Abstract
Given the importance of meiotic recombination in biology, there is a need to develop robust methods to estimate meiotic recombination rates. A popular approach, called the Marey map approach, relies on comparing genetic and physical maps of a chromosome to estimate local recombination rates. In the past, we have implemented this approach in an R package called MareyMap, which includes many functionalities useful to get reliable recombination rate estimates in a semi-automated way. MareyMap has been used repeatedly in studies looking at the effect of recombination on genome evolution. Here, we propose a simpler user-friendly web service version of MareyMap, called MareyMap Online, which allows a user to get recombination rates from her/his own data or from a publicly available database that we offer in a few clicks. When the analysis is done, the user is asked whether her/his curated data can be placed in the database and shared with other users, which we hope will make meta-analysis on recombination rates including many species easy in the future.
Keywords: crossover rates, Marey map approach, R package, Shiny, web applications
Meiotic recombination rates can vary quite a lot along chromosomes (Nachman 2002) and a good description of this variation in model organisms (humans, Drosophila, Caenorhabditis elegans, Arabidopsis) and in nonmodel organisms is important to understand both the process of meiosis and its evolutionary implications. In particular, recombination is considered one of the main factors influencing genome architecture (e.g., Gaut etal. 2007; Lynch 2007; Charlesworth and Campos 2014; Ellegren 2014; Melamed-Bessudo etal. 2016) and studying the influence of recombination on genome evolution is a hot topics in molecular evolution and genomics.
A key methodological point is of course the estimation of the local meiotic recombination rates. Several approaches have been developed to obtain such estimates. Sperm-typing gives very fine-scale and reliable estimates but this approach is costly, difficult to apply on a large-scale and only provides estimates in males (Kauppi etal. 2004, 2009). Studying SNPs and linkage disequilibrium from natural population data can give population-genetic-based estimates of recombination (Stumpf and McVean 2003; Coop and Przeworski 2007). If dense and genome-wide SNP data are available in a species, as for example in humans, this approach will return fine-scale estimates for the entire genome, and it has been very successful in identifying hot spots in humans and in other well-studied organisms (e.g., Ptak etal. 2004; Myers etal. 2005). Even more recently, genome-wide SNP data have been obtained for pedigrees in humans, yeast, Drosophila and Arabidopsis, which have allowed the direct and very precise study of recombination events that occurred in those pedigrees and have resulted in arguably the best estimates so far (Coop etal. 2008; Mancera etal. 2008; Comeron etal. 2012; Yang etal. 2012). With next-generation sequencing, obtaining such genome-wide SNP data is becoming easier, but at present this kind of data are not available for many organisms. Another approach, called the Marey map approach, relies on comparing physical and genetic maps of a given chromosome to estimate local recombination rates, which are given by the local slope of the curve adjusted to the datapoints (Chakravarti 1991). Historically, many studies using local recombination rate estimates have used this approach. Also, all methods have pros and cons (e.g., Buard and de Massy 2007) and the Marey map approach is still very popular as there are many more species for which both physical and genetic maps are available (the raw material for the Marey map approach) compared with species for which genome-wide SNP data are available.
Several tools implementing the Marey map approach have been developed but they focus on particular organisms: Drosophila (Fiston-Lavier etal. 2010) or mice and rat (Voigt etal. 2004) for example. In these tools, the users cannot work on their own data. The only tool for inferring local recombination rates in any organisms in a semiautomated way is a tool called MareyMap that we developed a few years ago (Rezvoy etal. 2007) and have updated recently (Siberchicot etal. 2015). MareyMap requires the user to have files for physical and genetic maps for each chromosome in a given species. The files are then uploaded and a number of functionalities allow the user to build a Marey map plot, to clean aberrant points from the map, to infer local estimates using different methods and to export estimates in different formats (Rezvoy etal. 2007). MareyMap is an R package with a tcl/tk graphical interface, which works well but requires the user to have R (R Core Team 2016) installed and to be familiar with installing R packages. Despite R being free and open-source, we have noticed that a significant number of users have experienced difficulties in installing the MareyMap R package when they are not frequent R users. Moreover, the many options proposed by MareyMap are useful for experienced users but can be confusing for beginners with the package and the Marey map approach.
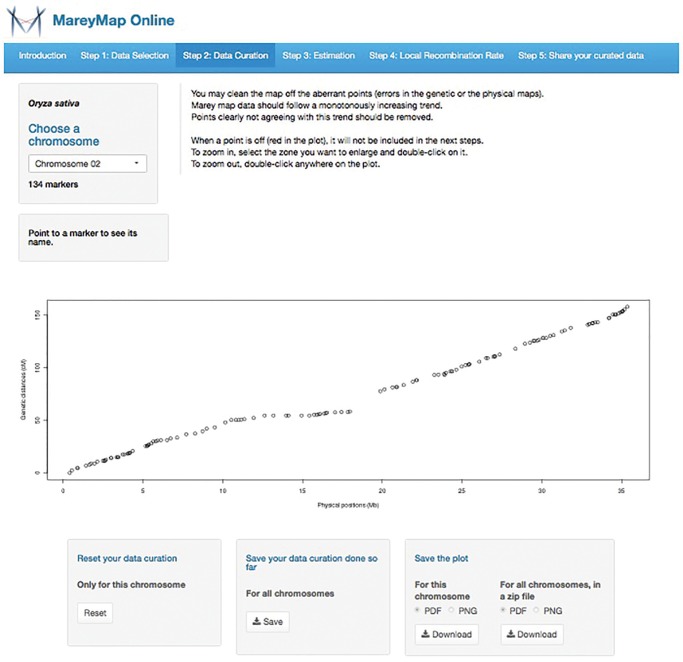
Here, we describe a web service called MareyMap Online that we have developed as a user-friendly version of MareyMap. MareyMap Online proposes a simplified version of MareyMap with five main steps: data uploading, data cleaning, recombination rate estimation, export and data sharing (see menu bar in fig. 1), which are described below. The user is guided through these steps with only essential options proposed and clear guidelines when there are choices to make. An important feature offered by MareyMap Online is to be able to pick some Marey map data from a publicly accessible database that we provide. This database currently includes 8 organisms: Arabidopsis thaliana, Caenorhabditis elegans, Danio rerio (zebrafish), Drosophila melanogaster, Homo sapiens, Oryza sativa (rice), Oryzias latipes (medaka), and Saccharomyces cerevisiae. MareyMap Online will ask each user the permission to keep her/his curated Marey map data once the analysis is done, which will then be stored in our database. We thus expect the database to include many more species in the future, which can be useful for meta-analysis on recombination and genome evolution for instance. Such studies are currently difficult as this kind of database does not exist and one needs to collect the Marey map data through literature search.
Fig. 1.
—Screenshot of a Marey map plot (step 2) in the MareyMap Online web service. The five steps are shown in the menu bar.
MareyMap Online has been implemented from the MareyMap R package with shiny, an R package which offers a framework to develop web applications in R (Chang etal. 2016). As explained above, the user follows a pipeline step by step and is asked first to pick a Marey map dataset from the database or to upload her/his own dataset. The format required is quite simple and an example is provided. In step 2, a plot comparing physical (X axis) and genetic (Y axis) map can be seen for each chromosome (fig. 1). Maps often contain errors. These can be easily spotted on a Marey map as they will clearly disrupt the expected overall monotonously increasing relationship between physical and genetic positions. Such erroneous datapoints need to be removed before further analysis and an interactive cleaning tool is proposed to the user in order to do it. Step 3 consists in selecting an estimation method to get local recombination rates. This is a very sensitive step. Users are proposed three different methods already available in the MareyMap R package: sliding windows, loess and cubic splines (Rezvoy etal. 2007; Siberchicot etal. 2015). Cubic splines are the best method to capture relatively fine-scale variations whereas sliding large windows will return large-scale estimates. Which is the right method for an analysis will depend on the features of the data. Key points are the density of the markers and the noise in the data. We offer some guidelines to help the users in picking up the best method for her/his data. In particular, a good criterion to check if a method worked well is to see whether no negative estimates have been obtained; negative estimates are indeed clear signs that another method with more “smoothing” effect is required. At step 4, the user will view the estimates and automatically generated warnings will tell her/him when negative values are present among the estimates. Back-and-forth adjustments between steps 3 and 4 may be needed to pick the estimation method most adapted to the user data. Then, export options are proposed. A useful one is the possibility to upload a list of physical positions and retrieve the estimates for all these positions, which is very useful for a downstream analysis of all the genes in a genome for example. The user can also query some positions to get estimates. At step 5, the user will be asked whether she/he is willing to authorise storage of her/his curated Marey map data in our public database.
In conclusion, MareyMap Online provides a simplified user-friendly pipeline for getting local recombination rate estimates. It is available online and requires no installation. We also offer a database with curated Marey map data, which we hope will grow in the future thanks to the MareyMap Online users. Importantly, for well-experienced users the R package will still be maintained and will offer the full set of options included in MareyMap.
Acknowledgments
We thank Stéphane Delmotte and Bruno Spataro who maintain the LBBE shiny server used for that project.
Literature Cited
- Buard J, de Massy B.. 2007. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet. 23(6): 301–309. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. 1991. A graphical representation of genetic and physical maps: the Marey map. Genomics 11(1): 219–222. [DOI] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire J, Xie Y, McPherson J.. 2016. shiny: Web application framework for R. R package version 0.14.2. Available from: https://cran.r-project.org/web/packages/shiny/index.html, last accessed September 11, 2017.
- Charlesworth B, Campos JL.. 2014. The relations between recombination rate and patterns of molecular variation and evolution in Drosophila. Annu Rev Genet. 48: 383–403. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S.. 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8(10): e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M.. 2007. An evolutionary view of human recombination. Nat Rev Genet. 8(1): 23–34. [DOI] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M.. 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319(5868): 1395–1398. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1): 51–63. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA.. 2010. Drosophila melanogaster recombination rate calculator. Gene 463(1–2): 18–20. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK.. 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 8(1): 77–84. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S.. 2004. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 5(6): 413–424. [DOI] [PubMed] [Google Scholar]
- Kauppi L, May CA, Jeffreys AJ.. 2009. Analysis of meiotic recombination products from human sperm. Methods Mol Biol. 557: 323–355. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sundarland (MA): Sinauer Associates Inc.
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM.. 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454(7203): 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Shilo S, Levy AA.. 2016. Meiotic recombination and genome evolution in plants. Curr Opin Plant Biol. 30: 82–87. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P.. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310(5746): 321–324. [DOI] [PubMed] [Google Scholar]
- Nachman MW. 2002. Variation in recombination rate across the genome: evidence and implications. Curr Opin Genet Dev. 12(6): 657–663. [DOI] [PubMed] [Google Scholar]
- Ptak SE, et al. 2004. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2: 0849–0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Available from: https://www. R-project. org/, last accessed September 11, 2017. [Google Scholar]
- Rezvoy C, Charif D, Gueguen L, Marais GA.. 2007. MareyMap: an R-based tool with graphical interface for estimating recombination rates. Bioinformatics 23(16): 2188–2189. [DOI] [PubMed] [Google Scholar]
- Siberchicot A, Rezvoy C, Charif D, Gueguen L, Marais G.. 2015. The MareyMap package. Version 1.3.3. CRAN. Available from: https://cran.r-project.org/web/packages/MareyMap/index.html, last accessed September 11, 2017.
- Stumpf MPH, McVean GA.. 2003. Estimating recombination rates from population-genetic data. Nat Rev Genet. 4(12): 959–968. [DOI] [PubMed] [Google Scholar]
- Voigt C, Moller S, Ibrahim SM, Serrano-Fernandez P.. 2004. Non-linear conversion between genetic and physical chromosomal distances. Bioinformatics 20(12): 1966–1967. [DOI] [PubMed] [Google Scholar]
- Yang S, et al. 2012. Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci U S A. 109(51): 20992–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]