Abstract
Ribosome assembly requires the concerted expression of hundreds of genes, which are transcribed by all three nuclear RNA polymerases. Transcription elongation involves dynamic interactions between RNA polymerases and chromatin. We performed a synthetic lethal screening in Saccharomyces cerevisiae with a conditional allele of SPT6, which encodes one of the factors that facilitates this process. Some of these synthetic mutants corresponded to factors that facilitate pre-rRNA processing and ribosome biogenesis. We found that the in vivo depletion of one of these factors, Arb1, activated transcription elongation in the set of genes involved directly in ribosome assembly. Under these depletion conditions, Spt6 was physically targeted to the up-regulated genes, where it helped maintain their chromatin integrity and the synthesis of properly stable mRNAs. The mRNA profiles of a large set of ribosome biogenesis mutants confirmed the existence of a feedback regulatory network among ribosome assembly genes. The transcriptional response in this network depended on both the specific malfunction and the role of the regulated gene. In accordance with our screening, Spt6 positively contributed to the optimal operation of this global network. On the whole, this work uncovers a feedback control of ribosome biogenesis by fine-tuning transcription elongation in ribosome assembly factor-coding genes.
INTRODUCTION
Transcription is the main target of gene regulation. Those genes that undergo tight control can switch transcription on or off in response to regulatory stimuli. Most of this control takes place during the assembly of pre-initiation complexes, but gene transcription can also be regulated during the transition from initiation to productive elongation. The so-called promoter-proximal pausing is in fact a critical regulatory step for many genes in most eukaryotes (1). In addition to these on/off regulation mechanisms, transcription elongation in gene bodies can be modulated when it becomes a rate-limiting step (2).
The main limitation to transcription elongation in eukaryotes results from the organisation of the DNA template into chromatin. Nucleosomes have to be evicted or extensively rearranged in order to facilitate RNA polymerase II (RNA pol II) progression (3,4). This highly dynamic process requires the functional contribution of a set of auxiliary factors that facilitate access to DNA in the chromatin template, modify histones covalently or catalyse the disassembly and reassembly of dynamic nucleosomes. One of these factors is Spt6, which plays an important role in transcription elongation from yeast to humans (5–10). Spt6 binds different phosphorylated forms of the RNA pol II C-terminal domain (CTD) through its SH2 domain (11–13), promotes H3K36 trimethylation (14), and acts as a histone chaperone during nucleosome reassembly after the passage of elongating RNA pol II (15–21). A contribution of Spt6 to rDNA transcription by RNA pol I has also been described (22).
In the present work, we screened for new genetic interactions of SPT6. Some of them link Spt6 with the genes that promote ribosome biosynthesis. These genes encode trans-acting factors that are required for pre-rRNA processing and/or the loading of ribosomal proteins into mature ribosomal subunits (23,24). The expression of these ribosome assembly factors is co-regulated with other groups (RNA pol I and II subunits, tRNA factors, translation factors, NTP synthesis) during cell growth and proliferation, which have all together been defined as the RiBi regulon (25,26). The genes that encode ribosomal proteins (the RP regulon) show higher expression levels than the RiBi regulon, and exhibit some regulatory differences, but are similarly regulated in response to, for instance, nutritional signals (25).
The transcriptional regulation of all the ribosome-related genes in response to growth-associated stimuli is mediated by the Ras-PKA pathway and the TOR kinase (25,27,28). On top of all this, RP genes respond to pre-rRNA processing by a direct interaction between the CURI complex, which activates RP transcription together with Rap1 and Fhl1 (29,30). However, no regulatory connection has been established between pre-RNA processing or ribosomal subunits assembly and the genes that encode the auxiliary factors that facilitate the whole process. We show herein that this regulatory connection exists and involves the differential regulation of specific genes according to their functional role on the ribosome biosynthesis pathway. We also show that this feedback control of ribosome biogenesis involves modulating the transcription elongation of the target genes.
MATERIALS AND METHODS
Strains, media and culture reagents
All the yeast strains used in this study are described in Supplementary Table S1. The strains for the transcriptomic analysis of the mutants that affected ribosome biogenesis were derivatives of BY4741. The concentrations of the translation drugs in the growth assays were: Cycloheximide (0.05 μg/ml), Anisomycin (15 μg/ml). Doxycycline (Sigma) was dissolved in distilled water in a concentrated stock and was added at a final concentration of 5 μg/ml. To test growth, yeast cultures were diluted to the an OD600 of 0.05 and serial dilutions (1:10) were spotted onto plates. At least three independent experiments were carried out in all cases.
Synthetic lethal screening
Screening was carried out following the plasmid-based strategy described in Supplementary Figure S1A. Sectoring colonies were able to lose the URA3-ADE3-SPT6 centromeric plasmid present in the strain that carried chromosomal alleles ura3, ade2, ade3 and spt6–140. Consequently, these cells were resistant to 5-fluororotic acid (5-FOA), a drug that requires the Ura3 protein for toxicity. In contrast, the colonies that exhibited only the characteristic red phenotype of ade2 were unable to lose the plasmid and displayed sensitivity to 5-FOA. This scenario suggests the existence of an additional mutation that conferred lethality in the absence of a wild-type SPT6 allele (Supplementary Figure S1B). Fifty-six candidates were identified from about 292 000 colonies screened in four different mutagenesis experiments. These candidates were tested to exclude the mutations that affected the following: plasmidic SPT6 allele; integration of the plasmid into a chromosome; presence of a second mutation in the spt6–140 allele; the polygenic nature of synthetic lethality. They were all backcrossed three times with the original strain to exclude the influence of any secondary mutation on the suppressor phenotype. SPT4 was cloned by complementation. The other eight complementation groups were transformed with a plasmidic DNA library and were screened for either complementation of their temperature-related phenotypes or restoration of their colony-sectoring ability.
Sucrose gradient centrifugation
Polysome preparations and analyses were performed as previously described (31) with an ISCO UA-6 system equipped to continuously monitor absorbance at 254 nm. A representative image of at least two replicates is shown.
Protein extractions and western blot analyses
Total yeast protein extracts were prepared and analysed by Western blotting following standard procedures. The following primary antibodies were used: mouse monoclonal anti-Myc (Santa Cruz Biotechnology), rabbit polyclonal anti-hexokinase (Abcam).
RNA extractions and northern hybridization
The RNA extraction and Northern hybridization analyses were carried out according to standard procedures. In all the experiments, RNA was extracted from exponentially growing cells. Equal amounts of total RNA were loaded onto gels. The oligonucleotides used for hybridizations have been previously described (32). The phosphorimaging analyses were performed in an FLA-5100 imaging system (Fujifilm).
Pulse-chase labeling of pre-rRNA
Pulse-chase labeling of pre-rRNA was performed as described elsewhere (33), using 100 μCi of [5,6–3H]uracil (45–50 Ci/mmol; Perkin Elmer) per 40 OD600 units of yeast cells. Cells were first transformed with an empty YCplac33 plasmid (CEN URA3) to make them prototrophic for uracil. A representative transformant was grown in liquid SD-Ura medium to the exponential growth phase at 30°C, pulse-labelled for 2 min, and chased for 5, 15, 30 and 60 min with SD medium that cotained an excess cold uracil. Total RNA was extracted by the acid–phenol method. Radioactive incorporation was measured by scintillation counting and ∼20 000 cpm per RNA sample were analysed by electrophoresis on 1.2% agarose–6% formaldehyde gel. RNA then was transferred to a nylon membrane and visualized using a BAS-TR2040 tritium screen (Fujifilm). Signal intensities were measured by a Typhoon 9400 imaging system (GE Healthcare) and the GelQuant.NET software (biochemlabsolutions.com).
Chromatin immunoprecipitation
The ChIP experiments were performed as previously described (34). Immunoprecipitations were performed with magnetic beads (Dynal) using the following antibodies: Myc (Santa Cruz Biotechnology), HA (clone 3F10, Roche) Rpb3 (ab81859; Abcam), H3 (ab1791, Abcam). DNAs were analysed by real-time quantitative PCR with SYBR Green Premix Ex Taq (Takara) in a Light Cycler 480 II (Roche). The amount of IP was represented in relation to input. Primer sequences are available upon request.
Genomic run-on and RPCC
The GRO and RPCC data shown in Figure 3 are stored in the Valencia Yeast database (VYdBase; http://vydbase.uv.es/) and in the GEO repository (accession number GSE14060). Detailed protocols for these methods have been previously described (2,35) and can also be found in the above-mentioned submission. Briefly for GRO, cells were washed twice in 0.5% of N-lauryl sarcosine sodium sulphate (sarkosyl) for permeabilization, chromatin clearance and transcription ternary complexes fixation. In vivo transcription was performed for 5 min at 30°C by re-suspending cells in an appropriate transcription buffer in the presence of [α33P]UTP. Cells were recovered, and the total radioactive UTP in vivo-labeled RNA was isolated by standard procedures and used for hybridization. The chromatin immunoprecipitation of RNA pol II (RPCC) was performed with an 8WG16 antibody (Santa Cruz, sc-56767) following standard procedures. Ligation-mediated PCR was used for the DNA amplification of the input and immunoprecipitated samples. The PCR product was used as a template for a radioactive labeling reaction and subsequent array hybridization.
Figure 3.
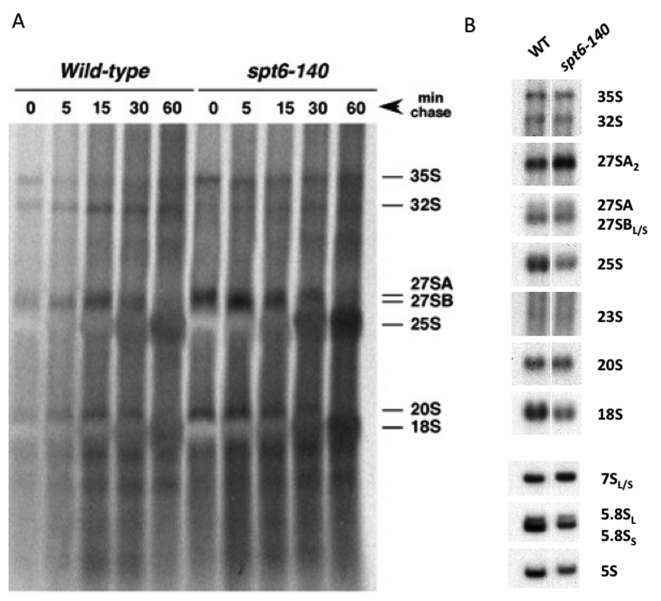
spt6–140 shows limited effects on rRNA synthesis. (A) Pulse-chase labeling of pre-rRNA in wild-type and isogenic spt6–140 cells growing at 30°C. Samples were taken at the indicated times. The position of the main pre-rRNAs and mature rRNAs is indicated on the right. (B) The high-molecular-mass pre-rRNAs and mature rRNAs of spt6–140 were compared with its isogenic wild type by northern analysis. Equal total RNA quantities from the cells grown exponentially at 30°C were loaded in gels.
Three biological replicates of the GRO and RPCC experiments were performed. The filters in each replicate were swapped among the different sampling times. Images were quantified by the ArrayVision 7.0 software (Imaging Research, Inc.). The signal intensity for each spot was background subtracted ARM Density (Artefact Removed Median). Only those values that were 1.35-fold higher than the corresponding background were taken as valid measurements. To compare the RPCC data between experiments, the median binding ratios of the 32 rDNA spots were arbitrarily set as a background. Reproducibility of replicates was checked by the ArrayStat software (Imaging Research, Inc.). Normalization between conditions was done by the global median method, as described elsewhere (35). The ratio between the immunoprecipitate and input in each experiment (or non antibody and input) after normalization was taken as the binding ratio. In order to compare RPCC and GRO, log data were standardized using z-scores (subtracting the mean and dividing by the standard deviation), as formerly described (2).
Measurement of mRNA levels and half-lives
Total RNA was purified by hot phenol-chloroform extraction and was reverse-transcribed to cDNA using M-MLV Reverse Transcriptase (Promega). A relative qPCR was then carried out for all the genes studied against SCR1. To determine the mRNA half-lives, we used the transcription shut-off assay, as previously described (36). Briefly, transcription was stopped by adding thiolutin at 5 μg/ml to log-phase cultures grown in YPD. Then culture samples were taken at different time points by a quick 2-minute centrifugation and the immediate freezing of cell pellets. mRNA levels were then determined and half-lives were estimated by the decay of signal over time.
Microarray gene expression profiling
The microarray data shown were submitted to the public ArrayExpress microarray database (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-3189, for the initial set of single mutants expression profiling (Figure 5), and E-MTAB-4574 for the spt6–140 single and double mutants (Figure 6), as well as the reprofiled RiBi single mutants. Detailed protocols can also be found in the above-mentioned submission. Yeast growth, RNA isolation, amplification, labeling, hybridization and data analysis were done as described elsewhere (37), but with the following modifications: the focal point was on genes RP and RiBi instead of on all the genes. Those genes with a very high intensity (proxy for expression levels) were excluded for saturation reasons, as were the genes that did not change significantly (fold change of >1.5, multiple test-adjusted P-value of <0.01) in at least two of the deletions, which left 56 genes. Of the 59 deletion analysed profiles, 37 had three significantly changing genes or more, or had a major overall effect on the 56 genes in question.
Figure 5.
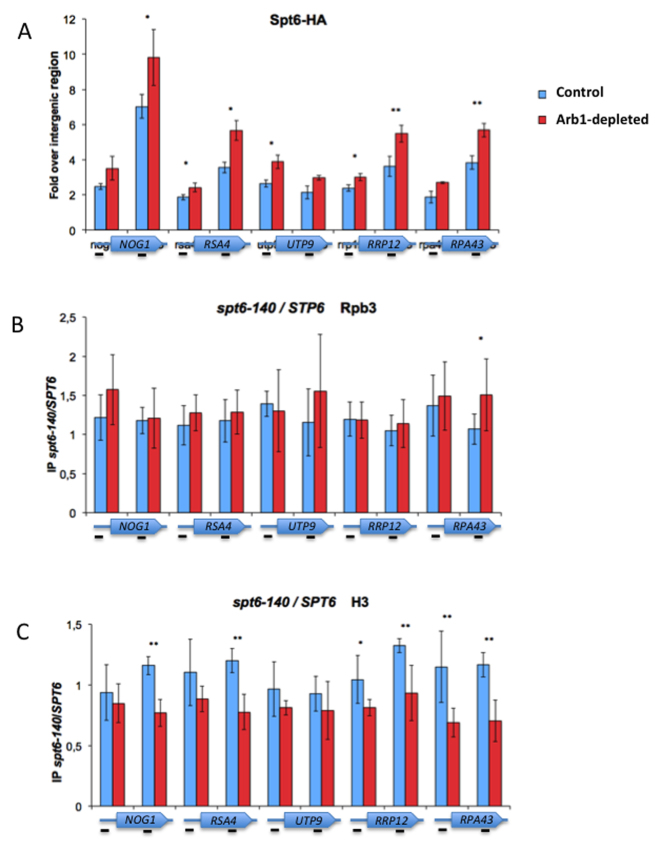
Spt6 recruitment upon Arb1 depletion prevents histone loss. (A) The arb1Δ TEToff:ARB1:MYC strain was transformed with an Spt6-HA-expressing centromeric plasmid to measure Spt6 occupancy during Arb1 depletion. The DNA regions located upstream of the transcription start sites and inside the ORFs of various RiBi genes were tested. (B and C) The occupancy of histone H3 (B) and RNA pol II-specific subunit Rpb3 (C) was measured by ChIP in the arb1Δ TEToff::ARB1::MYC strains (that contained either the SPT6 or the spt6–140 allele) at the indicated conditions. Both IPs came from the same extracts. ChIP values in relation to input. The spt6–140/SPT6 ratios are represented for Arb1-depleted and control conditions. The average and standard deviation of three biological replicates are shown. Two-tailed t-test; *P < 0.05, **P <0.005.
Figure 6.
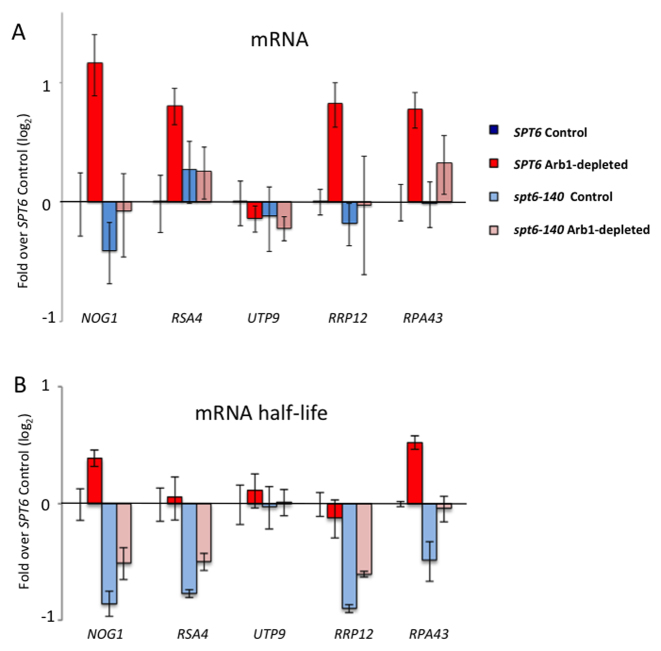
Spt6 contributes to the synthesis of stable mRNA of ribosome biogenesis genes upon Arb1 depletion. (A) Fold-change of the mRNA levels of the five genes tested in Figure 5 upon Arb1 depletion, mRNA levels were measured by RT-PCR. (B) Fold-change of the mRNA half-lives of the same set of genes, upon Arb1 depletion. The mRNA half-lives were calculated after performing transcription shut-off experiments (see Materials and Methods). All the values represented as a fold over the SPT6 non-depleted control. The average and standard deviation of three biological replicates are shown. See Supplementary Figure S10 for the spt6–140/SPT6 ratios under the Arb1-depleted and control conditions.
RESULTS
Ribosome assembly mutants display synthetic interactions with spt6-140
In order to study the functional implications of chromatin dynamics during transcription elongation, we screened for yeast mutations that cause lethality in combination with spt6–140, a thermosensitive allele of this chromatin factor that confers minor transcriptional defects at a permissive temperature (38). Fifteen bona fide candidates distributed in nine complementation groups were isolated (details in Materials and Methods, and Supplementary Figure S1). Seven genes have been identified to date; FUR1, SPT4, BUR2, SPN1/IWS1, PSA1, ARB1 and DBP6 (Table 1). FUR1 has been previously reported to be a common false-positive of the adopted screening strategy (39). SPT4, BUR2 and SPN1/IWS1 encode well-known transcriptional machinery elements. Spt4 is one of the subunits of elongation factor Spt4/Spt5 (9) that is homologous to metazoan DSIF (40) and is a well-known synthetic lethal of SPT6 (41). Bur2 is the regulatory subunit of Bur1/Bur2 cyclin-kinase (42), which is involved in the phosphorylation of Spt4-Spt5 and the elongating form of RNA pol II (43). Spn1/Iws1 interacts physically with Spt6 (44) and has been described to participate in the regulation of chromatin dynamics at the CYC1 promoter (45).
Table 1. Synthetic lethal mutants isolated in the screen.
| Complementation group | Growth | Mutated gene | ||
|---|---|---|---|---|
| 30°C | 37°C | 16°C | ||
| ssl1 | + | + | + | FUR1 |
| ssl2 | ssg | ssg | ssg | ARB1 |
| ssl3 | sg | sg | sg | DBP6 |
| ssl4 | sg | sg | – | |
| ssl5 | + | – | + | |
| ssl6 | sg | sg | - | BUR2 |
| ssl7 | sg | – | ssg | SPT4 |
| ssl8 | + | + | + | SPN1/IWS1 |
| ssl9 | ssg | ssg | sg | PSA1 |
Growth phenotypes are indicated. + Wild-type growth. sg slow growth, ssg severe slow growth, – lethality. The gene mutated in each complementation group is indicated. Our attempts to identify the genes that corresponded to ssl4 and ssl5 were unsuccessful. For numerical details of the screening, please check the Materials and Methods and Supplementary Figure S1C.
The above-described genetic interactors of Spt6 were either known or predictable according to the chromatin-related role of Spt6 in RNA pol II-dependent transcription. The other three genes detected in our screening did not encode transcriptional machinery components. PSA1 encodes the GDP-mannose pyrophosphorylase, an enzyme involved in fungal cell wall biosynthesis (46), which is particularly important for cell growth and septum formation after mitosis (47). DBP6 encodes a putative ATP-dependent RNA helicase of the DEAD-box family that participates in the early 60S ribosomal subunits assembly steps (32,33). ARB1 encodes an ABC-type factor involved in several aspects of the biogenesis of both the 40S and 60S ribosomal subunits (48). Since we found that two genes were involved in ribosome biosynthesis (DBP6 and ARB1), and the latter was isolated in two independent rounds of our screening, we decided to explore this genetic connection.
We firstly checked whether these two genes could play a role in RNA pol II-dependent transcription, as the other synthetic lethal genes do, but we did not find any evidence for this. For instance, we detect no recruitment of Arb1 to transcribed chromatin (Supplementary Figure S2A), no mRNA biogenesis defect (GLAM assay (49)) in the arb1 mutant (Supplementary Figure S2B), nor any sensitivity to NTP-depleting drugs (Supplementary Figure S2C). Instead we found mild resistance, similarly to what we previously described for other mutants that impair ribosome biogenesis (50).
Then we identified the mutant alleles of dbp6 and arb1. Both carried non-sense mutations (c.605C>A and c.1215delC, respectively). These new alleles, dbp6–100 and arb1–100, likely caused the partial lack of function of these two essential proteins. To confirm that this was indeed the case, we analysed their impact on ribosome biogenesis. We firstly performed a polysome profile analysis of these mutants in a wild-type SPT6 background. In agreement with previous reports (33,51), the dbp6–100 allele lowered the free 60S/40S ratio and led to half-mer polysomes to accumulate, and thus to a net deficit in the 60S ribosomal subunits (Figure 1A). The analysis of the steady-state levels of the pre- and mature rRNAs species in dbp6–100 indicated a slight delay in 35S pre-rRNA processing, as revealed by the reduction in pre-rRNAs 32S, 27SA2 and 20S, and a significant drop in the levels of both 27SB and 7S pre-rRNAs (Figure 1B).
Figure 1.
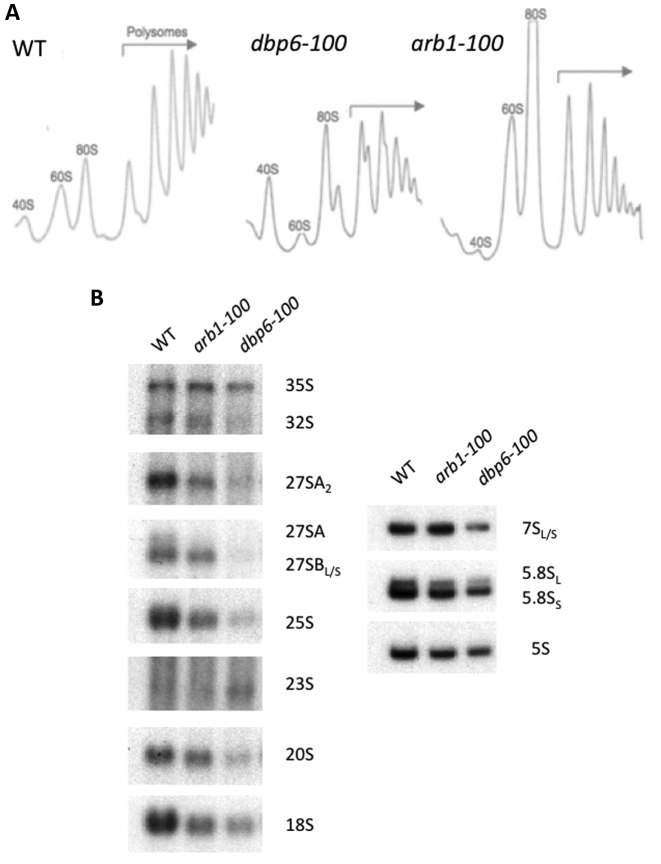
The spt6–140 synthetic lethal mutations in DBP6 and ARB1 affect ribosome biogenesis. (A) Polyribosome profiles. Strains were grown in YPD at 30°C and harvested at an OD600 of 0.8. Cell extracts were prepared and 10 A260 of each extract were resolved in 7–50% sucrose gradients. A254 was continuously measured. Sedimentation is from left to right. The peaks of free 40S and 60S ribosomal subunits, the 80S free couples/monosomes and polysomes, are indicated. (B) Northern analysis of the high-molecular-mass (left) and low-molecular-mass (right) pre-rRNAs and mature rRNAs. Equal total RNA quantities were loaded in gels.
Depletion of Arb1 has been reported to lead to a deficit of 40S ribosomal subunits (48); consistently, the polysome profile analysis of arb1–100 revealed a clear deficit of the 40S ribosomal subunits, as evidenced by an increase in the peak of the free 60S ribosomal subunits (Figure 1A). The Northern analysis showed that the arb1–100 allele also delayed 35S pre-rRNA processing, but had no impact on the other pre-rRNA precursors (Figure 1B). All these results are fully consistent with the published roles of Dbp6 and Arb1 (33,48,51).
In addition to dbp6 and arb1, a third gene in our screening seemed to be related to ribosome biogenesis. The thermo-sensitive mutation that defined the ssl5 complementation group, which has so far escaped our identification attempts, also exhibited a deficit of 40S subunits by a polysome profile analysis. This phenotype was enhanced at 37°C (Supplementary Figure S3A). We also found lowered levels of pre-rRNAs 32S, 27SA2, 27SB and 20S after transferring the ssl5 mutant cells at 37°C for 2 h. The aberrant 23S pre-rRNA (31) was seen to accumulate (Supplementary Figure S3B). These results indicated that the ssl5 mutant was impaired in ribosome biogenesis, and more specifically in early pre-rRNA processing steps.
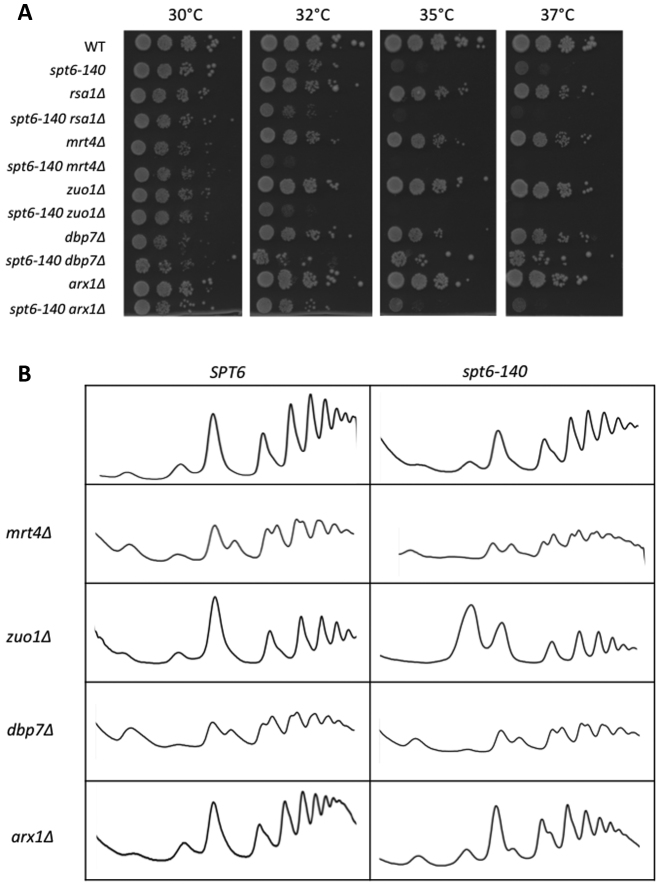
In order to check whether this synthetic genetic interaction between SPT6 and these three genes connected to ribosome biogenesis reflected a general functional link of Spt6 to the factors involved in this process, we combined spt6–140 and a set of viable deletions of the genes involved in different ribosome assembly steps (RSA1, MRT4, ZUO1, DBP7 and ARX1). Our screening revealed that four of these double mutants exhibited increased thermosensitivity (Figure 2A). A subpopulation of the double dbp7Δ spt6–140 mutant, which contained a duplication of chromosome VIII, was reproducibly able to grow at restrictive temperatures for spt6–140 (Figure 2A), which suggests that the strong synthetic phenotype of spt6–140 dbp7Δ can be suppressed by the overexpression of one of the genes, or more, located in chromosome VIII (see below). The polysome profiles of some double mutants were also analysed and confirmed these genetic interactions. The double arx1Δ spt6–140 mutant merely showed a moderately aggravated polysome profile, which is in line with the mild synergy that the two mutations produced in the thermosensitivity phenotype (Figure 2A and B). The combination of mrt4Δ, zuo1Δ and dbp7Δ, with spt6–140 showed worsened profiles compared with the single counterparts (Figure 2B). One particularly striking case was zuo1Δ, which, as a single mutant, showed a mild 60S defect, and exhibited a marked accumulation of the free 60S particles in combination with spt6–140 (Figure 2B).
Figure 2.
Spt6 is functionally linked to ribosome biogenesis. (A) Growth assay of the double mutants that combined spt6–140 and five different deletions of the genes involved in ribosome biogenesis. Isogenic wild-type and single mutants are also shown. Serial dilutions of the cultures exponentially grown at 30°C were spotted on YPD plates and incubated for 2 days at the indicated temperatures. (B) Polyribosome profiling of spt6–140, an isogenic wild type, mrt4Δ, zuo1Δ and dbp7Δ and the double mutants of these deletions in combination with spt6–140. Cells were grown at 30°C. Details are as mentioned in Figure 1A.
Taken altogether, the above-described results indicate a broad set of functional connections between SPT6 and the genes that encode the factors that facilitate ribosome biogenesis.
The functional connection of SPT6 to ribosome assembly genes is specific and not directly related to rDNA transcription
The functional connection of Spt6 to ribosome biogenesis might be explained through a direct role of Spt6 in rDNA transcription. It has been described that Spt6 binds rDNA and that the thermal inactivation (39°C) of the spt6–1004 thermosensitive allele almost abolishes rRNA synthesis (22). Although our screening was done with a different spt6 allele (spt6–140) and under non-restrictive thermal conditions (30°C) we decided to analyse its impact in rRNA synthesis. We performed pulse-chase labeling experiments to investigate this. As a result, we did not detect any deficit in rRNA labeling in spt6–140. Similar levels of labelled rRNA precursors were found in the wt and spt6–140 cells, including 35S pre-rRNA (see Figure 3A, time 0), which indicated no significant defect in overall rDNA transcription. Chase times suggested a minimum delay in 27S maturation to 25S rRNA (Figure 3A). This delay was consistent with the slight accumulation of the 27SA2 pre-rRNA that we observed when steady-state levels of pre-rRNAs were analysed by northern hybridization (Figure 3B). In agreement with this very weak processing phenotype, the single spt6–140 mutant exhibited a rather wild-type polysome profile, with very subtle half-mer polysomes (Figure 2B). Therefore, we concluded that the spt6–140 mutation did not lead to a significant defect in rRNA synthesis at this permissive temperature, and that its synthetic interactions with ribosome biogenesis genes were likely the result of defective ribosome assembly rather than impaired rDNA transcription.
Next we analyzed if transcription factors that participate in rDNA transcription elongation generated a similar synthetic phenotype than those observed with spt6–140. The complex formed by Spt4 and Spt5, which is homologous to the mammalian DSIF factor, plays a role in RNA pol I-dependent transcription and influences the coupling of rDNA transcription to ribosome assembly (52,53). We constructed double mutants by combining spt4Δ with the set of ribosome biogenesis genes that were shown to interact synthetically with spt6–140, including the alleles found in the synthetic lethal screening. None of the ribosome biogenesis factors that genetically interacted with spt6–140 displayed a synthetic phenotype when combined with spt4Δ, except for dbp6–100, which showed a very mild synthetic enhancement phenotype (Supplementary Figure S4). Accordingly to our previous results, the synthetic interaction between spt6–140 and the ribosome assembly genes are not simply the consequence of interfering with rDNA transcription.
We also investigated if lethality was simply caused by the simultaneous impairment of the two sequential processes that enable protein-coding gene expression: RNA pol II-dependent transcription, which is weakened by spt6–140, and mRNA translation, indirectly impaired by the mutation of ribosome assembly factors. We checked the sensitivity of spt6–140 and the double mutants described above to cycloheximide and anisomycin, two translation-inhibiting drugs. spt6–140 was resistant to both drugs and did not increase the sensitivity of the other single mutations. With rsa1Δ and dbp7Δ, the effect of spt6–140 was even suppressive, since their sensitivity to anisomycin was reduced (Supplementary Figure S5). We concluded that the genetic interaction of SPT6 to the ribosome biogenesis genes was not just the additive consequences of impaired transcription and translation.
Altogether, these results suggest a specific relationship between SPT6 and the genes that encode ribosome assembly factors, and situate this functional interaction outside the rDNA transcription context.
Transcription elongation of ribosome-related genes is altered in response to Arb1 depletion
The above results suggest that in addition to the direct presence of Spt6 during rDNA transcription (22), SPT6 may favor ribosome assembly by contributing to the function of the genes that encode ribosome assembly factors. Since Spt6 is a well-known chromatin factor during transcription elongation, we wondered whether the malfunction of Spt6 might hinder the correct expression and regulation of ribosome assembly genes. We previously described that the RiBi and RP regulons are specially regulated at the transcription elongation level (2,34,50). After considering this, we hypothesized that the transcription of the ribosome-related genes would be regulated in response to ribosome biogenesis impairment, and such regulation would be positively influenced by Spt6.
In order to test the first component of this hypothesis, we investigated the transcriptional consequences of ribosome assembly impairment. We set up an experimental system based on the depletion of Arb1. Given the strong transcriptional response of ribosome-related genes to any kind of stress (54,55), we decided to avoid those depletion systems that involve a sudden growth stop. Instead we placed the ARB1 gene under the control of a repressible Tet-off promoter. By doing so, we managed to achieve the progressive depletion of Arb1 after switching the promoter off by addition of doxycyclin (Supplementary Figure S6B). As expected, in the presence of sublethal doses of doxycycline, the TET::ARB1 allele conferred a synthetic growth phenotype in combination with spt6–140, which recapitulated the genetic interaction between ARB1 and SPT6 (Supplementary Figure S6A). In the absence of doxycycline the levels of Arb1 became slightly higher than in the non-engineered wild-type strain (Supplementary Figure S6B, time 0h). So after 4 h of doxycycline, wild-type levels of Arb1 protein, free ribosomal subunits and pre-rRNA precursors were still observed (Supplementary Figure S6B, D and E). After 8h, these levels had significantly lowered (Supplementary Figure S6B) without producing a significant effect on growth yet (Supplementary Figure S6C). At this time, cells started to accumulate free 60S ribosomal subunits (Supplementary Figure S6D) and abnormal levels of 35S pre-rRNA, which were followed by additional rRNA processing alterations at longer times (Supplementary Figure S6E). So, we decided to compare the transcriptional state of the genome in the samples treated with doxycyclin for 4 h (control) and 8 h (Arb1-depleted). We considered that under these mild conditions this system would be optimal for exploring the direct consequences of ribosome biogenesis impairment on genome-wide gene transcription, without the indirect effects produced by growth impairment.
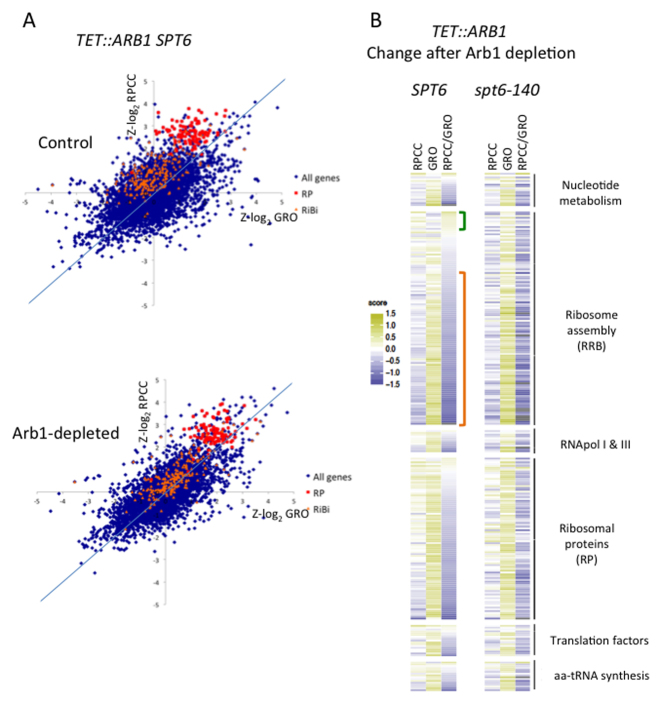
We firstly detected all the RNA pol II forms present in the genes by performing RNA pol II ChIP-on-chip (RPCC). Secondly using the same type of arrays, we detected transcriptionally active RNA polymerases by performing genomic run-on (GRO) assays (Supplementary Figure S7). This double strategy provides information about the amount of RNA pol II recruited to genes and the transcriptional activity of these recruited polymerases (2). As shown in a previous study (2), the genes that encode ribosomal proteins (RP) displayed very high levels of RNA pol II (RPCC) upon them compared with the rest of the genome, whereas the RiBi regulon genes exhibited intermediate levels (Figure 4A, top panel, Y-axis). These relative levels of RNA pol II did not increase (RP) and only slightly lowered (RiBi) when Arb1 was depleted (Figure 4A, compare the two panels, Y-axes). In contrast, we found an overall increase in the relative levels of transcriptionally active RNA pol II (GRO signals) in the RP and RiBi genes upon Arb1 depletion (Figure 4A, compare the two panels, X-axes). Changes in the RPCC and GRO signals after Arb1 depletion relocated the RP and RiBi genes in relation to the rest of the genome. Under the control conditions, both groups of genes occupied a biased position in the plot as they exhibited lower GRO levels than those expected for their RPCC (Figure 4A, top panel). After Arb1 depletion, both groups, and particularly the RiBi genes, moved to a more central location in the global genome context (Figure 4A, compare the two panels). We previously demonstrated that the RPCC/GRO ratios were indicative of the level of RNA pol II backtracking during transcription elongation (2,34,50). As a result of backtracking, the 3’ end of nascent RNA relocates out of the active site (56) and impedes backtracked RNA pol II to undergo a run-on reaction (57). This overall reduction in RNA pol II backtracking indicated that Arb1-depletion had provoked the activation of transcription elongation in ribosome-related genes.
Figure 4.
Arb1 depletion induces transcriptional elongation changes in the ribosome-related genes. (A) Comparison made between the standardised total RNA pol II occupancy (RPCC) and transcriptionally active RNA pol II (GRO) values (see the Materials and Methods) during Arb1 depletion. Arb1-depletion experiments were performed under the conditions defined in Supplementary Figure S6 using the arb1Δ TEToff::ARB1::MYC SPT6 strain. The samples taken before and after effective Arb1 depletion were compared. All the genes (blue), the RP regulon (red) and the RiBi regulon (orange) are indicated. (B) Heat map of the RPCC and GRO change during depletion for the RP genes and for the different subgroups present in the RiBi regulon. Fold-change of the z-scores (Arb1-depleted versus control) is indicated by the color bar as log2 values. Data are from the experiments described in A (arb1Δ SPT6 TEToff::ARB1::MYC strain) and in Supplementary Figure S8C (arb1Δ TEToff::ARB1::MYC spt6–140 strain). Genes were ranked according to the RPCC/GRO ratio variation in SPT6 to visualize the impact of spt6–140.
In order to check how general this phenomenon was, ribosome-related genes were distributed in functional categories (nucleotide metabolism, ribosome assembly, RNA pol I and III subunits, ribosomal proteins, translation factors and genes related to aminoacyl-tRNA synthesis) and ranked according to their RPCC/GRO ratio after Arb1 depletion. While the GRO signals of most genes in all the subsets increased, we found a more variable tendency in RPCC (Figure 4B).
When we focused on those genes directly involved in ribosome assembly (the more restricted RRB regulon defined by Wade et al. (26); see the Discussion) we found that changes in GRO and RPCC were reciprocal. In most of them increased GRO values were in parallel to lower RPCC signals (Figure 4B, red bracket). Consequently, their RPCC/GRO ratios predominantly lowered after Arb1 depletion (Figure 4B; Supplementary Figure S8A). In a few RBB genes with lowered GRO values, RPCC signals stay unchanged or even increased, which resulted in higher RPCC/GRO ratios (Figure 4B, green bracket). These reciprocal changes in the RPCC and GRO signals reinforced our conclusion of RNA pol II elongation regulation. Less RNA pol II backtracking should involve a higher proportion of active RNA pol II (higher GRO) and a shorter residence time (lower RPCC), and vice versa. This reciprocal behavior of the GRO and RPCC signals was less evident in the other categories (Figure 4B). In fact the overall change in the RPCC/GRO ratios after Arb1 depletion was weaker in the RP regulon than in the RBB genes (Supplementary Figure S8A-B). We concluded that the transcription elongation of RBB genes was particularly responsive to Arb1 depletion.
Spt6 recruitment to ribosome biogenesis genes after Arb1 depletion favours the productive synthesis of stable mRNAs
We also performed RPCC and GRO in spt6–140 cells upon depletion of Arb1. Under the control condition, we found the same pattern as in the wild type; most ribosome-related genes exhibited more total RNA pol II than expected from their GRO values (Supplementary Figure S8C left panel). However, upon Arb1 depletion we also detected a global increase in the relative levels of the GRO signals of all the ribosome-related genes, and a drop in the RPCC levels of many RiBi genes (Figure 4B and Supplementary Figure S8C). Moreover, the RBB genes with decreasing RPCC levels and increasing GRO signals were significantly more abundant in spt6–140 than in the wild type (Figure 4B and Supplementary Figure S9). These results indicate that spt6–140 does not impair the activation of transcription elongation in the RBB genes provoked by Arb1 depletion, and suggest that Spt6 is not an integral mediator of the feedback mechanism.
Spt6 is a chromatin factor that is recruited to the transcribed genes through its physical interaction with RNA pol II during transcription elongation. To test if Spt6 was recruited to responsive genes upon Arb1 depletion, we analysed five RiBi genes that exhibited an increased GRO signal. We found enhanced Spt6 binding to the gene body of four of them (all except UTP9) (Figure 5A). This increased binding of Spt6 after Arb1 depletion was consistent with a potential role of this factor in the up-regulated genes after their transcriptional activation. We then analysed the functional consequences of spt6–140 in the same set of genes by studying histone H3 and RNA pol II occupancy. We found that spt6–140 significantly decreased H3 occupancy after Arb1 depletion in the same four gene bodies that became enriched in Spt6 binding (Figure 5B). No significant impact of spt6–140 was observed in the occupancy of these genes by RNA pol II after Arb1 depletion (Figure 5C), which suggests that histone H3 changes were not due to altered RNA pol II levels. In contrast, we found a significant deficit in the mRNA levels after Arb1 depletion in spt6–140 in the same four genes (once again, all except UTP9) (Figure 6A, Supplementary Figure S10A). Since the GRO analysis showed no reduced transcription of these genes in spt6–140, lack of mRNA induction should be due to unproductive transcription (see the Discussion).
It has been recently described that ribosome-related genes are co-transcriptionally imprinted in response to transcriptional elongation stress, which provokes increased mRNA translatability and degradation (58). We wondered whether the observed deficit in the mRNA levels after Arb1 depletion caused by spt6–140 might result from an alteration of the mRNA half-lives of the target genes. We measured the mRNA stability of the five genes analysed above under the control and Arb1-depleted conditions in SPT6 and spt6–140, by a transcription shut-off assay (Supplementary Figure S10B). A comparison of the mRNA levels and half-lives in the SPT6 background showed that increased mRNA stability contributed to mRNA induction in response to Arb1-depletion. However, mRNA stabilization alone cannot explain all the mRNA changes detected (Figure 6A and B). For instance, increased expression of RSA4 and RRP12 was observed in the SPT6 background upon Arb1-depletion with no parallel change in mRNA half-life (Figure 6A–B).
When we focused in spt6–140, we detected changes in mRNA stability in the four genes targeted by Spt6 (all except UTP9 once more; Figure 6B). In these genes spt6–140 caused a significant decreased in mRNA stability under both the control and Arb1-depleted conditions (Figure 6B, Supplementary Figure S10C). Similarly, although the mRNA half-lives of all four responsive genes increased in spt6–140 after Arb1-depletion, none was able to reach the induced values observed in the SPT6 background (Figure 6B). Therefore, low mRNA stability is contributing to the limited mRNA levels exhibited by the four responsive genes upon Arb1-depletion in spt6–140 (compare Figure 6A and B). We conclude that the failure of spt6–140 to properly induce the RiBi genes in response to Arb1-depletion should be a combination of aberrant transcription and reduced mRNA stability.
Altogether, the results shown in Figures 5 and 6 indicate that the recruitment of Spt6 during the transcriptional activation provoked by Arb1 depletion contributes to an optimal chromatin dynamics in the target genes and ensures the synthesis of stable mRNAs from them.
The mutational impairment of ribosome biogenesis produces mutant-specific transcriptomic responses
The transcriptional changes of the ribosome-related genes in response to Arb1 depletion suggested the existence of specific regulatory pathways that operated when ribosome biogenesis was impaired. To investigate the validity and extension of this hypothesis, we performed a detailed dissection of the transcriptomes of the ribosome-related genes in response to a wide set of mutants defective in the ribosome biogenesis pathway. We analysed 59 null mutants that lacked global regulators of ribosome biogenesis, structural ribosome components (RP), RNA pol I subunits, and factors involved in pre-rRNA processing and ribosome assembly. We checked the changes in the expression of the RP and the different subsets of the RiBi regulons in all these deletions (see the Materials and Methods). Thirty-seven of the 59 mutants showed changes in three genes or more and were selected for further analyses.
Half the mutants induced the down-regulation of most genes (Supplementary Figure S11, right lanes), while the other half induced a complete or partial up-regulation (Supplementary Figure S11, left lanes). This indicates the existence of more than one alternative transcriptomic response to ribosome assembly impairment. sfp1Δ and sch9Δ, two slow-growth and small-cell mutants that lack the TORC1-dependent positive regulators of regulons RP and RiBi (25,59–61), clustered together and exhibited a general repression of all the ribosome-related genes (Supplementary Figure S11). In contrast, the mutant that lacked Hmo1, a high mobility group protein involved in the coordination between RP and rDNA transcription (62,63), exhibited a general up-regulation (Supplementary Figure S11). The transcriptomic patterns of these three regulators were fully expected, according to their well-known roles, and they validated this analysis.
When we examined the hierarchical clustering of the other analysed mutants, deletions of structural ribosomal proteins formed the best-defined cluster (Supplementary Figure S11). These mutants up-regulated a large subset of the genes that encode ribosome assembly factors (RBB genes), without affecting the RP genes themselves (Supplementary Figure S11). Four other analysed mutants that clustered together, rpa14Δ, tom1Δ, utp30Δ and rsa1Δ, showed an even more general up-regulation of the RBB group (Supplementary Figure S11). No clear functional connection has been established among these four mutants, which respectively lacked an RNA pol I subunit, a ubiquitin ligase involved in pre-40S assembly, a 90S-acting factor and a pre-60S factor related to snoRNA stability.
We found that some specific deletions changed the expression of a very specific subset of ribosome assembly factors; e.g., only some exhibited an increased expression in dbp7Δ, while the rest were mostly unaffected or repressed (Supplementary Figure S11). These up-regulated genes encode factors that act upon the assembly of large ribosomal subunits, and some maintain a direct functional relation with Dbp7 (32). A different subset of ribosome assembly factors was up-regulated in zuo1Δ (Supplementary Figure S11). Other mutations, like tsr2Δ and ltv1Δ, provoked the induction of RP genes and caused neutral, or even negative, effects on the assembly category (Supplementary Figure S11). These results reveal the existence of regulatory transcriptomic responses to ribosome biogenesis impairment that are highly mutant-specific.
The transcriptomic responses to ribosome biogenesis perturbation uncover an internal regulatory structure within RBB genes
The hierarchical clustering of the gene expression changes caused by the above-mentioned deletions showed that most of the genes directly involved in ribosome assembly (the RRB regulon; see the Discussion) segregated apart from the RP genes (Supplementary Figure S11). Save the RNA pol I and III subunits, which were grouped mostly with the assembly factors, the other functional categories present in the RiBi regulon also segregated apart. For instance, most of the genes that encode aminoacyl-tRNA synthetases and nucleotide metabolism enzymes correlated as a group and displayed a clearly different behavior compared to the RBB genes. While the latter were repressed in some mutants and were up-regulated in others, the expression of most nucleotide metabolism, aminoacyl-tRNA synthetases and translation factors was repressed in almost all the tested deletions (Supplementary Figure S11). We detected no independent clustering between the large subunit and the small subunit In the RP genes (Supplementary Figure S11). These results suggest that the large and functionally complex RiBi regulon, originally defined by its co-regulation under nutritional and environmental conditions (25), is less consistent than the more specific RRB regulon in its response to ribosome assembly impairment.
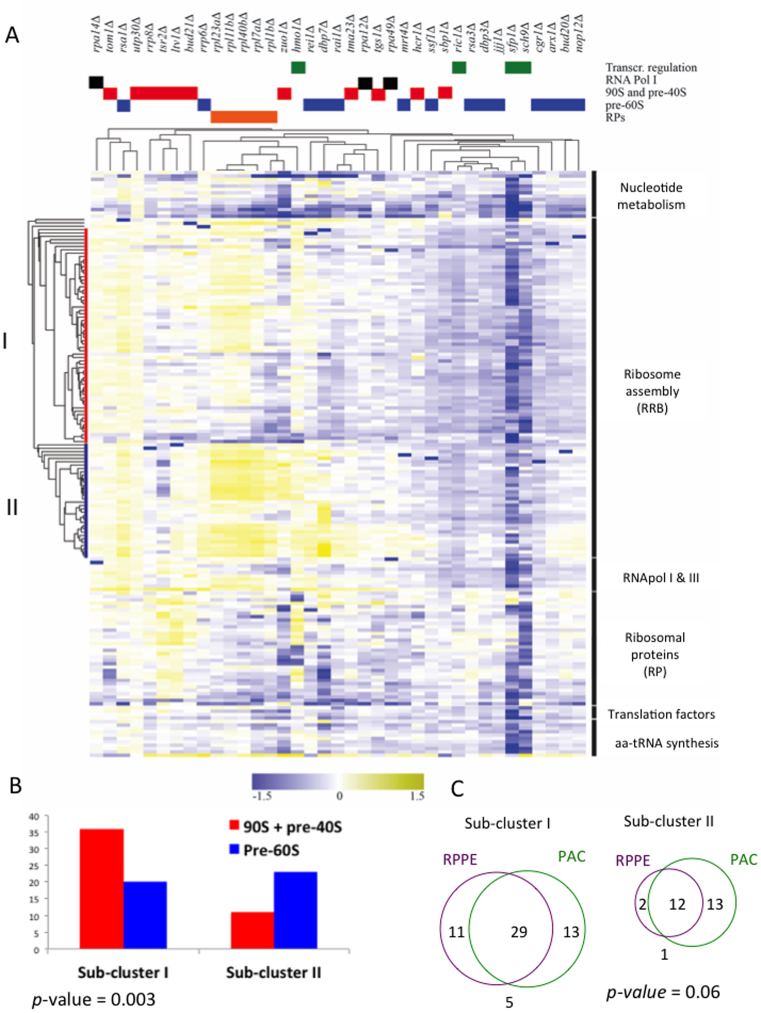
To further clarify this observation, we repeated the clustering analysis of each RiBi subgroup independently (Figure 7A). The differential transcriptomic patterns of the mutants displayed RBB genes in four subclusters (see the vertical gene tree in Figure 7A). We analysed the composition of the two subclusters that comprised more than two genes. Subcluster I (Figure 7A, in red on the left) was significantly enriched in the factors involved in early ribosome assembly steps (90S factors) or in the factors related to the maturation of the small ribosomal subunit (pre-40S factors) (Figure 7B and Supplementary Figure S12A). Subcluster II (Figure 7A, in blue on the left) was enriched in the factors specifically involved in the assembly and maturation of the large subunit (pre-60S factors) (Figure 7B and Supplementary Figure S12A). This difference revealed the existence of a regulatory structure within ribosome assembly genes, which was related to their functional roles. The genes in Subcluster II were specifically up-regulated by some deletions (rrp6Δ, rpl7AΔ, rpl1BΔ, zuo1Δ, dbp7Δ and rei1Δ), although their induction patterns were not identical (Figure 7A). In contrast, the genes of Subcluster I were negatively affected by these deletions and were never differentially induced from Subcluster II. In those mutants where Subcluster I was up-regulated (as in rpa14Δ, tom1Δ, rsa1Δ and utp30Δ), the expression of Subcluster II also increased (Figure 7A). This suggests the existence of a general control of ribosome assembly factors capable of enhancing the expression of the two subclusters in parallel, and a second switch capable of specifically regulating the genes within Subcluster II, which would mainly cause a specific induction of the factors involved in pre-60S processing. We considered it meaningful that most of the mutations that specifically induced the genes of Subcluster II (rrp6Δ, rpl7AΔ, rpl1BΔ, dbp7Δ and rei1Δ) were linked to the assembly of the 60S subunit. We realised that Subcluster II was enriched in the genes located in chromosome VIII (10 out of 34). This would explain the tendency to duplicate this chromosome shown by dbp7Δ, especially in the context of spt6–140 (see above). However, the enrichment of cluster II in the pre-60S factors persisted after removing the ten chromosome VIII-located genes from the analysis (Supplementary Figure S12B).
Figure 7.
The impairment of ribosome biogenesis changes the general expression pattern of the ribosome-related genes. (A) Hierarchical clustering of the changes in the mRNA levels upon single gene deletions (top list) on the significantly changing genes in the different functional categories of ribosome-related genes (right). The RRB regulon, split into four subclusters, is represented on the left (Subcluster I in red and Subcluster II in blue). The fold change of the mRNA expression in the mutant versus the corresponding wild type is indicated by the colour bar as log2 values. (B) Distribution of pre-60S and the combination of the 90S and pre-40S factors between the two main subclusters of RBB genes. (C) Venn diagrams showing the balanced (Subcluster I) and biased (Subcluster II) presence of PAC and RRPE elements in the two main subclusters within the RBB group. P-values correspond to the Chi-square test.
Two regulatory sequences have been described as being usually present in the promoter regions of RBB genes: the PAC and RRPE elements (26,64) bound by transcriptional repressors Dot6 and Tod6, in the former (65), and by Stb3 in the latter (66). We analysed the distribution of the PAC and RRPE elements in the genes of Subclusters I and II to find that the RRPE element was less frequently present in Subcluster II than in I (Figure 7C). This bias was also conserved after removing the chromosome VIII-located genes from Subcluster II (Supplementary Figure S12C). In contrast, the RRPE element was not significantly less abundant in the genes that encoded pre-60S-specific factors than in those involved in 90S and pre-40S maturation (Supplementary Figure S12D). This suggests that the lower frequency of the RRPE elements in Subcluster II is not the indirect result of the enrichment of this subcluster in the pre-60S factors. In Subcluster II, the RRPE elements were particularly scarce in those genes located in the bottom half (Supplementary Figure S12A). Interestingly, these genes were most frequently up-regulated by the tested deletion (Figure 7A). We concluded that an internal regulatory structure exists in the group of genes that encoded ribosome assembly factors, which is related to the function of each gene in the assembly process. This regulation would consist in several components: a general control of all the assembly factors, including the 90S, pre-40S and pre-60S factors, and additional mechanisms that control specific subsets of genes, especially one that is enriched in the genes that lacked RRPE elements and preferentially encoded pre-60S factors.
spt6-140 impairs the feedback up-regulation of ribosome assembly genes
We showed above that spt6–140 did not interfere with the transcriptional activation provoked by the depletion of Arb1 (Figure 4B), although it destabilized the chromatin of some activated ribosome assembly genes and impaired the productive biosynthesis of stable mRNAs from them (Figures 5C and 6A). We also demonstrated that other mutational perturbations often caused a feedback up-regulation of the mRNA levels of the RBB genes (Figure 7A). In order to clarify how extensive the contribution of Spt6 was to the feedback regulation of ribosome biogenesis, we analysed the mRNA transcriptome of spt6–140. We found that this mutant has mild overall effects on the mRNA levels of some genes related to mating, cell wall, oxidative stress and chemical stimuli. This was consistent with the known phenotypes of spt6 mutants, like loss of invasive growth and increased heat sensitivity (67,68). This transcriptomic pattern, compared to the database of chromatin-related mutants (69), grouped spt6–140 together with bre1Δ, rtf1Δ, lge1Δ, bur2Δ, ctr9Δ, paf1Δ and rad6Δ, which are all related to transcription elongation and co-transcriptional histone modification (70) (Supplementary Figure S13).
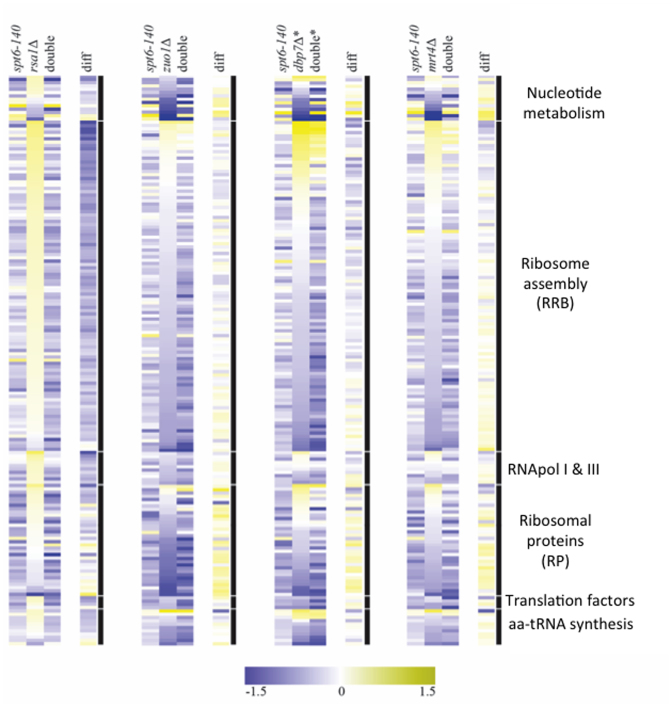
We then analysed the effect of spt6–140 on the transcriptomic profiles generated by four representative mutants (Figure 8). We showed above that rsa1Δ produced a general increase in the expression of the RBB genes (Figures 7A and 8). This up-regulation was completely abolished by spt6–140. The double mutant exhibited almost the same profile as the single spt6–140, which had a mild negative effect on the expression of all the ribosome-related genes (Figure 8, first panel). The difference (dif) between the transcriptomic pattern of the double mutant (double) and the expected additive combination of the two single mutants suggested a very strong epistasis of spt6–140 over rsa1Δ (Figure 8, first panel).
Figure 8.
spt6–140 modifies the transcriptomic profiles of ribosome assembly mutants. Heat maps of the changes in the mRNA levels in spt6–140, ribosome biogenesis mutants and double mutants are shown. The difference between the observed changes in the double mutant (with respect to the wild type) and the additive combination of the single mutants (diff) is also presented. Fold change of the mRNA expression in the mutant versus the corresponding wild type is indicated by the color bar as log2 values. Genes are grouped by functional categories and are ordered by the relative expression changes upon the ribosome biogenesis mutant. An asterisk (*) in the two strains that contain dbp7Δ signifies chromosome VIII duplication due to the marked tendency of these strains to accumulate this chromosome anomaly. The chromosome VIII duplication effect was removed from the diff column as it is present in both the dbp7Δ and spt6–140 dbp7Δ strains.
The epistasis of spt6–140 over dbp7Δ and mrt4Δ was also clear but, in this case, the double mutants retained some induced expression of those RBB genes which markedly up-regulated in the single deletions (Figure 8, third and fourth panels). This epistasis is consistent with the enhancement of the polysome profile defects of the single dbp7Δ and mrt4Δ deletions when combined with spt6–140 (Figure 3B). With zuo1Δ, the profile of the double mutant was more additive than epistatic (Figure 8, second panel). This is consistent with the drastic change in the polysome profile observed from the single zuo1Δ mutant to the double spt6–140 zuo1Δ (Figure 3B).
These relevant effects of spt6–140 on the transcriptomic phenotype of the four tested mutants indicate a substantial contribution of Spt6 to ribosome biogenesis regulation, especially in those cases that involve enhanced expression. In combination with the Arb1-depletion experiments shown above, these results also suggest that regulation of transcription elongation and its associated chromatin dynamics are relevant for the feedback control of ribosome biogenesis.
DISCUSSION
Screening synthetic lethal mutations for spt6–140 allowed us to dissect the network of the functional relationships of SPT6 within the yeast genome. A subset of the identified genes confirmed the cooperation of Spt6 with two other transcriptional machinery elements, which have already been described: Spt4-Spt5/DSIF and Spn1/Iws1. In both cases, the physical and genetic interactions with Spt6 have been previously reported (41,44,71,72). We also found Bur2, one of the BUR kinase components involved in the recruitment of Spt6 to RNA pol II CTD upon transcription initiation (73). This functional relationship between SPT6 and BUR2 was also confirmed by the comparison of the spt6–140 and bur2Δ transcriptomes and their highest overall correlation (Supplementary Figure S13). The fact that these genes appeared throughout our genetic screening was a quality sign.
The genetic interactions between non-essential cell components have been extensively mapped (74,75). However, SPT6 have not been systematically analysed. Hence some relevant aspects of its functional role may remain unknown. The unexpected isolation of four synthetic mutants with clear defects in ribosome biogenesis is a good example. Four additional deletions in different ribosome assembly factors also exhibited synthetic phenotypes when combined with spt6–140 (Figure 2). All these data indicate a functional connection between Spt6 and ribosome biogenesis. Spt6 has been shown to bind rDNA and its thermal inactivation provokes the severe inhibition of rRNA synthesis (22). Two-hybrid evidence has also suggested a direct contact between Spt6 and RNA pol I (76). According to thse reports, our results might be explained as an indirect result of the contribution of Spt6 to rDNA transcription. However, we did not find any evidence for suboptimal rRNA transcription in spt6–140 at 30°C, which was the permissive temperature that we used for our screening and the subsequent genetic interaction analyses (Figure 3A), nor did the rDNA transcription factor Spt4/Spt5 show the extensive set of synthetic interactions with the ribosome assembly factors that we detected for Spt6 (Supplementary Figure S4). Moreover, the subtle variation in the levels of the rRNA precursors shown by spt6–140 seems the consequence of the posttranscriptional defects that are usually associated with the impairment of the assembly process (e.g. (51)). We think that all these results indicate that, in addition to its proposed function in rDNA transcription, Spt6 must contributes to ribosome assembly in a second manner, likely related to its role in the RNA pol II-dependent transcription context.
Feedback regulation in the ribosome assembly gene network
It has been extensively demonstrated that Spt6 facilitates chromatin dynamics during transcription by RNA pol II (6,7,9,73). Therefore, in order to find an alternative scenario for a contribution of Spt6 to ribosome biogenesis we focused on those genes protein-coding genes that are directly related to ribosome biogenesis. We hypothesised that Spt6 would play a role in the context of RiBi gene regulation. This role would be particularly important according to the regulatory changes in RiBi expression that the malfunction of the ribosome assembly process might cause. This hypothesis assumed that Spt6 played an important function in the regulated expression of RiBi genes, but did not necessarily involve the role of Spt6 as a mediator of the regulatory signal itself (see later).
Two independent sets of experiments confirmed that RiBi genes undergo extensive regulation upon ribosome biogenesis impairment. In the first one we found that the depletion of an essential ribosome assembly factor, Arb1, under the experimental conditions that did not affect growth, provoked the transcriptional activation of ribosome-related genes (Figure 4A–B). In a second set of experiments we measured the transcriptomic pattern of 59 viable deletion mutants, defective for the ribosome biogenesis pathway. Thirty-seven of these deletions exhibited a substantial alteration in the mRNA levels of the ribosome-related genes, and this alteration in half of them involved an up-regulated expression (Figure 7A). In these two different experimental systems we found that the production of increased mRNA levels by the up-regulated RiBi genes was significantly interfered with, and in some cases abolished, by spt6–140 (Figures 6A and 8).
When we compared the transcriptomic patterns of all the analysed mutants we found considerable heterogeneity within the RiBi genes. This variance led to a segregated clustering of the different groups present in this large regulon, which encompasses nucleotide synthesis enzymes, RNA polymerase I and III subunits, and the factors involved in translation, tRNA metabolism and ribosome assembly (Supplementary Figure S11). This complex group of genes displays a homogeneous regulation under nutritional and environmental conditions (25), but is less distinguishable in response to ribosome assembly perturbation. In the present work, the transcriptomic patterns of those genes specifically devoted to ribosome assembly (the RRB regulon defined by (26)) provided more useful information. For instance, the RBB and RP genes displayed reciprocal patterns in several analyzed mutants (compare, e.g. rsa1Δ and tsr2Δ in Figure 7A), while the other RiBi subgroups were never differentially up-regulated.
This independent regulation of RP and RRB in response to specific alterations of ribosome assembly is consistent with their different promoter architecture and the distinct sets of transcription factors that drive their expression (77). Most RP genes are controlled by Rap1, Fhl1, Ifh1 and Hmo1 (30,78,79). In contrast, RRB genes are driven mostly by Stb3, which recognises the RRPE motif (66), and also by Dot6 and Tod6, which bind the PAC sequence element (80). These different constellations of transcription factors involve substantial differences in the mechanisms that regulate the transcription of RP and RRB. For example, both regulons share some important chromatin marks, like H3K4 methylation, when they respond to oxidative stress but differ for others, e.g. histone deacetylation (81).
Taken altogether, our transcriptomic results suggest a model in which the genes that encode the RP and ribosome assembly factors could respond independently to ribosome biogenesis impairment (Supplementary Figure S14). The regulation of the RP regulon in response to ribosome biogenesis is mediated by transcription factor Ifh1 and its capacity to physically interact with some rRNA processing factors in the CURI complex (29). RRB genes also responded globally to some mutational perturbations, as in rpa14Δ, tom1Δ, rsa1Δ and utp30Δ (Figure 7A). Since Ifh1 does not drive the transcription of RRB genes, another mechanism, independent of the CURI complex, should support this feedback control. How this regulation occurs is still unclear and requires the further dissection of global regulators, including those that bind to PAC and RRPE elements.
This global control of RRB genes would co-exist with specific regulations of more restricted subsets of assembly genes, as in dbp7Δ (Figure 7A). In these situations, the control of a set of assembly genes, enriched in pre-60S factors, would uncouple from a second group composed preferentially of 90S and pre-40S factors (Supplementary Figure S14). The clustering analysis helped us to visualise this phenomenon (Figure 7A and B). The differential presence of the RRPE elements in these two groups of genes might facilitate this uncoupling (Figure 7C). Presence of additional regulatory motifs in the promoter regions of some RRB genes could also contribute to the differential regulation of more restricted gene subsets (77).
Control of transcription elongation in the ribosome assembly gene network
The use of a carefully controlled Arb1-depletion system allowed us to show the marked alteration of transcription elongation that the RRB genes underwent as soon as ribosome assembly was impaired (Figure 4A and B). In most RRB genes these changes involved increased transcriptional activity, measured by transcriptional run-on, with no simultaneous increase in the amount of RNA pol II present (Figure 4A). This phenomenon suggests reduced RNA pol II backtracking frequency. These results not only confirmed our previous reports about the regulation of ribosome-related genes at the transcription elongation level (2,34,50), but also reinforced RNA pol II backtracking as a physiologically regulated process (reviewed in (57)). While transcription initiation regulation is the simplest way to switch a gene expression on or off, it is conceivable that backtracking provides a mechanism to fine-tune ribosome biogenesis, a phenomenon that requires the accurate stoichiometry of structural components and the coordinated action of a large number of assembly factors.
At constant initiation rates, decreased backtracking predicts lower RNA pol II occupancy. This is what we observed as a general tendency in the subgroup of RRB genes (Figure 4B). However, in both this subgroup and other ribosome-related genes, we found genes with increased GRO signals and no significant change in RNA pol II occupancy (Figure 4B). This indicates that the observed transcription up-regulation also involves an increase in initiation rates to some extent.
The feedback control of the RRB genes was severely interfered with by spt6–140 (Figure 8). This defective regulation might be an indirect result of the proposed participation of Spt6 in rRNA transcription by RNA pol I (22). However, we detected the recruitment of Spt6 to the bodies of the up-regulated ribosome assembly genes, and a correlation between this recruitment and its positive effect at the mRNA level (Figure 5). These results support a direct role of Spt6 in the expression of ribosome-assembly genes. Moreover, the regulation of ribosome assembly genes at the transcription elongation level well matches the known function of Spt6, which has been physically and functionally linked to this specific RNA pol II-dependent transcription step in several biological systems (6–10,73).
This positive contribution of Spt6 to the regulation of RRB genes does not involve a role of this factor as a direct mediator of the regulatory signal. Spt6 did not seem to mediate the transcriptional up-regulation of ribosome assembly genes since spt6–140 did not negatively interfere with this activation (Figure 4B). Instead Spt6 was recruited to the transcriptionally activated genes, where it acted locally to maintain its chromatin state (Figure 5C). These results support a chromatin-preserving role of Spt6 during the transcriptional up-regulation of RRB genes. The specific genetic interactions of spt6–140 with ribosome assembly mutants, not shared by spt4Δ (Figure 2 and Supplementary Figure S4) also support this chromatin connection. Both Spt6 and DSIF (Spt4/Spt5) have a comparable link to RNA pol II-dependent transcription elongation (8,9). Unlike DSIF, which directly acts on the elongating form of RNA pol II (82), Spt6 operates in the chromatin transactions that take place during transcription elongation to favour nucleosome reassembly and to maintain the proper co-transcriptional dynamics of chromatin (13–18,83,84).
In our model, the feedback up-regulation of the RRB genes would involve the transition between a scenario dominated by backtracked RNA polymerases to a different one dominated by actively elongating enzymes (Supplementary Figure S15). This regulatory transition would be coupled to a parallel change in the dynamics of transcribed chromatin, where Spt6 would favour the maintenance of transcribed nucleosomes and would, therefore, prevent aberrant histone eviction. The malfunction of Spt6 during transcription elongation provokes these chromatin disorders, which are detrimental for the production of functional mRNAs, but do not impair transcription itself (85). This chromatin-preserving function of Spt6 during transcription elongation would be most relevant for completing the correct up-regulation of RRB genes by allowing the increased synthesis of their mRNAs. In spt6–140, those genes that are transcriptionally activated would fail to preserve chromatin and, therefore, to sustain enhanced mRNA production (Supplementary Figure S15). Aberrant and prematurely terminated transcripts are likely degraded by nuclear RNA decay machinery, which has been shown to be efficiently recruited to ribosome-related genes (86). This model is consistent with the effect of spt6–140 on the transcriptomic patterns of the analysed ribosome assembly mutants, where those mRNAs that became induced by the single deletion failed to be expressed or remained only partially induced in the double mutants (Figure 8).
Alterations to mRNA levels result from changes in gene transcription, mRNA stability or both (87). In addition to the aforementioned chromatin effect, spt6–140 provoked a decrease in mRNA stability in the analysed target genes, which helped explain their poor response to Arb1 depletion at the mRNA level (Figure 6). This influence of Spt6 on mRNA stability can be understood in the context of the different forms of transcription/mRNA decay crosstalk that have been described (Reviewed in (88), including the co-transcriptional imprinting of ribosome-related mRNAs for their cytoplasmic fate (58). Other factors that cotranscriptionally bind RNA pol II, like the Paf1 factor, has been shown to influence mRNA fate in addition to foster transcription elongation (89). Moreover, direct mechanistic links between Spt6 and post-transcriptional mRNA phenomena (mRNA splicing and export) have been described in mammalian cells (12,90). In our model, in spite of non-directly mediating the transcriptional activation of RRB genes, Spt6 would favour that the up-regulated genes actually produce enhanced levels of properly stable mRNAs.
The results discussed above enable us to conclude that the ribosome assembly network requires feedback regulation to ensure the accurate expression of all its components, and that this feedback involves a fine control of transcription elongation and the co-transcriptional events that lead to the synthesis of stable mRNAs.
ACCESSION NUMBER
PDB ID: GSE14060.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all the members of our laboratories for discussion and support. We thank Helen Warburton for correcting the English.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness, and European Union funds (FEDER) [BFU2013-48643-C3-1-P and BFU2016-77728-C3-1-P to S.C., BFU2013-48643-C3-3-P to J.E.P.-O., BFU2013-42958-P to J.d.l.C, RYC-2014-16665 to F.G-R.]; FPI and FPU grants from the Spanish Government [to V.B. and G.M.-Z.]; Regional Andalusian Government [P12-BIO1938MO to S.C. and P08-CVI-03508 to J.d.l.C.]; Regional Valencian Government [PROMETEO II 2015/006 to J.E.P-O.]. Funding for open access charge: Consejería de Economía, Innovación, Ciencia y Empleo, Junta de Andalucía [P12-BIO1938MO].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jonkers I., Lis J.T.. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pelechano V., Jimeno-Gonzalez S., Rodriguez-Gil A., Garcia-Martinez J., Perez-Ortin J.E., Chavez S.. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009; 5:e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwabish M.A., Struhl K.. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004; 24:10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang H.W., Kulaeva O.I., Shaytan A.K., Kibanov M., Kuznedelov K., Severinov K.V., Kirpichnikov M.P., Clark D.J., Studitsky V.M.. Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 2014; 42:1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winston F., Chaleff D.T., Valent B., Fink G.R.. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984; 107:179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ardehali M.B., Yao J., Adelman K., Fuda N.J., Petesch S.J., Webb W.W., Lis J.T.. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009; 28:1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Endoh M., Zhu W., Hasegawa J., Watanabe H., Kim D.K., Aida M., Inukai N., Narita T., Yamada T., Furuya A. et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 2004; 24:3324–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaplan C.D., Morris J.R., Wu C., Winston F.. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000; 14:2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartzog G.A., Wada T., Handa H., Winston F.. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998; 12:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andrulis E.D., Guzman E., Doring P., Werner J., Lis J.T.. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000; 14:2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun M., Lariviere L., Dengl S., Mayer A., Cramer P.. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J. Biol. Chem. 2010; 285:41597–41603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoh S.M., Cho H., Pickle L., Evans R.M., Jones K.A.. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007; 21:160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burugula B.B., Jeronimo C., Pathak R., Jones J.W., Robert F., Govind C.K.. Histone deacetylases and phosphorylated polymerase II C-terminal domain recruit Spt6 for cotranscriptional histone reassembly. Mol. Cell. Biol. 2014; 34:4115–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Youdell M.L., Kizer K.O., Kisseleva-Romanova E., Fuchs S.M., Duro E., Strahl B.D., Mellor J.. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 2008; 28:4915–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bortvin A., Winston F.. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996; 272:1473–1476. [DOI] [PubMed] [Google Scholar]
- 16. Ivanovska I., Jacques P.E., Rando O.J., Robert F., Winston F.. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 2011; 31:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeronimo C., Watanabe S., Kaplan C.D., Peterson C.L., Robert F.. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol. Cell. 2015; 58:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morillo-Huesca M., Maya D., Munoz-Centeno M.C., Singh R.K., Oreal V., Reddy G.U., Liang D., Geli V., Gunjan A., Chavez S.. FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 2010; 6:e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallastegui E., Millan-Zambrano G., Terme J.M., Chavez S., Jordan A.. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 2011; 85:3187–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanti M., Gallastegui E., Respaldiza I., Rodriguez-Gil A., Gomez-Herreros F., Jimeno-Gonzalez S., Jordan A., Chavez S.. Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLoS Genet. 2009; 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerard A., Segeral E., Naughtin M., Abdouni A., Charmeteau B., Cheynier R., Rain J.C., Emiliani S.. The integrase cofactor LEDGF/p75 associates with Iws1 and Spt6 for postintegration silencing of HIV-1 gene expression in latently infected cells. Cell Host Microbe. 2015; 17:107–117. [DOI] [PubMed] [Google Scholar]
- 22. Engel K.L., French S.L., Viktorovskaya O.V., Beyer A.L., Schneider D.A.. Spt6 is essential for rRNA synthesis by RNA polymerase I. Mol. Cell. Biol. 2015; 35:2321–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Woolford J.L. Jr, Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de la Cruz J., Karbstein K., Woolford J.L. Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015; 84:93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M.. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004; 18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wade C.H., Umbarger M.A., McAlear M.A.. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006; 23:293–306. [DOI] [PubMed] [Google Scholar]
- 27. Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., Shore D.. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004; 432:1058–1061. [DOI] [PubMed] [Google Scholar]
- 28. Martin D.E., Soulard A., Hall M.N.. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004; 119:969–979. [DOI] [PubMed] [Google Scholar]
- 29. Rudra D., Mallick J., Zhao Y., Warner J.R.. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 2007; 27:4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Downey M., Knight B., Vashisht A.A., Seller C.A., Wohlschlegel J.A., Shore D., Toczyski D.P.. Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor Ifh1. Curr. Biol.: CB. 2013; 23:1638–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kressler D., Rojo M., Linder P., Cruz J.. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 1999; 27:4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosado I.V., Dez C., Lebaron S., Caizergues-Ferrer M., Henry Y., de la Cruz J.. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 2007; 27:1207–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kressler D., de la Cruz J., Rojo M., Linder P.. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998; 18:1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez-Gil A., Garcia-Martinez J., Pelechano V., Munoz-Centeno Mde L., Geli V., Perez-Ortin J.E., Chavez S.. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 2010; 38:4651–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garcia-Martinez J., Aranda A., Perez-Ortin J.E.. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004; 15:303–313. [DOI] [PubMed] [Google Scholar]
- 36. Grigull J., Mnaimneh S., Pootoolal J., Robinson M.D., Hughes T.R.. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004; 24:5534–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Wageningen S., Kemmeren P., Lijnzaad P., Margaritis T., Benschop J.J., de Castro I.J., van Leenen D., Groot Koerkamp M.J., Ko C.W., Miles A.J. et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010; 143:991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clark-Adams C.D., Winston F.. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987; 7:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koren A., Ben-Aroya S., Steinlauf R., Kupiec M.. Pitfalls of the synthetic lethality screen in Saccharomyces cerevisiae: an improved design. Curr. Genet. 2003; 43:62–69. [DOI] [PubMed] [Google Scholar]
- 40. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998; 12:343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swanson M.S., Winston F.. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992; 132:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yao S., Neiman A., Prelich G.. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 2000; 20:7080–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wood A., Shilatifard A.. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle. 2006; 5:1066–1068. [DOI] [PubMed] [Google Scholar]
- 44. Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F.. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002; 22:6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang L., Fletcher A.G., Cheung V., Winston F., Stargell L.A.. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell. Biol. 2008; 28:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoda K., Kawada T., Kaibara C., Fujie A., Abe M., Hitoshi Hashimoto, Shimizu J., Tomishige N., Noda Y. et al. Defect in cell wall integrity of the yeast saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene VIG9. Biosci. Biotechnol. Biochem. 2000; 64:1937–1941. [DOI] [PubMed] [Google Scholar]
- 47. Munoz-Centeno M.C., Martin-Guevara C., Flores A., Perez-Pulido A.J., Antunez-Rodriguez C., Castillo A.G., Sanchez-Duran M., Mier P., Bejarano E.R.. Mpg2 interacts and cooperates with Mpg1 to maintain yeast glycosylation. FEMS Yeast Res. 2012; 12:511–520. [DOI] [PubMed] [Google Scholar]
- 48. Dong J., Lai R., Jennings J.L., Link A.J., Hinnebusch A.G.. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol. Cell. Biol. 2005; 25:9859–9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morillo-Huesca M., Vanti M., Chavez S.. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 2006; 273:756–769. [DOI] [PubMed] [Google Scholar]
- 50. Gomez-Herreros F., de Miguel-Jimenez L., Morillo-Huesca M., Delgado-Ramos L., Munoz-Centeno M.C., Chavez S.. TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res. 2012; 40:6508–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de la Cruz J., Lacombe T., Deloche O., Linder P., Kressler D.. The putative RNA helicase Dbp6p functionally interacts with Rpl3p, Nop8p and the novel trans-acting Factor Rsa3p during biogenesis of 60S ribosomal subunits in Saccharomyces cerevisiae. Genetics. 2004; 166:1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schneider D.A., French S.L., Osheim Y.N., Bailey A.O., Vu L., Dodd J., Yates J.R., Beyer A.L., Nomura M.. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:12707–12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson S.J., Sikes M.L., Zhang Y., French S.L., Salgia S., Beyer A.L., Nomura M., Schneider D.A.. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J. Biol. Chem. 2011; 286:18816–18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marion R.M., Regev A., Segal E., Barash Y., Koller D., Friedman N., O'Shea E.K.. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14315–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miyoshi K., Shirai C., Mizuta K.. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003; 31:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheung A.C., Cramer P.. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011; 471:249–253. [DOI] [PubMed] [Google Scholar]
- 57. Gomez-Herreros F., de Miguel-Jimenez L., Millan-Zambrano G., Penate X., Delgado-Ramos L., Munoz-Centeno M.C., Chavez S.. One step back before moving forward: regulation of transcription elongation by arrest and backtracking. FEBS Lett. 2012; 586:2820–2825. [DOI] [PubMed] [Google Scholar]
- 58. Gupta I., Villanyi Z., Kassem S., Hughes C., Panasenko O.O., Steinmetz L.M., Collart M.A.. Translational capacity of a cell is determined during transcription elongation via the Ccr4-Not complex. Cell Rep. 2016; 15:1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jorgensen P., Nishikawa J.L., Breitkreutz B.J., Tyers M.. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002; 297:395–400. [DOI] [PubMed] [Google Scholar]
- 60. Singh J., Tyers M.. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev. 2009; 23:1944–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lempiainen H., Uotila A., Urban J., Dohnal I., Ammerer G., Loewith R., Shore D.. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell. 2009; 33:704–716. [DOI] [PubMed] [Google Scholar]
- 62. Merz K., Hondele M., Goetze H., Gmelch K., Stoeckl U., Griesenbeck J.. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008; 22:1190–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Berger A.B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O.. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell. Biol. 2007; 27:8015–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hughes J.D., Estep P.W., Tavazoie S., Church G.M.. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000; 296:1205–1214. [DOI] [PubMed] [Google Scholar]
- 65. Lippman S.I., Broach J.R.. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liko D., Slattery M.G., Heideman W.. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007; 282:26623–26628. [DOI] [PubMed] [Google Scholar]
- 67. Jin R., Dobry C.J., McCown P.J., Kumar A.. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell. 2008; 19:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaplan C.D., Holland M.J., Winston F.. Interaction between transcription elongation factors and mRNA 3΄-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 2005; 280:913–922. [DOI] [PubMed] [Google Scholar]
- 69. Lenstra T.L., Benschop J.J., Kim T., Schulze J.M., Brabers N.A., Margaritis T., van de Pasch L.A., van Heesch S.A., Brok M.O., Groot Koerkamp M.J. et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011; 42:536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Laribee R.N., Krogan N.J., Xiao T., Shibata Y., Hughes T.R., Greenblatt J.F., Strahl B.D.. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol.: CB. 2005; 15:1487–1493. [DOI] [PubMed] [Google Scholar]
- 71. Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R. 3rd, Hartzog G.A.. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003; 23:1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McDonald S.M., Close D., Xin H., Formosa T., Hill C.P.. Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Mol. Cell. 2010; 40:725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dronamraju R., Strahl B.D.. A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation. Nucleic Acids Res. 2014; 42:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zheng J., Benschop J.J., Shales M., Kemmeren P., Greenblatt J., Cagney G., Holstege F., Li H., Krogan N.J.. Epistatic relationships reveal the functional organization of yeast transcription factors. Mol. Syst. Biol. 2010; 6:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M. et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007; 446:806–810. [DOI] [PubMed] [Google Scholar]
- 76. Beckouet F., Mariotte-Labarre S., Peyroche G., Nogi Y., Thuriaux P.. Rpa43 and its partners in the yeast RNA polymerase I transcription complex. FEBS Lett. 2011; 585:3355–3359. [DOI] [PubMed] [Google Scholar]
- 77. Bosio M.C., Negri R., Dieci G.. Promoter architectures in the yeast ribosomal expression program. Transcription. 2011; 2:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao Y., McIntosh K.B., Rudra D., Schawalder S., Shore D., Warner J.R.. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 2006; 26:4853–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Knight B., Kubik S., Ghosh B., Bruzzone M.J., Geertz M., Martin V., Denervaud N., Jacquet P., Ozkan B., Rougemont J. et al. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev. 2014; 28:1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R.. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011; 30:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S. et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012; 10:e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bernecky C., Herzog F., Baumeister W., Plitzko J.M., Cramer P.. Structure of transcribing mammalian RNA polymerase II. Nature. 2016; 529:551–554. [DOI] [PubMed] [Google Scholar]
- 83. Adkins M.W., Tyler J.K.. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006; 21:405–416. [DOI] [PubMed] [Google Scholar]
- 84. McCullough L., Connell Z., Petersen C., Formosa T.. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics. 2015; 201:1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kaplan C.D., Laprade L., Winston F.. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003; 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- 86. Bresson S., Tuck A., Staneva D., Tollervey D.. Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Mol. Cell. 2017; 65:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Garcia-Martinez J., Ayala G., Pelechano V., Chavez S., Herrero E., Perez-Ortin J.E.. The relative importance of transcription rate, cryptic transcription and mRNA stability on shaping stress responses in yeast. Transcription. 2012; 3:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Perez-Ortin J.E., Alepuz P., Chavez S., Choder M.. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013; 425:3750–3775. [DOI] [PubMed] [Google Scholar]
- 89. Fischl H., Howe F.S., Furger A., Mellor J.. Paf1 has distinct roles in transcription elongation and differential transcript fate. Mol. Cell. 2017; 65:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yoh S.M., Lucas J.S., Jones K.A.. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008; 22:3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.