etal. 2005). Primers for PCR have been designed based on 16 GenBank entries for rdpA and 3 for sdpA. These primers were used to determine the presence of these genes in agricultural soil and to evaluate the expression of them in soils amended with D. acidovorans MC1 during DCPP degradation (Paulin etal. 2010). The soil used in that study contained no rdpA genes, but had 103 copies of tfdA gene per gram of soil. The tfdA genes also displayed some variation (four clusters) in the nucleotide sequences. In another soil-based study, the expression and abundance of the sdpA, rdpA, and tfdA genes were monitored following a DCPP amendment by using the same primers as above for qPCR. The diversity of both rdpA and sdpA genes and transcripts was very low with only a single transcript sequence detected for each gene (Paulin etal. 2011). The rdpA and sdpA transcripts were identical to those of D. acidovorans MC1 and S. herbicidovorans MH, respectively. Low diversity among rdpA/sdpA observed in soil was later confirmed in other settings (Liu etal. 2013), suggesting that either the primers are not targeting many natural variants of these genes or that the variability of rdpA and sdpA is indeed low. The latter study also showed that gene expression of tfdA, rdpA, sdpA, and cadA was higher in microcosms exposed to MCPA, highlighting the significance of these genes in natural settings. This is supported by a recent study on bacterial communities in a sand filter that degraded DCPP and MCPP during groundwater treatment, where the abundance of rdpA and sdpA genes significantly increased during pesticide degradation (Feld etal. 2015). In addition to strains MH and MC1, two exogenously isolated plasmids (pAKD16 and pAKD34) that have identical plasmid backbones are reported to harbor rdpA and sdpA (Drønen etal. 1998, 1999; Sen etal. 2011). Likewise, cadA was found on the large conjugative plasmid pCADAB1 in Sphingomonas sp. ERG5 (Nielsen etal. 2013).
This study presents the complete genome of Sphingobium herbicidovorans MH and an in-depth analysis of the complex genetic context of its sdpA, rdpA, and cadA genes. The gene clusters containing sdpA and cadA display complicated structures that appeared to have undergone major genomic rearrangements, which would have facilitated gene transfer between chromosomes and plasmids, horizontal gene transfer (HGT), and the adaptation for chlorophenoxy pesticide degradation. These gene clusters were compared with their homologs and paralogs in the exogenously isolated plasmids pAKD34, pAKD16, the canonical Delftia acidovorans MC1, the MCPA-degrading Sphingomonas sp. ERG5, and the draft genome of Sphingomonas sp. TFD44 which was produced for this study. Finally, through single-strain mobilomic sequencing of exonuclease treated DNA from strain MH, we observed several circular elements, including a 10-kb mobilizable plasmid and an 11-kb circular intermediate state of an IS91-transposon harboring sdpA, along with several circular intermediates of insertion sequence elements (IS) and integrated prophages. We here set out to document the high genomic plasticity of sphingomonads that can help to explain the very recent evolution of other degradation pathways of xenobiotics such as pentachlorophenol and hexachlorocyclohexane, which were introduced <100 years ago, that have been suggested to have evolved in this group of bacteria (Mohn etal. 2006; Copley etal. 2012; Verma etal. 2014; Pearce etal. 2015; Tabata etal. 2016). This study provides insight into the specific evolution of chlorophenoxy catabolic gene clusters and the general sphingomonad evolution as xenobiotic degraders.
Materials and Methods
Preparation of DNA and Sequencing
For complete genome assembly and mobilomic sequencing of Sphingobium herbicidovorans MH and draft genome assembly of strain TFD44, five separate sequencing libraries were prepared for these strains (supplementary table S1, Supplementary Material online) using Illumina, PacBio, and Nanopore platforms. A flowgram of the DNA preparations, sequencing, and genome assemblies is shown in supplementary figure S1, Supplementary Material online.
Sphingobium herbicidovorans MH was inoculated in R2B medium, incubated at 30 °C, and shaken for 48 h. High molecular weight (HMW) DNA was extracted using the Gram-negative protocol of the QIAGEN Puregene DNA kit (QIAGEN, Hilden, Germany), starting with 6× 1.2 ml of a fully grown culture that had been centrifuged at 14,100×g for 3 min to pellet bacterial cells. After DNA extraction and elution according to manufacturer instructions, tubes with eluted DNA were shaken overnight at ambient temperature in order to rehydrate the DNA. An Illumina Nextera XT (Illumina, San Diego, CA) library was prepared for paired-end sequencing using the HMW DNA as template and was sequenced on the Illumina MiSeq with a V2 kit, yielding 2 ×250 bp reads. Sphingomonas sp. TFD44 was kindly provided by Prof. Dr Michael Schlömann, Technische Universität Bergakademie Freiberg, Germany. Sphingomonas sp. TFD44 DNA was prepared from the R2B culture using the MOBIO UltraClean Microbial DNA Isolation Kit (QIAGEN, Hilden, Germany) and a sequencing library for the Illumina MiSeq was likewise prepared with the Illumina Nextera XT kit and was sequenced on the Illumina MiSeq with a V2 kit, yielding 2 ×250 bp reads. For Nanopore sequencing, two libraries were built from HMW DNA from strain MH. In order to get as long reads as possible, one library was built without shearing of DNA, whereas the other was sheared to 8 kb using G-tubes (Covaris, Brighton, UK). Both libraries were prepared with the SQK-MAP006 kit according to the manufacturer instructions (Oxford Nanopore Technologies, Oxford, UK) and sequenced on the same MinION flow cell. For PacBio sequencing (Pacific Biosciences, Menlo Park, CA), 10 µg HMW DNA from strain MH was diluted in 150 µl and sheared on a Covaris g-tube spinning 2× 1 min at 4,600 rpm on an Eppendorf 5424, targeting an insert size of 15–20 kb. Sheared DNA was size selected on a Sage Pippin (Sage Science, Beverly, MA) with an S1 ladder and using a 15- to 20-kb v3 protocol. PacBio protocol for 10-kb inserts was used and ligation run overnight. Final library was loaded on two SMRTcells with either 0.02 or 0.04 nM on plate concentration.
For mobilomic sequencing of circular genetic elements from strain MH, DNA was extracted from culture grown in R2B for 48 h using Plasmid AX mini kit (A&A Biotechnology, Gdynia, Poland) with the protocol for large plasmids and/or low copy number. For exonuclease-based removal of chromosomal DNA and fragmented plasmids, 20 µl of DNA was treated with 0.5 µl (5 U) Plasmid-Safe ATP-dependent exonuclease (Epicenter Technologies, Madison), as described previously (Jørgensen etal. 2014). The reaction was incubated at 37 °C for 30 min and then 70 °C for 30 min to inactivate the exonuclease. An Illumina Nextera XT library was built on digested DNA and was sequenced on the Illumina NextSeq platform with the V2 kit, yielding 2 ×250 bp paired-end reads.
Bioinformatic Analyses
PacBio reads were used for assembly using the Canu assembler (Koren etal. 2017) (version 1.3) with default settings and genome size set to 4.5 Mb which resulted in seven contigs. For error correction using high-quality Illumina reads from the HMW DNA library, paired-end reads were trimmed for contaminating sequences (adapters and barcodes) using Cutadapt (version 1.8.3) (Martin 2011) and were quality trimmed at the ends for sequences with quality below Q20. The seven Canu-assembled contigs were imported to CLC Genomics Workbench (version 9) (QIAGEN, Hilden, Germany), where high quality Illumina reads were mapped to the contigs and used for correcting errors from the PacBio assembly. Uncorrected 1-D Nanopore reads and PacBio reads were mapped with BLASTN (Altschul etal. 1990) to the corrected seven contigs and were used to manually merge contigs to four final contigs. All three types of sequencing reads were mapped (Illumina reads were mapped with the “Map Reads To Contigs” tool from CLC and long reads mapped with BLASTN) to the assembled contigs and manually inspected for sequencing errors, misassemblies, areas with low coverage, and circularity of assembled contigs by mapping long reads to ends of contigs and checking for overlap with the other end of the same contig. Paired-end Illumina reads from Sphingomonas sp. TFD44 were assembled using the SPAdes assembler (version 3.6.0) (Bankevich etal. 2012).
The final assembly of strain MH was automatically annotated with PROKKA (Seemann 2014) (v. 1.11), whereas features highlighted in this paper were manually verified and curated using BLASTX against the NR database and associated Conserved Domain Database (CDD) for conserved protein domains (Altschul etal. 1990; Marchler-Bauer etal. 2015). IS elements were predicted with ISSaga (Varani etal. 2011) and those in proximity to genes of special interest were checked with ISfinder (Siguier etal. 2006) and BLASTX (supplementary fig. S1, Supplementary Material online). Previously studied genetic fragments from strain MH (Müller etal. 2004; Schleinitz etal. 2004) were downloaded from NCBI: tnpA-sdpA [AJ628860.1], rdpA [AF516752.2], tfdK-ORFB-dccAII-dccDII-tnp A[AJ628863.1], cadR-tfdB [AJ628861.1], chloromuconate cycloisomerase [DQ146618.1], dccE-dccAI-dccDI [AJ628862.1], and tnpA-rdpA [AJ628859.1]. These nucleotide sequences were mapped to the MH genome using BLASTN (all matches except DQ146618.1 had >99% similarity). Genetic regions surrounding identified sdpA, rdpA, and cadA genes were used as query for BLASTN, in order to find similar regions for comparative genomics. Identified similar regions from pAKD16 [JN106167.1], pAKD34 [JN106175.1], pMC1 [AY327575.2], pCADAB1 [KF494257.1], Sphingomonas sp. 58-1 [AB353895.1], and Sphingomonas sp. TFD44 (draft genome from this study produced from MiSeq data) were aligned using Clustal Omega (Sievers etal. 2011) in CLC. Proposed hypothetical evolutionary states of genetic clusters were manually edited using CLC. A genome map of replicons was made in Circos (Krzywinski etal. 2009), whereas remaining graphics were produced in CLC. Mobilome sequencing coverage of replicons was derived from alignment of reads using the BWA aligner (v. 0.7.12-r1039) (Li and Durbin 2009) with the “mem” algorithm and default parameters.
The previously identified minimal core 206 gene set for bacteria (Gil etal. 2004) was used to separate chromosomal replicons from plasmid ones. These are referred to as housekeeping genes. A manual subset of the ACLAME (Leplae etal. 2010) plasmid database (4,721 genes; supplementary file 1) was used to identify genes involved in plasmid transfer, replication, and partitioning using BLASTP.
Mobilome Analyses
Sequencing reads from the exonuclease-treated MH DNA were quality checked as done for the Illumina data above and were de novo assembled using the SPAdes assembler (version 3.6.0) (Bankevich etal. 2012) with “careful” option enabled. Cleaned paired-end reads from the mobilomic MH DNA were mapped onto the final assembly of strain MH using the “Map reads to contigs” function in CLC Genomics Workbench with default parameters except for setting the Similarity fraction option to 0.95, corresponding to a minimum of 95% similarity for accepting a mapped read. A log–log linear model was fitted on mobilome sequencing coverage against replicon size in R. The model was tested with ANOVA. Subsequently, all replicons were manually inspected for regions of increased mobilome read coverage, indicating potential extrachromosomal and extraplasmidal circular states of DNA molecules. In order to obtain average mobilome sequencing coverages of all replicons, identified regions with increased coverage were masked in a second read mapping of mobilome reads to the assembled genome. A third read mapping of mobilome reads to only the identified regions of increased coverage was performed. The background read coverage of the replicons were subtracted from the coverage of the regions of increased coverage, in order to account for the baseline coverage that contributes to the coverage of potential small circular DNA molecules originating from their incorporated positions on the bacterial replicons (supplementary fig. S2, Supplementary Material online). Regions with elevated coverage of mobilome reads were extracted from their positions in the original replicons and investigated for circularity by manually verifying that paired-end reads spanned the terminal start and end regions of the sequences, as previously described (Jørgensen etal. 2014). The copy number per bacterial cell of circular intermediate molecules were calculated relative to the mobilome coverage of plasmid pSHV2, since this is the smallest of the main replicons and thus the most stable during exonuclease treatment. Calculation of copy numbers per cell of regions with elevated mobilome coverage were in summary done as following:
Where Nix is copy number of sequence i on replicon x and BCxis the background coverage of replicon x. Subsequently, the copy number per cell of region i with elevated mobilome coverage was calculated as:
Where OCi is the observed coverage of region i and CNpSHV2 is the copy number of replicon pSHV2 as defined from the nonmobilome MiSeq sequencing to be 1.46.
Ten control genomic regions with no obvious increase in mobilome sequencing coverage were used to calculate the background level of DNA that does not form free circular molecules (supplementary table S2, Supplementary Material online).
Results and Discussion
Genome Assemblies and General Features
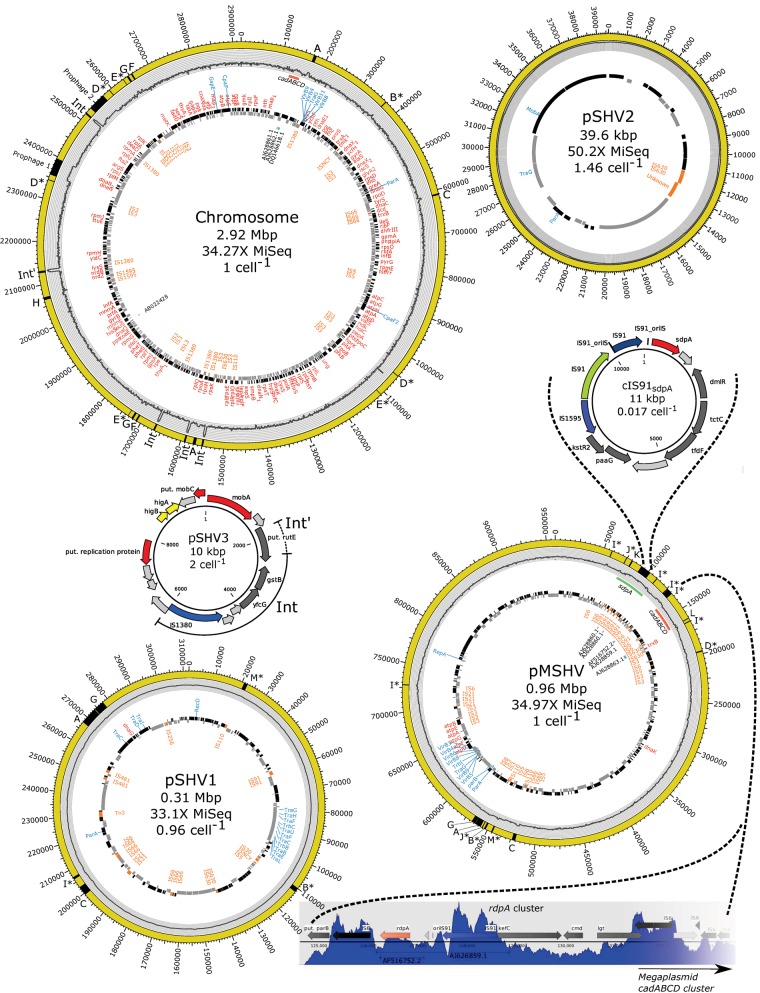
Assembly and subsequent manual curation, including joining of contigs based on overlapping long reads (Nanopore, MiSeq, and PacBio) and assembly error correction with high quality Illumina data, resulted in a 4.24-Mb genome consisting of a 2.92-Mb chromosome with 162 identified housekeeping genes, a 956-kb megaplasmid (pMSHV) with eight identified housekeeping genes, a 312-kb plasmid with one identified housekeeping gene (pSHV1) and a 40-kb plasmid (pSHV2) (fig. 1). The nine housekeeping genes on plasmids pMSHV and pSHV1 were also identified on the chromosome, so pMSHV and pSHV1 can be considered plasmids and not secondary chromosomes. In addition, a 10-kb mobilizable plasmid (pSHV3) and an 11-kb circular intermediate of an IS91-transposon harboring sdpA (cIS91sdpA) were derived from the single-strain mobilome data. The mobilome data from MiSeq sequencing showed low coverage of most of the chromosome and pMSHV (∼1.66–4.28×), whereas pSHV1 had a coverage of 33.66× and pSHV2 and pSHV3 had much higher coverages at 31,038× and 42,469×, respectively. From assembly of mobilome sequencing reads, the completeness and circularity of replicons pSHV1, pSHV2, and pSHV3 were confirmed. The larger replicons pMSHV and the chromosome did not have high mobilome sequencing coverage and were therefore very fragmented in the SPAdes mobilome assembly. The relationship between replicon size and mobilome sequencing coverage fits a log–log linear model, showing that increasing DNA molecule size leads to decrease in coverage from the exonuclease-treated DNA sequencing (P value < 0.01, adjusted R2 = 0.9379; supplementary table S3 and fig. S3, Supplementary Material online). This most likely reflects the probability that a circular DNA molecule of a given size will fragment into linear segments by shear forces during DNA extraction and subsequent handling, leading to digestion by exonuclease treatment. Several factors will size-dependently influence the probability of fragmentation of circular DNA molecules, such as vortexing, pipetting, and bead-beating (Santiago etal. 2014) during DNA extraction which was not done in the Puregene DNA extraction in this study. Replicon copy numbers for chromosome, pMSHV, pSHV1, pSHV2, and pSHV3 were estimated from the nonmobilome MiSeq data to be 1, 1, 1, and 1.46, respectively. The possibly mobilizable plasmid pSHV3 was only fully represented in the mobilome assembly and is therefore not considered a main replicon. As the smallest replicon in the nonmobilome data, the 39,633-bp plasmid pSHV2 is assumed to be the most stable of the major replicons during exonuclease treatment and was, therefore, used to estimate the copy numbers of pSHV3 and cIS91sdpA to be 2 and 0.017, respectively (supplementary table S3, Supplementary Material online).
Fig. 1.
—The four major replicons of Sphingobium herbicidovorans MH (yellow circles: Chromosome, Megaplasmid pMSHV, pSHV1, and pSHV2) and minor circular elements (10-kb mobilizable plasmid pSHV3 and 11-kb IS91-mediated circular element cIS91sdpA) determined from sequencing of exonuclease treated DNA (mobilome data). Relative sizes of circles do not represent their actual scale. Black and gray tiles represent CDS features, as predicted by PROKKA, on the forward and reverse strand, respectively. Identified housekeeping genes are marked in red, plasmid-backbone genes are in blue, and IS elements are in orange on the inside of the CDS features. Previously studied genes and PCR clones are marked as their GenBank accession numbers in black. The sdpA (green) and cadABCD (red) gene clusters on major replicons are highlighted just under the mobilome histogram. The histogram on the gray background shows the mapping of paired-end reads from the exonuclease-treated mobilome sequencing. The mobilome coverage was log-transformed and histograms for all replicons have the same scale for direct comparison (e.g., the average mobilome coverage of the chromosome is much lower than that of pSHV2). Putative circular elements from the mobilome data are highlighted in black on the outer yellow diagram (A–M) and are shown in further detail in figure 4. The mobilizable plasmid pSHV3 has been partially integrated five separate times into the chromosome (Int) of which one integration is longer than the other four and contains the full rutE ORF, encoding a putative nitroreductase-like family protein, besides the yfcG and gstB genes encoding putative a GSH-dependent disulfide bond oxidoreductase and a putative glutathione S-transferase. For pSHV3, cIS91sdpA, and rdpA cluster arrows indicate hypothetical genes (light gray), miscellaneous genes (dark gray), toxin–antitoxin genes (yellow), and genes of special interest (other colors).
Gene Clusters and Transposons Define Genome Structure and Metabolic Arsenal
All previously identified PCR products were mapped onto the genome with ∼99% similarity with the exception of the chloromuconate cycloisomerase [DQ146618.1], which was only 89% similar (fig. 1). Based on BLASTN comparisons with previously mentioned PCR fragments from strain MH and with pCADAB1, pAKD16, pAKD34, pMC1, and strain TFD44, three gene clusters of catabolic interest were identified. A 27.4-kb cluster is situated on the chromosome with high structural and sequence similarity to the 33.7-kb composite transposon Tn6228 of pCADAB1 [KF494257.1], although the MH chromosomal gene cluster lacks the essential cadABCD genes. A similar 33.8-kb gene cluster was identified on megaplasmid pMSHV, complete with the cadABCD genes and flanked by identical IS6 elements, suggesting that this cluster was transferred as a composite transposon. Furthermore, a previously described 10.5-kb PCR fragment of the Sphingomonas sp. TFD44 cadABCD gene cluster [AY598949.1] was identified in the draft genome of this strain. The two cadABCD-associated gene clusters from the chromosome and pMSHV of strain MH were aligned to the similar clusters of pCADAB1, strain TFD44, and a reference cadABCD genetic fragment from Sphingomonas sp. 58-1 (Shimojo etal. 2009) [AB353895.1] (supplementary fig. S4, Supplementary Material online). Besides the essential cadABCD genes, the genes encoding the enantiomer-specific SdpA and RdpA DCPP degradation enzymes are furthermore situated on the megaplasmid pMSHV, supporting the previous proposal that sphingomonad megaplasmids may act as reservoirs for catabolic genes acquired via HGT, since acquisition of new genes will not disrupt important housekeeping genes on this replicon (Copley etal. 2012). As discussed in further detail below, the three individual gene clusters harboring cadABCD, sdpA, and rdpA are all tightly associated with IS elements and the high sequence similarity to other strains suggests that these gene clusters are actually genomic islands that are frequently transferred horizontally between bacteria.
Evolution of the cadABCD Gene Cluster in Sphingomonad Strains
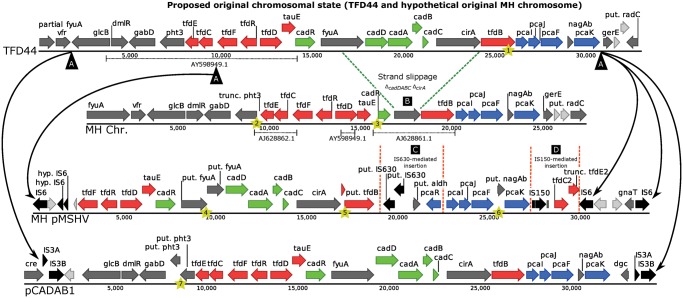
Since the role of the chlorophenoxy herbicide degradation genes in these clusters have been studied previously (Don etal. 1985; Filer and Harker 1997; Leveau etal. 1998; Kitagawa etal. 2002; Müller etal. 2004; Thiel etal. 2005; Shimojo etal. 2009; Nielsen etal. 2013), this study focused on the evolution and arrangement of these features. Based on multiple alignments of the genetic clusters, a model was proposed that describes the most parsimonious chain of events leading to the currently observed states of the cadABCD cluster in sphingomonads (fig. 2). They have been isolated from geographically disparate locations (Austria, Denmark, North America, and Japan) for strains MH, ERG5, TFD44, and 58-1, respectively (Horvath etal. 1990; Fulthorpe etal. 1995; Shimojo etal. 2009; Gözdereliler etal. 2013). Therefore, this cluster is highly conserved in sphingomonads worldwide.
Fig. 2.
—Proposed most parsimonious model of genomic rearrangements leading to the observed states of the cadABCD gene cluster. The top sequence was observed in Sphingomonas sp. TFD44 and is a hypothetical original state in the chromosome of strain MH. Previously described PCR products are marked with dotted lines and accession numbers under their respective sequences. Major events leading to all observed states were (A) the insertion of IS elements (IS6 for strain MH megaplasmid pMSHV and IS3 for strain ERG5 pCADAB1) that led to the formation of composite transposons and subsequent transfer to plasmids, (B) strand slippage between two homologous sequences (fyuA and cirA) that led to the deletion of the cadABCD genes on the MH chromosome, (C) the insertion of aldehyde dehydrogenase and pcaR genes mediated by an IS630 element and (D) the insertion of tfdC2 and tfdE2 mediated by an IS150 element of the IS3 family. Minor mutations are marked with yellow stars showing 1) “A” insertion causing elongation of the tfdB reading frame, 2) G-to-A mutation that led to a shortened pht3 reading frame, 3) “G”-deletion and truncation of cadR gene, 4) “G”-deletion and disruption of the reading frame in fyuA gene, 5) “C”-deletion and disruption of the reading frame in the tfdB gene, 6) a 42-bp deletion in the nagAB gene, and 7) “A”-deletion and disruption of reading frame in the pht3 gene.
We propose that the cadABCD cluster from strain TFD44 constitutes the most ancestral state, since it contains most of the genes that were shared across all other observed clusters and did not appear to be flanked by IS elements. Furthermore, we propose that strain MH has had a similar original state on the chromosome (fig. 2 0), but that the cadABCD and tonB-dependent receptor cirA have been lost through illegitimate recombination between the homologous stretches between fyuA and cirA genes (event B in fig. 2). For all clusters containing a full length fyuA (∼2,337 bp) and cirA (∼2,421 bp), there was 40% nucleotide similarity with ∼1,000 identical nucleotides shared between the two genes (supplementary fig. S5, Supplementary Material online). This similarity between the two genes could have formed the basis for a strand slippage event (Darmon and Leach 2014) that led to previously described deletion in the chromosomal cadABCD cluster in MH. The second cadABCD gene cluster on pMSHV in MH has likely been transferred to the plasmid from the chromosome prior to the strand slippage-mediated deletion through the insertion of identical IS6 elements on either side of the cluster that created a composite transposon on the chromosome and was then transferred to pMSHV (event A in fig. 2). Two or more IS6-related events have likely occurred later and led to the formation of an additional broken IS6 element in the left part of pMSHV and an additional IS6 element on the opposite strand in the right part. Analogously to this event, IS3 elements have formed the Tn6228 composite transposon on pCADAB1 by inserting further upstream than the left-most IS6 element in pMSHV, and therefore, introduced the additional genes glcB, dmlR, gabD, pht3, tfdE, and tfdC in Tn6228. The right-most site of insertion of an IS3 element in Tn6228 seems to be close to the same as that forming the cluster on pMSHV through IS6 elements, which indicates that this transposon contains the core genes of chlorophenoxy herbicide degradation that have been mobilized multiple times via IS elements. Interestingly, the cadABCD cluster is not present in the remaining genome of Sphingomonas sp. ERG5 (that harbors pCADAB1), so this strain might have acquired the cluster via HGT or a complete deletion from the chromosome following copying to plasmid pCADAB1. After the IS6-mediated transfer to pMSHV, two insertion events have occurred in this gene cluster: the introduction of IS630, a putative aldehyde dehydrogenase, and the regulatory gene pcaR (event C, fig. 2) and the introduction of IS150, tfdC2 and a possibly truncated tfdE2 (event D, fig. 2) that are each ∼90% similar to the other tfdC and tfdE in the same gene cluster. Several minor mutations have caused gene disruptions that likely led to nonfunctional variants of tfdB, pht3, cadR, fyuA, and nagAb, demonstrated by multiple sequence alignment (supplementary fig. S4, Supplementary Material online). It is possible that other genetic rearrangements have led to the observed states of the cadABCD cluster, but the hereby proposed evolutionary model requires the fewest changes and is thus the most likely model.
The MH sdpA Gene Cluster Displays IS Elements and Sequence Duplications
A 36.7-kb gene cluster on pMSHV, containing sdpA, was aligned to similar regions on plasmids pAKD16 [JN106167.1] and pAKD34 [JN106175.1], both isolated from an agricultural soil, and on Delftia acidovorans plasmid pMC1 [AY327575.2]. However, the alignable part of the MH sequence occurred in three consecutive, imperfect, ∼10-kb repeats, requiring that the pAKD16, pAKD34, and pMC1 clusters were copied 3 times manually for multiple sequence alignment (supplementary fig. S6, Supplementary Material online).
The pAKD16 and pAKD34 clusters were very similar to the first of the three replicated clusters in MH, which suggests that the initial replicate is the original state in MH. pAKD16 and pAKD34 had been exogenously isolated from the same soil, so there is no information on the identity of the original host, although they do share similarity to plasmid p712 from Ralstonia pickettii [JQ436722.1] as revealed by BLASTN. However, both plasmids were shown to have broad-host ranges (Drønen etal. 1998, 1999), which indicates that this gene cluster was potentially spread across a wider phylogenetic range than the sphingomonad cadABCD cluster. Furthermore, there is also similarity to a genetic region on Delftia acidovorans MC1 plasmid pMC1 (supplementary fig. S6, Supplementary Material online).
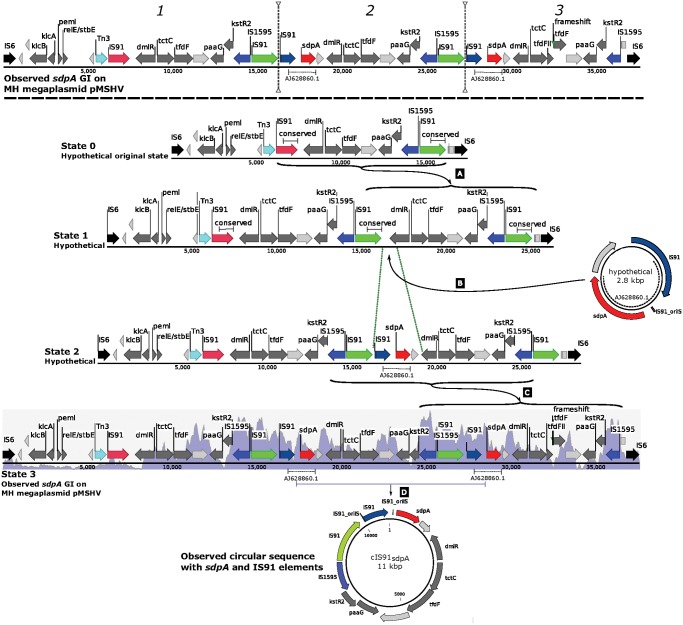
An evolutionary model is proposed to account for the intricate arrangement of the sdpA cluster on megaplasmid pMSHV (fig. 3). The most conserved part (fig. 3 and supplementary fig. S6, Supplementary Material online: leftmost IS6 to first green IS91) of the MH sdpA gene cluster is shown as the hypothetically original “State 0”. The entire current (observed) state is flanked by identical IS6 elements, which indicates that the entire gene cluster can appear as a transferrable composite transposon. In the hypothetical ancestral State 0, the flanking IS6 elements were already incorporated, since it is likely that this region was transferred from a chromosome, although the IS6 elements could have been inserted at any step in the proposed model. Two subtypes of IS91 elements are present in the hypothetical ancestral State 0 that share a conserved 872-bp terminal region that could serve as the basis for a tandem repeat duplication event (Darmon and Leach 2014), leading to duplication of the entire region between the two 872-bp repeats (hypothetical State 1). However, of the three observed imperfectly replicated regions, the two right-most ones carry an IS91 element, the sdpA gene, and a hypothetical protein each. This 2.8-kb sequence was neither present in pAKD16, pAKD34, nor pMC1, which is consistent with a feature that could have been introduced after the first copy event, since it was present in the second and third copy but not in the first one. IS91 elements are known to form gene cassettes with accessory genes and to have circular intermediates (Garcillán-Barcia etal. 2002; Toleman etal. 2006) that can be inserted at new sites, as is also observed in the single-strain mobilome in this study. We propose that this is the mode of entry for the essential sdpA gene, the IS91 element, and the adjacent hypothetical gene into the second cluster copy in hypothetical State 1 between the already present IS91 and the dmlR gene (hypothetical State 2). Subsequently, another tandem repeat duplication event was predicted with two identical IS1595 elements of the ISSod11 subgroup forming the basis for duplication (observed State 3). This duplication event could have involved the two identical IS91 elements (Green arrows in fig. 3), which would, however, have required an additional excision of the IS91 element that would be placed next to the terminal IS6 element, since that was not present in the observed state. This final tandem duplication event led to the formation of the observed state of the sdpA cluster on pMSHV, where there are copies of the sdpA gene in the second and third cluster copies, but not in the first one.
Fig. 3.
—Proposed most parsimonious model of the events leading to the observed state of the sdpA gene cluster on MH megaplasmid pMSHV (top, above dashed line). State 0 (top sequence below dashed line) represents the proposed hypothetical state flanked by IS6 elements. Vertical dotted lines indicate the three imperfect repeated sequences in the observed gene cluster. Proposed events leading from State 0 to the currently observed State 3 are (A) tandem repeat duplication of conserved IS91 sequences (green and pink IS91 genes), (B) IS91-mediated insertion of a 2,790-bp sequence encoding sdpA (red), a hypothetical gene (light gray), and an IS91 element (purple), (C) tandem repeat duplication between two identical IS1595 elements of the ISSod11 subgroup (blue), and (D) the 11-kb circular sequence cIS91sdpA is observed from the assembled mobilome data, encoding sdpA, two IS91 elements, the IS1595 element, and seven other miscellaneous CDS features. Dark gray arrows represent miscellaneous genes with predicted functions, light gray arrows represent hypothetical genes, and colored arrows represent genes of importance to the model or general genetic mobilization function. The blue histogram in the background of State 3 shows sequence coverage of the mobilome data, displaying regions of elevated mobilome sequencing coverage and thus existence of a circular genetic element.
The rdpA Cluster Is Likely Introduced into MH through One-Ended Transposition
The rdpA cluster (fig. 1) was located between the sdpA and cadABCD gene clusters on pMSHV, and therefore, its origin and evolution were difficult to determine, since there are no good contiguous BLASTN hits in the NR database similar to another study that showed this gene to be less abundant in the environment (Paulin etal. 2010). We hypothesize that rdpA, which was flanked by an IS91, has been introduced to strain MH through a one-ended transposition event similar to that for sdpA (fig. 3). There are 21 ORFs over ∼14 kb between rdpA and flanking IS6 gene and the sdpA gene cluster (fig. 1) that encode a putative plasmid partitioning parB protein, putative chromate resistance proteins (chrC, chrA1, and chrB), subunits of DNA polymerase V (umuD, umuC, and lmbB), and some hypothetical proteins. Due to the lack of genes related to catabolism and an absence of IS elements, this 14-kb stretch of DNA could represent the ancestral DNA of pMSHV that was the site of multiple insertion events of IS elements and composite transposons harboring genes related to degradation of xenobiotics.
Single-Strain Mobilomics Shows a Plastic Genome in Flux
Manual inspection of sequence coverage from the single-strain mobilome, coupled with the automated Coverage Analyses implemented in CLC Genomics Workbench, revealed elevated mobilome coverage of 44 genetic regions on the chromosome and plasmids pMSHV and pSHV1 (fig. 1). Many of the 44 regions with increased mobilome coverage are duplicates occurring multiple times on the main replicons and are represented by 13 unique short sequences ranging from 855 to 6,229 bp and four longer regions representing two prophages and the sdpA- and rdpA clusters (figs. 1 and 4;supplementary table S2, Supplementary Material online).
Long reads from PacBio and Nanopore were used to verify that regions of higher mobilome coverage were not repeated sequences that were misassembled. Sequences of the single-strain mobilome were assembled with SPAdes and revealed an 11-kb segment of the sdpA cluster that has a circular state (cIS91sdpA) and includes the sdpA gene, some IS elements, and seven other genes (figs. 1 and 3). This circular molecule was likely generated by either of the two IS91 elements which both display a conserved 17-bp origin of replication (oriIS) that is 167-bp downstream of the CDS feature consistent with previous findings (Schleinitz etal. 2010). Several IS91 termination sites (terIS) were also predicted throughout the sdpA gene cluster (not shown) and as discussed by Toleman etal. (2006) IS91 elements might utilize several different terIS sites or might terminate replication at random sites. The case of transposition without the terIS site is termed one-ended transposition and requires only the conserved oriIS and a tetranucleotide (5′-CTTG or 5′-GTTC) in proximity and results in translocation of IS91 and variable lengths of adjacent sequence (Mendiola etal. 1994; Garcillán-Barcia and De La Cruz 2002). This could explain the observation that coverage of the mobilome sequence data extended beyond the boundaries of the SPAdes-assembled circular molecule, as is evident in the sequence coverage in the observed State 3 (fig. 3). Supported by the single-strain mobilome sequencing, we propose that sdpA was inserted via IS91-mediated one-ended transposition and subsequently formed the 11-kb circular cIS91sdpA which could lead to propagation of sdpA in a microbial community via its insertion into conjugative plasmids.
Although the exact circular state of cIS91sdpA could be determined through mapping of mobilome reads, a circular intermediate molecule of the rdpA cluster could not be established, although the mobilome sequencing coverage is higher in this region than the background coverage of pMSHV and indicates that every 580th bacterial cell would have a free circular copy of the rdpA cluster, corresponding to 0.0017 copies per cell (supplementary fig. S7, Supplementary Material online).
There is no data indicating circular molecules originating from the cadABCD clusters on pMSHV and the chromosome (fig. 1). However, pSHV3 contains an IS1380 element that seems to have inserted four identical copies of the IS element and adjacent genes, yfcG, gstB, and partial rutE into the MH chromosome and in a single case including a complete rutE gene (fig. 1).
Short, Circular DNA Molecules from Various Genomic Locations
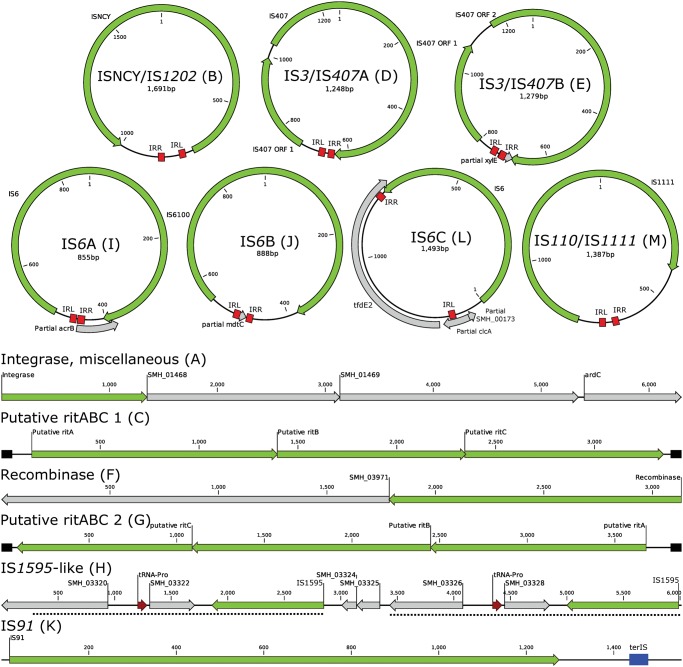
Through read mapping of mobilome reads to the 13 short unique sequences, it was possible to determine configurations of the exact circular intermediates of seven DNA molecules, whereas six regions with elevated mobilome coverage could not be definitively determined to have circular intermediates (fig. 4). For all seven sequences with determined circular intermediate molecules, it was possible to identify inverted repeats in the size range 14–26 bp flanking the IS elements, corresponding largely to the inverted repeats of their closest homologs in the ISfinder database (Siguier etal. 2006). The seven small circular sequences harbor IS elements from the families ISNCY, IS3, IS6, and IS110. Two different IS3 elements, both of the IS407 group, are present with each element being constituted by two ORFs. Three different variants of IS6-family elements occur, where one of the small circular elements harbors the possibly truncated variant of the tfdE gene from the cadABCD gene cluster (fig. 2; tfdE2) besides the IS6 ORF.
Fig. 4.
—Regions with elevated mobilome coverage from the genome of Sphingobium herbicidovorans MH. Families and groups of IS elements were assigned from ISfinder and are denoted as family/group for those that could be assigned to a group (e.g., ISNCY/IS1202) and simply as the IS family if no group could be assigned (e.g., IS91). Two unique IS3/IS407 elements appeared as circular elements and are named IS3/IS407A and IS3/IS407B. Three unique IS6 elements were identified and are named IS6A, IS6B, and IS6C. Two unique putative ritABC elements are shown as ritABC 1 and ritABC 2. Length of inverted repeats (IRL and IRR) are B: 18 bp, D: 25/26 bp, E: 24/25 bp, I: 14/16 bp, J: 14 bp, L: 20/21 bp, M: 15 bp. Furthermore, ritABC 1 and ritABC 2 have direct repeats flanking the ORFs consisting of a 42-bp perfect repeat and a 51-bp imperfect, respectively. Finally, a 40-bp terminator sequence (terIS) could be identified for the IS91 (K) element, but no origin of replication. Letters in parentheses were given in alphabetical order based on the order in which they first occur in the genome shown in figure 1. Many of the sequences occur in multiple copies in the genome (fig. 1 and supplementary table S2, Supplementary Material online). Sequences are not drawn to scale.
Intriguingly, several genes encoding recombinases/integrases with the XerDC protein domains COG4973 and COG4974, including the newly described recombinase in trio (RIT) elements (Ricker etal. 2013) show elevated mobilome coverage, but circular intermediates of these sequences could not be determined by read mapping of paired-end reads (fig. 4; A, C, F, and G). It was previously shown that RIT elements are likely mobilized intracellularly as a unit, although the mechanism of mobilization has yet to be deduced (Van Houdt etal. 2012; Ricker etal. 2013). The mobilome data presented here indicates that RIT elements, along with other integrase/recombinase genes form free intermediate DNA molecules that are protected from exonuclease digestion, possibly as a stage in their mobilization. The presence of respectively 47- and 51-bp long imperfect palindromic repeats in the ends of both putative RIT elements suggests that the RIT elements form intermediate hairpin-structured molecules during their mobilization that may be protected from exonuclease digestion. Furthermore, it was not possible to determine the complete circular structure of two regions with elevated mobilome coverage containing IS1595-like IS elements and an IS91 element, respectively (fig. 4: H, K). One of these contains a 2,580-bp perfect repeat encoding a IS1595-like element, a tRNA, and two hypothetical genes of which one might be in a truncated form. Putative circular DNA molecules originating from this region is estimated to have an only slightly higher copy number than the mean of ten control regions with no increased mobilome coverage and less than the +2SD limit (supplementary fig. S7, Supplementary Material online). It is thus not expected that any circular DNA molecules originate from this region with IS1595-like elements, although manual inspection of mobilome read mapping indicated a slightly elevated coverage. Finally, a region with elevated mobilome coverage encode an IS91-like element that is ∼83% similar to the large IS91 element in cIS91sdpA and 97% similar to the ISPps1 transposase from Pseudomonas huttiensis. A putative 40-bp terIS could be identified downstream of the IS91 ORF, but no oriIS was found (fig. 4). This suggests that this IS91 may have lost the ability to form circular intermediates during mobilization or that this IS element uses a novel oriIS.
The small circular DNA molecules range in copy number per cell from 0.0004 for the 1,248-bp region with an IS407-like element to 0.0131 for the 855-bp IS6A element (supplementary fig. S7, Supplementary Material online), corresponding to a free circular molecule occurring in every 2,364th and 76th cell, respectively. The mobilome sequencing approach can therefore be used to study the dynamics mobile genetic elements (MGEs) with circular stages, or other tertiary DNA structures that are protected from exonuclease digestion such as hairpin loops, and their effect on genome evolution and adaptation under various conditions.
Excised Prophage Genomes Revealed by Mobilome Sequencing
From the mobilome data, it is possible to not only investigate the circular intermediates of IS elements and recombinases/integrases during mobilization but also of prophages that are integrated into the chromosome (fig. 1). Two such prophages in the chromosome, both ∼40 kb in size, were identified in their excised state from the mobilome data and confirmed with mapping of paired-end mobilome reads to be circular (supplementary fig. S8, Supplementary Material online). Although circular excised genomes of Prophage 1 had a copy a number of 0.00087 per cell (corresponding to a single copy in every 1,150th cell), and thus higher than the defined limit of mean of background mobilome coverage of ten noncircular regions +2 SD (0.00042), Prophage 2 had a copy number of 0.00021 (one copy in every 4,762nd cell) which was lower than this limit. However, the presence of excised circular genomes of both prophages is proven by the mapping of mobilome reads that overlap ends of both prophage sequences. The mobilome sequencing approach will be very useful in future investigations of the dynamics of prophage excision rates and what factors might affect these.
Replicons, MGEs, and Genome Plasticity in Sphingomonads Drive the Formation of Catabolic Traits
From single-strain mobilomic sequencing, circular states were observed of the replicons pSHV1, pSHV2, and pSHV3, as well as transposon cIS91sdpA, two prophages, several integrases/recombinases, ritABC elements, along with IS elements of the families IS3, IS1595, IS6, IS91, and IS110 (figs. 1 and 4). The rate at which genetic elements excise from the their genomic positions and form free circular intermediate states, ready to integrated into new locations and cause genetic rearrangements, was moreover calculated (supplementary fig. S7, Supplementary Material online).
A few models describing the formation of gene clusters with specific pathways occur have been proposed, including the selfish operon model that dictates that specialized gene clusters are more likely to be mobilized in HGT (Lawrence and Roth 1996; Ballouz etal. 2010). It is furthermore commonly understood that sphingomonads harbor a large array of genes encoding catabolic enzymes and have generally specialized in degradation of recalcitrant hydrocarbon compounds (Stolz 2009; Aylward 2013). The present study adds insight to the mechanisms through which catabolic gene clusters arise and mobilize. Specifically, the essential sdpA gene was shown to be mobilized on an 11-kb circular DNA element that excise from its original location on the pMSHV megaplasmid approximately once per 59th cell and can thus be transferred to new replicons and subsequently propagate through a bacterial population. Furthermore, short DNA sequences encoding various IS elements was shown to form circular intermediates in high copy numbers, that can lead to the formation of gene clusters such as the cadABCD cluster that is flanked by IS6 elements that was likely copied from the original chromosomal location to the megaplasmid. These results demonstrate the remarkable plasticity of the Sphingobium herbicidovorans MH genome and continuous evolution of xenobiotics degradation pathways. Application of the single-strain mobilome sequencing furthermore provides a unique tool to study the extent of genomic rearrangements mediated by MGEs such as plasmids, prophages, IS- and RIT elements and should be considered as a new standard approach to studying microbial genomes in detail.
Conclusion
The complete genome and single-strain mobilome of chlorophenoxy herbicide-degrading Sphingobium herbicidovorans MH shows how clusters of xenobiotics catabolic genes were formed through interplay of genomic rearrangements and IS element-related alterations. The two cadABCD clusters on chromosome and megaplasmid pMSHV led to divergence and natural variations of genetic arrangements that in turn may include more efficient configurations for improved xenobiotic degradation. The hereby proposed model for evolution of the cadABCD cluster accounts for the currently observed genetic diversity in sphingomonads and supports the previously hypothesized restricted spread of this cluster outside the sphingomonad group and that the evolution of these clusters have occurred in a phylogenetically isolated manner in the sphingomonads (Itoh etal. 2004; Nielsen etal. 2013), since we did not find any similar sequences in nonsphingomonads.
The sdpA gene cluster showed an expansion and gene redundancy through the two tandem repeat duplications. Furthermore, the mobilome data revealed that sdpA is part of the highly active IS91 element-associated circular molecule cIS91sdpA that has potential for spreading to new plasmids and subsequent dissemination of the essential sdpA gene to new bacterial hosts. A total of 44 genetic regions displayed elevated sequence coverage from the mobilome data set, leading to the identification of several unique short, circular, intermediate DNA molecules consisting of IS elements and other MGEs that can mobilize between replicons. Furthermore, the formation rates of these intermediate DNA molecules could be estimated to provide insight into the dynamics of their mobilization. Finally, two chromosomally integrated prophages were shown, based on the mobilome data, to excise and form free circular DNA molecules. The single-strain mobilome method does not only produce plasmid sequences (a plasmidome) or a “circularome” (circular DNA molecules), but diverse genetic information in transit. The application of single-strain mobilomics holds great potential for studying dynamics of bacterial adaptation in microbial ecology rather than the static insight provided by traditional whole genome sequencing.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported partly by a Villum foundation Block Stipend and MEM2BIO (Innovation Fund Denmark, contract number 5157-00004B) awarded to LHH and ORIGENE (AUFF NOVA) awarded to LHH and TKN.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aylward FO. 2013. Comparison of 26 sphingomonad genomes reveals diverse environmental adaptations and biodegradative capabilities. Appl Environ Microbiol. 79(12):3724–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballouz S, Francis AR, Lan R, Tanaka MM, Eisen JA.. 2010. Conditions for the evolution of gene clusters in bacterial genomes. PLoS Comput Biol. 6(2):e1000672.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf D, Davidson I, Babel W.. 2004. Regulation of catabolic enzymes during long-term exposure of Delftia acidovorans MC1 to chlorophenoxy herbicides. Microbiology 150(Pt 4):1005–1014. [DOI] [PubMed] [Google Scholar]
- Copley SD, et al. 2012. The whole genome sequence of Sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol Evol. 4(2):184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E, Leach DRF.. 2014. Bacterial genome instability. Microbiol Mol Biol Rev. 78(1):1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don RH, Weightman AJ, Knackmuss HJ, Timmis KN.. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2, 4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcalienes eutrophus JMP134(pJP4). J Bacteriol. 161:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drønen AK, Torsvik V, Goksøyr J, Top EM.. 1998. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol Ecol. 27(4):381–394. [Google Scholar]
- Drønen AK, Torsvik V, Top EM.. 1999. Comparison of the plasmid types obtained by two distantly related recipients in biparental exogenous plasmid isolations from soil. FEMS Microbiol Lett. 176(1):105–110. [Google Scholar]
- Feld L, et al. 2015. Establishment of bacterial herbicide degraders in a rapid sand filter for bioremediation of phenoxypropionate-polluted groundwater. Appl Environ Microbiol. 82(3):878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer K, Harker AR.. 1997. Identification of the inducing agent of the 2,4-dichlorophenoxyacetic acid pathway encoded by plasmid pJP4. Appl Environ Microbiol. 63(1):317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori F, Hausinger RP.. 1993. Alcaligenes eutrophus JMP134 ‘2, 4-dichlorophenoxyacetate monooxygenase’ is an α-ketoglutarate-dependent dioxygenase. J Bacteriol. 175(7):2083–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe RR, McGowan C, Maltseva OV, Holben WE, Tiedje JM.. 1995. 2, 4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 61(9):3274–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillán-Barcia M, Bernales I, Mendiola M, De La Cruz F.. 2002. IS91 rolling-circle transposition. In Mobile DNA II (eds), Craig N, Craigie R, Gellert M, Lambowitz A, 891–904. Washington, DC: ASM Press.
- Garcillán-Barcia MP, De La Cruz F.. 2002. Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. FEMS Microbiol Ecol. 42(2):303–313. [DOI] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Pereto J, Moya A.. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 68(3):518–537. Table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gözdereliler E, et al. 2013. Comparing metabolic functionalities, community structures, and dynamics of herbicide-degrading communities cultivated with different substrate concentrations. Appl Environ Microbiol. 79(1):367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann D, Müller RH.. 2006. 2, 4-Dichlorophenoxyacetic acid (2, 4-D) utilization by Delftia acidovorans MC1 at alkaline pH and in the presence of dichlorprop is improved by introduction of the tfdK gene. Biodegradation 17(3):263–273. [DOI] [PubMed] [Google Scholar]
- Horvath M, Ditzelmüller G, Loidl M, Streichsbier F.. 1990. Isolation and characterization of a 2-(2, 4-dichlorophenoxy) propionic acid-degrading soil bacterium. Appl Microbiol Biotechnol. 33(2):213–216. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. 2016. Microbial catabolism of chemical herbicides: microbial resources, metabolic pathways and catabolic genes. Pestic Biochem Physiol. doi: 10.1016/j.pestbp.2016.11.010. [DOI] [PubMed] [Google Scholar]
- Itoh K, et al. 2004. Root nodule Bradyrhizobium spp. harbor tfdAα and cadA, homologous with genes encoding 2, 4-dichlorophenoxyacetic acid-degrading proteins. Appl Environ Microbiol. 70(4):2110–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen TS, Xu Z, Hansen MA, Sørensen SJ, Hansen LH.. 2014. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS One 9(2):e87924.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa W, et al. 2002. Novel 2, 4-dichlorophenoxyacetic acid degradation genes from oligotrophic bradyrhizobium sp. strain HW13 isolated from a pristine environment. J Bacteriol. 184(2):509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler HPE. 1999. Sphingomonas herbicidovorans MH: a versatile phenoxyalkanoic acid herbicide degrader. J Ind Microbiol Biotechnol. 23(4–5):336–340. [DOI] [PubMed] [Google Scholar]
- Koren S, et al. 2017. Canu: Scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR.. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143(4):1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibeling S, et al. 2010. Declining capacity of starving delftia acidovorans MC1 to degrade phenoxypropionate herbicides correlates with oxidative modification of the initial enzyme. Environ Sci Technol. 44(10):3793–3799. [DOI] [PubMed] [Google Scholar]
- Leplae R, Lima-Mendez G, Toussaint A.. 2010. ACLAME: a CLAssification of mobile genetic elements, update 2010. Nucleic Acids Res. 38(Suppl 1):D57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau JHJ, Zehnder AJB, Van Der Meer JR.. 1998. The tfdK gene product facilitates uptake of 2, 4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J Bacteriol. 180(8):2237–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Liu SJ, Drake HL, Horn MA.. 2013. Consumers of 4-chloro-2-methylphenoxyacetic acid from agricultural soil and drilosphere harbor cadA, r/sdpA, and tfdA-like gene encoding oxygenases. FEMS Microbiol Ecol. 86(1):114–129. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43(Database issue):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10–12. [Google Scholar]
- McManus SL, Richards KG, Grant J, Mannix A, Coxon CE.. 2014. Pesticide occurrence in groundwater and the physical characteristics in association with these detections in Ireland. Environ Monit Assess. 186(11):7819–7836. [DOI] [PubMed] [Google Scholar]
- Mendiola MV, Bernales I, de la Cruz F.. 1994. Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci U S A. 91(5):1922–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn WW, Mertens B, Neufeld JD, Verstraete W, De Lorenzo V.. 2006. Distribution and phylogeny of hexachlorocyclohexane-degrading bacteria in soils from Spain. Environ Microbiol. 8(1):60–68. [DOI] [PubMed] [Google Scholar]
- Müller RH, Hoffmann D.. 2006. Uptake kinetics of 2, 4-dichlorophenoxyacetate by Delftia acidovorans MC1 and derivative strains: complex characteristics in response to pH and growth substrate. Biosci Biotechnol Biochem. 70(7):1642–1654. [DOI] [PubMed] [Google Scholar]
- Müller RH, Jorks S, Kleinsteuber S, Babel W.. 1999. Comamonas acidovorans strain MC1: a new isolate capable of degrading the chiral herbicides dichlorprop and mecoprop and the herbicides 2, 4-D and MCPA. Microbiol Res. 154(3):241–246. [DOI] [PubMed] [Google Scholar]
- Müller TA, Byrde SM, Werlen C, Van Der Meer JR, Kohler HPE.. 2004. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl Environ Microbiol. 70(10):6066–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TA, Fleischmann T, van der Meer JR, Kohler H-PE.. 2006. Purification and characterization of two enantioselective alpha-ketoglutarate-dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl Environ Microbiol. 72(7):4853–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel K, Suter MJ, Kohler HP.. 1997. Involvement of two alpha-ketoglutarate-dependent dioxygenases in enantioselective degradation of (R)- and (S)-mecoprop by Sphingomonas herbicidovorans MH. J Bacteriol. 179(21):6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TK, et al. 2013. Novel insight into the genetic context of the cadAB genes from a 4-chloro-2-methylphenoxyacetic acid-degrading Sphingomonas. PLoS One 8(12):e83346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin MM, Nicolaisen MH, Sørensen J.. 2011. (R, S)-dichlorprop herbicide in agricultural soil induces proliferation and expression of multiple dioxygenase-encoding genes in the indigenous microbial community. Environ Microbiol. 13(6):1513–1523. [DOI] [PubMed] [Google Scholar]
- Paulin MM, Nicolaisen MH, Sørensen J.. 2010. Abundance and expression of enantioselective rdpA and sdpA dioxygenase genes during degradation of the racemic herbicide (R, S)-2-(2, 4-dichlorophenoxy)propionate in soil. Appl Environ Microbiol. 76(9):2873–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SL, Oakeshott JG, Pandey G.. 2015. Insights into ongoing evolution of the hexachlorocyclohexane catabolic pathway from comparative genomics of ten Sphingomonadaceae strains. G3 Genes Genomes Genet 5:1081 LP–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker N, Qian H, Fulthorpe RR.. 2013. Phylogeny and organization of recombinase in trio (RIT) elements. Plasmid 70(2):226–239. [DOI] [PubMed] [Google Scholar]
- Santiago A, et al. 2014. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 14:112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz KM, Kleinsteuber S, Vallaeys T, Babel W.. 2004. Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2, 4-dichlorophenoxy)propionate cleavage in Delftia acidovorans MC1. Appl Environ Microbiol. 70(9):5357–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz KM, Vallaeys T, Kleinsteuber S.. 2010. Structural characterization of ISCR8, ISCR22, and ISCR23, subgroups of IS91-like insertion elements. Antimicrob Agents Chemother. 54(10):4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- Sen D, et al. 2011. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl Environ Microbiol. 77(22):7975–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Kawakami M, Amada K.. 2009. Analysis of genes encoding the 2,4-dichlorophenoxyacetic acid-degrading enzyme from Sphingomonas agrestis 58-1. J Biosci Bioeng. 108(1):56–59. [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M.. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(Database issue):D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A. 2009. Molecular characteristics of xenobiotic-degrading sphingomonads. Appl Microbiol Biotechnol. 81(5):793–811. [DOI] [PubMed] [Google Scholar]
- Tabata M, et al. 2016. Comparison of the complete genome sequences of four γ-hexachlorocyclohexane-degrading bacterial strains: insights into the evolution of bacteria able to degrade a recalcitrant man-made pesticide. DNA Res. 23:581–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel M, Kaschabek SR, Gröning J, Mau M, Schlömann M.. 2005. Two unusual chlorocatechol catabolic gene clusters in Sphingomonas sp. TFD44. Arch Microbiol. 183(2):80–94. [DOI] [PubMed] [Google Scholar]
- Toleman MA, Bennett PM, Walsh TR.. 2006. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J Antimicrob Chemother. 58(1):1–6. [DOI] [PubMed] [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M.. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12(3):R30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R, Leplae R, Lima-Mendez G, Mergeay M, Toussaint A.. 2012. Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes. Mob DNA 3(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma H, et al. 2014. Comparative genomic analysis of nine Sphingobium strains: insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genomics 15:1014.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf A, Benndorf D, Müller RH, Babel W.. 2002. The two enantiospecific dichlorprop/alpha-ketoglutarate-dioxygenases from Delftia acidovorans MC1–protein and sequence data of RdpA and SdpA. Microbiol Res. 157(4):317–322. [DOI] [PubMed] [Google Scholar]
- Westendorf A, Benndorf D, Pribyl T, Harms H, Müller RH.. 2006. Kinetic traits and enzyme form patterns of (R)-2-(2, 4-dichlorophenoxy) propionate/alpha-ketoglutarate dioxygenase (RdpA) after expression in different bacterial strains. Eng Life Sci. 6(6):552–559. [Google Scholar]
- Zipper C, Bunk M, Zehnder AJB, Kohler H-PE.. 1998. Enantioselective uptake and degradation of the chiral herbicide dichlorprop [(RS)-2-(2, 4-dichlorophenoxy)propanoic acid] by Sphingomonas herbicidovorans MH. J Bacteriol. 180:3368–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper C, Nickel K, Angst W, Kohler H.. 1996. Complete microbial degradation of both enantiomers of the chiral herbicide mecoprop [(RS)-2-(4-chloro-2-methylphenoxy)propionic acid] in an enantioselective manner by Sphingomonas herbicidovorans sp. nov. Appl Environ Microbiol. 62:4318–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.