Abstract
The nematode family Strongyloididae is of particular interest because it contains important parasites of medical and veterinary relevance. In addition, species of this family can form parasitic and free-living generations and it also occupies an interesting phylogenetic position within the nematodes. Nematodes differ in several ways from other taxa with respect to their small noncoding RNAs. Recent comparative studies revealed that there is also considerable variability within the nematodes. However, no Strongyloididae species or close relative was included in these studies. We characterized the small RNAs of two developmental stages of three different Strongyloididae species and compared them with the well-studied free-living nematodes Caenorhabditis elegans and Pristionchus pacificus. Strongyloididae have conserved and taxon-specific microRNAs, many of which are differentially regulated between the two developmental stages. We identified a novel class of around 27-nucleotide-long RNAs starting with 5′G or A, of which a large fraction have the potential to target transposable elements. These RNAs most likely have triphosphates at their 5′ ends and are therefore presumably synthesized by RNA-dependent RNA polymerases. In contrast to C. elegans but similarly to some other nematode taxa, Strongyloididae have no Piwi-interacting RNAs, nor do their genomes encode Argonaute proteins of the Piwi family. Finally, we attempted but failed to detect circulating parasite small RNAs in the blood of hosts.
Keywords: nematodes; parasites, piRNAs; miRNAs; secondary siRNAs; TAP-treatment
Introduction
Nematodes exhibit a wide variety of lifestyles ranging from soil-dwelling or marine free-living species feeding on bacteria, fungi, and/or other nematodes, to parasitic species infecting all kinds of animals and plants through different mechanisms (Lee 2002; Perry and Wharton 2011). Reproductive modes vary across the nematode clades, with gonochoristic, hermaphroditic, and parthenogenetically reproducing species found amongst the more than 23,000 nematode species that have been described so far. Phylogenetic analysis of free-living and parasitic nematodes (Blaxter et al. 1998; Holterman et al. 2006) led to the conclusion that parasitism must have evolved independently several times within this phylum (Blaxter et al. 1998; Dorris et al. 1999; Blaxter 2011). This diversity of lifestyles must have arisen from evolutionary adaptations in conserved and novel molecular, developmental and physiological processes, all of which are associated with changes in the genome organization and in gene expression. Small noncoding RNAs (sRNAs), produced through multiple regulatory pathways, play important roles in the regulation of these processes, in particular the fine-tuning of the expression of a wide variety of genes (Ghildiyal and Zamore 2009). They are therefore likely targets for evolution to act on during the adaptation to the different lifestyles. Indeed, some interesting differences in the sRNA complements between nematodes and other phyla, and between different nematode taxa have been described (Czech and Hannon 2011; Wang et al. 2011; Sarkies and Miska 2014; Sarkies et al. 2015). Within the animal kingdom, three major conserved classes of sRNAs (18–35 nucleotides [nt]) have been described: microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), and small-interfering RNAs (siRNAs) (Czech and Hannon 2011). These different sRNA classes are defined by their biogenesis and their interaction with different proteins of the Argonaute family. They play important roles in transcriptional and posttranscriptional gene regulation and genome maintenance (Carthew and Sontheimer 2009; Ghildiyal and Zamore 2009; Moazed 2009; Sabin et al. 2013). The nematode Caenorhabditis elegans is one of the best-studied organisms with respect to sRNA families and their Argonaute partners (Buck and Blaxter 2013; Billi et al. 2014; Sarkies and Miska 2014). Known classes of small RNAs in C. elegans include the conserved miRNAs (20–23 nt), which are transcribed as single-stranded precursor RNAs with a secondary hairpin structure. This precursor is then processed by Drosha and Dicer resulting in the mature miRNA, which binds to the Ago subfamily of Argonautes (Sarkies and Miska 2014). Many miRNAs from C. elegans are conserved even in humans. They repress translation or promote degradation of mRNAs, with many of them involved in controlling important developmental transitions (Kaufman and Miska 2010). Another conserved sRNA class in C. elegans is the piRNAs, also called 21 U RNAs because they are 21 nt long and start with a 5′U. They target and silence transposable elements in the germline (Siomi et al. 2011; Ku and Lin 2014). 21 U RNAs are transcribed as short (26–30 nt) precursors, which are processed to give rise to mature 21 U RNAs (Ruby et al. 2006; de Wit et al. 2009; Gu et al. 2012; Weick et al. 2014). Each 21 U RNA is transcribed from its own transcription unit and many of them contain a conserved ∼8 nt upstream motif (Ruby motif). Most 21 U RNA coding units are located in one of two large clusters on chromosome IV.
A third class of sRNAs in C. elegans is the primary siRNAs. Endogenous primary siRNAs are around 26 nt in length and have a strong bias toward a 5′ G (hence 26 G RNAs) and their biogenesis is dependent on an RNA-dependent RNA polymerase (RdRP) and Dicer (Han et al. 2009; Thivierge et al. 2011; Billi et al. 2014). 26 G RNAs are enriched in the gonad (Han et al. 2009; Thivierge et al. 2011; Billi et al. 2014). Exogenous primary siRNAs of 21–22 nt in length are formed upon challenge with exogenous double-stranded RNA through the action of Dicer (Sarkies and Miska 2014).
Both the primary siRNAs (exogenous and endogenous) and the piRNAs (21 U RNAs) can trigger the production of secondary siRNAs, thereby amplifying and enhancing their action. Secondary siRNAs in C. elegans are around 22 nt long and have a 5′ G (22 G RNAs) (Pak and Fire 2007; Sijen et al. 2007; Gu et al. 2009; Vasale et al. 2010; Ashe et al. 2012; Lee et al. 2012; Shirayama et al. 2012; ). According to their differing biogenesis, the different sRNA classes vary in their lengths, their 5′-nucleotide biases toward certain nucleotides and in their 5′- and 3′-end modifications. miRNAs, 21 U and 26 G RNAs have 5′-monophosphates, due to the fact that their mature 5′ ends are generated through endonucleolytic cleavage. In contrast, the secondary siRNAs (22 G RNAs) are produced by nematode-specific RdRPs without 5′ endonucleolytic processing, resulting in 5′-triphosphates (Pak and Fire 2007; Sijen et al. 2007; Billi et al. 2014).
Although no other nematode has been studied as well as C. elegans, it has recently emerged that both nematodes small RNAs and their resulting biological functions are surprisingly diverse. For example, PIWI proteins and piRNAs have apparently been lost independently in multiple branches of the nematode phylogenetic tree, and 5′-triphosphorylated small RNAs were found in nematodes of clades III–V but not clade I (Wang et al. 2011; Sarkies et al. 2015).
The phylogenetic clade IV of nematodes (according to Blaxter et al. 1998) is of particular interest because it contains free-living, entomopathogenic, plant parasitic, and animal parasitic species. The animal parasitic clade IV family Strongyloididae consists of the two fairly closely related genera: Strongyloides and Parastrongyloides. Strongyloides spp. are obligate small intestinal parasites of vertebrates which can undergo facultative single nonparasitic generations with males and females between parthenogenetic parasitic generations (Viney and Lok 2015). Parastrongyloides spp. are facultative parasites of various marsupials with their best-studied representative, Parastrongyloides trichosuri, parasitizing Australian Brush-tailed Possums (Grant et al. 2006). The Strongyloididae have enjoyed a dramatic increase in scientific attention over the last few years, mainly because of two reasons. Firstly, this family contains Strongyloides stercoralis, one of the most prevalent nematode parasites of humans. Although its medical relevance had been underestimated for a long time, it is now increasingly appreciated as an important pathogen in poor communities with unsatisfactory sanitary conditions, and as an important complication in the context of immunosuppressive treatments for cancer and organ transplantation patients (Bisoffi et al. 2013; Nutman 2017). Secondly, with its interesting life cycle (supplementary fig. 1A–C, Supplementary Material online) at the interface of free-living and parasitic lifestyles, this family represents a highly interesting set of model species for basic biological and translational research (Lok 2007; Viney 2017; Baskaran et al. 2017). Recently, high-quality draft genomes of Strongyloides ratti, Strongyloides venezuelensis (which are both parasites of rats), S. stercoralis, Strongyloides papillosus (parasite of sheep), and P. trichosuri have been published (Hunt et al. 2016). The authors of this article also analyzed and compared the gene expression between different developmental stages and different species. However, this analysis is limited to mRNAs and protein coding genes and does not include data for sRNAs. To our knowledge, only a single analysis of the small RNAs of S. ratti has been published (Ahmed et al. 2013). However, this study focused on miRNAs and used methodology that included only small RNAs with 5′ monophosphates and therefore would have missed 5′ triphosphorylated sRNAs, which in other nematodes form a large portion of the sRNAs. In a recent extensive analysis of all known classes of small RNAs of numerous nematodes neither representatives of the family Strongyloididae nor of any other animal parasitic species of clade IV were included (Sarkies et al. 2015).
In order to compare the sRNA complement of the Strongyloididae, which are not only important parasites but also occupy an interesting phylogenetic position, with the sRNAs of other nematodes, we characterized the small RNA contents of two developmental stages (free-living adults and infective larvae) of three different species of Strongyloididae (S. ratti, S. papillosus, and P. trichosuri). For comparison, we also reanalyzed the sRNAs of adult hermaphrodites and dauer larvae of the two well-studied free-living nematodes C. elegans and Pristionchus pacificus. Dauer larvae in C. elegans and P. pacificus and infective larvae in Strongyloididae are believed to be evolutionary equivalents (Ogawa et al. 2009; Wang et al. 2009; Crook 2014). Like C. elegans and P. pacificus, Strongyloididae have a prominent class of small RNAs with 5′ triphosphates. However, different from C. elegans, P. pacificus, and any other nematode analyzed so far, these RNAs are longer and do not show the strong bias toward a G nucleotide at their 5′-end. Further, Strongyloididae possess no class of small RNAs that show the typical features of piRNAs and, consistently, there are no recognizable orthologs of PIWI protein genes present in their genomes.
Materials and Methods
Nematode Strains, Culture Conditions, and Sample Collection
Wild-type strains of five different nematode species were used for this study: C. elegans N2 (received from the C. elegans Genetics Center at the University of Minnesota), P. pacificus RS2333 (received from Ralf J. Sommer), S. ratti ED321 (Viney 1996), S. papillosus LIN (Eberhardt et al. 2007), P. trichosuri (received from Warwick N. Grant, for description of isolate, see Kulkarni et al. (2013)).
Caenorhabditis elegans and P. pacificus were maintained on 6 cm plates with Nematode Growth Medium (NGM) agar on a lawn of Escherichia coli OP50 at 20 °C as previously described (Brenner 1974; Stiernagle 2006). For mixed-stage P. pacificus cultures, three adults per plate were picked onto 6 cm NGM plates and grown at 20 °C for 7 days. For mixed-stage C. elegans cultures, ∼50 L1/L2 larvae were picked onto 6 cm NGM plates and grown at 20 °C for 3 days. To obtain adult hermaphrodites, mixed-stage worms from twenty 6 cm plates were washed off with 0.1 M NaCl into a 50 ml Falcon tube, and gravid adult worms were allowed to sink to the bottom of the tube for 3–5 min. The supernatant (with young larvae) was removed and the remaining worm pellet was resuspended in 50 ml 0.1 M NaCl. The washing and settling was repeated three times. After the washing, the supernatant was removed and the pellet of worms transferred into a 1.5 ml Eppendorf tube and filled with 0.1 M NaCl. The adult worms were finally pelleted by centrifuging for 30 s and as much supernatant removed as possible. Caenorhabditis elegans and P. pacificus dauer larvae were obtained from liquid culture (Lewis and Fleming 1995): Mixed-stage worms from thirty 6 cm NGM plates were inoculated into 150 ml S-medium without Cholesterol containing 1% (w/v) OP50, 50 µg/ml Streptomycin and 50 µg/ml Nystatin and grown at 22 °C and 180 rpm for 8 days. For the first replicate of dauer cultures, clean dauer larvae were obtained by washing with 0.1 M NaCl as mentioned before. For the second and third replicate of dauer cultures, dauer larvae were purified by sucrose flotation and Ficoll precipitation (http://diamond.tuebingen.mpg.de/wiki/index.php/Dauer_ purification_by_Sucrose_Floatation_and_Ficoll_precipitation (last accessed October 2, 2017)).
Strongyloides ratti and S. papillosus were maintained under laboratory conditions in female Wistar rats or female New Zealand White rabbits, respectively, as previously described (Viney et al. 1992; Eberhardt et al. 2007). All relevant national and international animal welfare regulations and guidelines were followed. The experiments were approved by the Regierungspräsidium Tübingen (AZ: 35/9185.82-5/15.07.2015). Strongyloides ratti and S. papillosus adults (males and females) were obtained from 48 h fecal cultures at 19 °C for S. ratti and 25 °C for S. papillosus. Adult free-living worms were isolated from fecal cultures using the Baermann technique (2–3 h) (Lok 2007) and washed with tap water. Young larvae were removed by letting worms sink down for 3–5 min in 50 ml Falcon tubes filled with tap water. The supernatant (with young larvae) was removed and the remaining worm pellet was resuspended in 50 ml of tap water. The washing and settling was repeated three times. After the washing the supernatant was removed and the pellet of worms transferred into a 1.5 ml Eppendorf tube and filled with water. The adult worms were finally pelleted by centrifuging for 30 s and as much supernatant removed as possible. Strongyloides ratti infective larvae were obtained from 7-day fecal cultures using the Baermann technique (overnight) and washed three times with tap water. Strongyloides papillosus infective larvae were obtained from 7-day cultures as described (Eberhardt et al. 2008). In brief, 48 h fecal culture petri dishes were placed in a larger petri dishes with tap water. The infective larvae that accumulated in the water were collected using the Baermann technique (overnight) and washed three times with tap water. Parastrongyloides trichosuri was maintained in continuous free-living cycles on 6 cm NGM plates with a piece of autoclaved rabbit feces on a lawn of E. coli OP50 at 20 °C (Grant et al. 2006; Kulkarni et al. 2013). For mixed-stage P. trichosuri cultures, ∼100–200 adults were picked onto NGM plates and grown at 20 °C for 8 days. As the worms tend to burrow into the agar, the agar of 5–10 plates was chopped into smaller pieces and the worms isolated using the Baermann technique (2 h) with M9 buffer. The worms were transferred to 50 ml Falcon tubes and adult worms were allowed to sink to the bottom of the tube for 3–5 min. The supernatant (with young larvae) was removed and the remaining worm pellet was resuspended in 50 ml M9 buffer. The washing and settling was repeated three times and the final worm pellets obtained as in C. elegans. Parastrongyloides trichosuri infective larvae were obtained by picking ∼100–200 adults onto NGM plates and grown at 20 °C for 15 days. For each replicate, 10–20 plates were used and infective larvae isolated and washed like the adults.
Collection of Whole Blood from Rats Infected and Uninfected with S. ratti
Three 4-week-old female Wistar rats (Charles River) were infected with ∼500 infective larvae from the S. ratti strain ED231 in 200 µl PBS as previously described (Viney et al. 1992). At the same time, two 4-week-old rats were mock-infected with 200 µl PBS. On the 8th day postinfection, the rats were first sedated with CO2 for ∼3 min and then killed by cervical dislocation. The thoracic cavity of the rats was opened, the heart was cut with a clean scalpel blade and whole blood obtained from the blood puddles forming in the cavity. The blood was collected in RNAprotect Animal Blood Tubes (500 µl Qiagen) and incubated for 5 h at room temperature. All relevant national and international animal welfare regulations and guidelines were followed. The experiments were approved by the Regierungspräsidium Tübingen (AZ: 35/9185.82-5/15.07.2015, Anzeige vom 13.10.2016).
RNA Extraction
For the nematodes, 1 ml TRIzol (Thermo Fisher Scientific) was added to each worm pellet and briefly grinded with a pestle. The mixture was then frozen in liquid nitrogen and thawed at room temperature three times. RNA of adult worms was extracted with the standard TRIzol protocol followed by a separation of large and small RNAs with the PureLink miRNA Isolation Kit (Thermo Fisher Scientific). RNA of dauer/infective larvae was extracted the same as adult RNA but the small RNA fraction contained a lot of large RNAs and was reseparated into large and small RNA fractions using the RNA Clean & Concentrator kit (Zymo Research). For the mouse blood, the total RNA (including short RNAs) from whole blood of infected and uninfected rats was extracted with the RNeasy Protect Animal Blood kit (Qiagen) according to the manufacturer’s protocol. Subsequently, the large RNA and small RNA fractions were obtained with the RNA Clean & Concentrator kit (Zymo Research). The quality and quantity of the extracted RNA fractions was assessed using 1% Agarose gels, 2100 Bioanalyzer RNA 6000 Nano chips (Agilent Technologies), and Qubit RNA BR Assays (ThermoFisher Scientific) according to the manufacturer’s protocol. The RNA was stored at –80 °C until library preparation.
Library Preparation
Following RNA isolation, each small RNA replicate was divided, and one part was treated with 25 U tobacco acid pyrophosphatase (Epicenter) or Cap-Clip acid pyrophosphatase (Biozym) in 50 µl volume for 2 h at 37 °C to remove 5′-triphosphates and allow 5′-independent library preparation. The other part was mock treated under the same conditions (5′-dependent library preparation). From those treated or untreated small RNA fractions, libraries were prepared with the TruSeq Small RNA Library Preparation Kit (Illumina) according to the manufacturer’s protocol. cDNA constructs containing both adapters were cut from a gel by hand in the range of ∼140–160 base pairs (bp) corresponding to sRNAs with lengths in the range of ∼17–37 nt. Every library was validated with 2100 Bioanalyzer DNA 1000 chips.
Small RNA Sequencing and Read Preprocessing
Forty-eight samples were sequenced on the same flowcell on a HiSeq 3000 (Illumina; 150 bp single-end). Eighteen samples were sequenced in four batches on a MiSeq (Illumina; 50 bp single-end). See supplementary table 1, Supplementary Material online for more detailed information on the nematode data set and supplementary table 2, Supplementary Material online for information on the whole blood samples from infected and uninfected rats. Samples were demultiplexed, adapter sequences were removed with flexbar (v2.5; parameters ‘-u 100 -m 14 -at 1 -n 8 -ag -7 -ao 3’) (Dodt et al. 2012) and only reads with 14 nt or longer retained. Fastq files were converted to fasta files using FASTX Toolkit (v0.0.14). All read data will be made publically available through appropriate databases upon acceptance of the manuscript.
miRNA Prediction and Quantification
miRDeep2 (v0.0.7) (Friedlander et al. 2012) was used for novel miRNA prediction and quantification of known miRNAs. The full initial list of predicted novel miRNAs was filtered against a custom database containing tRNAs, rRNAs, snoRNAs, snRNAs, 21 U RNAs, and repeats. All sequences with a miRDeep2 score <2 and/or matching the database and/or showing a ratio of 5′-all-phosphate to 5′-monophosphate read counts ≥1 were excluded. miRBase release 21 was used for reference precursors and mature miRNAs. A 10 nt genomic sequence on the 5′ and 3′ end of all precursors was added to ensure complete capture of miRNA-related reads. Mature miRNAs arms were defined by calculating the ratio of read counts matching to the 5p- or 3p-arm (all sample counts combined, 1 pseudocount was added to avoid division by zero). For ratios >1, the 5p-arm was considered to be the mature miRNA; for ratios <1, the 3p-arm (supplementary table 3, Supplementary Material online).
miRNA Classification into Seed-Families and Inference of Phylogenetic Trees
The known and novel miRNAs of all species were grouped into families based on the identity of their seed sequences (positions 2–8 of the mature miRNA; see supplementary table 4, Supplementary Material online). For the conserved miRNA seed families, seed-constrained multiple sequence alignments were constructed using LocARNA (Will et al. 2007, 2012), which takes the predicted secondary structure of the miRNAs into account. For each miRNA group, the similarity matrix (LocARNA scores) was transformed into a distance matrix as described in Will et al. (2007), and used to infer phylogenetic trees with the Unweighted Pair Group Method with Arithmetic Mean hierachical clustering method (upgma, R package phangorn v1.99-7 (Schliep 2011)). The resulting phylogenetic trees were combined with our miRNA expression fold changes between dauers/infectives and adults, and the 5p/3p ratio information of miRNA arm usage, into plots with R.
sRNA Classification
All sRNA reads were classified into four classes: tRNA-, rRNA-, miRNA-derived, or other. For this classification, all reads were aligned to defined tRNA, rRNA, miRNA precursors, and finally to the respective genome assemblies (see supplementary table 5, Supplementary Material online). tRNAs were predicted for all species genomes using tRNAscan-SE (v1.3.1; default parameters) (Lowe and Eddy 1997). For the alignments, bowtie was used with no allowance for mismatches (v1.0.0; parameters ‘-v 0 –k 1 -a –best –strata’) (Langmead et al. 2009).
Length Distribution and 5′ Nucleotides
Custom Shell and Perl scripts were used to get the length and first nucleotide of all reads. The output was read into R to calculate means and generate barplots.
Differential Expression Analysis
MicroRNAs
The miRDeep2 quantifier module was used to quantify reads from untreated (5′-monophosphate) libraries representing either dauers/infectives or free-living stages in triplicates for each of the five species. The sum of read-counts on the 3p-arm and 5p-arm of each miRNA was calculated for each sample. For the differential expression analysis of miRNAs, the R package edgeR (v 3.14.0) was used (Robinson et al. 2010). For this analysis, only miRNAs that have at least 1 read per million (miRNA mapping reads) in at least three samples were kept. The data were TMM (trimmed mean of M values) normalized (Robinson and Oshlack 2010), and differential expression determined using the generalized linear model (GLM) quasi-likelihood (QL) F-test (Lund et al. 2012). Up- and downregulated miRNAs are defined as significantly differentially expressed if they have a log2 fold change of ≥ 1 and ≤–1, respectively and a false discovery rate (FDR) <5% (supplementary table 3, Supplementary Material online).
Other-Mapped Reads
All other-mapped reads from all samples were filtered, and only reads with a single unambiguous match in the respective genomes were kept. Other-mapped reads falling on annotated mRNAs were then counted with htseq-count from the HTSeq framework (v0.6.0; parameters ‘-s no -m union’) (Anders et al. 2015). For the differential expression analysis of the other-mapped reads, R package edgeR (v 3.14.0) (Robinson et al. 2010) was used. For the analysis, only those genes that have at least 1 read per million in at least three samples were kept and 5′-all-phosphate and 5′-monophosphate samples were compared. The data were TMM normalized (Robinson and Oshlack 2010), and differential expression determined using the GLM QL F-test (Lund et al. 2012). Up- and downregulated target genes are defined as significantly differentially expressed if they have a log2 fold change of ≥ 1 and ≤–1, respectively and an FDR <5%. In order to investigate if tobacco acid pyrophosphatase (TAP)-treatment enriches sRNAs specifically matching to certain protein families containing specific protein domains (PFAM v.29), an hmmsearch of annotated proteins (supplementary table 5, Supplementary Material online) against the PFAM v.29 HMM library was performed. Gene set enrichment analysis was done in R with camera (Competitive Gene Set Test Accounting for Intergene Correlation (Wu and Smyth 2012)) from the package limma (v. 3.28.14). Significantly (FDR < 5%), enriched protein families that are up- or downregulated between 5′-all-phosphate and 5′-monophosphate samples were compared bewteen all five nematode species.
Investigation of Other-Mapped Reads and Their Abundance on Annotated Genes
All other-mapped reads from all samples were filtered, and only reads with a single unambiguous match in the respective genomes were kept. Other-mapped reads mapping sense or antisense on annotated mRNAs (in case of C. elegans also transposons) were counted using htseq-count from the HTSeq framework (v0.6.0; parameters ‘-s yes/reverse -m union’) (Anders et al. 2015). The percentage of alignments being sense, antisense, or not aligned to an annotated gene was then calculated.
Comparison of Our miRNA Expression Changes to Published Data from Illumina and SOLiD Sequencing
miRNA expression changes was compared with published data obtained by Illumina and SOLiD sequencing of dauers/infectives and mixed-stages of C. elegans, P. pacificus, and S. ratti (Ahmed et al. 2013). For all miRNA genes that were shared and highly enough expressed in both data sets (at least three reads per million in three samples), the expression changes were calculated into up-, downregulated, and unaffected. The miRNA genes were then grouped into a contigency table. A χ2 test was then performed in R to assess statistical independence of the two data sets.
Calculation of Genome Coverage of Potential piRNAs
All other-mapped reads from all samples were filtered and only reads with a single unambiguous match in the respective genomes, a 5′U and 21 (C. elegans) or 20–24 nt (S. ratti and S. papillosus) length were considered. samtools (v. 0.1.19 (Li et al. 2009)) and bedtools (v. 2.17.0 (Quinlan and Hall 2010)) were used to calculate the number of reads matching 100 kb bins of the genomes (considering only scaffolds with >100 kb length). The number of reads was normalized to reads per million in relation to all mapped reads (tRNA-, rRNA-, miRNA-, and other-mapped). The coverage was plotted with R.
piRNA Motif Detection
All other-mapped reads from all samples were filtered and only unique sequences with a single unambiguous match in the respective genomes, a 5′U and 21 (C. elegans and P. pacificus) or 20–24 nt (Strongyloididae) length were kept. The filtered sequences from adults were merged, and the 60 nt upstream and the first 2 nt of each location of these sequences were taken. meme (v. 4.11.1 (Bailey and Elkan 1994; Bailey et al. 2015), -dna -oc -maxsize 3000000 -mod zoops -nmotifs 1 -minw 4 -maxw 62 -p 64) was run on a random subset of 5,000 upstream sequences for each species to discover a common motif. fimo (v. 4.11.1 (Bailey et al. 2015; Grant et al. 2011), –oc –verbosity 1 –thresh 1.0E-4 –norc) was used to detect the predicted motifs in the entire sets of upstream sequences, and those which have at least one hit with a P-value of ≤1 × 10−4 were counted.
Phylogenetic Analysis of Argonautes and RdRPs
To obtain a list of RdRPs and Argonaute proteins in C. elegans, P. pacificus, S. ratti, S. papillosus, and P. trichosuri, a jackhmmer (HMMER 3.1b1 (Eddy 2011)) search with five iterations (default options) against the accessions in supplementary table 5, Supplementary Material online using the protein sequences of C. elegans rrf-1 (Wormbase ID CE27141) and alg-1 (Wormbase ID CE31525) was performed. For Argonautes, the same search was also performed in Homo sapiens, Mus musculus, and Drosophila melanogaster. This resulted in RDRP orthologs with E-values <2 × 10−46, and Argonaute orthologs with <2 × 10−8. Multiple sequence alignments were then performed using MUSCLE with default settings (Edgar 2004). As outgroups for RdRPs, Arabidopsis thaliana (RDR1-6; GenBank accessions OAP18817.1, AEE82976.1, O82190.2, O82189.2, O82188.2, AEE78550.1) and Saccharomyces pombe (RDP1, GenBank accession CAB11093.1) RdRPs were used. As outgroups for the PIWI Argonautes, H. sapiens (PIWIL1 and PIWIL4; GenBank accessions Q96J94.1 and AAH31060.1), M. musculus (PIWIL1-2; GenBank accessions AAI29859.1 and AAK31965.1), and D. melanogaster (PIWI and AUBERGINE; GenBank accessions AGL81535.1 and CAA64320.1) PIWIs and the Argonaute protein from the Archaea Pyrococcus furiosus (PDB: 1Z25) were used. Unrooted trees were constructed with the outputs of MUSCLE, using the R package phangorn (v1.99-7 neighbour joining, maximum likelihood optim.pml, LG model, 100 bootstraps (Schliep 2011)).
Results
We isolated and sequenced sRNAs from three biological replicates for two developmental stages (free-living males and females/hermaphrodites, and infective/dauer larvae) of three species of Strongyloididae (S. ratti, S. papillosus, and P. trichosuri), and for comparison, of the two well-characterized clade V free-living nematodes C. elegans and P. pacificus. In order to obtain additional information about the chemical nature of the 5′ ends of the different sRNAs, from each RNA preparation, we prepared sequencing libraries with (5′-all-phosphate) and without (5′-monophosphate) prior treatment with TAP. 5′-Monophosphate libraries only include sRNAs with a 5′-monophosphate, whereas 5′-all-phosphate libraries also represent sRNAs that have 5′-di-, -triphosphates or cap-structures in vivo. After preprocessing and filtering (see Materials and Methods), the sRNA reads were classified into four groups: miRNAs (aligning to identified miRNA precursors), tRNA-derived (aligning to tRNA genes), rRNA-derived (aligning to ribosomal RNA genes), or “other” (aligning to any other part of the respective genome). See supplementary figure 1, Supplementary Material online for a graphical overview over the samples, treatments, and the analysis pipeline, and supplementary table 1, Supplementary Material online for detailed information on all samples and treatments.
Improvement of the miRNA Annotations
In our attempt to classify the sRNA reads, we had to consider the fact that the quality and the quantity of the miRNA precursor annotations varied dramatically between the five species under consideration. While C. elegans has been analyzed extensively and miRBase (release 21) contains a close to complete list of miRNA precursors for this species; for S. papillosus and P. trichosuri, no miRNAs have been annotated yet. Therefore, we used miRDeep2 (Friedlander et al. 2012) to predict novel miRNAs based on our 5′-monophosphate sequencing reads and the available genome information (for details see Material and Methods). We then compared our predictions with the entries in miRBase release 21 (table 1).
Table 1.
Number of Expressed miRNA Precursors
| C. elegans | P. pacificus | S. ratti | S. papillosus | P. trichosuri | |
|---|---|---|---|---|---|
| miRBase listed miRNA precursorsa | 250 | 354 | 106 | — | — |
| expressed miRBase listed miRNAsb | 218 | 344 | 106 | — | — |
| newly predicted miRNA precursorsc | 7 | 37 | 33 | 140 | 163 |
| Total expressed miRNAs | 225 | 381 | 139 | 140 | 163 |
miRNA precursors listed in miRBase (http://www.mirbase.org; last accessed October, 2 2017) release 21 for this particular species.
Number of miRBase listed miRNA precursors for which we found mature miRNAs in any of our samples from the particular species.
miRNA precursors not previously described for this particular species (new predictions were only made if the corresponding miRNA was detected in our samples).
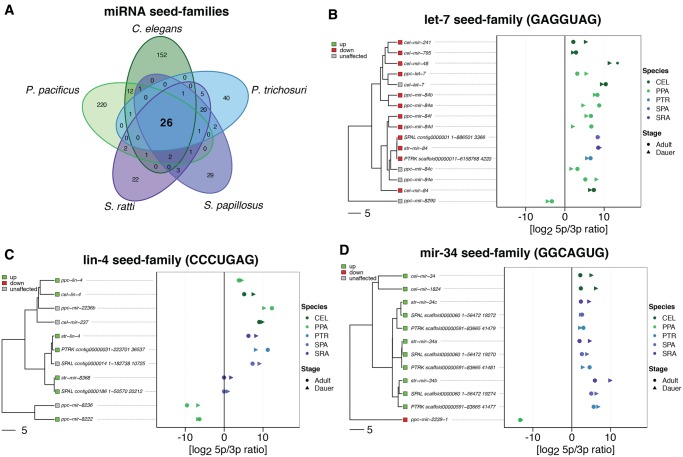
We found evidence for expression of the vast majority of the listed miRNAs in all three species where annotations were available. We identified only seven novel miRNAs in C. elegans, the species where the most extensive prior analysis has been conducted. Furthermore, 37 novel miRNA predictions for P. pacificus, 33 for S. ratti, 140 for S. papillosus, and 163 for P. trichosuri fulfilled our criteria for inclusion in the analysis (see Materials and Methods and supplementary table 6, Supplementary Material online). In total, we found 225 (C. elegans), 381 (P. pacificus), 139 (S. ratti), 140 (S. papillous), and 163 (P. trichosuri) expressed miRNA precursors and used those as basis for further analysis. The differences in the numbers of miRNA precursors may reflect the differences in the genome sizes as was proposed by Ahmed et al. (2013). For each known or novel miRNA precursor, the mature miRNA arm was predicted by calculating the ratio of read counts matching to the 5p- or 3p-arm. For ratios >1, the 5p-arm was considered to be the mature miRNA, and for ratios <1, the 3p-arm (see supplementary table 3, Supplementary Material online). Although there were cases with ratios relatively close to 1, in particular for weakly expressed miRNAs, in the vast majority of the cases the preference for one arm was strong. Next, all miRNAs were grouped into families based on their seed sequences (perfect match at positions 2–8 of the mature miRNA, see supplementary table 4, Supplementary Material online). This procedure is common (Wheeler et al. 2009) because the seed sequence is the main determinant of the target specificity of miRNAs. In total, we found 541 different seeds of which only 26 are shared among all five species (fig. 1A). However, members of these 26 seed families amount to a considerable fraction of the total miRNAs, namely 62/224 (28%) in C. elegans, 86/381 (23%) in P. pacificus, 48/139 (35%) in S. ratti, 44/140 (31%) in S. papillosus, and 42/163 (26%) in P. trichosuri. Further we identified 20 seed families that are shared only between Strongyloididae and 12 seeds that are shared only between the two clade V nematodes.
Fig. 1.
—Comparative analysis of miRNA seed-families. (A) Venn diagram showing the number of miRNA seed families and their overlaps between the five nematodes species under study. (B–D) Phylogenetic trees based on multiple sequence alignments of all members of three miRNA seed-families (left) and arm preference (right) represented as ratio of reads derived from the 5p arm and the 3p arm of the miRNA precursor. The species are color coded, filled circles represent reads from libraries derived from adults, filled arrow heads represent reads from dauer/infective larvae derived libraries. The seed sequence is given in parentheses. (B) One example of a conserved seed family (present in all five species) that is consistently downregulated in dauer/infective larvae compared with adults (red squares). (C) One example for a conserved seed family that is consistently upregulated in dauer/infective larvae compared with adults (green squares). For the other conserved differentially expressed miRNAs see supplementary figure 7, Supplementary Material online. (D) The mir-34 seed family. Notice that P. pacificus mir-34 is differentially expressed in the opposite direction than the rest and its gene phylogeny is not in agreement with the species phylogeny, suggesting that it has acquired the seed by convergence.
Results in C. elegans and P. pacificus and Comparison with Previously Published Literature
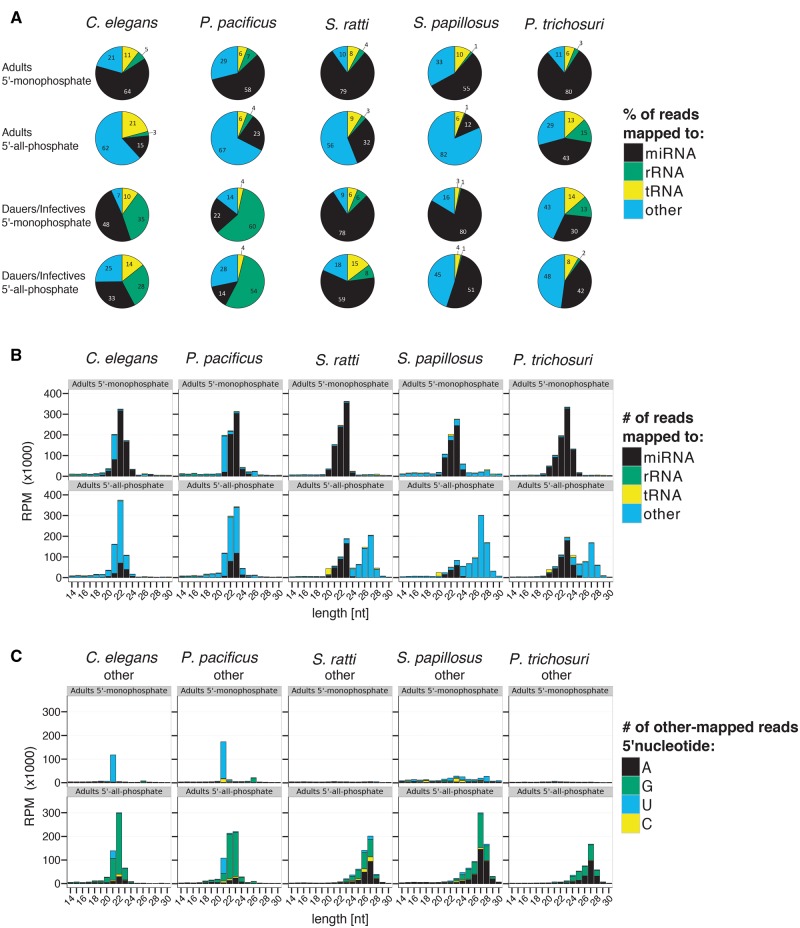
In C. elegans and P. pacificus adult hermaphrodites, miRNAs were the most abundant class in 5′-monophosphate samples, representing on average 64% and 58% of all mapped reads with a size distribution around 22 nt length and either a 5′U or 5′A start (fig. 2A and B and supplementary fig. 2, Supplementary Material online). The second most abundant class of sRNAs in the 5′-monophosphate libraries were other-mapped reads, representing on average 21% and 29% of all mapped reads in C. elegans and P. pacificus adults. Most of these other-mapped reads included in the 5′-monophosphate libraries have a distinct length of 21 nt and a 5′U representing 21 U RNAs (fig. 2A–C). These 21 U RNAs are known to be the piRNAs (Ruby et al. 2006; de Wit et al. 2009; Gu et al. 2012; Weick et al. 2014). Additionally there is a small other-mapped peak in both species at 26 nt with 5′Gs representing the primary (endo-)siRNA class of 26 G RNAs (fig. 2B and C) (Han et al. 2009; Thivierge et al. 2011; Billi et al. 2014). In the 5′-all-phosphate adult samples, miRNA reads amounted to only 15% and 23% of all mapped reads in C. elegans and P. pacificus, respectively, whereas other-mapped reads accounted for the majority of all mapped reads (62% and 67%, respectively). The TAP-enriched other-mapped reads have a size distribution around 22 nt of length and a very strong 5′G bias (22 G RNAs) (fig. 2A–C). In the dauer samples, the same trend as in adults can be observed, but a larger proportion (25–60%) of the reads are rRNA derived. Because most of them are in sense orientation, they probably represent degradation products (fig. 2A). The 22 G RNAs have been described to be secondary siRNAs, which are produced by the RdRPs RRF-1 and EGO-1 (Gu et al. 2009) without 5′ processing and therefore have 5′-triphosphates (Pak and Fire 2007; Sijen et al. 2007; Gu et al. 2009; Vasale et al. 2010; Ashe et al. 2012; Lee et al. 2012; Shirayama et al. 2012; ). For the length distribution and 5′ nucleotides of dauer samples and other sRNA classes, see supplementary figure 2, Supplementary Material online. In order to also have a quantitative comparison with published data, we reclassified the sRNA reads from the previously published C. elegans and P. pacificus data sets from Weick et al. (2014) in the same way as our own data set and compared them with our data (supplementary table 7, Supplementary Material online). We found that in the published data roughly the same percentage of reads from 5′-monophosphate samples were miRNAs (69% and 54% in C. elegans and P. pacificus, respectively, compared with 64% and 58% in our sample) or other-mapped (12% and 33% compared with 21% and 29% in our sample). Also in the published data set, the proportion of miRNA reads was lower after TAP-treatment (21% and 19% compared with 15% and 23% in our sample) and the majority of reads fell in the class of other-mapped reads (72% and 75% compared with 62% and 67% in our sample). Overall, our results in C. elegans and P. pacificus are in excellent agreement with previously published data (Ahmed et al. 2013; Shi et al. 2013; Sarkies et al. 2015). From this we conclude that our experimental design was suitable to reproduce earlier findings, and therefore our new data on the Strongyloididae species are comparable with the existing literature on other systems.
Fig. 2.
—sRNA profiles. (A) Piecharts representing the percentage of all mapped reads classified as miRNAs (black), rRNA-derived (green), tRNA-derived (yellow), or other (blue). (B) Barplots showing the length distribution of small RNA classes. The color code is the same as in (A). (C) Barplots of the length distribution and 5′ nucleotide of sRNAs classified as other. Reads starting with A, G, U, or C are represented in black, green, blue, and yellow, respectively. In all cases, the average of the three biological replicates is given. In B and C only the results for adults and in C only the results for the class “other” are given. For the corresponding plots for dauers/infectives and for the other sRNA classes see supplementary figure 2, Supplementary Material online. 5′-Monophosphate RNA not treated with TAP before library construction; 5′-all-phosphate RNA treated with TAP before library construction; RPM: reads per million; length[nt]: read length in nucleotides.
Classes of Small RNAs Identified in Strongyloididae
In free-living adults of all three Strongyloididae species, most of the reads from 5′-monophosphate samples are derived from miRNAs (on average 79%, 55%, and 80% of all mapped reads for S. ratti, S. papillosus, and P. trichosuri, respectively; fig. 2A), which is comparable with C. elegans and P. pacificus. As in C. elegans and P. pacificus, the length of miRNAs peaks at 22–23 nt in all three species (fig. 2B) and they have a very strong 5′U bias (supplementary fig. 2, Supplementary Material online). The proportion of reads mapping to rRNA or tRNA sequences was comparable in all five species tested. However, unlike in C. elegans and P. pacificus, we found no peaks corresponding to the 21 U and the 26 G RNAs among the reads mapping to “other” parts of the genome in Strongyloididae (fig. 2A–C). These peaks were, however, clearly present in our C. elegans and P. pacificus data sets.
In adults of all five species tested, reads mapping to “other” parts of the genome make up a much greater proportion in the 5′-all-phosphate samples than in the 5′-monophosphate data sets. In C. elegans and in P. pacificus, the TAP-enriched fraction consists predominantly of the 22 G RNAs. In contrast, the TAP-enriched sRNAs of all three Strongyloididae species form a novel class of sRNAs, with a length peak at 27 nt (fig. 2B) and 5′G and 5′A being similarly abundant (27GA RNAs) (fig. 2C). Qualitatively, all findings in the adults described above also hold true for dauer/infective larvae (fig. 2A and supplementary fig. 2, Supplementary Material online). However, the enrichment of other-mapped reads in the 5′-all-phosphate samples is less pronounced in this developmental stage. The biological replicates of all species and developmental stages are consistent. The sRNA profiles are presented separately and with full size range (up to 50 nt) for each biological replicate in supplementary figure 3, Supplementary Material online.
Putative Targets of the TAP-Enriched 27GA RNAs
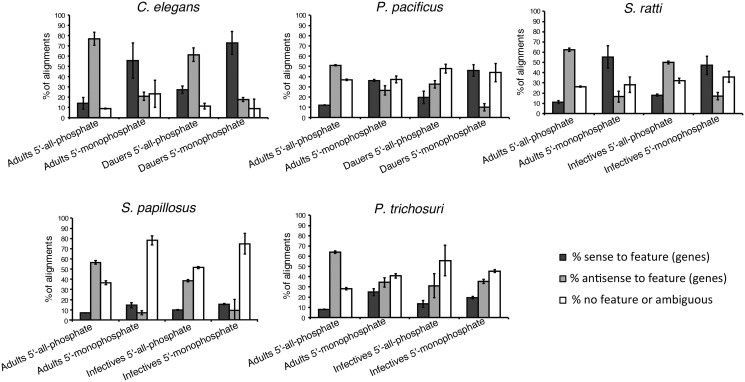
In C. elegans, the 22 G RNAs have been shown to be secondary siRNAs mapping mainly antisense to transposons and other genes. We speculated that the TAP-enriched 27GA RNAs are the Strongyloididae equivalent of the 22 G RNAs of C. elegans. In that case, one would predict that these RNAs correspond to the noncoding strand of genes, such that they could target the corresponding mRNAs by base pairing. To test this, we counted reads mapping sense or antisense or not mapping to annotated genes with htseq (Anders et al. 2015). We then compared the differences between 5′-all-phosphate and 5′-monophosphate libraries (fig. 3). In all five species analyzed, TAP-treatment led to a strong increase in the proportion of “antisense to gene” reads in adults. In fact, the majority of other-mapped reads (51–77% of unambiguously other-mapped reads) in 5′-all-phosphate adult samples map antisense to annotated genes. The same trend was observed in dauer/infective samples except for P. trichosuri (fig. 3). Next we asked what kind of genes are targeted by the TAP-enriched RNAs and performed a differential expression analysis, comparing other-mapped reads from 5′-all-phosphate libraries versus 5′-monophosphate libraries. We then identified the gene predictions targeted by the TAP-enriched RNAs and performed a gene set enrichment analysis. We found that a good portion of the TAP-enriched RNAs target gene predictions containing transposable element-related protein domains in all five nematode species, but also other protein coding genes are putative targets (table 2 and supplementary table 8, Supplementary Material online). Given the limited quality of the annotations of the Strongyloididae genomes, it is difficult to make a more quantitative statement. However, estimates based on two different methods of predicting transposons are provided in supplementary figure 4, Supplementary Material online.
Fig. 3.
—TAP-treatment enriches for reads that align antisense to annotated genes in all five species studied. Barplots showing the percentage of other mapped reads that map to annotated features (genes) in either sense (dark gray) or antisense (light gray) orientation or reads that map ambiguously or to regions of the genome without annotated features (white). Notice that the proportion of reads that does not map to annotated parts of the genome is highly dependent on the quality of the genome annotations, which varies strongly between the five species under study. Therefore only the ratio of sense mapped reads to antisense mapped reads is relevant. Given are the means of the three biological replicates. The errorbars indicate the standard deviations.
Table 2.
TAP-Enriched Other-Mapped sRNAs Target Genes with Protein Domains Related to Transposable Elements in all Five Nematode Species
| PFAM Domain | Function | C. elegansAdults 5′-all-phosphate | P. pacificusAdults 5′-all-phosphate | S. rattiAdults 5′-all-phosphate | S. papillosusAdults 5′-all-phosphate | P. trichosuriAdults 5′-all-phosphate |
|---|---|---|---|---|---|---|
| transposable_element | up | not annotated | not annotated | not annotated | not annotated | |
| Rve | Retroviral integrase | — | up | up | up | up |
| gag-asp_protease | gag-polyprotein putative aspartyl protease | — | up | up | up | up |
| rve_3 | Integrase core domain | — | — | up | up | up |
| zf-H2C2 | binds to histone upstream activating sequence elements that are found in histone gene promoters | — | up | up | up | up |
| zf-CCHC | Zinc finger | up | up | up | up | — |
| RVT_1 | Reverse transcriptase | — | — | up | up | up |
| DDE_Tnp_IS1595 | ISXO2-like transposase domain | — | — | up | up | up |
| Asp_protease_2 | Aspartyl protease | — | up | up | — | |
| RVP | Retroviral aspartyl protease | — | — | up | up | — |
| RVP_2 | Retroviral aspartyl protease | — | — | up | up | — |
| DDE_3 | DDE superfamily endonuclease | — | — | up | up | up |
| Asp_protease | Aspartyl protease | — | — | up | up | — |
| AT_hook | DNA-binding motif present in many proteins | up | — | up | up | — |
| gag_preintegrs | associated with retroviral insertion elements, lies just upstream of the integrase region | — | — | up | up | — |
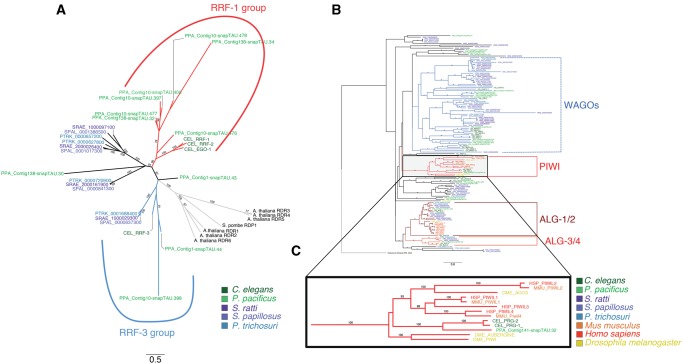
Based on these findings, we hypothesize that these TAP-enriched sRNAs in Strongyloididae represent a class of secondary siRNAs which are produced by RdRPs without 5′ processing, comparable with the 22 G RNAs in C. elegans. Indeed, multiple putative RdRP genes are present in the genomes of all three Strongyloididae species (fig. 4A).
Fig. 4.
—Phylogenetic gene trees for RdRPs and Argonautes. (A) Unrooted neighbour joining tree of the RdRP genes from the five different nematodes. As outgroups we added RdRPs from Arabidopsis thaliana and Saccharomyces pombe. The different species are color-coded, and the two previously described subfamilies (RRF-1 and RRF-3) are indicated by colored branches. Only branch support values ≥ 70 from 100 bootstraps are shown. Notice that the RRF-3 group is not supported by a high bootstrap value. (B) Neighbour joining tree of the Argonaute genes from the five nematodes under study and Drosophila melanogaster, Homo sapiens, Mus musculus and one argonaute gene from the archean species Pyrococcus furiosus. For graphical representation we used FigTree to reroot the tree to the outgroup Argonaute from P. furiosus. The different species are color-coded. The conserved subfamilies that contain nematode, mammalian and Drosophila members are indicated by colored solid branches (ALG-1/2, ALG3/4, PIWI). The previously described group of nematode (worm) specific Argonautes (WAGOs) are indicated by dashed lines. Notice that monophyly of WAGOs is not supported by high bootstrap values in our analysis. For a high resolution graph of the complete tree see supplementary figure 5, Supplementary Material online. (C) Zoom in of the Piwi branch of Argonautes, which is lacking Strongyloididae representatives. Only branch support values ≥70 from 100 bootstraps are shown.
piRNAs Are Lost in Strongyloididae Nematodes
In C. elegans and P. pacificus, piRNAs are also known as 21 U RNAs. As described above, we failed to detect a prominent group of “other” mapped TAP insensitive (5′ monophosphorylated) RNAs of 21 or a similar defined number of nucleotides in length with a 5′U in any of the Strongyloididae, although this class of RNAs was clearly present in our C. elegans and P. pacificus samples (fig. 2C). This indicated that these worms either do not possess piRNAs, or that the piRNAs vary in length. In order to investigate this, we asked if we could identify a class of RNAs that showed any of the general properties of piRNAs.
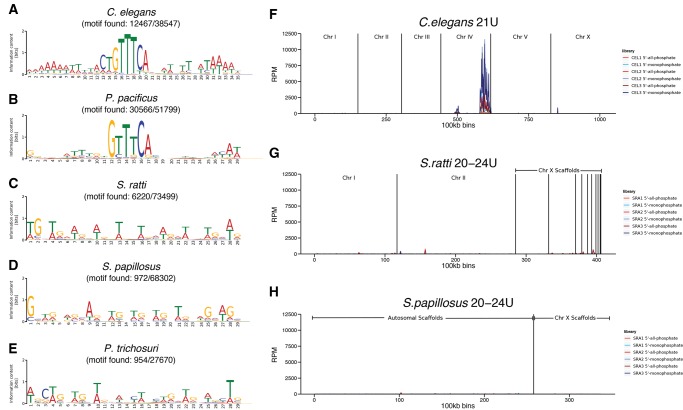
The 21 U RNA genes in C. elegans are located in two big clusters on chromosome IV (Ruby et al. 2006), whereas in P. pacificus, the 21 U RNA genes are organized in multiple smaller clusters, which are disseminated throughout the genome (de Wit et al. 2009). The tendency to start with a 5′U and to be clustered in the genome is a widespread feature of piRNAs far beyond nematodes (Grimson et al. 2008; Malone et al. 2009; Juliano et al. 2014). Therefore, we analyzed the genomic distribution of all uniquely mapped “other” 20–24 U RNAs in S. ratti and in S. papillosus. We found no indication for clustering of the genomic regions from which the 20–24 U RNAs originate (fig. 5F–H).
Fig. 5.
—Absence of a Ruby-type motif and no genomic clustering of 20–24 U RNAs in Strongyloididae. (A–E) Sequence logos of de novo predicted motifs based on 5,000 randomly selected 60 bp regions immediately upstream of aligned 21 U (A, B) or 20–24 U (C–E) other-mapped sequences. The numbers beneath the species indicate the number of places in the genome to which 21 U or 20–24 U reads align that do have the motif in the upstream region/total number of places to which 21 U or 20–24 U reads align. (F–H) Plots representing the genome coverage by 21 U or 20–24 U other-mapped reads from adult worm replicates along the chromosomes/scaffolds (100kb bins) for C. elegans (F), S. ratti (G), and S. papillosus (H). The different biological replicates and treatments are color coded. The two strongest peaks in C. elegans correspond to the known major and minor piRNA gene clusters. RPM reads per million.
In C. elegans, each 21 U RNA is transcribed from its own gene and many of these 21 U RNA genes contain a conserved motif (Ruby motif), about 40 nt upstream of the position corresponding to the 5′U of the mature 21 U RNA (Ruby et al. 2006; de Wit et al. 2009; Gu et al. 2012; Weick et al. 2014). A similar conserved motif is also found in P. pacificus and the clade V parasitic nematodes Haemonchus contortus and Nippostrongylus brasiliensis (de Wit et al. 2009; Sarkies et al. 2015). In order to find evidence for a Ruby-like motif in Strongyloididae, we randomly selected 5,000 genomic positions from where “other” mapped 20–24 U RNAs (21 U RNAs for C. elegans and P. pacificus) originated. We then used the MEME Suite (Bailey et al. 2015) to search for the presence of a conserved motif in the upstream regions. We determined in what proportion of all “other” mapped 20–24 U RNAs the identified motif occurred within 60 bp upstream of the position corresponding to the 5′U. Although we were able to rederive the published Ruby motif from the C. elegans and the P. pacificus data, we did not find a comparable motif in any of the three Strongyloididae species. Even if we accepted the best identified consensus sequence as a motif, it was present in the upstream region of only a small fraction of all 20–24 U RNA coding regions (fig. 5A–E andsupplementary table 9, Supplementary Material online).
In other systems, piRNAs specifically interact with PIWI proteins, which are a class of Argonaute proteins that are highly conserved within the animal kingdom (Czech and Hannon 2011). Therefore as a final line of evidence, we asked if genes encoding PIWI proteins are present in the Strongyloididae genomes. To obtain a list of Argonaute proteins in the five species analyzed, we performed a jackhmmer search (HMMER 3.1b1 (Eddy 2011)) on the (predicted) proteins using the protein sequence of C. elegans ALG-1 as bait. We accepted Argonaute homologs with an e-value <2 × 10−8 and aligned the protein sequences using MUSCLE with default settings (Edgar 2004). As outgroups for the PIWI Argonautes, we used PIWI proteins from H. sapiens (PIWIL1 and PIWIL4), M. musculus (PIWIL1-2), and D. melanogaster (PIWI and AUBERGINE). We then reconstructed unrooted trees with the R package phangorn (Schliep 2011) (fig. 4B and C and supplementary fig. 5, Supplementary Material online). We identified 25 different Argonaute genes in S. ratti. It should be noted that the somewhat higher number in S. papillosus is likely an overestimate due to assembly problems in the S. papillosus draft genome, which was derived from an outbred population with rather high genetic variability. The number of Argonaute genes in S. ratti is very comparable with the number in C. elegans (both 25) and P. pacificus (23). Several of the major groups contain genes from the two clade V nematodes (C. elegans and P. pacificus) and the Strongyloididae but cases of clear one to one orthology are rare. This indicates that the Argonaute gene families have independently expanded in the two clades. There are also phylogenetic groups of argonaute genes that appear entirely absent from either of the two clade V species or from the Strongyloididae. Interestingly, one of the two groups not present in Strongyloididae are the PIWI coding genes. Overall, from these results we conclude that “standard” piRNAs are absent from Strongyloididae as is the case for a number of other nematode species (Sarkies et al. 2015).
A Conserved Set of miRNA Families Is Differentially Expressed in Dauers/Infectives versus Adults
In order to identify developmentally regulated miRNAs, we quantified the miRNA expression (as described in Materials and Methods) and compared the miRNA expression levels in dauer/infective larvae and in free-living adults. Only those miRNAs that had at least 1 read per million (miRNA mapping reads) in at least three samples were included in the analysis. Up- and downregulated miRNAs were considered significantly differentially expressed if they showed at least a 2-fold change in expression (log2 fold change (log2FC) of ≥1 or ≤–1) and an FDR <5%. Of all expressed miRNAs, we found 49% (109/224) in C. elegans, 44% (167/381) in P. pacificus, 47% (65/139) in S. ratti, 52% (73/140) in S. papillosus, and 61% (99/163) in P. trichosuri to be significantly differentially expressed (supplementary table 3, Supplementary Material online). Roughly equal numbers of miRNAs were up- and downregulated in dauers/infectives compared with adults across all species, with 54 up and 55 down in C. elegans, 75 up and 92 down in P. pacificus, 37 up and 28 down in S. ratti, 41 up and 32 down in S. papillosus, and 43 up and 56 down in P. trichosuri.
For some of the species, we investigated (C. elegans, P. pacificus, and S. ratti), Ahmed et al. (2013) had already compared the expression of miRNAs in dauers/infectives and in mixed stages. In total, these authors had looked at 177 miRNAs (of which 152 were also present at high enough levels to be included in our analysis) in C. elegans, 331 (of which 291 are also included in our data set) in P. pacificus, and 106 (of which 100 are also included in our data set) in S. ratti (supplementary fig. 6 and table 10, Supplementary Material online). In order to compare our data with Ahmed et al. (2013), we categorized the miRNA expression changes into upregulated, downregulated, and unaffected, made a contingency table and performed a χ2 test. The two data sets show a highly significant correlation of the categorized expression changes in dauers versus mixed/adult stages for all three species (, P and P for C. elegans, P. pacificus, and S. ratti, respectively, supplementary fig. 6A, C and E, Supplementary Material online). We plotted the log2FC values of dauers/infectives versus mixed stage/adults of the two studies against each other and performed a linear regression (supplementary fig. 6B, D and F, Supplementary Material online). In general, the expression changes of miRNAs detected in both data sets are comparable and only very few miRNAs defined as up- or downregulated in one of the data sets show the opposite trend in the other data set. This again demonstrates that our findings are consistent with previously published data as far as such data exists, adding credibility to our data about not previously analyzed species.
Among the 26 miRNA seed families that are shared between all five nematode species, we found ten that showed consistent differential expression in all five nematode species. We considered differential expression to be consistent if at least one seed family member from each species is differentially expressed in the same direction (up- or downregulated in dauers/infectives), as the majority of the miRNAs that are members of the same seed family. Eight seed families were upregulated (lin-4, mir-124, mir-231, mir-232, mir-251, mir-50, mir-51, and mir-86) and two families were downregulated (let-7 and mir-35) in dauers/infectives versus adults in all five nematode species (fig. 1B and C and supplementary fig. 7, Supplementary Material online). For each of the conserved miRNA seed families, we constructed seed-constrained multiple sequence alignments with LocARNA (Will et al. 2007, 2012), which takes the predicted secondary structure of the miRNAs into account. These alignments were then used to infer gene trees shown in figure 1. Furthermore we analyzed the conservation of the preference for the 5p- or 3p-arm giving rise to the mature miRNA for each seed family, and found that most of the miRNAs members of the different seed families share the origin (5p- or 3p-arm) of their mature miRNA (fig. 1B–D and supplementary fig. 7, Supplementary Material online). Almost all exceptions were found for miRNAs that were phylogenetically more distant from the rest of the members in a seed family and had possibly acquired the same seed by convergence. These findings indicate that certain miRNA seed families perform stage specific functions that are conserved over large evolutionary distance. An interesting case among the 26 seed families is the mir-34 family (fig. 1D). Against the general trend that Strongyloididae have fewer miRNAs, we found three paralogs in the Strongyloididae, two in C. elegans and only one in P. pacificus. Further, the C. elegans and the Strongyloididae mir-34 family genes are more closely related to each other than to the single P. pacificus gene, and in P. pacificus the mature miRNA is on the 5p-arm, whereas in the others the mature miRNA is on the 3p-arm. All of this indicates that the P. pacificus mir-34 is of different phylogenetic origin (convergent acquisition of the seed). Fittingly, the P. pacificus mir-34 is differentially expressed in the other direction suggesting a different function.
No Signs of Circulating Parasite-Derived sRNAs in the Host Blood
There are more and more reports of parasite-derived sRNAs circulating in the blood or other tissues of animal hosts. Circulating miRNAs were found in the blood of hosts infected with the filarial nematodes Dirofilaria immitis and Onchocerca volvulus (Tritten et al. 2014) and in hosts infected with trematodes of the genus Schistosoma (Cai et al. 2015). For the intestinal parasitic nematode Heligmosomoides polygyrus, it has been shown that it secretes vesicles containing miRNAs as well as a nematode Argonaute protein (Buck et al. 2014). We sequenced sRNAs from whole blood of three rats that were infected with S. ratti, and compared their sRNA expression to two sRNA data sets from uninfected rats, to see if any sRNAs from S. ratti are circulating in the blood of their host animal (supplementary table 2, Supplementary Material online). We could not find any S. ratti-specific miRNAs (or any other sRNAs) that were present in the infected rats but not the uninfected ones. Either there are no circulating sRNAs that are secreted from S. ratti, they are identical in sequence to host sRNAs, or they are present in such low amounts that our method was not sensitive enough to detect them. As expected, TAP-treatment made no difference for rat-derived sRNAs except for a strong TAP-dependent peak of 39 nt entirely derived from 5 S rRNA and a slight enrichment of tRNA fragments (supplementary fig. 8, Supplementary Material online).
Discussion
The nematodes are probably the most species rich animal phylum and differ in many ways not only from other phyla but also from each other (Lee 2002). One example are various aspects about their noncoding small RNAs. Here, we present the first global characterization of the sRNA profiles of three members of the parasitic nematode family Strongyloididae. In a previous study that compared sRNAs in various nematodes (Sarkies et al. 2015), the closest relative of the Strongyloididae that had been included was Globodera pallida, a plant parasite that although part of the same major clade (clade IV) of nematodes (Blaxter et al. 1998), is as phylogenetically distant as is possible within a clade. This is the first global characterization of sRNAs in any animal parasite of clade IV. Therefore, the Strongyloididae represent almost certainly an independent transition to parasitic life style from the few other parasitic nematodes for which such an analysis had been done.
Like in other nematodes, the Strongyloididae sRNA libraries that were limited to RNAs with 5′ monophosphates were dominated by miRNA-derived sequences. Comparable with other systems, Strongyloididae have conserved and taxon-specific miRNAs. Among the 157 different miRNA seeds identified in Strongyloididae, only 48 were shared amongst all three species tested. Interestingly, more than half of them (26) were also found in C. elegans and P. pacificus. Also for these two clade V species, the number of seeds shared between them but not with the Strongyloididae was smaller than the number of seeds shared among all five species of the two clades. A substantial fraction (10/26) of the miRNAs with conserved seeds, showed conserved differential expression between free-living adults and infective/dauer larvae, indicating that in spite of the large phylogenetic distance, these miRNAs might still fulfill similar functions. Consistent with a largely conserved miRNA system, we identified Argonaute proteins in all three species which clearly belong to the families of Argonautes involved in miRNA function in other systems.
Similarly to other nematodes of clades III–V (Wang et al. 2011; Sarkies et al. 2015), we identified a population of RNAs in Strongyloididae only detectable upon TAP treatment. However, differently from all other previously characterized nematodes where these RNAs are around 22 nt long (24 nt in the clade IV representative G. pallida) and in their overwhelming majority start with 5′ G (hence called 22 G RNAs), in Strongyloididae the TAP-enriched RNAs are longer, around 27 nt, and are about as likely to have 5′ As as 5′ Gs (27GA RNAs). This difference suggests that Strongyloididae also have different RdRPs for their biosynthesis and different Argonautes capable of binding these longer RNAs. Consistently, we identified four putative RdRP genes in each of the three species of Strongyloididae. For two of them, clear one to one orthologs were present in all three species analyzed. For the other two, there are one to one orthologs in the two Strongyloides species, whereas both genes in P. trichosuri are more closely related to one of the Strongyloides paralogs. For none of the Strongyloididae RdRPs, a clear orthology relationship to any of the four C. elegans RdRPs was detectable (fig. 4A). We found about the same number of Argonaute genes in Strongyloididae as in C. elegans. However, cases of clear orthology are rare and largely limited to the two conserved families (ALG-1/2 and ALG-3/4, involved in miRNA and primary endogenous siRNA function, respectively) (fig. 4B) (Conine et al. 2010; Buck and Blaxter 2013). This indicates that most of the expansion of the Argonaute family occurred after the phylogenetic separation of C. elegans and Strongyloididae. We showed that a large portion of the 27GA RNAs has the potential to target transposable elements (supplementary fig. 4, Supplementary Material online), like 22 G RNAs do in C. elegans (Gu et al. 2009). The fact that they appear to be made by RdRPs, comparable with 22 G RNAs, leads us to hypothesize that the 27GA RNAs correspond to the 22 G secondary siRNAs in C. elegans. The 22 G RNAs in C. elegans are secondary siRNAs triggered by the primary endogenous siRNAs (26 G RNAs) and 21 U RNAs (Pak and Fire 2007; Sijen et al. 2007; Gu et al. 2009; Vasale et al. 2010; Ashe et al. 2012; Lee et al. 2012; Shirayama et al. 2012; ). Although in our data 26G RNAs are visible as small peaks in the C. elegans and P. pacificus samples (fig. 2C), no such peaks are visible in the samples derived from Strongyloididae. Also in Sarkies et al. (2015) no 26 G peaks are visible in the samples from Brugia malayi (clade III) and G. pallida (clade IV). However, 26 G RNAs have been found in the clade III parasitic nematode Ascaris suum, where they are strongly enriched in the male gonad (Wang et al. 2011). We speculate that primary endogenous siRNAs, which trigger the production of the 27GA RNAs do exist in Strongyloididae but they might be more heterogeneous in length compared with clade V nematodes and A. suum. We cannot exclude the presence of an unknown chemical modification that prevents certain sRNAs from being included in the sequencing libraries. In C. elegans portions of the 26 G and the 21 U RNAs have been shown to be 3′-end methylated (Kamminga et al. 2012). We do not think that this particular modification is present in the Strongyloididae because in none of the genomes we found a homolog of HENN-1, the methyltransferase responsible for this modification. This is consistent with earlier studies in which neither the gene nor 3′-end methylation have been found in nematodes outside the clade V (Wang et al. 2011; Sarkies et al. 2015).
We found no indication of the presence of piRNAs or Piwi proteins in Strongyloididae. This suggests that this taxon lacks this widely conserved class of small RNAs normally involved in silencing transposable elements in the germline. The absence of piRNAs had already been postulated for various other nematodes of multiple clades based on the absence of recognizable Piwi genes in the genomes (Wang et al. 2011; Sarkies et al. 2015). Among them is Panagrellus redivivus, a relatively closely related free-living nematode (Srinivasan et al. 2013). In fact, although piRNAs are widely conserved, within nematodes they have so far only been found in one of the major clades (clade V), which contains C. elegans. Although the exact phylogeny of nematodes is under debate, all phylogenetic studies we are aware of Blaxter (2011, 2007), Holterman et al. (2006), and Meldal et al. (2007) agree that Strongyloididae are more closely related to C. elegans than to some of the nematode species shown to lack piRNAs. Given the wide conservation of piRNAs among animals, the assumption that the last common ancestor of all nematodes had piRNAs appears reasonable. If we accept that the last common ancestor of all nematodes had piRNAs, the absence of piRNAs in all nematodes outside of clade V studied so far can only by explained by independent losses during evolution in multiple branches of nematodes, as proposed by Sarkies et al. (2015). However, maybe one should not prematurely exclude an early loss followed by a regain of piRNAs in clade V, possibly through horizontal gene transfer.
For some parasitic nematodes, it has been recently described that they secrete small RNAs and in a few cases, parasite-derived sRNAs were found circulating in the host blood (Cai et al. 2015; Tritten et al. 2014). Therefore, we sequenced 5′-all-phosphate and 5′-monophosphate sRNAs isolated from blood of rats which were infected or mock infected with S. ratti. We failed to detect S. ratti-derived sRNAs, indicating, that if such RNAs are present in the blood, they are either extremely low abundant or they are identical with host sRNAs. This finding does not exclude that parasite-derived sRNAs are present locally in the small intestine of the host and might play a role in the host–parasite interaction.
Taken together, we identified a novel class of small RNAs termed 27GA RNAs in the Strongyloididae, a family of parasitic nematodes, which includes the human pathogen S. stercoralis. This class of RNAs is only detectable in RNA seq experiments if the RNA is treated with tobacco acid pyrophosphatase, indicating that these RNAs carry triphosphates at their 5′ ends and are made by RdRPs. Correspondingly, Strongyloididae possess different subfamilies of RdRPs and Argonaute proteins. A large fraction of the 27GA RNAs have the potential to target transposable elements. We hypothesize that 27GA RNAs correspond to the 22 G secondary siRNAs in C. elegans. Further, Strongyloididae lack piRNAs and Piwi-type Argonaute proteins.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Declarations
Animal Experiments
For animal experiments, all relevant national and international animal welfare regulations and guidelines were followed. The experiments were approved by the Regierungspräsidium Tübingen (AZ: 35/9185.82-5/15.07.2015).
Availability of Data and Materials
All data sets generated and analysed in this study are available at the European Nucleotide archive (ENA) under the accession PRJEB21191. It is policy of miRBase to only assign definite miRNA names after acceptance of a publication. Therefore, the final names for the newly described miRNAs will be assigned by miRBase according to their schedule.
Funding
This work was supported by the Max Planck Society.
Authors’ Contributions
A.H. and A.S. designed the experiments. A.H. collected the samples and constructed the sRNA libraries. A.H. performed the analysis pipeline. A.H. and A.S. interpreted the data. A.H. prepared the figures and tables with input from A.S. A.H. and A.S. cowrote the manuscript. Both authors approved the final draft.
Supplementary Material
Acknowledgments
We thank Dorothee Harbecke, Christa Lanz, and the in-house sequencing facility for their technical support. We thank Christian Rödelsperger, Peter Sarkies, and members of our department for valuable discussions, and Christian Rödelsperger and Alex Dulovic for critically reading the manuscript. Caenorhabditis elegans N2 was provided by the C. elegans Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Pristionchus pacificus was provided by Ralf J. Sommer.
Literature Cited
- Ahmed R, et al. 2013. Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes. Genome Biol Evol. 5(7): 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2): 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150(1): 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS.. 2015. The MEME Suite. Nucleic Acids Res. 43(W1): W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran P, Jaleta TG, Streit A, Rodelsperger C.. 2017. Duplications and positive selection drive the evolution of parasitism-associated gene families in the nematode Strongyloides papillosus. Genome Biol Evol. 9(3): 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SE, Kim JK.. 2014. Endogenous RNAi pathways in C. elegans. (May 7, 2014), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.170.1, http://www.wormbook.org (last accessed October 2, 2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisoffi Z, et al. 2013. Strongyloides stercoralis: a plea for action. PLoS Negl Trop Dis. 7(5): e2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. 2011. Nematodes: the worm and its relatives. PLoS Biol. 9(4): e1001050.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. 2007. Symbiont genes in host genomes: fragments with a future? Cell Host Microbe 2(4): 211–213. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, et al. 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392(6671): 71–75. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77(1): 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Blaxter M.. 2013. Functional diversification of Argonautes in nematodes: an expanding universe. Biochem Soc Trans. 41(4): 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, et al. 2014. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 5: 5488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Gobert GN, You H, Duke M, McManus DP.. 2015. Circulating miRNAs: potential novel biomarkers for hepatopathology progression and diagnosis of Schistosomiasis japonica in two murine models. PLoS Negl Trop Dis. 9(7): e0003965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ.. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136(4): 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, et al. 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A 107(8): 3588–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook M. 2014. The dauer hypothesis and the evolution of parasitism: 20 years on and still going strong. Int J Parasitol. 44(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ.. 2011. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 12(1): 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SE, Cuppen E, Berezikov E.. 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19(11): 2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C.. 2012. FLEXBAR—Flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1(3): 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris M, De Ley P, Blaxter ML.. 1999. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol Today 15(5): 188–193. [DOI] [PubMed] [Google Scholar]
- Eberhardt AG, Mayer WE, Bonfoh B, Streit A.. 2008. The Strongyloides (Nematoda) of sheep and the predominant Strongyloides of cattle form at least two different, genetically isolated populations. Vet Parasitol. 157(1-2): 89–99. [DOI] [PubMed] [Google Scholar]
- Eberhardt AG, Mayer WE, Streit A.. 2007. The free-living generation of the nematode Strongyloides papillosus undergoes sexual reproduction. Int J Parasitol. 37(8-9): 989–1000. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10): e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N.. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40(1): 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD.. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet. 10(2): 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7): 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant WN, et al. 2006. Parastrongyloides trichosuri, a nematode parasite of mammals that is uniquely suited to genetic analysis. Int J Parasitol. 36(4): 453–466. [DOI] [PubMed] [Google Scholar]
- Grimson A, et al. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455(7217): 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, et al. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151(7): 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36(2): 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, et al. 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A 106(44): 18674–18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman M, et al. 2006. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol Biol Evol. 23(9): 1792–1800. [DOI] [PubMed] [Google Scholar]
- Hunt VL, et al. 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet 48: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, et al. 2014. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci U S A 111(1): 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, et al. 2012. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 8(7): e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman EJ, Miska EA.. 2010. The microRNAs of Caenorhabditis elegans. Semin Cell Dev Biol. 21(7): 728–737. [DOI] [PubMed] [Google Scholar]
- Ku HY, Lin H.. 2014. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev. 1(2): 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Dyka A, Nemetschke L, Grant WN, Streit A.. 2013. Parastrongyloides trichosuri suggests that XX/XO sex determination is ancestral in Strongyloididae (Nematoda). Parasitology 140(14): 1822–1830. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3): R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DL. 2002. The Biology of Nematodes: CRC Press. [Google Scholar]
- Lee HC, et al. 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150(1): 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT.. 1995. Basic culture method In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego: Academic Press; p. 3–29. [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok JB. 2007. Strongyloides stercoralis: a model for translational research on parasitic nematode biology (February 17, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.134.1, http://www.wormbook.org (last accessed October 2, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5): 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund SP, Nettleton D, McCarthy DJ, Smyth GK.. 2012. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol, ISSN (Online) 1544-6115, DOI: https://doi.org/10.1515/1544-6115.1826 (last accessed October 2, 2017) [DOI] [PubMed] [Google Scholar]
- Malone CD, et al. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137(3): 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal BH, et al. 2007. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol. 42(3): 622–636. [DOI] [PubMed] [Google Scholar]
- Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457(7228): 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman TB. 2017. Human infection with Strongyloides stercoralis and other related Strongyloides species. Parasitology 144:263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Streit A, Antebi A, Sommer RJ.. 2009. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol. 19(1): 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A.. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315(5809): 241–244. [DOI] [PubMed] [Google Scholar]
- Perry RN, Wharton DAeds.. 2011. Molecular and physiological basis of nematode survival: CAB International, Oxfordshire, ISBN 978-1-84593-687-7. [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6): 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1): 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11(3): R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, et al. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127(6): 1193–1207. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Delas MJ, Hannon GJ.. 2013. Dogma derailed: the many influences of RNA on the genome. Mol Cell 49(5): 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA.. 2014. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol. 15(8): 525–535. [DOI] [PubMed] [Google Scholar]
- Sarkies P, et al. 2015. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol. 13(2): e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27(4): 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y, Ruvkun G.. 2013. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 23(3): 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, et al. 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150(1): 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH.. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315(5809): 244–247. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA.. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 12(4): 246–258. [DOI] [PubMed] [Google Scholar]
- Srinivasan J, et al. 2013. The draft genome and transcriptome of Panagrellus redivivus are shaped by the harsh demands of a free-living lifestyle. Genetics 193(4): 1279–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org (last accessed October 2, 2017). [Google Scholar]
- Thivierge C, et al. 2011. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol. 19(1): 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritten L, et al. 2014. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl Trop Dis. 8(7): e2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, et al. 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A 107(8): 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney M. 2017. Strongyloides. Parasitology 144:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney ME. 1996. Developmental switching in the parasitic nematode Strongyloides ratti. Proc Biol Sci. 263(1367): 201–208. [DOI] [PubMed] [Google Scholar]
- Viney ME, Lok JB.. 2015. The biology of Strongyloides spp. (May 23, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.141.1, http://www.wormbook.org (last accessed October 2, 2017). [Google Scholar]
- Viney ME, Matthews BE, Walliker D.. 1992. On the biological and biochemical nature of cloned populations of Strongyloides ratti. J Helminthol. 66(1): 45–52. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. 2011. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 21(9): 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2009. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc Natl Acad Sci U S A 106(23): 9138–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick EM, et al. 2014. PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev. 28(7): 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BM, et al. 2009. The deep evolution of metazoan microRNAs. Evol Dev. 11(1): 50–68. [DOI] [PubMed] [Google Scholar]
- Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R.. 2012. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA 18(5): 900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R.. 2007. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol. 3(4): e65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Smyth GK.. 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40(17): e133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets generated and analysed in this study are available at the European Nucleotide archive (ENA) under the accession PRJEB21191. It is policy of miRBase to only assign definite miRNA names after acceptance of a publication. Therefore, the final names for the newly described miRNAs will be assigned by miRBase according to their schedule.