Abstract
Background
Tumor infiltrating lymphocytes (TILs) and programmed death ligand 1 (PD-L1) are targets of immune checkpoint inhibitors.
Methods
Forty-three World Health Organization (WHO) grade II/III gliomas (39 IDH-mutant [mut], 4 IDH-wildtype [wt]) and 14 IDH-mut glioblastomas (GBM) were analyzed for TIL (CD3+; PD1+) infiltration and PD-L1 expression. Results were compared with the data of a previously published series of 117 IDH-wt glioblastomas. PD-L1 gene expression levels were evaluated in 677 diffuse gliomas grades II–IV from The Cancer Genome Atlas (TCGA) database.
Results
TIL and PD-L1 expression were observed in approximately half of WHO grade II/III gliomas. IDH-wt status was associated with significantly higher TIL infiltration and PD-L1 expression among all (grades II–IV) cases (n = 174, P < 0.001) and within the cohort of glioblastomas (n = 131, P < 0.001). In low-grade glioma (LGG) and glioblastoma cohorts of TCGA, significantly higher PD-L1 gene expression levels were evident in IDH-wt compared with IDH-mut samples (LGG: N = 516; P = 1.933e-11, GBM: N = 161; P < 0.009). Lower PD-L1 gene expression was associated with increased promoter methylation (Spearman correlation coefficient −0.36; P < 0.01) in the LGG cohort of TCGA. IDH-mut gliomas had higher PD-L1 gene promoter methylation levels than IDH-wt gliomas (P < 0.01).
Conclusions
The immunological tumor microenvironment of diffuse gliomas differs in association with IDH mutation status. IDH-wt gliomas display a more prominent TIL infiltration and higher PD-L1 expression than IDH-mut cases. Mechanistically this may be at least in part due to differential PD-L1 gene promoter methylation levels. Our findings may be relevant for immune modulatory treatment strategies in glioma patients.
Keywords: glioblastoma, IDH mutation, immune microenvironment, low grade glioma, PD-L1, promoter methylation
Importance of the study.
Here, we show that the immunological tumor microenvironment of diffuse gliomas differs in dependence of the molecular tumor status, with IDH-wt cases showing more prominent infiltration by lymphocytes and higher expression of the immune checkpoint molecule PD-L1 than IDH-mut cases. The difference in PD-L1 expression was evident not only at the protein level as assessed by immunohistochemistry, but also at the gene expression level, as confirmed in a large series of diffuse gliomas from the database of TCGA. As a potential mechanistic link between PD-L1 gene expression and IDH mutations we identified increased PD-L1 gene promoter methylation in the IDH-mut subpopulation. Given the possible predictive value of TIL infiltration and PD-L1 expression as biomarkers for response to immune checkpoint inhibitors, our findings may be relevant for immune modulatory treatment strategies in glioma patients.
Diffuse gliomas are the most common primary brain tumors of adults and comprise a heterogeneous group of neoplasms that differ with regard to their natural course and sensitivity to chemo- and radiotherapy. Traditionally, diffuse gliomas have been separated in astrocytic versus oligodendroglial neoplasms and in 3 tumor grades based on histological features.1 However, in recent years, distinct molecular classes of diffuse gliomas have been identified and the revised fourth edition of the internationally accepted World Health Organization (WHO) Classification of CNS Tumors published in 2016 has incorporated important molecular features as an integral part of glioma subclassification.2 These include the mutational status of the isocitrate dehydrogenase 1 and 2 genes (IDH1/2), the codeletion status of chromosome arms 1p and 19q, and histone 3 mutational status, which have been shown to distinguish biologically and clinically distinct diffuse glioma types.3,4 Although neurosurgical resection and adjuvant radio- and chemotherapy may prolong patients’ survival times, most diffuse gliomas recur and limit life expectancy. So far, novel treatments based on biological insights have not been able to improve patient outcomes, and new treatment modalities are needed for patients with diffuse gliomas.
Immunotherapies blocking specific immunomodulatory molecules, so-called immune checkpoint inhibitors, have shown clinically relevant efficacy in a number of tumor types and have emerged as a novel treatment paradigm in clinical oncology. Monoclonal antibodies targeting the immunosuppressive molecule programmed death 1 (PD1) and programmed death ligand 1 (PD-L1) have proven particularly successful and have been approved in melanoma, lung cancer, renal cell cancer, and other tumor types. Among diffuse gliomas, glioblastoma (GBM) has repeatedly been described to overexpress PD-L1 and contain tumor-infiltrating lymphocytes (TILs).5–7 Moreover, efficacy of PD1/PD-L1 inhibitors has been observed in preclinical glioma models and in individual human cases.8–11 Several clinical trials, including large international randomized studies, are enrolling glioblastoma patients and will report efficacy data in this glioma type in the near future. However, so far little data on the immune composition of the tumor microenvironment of other diffuse gliomas are available. Intriguingly, a recent paper reported some differences in the frequency of TILs and PD-L1 expression between tumor types and tumor grades as defined by histological features and the WHO 2007 classification.12,13 Associations of molecular subtypes with tumor immunogenicity and the capacity for immune evasion have been reported for several tumor types. We hypothesized that the recently defined molecular glioma subtypes may also associate with distinct immunological tumor characteristics.14–16 Therefore, we compiled a large series of cases across all diffuse glioma types as defined by the WHO 2016 classification and characterized them with regard to infiltration by TIL subsets and expression of PD-L1. To validate our findings, we performed an analysis of PD-L1 gene expression levels in diffuse gliomas derived from The Cancer Genome Atlas (TCGA) database.
Methods
Vienna WHO II/III Glioma Cohort
Patients treated for diffuse astrocytoma, anaplastic astrocytoma, oligodendroglioma, or anaplastic oligodendroglioma at the Medical University of Vienna were identified from the Neuro-Biobank of the Institute of Neurology, Medical University of Vienna. Diagnosis was performed according to the WHO 2016 classification by a board certified neuropathologist.2 The ethics committee of the Medical University of Vienna approved the study (Vote 078/2004).
Vienna Glioblastoma Cohort
Data on TIL infiltration and PD-L1 expression in 117 IDH-R132H negative newly diagnosed glioblastomas were available from a previous study.6 For the present study, we expanded this cohort by 14 newly diagnosed glioblastoma cases harboring an immunohistochemically detected IDH-R132H. The overall glioblastoma cohort analyzed in this study therefore encompassed 131 cases.
Immunohistochemistry and Molecular Pathology
Tumor tissue was formalin fixed and paraffin embedded according to standard laboratory practice. Specimens presenting with a specific anti–IDH-R132H immunohistochemical signal were scored as IDH mutated (IDH-mut). Anti–IDH-R132H negative WHO II/III glioma cases underwent genetic sequencing of IDH1 and IDH2 genes to detect less common forms of IDH mutations.17 Only WHO II/III glioma specimens with no evidence of IDH1/2 mutations at immunohistochemistry and gene sequencing were classified as IDH wild-type (IDH-wt).18 Immunohistochemistry for cluster of differentiation 3 (CD3) and PD1 was performed as previously published on a Ventana Benchmark Ultra immunostaining system.6 Tissue of human nonmalignant lymph nodes was used as positive control and omission of primary antibody was used as negative control. Immunohistochemistry for PD-L1 was performed using a Dako AutostainerPlusLink immunostaining system and the monoclonal mouse antibody clone 5H1 (dilution 1:400; kindly provided by Dr Lieping Chen), which was used in several studies correlating PD-L1 expression and response to immune checkpoint inhibitors as well as in previous studies of our group investigating PD-L1 expression.6,19–23 In brief, antigen retrieval was performed using the Target Retrieval Solution high pH9 buffer (Dako) followed by antibody incubation using a 1:100 dilution and detection using the EnVision Flex and visualization system (Dako). Human placenta served as positive control and omission of the primary antibody as negative control. The 1p/19q status (1p/19q codeleted [codel] vs 1p/19q non-codeleted [non-codel]) was evaluated in all WHO grade II/III gliomas using fluorescence in situ hybridization as described previously.24
Evaluation of Immunohistochemistry
Tumor infiltrating lymphocytes
Density of CD3+ and PD1+ TILs was evaluated semiquantitatively by overall impression at low microscopic magnification (100x) and scored absent (less than 4 TILs in the entire specimen), sparse (more than 4 but no accumulation), moderate (single areas with accumulation of TILs), dense (TILs throughout the tumor section), or very dense (high frequency of TILs throughout the entire tumor section) according to previously published criteria.6,25 Further, accumulation of TILs in predefined areas (within the viable tumor tissue, in the perivascular region, and, if applicable, in the invasion zone to the surrounding brain parenchyma) was analyzed at higher magnification (200x–400x).
PD-L1 expression
PD-L1 expression was evaluated according to a previously published algorithm: diffuse/fibrillary PD-L1 expression was semiquantitatively assessed according to the following criteria: (i) no positive tumor areas; (ii) expression in <25% of nonnecrotic tumor area; (iii) expression in >25% and <50% of nonnecrotic tumor area; (iv) expression in >50% and <75% of nonnecrotic tumor area; (v) expression in >75% of nonnecrotic tumor area.6 Further, membranous PD-L1 labeling was recorded as percentage of tumor cells presenting with strong, complete, membranous PD-L1 staining. For subsequent statistical analysis, specimens with fibrillary/diffuse PD-L1 expression in >25% of viable tumor tissue or membranous PD-L1 expression in at least 1% of tumor cells were defined as PD-L1 positive.
The Cancer Genome Atlas Dataset
TCGA RNA-Seq level 3 (normalized) data for WHO grade II/III diffuse glioma (TCGA LGG dataset) and glioblastoma (WHO grade IV, TCGA GBM dataset) samples were obtained through the National Cancer Institute Genomic Data Commons portal and preprocessed to produce single data frames of expression values with sample annotations (sample type, IDH mutation status) based on Supplementary Tables S2 and S3 from Ceccarelli et al.3 For the PD-L1 gene expression analysis, TCGA WHO grade II/III gliomas and glioblastoma samples were selected for which RNA-Seq data and annotation information were available (N = 516). For the correlation analysis of PD-L1 gene expression and promoter cytosine-phosphate-guanine island (CGI) methylation levels, a smaller subset of samples was selected due to the lack of glioblastoma samples which have both RNA-Seq and Infinium 450k data available (N = 51 IDH-wt and 4 IDH-mut). Infinium 450k probes measuring DNA methylation at the PD-L1 promoter were selected for analysis based on the previously described methodology.26 Briefly, PD-L1 promoter probes exhibiting functional methylation were determined based on their (i) annotated location and (ii) exhibition of a significant negative correlation methylation/PD-L1 expression (Spearman correlation) in the TCGA LGG dataset. TCGA data were downloaded between November 2015 and January 2016 and have been preprocessed to produce single data frames of expression values for each dataset. Compiled molecular information, comprising IDH1/2 mutation status, 1p/19q codeletion status, and CGI methylated phenotype status were kindly made available by Pierre Bady.27
Statistical Analysis
All pairwise comparisons with 2 groups were performed using Student’s t-test or chi-square test as appropriate. Comparisons with more than 2 groups were performed using ANOVA, with pairwise comparisons performed using Tukey’s honestly significant difference post-hoc analysis. P-values < 0.05 were considered statistically significant. No survival analysis was performed due to the high percentage of censored patients (>90%). Due to the exploratory and hypothesis-generating design of the present study, no adjustment for multiple testing was applied.28 All statistical analyses were performed with SPSS 20.0 software or in R.29
Results
TIL Density and PD-L1 Expression in the Vienna WHO Grade II/III Glioma Cohort
Overall, tissue samples of 43 patients with WHO grade II and III diffuse gliomas were available. Tumor typing according to WHO 2016 classification and patient characteristics are given in Table 1. Infiltration by TILs was present in 22/43 (51.2%) specimens. The TIL density overall was only sparse to moderate, and none of the investigated specimens presented with dense or very dense infiltration by any TIL subtype (Fig. 1). Infiltration was observed diffusely throughout the viable tumor tissue. TILs were only infrequently observed in the invasion zone to the surrounding brain parenchyma, and if present only at sparse density. PD1+ TILs were not detected in any of the investigated WHO grade II/III glioma samples (Supplementary Table S1).
Table 1.
Patients’ characteristic Vienna WHO grade II/III glioma cohort
| Entire Population (n = 43) | ||
|---|---|---|
| N | % | |
| Median age at diagnosis of glioma, y (range) |
38 (20–74) |
|
| WHO 2016 grade | ||
| WHO grade II | 25 | 58.1 |
| WHO grade III | 18 | 41.9 |
| Tumor type | ||
| Diffuse astrocytoma, IDH-mut‒1p/19q non-codel | 15 | 34.9 |
| Anaplastic astrocytoma, IDH- mut‒1p/19q non-codel | 7 | 16.3 |
| Oligodendroglioma, IDH- mut‒1p/19q codel | 8 | 18.6 |
| Anaplastic oligodendroglioma, IDH-mut‒1p/19q codel | 9 | 20.9 |
| Diffuse astrocytoma, IDH-wt‒1p/19q non-codel | 2 | 4.7 |
| Anaplastic astrocytoma, IDH- wt‒1p/19q non-codel | 2 | 4.7 |
| IDH mutation status | ||
| IDH-R132H mutation present | 36 | 83.7 |
| IDH-R132C mutation present | 3 | 7.0 |
| No IDH-R132 mutation present | 4 | 9.3 |
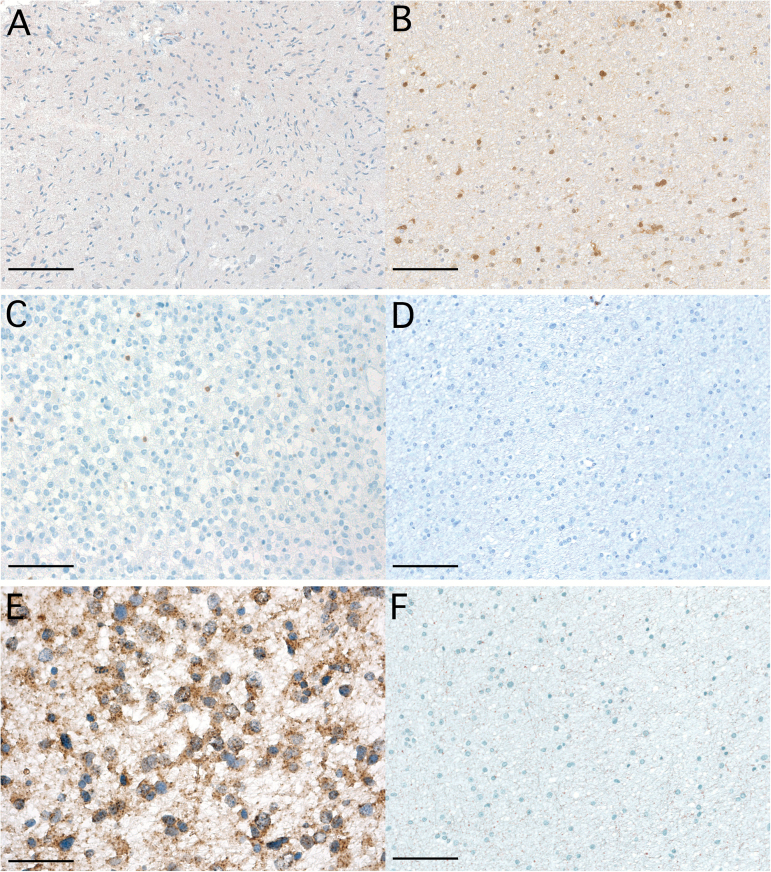
Fig. 1.
Difference in TIL density and PD-L1 expression in IDH-wt and IDH-mut glioma. (A, C, E) IDH-wt glioma WHO grade III without immunohistochemical staining for IDH-R132H mutant and lack of IDH gene 1 or 2 mutations based on gene sequencing (magnification × 100; A; scale bar 250 μm), scattered infiltration with CD3+ TILs (magnification × 200; C; scale bar 100 μm), fibrillary expression of PD-L1 (magnification × 400; E). (B, D, F) IDH-mut WHO grade II glioma presenting with anti–IDH-R132H immunostaining (magnification × 100; B), absence of CD3+ TILs (magnification × 200; D; scale bar 100 μm), lack of PD-L1 expression (magnification × 200; F; scale bar 100 μm).
No correlation between histology (IDH-mut‒1p/19q codel vs IDH-mut‒1p/19q non-codel) and CD3+ TIL density was detected (P = 0.408; chi-square test). Correlation of TIL infiltration and IDH status was not performed due to limited statistical power (only 4/43 IDH-wt specimens).
Diffuse/fibrillary PD-L1 expression in tumor tissue was observed in 22/43 (51.2%) specimens (Fig. 1E). Membranous PD-L1 expression of individual tumor cells was evident in 3/43 (7.0%) specimens (Supplementary Fig S1; Supplementary Table S1). Only 1/43 (2.3%) case, an anaplastic astrocytoma IDH-mut, displayed membranous PD-L1 expression in approximately 10% of viable glioma tumor cells. No statistical difference in frequency of PD-L1 expression according to molecular subtype (IDH-mut‒1p/19q codel vs IDH-mut‒1p/19q non-codel) was observed (P = 0.855 chi-square test). Correlation of PD-L1 expression and IDH mutation was not performed due to limited statistical power (only 4/43 IDH-wt specimens).
TIL Density and PD-L1 Expression in the Vienna Glioblastoma Cohort (WHO Grade IV)
In the 14 cases of IDH-mut glioblastoma we found CD3+ TILs in 3/14 (21.4%) cases. PD1+ TILs were absent in all 14 IDH-mut glioblastoma cases. Diffuse/fibrillary and membranous PD-L1 expression was evident in 1/14 (7.1%) of these specimens, while none of the cases showed membranous PD-L1 expression. Comparing the results from this series of 14 IDH-R132H positive glioblastomas with the data from our previously reported series of 117 IDH-R132H negative glioblastomas,6 we found a strong correlation of IDH status with characteristics of the inflammatory microenvironment: IDH-wt glioblastoma presented significantly more frequently with CD3+ TILs (66.7% vs 21.4%; P = 0.001; chi-square test), PD1+ TILs (17.1% vs 0.0%; P = 0.002; chi-square test), fibrillary/diffuse PD-L1 expression (84.6% vs 7.1%; P < 0.001; chi-square test), and membranous PD-L1 expression (58.1% vs 0%; P < 0.001; chi-square test) than IDH-mut specimens (Supplementary Table S1).
TIL Density and PD-L1 Expression in the Overall Vienna Glioma Cohort (WHO Grades II, III, and IV)
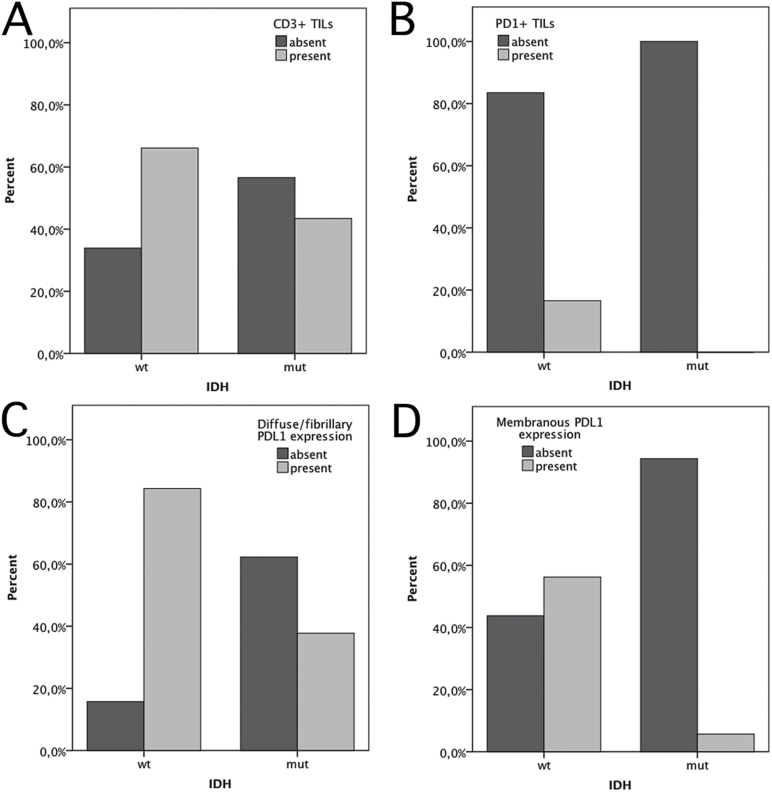
IDH mutation status also correlated with characteristics of the inflammatory microenvironment in the entire Vienna glioma cohort containing 43 WHO grade II/III gliomas and 131 glioblastomas (total n = 174). IDH-wt glioma presented significantly more frequently with CD3+ TILs (66.1% vs 43.4%; P = 0.005; chi-square test; Fig. 2A; Table 2), with PD1+ TILs (16.5% vs 0%; P = 0.002; chi-square test; Fig. 2B; Table 2), with fibrillary/diffuse PD-L1 expression (56.2% vs 5.7%; P < 0.001; chi-square test; Fig. 2C; Table 2), and with membranous PD-L1 expression (56.2% vs 5.7%; P < 0.001; chi-square test; Fig. 2D; Table 2) compared with IDH-mut glioma.
Fig. 2.
Bar graphs illustrating the correlation of TIL infiltration and PD-L1 expression with IDH status in the overall Vienna cohort of WHO grades II–IV diffuse glioma (n = 174). (A) CD3+ TILs in IDH-mut and IDH-wt glioma. (B) Fibrillary/diffuse PD-L1 expression in IDH-mut and IDH-wt glioma. (C) Membranous PD-L1 expression in IDH-mut and IDH-wt glioma.
Table 2.
Presence of TIL infiltration and PD-L1 expression in IDH-wt and IDH-mut gliomas
|
IDH-wt Glioma (n = 121) |
IDH-mut Glioma (n = 53) |
P-value | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| CD3+ TIL | 0.005 | ||||
| Present | 80 | 66.1 | 23 | 43.4 | |
| Absent | 41 | 33.9 | 30 | 56.6 | |
| PD1+ TIL | 0.002 | ||||
| Present | 20 | 16.5 | 0 | 0.0 | |
| Absent | 101 | 83.5 | 53 | 100.0 | |
| Diffuse/fibrillary PD-L1 expression | <0.001 | ||||
| Present | 102 | 84.3 | 20 | 37.7 | |
| Absent | 19 | 15.7 | 33 | 63.3 | |
| Membranous PD-L1 expression | <0.001 | ||||
| Present | 68 | 56.2 | 3 | 5.7 | |
| Absent | 53 | 43.8 | 50 | 94.3 | |
PD-L1 Gene Expression and PD-L1 Gene Promoter Methylation in TCGA Dataset
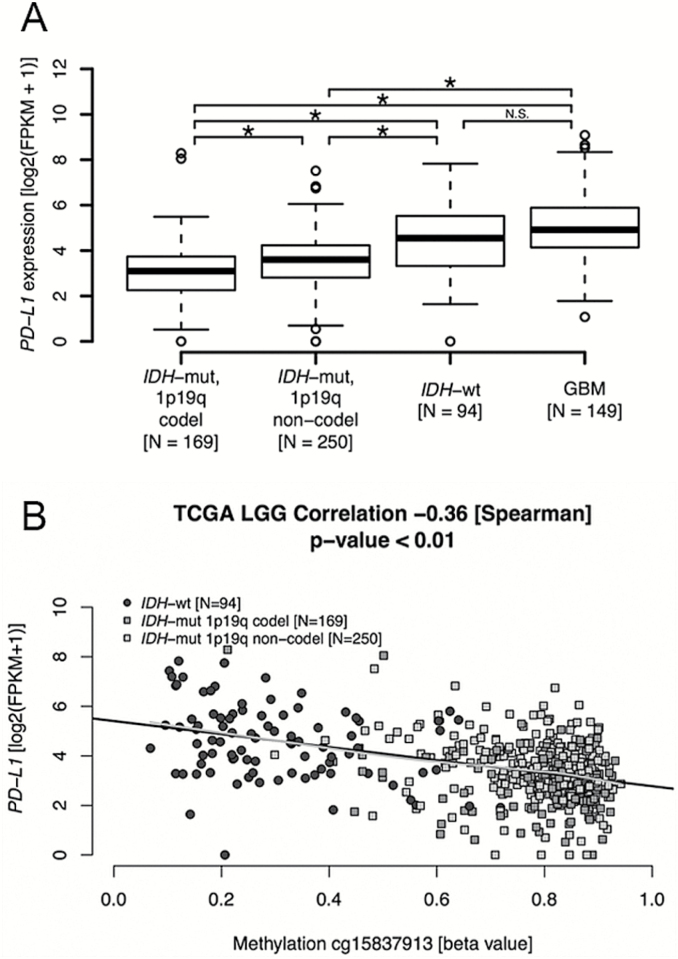
PD-L1 expression differed significantly between molecular glioma subtypes. The lowest expression was observed in IDH-mut‒1p/19q codel gliomas, followed by IDH-mut‒1p/19q non-codel, IDH-wt, and glioblastoma (P < 0.001; Fig. 3A). PD-L1 gene expression was statistically significantly higher in IDH-wt WHO grade II/III gliomas compared with IDH-mut WHO grade II/III gliomas (P = 1.933e-11). However, there was no statistical difference in PD-L1 gene expression levels between IDH-wt WHO grade II/III glioma and glioblastoma cases.
Fig. 3.
PD-L1 gene expression and gene promoter methylation in diffuse glioma. (A) PD-L1 gene expression is significantly lower in IDH-mut WHO II/III glioma than in IDH-wt WHO II/III glioma of TCGA WHO LGG cohort, while IDH-wt WHO II/III gliomas are not different from the cohort of TCGA GBM (for which RNA-Seq data are available). (B) PD-L1 gene expression levels show a significant negative correlation with PD-L1 gene promoter methylation, illustrated for a representative functional CpG (probe cg15837913, −0.36 [Spearman], P < 0.01) in WHO II/III glioma of the dataset of TCGA LGG. Black lines in the plot show the fit of linear regression, in gray local regression using Lowess smoothing. Methylation of the PD-L1 promoter is significantly higher in IDH-mut WHO II/III glioma than IDH-wt WHO II/III glioma (beta-values, P < 0.01, 2-sided Student’s t-test).
PD-L1 gene promoter methylation was studied as a possible explanation for these differing results. A negative correlation of PD-L1 gene expression with PD-L1 gene promoter methylation was observed (cg15837913, −0.36 [Spearman correlation coefficient], P < 0.01, Fig. 3B; cg19724470, −0.27, P < 0.01; Supplementary Fig S1). In line, PD-L1 gene promoter methylation levels were higher in IDH-mut compared with IDH-wt samples, supporting the notion that PD-L1 gene promoter methylation may be causally linked to the lower PD-L1 expression levels in IDH-mut versus IDH-wt samples (P < 0.01; Fig. 3B). In the glioblastoma cohort of TCGA, the number of IDH-mut cases with 450k and RNA-Seq data was too small for meaningful analysis (4/55).
Discussion
In this project, we investigated TIL infiltration and PD-L1 expression in diffuse gliomas and report a significant association of these immunological parameters with their molecular tumor subtype. A previous paper by Garber et al has already documented some difference in TIL infiltration and PD-L1 expression among histological glioma types.12 Our study indicates that the main factor influencing the extent of TIL infiltration and presence of PD-L1 expression in diffuse gliomas is IDH mutational status. IDH-wt cases had more TIL and PD-L1 expression and may be considered more immunologically activated than IDH-mut cases. The association of PD-L1 expression with IDH status was evident both in protein-based analysis using immunohistochemistry in our series and at the gene expression level in the dataset of TCGA.
The mechanistic basis for the association of IDH mutation with the immunologic makeup of the tumor microenvironment remains to be determined. However, the lower PD-L1 gene expression was associated with increased promoter methylation in the IDH-mut gliomas. Based on our data, we hypothesize that the higher PD-L1 promoter methylation is associated with the characteristic hypermethylator phenotype of IDH-mut gliomas that has been shown to be induced by the oncometabolite 2-hydroxglutarate.30–32 Further factors influencing the different immune phenotypes of IDH-wt and IDH-mut gliomas may include epigenetic alterations in other immune-relevant signaling pathways and reprogramming of the metabolism.33,34 In addition, the effects of 2-hydroxglutarate on the tumor microenvironment including TILs will need to be considered.31,35
Clinical trials are currently evaluating the role of PD1/PD-L1 inhibitors in newly diagnosed and recurrent primary glioblastoma. Should these trial efforts show a positive therapeutic effect of these drugs in this tumor type, expansion of the subset of nonglioblastoma diffuse gliomas without IDH mutation should be considered. Such cases (ie, diffuse and anaplastic astrocytoma with IDH-wt status) have poor clinical outcome and limited treatment options and are in need of new therapies.2 Our data and the data from TCGA may suggest that these cases are amenable to immune checkpoint inhibition.
Preclinical studies suggest that IDH mutations may serve as a specific target for vaccination approaches in glioma, and a clinical trial evaluating this approach is currently recruiting patients with IDH-mut diffuse gliomas (NCT02454634, NOA-16).35 Our findings show a low baseline infiltration by TILs in IDH-mut gliomas, and combination of an IDH1-R132H–specific vaccine with other immune-stimulatory agents boosting immune cell migration into the tumor microenvironment may be useful to facilitate an efficient antitumor response.10 Given the low PD-L1 expression found in IDH-mut cases, likely silenced through methylation of the PD-L1 gene promoter, PD1/PD-L1 immune checkpoint inhibitors are not indicated, and other strategies need to be employed—for instance, agonists of co-stimulatory checkpoint molecules. Further, high mutational load is considered an emerging biomarker for response to immune checkpoint inhibitors and pediatric hypermutant glioblastoma resulting from a germline mismatch repair deficiency presented with response to immune checkpoint inhibitors.36,37 However, LGGs present with a rather low mutational load (0.77 mutations/Mb) in comparison to glioblastoma (2.2 mutations/Mb) and especially other tumor entities with high response rates to immune checkpoint inhibitors like melanoma (12.9 mutations/Mb) and lung cancer (9.9 mutations/Mb).38,39 A subset of IDH-mut‒1p/19q non-codel LGG and glioblastoma have been reported to present with a hypermutator phenotype after temozolomide-based therapy.40,41 The hypermutator phenotype has been found in MGMT methylated glioma at recurrence, usually associated with the acquisition of a mutation in MSH6 or another gene of the mismatch repair pathway that provide resistance to temozolomide treatment.41,42 The plethora of acquired mutations may yield neo-epitopes and render the tumors sensitive to immunotherapies. However, the frequency of the hypermutator phenotype is unknown, as only small series have been published so far.40–42 It has been proposed that MSH6 mutations and mutations in other mismatch repair genes may serve as biomarkers to detect the hypermethylation phenotype in temozolomide-treated patients with MGMT methylated glioma. The latter is common in IDH-mut LGG or GBM (>90%), and close to 50% in IDH-wt glioma (WHO grades II to IV), and most importantly these patients are usually treated with an alkylating agent.27
Therefore, combinational strategies taking into account the specific characteristics of the immune microenvironment like TIL density and PD-L1 expression, as well as mutational characteristics and previously applied therapies, might in the future define the immune modulatory therapy approach.
Several different antibodies and protocols have been used for the detection on PD-L1 by immunohistochemistry. Importantly, the resulting signal can vary according to the main targeted domain as antibodies targeting the extracellular domain produce a rather cytoplasmatic signal and antibodies targeting the cytoplasmic domain a rather membranous signal.43,44 In the current study, we used the antibody clone 5H1, which has been used previously to study PD-L1 expression in glioma and other tumor types by our group and others to correlate PD-L1 expression likelihood of response to immune checkpoint inhibitors.6,20,21,23 Here, we observed a diffuse/fibrillary as well as a membranous staining pattern, both of which have been described previously.45 We believe that these staining patterns resemble the heterogeneous microarchitecture of glioblastoma, with the membranous labeling being visible on only epithelioid cancer cells and the diffuse/fibrillary staining reflecting membrane staining on the delicate tumor cell process forming the pathognomic “neurofibrillary matrix” of glial tumors. Most likely only studies on ultrastructure level (eg, using electron microscopy) can answer to which component the PD-L1 is bound in case of the fibrillary staining patterns. Importantly, so far no standard protocol has been published for the detection of PD-L1 expression in glioma, and the optimal method for routine clinical use as well as the cutoff values need to be defined.13 A further interesting parameter could be PD1 expression on TILs besides the proposed predictive value of PD-L1 expression on tumor cells, macrophages, or TILs.23,46,47 Indeed, a retrospective study suggests an increased likelihood of response to PD1 axis targeting immune checkpoint inhibitors in patients with dense infiltration by PD1+ TILs.48 The currently ongoing clinical trials on immune checkpoint inhibitors in glioma patients will provide deeper insights into which characteristics of the inflammatory microenvironment might be of predictive value.
In conclusion, our data show that the immunological tumor microenvironment of diffuse gliomas differs in association with their IDH mutation or CGI methylated phenotype status, respectively, although the mechanistic basis of the observed relationship remains to be elucidated. Our findings suggest that the characteristics of the inflammatory microenvironment may differ according to the genetic glioma subtype and may be relevant for the further conduct and planning of clinical trials investigating the therapeutic value of immune modulatory treatment strategies in glioma patients.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was funded by the research budget of the Medical University of Vienna and a grant by the “Hochschuljubuläumsstiftung” with the project title “Das Immunsystem im Kampf gegen Krebs” and the Swiss National Science Foundation 31003A_163297.
Conflict of interest statement. A.S.B. has received travel support from Roche and Bristol-Myers Squibb (BMS), as well as honoraria from Roche. M.P. has received research support from Böhringer-Ingelheim, GlaxoSmithKline, Merck Sharp & Dohme, and Roche and honoraria for lectures, consultation, or advisory board participation from BMS, Novartis, Gerson Lehrman Group, CMC Contrast, GlaxoSmithKline, Mundipharma, Roche, and AstraZeneca. M.E.H. is a consultant for BMS. All other authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgment
The results published here are in part based upon data generated by TCGA Network, http://cancergenome.nih.gov/
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reuss DE, Kratz A, Sahm F, et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015;130(3):407–417. [DOI] [PubMed] [Google Scholar]
- 5. Wintterle S, Schreiner B, Mitsdoerffer M, et al. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res. 2003;63(21):7462–7467. [PubMed] [Google Scholar]
- 6. Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nduom EK, Wei J, Yaghi NK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18(2):195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reardon DA, Gokhale PC, Klein SR, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res. 2016;4(2):124–135. [DOI] [PubMed] [Google Scholar]
- 9. Kim JE, Patel MA, Mangraviti A, et al. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res. 2017;23(1):124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11(9):504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 12. Garber S, Hashimoto Y, Weathers S, et al. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol. October 2016;18(10):1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berghoff AS, Preusser M. In search of a target: PD-1 and PD-L1 profiling across glioma types. Neuro Oncol. 2016;18(10):1331–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huynh TG, Morales-Oyarvide V, Campo MJ, et al. Programmed cell death ligand 1 expression in resected lung adenocarcinomas: association with immune microenvironment. J Thorac Oncol. 2016;11(11):1869–1878. [DOI] [PubMed] [Google Scholar]
- 15. Koh J, Go H, Keam B, et al. Clinicopathologic analysis of programmed cell death-1 and programmed cell death-ligand 1 and 2 expressions in pulmonary adenocarcinoma: comparison with histology and driver oncogenic alteration status. Mod Pathol. 2015;28(9):1154–1166. [DOI] [PubMed] [Google Scholar]
- 16. Kim JH, Park HE, Cho NY, Lee HS, Kang GH. Characterisation of PD-L1-positive subsets of microsatellite-unstable colorectal cancers. Br J Cancer. 2016;115(4):490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Preusser M, Capper D, Hartmann C; Euro-CNS Research Committee IDH testing in diagnostic neuropathology: review and practical guideline article invited by the Euro-CNS research committee. Clin Neuropathol. 2011;30(5):217–230. [DOI] [PubMed] [Google Scholar]
- 18. Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20(1):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berghoff AS, Ricken G, Wilhelm D, et al. Tumor infiltrating lymphocytes and PD-L1 expression in brain metastases of small cell lung cancer (SCLC). J Neurooncol. 2016;130(1):19–29. [DOI] [PubMed] [Google Scholar]
- 20. Berghoff AS, Ricken G, Widhalm G, et al. Tumour-infiltrating lymphocytes and expression of programmed death ligand 1 (PD-L1) in melanoma brain metastases. Histopathology. 2015;66(2):289–299. [DOI] [PubMed] [Google Scholar]
- 21. Berghoff AS, Fuchs E, Ricken G, et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology. 2016;5(1):e1057388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berghoff AS, Ricken G, Widhalm G, et al. PD1 (CD279) and PD-L1 (CD274, B7H1) expression in primary central nervous system lymphomas (PCNSL). Clin Neuropathol. 2014;33(1):42–49. [DOI] [PubMed] [Google Scholar]
- 23. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woehrer A, Hainfellner JA. Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncol. 2015;4(5):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dahlin AM, Henriksson ML, Van Guelpen B, et al. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24(5):671–682. [DOI] [PubMed] [Google Scholar]
- 26. Kurscheid S, Bady P, Sciuscio D, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. [DOI] [PubMed] [Google Scholar]
- 28. Bender R, Lange S. Adjusting for multiple testing—when and how? J Clin Epidemiol. 2001;54(4):343–349. [DOI] [PubMed] [Google Scholar]
- 29. R: A Language and Environment for Statistical Computing [computer program] Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 30. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tateishi K, Wakimoto H, Iafrate AJ, et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell. 2015;28(6):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schumacher T, Bunse L, Pusch S, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 36. Yuan J, Hegde PS, Clynes R, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer. 2016;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22(8):1865–1874. [DOI] [PubMed] [Google Scholar]
- 38. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. [DOI] [PubMed] [Google Scholar]
- 39. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mahoney KM, Sun H, Liao X, et al. PD-L1 antibodies to its cytoplasmic domain most clearly delineate cell membranes in immunohistochemical staining of tumor cells. Cancer Immunol Res. 2015;3(12):1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Igarashi T, Teramoto K, Ishida M, Hanaoka J, Daigo Y. Scoring of PD-L1 expression intensity on pulmonary adenocarcinomas and the correlations with clinicopathological factors. ESMO Open. 2016;1(4):e000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rittmeyer A, Barlesi F, Waterkamp D, et al. ; OAK Study Group. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest. 2016;126(9):3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.