Abstract
Firmicutes have two distinct replicative DNA polymerases, the PolC leading strand polymerase, and PolC and DnaE synthesizing the lagging strand. We have reconstituted in vitro Bacillus subtilis bacteriophage SPP1 θ-type DNA replication, which initiates unidirectionally at oriL. With this system we show that DnaE is not only restricted to lagging strand synthesis as previously suggested. DnaG primase and DnaE polymerase are required for initiation of DNA replication on both strands. DnaE and DnaG synthesize in concert a hybrid RNA/DNA ‘initiation primer’ on both leading and lagging strands at the SPP1 oriL region, as it does the eukaryotic Pol α complex. DnaE, as a RNA-primed DNA polymerase, extends this initial primer in a reaction modulated by DnaG and one single-strand binding protein (SSB, SsbA or G36P), and hands off the initiation primer to PolC, a DNA-primed DNA polymerase. Then, PolC, stimulated by DnaG and the SSBs, performs the bulk of DNA chain elongation at both leading and lagging strands. Overall, these modulations by the SSBs and DnaG may contribute to the mechanism of polymerase switch at Firmicutes replisomes.
INTRODUCTION
Replicative DNA polymerases are unable to initiate DNA synthesis de novo. As a consequence, initiation of DNA replication requires a previous step to generate a primer, which contains the essential 3’-hydroxyl moiety that is requisite for polymerase dependent DNA synthesis. Initiation of DNA replication is best understood in γ-Proteobacteria, being Escherichia coli the best characterized member of the class. The first in vitro θ-type replication assays, performed with purified proteins and with plasmids containing the unique origin of replication of this bacterium, oriC, provided an important tool to understand the basic steps of initiation of DNA replication. Here, oriC-dependent θ-type (circle-to-circle) replication could be initiated with any of the three priming systems: primase (DnaG), RNA polymerase, or both combined (1), but later experiments suggested that RNA polymerase is important to transcriptionally activate oriC (2). In bacteria of the Firmicutes phylum, as Bacillus subtilis, the direct involvement of RNA synthesis in the initiation of chromosome replication was early demonstrated using thermosensitive mutants (3), but the in vitro replication system developed with a B. subtilis plasmid-borne oriC and cell extracts showed a high variability in the preparation of protein extracts, and the contribution of DnaG, RNA polymerase, or both to the initiation of DNA replication in this bacterium could not be addressed (4).
Firmicutes possess three conspicuous genome features: purine asymmetry across the two strands of replication, a marked strand-biased gene distribution (5–8), and presence of two essential replicative C-family DNA polymerases, PolC and DnaE (also called DnaE3) (9–11). In contrast, E. coli codes for only one replicative C-family DNA polymerase, the Pol III-α subunit (also termed DnaE1, 11). Escherichia coli Pol III-α extends the RNA primers and catalyses leading and lagging strand DNA synthesis (2). Firmicutes PolC contains both DNA polymerase and proofreading 3′→5′ exonuclease activities in one polypeptide chain (12–14), whereas DnaE has no proofreading activity (15,16). DnaE presents homology to E. coli Pol III-α, and to a lesser extent to PolC (17). In B. subtilis, genes positioned on the lagging strand evolve at a significantly higher rate than those encoded on the leading strand (18). All these features raise the question whether there is a division of labor between the two replicative DNA polymerases, each one devoted to one strand.
Initial genetic analysis suggested a division of labor between these two DNA polymerases in Firmicutes, DnaE synthesizing the lagging and PolC the leading strand (19). Later, a biochemical analysis was performed with a substrate, which represented an advancing replication fork (i.e. a ‘synthetic nicked mini-circle’ that contains a 3’-OH DNA end for leading strand extension and a 5’-unpaired flap for initiation of lagging strand synthesis) (20). These assays revealed that PolC is the leading strand polymerase that initiated synthesis at the pre-existing 3’-OH DNA end, independently of DnaE (20). In the lagging strand, however, DnaE extended the RNA primer synthesized by DnaG before PolC rapidly displaced it to synthesize the bulk of DNA on both lagging and leading strands (20). These results showed a role for PolC in leading and lagging strand synthesis, whereas DnaE seemed to be restricted to the lagging strand, and suggested that B. subtilis DnaE is the bacterial homolog of eukaryotic Pol α for lagging strand synthesis. This was further supported by recent results that showed that B. subtilis DnaE interacts directly with the replicative helicase DnaC, and DnaG, forming a ‘primosome’ complex (21). These previous in vivo (19) and in vitro (20) studies were not designed to study de novo DNA synthesis at the leading strand, as it occurs during initiation of θ-type DNA replication in Firmicutes. To gain insight into the initiation of θ-type DNA replication in bacteria having both DNA polymerases, PolC and DnaE, we have reconstituted for the first time an in vitro origin-dependent θ-type DNA replication system using the B subtilis SPP1 bacteriophage (phage) replisome. It was previously shown that phage SPP1 codes for the replisome organizer, G38P, the replicative DNA helicase, G40P, its helicase loader, G39P, and a single-stranded binding protein (SSB), G36P, and it recruits the rest of the replisome components from its host for an efficient viral replication (22–24). In vivo SPP1 DNA replication starts by the θ-mechanism from a replication origin (oriL), but after one or a few rounds, it switches to concatemeric replication (termed also σ-type replication) by a process driven by viral recombination proteins (25–27). Previously the SPP1 σ-type DNA replication was established in vitro with a synthetic nicked mini-circle template, and it was found to require eleven purified proteins. Four are viral-encoded (G38P, G40P, G39P, G36P), and seven host-encoded: β clamp, clamp loader (τ complex, composed by the τ, δ and δ′ subunits), primase (DnaG), PolC and DnaE (28). In this system, leading strand synthesis initiated at the pre-existing 3’-OH DNA end present in the synthetic nicked mini-circle template. The recruitment of the host replication proteins occurred presumably by protein-protein interactions with viral proteins, as it had been previously demonstrated in vitro that G40P interacts with DnaG, and with the τ subunit of the clamp loader complex (29,30). For in vitro σ-type SPP1 replication, DnaG and DnaE were necessary for lagging strand DNA synthesis, and PolC was involved in leading and lagging strand synthesis, as previously observed with B. subtilis (20,28).
In this work we have reconstituted for the first time SPP1 oriL-dependent θ-type DNA replication. We show that during θ-type DNA replication DnaE plays a crucial role in both DNA strands. Our results show that DnaG is required to prime the leading-strand at oriL, but this primer is not sufficient for PolC initiation of leading-strand synthesis, although DnaG stimulates PolC-mediated leading strand replication. DnaE is essential for extending the leading strand primer at oriL, and then PolC continues leading strand synthesis to give full replication products. Our results suggest that this division of labor can occur in all bacteria having the two replicative polymerases, and that primase and the SSBs may contribute to the mechanism of polymerase switch.
MATERIALS AND METHODS
Plasmids
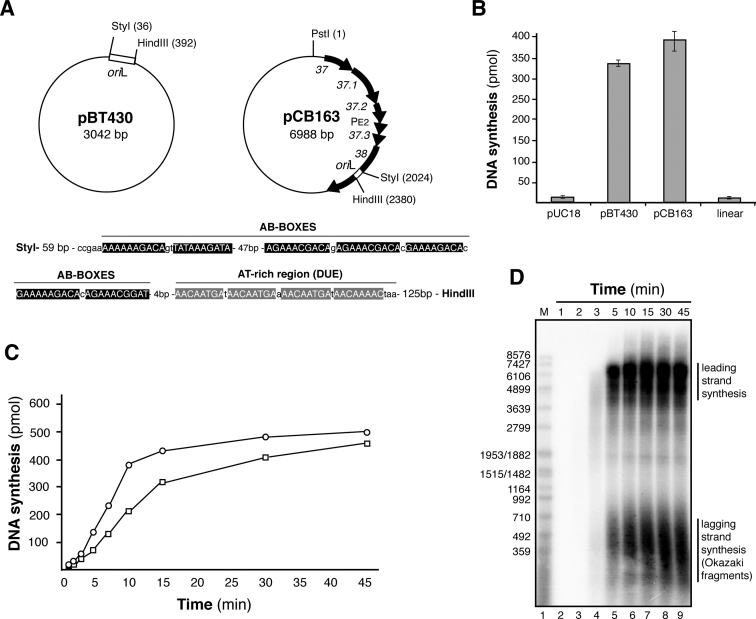
Three plasmids were used as DNA templates for in vitro θ replication: oriL− (pUC18 2686 bp, used as negative control); a short oriL+ (pBT430, a pUC18 plasmid derivative containing the 356 bp HindIII–StyI SPP1 oriL DNA segment, see (23)); and long oriL+ (pCB163, derived from pUC18-cat, it contains a SPP1 2.7 kb PvuII–SmaI DNA segment encompassing gene 37 to gene 38 (23)). A brief scheme of the two oriL+ plasmids is depicted in Figure 1A.
Figure 1.
Reconstitution of SPP1 oriL-dependent θ-type DNA replication with B. subtilis and SPP1 purified proteins. (A) Diagram of the oriL-borne plasmid DNA templates used. Plasmid pBT430 only contains the 356-bp oriL region (StyI–HindIII fragment). Plasmid pCB163 has a 2.7-kb SPP1 region cloned (from gene 37 to gene 38). In the bottom, the oriL region is enlarged. Its is composed by a series of repeated elements called AB boxes where G38P specifically binds, and the AT-rich adjacent region known as DUE. (B) Quantification of total DNA synthesis obtained after 15 min incubation using the DNA templates: pUC18 as negative control, and pBT430 and pCB163, both carrying oriL. In lane 4, DNA synthesis obtained with PstI-linearized pCB163. The values represented are the mean of three independent experiments. (C) Time course of DNA synthesis obtained using pBT430 (□) or pCB163 (○) plasmids (both carrying oriL), quantified by incorporation of [α-32P]dATP. (D) The DNA products generated in the replication reaction performed with pCB163 as DNA template were separated on a 0.7% alkaline agarose gel to visualize leading and lagging strand synthesis. M: 3′-labeled EcoRI-digested SPP1 DNA marker.
Non-denaturing supercoiled plasmid DNA isolation
Plasmid DNA was isolated from 1 liter of an E. coli culture using a non-denaturing protocol (31) with some modifications. Cells were harvested, washed with T10E1N10 buffer (10 mM Tris–HCl pH7.5, 1 mM EDTA, 10 mM NaCl) and resuspended in 30 ml of T10E1N10 buffer. EDTA up to 25 mM and 1.2 ml of 1% lysozyme were added. After incubation at 4°C for 10 min, 1.8 ml of 10% Triton X-100 was added. After further incubation for 40 min at room temperature, ultracentrifugation at 30 000 rpm in a 50.2Ti rotor at 4°C for 60 min was performed. The supernatant was extracted three times with phenol:chloroform:isoamyl solution (50:50:1). DNA was precipitated by adding 1/10 volume of 3M KOAc and a volume of 2-propanol, and resuspended in 5 ml of TE buffer carrying 20 μg/ml RNAse. After incubation for 60 min, plasmid DNA was extracted again with phenol:chloroform:isoamyl (50:50:1), and after ethanol precipitation, it was finally resuspended in 8 ml of T10E1N10 buffer. Eight gram of CsCl and ethidium bromide (200 μg/ml) were added for a further purification by CsCl gradient centrifugation. The gradient was performed in Beckman 13 × 51 mm tubes. The mixture was centrifuged in a NVT90 rotor (55 000 rpm, 15°C) for 16 h. The UV-visualized plasmid DNA band was collected and ethidium bromide was removed by extraction with 2-propanol saturated with cesium chloride and the DNA was precipitated. The pellet was resuspended in 0.5 ml of T50E1N10 buffer and Form I DNA was finally purified by sucrose gradient centrifugation. The sucrose gradient (15–40%) was formed in Beckman 14 × 89 mm tubes using a Gradient Station. The sample was loaded on the top of the tubes, and centrifuged in a SW41 rotor (35 000 rpm, 4°C, 16 h). Fractions of 0.5 ml were collected and analyzed in a 0.8% agarose gel. Fractions carrying pure Form I DNA were pooled and precipitated. The pellet was washed, resuspended in 0.5 ml of TE buffer and dialyzed. Supercoiled plasmids were stored at 4°C. DNA concentrations are indicated in mols of nucleotides.
Proteins
SPP1 G40P, G39P, G38P and G36P were purified as described (28). B. subtilis DnaG primase, DnaE, PolC, τ, δ, δ′, β, SsbA and DNA gyrase subunits GyrA and GyrB were purified as described (20,32). B. subtilis RNA polymerase and DNA gyrase were a gift from Begoña Carrasco and Dagmar Klostermeier, respectively, and E. coli Topoisomerase III was a gift from Kenn Marians. DNA polymerase I (Pol I), and T4 DNA ligase were from NEB Biolabs.
DNA replication assays
Standard oriL-dependent SPP1 θ-type DNA replication reactions consisted of 30 nM G40P6, 120 nM G39P, 180 nM G38P, 40 nM DnaG, 10 nM DnaE, 20 nM PolC, 25 nM τ4, 25 nM δ, 25 nM δ′, 24 nM β2, 50 nM G36P4, 30 nM gyrase, DNA template (400 pmol in nucleotides), 350 μM ATP, 100 μM CTP, GTP and UTP, 48 μM dNTPs (except 18 μM dATP). α-[32P]dATP was added in a 1*:2500 ratio. The reactions were carried out in 25 μl of buffer BsRC (20) that additionally contained 4% of glycerol and 19 mM NaCl contributed by the proteins buffers. An enzyme mix containing all protein components except the SSB (G36P, or SsbA, as indicated) was prepared in buffer BsRC. The substrate mix contained template DNA, rNTPs, dNTPs and the SSB protein. Reactions were initiated by mixing the enzyme mix with the substrate mix, and incubations were conducted for 15 min at 37°C. After incubation, reactions were stopped by addition of an equal volume of stop mix composed of 40 mM Tris–HCl (pH 8.0), 0.2% SDS, 100 mM EDTA and 0.5 mg/ml Proteinase K. Samples were treated for 20 min at 37 °C, then applied into Sephadex G-50 packed cartridges to eliminate the non-incorporated dNTPs. The extent of DNA synthesis was measured by scintillation counting.
Replication reactions were also performed with the synthetic nicked mini-circle substrate. This DNA template is a 409 nt nicked circle containing a 396 nt tail (20). It has a asymmetric 50:1 G:C distribution, so that in the presence of α-[32P]dCTP leading strand synthesis is observed, and with α-[32P]dGTP lagging strand synthesis. With this substrate σ-type concatemeric DNA replication is observed. Reactions were performed as published (28) with some minor modifications. Briefly they contained 80 pmol of DNA template (in nt), 30 nM G40P6, 180 nM G39P, 200 nM G38P, 20 nM DnaG, 20 nM DnaE, 20 nM PolC, 25 nM τ4, 25 nM δ, 25 nM δ′, 24 nM β2, and the indicated concentrations of the SSB (G36P or SsbA). Radiolabeled nucleotides were used in a 1*:2000 ratio with respect to cold nucleotide. When DnaE was the only DNA polymerase present, reactions were incubated for 40 min at 37°C. Reactions with the full replicase (PolC + DnaE) were incubated for 2 min at 37°C, and reactions having only PolC were incubated for 2 min at 37°C.
Gel electrophoresis of DNA replication products
Leading and lagging strand synthesis were separated by alkaline agarose gel electrophoresis. After replication assays, reactions were adjusted to 50 mM NaOH, 5% (v/v) glycerol and 0.05% bromophenol blue and fractionated on alkaline 0.7% agarose gels for approximately 4 h at 70 V. Gels were fixed in 7% trichloroacetic acid, dried, auto-radiographed on storage phosphor screens and analyzed with Quantity One software (BioRad). Total DNA synthesis was also analyzed in neutral gels. For that, the reaction mixtures were mixed with one-tenth volume of a dye mixture containing 30% glycerol, 10 mM EDTA, 0.1% xylene cyanol and 0.1% bromphenol blue. Samples were separated on 0.8% agarose gels, run at 25 V for ∼16 h in TBE buffer (89 mM Tris Base, 89 mM boric acid, 2 mM EDTA pH 8.0) in the absence of ethidium bromide. Gels were stained in 1 μg/ml ethidium bromide, fixed in 7% trichloroacetic acid, dried and autoradiographed.
RESULTS
Reconstitution of oriL-dependent SPP1 DNA replication
Previously, it was shown that G38P, the SPP1 replisome organizer, specifically binds in vitro to two short discrete regions of the phage genome (oriL and oriR) with similar affinity (22,23). Plasmid pCB163, a pUC18 derivative containing a 2.7-kb region of SPP1 DNA (including genes 37 to 38, the PE2 promoter, and the oriL region embedded in gene 38, Figure 1A) replicates in vivo in B. subtilis, if genes 39 and 40 are provided in trans (23). The oriL region has the classic signature of a bacterial replication origin (reviewed in 33,34), with DNA repeats (the iterons, termed here AB boxes), which are specifically bound by G38P (23), and adjacent to them a DNA unwinding element (DUE, an A-T rich region, see Figure 1A). In order to test if this region is necessary and sufficient to drive DNA replication we cloned the 356-base pair (bp) SPP1 region that includes the oriL region onto pUC18, leading to pBT430. We used these three DNA substrates as templates for analyzing in vitro θ-type DNA replication: oriL− (pUC18, negative control); the in vivo analyzed oriL+ (pCB163) and a shorter version containing only oriL (pBT430).
First, the concentrations of the required proteins were individually titrated with pCB163 as the DNA template to optimize the SPP1 oriL-dependent DNA replication (Supplementary Figure S1). In vitro DNA replication with this template required the four viral G40P, G39P, G38P, G36P proteins previously defined by in vivo and in vitro studies to be essential for DNA replication (22,24,28). Host proteins DnaG, PolC, DnaE, β and τ-complex were necessary for replication in addition to DNA gyrase (Supplementary Figures S1 and S2). Addition of purified B. subtilis RNA polymerase neither replaced DnaG nor enhanced the level of DNA synthesis obtained with the pCB163 template. This is consistent with genetic analysis that showed that SPP1 DNA replication is independent on RNA polymerase (22).
In vitro DNA replication was not detected with the supercoiled oriL− substrate (pUC18 control, Figure 1B), while it was obtained using the long oriL+ template (plasmid pCB163). No DNA synthesis was observed when this oriL+ template was Form III DNA (i.e. linearized DNA) (Figure 1B, lane 4), showing that DNA supercoiling is required for oriL-dependent DNA replication. When plasmid pBT430 (containing the 356 bp oriL region) was used, a similar level of DNA synthesis was observed, showing that DNA replication is dependent on oriL, and that this 356-bp region is necessary and sufficient for efficient DNA synthesis in vitro.
As occurred in σ-type DNA replication, the oriL-assembled replisome could use both viral- (G36P) or host-encoded (SsbA) SSB proteins (Supplementary Figures S1C and S1D), and the quantification showed that the presence of an SSB is essential to have DNA synthesis. Both SSBs support similar levels of DNA synthesis. High concentrations of SsbA slightly inhibited oriL-dependent DNA replication, whereas this inhibition was not observed increasing G36P. Conversely, high G36P concentrations were inhibitory with the ‘synthetic nicked mini-circle’ substrate (which mimics the σ-type concatemeric mode of SPP1 replication) and increasing SsbA stimulated its replication (28). The nature of these inhibition discrepancies remains unknown, but it may be due to an effect on the activity of the DnaE enzyme (see below).
A time course of oriL-dependent DNA replication with the two oriL+ templates (pCB163 and pBT430 plasmids) showed an initial lag phase longer than 3 min. Then, DNA synthesis was linear during ∼10 min (Figure 1C). Two populations of DNA products were detected in an alkaline agarose gel (Figure 1D and Supplementary Figure S3): nascent leading strands, which corresponded to full-length plasmid DNA and suggested unidirectional replication from oriL (see Supplementary text and Supplementary Figure S4), and nascent lagging strands composed by Okazaki fragments of ∼500 bp, because there was neither DNA Polymerase I (Pol I) nor DNA ligase in the reaction mixture. A quantitative analysis by densitometry of the gels showed similar amounts of leading and lagging strands over time, as expected for a coordinated leading and lagging strand DNA synthesis from the oriL-containing templates. Replication products larger than the original plasmid were not observed (Figure 1D), suggesting that no over-replication due to strand switching or SPP1 σ-type DNA replication takes place.
Analysis of oriL-dependent DNA replication
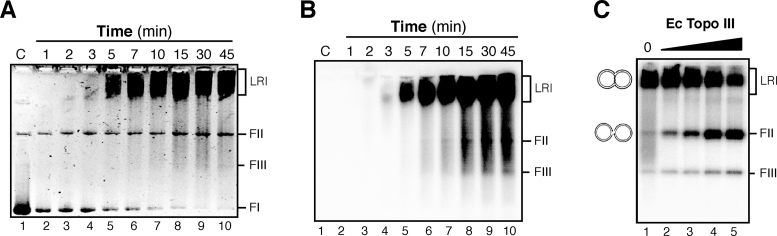
To obtain more information about the type of DNA replication initiated from oriL, replication products obtained with pCB163 were analyzed in native gels (Figure 2). The ethidium bromide-stained neutral gel confirmed that Form II (i.e. nicked or gapped circular) DNA, which was present in a small amount in our plasmid preparation, is not substrate for oriL-dependent DNA replication. Form I of the plasmid template (i.e. supercoiled DNA) was the substrate from the beginning of the reaction, and it was transformed over time to the late replication intermediate (LRI, Figure 2A).
Figure 2.
oriL-dependent DNA replication products are of the θ-type. (A) The replication products that accumulate over the time using supercoiled pCB163 plasmid as the DNA template were analyzed in a native agarose gel stained with ethidium bromide. (B) Autoradiogram of the same gel. (C) The late replication intermediates formed are decatenated by the E. coli Topoisomerase III enzyme. Standard replication reactions with plasmid pCB163 as DNA template were performed for 15 min and then E. coli Topoisomerase III was added (varied in a 2-fold dilution series from 400 nM down to 50 nM), and reaction continued for 15 min. The deproteinized products were fractionated on a native agarose gel and autoradiographed. The abbreviations used are: C, control, pCB163 plasmid; LRI, late replication intermediates; FI, FII, FIII: Forms I, II and III of plasmid DNA, respectively.
Late replication intermediates, which run close to the top of the gel, accumulated when DNA gyrase was the only topoisoimerase present, suggesting θ-type of DNA replication (Figure 2B). Form II is the expected decatenated replication product, because as stated, reactions lacked Pol I and DNA ligase. The accumulation over time of a radiolabelled faint band that migrated as Form II DNA was detected in the autoradiogram. This band is probably due to the poor decatenase activity of B. subtilis DNA gyrase (Figure 2B, see 35). To confirm that the products that run close to the top of the gel are genuine late replication intermediates, which accumulated in a θ-type replication, we used E. coli Topoisomerase III, a potent decatenase on DNA rings that have small gaps (36). Standard SPP1 replication assays were performed with plasmid pCB163, and then E. coli Topoisomerase III was added, and the reactions were incubated for additional 15 min. After deproteinization, the replication products were resolved in native agarose gels, and the accumulation of Form II DNA, due to Topoisomerase III-mediated decatenation of the late replication intermediates, was observed (Figure 2C).
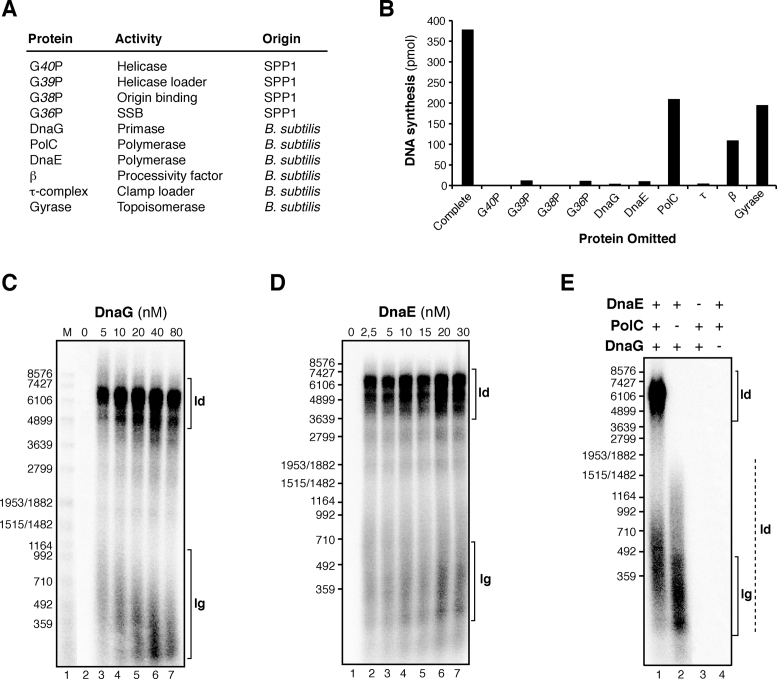
DnaG and DnaE are required for leading strand synthesis
In assays where single proteins were omitted from the optimized oriL-dependent θ-type DNA replication reactions, we observed that all proteins where required, but with some differences (Figure 3). Significant levels of DNA synthesis could be observed in the absence of DNA gyrase, because in the absence of a topoisomerase, θ-type DNA replication starts but stalls due to accumulation positive supercoiling (36,37). When analyzed on an alkaline gel, early replication intermediates of around 1–2 kb were observed in the absence of DNA gyrase (Supplementary Figure S2). No DNA synthesis was observed in the absence of DnaG, indicating that the initiation of leading and lagging strand synthesis is DnaG primase dependent. The size of Okazaki fragments decreased with increasing DnaG concentrations, although here the Okazaki fragment length was not as sensitive to DnaG concentration as it was observed in σ-type concatemeric SPP1 replication (Figure 3C and (28)).
Figure 3.
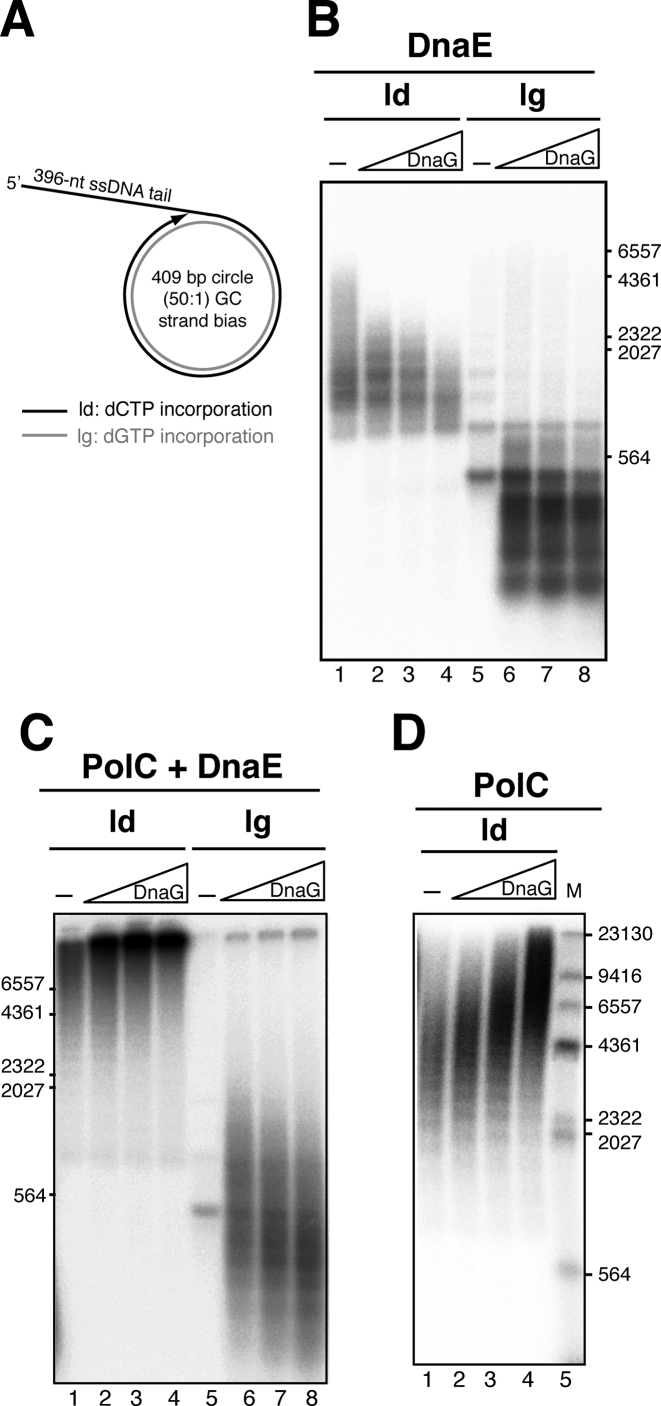
DnaG and DnaE are required for initiation of leading strand synthesis in SPP1 θ-type DNA replication. (A) Activity and source of proteins involved. (B) Protein requirements for oriL-dependent θ-type DNA replication. The level of DNA synthesis obtained with plasmid pCB163 as DNA template in reactions omitting just one protein component was quantified by [α-32P]dATP incorporation. The values shown are the mean of three independent experiments. (C and D) Alkaline agarose gel of oriL-dependent replication products obtained from reactions omitting or varying DnaG (C) or DnaE (D). The protein concentration varied in a 2-fold dilution series as indicated. (E) Replication products obtained in 15 min reactions omitting PolC, DnaE and DnaG as indicated. Samples were fractionated on alkaline agarose gels. Abbreviations: ld, leading strand synthesis; lg, lagging strand synthesis. The dashed line highlights the leading strand product putatively synthesized by the DnaE enzyme.
DnaE was strictly required to have oriL-dependent θ-type DNA replication (Figure 3B and 3D). In contrast, DnaE and DnaG were dispensable for leading strand DNA synthesis in SPP1 σ-DNA replication (28). These results suggested that DnaE may be strictly required for extending the leading strand RNA primer synthesized by DnaG at the oriL region. This is consistent with the previous observation that DnaE, but not PolC, efficiently uses RNA primers in primer extension assays (20).
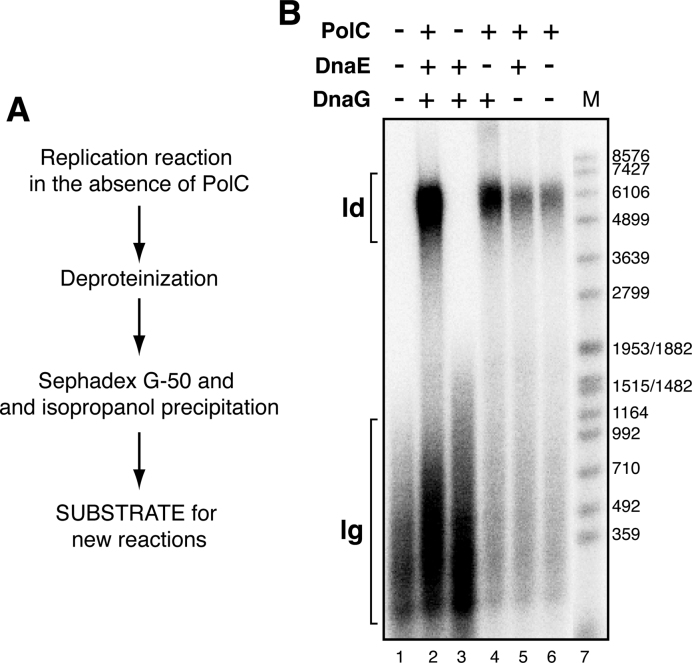
A significant level of DNA synthesis was observed when just the processive PolC polymerase was omitted (i.e. DnaE was the only polymerase, Figure 3B). When these replication products were analyzed in an alkaline gel, we observed that the size of the DNA synthesized was broad, from DNA replication products with the expected length for Okazaki fragments (300–500 bp) to longer DNA fragments (up to 1.5 kb) (Figure 3E, lane 2). This result suggested that in the absence of PolC, DnaE could support significant replication of the leading strand, producing the 1.5 kb DNA fragments in a DnaG-dependent manner (Figure 3E, lane 4). To test this hypothesis, the replication products generated with DnaE as the only DNA polymerase present were deproteinized, purified and this pre-initiated substrate was used for another round of in vitro DNA replication (Figure 4A). As revealed in Figure 4B (lanes 4 and 6), the addition of PolC as the unique polymerase to this pre-initiated substrate, which contains RNA-DNA hybrid primers of variable length, was necessary and sufficient to yield full-length leading strand replication products (i.e. a 6.9 kb DNA). Higher levels of complete leading strand synthesis were obtained when DnaG was added in addition to PolC (Figure 4B, lane 4 in comparison with lane 6). This stimulation is further analyzed below.
Figure 4.
DnaE and DnaG prime oriL-dependent leading strand synthesis. (A) Scheme of the assay. Six standard replication reactions with all components except PolC were performed with pCB163 as the DNA template and pooled. After deproteinization the replication products were purified. The DNA pellets were resuspended in MilliQ water and this partially replicated substrates were used as templates for new replication reactions. (B) Denaturing gel showing the final replication products obtained when these purified DnaG-DnaE-primed templates were used in a new in vitro replication reaction with all the components except the indicated enzyme. Abbreviations: ld: leading strand, lg: lagging strand, M: size marker (EcoRI-labelled SPP1-DNA).
DnaG stimulates PolC, but inhibits DnaE
To further explore the extent of DnaE-mediated DNA replication of the leading strand in the plasmid oriL system, we performed reactions where all components except PolC were present (i.e DnaE was the only DNA polymerase) and longer times of incubation (Supplementary Figure S5). Full-length replication products were not observed, even after 90 min of incubation. The maximal length obtained was ∼2500 bp instead of the expected 6.9 kb, which is the plasmid size. In contrast, Streptococcus pyogenes DnaE, which has 34% sequence identity with the B. subtilis DnaE enzyme, completed a M13mp18-primed ssDNA template (7249 bp) in a reaction having, in addition to DnaE, the τ-complex, β and the SSB protein. The rate of this reaction was 60 nt/s (38). Similarly, earlier studies showed that the rate of DNA synthesis for DnaE with a single-stranded DNA template was 16 nt/s and raised to 75 nt/s if the τ-complex and β proteins were also present (20). In these two reports, the reactions were done with DnaE, τ-complex and β, but in the absence of the other components of the primosome (primase and helicase), which were present in our experiment. We wondered whether the low extent of leading strand synthesis observed by us in reactions whith DnaE as the only DNA polymerase could be due to an inhibitory effect of any component of the primosome (DnaG or G40P). In addition, since in the previous experiment we had observed that DnaG stimulated PolC (Figure 4B), we decided to analyze more carefully the effect of primase in replication reactions having only DnaE or only PolC as the DNA polymerases.
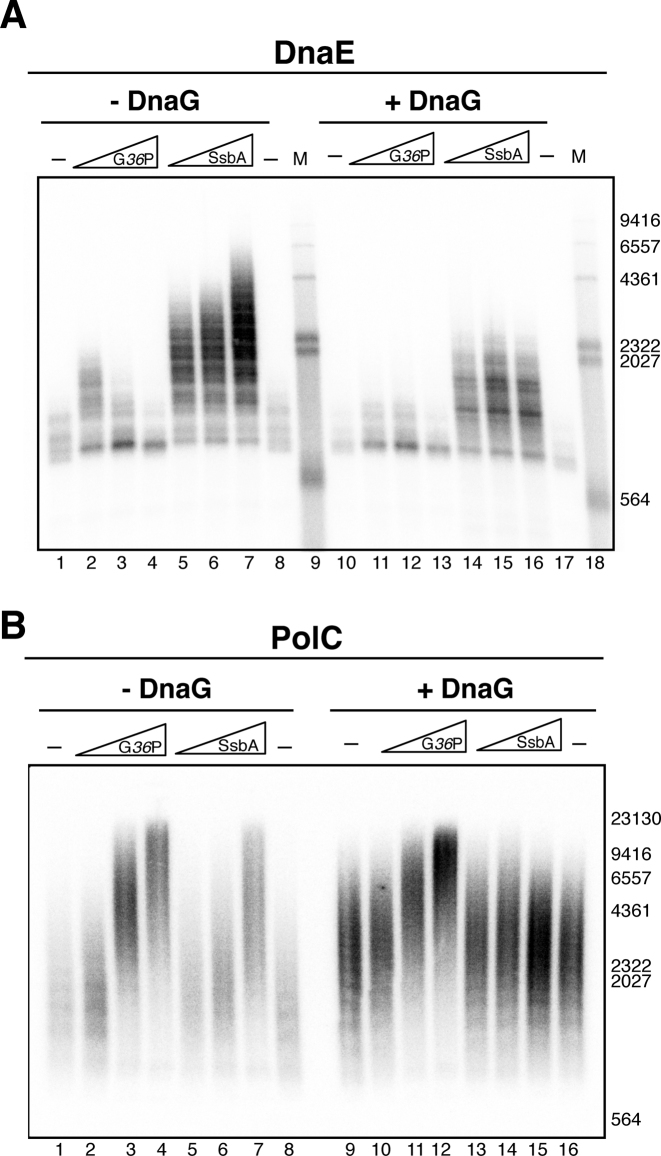
To analyze this, we used the ‘synthetic nicked mini-circle’ substrate as DNA template. This substrate is primed by a pre-existing 3’-OH DNA end (see Figure 5A), therefore DnaG is only required to prime the lagging strand. In addition, this DNA template has a 50:1 GC strand bias, so that radiolabeled dCTP is almost only incorporated in the leading strand, and radiolabelled dGTP is incorporated in the lagging strand. Reactions contained all replisome components, except the PolC polymerase (i.e. they contained G38P, G39P, G40P, DnaG, SsbA, τ-complex, β and DnaE, Figure 5A). In the absence of DnaG (only leading strand synthesis is permitted), DnaE-synthesized replication products up to 5-kb were observed with the SsbA concentration used in these reactions (100 nM), in agreement with the previous results obtained by the other authors (20,38). Interestingly, increasing concentrations of DnaG resulted in an inhibition of DnaE-mediated leading strand DNA synthesis, and the accumulation of shorter leading strand products was observed. However, this effect was not observed on the lagging strand. Varying DnaG did not have any effect on the size of the Okazaki fragments synthesized by the DnaE enzyme (Figure 5B). In contrast, when reactions also had PolC (i.e. the full replisome was present, Figure 5C), increasing concentrations of DnaG stimulated leading strand synthesis (Figure 5C, lanes 2–4). With the full replisome (DnaE+PolC present), the size of Okazaki fragments decreased with increasing DnaG concentrations as previously reported (Figure 5C, lanes 6–8, and (28)).
Figure 5.
DnaG differentially modulates DNA synthesis mediated by PolC or by DnaE. (A) Scheme of the synthetic nicked mini-circle substrate used in these assays; leading strand synthesis is primed by the pre-existing 3’-OH DNA end, and with this substrate σ-type concatemeric DNA replication is obtained. (B) DnaG inhibits leading strand synthesis catalyzed by DnaE. DNA replication reactions were performed with the synthetic nicked mini-circle as DNA template in the presence of [α-32P]dCTP or [α-32P]dGTP to visualize leading or lagging strand synthesis respectively. In addition to DnaE, reactions had G38P, G39P, G40P, τ complex, β and 100 nM SsbA as the SSB protein. As indicated, reactions were performed in the absence or presence of increasing DnaG concentrations (8, 16 and 32 nM), and were incubated for 40 min at 37 °C. (C) DnaG stimulates leading strand synthesis catalyzed by a complete replisome. Increasing concentrations of DnaG (0, 8, 16, and 32 nM) were added to the full SPP1 replisome (PolC plus DnaE and all replisome components). Reactions contained [α-32P]dCTP or [α-32P]dGTP to visualize leading or lagging strand synthesis and were incubated for 2 min at 37°C. (D) DnaG stimulates leading strand synthesis catalyzed by PolC. Reactions were performed similarly but here [α-32P]dCTP was used, and PolC was the sole DNA polymerase present. ld: leading strand, lg: lagging strand; M: DNA ladder used, HindIII labelled λ DNA.
To confirm that this stimulation of leading strand synthesis is due to a positive effect of DnaG over the DNA synthesis performed by the PolC enzyme, we analyzed with this synthetic nicked mini-circle’ substrate the effect of DnaG on reactions having only PolC as the DNA polymerase (Figure 5D). With this substrate, leading strand synthesis does not require DnaG priming. In the absence of DnaG, leading strand replication products synthesized by PolC in the presence of all replisome components except DnaE (i.e, reactions contained G38P, G39P, G40P, SsbA, τ-complex, β and PolC) reached a maximum size of 6.5 kb in a 2 min reaction. Addition of increasing concentrations of DnaG stimulated leading strand synthesis mediated by PolC, and the size of the DNA products obtained was up to 20 kb at high primase concentrations (Figure 5D). In contrast, increasing DnaG concentrations decreased the size of the Okazaki fragments poorly synthesized by the PolC enzyme in the absence of DnaE (Supplementary Figure S6A). These experiments showed that independently of the plasmid replication mechanism (θ-type or σ-type), DnaG stimulated PolC-mediated leading strand synthesis (Figures 4B and 5D respectively). In the synthetic nicked mini-circle substrate, the leading strand is DNA primed (Figure 5A), but DnaG stimulated leading strand synthesis. One hypothesis is that DNA synthesis is discontinous, and that DnaG may catalyze the synthesis of new RNA primers at the leading strand, which are further extended by PolC. To evaluate this, we analyzed if ribonucleotides are incorporated into the leading strand in replication reactions having DnaG, and PolC (i.e, reactions contained G38P, G39P, G40P, DnaG, SsbA, τ-complex, β and PolC). Previous studies showed that B. subtilis DnaG initiates primer synthesis from 5′-d(CTA), 5′-d(TTA) and 5′-d(TTT) (21), hence we analyzed if [α-32P]ATP is incorporated. We only observed incorporation of the radiolabeled ATP into the lagging strand (Supplementary Figure S6B). The reaction was very poor, confirming that in the absence of DnaE, PolC performs limited lagging strand synthesis (see also Supplementary Figure S6A). These results rule out the hypothesis that DnaG stimulates leading strand synthesis by primer formation at the leading strand. Another possible mechanism for the increment in DNA synthesis involves a direct PolC–DnaG interaction. To test this, we performed pulldown experiments with a N-terminal His-tagged variant of DnaG, which is fully active in DNA replication, and native PolC. Proteins were preincubated as indicated in Supplementary methods, followed by treatment of the reaction mixtures with Ni-magnetic beads to capture any protein complex that might have formed. We found no specific retention of PolC, which suggests that both proteins do not form stable complexes (Supplementary Figure S7A).
Effect of the SSBs in DnaE-mediated and PolC-mediated DNA synthesis
We further analyzed if other proteins may contribute to the polymerase exchange at the SPP1 replication fork. Previous experiments in other systems have shown that the SSBs may control the mechanism of primase polymerase switch (39,40). To test if any of the two SSBs that are active at the SPP1 replication fork (G36P and SsbA) may contribute to the polymerase switch, we analyzed if the presence of a SSB protein has an effect on DnaE or PolC catalyzed DNA synthesis. This question can only be tested with the synthetic nicked mini-circle substrate (σ-type replication), because oriL-dependent θ-type DNA replication strictly requires a SSB protein (Supplementary Figure S1). First we set up reactions containing DnaE, τ-complex, β, the replicative helicase and its loaders (G38P, G39P, G40P). The reactions additionally contained primase when indicated, and different concentrations of the two SSBs (G36P or SsbA, from 25 to 100 nM). In the absence of DnaG (i.e, only leading strand synthesis, primed from the pre-existing 3’-OH DNA end, is permitted), DNA synthesis mediated by DnaE was stimulated by the presence of a SSB, albeit to a different extent. DNA synthesis was poor in reactions having the viral SSB (G36P) in comparison with reactions having SsbA (Figure 6A, lanes 2–4 versus 5–7). This strong stimulation by SsbA could be due to the interaction of DnaE with SsbA (41). However G36P also interacts with DnaE (Supplementary Figure S7B). The amount of SsbA present in the reaction modulated the length of leading strand products synthesized by DnaE. When lagging strand synthesis was also allowed (i.e. reactions additionally contained DnaG), leading strand DNA synthesis with DnaE as the sole polymerase was inhibited by the presence of DnaG, and this effect was observed with both SSBs, albeit in a different extent (Figure 6A, lanes 11–13 and 14–16). We then tested the effect of the SSBs in reactions having PolC as the only polymerase (i.e, reactions had the full replisome except DnaG and DnaE, Figure 6B, lanes 1–8). We found that in contrast to the effect observed in DnaE reactions, increasing concentrations of the SSBs stimulated leading strand DNA synthesis mediated by PolC, although the SSBs are thought to function only on the lagging strand. Here, no difference was observed between SsbA and G36P, and their stimulatory effect was also observed when reactions additionally had DnaG (Figure 6B, lanes 9–16).
Figure 6.
DNA synthesis is modulated by the SSBs. (A) Effect of the SSBs in DnaE-mediated leading strand synthesis. DNA replication reactions were performed with the synthetic nicked mini-circle in the presence of [α-32P]dCTP and contained G38P, G39P, G40P, τ complex, β and DnaE as the only DNA polymerase. As indicated, reactions additionally contained or not DnaG (20 nM), and increasing concentrations of G36P, or SsbA (25, 50 and 100 nM). After 40 min of incubation, the reactions were stopped, and the leading strand replication products were analyzed by denaturing agarose gel electrophoresis and autoradiography. M is HindIII labelled λ DNA. (B) Effect of the SSBs in PolC-mediated leading strand synthesis. Reactions were performed as in A, but contained PolC instead of DnaE. After 2 min of incubation, the reactions were stopped, and the leading strand replication products were analyzed by denaturing gel electrophoresis and autoradiography.
DISCUSSION
We have succeeded in establishing for the first time an in vitro origin-dependent θ-type DNA replication system from Firmicutes using the SPP1 replisome, which consists of SPP1 and B. subtilis proteins. Initiation of SPP1 oriL-dependent θ-type DNA replication relied on thirteen proteins: four viral proteins (G38P, G39P, G40P and G36P) and nine host proteins (DnaG, PolC, DnaE, β, τ-complex (τ, δ and δ’) and DNA gyrase, composed by GyrA and GyrB subunits). Our in vitro data are in agreement with previous genetic data which showed that SPP1 replication is independent of host encoded DnaA initiator, chaperones (DnaK and DnaJ) and RNA polymerase (22). This is in contrast to other bacterial or phage systems, where other proteins, in addition to the origin binding protein, are required, either to help in origin opening, or to remodel and activate proteins at the origin of replication (42–45). Unlike SPP1 σ-type replication (28), SPP1 θ-type replication cannot proceed without supercoiling. The strict requirement of a supercoiled substrate seems to be a common feature of DNA replication by the θ mechanism. It has been reported for systems initiated by DnaA (46), or by the λO protein (47), and here we observe this in SPP1 θ-type replication, which is initiated by the binding of G38P to the oriL region. In contrast to DnaA and many other replication initiators, G38P (pfam09524 protein family) does not contain an AAA+ domain (48). In addition, a SSB protein was required to have in vitro DNA replication in the oriL system. Earlier studies showed that E. coli SSB is required for optimal unwinding of ori λ plasmid DNA by the DnaB helicase (43,49). Our previous assays showed that a ssDNA region is necessary for helicase G40P loading and assembly of the replisome (50). We can envision that DNA supercoiling and the SSB proteins (G36P or SsbA) may facilitate the G38P-mediated unwinding of the DUE element (AT-rich region) present in the oriL region, probably by stabilizing the single stranded replication bubble, as it occurs in other bacterial systems (51).
The in vitro reconstituted oriL-dependent system has allowed us to study the mechanism of priming during initiation of DNA replication in a bacterium that has two replicative C-family DNA polymerases, PolC and DnaE. In all organisms, initiation of DNA replication requires de novo priming to provide with a free 3’-OH end, and this may occur by several different mechanisms. Assays using purified proteins from E. coli (γ-Proteobacteria class) confirmed that the only C-family replicative polymerase present in this bacterium (i.e, Pol III-α subunit, DnaE1) extends RNA primers, synthesized either by DnaG or by the RNA polymerase, to initiate leading and lagging strand synthesis (52). However, ∼60% of all sequenced bacteria have two, three or even four putative C-family polymerases (11). Bacteria of the Bacilli class have PolC and DnaE (also termed DnaE3). Initial genetic studies proposed that B. subtilis DnaE is restricted to lagging strand replication, based on results showing that in dnaE mutant cells the synthesis of plasmid pAMβ1 lagging strand, but not leading strand was deficient (19). Plasmid pAMβ1 replicates by the θ mechanism, but the initiation of DNA replication is not performed by DnaG priming, followed by extension of this primer by PolC and/or by DnaE (53). Here, RNA polymerase, aided by the plasmid-encoded initiator protein RepE, generates an RNA at the plasmid origin of replication (53,54). Then, DNA Pol I uses this leading strand RNA primer to generate a ∼200-nt long displaced loop structure, which is used as a signal for entry of the host PriA-dependent primosome, and the subsequent assembly of the full replisome (53,55). This replication restart reaction is already DNA primed, because the displaced loop structures contain the nascent leading strand paired to its template, and DnaG and DnaE will not be required for leading strand synthesis. Similarly, in the previous σ-type replication assays, done in vitro with B. subtilis and SPP1 replisomes, leading strand synthesis initiated at a 3’-OH end, pre-existing in the DNA template, and relied only on PolC (20,28). We show with our in vitro θ-type DNA replication system that the processive PolC enzyme cannot initiate leading strand synthesis in the absence of DnaE. Initiation requires DnaG and DnaE, which synthesize the hybrid RNA/DNA initiator primer, used by the DNA-dependent PolC polymerase to perform leading strand synthesis at oriL. These results show that the two replicative polymerases present in B. subtilis are not restricted to one strand.
We propose that in bacteria having two replicative polymerases of the C-family, as in Firmicutes, there is a division of labor between the two polymerases (Supplementary Figure S8). In initiation of unidirectional DNA replication, or when the 3΄-OH of the nascent leading strand is unavailable (for example when DNA replication needs to be restarted downstream of an unrepaired block) only one replisome is present. In SPP1, the replisome organizer, G38P, in concert with the helicase laoder, G39P, loads the hexameric replicative helicase, G40P, to the DNA (24). Here, we propose that one replicative helicase (G40P in the figure, but the same may occur with the host DnaC helicase) assembles two DnaG-DnaE complexes, enabling the priming of both leading and lagging strands for initiation of unidirectional replication (Supplementary Figure S8A). This is in agreement with previous studies that showed that DnaC, DnaE and DnaG form a ternary complex (21), and that a single hexamer of the E. coli replicative helicase was sufficient to coordinate priming by DnaG of both the leading and lagging strands (56). Similarly, in eukaryotes, Pol α is able to prime both strands in vitro (57), and Ctf4 can couple two molecules of Pol α to one CMG helicase (58). Once DnaG synthesizes the RNA primers, DnaE as an RNA-dependent DNA polymerase will extend these primers, and PolC will elongate the hybrid RNA–DNA primers at both, the leading and the lagging strand. However, when there is a free 3’-OH end, as once DNA replication has started, or at pausing sites, DnaG and DnaE will be devoted to lagging strand synthesis. In bidirectional replication, as during initiation of DNA replication in B. subtilis, two complete replisomes are loaded at the origin of replication (Supplementary Figure S8B), so that two DnaC-DnaG-DnaE complexes can be loaded, each on one strand. Then the first primer synthesized by a DnaE–DnaG complex moving in the clockwise direction away from oriC may become the leading strand for the counterclockwise moving replication fork and viceversa (52) (Supplementary Figure S8B).
An interesting open question in the field is how the DnaG–DnaE complex hands off the initiator hybrid RNA/DNA primer to PolC. DnaE is displaced by PolC, and this must occur early in vivo, because DnaE is error-prone and efficient at lesion bypass (15,16). Previous studies showed that PolC early gains access to primers synthesized by the DnaG–DnaE complex on the lagging-strand (20,59). However, the exact mechanism of polymerase switch is unknown. Experiments performed with the eukaryotic DnaG–DnaE counterpart, the Pol α enzyme, showed that its polymerase activity is inhibited by RFC/PCNA, the eukaryotic counterparts of the τ-complex and β respectively (60), and this inhibition may contribute to the mechanism of primer handhoff. However, the polymerase activity of DnaE is stimulated by β and the τ-complex (20,38). In this work, we analyzed whether other proteins located on the lagging strand (DnaG, and the SSBs) may contribute to polymerase hand-off. We found that DnaG stimulated PolC-mediated but inhibited DnaE-mediated leading strand synthesis. The inhibition by DnaG of DnaE-mediated DNA synthesis was not observed before, because the previous experiments were done with reactions having DnaE in the presence of DnaG, the DnaC helicase and its loader DnaI only, and not in the context of a full replisome (21). This inhibition by the primase was observed independently of which SSB (the bacterial SsbA or the viral G36P) was present in the reaction, and only on the leading strand. We propose that DnaG may contribute to removal of DnaE from the leading strand. Our results with DnaE are similar with results obtained with an E. coli in vitro system (which also has a DnaE enzyme, although from another family). They showed that primase lowered DNA synthesis in vitro (61). The stimulation of PolC-mediated leading strand synthesis by DnaG was not previously analyzed. We do not believe that this stimulation of DNA synthesis is just because in the presence of DnaG, G40P has higher helicase activity (29), allowing the DNA polymerases to move faster. The contrary result is observed in reactions with DnaG and DnaE as the only DNA polymerase. Besides, we did not detect any specific interaction between PolC and DnaG. Therefore, we favor the hypothesis that this stimulation could be mediated by an interaction of DnaG with PolC through other proteins of the replisome. One candidate is a four-protein interaction between the τ-complex, the SSB protein, DnaG and PolC.
Initiation of DNA replication requires priming of both strands, and in this work we have observed that the DnaG primase may actively contribute to DNA polymerase switch at the leading strand, by inhibiting DnaE and stimulating PolC leading strand synthesis. In addition, we found that the viral SSB, also stimulates PolC but inhibits DnaE at the leading strand, suggesting that the SSB may also contribute to the mechanism of polymerase switch. This result is different to our results previously published (28), because the DNA templates and conditions used were different. Here, we used a more physiological substrate, the synthetic nicked mini-circle, where coordinated leading and lagging strand synthesis is obtained, and the reactions were performed with one DNA polymerase, in the presence of the other replisome components, dNTPs and rNTPs. These reactions also showed that both SSBS (SsbA, and G36P), bound to the ssDNA region present at the lagging strand of the synthetic nicked mini-circle, stimulated leading strand synthesis mediated by PolC. Similarly, SSB bound to the lagging strand stimulated leading strand synthesis catalyzed by the E. coli DnaE1 enzyme (61). Our results suggest that a network of reactions collectively regulate the polymerase switch and speed and processivity of the Firmicutes replisomes.
In bacteria, the C-family of DNA polymerases comes in two major forms, PolC and DnaE. While PolC represents an evolutionary compact group, DnaE can be further subdivided into at least three groups (DnaE1–3) (62). In Firmicutes, PolC is accompanied by a DnaE-type polymerase, which, according to phylogenetic analysis can be either DnaE1 or DnaE3 (11). DnaE3 polymerases combined with PolC are found in class Bacilli (11,62). Our results and previous suggest that in bacteria having only one replicative polymerase of the DnaE1 family, as in E. coli the Pol III-α polymerase, this enzyme evolved to have fidelity, and extend both DNA and RNA primers (63). In contrast, B. subtilis DnaE (DnaE3), evolved to be error-prone (15,16), and specifically extend RNA primers. The inhibition of DnaE-mediated leading strand synthesis by DnaG primase is conserved in the two families (this work, and (61)). Firmicutes PolC polymerase evolved to have fidelity, to extend only DNA primers, and to be stimulated by DnaG on the leading strand. It will be of significant interest to determine which polymerase will extend DNA primers and perform leading strand-synthesis in classes Clostridia and Negativicutes of Firmicutes, which have DnaE1 in addition to PolC (11).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Charles S. McHenry (Univ. Boulder, USA), Begoña Carrasco (CNB-CSIC, Spain), Dagmar Klostermeier (University of Muenster, Germany) and Kenn Marians (Memorial Sloan Kettering Cancer Center, USA) for providing us with proteins. We thank María López for excellent technical assistance; and Juan C. Alonso and Charles S. McHenry for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitvidad (MINECO) [BFU2012–39879-C02-02, BFU2015–67065-P]. Funding for open access charge: MINECO.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ogawa T., Baker T.A., van der Ende A., Kornberg A.. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: contributions of RNA polymerase and primase. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:3562–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker T.A., Kornberg A.. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988; 55:113–123. [DOI] [PubMed] [Google Scholar]
- 3. Murakami S., Inuzuka N., Yamaguchi M., Yamaguchi K., Yoshikawa H.. Initiation of DNA replication in Bacillus subtilis. III. Analysis of molecular events involved in the initiation using a temperature-sensitive dna mutant. J. Mol. Biol. 1976; 108:683–704. [DOI] [PubMed] [Google Scholar]
- 4. Moriya S., Firshein W., Yoshikawa H., Ogasawara N.. Replication of a Bacillus subtilis oriC plasmid in vitro. Mol. Micro. 1994; 12:469–478. [DOI] [PubMed] [Google Scholar]
- 5. Rocha E.P. The replication-related organization of bacterial genomes. Microbiology. 2004; 150:1609–1627. [DOI] [PubMed] [Google Scholar]
- 6. Hu J., Zhao X., Yu J.. Replication-associated purine asymmetry may contribute to strand-biased gene distribution. Genomics. 2007; 90:186–194. [DOI] [PubMed] [Google Scholar]
- 7. Saha S.K., Goswami A., Dutta C.. Association of purine asymmetry, strand-biased gene distribution and PolC within Firmicutes and beyond: a new appraisal. BMC Genomics. 2014; 15:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goswami A., Roy Chowdhury A., Sarkar M., Saha S.K., Paul S., Dutta C.. Strand-biased gene distribution, purine assymetry and environmental factors influence protein evolution in Bacillus. FEBS Lett. 2015; 589:629–638. [DOI] [PubMed] [Google Scholar]
- 9. Inoue R., Kaito C., Tanabe M., Kamura K., Akimitsu N., Sekimizu K.. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet. Genom.: MGG. 2001; 266:564–571. [DOI] [PubMed] [Google Scholar]
- 10. Zhang G., Gao F.. Quantitative analysis of correlation between AT and GC biases among bacterial genomes. PloS One. 2017; 12:e0171408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Timinskas K., Balvociute M., Timinskas A., Venclovas C.. Comprehensive analysis of DNA polymerase III alpha subunits and their homologs in bacterial genomes. Nucleic Acids Res. 2014; 42:1393–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammond R.A., Barnes M.H., Mack S.L., Mitchener J.A., Brown N.C.. Bacillus subtilis DNA polymerase III: complete sequence, overexpression, and characterization of the polC gene. Gene. 1991; 98:29–36. [DOI] [PubMed] [Google Scholar]
- 13. Low R.L., Rashbaum S.A., Cozzarelli N.R.. Purification and characterization of DNA polymerase III from Bacillus subtilis. J. Biol. Chem. 1976; 251:1311–1325. [PubMed] [Google Scholar]
- 14. Sanjanwala B., Ganesan A.T.. DNA polymerase III gene of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:4421–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Chatelier E., Becherel O.J., d’Alencon E., Canceill D., Ehrlich S.D., Fuchs R.P., Janniere L.. Involvement of DnaE, the second replicative DNA polymerase from Bacillus subtilis, in DNA mutagenesis. J. Biol. Chem. 2004; 279:1757–1767. [DOI] [PubMed] [Google Scholar]
- 16. Bruck I., Goodman M.F., O’Donnell M.. The essential C family DnaE polymerase is error-prone and efficient at lesion bypass. J. Biol. Chem. 2003; 278:44361–44368. [DOI] [PubMed] [Google Scholar]
- 17. Flett F., de Mello Jungmann-Campello D., Mersinias V., Koh S.L., Godden R., Smith C.P.. A ‘gram-negative-type’ DNA polymerase III is essential for replication of the linear chromosome of Streptomyces coelicolor A3(2). Mol. Micro. 1999; 31:949–958. [DOI] [PubMed] [Google Scholar]
- 18. Paul S., Million-Weaver S., Chattopadhyay S., Sokurenko E., Merrikh H.. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013; 495:512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dervyn E., Suski C., Daniel R., Bruand C., Chapuis J., Errington J., Janniere L., Ehrlich S.D.. Two essential DNA polymerases at the bacterial replication fork. Science. 2001; 294:1716–1719. [DOI] [PubMed] [Google Scholar]
- 20. Sanders G.M., Dallmann H.G., McHenry C.S.. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010; 37:273–281. [DOI] [PubMed] [Google Scholar]
- 21. Rannou O., Le Chatelier E., Larson M.A., Nouri H., Dalmais B., Laughton C., Janniere L., Soultanas P.. Functional interplay of DnaE polymerase, DnaG primase and DnaC helicase within a ternary complex, and primase to polymerase hand-off during lagging strand DNA replication in Bacillus subtilis. Nucleic Acids Res. 2013; 41:5303–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pedre X., Weise F., Chai S., Luder G., Alonso J.C.. Analysis of cis and trans acting elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 1994; 236:1324–1340. [DOI] [PubMed] [Google Scholar]
- 23. Missich R., Weise F., Chai S., Lurz R., Pedre X., Alonso J.C.. The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol. 1997; 270:50–64. [DOI] [PubMed] [Google Scholar]
- 24. Ayora S., Stasiak A., Alonso J.C.. The Bacillus subtilis bacteriophage SPP1 G39P delivers and activates the G40P DNA helicase upon interacting with the G38P-bound replication origin. J. Mol. Biol. 1999; 288:71–85. [DOI] [PubMed] [Google Scholar]
- 25. Ayora S., Missich R., Mesa P., Lurz R., Yang S., Egelman E.H., Alonso J.C.. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 2002; 277:35969–35979. [DOI] [PubMed] [Google Scholar]
- 26. Alonso J.C., Tavares P., Lurz R., Trautner T.A.. Bacteriophage SPP1. 2006; 2nd edn, Oxford; NY: Oxford University Press. [Google Scholar]
- 27. Lo Piano A., Martinez-Jimenez M.I., Zecchi L., Ayora S.. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011; 160:1–14. [DOI] [PubMed] [Google Scholar]
- 28. Seco E.M., Zinder J.C., Manhart C.M., Lo Piano A., McHenry C.S., Ayora S.. Bacteriophage SPP1 DNA replication strategies promote viral and disable host replication in vitro. Nucleic Acids Res. 2013; 41:1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ayora S., Langer U., Alonso J.C.. Bacillus subtilis DnaG primase stabilises the bacteriophage SPP1 G40P helicase-ssDNA complex. FEBS Lett. 1998; 439:59–62. [DOI] [PubMed] [Google Scholar]
- 30. Martínez-Jiménez M.I., Mesa P., Alonso J.C.. Bacillus subtilis tau subunit of DNA polymerase III interacts with bacteriophage SPP1 replicative DNA helicase G40P. Nucleic Acids Res. 2002; 30:5056–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marians K.J., Soeller W., Zipursky S.L.. Maximal limits of the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. J. Biol. Chem. 1982; 257:5656–5662. [PubMed] [Google Scholar]
- 32. Gottler T., Klostermeier D.. Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J. Mol. Biol. 2007; 367:1392–1404. [DOI] [PubMed] [Google Scholar]
- 33. Bramhill D., Kornberg A.. A model for initiation at origins of DNA replication. Cell. 1988; 54:915–918. [DOI] [PubMed] [Google Scholar]
- 34. Wolanski M., Donczew R., Zawilak-Pawlik A., Zakrzewska-Czerwinska J.. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Micro. 2014; 5:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lanz M.A., Klostermeier D.. The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 2012; 40:10893–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hiasa H., DiGate R.J., Marians K.J.. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994; 269:2093–2099. [PubMed] [Google Scholar]
- 37. Peng H., Marians K.J.. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:8571–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bruck I., O’Donnell M.. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 2000; 275:28971–28983. [DOI] [PubMed] [Google Scholar]
- 39. Yuzhakov A., Kelman Z., O’Donnell M.. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999; 96:153–163. [DOI] [PubMed] [Google Scholar]
- 40. Hernandez A.J., Lee S.J., Richardson C.C.. Primer release is the rate-limiting event in lagging-strand synthesis mediated by the T7 replisome. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Costes A., Lecointe F., McGovern S., Quevillon-Cheruel S., Polard P.. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 2010; 6:e1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Learn B., Karzai A.W., McMacken R.. Transcription stimulates the establishment of bidirectional lambda DNA replication in vitro. Cold Spring Harbor Symp. Quant. Biol. 1993; 58:389–402. [DOI] [PubMed] [Google Scholar]
- 43. Mensa-Wilmot K., Seaby R., Alfano C., Wold M.C., Gomes B., McMacken R.. Reconstitution of a nine-protein system that initiates bacteriophage lambda DNA replication. J. Biol. Chem. 1989; 264:2853–2861. [PubMed] [Google Scholar]
- 44. Zylicz M., Ang D., Liberek K., Georgopoulos C.. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989; 8:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alfano C., McMacken R.. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J. Biol. Chem. 1989; 264:10709–10718. [PubMed] [Google Scholar]
- 46. Bramhill D., Kornberg A.. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988; 52:743–755. [DOI] [PubMed] [Google Scholar]
- 47. Wold M.S., Mallory J.B., Roberts J.D., LeBowitz J.H., McMacken R.. Initiation of bacteriophage lambda DNA replication in vitro with purified lambda replication proteins. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:6176–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bleichert F., Botchan M.R., Berger J.M.. Mechanisms for initiating cellular DNA replication. Science. 2017; 355:eaah6317. [DOI] [PubMed] [Google Scholar]
- 49. Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J.D., McMacken R.. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:7638–7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ayora S., Weise F., Mesa P., Stasiak A., Alonso J.C.. Bacillus subtilis bacteriophage SPP1 hexameric DNA helicase, G40P, interacts with forked DNA. Nucleic Acids Res. 2002; 30:2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Magnan D., Bates D.. Regulation of DNA Replication Initiation by Chromosome Structure. J. Bact. 2015; 197:3370–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hiasa H., Marians K.J.. Primase couples leading- and lagging-strand DNA synthesis from oriC. J. Biol. Chem. 1994; 269:6058–6063. [PubMed] [Google Scholar]
- 53. Bruand C., Le Chatelier E., Ehrlich S.D., Janniere L.. A fourth class of theta-replicating plasmids: the pAMbeta 1 family from gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:11668–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Le Chatelier E., Janniere L., Ehrlich S.D., Canceill D.. The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMbeta 1 from Gram-positive bacteria. J. Biol. Chem. 2001; 276:10234–10246. [DOI] [PubMed] [Google Scholar]
- 55. Bruand C., Ehrlich S.D., Janniere L.. Primosome assembly site in Bacillus subtilis. EMBO J. 1995; 14:2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heller R.C., Marians K.J.. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006; 439:557–562. [DOI] [PubMed] [Google Scholar]
- 57. Georgescu R.E., Schauer G.D., Yao N.Y., Langston L.D., Yurieva O., Zhang D., Finkelstein J., O’Donnell M.E.. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015; 4:e04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simon A.C., Zhou J.C., Perera R.L., van Deursen F., Evrin C., Ivanova M.E., Kilkenny M.L., Renault L., Kjaer S., Matak-Vinkovic D. et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014; 510:293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan Q., Dohrmann P.R., Sutton M.D., McHenry C.S.. DNA Polymerase III, but not Polymerase IV, must be bound to a tau-containing DnaX complex to enable exchange into replication forks. J. Biol. Chem. 2016; 291:11727–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mossi R., Keller R.C., Ferrari E., Hubscher U.. DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J. Mol. Biol. 2000; 295:803–814. [DOI] [PubMed] [Google Scholar]
- 61. Georgescu R.E., Yao N., Indiani C., Yurieva O., O’Donnell M.E.. Replisome mechanics: lagging strand events that influence speed and processivity. Nucleic Acids Res. 2014; 42:6497–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao X.Q., Hu J.F., Yu J.. Comparative analysis of eubacterial DNA polymerase III alpha subunits. Genom. Proteom. Bioinf. 2006; 4:203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McHenry C.S. Breaking the rules: bacteria that use several DNA polymerase IIIs. EMBO Rep. 2011; 12:408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.