Abstract
Studies of regulatory activity and gene expression have revealed an intriguing dichotomy: There is substantial turnover in the regulatory activity of orthologous sequences between species; however, the expression level of orthologous genes is largely conserved. Understanding how distal regulatory elements, for example, enhancers, evolve and function is critical, as alterations in gene expression levels can drive the development of both complex disease and functional divergence between species. In this study, we investigated determinants of the conservation of regulatory enhancer activity for orthologous sequences across mammalian evolution. Using liver enhancers identified from genome-wide histone modification profiles in ten diverse mammalian species, we compared orthologous sequences that exhibited regulatory activity in all species (conserved-activity enhancers) to shared sequences active only in a single species (species-specific-activity enhancers). Conserved-activity enhancers have greater regulatory potential than species-specific-activity enhancers, as quantified by both the density and diversity of transcription factor binding motifs. Consistent with their greater regulatory potential, conserved-activity enhancers have greater regulatory activity in humans than species-specific-activity enhancers: They are active across more cellular contexts, and they regulate more genes than species-specific-activity enhancers. Furthermore, the genes regulated by conserved-activity enhancers are expressed in more tissues and are less tolerant of loss-of-function mutations than those targeted by species-specific-activity enhancers. These consistent results across various stages of gene regulation demonstrate that conserved-activity enhancers are more pleiotropic than their species-specific-activity counterparts. This suggests that pleiotropy is associated with the conservation of regulatory across mammalian evolution.
Keywords: enhancer, evolutionary conservation, pleiotropy
Introduction
Mammalian genomes harbor hundreds of thousands of regulatory enhancer sequences that are essential for directing spatiotemporal patterns of gene expression during development and differentiation (Shlyueva etal. 2014; Roadmap Epigenomics Consortium etal. 2015). Enhancers contain binding sites for transcription factors (TFs), the binding patterns of which regulate gene expression. Genetic variants that disrupt the functionality of enhancer sequences, and thereby alter gene expression levels, are major contributors to both speciation events (Romero etal. 2012) and risk for complex disease (Maurano etal. 2012; Corradin and Scacheri 2014). Given their functional importance, there is considerable interest in better understanding the evolutionary processes underlying both enhancer sequence conservation and, more importantly, enhancer activity conservation.
Much of the transcriptional machinery responsible for regulating gene expression levels is conserved across species. For example, TFs and the sequence motifs they recognize are often conserved between human and fly (Amoutzias etal. 2007; Wei etal. 2010; Cheng etal. 2014; Nitta etal. 2015). Consequently, a sequence’s TF binding profile across different species is typically similar (Wilson etal. 2008); however, the enhancer activity of orthologous sequences is less consistent. Ritter etal. (2010) examined the activity profiles of 41 pairs of conserved regulatory elements between human and zebrafish. Roughly a third of these pairs demonstrated consistent activity patterns between species, but the majority did not (Ritter etal. 2010). Villar etal. (2015) demonstrated that regulatory activity turnover is pervasive between even more closely related species; only 1% of human liver enhancers had conserved activity across 20 mammals. Thus, despite similarity in TFs and their binding motifs, orthologous sequences can have highly variable enhancer activity across species.
Pleiotropy—broadly defined as a single genetic locus influencing multiple traits (Paaby and Rockman 2013)—has been proposed to contribute to the evolutionary conservation of both genes and regulatory activity (Galis etal. 2002; He and Zhang 2006; Cheng etal. 2014; Papakostas etal. 2014; Chesmore etal. 2016; Huang etal. 2017). Mutations in pleiotropic regions face a trade-off: Variants potentially advantageous to one function may be deleterious for others (Guillaume and Otto 2012). Consequently, pleiotropic regions may be more likely to be constrained by selection than nonpleiotropic regions. The relationship between pleiotropy and conservation has been demonstrated on several scales. Highly pleiotropic genes are more likely to have conserved orthologs in other species (He and Zhang 2006) and are more likely to have constrained expression levels (Papakostas etal. 2014). In the context of regulatory functions, binding sites for transcription factors that are observed in multiple cellular contexts, and are therefore presumed to be more pleiotropic, are more likely to be conserved between human and mouse (Cheng etal. 2014). Thus, we predicted a positive relationship between pleiotropy and enhancer activity conservation across species.
In this study, we investigated whether enhancers with conserved regulatory activity between species were more likely to be pleiotropic than enhancers with similarly alignable sequences, but species-specific regulatory activity. We quantified pleiotropy at several stages of human gene regulation: Density and diversity of TF binding motifs, extent of regulatory activity across cellular contexts, and number of target genes. We investigated these measures of pleiotropy in liver enhancers recently identified from genome-wide histone modification profiles across ten diverse mammalian species. We compared two groups of sequences present and alignable across all ten species: 1) those with liver activity in all ten mammals considered (conserved-activity) and 2) those with liver enhancer activity in only one species (species-specific-activity). We found that the conserved-activity enhancers consistently had stronger evidence of more, and more diverse, regulatory functions than the species-specific-activity enhancers. We also demonstrated that machine learning classifiers can accurately distinguish these two classes of enhancers using these measures of functional potential and diversity. Overall, our results argue that conserved-activity enhancers are more pleiotropic than species-specific-activity enhancers with similar levels of sequence alignability. This suggests that more diverse functional activity contributes to conserved activity across species, and that conserved activity may facilitate acquisition of additional functions.
Materials and Methods
Identifying Enhancers and Cross-Species Alignments
Enhancers were previously identified by Villar etal. (2015) in primary liver tissues collected from 20 mammalian species. Using ChIP-seq, Villar etal. (2015) identified H3K27ac and H3K4me3 peaks across the entire genome; putative enhancers were defined as genomic regions exclusively containing H3K27ac peaks (i.e., H3K27ac peaks that did not overlap H3K4me3 peaks) found in at least two representatives of the species. We restricted our analysis to the following ten species with high quality genome builds: Human (Homo sapiens), macaque (Macaca mulatta), marmoset (Callithrix jacchus), mouse (Mus musculus), rat (Rattus norvegicus), rabbit (Oryctolagus cuniculus), cow (Bos taurus), pig (Sus scrofa), dog (Canis familiaris), and cat (Felis catus). Cross-species comparisons to identify whether or not a sequence was present and active in other species were performed in reference to the eutherian mammal EPO alignment. To determine enhancer activity in cellular contexts other than the liver, we used enhancers identified by CAGE by the FANTOM Consortium (http://enhancer.binf.ku.dk/presets/; last accessed September 21, 2017) (Andersson etal. 2014).
Standardizing Enhancer Length
To avoid confounding by length, we restricted enhancer sequences used in the majority of analyses to 5 kb centered on the middle of the enhancer. If a putative enhancer was shorter than 5 kb, we extended the enhancer boundaries symmetrically in both directions until it was 5 kb. The length of 5 kb was selected as it the intermediate point between the average length of the conserved-activity enhancers (7,895 bp) and species-specific-activity enhancers (2,545 bp). For sequences shorter than 5 kb, standardizing length could potentially dilute the density of TF binding motifs; however, it would increase the likelihood of overlapping enhancers in multiple cellular contexts or mapping to additional gene targets. In other words, it would have inconsistent effects on measures of pleiotropy; only in the TF binding motif analysis would the potential for pleiotropy for shorter sequences possibly be reduced. We demonstrated that the decreased density of TF binding motifs in human-specific-activity enhancers was not a product of length standardization (supplementary fig. 3, Supplementary Material online). Consequently, any influence of the length standardization in the subsequent analyses of breadth of activity and gene targets would only increase the likelihood of pleiotropic effects in species-specific-activity enhancers. This would reduce our power to detect increased evidence for pleiotropy in conserved-activity enhancers, but would not result in false positives.
Identification of TF Binding Motifs
We identified TF binding motifs using four databases derived across diverse sets of species and using different experimental approaches: Motifs derived from ChIP-Seq peaks in human by the ENCODE Project (n = 2,065) (Kapur etal. 2011); motifs derived from ChIP-Seq peaks and HT-SELEX in vertebrates by JASPAR (Core Vertebrates) (n = 519) (Mathelier etal. 2016); motifs derived from ChIP-Seq peaks in human and HT-SELEX by HOCOMOCOv9 (n = 426) (Kulakovskiy etal. 2016); and motifs derived from ChIP-Seq peaks and HT-SELEX from human and mouse (n = 843) (Jolma etal. 2013). For each of these data sets, we scanned the putative enhancer sequences for motif occurrences using FIMO (Grant etal. 2011), using the default settings and requiring a q-value of <0.1 to be considered a match.
SVM Classifiers
We trained SVM classifiers to distinguish between conserved-activity enhancers and species-specific-activity enhancers using three different kinds of features: TF binding motif frequencies, k-mer spectra, and functional genomics annotations. For the TF motif-based classifiers, each enhancer was associated with a feature vector that included the frequency of all possible TF motifs in its sequence. We then trained a linear SVM to distinguish the two classes of enhancers. The kernel was normalized using the square root diagonal kernel normalizer. All training and testing was done in the EnhancerFinder framework (Erwin etal. 2014).
K-mer spectra quantify sequence content with the frequency of each unique nucleotide combination of length k in the enhancer sequence. We determined the k-mer spectra of each enhancer sequence using EnhancerFinder (Erwin etal. 2014); the kernel was normalized using the square root diagonal kernel normalizer. The reverse complement of the sequence was considered (i.e., counts for ATG and CAT were combined). We examined various k (4, 5, 6, 7, 8) and found consistent results across settings (supplementary fig. 4, Supplementary Material online).
To investigate whether functional genomics annotations were predictive of enhancer activity conservation, we used data collected by the ENCODE Project. Specifically, we used DNase-Seq, histone modifications, and TFBS Peaks (SPP) curated by the ENCODE Analysis Hub at the European Bioinformatics Institute (https://genome.ucsc.edu/ENCODE/downloads.html; last accessed September 21, 2017). We considered each genome-wide annotation as a binary feature, and each enhancer was assigned 0 if it did not overlap an element of the annotation set or 1 if it did overlap. Training and testing of the functional genomics classifier was also carried out in the EnhancerFinder framework (Erwin etal. 2014).
Target Gene Mapping and Analysis of Gene Expression across Contexts
We used two methods to map the enhancers to their target genes: 1) GTEx eQTL association based target gene mapping. We first identified SNPs in the enhancer regions of interest and SNPs in high linkage disequilibrium with them (r2 > 0.9, based the 1000 Genomes EUR super population). Then, using expression data from GTEx, we considered genes for which these SNPs were eQTL to be putative target genes (The GTEx Consortium 2015). 2) FANTOM enhancer–TSS associations. The FANTOM consortium released a set of target predictions for each of their predicted transcribed enhancers based on the coexpression of the enhancer and genes across tissues (Andersson etal. 2014). We overlapped each liver enhancer of interest with the FANTOM enhancers. We then considered any genes associated with an overlapping FANTOM enhancer as putative target genes. To analyze the breadth of activity of target genes, we used the median Reads Per Kilobase of transcript per Million mapped reads (RPKMs) for genes from the GTEx v6 RNA-Seq data, which includes 53 types of tissue (The GTEx Consortium 2015).
Results
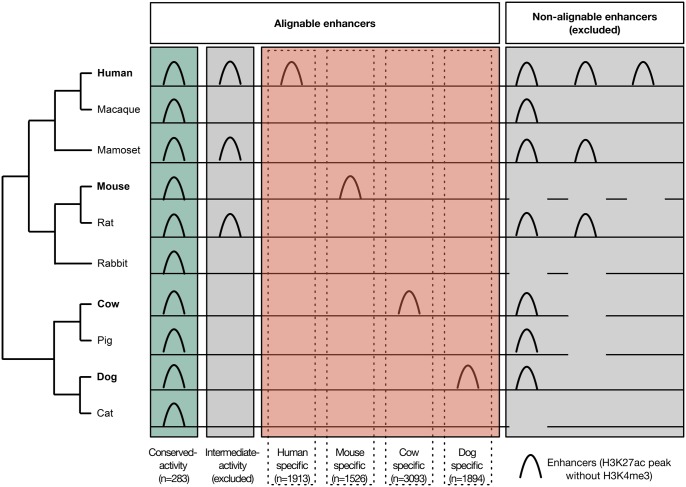
In this study, we explored attributes that distinguish genomic regions with both alignable sequence and regulatory activity across diverse mammals from those with similarly alignable sequences, but regulatory activity isolated to a single species. We analyzed genome-wide maps of histone modifications in primary liver tissue from ten mammals to quantify the regulatory activity conservation spectrum for liver regulatory sequences. Following Villar etal. (2015), we defined regulatory activity as peaks of H3K27ac histone modifications without the H3K4me3 modification. As histone modifications are correlated with enhancer activity in reporter assays (Creyghton etal. 2010; Nord etal. 2013; Villar etal. 2015), we refer to these sequences as enhancers for brevity. As illustrated in figure 1, we considered two enhancer sets of interest: Sequences that can be aligned across the genomes of ten mammalian species with evidence of enhancer activity in each species (conserved-activity enhancers; n = 283) and sequences that can be aligned across the ten species with evidence of enhancer activity exclusively in a single species (species-specific-activity enhancers). We examined species-specific-activity liver enhancer sets across four different mammalian species: Human (n = 1,913), mouse (n = 1,526), dog (n = 1,894), and cow (n = 3,093).
Fig. 1.
—Defining the enhancers analyzed in this study. We analyzed enhancers previously identified in primary liver tissue from ten mammals (Villar etal. 2015). Enhancers were defined on the basis of histone modifications (presence of H3K27ac peaks, absence of H3K4me3 peak). We analyzed sequences alignable across all ten species to disentangle sequence conservation from regulatory activity conservation across species. To identify trends that would hold across all mammals, we restricted our analysis to extremes of the activity conservation spectrum: conserved-activity enhancers (green), and species-specific-activity enhancers (red). We considered species-specific-activity enhancers from human, mouse, cow, and dog to represent a diverse array of clades.
Conserved-Activity Enhancers Have Greater Density and Diversity of TF Binding Motifs than Species-Specific-Activity Enhancers
The differential enhancer activity of alignable sequences may be attributable to differences in sequence properties that determine their regulatory potential, as quantified by both the density of TF binding motifs and the diversity of distinct TFs with binding motifs. Within a species, enhancers with a greater density of TF binding motifs are both stronger (Erceg etal. 2014) and more robust to disruptive genetic variation (Ludwig etal. 2011). We hypothesized that these principles generalize to enhancer conservation between species. To investigate this, we scanned all enhancer sequences for matches to a curated set of nonredundant TF binding motifs from the JASPAR database (Mathelier etal. 2016). Unless otherwise noted, enhancers were length standardized (Materials and Methods) to avoid confounding.
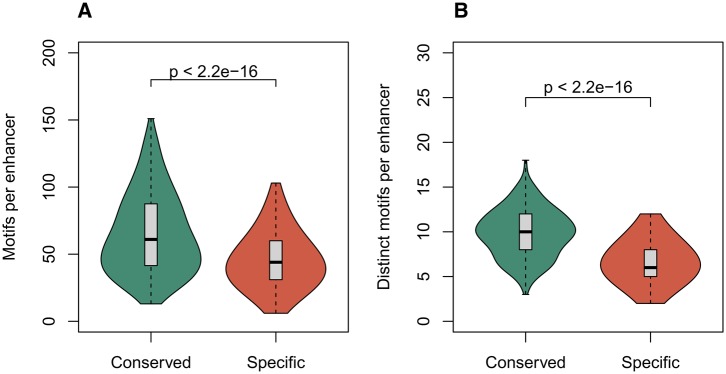
Conserved-activity liver enhancers have a greater density of TF binding motifs than human-specific-activity enhancers (fig. 2A; median: 61 versus 44 per enhancer; Mann–Whitney U (MWU) test, P < 2.2 × 10−16). Moreover, conserved-activity enhancers contain binding sites for almost double the number of distinct TFs (fig. 2B; median: 10 versus 6 per enhancer; MWU test, P < 2.2 × 10−16). This finding is robust across other databases of TF binding motifs, including motifs from the ENCODE Project (Kapur etal. 2011), HOCOMOCO (Kulakovskiy etal. 2016), and SELEX studies (Jolma etal. 2013) (supplementary fig. 1, Supplementary Material online). Furthermore, the other species-specific-activity (mouse, dog, and cow) enhancers also had both lower density and diversity of TF binding motifs relative to conserved-activity enhancers (supplementary fig. 2, Supplementary Material online). This trend also was consistent when enhancers were not length standardized (supplementary fig. 3, Supplementary Material online). Thus, conserved-activity enhancers have both a greater density and diversity of TF binding motifs than species-specific-activity enhancers, across multiple species and TF motif databases.
Fig. 2.
—Conserved-activity enhancers have greater TF binding motif density and diversity than species-specific-activity enhancers. We quantified the total number of TF binding motifs (JASPAR, Core Vertebrates [Mathelier etal. 2016]) and of distinct TF motifs within the enhancer sequences. The 283 conserved-activity enhancers have (A) significantly more TF binding motifs per enhancer (median of 61 vs. 44) and (B) binding motifs for significantly more distinct TFs (median of 10 vs. 6) than the 1,913 human-specific activity enhancers. Each box covers the first through third quartiles, and the whiskers extend to the most extreme data point within 1.5 times the interquartile range of the full distribution. The distributions are summarized using the R vioplot package with default parameters applied to each distribution with boxplot outliers removed. The enhancers were standardized by length and compared with the Mann–Whitney U test.
We next examined whether differences in TF binding motif profiles were sufficient to distinguish conserved-activity enhancers from species-specific-activity enhancers in a machine learning framework. First, we trained linear support vector machine (SVM) classifiers with conserved-activity enhancers as positives and species-specific-activity enhancers as negatives using the frequency of each distinct TF binding motif in the enhancer sequence as features. We performed 10-fold cross validation, computed receiver operator characteristic (ROC) curves, and evaluated classifier performance by the area under the ROC curve (auROC).
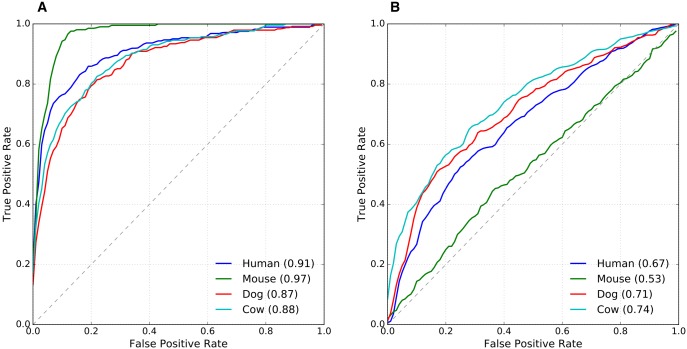
The classifiers accurately discriminated the conserved-activity enhancers from the species-specific-activity enhancers in each species (auROC: 0.88–0.97, fig. 3A). We hypothesize that the particularly strong performance of the mouse classifier may be due to rodent-specific differences in the genomic GC content distribution compared with other mammals (Romiguier etal. 2010). To benchmark the performance of the classifiers, we ranked enhancers by the density of TF binding motifs in the sequence and evaluated the predictive ability of this single feature (fig. 3B). Performance notably decreased when only considering the density of TF binding motifs (auROC: 0.53–0.74) for all species, especially mouse. Other approaches for quantifying enhancer sequence properties, such as k-mer spectra (Materials and Methods), were not as effective at predicting the conservation of regulatory activity (supplementary fig. 4, Supplementary Material online). These results indicate that not only do conserved-activity enhancers have a greater density and diversity of TF binding motifs than species-specific-activity enhancers, but the occurrence patterns of specific TF binding motifs are informative about the conservation of enhancer activity across species.
Fig. 3.
—TF binding motif patterns can distinguish between conserved-activity and species-specific-activity enhancers. (A) In each species (human, mouse, dog, and cow), we trained SVM classifiers to distinguish conserved-activity from species-specific-activity enhancers using the frequency of each TF binding motif individually as features. (B) For comparison, we attempted to distinguish the two classes of enhancers based only on the total number of TF binding motifs. The ROC curves display classifier performance, and the area under the curves is provided in the legend.
Conserved-Activity Enhancers Are Active in More Cellular Contexts than Human-Specific-Activity Enhancers
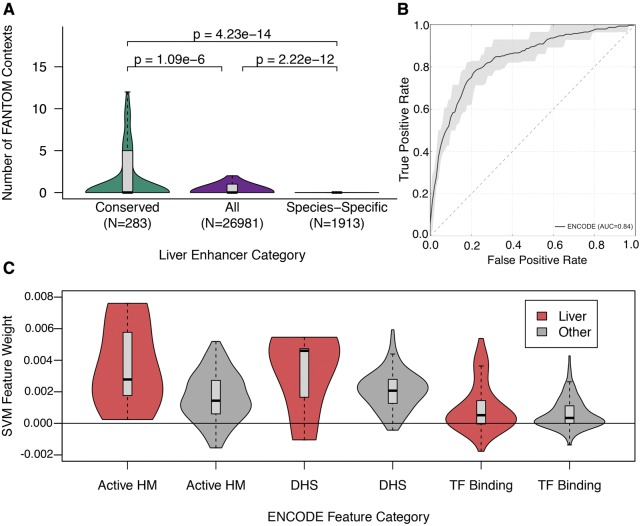
We next investigated whether the greater density and diversity of TF binding motifs of conserved-activity enhancers translated to increased regulatory activity across biological contexts within a species. We focused on human, as enhancers have been identified in a more diverse set of biological contexts for human than other species. We used enhancers in 108 cellular contexts identified by the FANTOM consortium, which used cap analysis of gene expression (CAGE) assays to identify bi-directionally transcribed “eRNA” transcripts (Andersson etal. 2014). On average, conserved-activity enhancers overlapped an active FANTOM enhancer in more than seven cellular contexts, which was double the number of cellular contexts expected from all human liver enhancers (mean: 7.2 vs. 3.6 per enhancer; P = 1.09 × 10−6, MWU test) and almost quadruple the number of active contexts for human-specific-activity enhancers (mean: 7.2 vs. 1.9 per enhancer; P = 2.22 × 10−12, MWU test) (fig. 4A). We next tested whether specific cellular contexts in FANTOM drove this enrichment by evaluating the overlap with enhancers from each FANTOM context separately. Conserved-activity enhancers were significantly more likely to overlap FANTOM enhancers relative to all human liver enhancers in 36 of 108 cellular contexts, and relative to human-specific-activity enhancers in 72 of 108 cellular contexts (P < 0.05 after Bonferroni correction, Fisher’s exact test) (supplementary table 1, Supplementary Material online). The converse—significant depletion of conserved-activity enhancers relative to all enhancers, or human-specific-activity enhancers—was never observed. These results demonstrate that conservation of activity across species within one cellular context (the liver) is positively correlated with the breadth of activity across other cellular contexts in humans.
Fig. 4.
—Breadth of activity across cellular contexts within species is positively associated with conservation of regulatory activity across species. (A) We identified the overlap of conserved-activity liver enhancers, all human liver enhancers (regardless of their conservation status), and human-specific-activity enhancers with enhancers identified across 108 human cellular contexts via CAGE by the FANTOM consortium. In addition to being less active across contexts than conserved-activity enhancers (P = 4.23 × 10−14, MWU test), the human-specific-activity enhancers were active in significantly fewer cellular contexts than expected based on all liver enhancers (P = 2.22 × 10−12). (B) We then trained a SVM on DNase I hypersensitivity sites (DHS), histone modifications, and TF binding profiles identified genome-wide in 125 cellular contexts by ENCODE in human to distinguish conserved-activity enhancers from human-specific-activity-enhancers (auROC = 0.84). We considered each genome-wide annotation as a binary feature, and each enhancer was assigned 0 if it did not overlap an element of the annotation set or 1 if it did overlap. Shaded areas are bounded by the max and min ROC obtained across 10-fold cross validation. (C) We examined the weights assigned to each feature by the classifier; the absolute value of a feature’s weight indicates its overall importance, and the sign indicates whether it is more associated with conserved-activity enhancers (positive) or human-specific-activity enhancers (negative). The weights for features associated with active genomic regions (i.e., active histone modifications (HM), DHS, and TF binding) are positively skewed regardless of the cellular context, indicating they are generally more associated conserved-activity enhancers. The weights for features from liver contexts (red) are consistently more positive than similar features from other contexts (gray). Distributions are summarized as described for figure 2.
We next examined whether patterns in functional genomics data indicative of regulatory activity (and inactivity) across diverse cellular contexts within a species could predict the activity conservation of liver enhancers across species. We used human data collected by the ENCODE Project for this component of the analysis (Bernstein etal. 2012; Sloan etal. 2016). In contrast to FANTOM, ENCODE performed a broad array of functional genomic assays, including DNase I hypersensitivity sites (DHS), histone modifications, and TF binding profiles genome-wide in 125 cellular contexts. In 10-fold cross-validation, the ENCODE classifier was able to distinguish many conserved-activity enhancers from human-specific-activity enhancers (auROC = 0.84; fig. 4B); however, it was not as accurate as the TF binding motif based classifier (fig. 3A; auROC = 0.91). As anticipated, the majority of the most predictive features for both conserved-activity and human-specific-activity enhancers were from liver contexts (supplementary tables 2 and 3, Supplementary Material online). Additionally, there was a general association between active functional annotations and conserved-activity enhancers (fig. 4C), regardless of the cellular context in which the annotation was identified. This association between conserved-activity enhancers and active annotations across contexts argues that they are active in a broader range of cellular contexts.
Motivated by these differences in the breadth of activity of the conserved-activity and human-specific liver enhancers, we analyzed the weights assigned to each TF motif by the trained human enhancer SVM classifier from the previous section. Three transcription factors’ motifs (SP1, SP2, and EWSR1-FLI) were assigned notably higher weights than others (supplementary fig. 5, Supplementary Material online). SP1 and SP2 are broadly expressed zinc finger TFs from the Sp/XKLF family that recognize common GC box motifs and carry out diverse functions across many tissues (Philipsen and Suske 1999). EWSR1-FLI1 is a fusion of EWSR1 and FLI1, an ETS family TF, that is involved in oncogenesis in Ewing tumors (Guillon etal. 2009). The ETS family is a diverse family of TFs with broad functions, but all members have a conserved DNA-binding domain that recognizes diverse motifs with a core GGA(A/T) sequence. While many specific ETS family TFs are present in the motif database, the EWSR1-FLI1 motif consists of GGAA repeats, so we believe that this motif is likely highly weighted as a proxy for this family of broadly active factors. Thus, motifs useful in distinguishing the conserved-activity from human-specific-activity enhancers can be bound by diverse, broadly expressed TFs, further supporting their potential for activity in many cellular contexts.
Conserved-Activity Enhancers Have More Target Genes and Their Target Genes Are More Broadly Active than Human-Specific-Activity Enhancers
Conserved-activity enhancers have a higher density and diversity of TF binding sites, and they exhibit regulatory activity in more genomic contexts than human-specific-activity enhancers. Given their greater regulatory potential and function, we hypothesized that conserved-activity enhancers may regulate more genes, genes with more diverse functions, or both, than do human-specific-activity enhancers. To investigate this, we mapped enhancers to target genes using two complementary approaches. First, we considered the enhancer–gene pairs predicted by the FANTOM project, which are derived from the coexpression patterns of eRNA and mRNA across many cellular contexts in human (Andersson etal. 2014). FANTOM target data are available for 89 of 283 (31.4%) conserved-activity enhancers and 317 of 1,913 (16.6%) human-specific-activity enhancers. Second, we mapped enhancers to genes using genotype and expression data from the GTEx project (The GTEx Consortium 2015). We identified genetic variants within the enhancer sequences that were significantly associated with gene expression levels, and we then used these expression quantitative trait loci (eQTLs) to match enhancers to potential target genes (Materials and Methods). Using this approach, 174 out of 283 (61.4%) conserved-activity enhancers and 1,250 of 1,913 (65.0%) human-specific-activity enhancers mapped to at least one target gene.
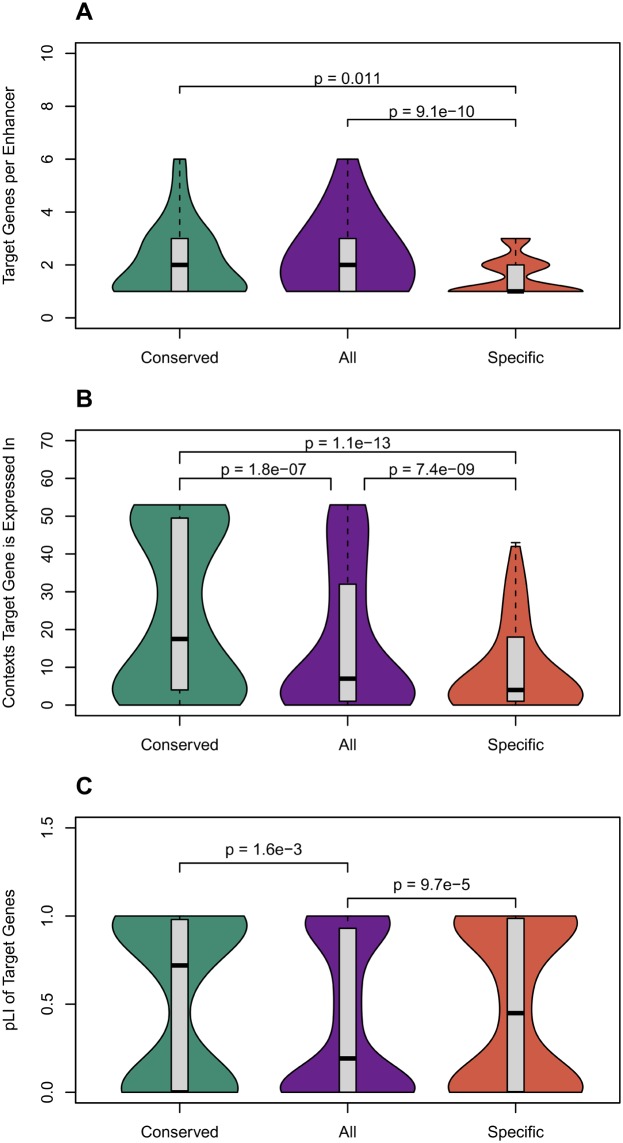
In both the FANTOM and GTEx target sets, conserved-activity enhancers map to significantly more gene targets than human-specific-activity enhancers (fig. 5A and supplementary fig. 6A, Supplementary Material online; mean FANTOM: 2.4 vs. 1.9, P = 0.01; mean GTEx: 3.9 vs. 3.0, P = 0.05; MWU test). Conserved-activity enhancers target a similar number of genes as human liver enhancers in general (mean FANTOM: 2.4 vs. 2.5, P = 0.6; mean GTEx: 3.9 vs. 3.7, P = 0.8), and thus the lower number of targets predicted for human-specific-activity enhancers suggests that human-specific-activity enhancers are depleted of targets. However, the gene targets of conserved-activity enhancers are expressed in a more diverse array of cellular contexts than both all liver enhancers (mean FANTOM: 23.6 vs. 16.4, P = 1.84 × 10−7; mean GTEx: 17.8 vs. 15.9, P = 0.02; MWU test) and human-specific-activity enhancers (mean FANTOM: 23.6 vs. 11.6, P = 1.14 × 10−13; mean GTEx: 17.8 vs. 14.7, P = 0.002; MWU test), for both FANTOM and GTEx mappings (fig. 5B and supplementary fig. 6B, Supplementary Material online). Ultimately, conserved-activity enhancers appear to regulate the expression of more genes than human-specific-activity enhancers, and their gene targets are more broadly expressed than those of human liver enhancers collectively.
Fig. 5.
—Conserved-activity enhancers have more target genes than human-specific-activity enhancers; their targets are expressed in more cellular contexts; and their targets are under stronger evolutionary constraint. (A) Conserved-activity enhancers have significantly more target genes than human-specific-activity enhancers, but a similar number as all human liver enhancers. (B) The gene targets of conserved-activity enhancers were active in significantly more contexts than either human-specific-activity or all liver enhancers. Target gene expression was determined from GTEx (The GTEx Consortium 2015). (C) The target genes for conserved-activity enhancers have significantly higher probabilities of being loss-of-function intolerant (pLI) according to ExAC than target genes for all human liver enhancers. Thus, the target genes of conserved-activity enhancers are under more evolutionary constraint. The targets of human-specific-activity enhancers were also less tolerant of loss of function than liver enhancers overall. Enhancers were mapped to target genes based on coexpression patterns by the FANTOM Consortium. Results were similar when identifying targets based on eQTL (supplementary fig. 6, Supplementary Material online ). The Mann–Whitney U test was used for all comparisons. Distributions are summarized as described for figure 2.
We next hypothesized that the gene targets of conserved-activity enhancers would be more likely to be evolutionarily constrained. To investigate this, we analyzed probability of loss-of-function intolerance (pLI) scores computed by the Exome Aggregation Consortium (ExAC) to quantify constraint on genes. Using the FANTOM enhancer-gene mappings, conserved-activity enhancers mapped to genes with significantly higher pLI scores than the target genes of all human liver enhancers (mean pLI: 0.53 vs. 0.41, P = 1.6 × 10−3; MWU test) (fig. 5C). This finding generalized to the GTEx enhancer-gene mappings (P = 2.0 × 10−3) (supplementary fig. 6C, Supplementary Material online). As expected, the human-specific-activity enhancer targets had lower median pLI than the conserved-activity targets from FANTOM, but surprisingly, the pLI of human-specific-activity enhancers was greater than for liver enhancers overall (fig. 5C). However, this was not true among the GTEx targets; the pLI scores for the human-specific-activity enhancer targets from GTEx were not significantly different from the targets of all human liver enhancers. This will require further study as enhancer–target mappings improve.
Overall, these results indicate that conserved-activity enhancers regulate the expression of more genes than human-specific-activity enhancers, and that these genes are both more broadly expressed and experience stronger constraint than the gene targets of all human liver enhancers.
Discussion
In this study, we demonstrated that liver enhancers with conserved activity across mammals have greater evidence for pleiotropy than similarly alignable sequences with only species-specific activity across three levels of regulatory function: TF binding potential, enhancer activity across tissues, and downstream gene targets. We first found that conserved-activity enhancers have both significantly more TF binding motifs and binding motifs for more distinct TFs, illustrating a greater potential for diverse regulatory activity. We then demonstrated that this increased potential is realized: Conserved-activity liver enhancers are active enhancers in significantly more cellular contexts than species-specific-activity liver enhancers. Furthermore, these differences in activity are also apparent in the attributes of their gene targets; conserved-activity enhancers have more gene targets, and their targets are both more broadly expressed and under greater levels of constraint than species-specific-activity enhancers. These overall differences are sufficiently large that we could accurately classify conserved-activity and human-specific-activity enhancers in a machine learning framework.
Several previous studies have suggested that pleiotropy may play a role in the conservation of regulatory activity across species, but the relationship between pleiotropy and regulatory conservation has not been comprehensively evaluated. For example, the conservation of TF binding at orthologous sequences was positively correlated with the number of cellular contexts in which the sequence had an open chromatin conformation (Cheng etal. 2014). Similarly, an enhancer’s breadth of activity across cellular contexts is positively correlated with the predicted deleteriousness of variants within the enhancer sequence, suggesting that breadth of enhancer activity across contexts is associated with stronger purifying selection (Huang etal. 2017). Our results significantly expand these previous findings beyond the breadth of enhancer activity to other dimensions of regulatory activity, including TF binding density and diversity and gene targets. Additionally, we demonstrate that these trends generalize across mammalian species. Thus, our results provide consistent evidence that enhancers with conserved activity are more pleiotropic than other enhancers.
Given the fast turnover of liver enhancers relative to species divergence (Villar etal. 2015), we anticipate that the majority of species-specific enhancers are young, rather than being remnants of ancestral enhancer elements lost in other lineages. Newly created enhancers likely vary in their regulatory potential. For example, an enhancer that first gains activity in a genomic region that is accessible in many cellular contexts or by gaining a binding site for a broadly expressed TF is likely to have greater pleiotropic potential than an enhancer that arises in a more context-specific region. Over time, the first enhancer would have an easier path to expanding its regulatory role, and thus its constraint. However, constrained activity in one context could also promote pleiotropy by providing a stable functional substrate for developing regulatory activity in additional contexts. Furthermore, enhancers are a diverse and heterogeneous assortment of DNA elements, and other factors likely contribute to their evolutionary dynamics. For instance, our results on the density and diversity of TF binding sites suggest that the robustness of enhancer sequences to disruptive genetic variation may influence activity conservation. More work on the interactions of pleiotropy, activity, and constraint is needed to shed light on the development and evolution of regulatory sequences. Comprehensive mapping of enhancer activity across multiple tissues and species will help resolve these questions.
Several technical limitations may impact the interpretation of our results. Genome-wide profiles of histone modifications have a limited resolution to identify the boundaries of enhancer elements (Shlyueva etal. 2014). As a consequence, it is possible that separate enhancers in close proximity to one another might not be distinguished as separate elements; if multiple enhancers were merged together, this could result in apparent signatures of pleiotropy. However, we demonstrate pleiotropy for these genomic regions at the finest resolution achievable using current, high-throughput techniques. Second, we focused on deeply alignable sequences with extreme differences in activity conservation—those active in all species versus those active only in one. We focused on these extremes to increase our likelihood of detecting differences and to identify patterns that hold across mammals. However, it is possible this may have obfuscated lineage-specific (e.g., primate-specific) patterns underlying conservation of regulatory activity in some clades. Third, mapping enhancers to target genes is a challenging problem, and our current knowledge of gene targets is incomplete. Many enhancers do not have any predicted target genes identified and those that do are likely to include false positives. To account for this uncertainty, we considered two independent mapping strategies and found consistently more targets for the conserved-activity enhancers compared with those with species-specific-activity. Despite these caveats, our findings are consistent across multiple methods of defining TF binding motifs, breadth of enhancer activity, and downstream gene targets.
Finally, the identification of enhancers is an imperfect process, and no genome-wide identification strategy is completely accurate. Histone modification profiles, in particular H3K27ac without H3K4me3, are strongly correlated with enhancer activity in reporter assays, and their use has enabled fundamental studies of enhancer activity genome-wide (Creyghton etal. 2010; Nord etal. 2013; Villar etal. 2015). Nonetheless, there is the possibility of both false positives and false negatives using this approach. False negatives could potentially result in some sequences with true enhancer activity in multiple species being considered species-specific-activity enhancers. Their inclusion would be unlikely to create spurious results as they would likely diminish differences between the species-specific and conserved-activity-enhancer categories. In contrast, false positives are more concerning as they could include nonenhancers in the species-specific-activity enhancer category. However, we demonstrate that all human enhancers, regardless of conservation, demonstrate reduced pleiotropy relative to conserved-activity enhancers, which suggests this finding is not a product of false positives within the species-specific-activity enhancer category. Furthermore, observation of the histone modification signature was required in two biological replicates to define an enhancer, decreasing the risk for false positives. Thus, while the use of histone modifications to identify putative enhancers has caveats, the difference in pleiotropy between enhancer categories is unlikely to be a product of false positives or negatives.
Overall, our work argues that pleiotropy influences the conservation of enhancer activity of noncoding sequences across mammalian evolution. The functional diversity of regulatory sequences must be integrated into models of their evolution. In addition to improving our theoretical understanding of evolutionary constraint on regulatory regions, better understanding the evolutionary forces acting upon the genomic regulatory landscape will also have practical benefits. For example, we demonstrate that machine-learning classifiers can be trained to distinguish conserved-activity from species-specific-activity enhancers using features that reflect their pleiotropy. In the future, these classifiers could be adapted to predict which enhancers will generalize between species, prioritize new tissues for genome-wide assays, and estimate the effects of mutations on enhancer activity.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Acknowledgment
This work was supported in part by US National Institutes of Health (NIH) grant (1R01GM115836 to J.A.C) and an Innovation Catalyst Award from the March of Dimes Prematurity Research Center Ohio Collaborative. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
Literature Cited
- Amoutzias GD, et al. 2007. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol. 24(3): 827–835. [DOI] [PubMed] [Google Scholar]
- Andersson R, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507(7493): 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414): 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, et al. 2014. Principles of regulatory information conservation between mouse and human. Nature 515(7527): 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesmore KN, Bartlett J, Cheng C, Williams SM.. 2016. Complex patterns of association between pleiotropy and transcription factor evolution. Genome Biol Evol. 8(10): 3159–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Scacheri PC.. 2014. Enhancer variants: evaluating functions in common disease. Genome Med. 6(10): 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 107(50): 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erceg J, et al. 2014. Subtle changes in motif positioning cause tissue-specific effects on robustness of an enhancer’s activity. PLoS Genet. 10(1): e1004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin GD, et al. 2014. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol. 10(6): e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galis F, van Dooren TJM, Metz JAJ.. 2002. Conservation of the segmented germband stage: robustness or pleiotropy?. Trends Genet. 18(10): 504–509. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7): 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume F, Otto SP.. 2012. Gene functional trade-offs and the evolution of pleiotropy. Genetics 192(4): 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon N, et al. 2009. The oncogenic EWS-FLI1 protein binds invivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One 4(3): e4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J.. 2006. Toward a molecular understanding of pleiotropy. Genetics 173(4): 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-F, Gulko B, Siepel A.. 2017. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 69682: doi: 10.1101/069682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152(1–2): 327–339. [DOI] [PubMed] [Google Scholar]
- Kapur K, Schüpbach T, Xenarios I, Kutalik Z, Bergmann S.. 2011. Comparison of strategies to detect epistasis from eQTL data. PLoS One 6(12): e28415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, et al. 2016. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 44(D1): D116–D125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Manu Kittler R, White KP, Kreitman M.. 2011. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLOS Genet. 7: e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, et al. 2016. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44(D1): D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337(6099): 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, et al. 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 4: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155(7): 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV.. 2013. The many faces of pleiotropy. Trends Genet. 29(2): 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papakostas S, et al. 2014. Gene pleiotropy constrains gene expression changes in fish adapted to different thermal conditions. Nat Commun. 5: 4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S, Suske G.. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27(15): 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter DI, et al. 2010. The importance of Being Cis: evolution of orthologous fish and mammalian enhancer activity. Mol Biol Evol. 27(10): 2322–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium RE, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518(7539): 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I, Ruvinsky I, Gilad Y.. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 13(7): 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N.. 2010. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 20(8): 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A.. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 15(4): 272–286. [DOI] [PubMed] [Google Scholar]
- Sloan CA, et al. 2016. ENCODE data at the ENCODE portal. Nucleic Acids Res. 44(D1): D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160(3): 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G-H, et al. 2010. Genome-wide analysis of ETS-family DNA-binding invitro and invivo. EMBO J. 29(13): 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, et al. 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322(5900): 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.