Abstract
Although oxidative stress has been shown to induce senescence and replication stress independently, no study has implicated unresolved replication stress as the driver for cellular senescence in response to oxidative stress. Using cells exposed to increasing concentrations of hydrogen peroxide, we show that sub-lethal amount of exogenous hydrogen peroxide induces two waves of DNA damage. The first wave is rapid and transient while the second wave coincides with the cells transition from the S to the G2/M phases of cell cycle. Subsequently, cells enter growth arrest accompanied by the acquisition of senescence-associated characteristics. Furthermore, a p53-dependent decrease in Rad51, which is associated with the formation of DNA segments with chromatin alterations reinforcing senescence, and Lamin B1 that is involved in chromatin remodeling, is observed during the establishment of the senescent phenotype. On the other hand, increase in senescence associated-β-Gal activity, a classical marker of senescence and HMGA2, a marker of the senescence-associated heterochromatin foci, is shown to be independent of p53. Together, our findings implicate replication stress-induced endogenous DNA damage as the driver for the establishment of cellular senescence upon sub-lethal oxidative stress, and implicate the role of p53 in some but not all hallmarks of the senescent phenotype.
INTRODUCTION
Cellular senescence is defined as a stable cell cycle arrest elicited in response to a variety of stressors. Intense oncogenic signaling, telomere loss, radiation, chemotherapeutic drugs, bacterial toxins and oxidative stress have all been linked to the induction of the senescent phenotype through direct DNA damage or replication stress-induced DNA damage (1–9). Interestingly, oxidative stress has been shown to induce cellular senescence (8,10,11) and replication stress independently (12,13). There is a lack of evidence to implicate replication stress-induced DNA damage as the driver for the initiation of cellular senescence in response to oxidative stress. The acquisition of cellular senescence is a dynamic process in which changes take place over an extended period of time (14–17). These changes are necessary for the permanent halt of proliferation, failing which cells might escape from senescence to a pro-oncogenic state (5,18). The senescent phenotype is associated with the activation of the tumor suppressor p53 through its phosphorylation at Ser15 residue, which prevents cells carrying genomic lesions from progressing through the cell cycle (19–22) and the acquisition of persistent DNA damage foci or DNA segments with chromatin alterations reinforcing senescence (DNA-SCARS) (5,16,21). DNA-SCARS contain mediators of the DNA damage response (DDR) such as CHK2 and p53, but lack DNA repair proteins and single-stranded DNA (ssDNA)-binding proteins such as Rad51 and the replication protein A (RPA) (21). The absence of DNA repair proteins such as Rad51 in DNA-SCARS has led to the proposal that DNA-SCARS formation is due to an ineffective DNA repair process, which is accelerated in cells deficient in DNA repair proteins of the homologous recombination (HR) repair system, such as Rad51 (21). However, induction of persistent foci and cell growth arrest is insufficient to complete the acquisition of the senescent phenotype. After the establishment of growth arrest, senescent cells undergo extensive changes in chromatin, which contribute to the progression of senescence into a deep senescent state (23). Among these important changes is the formation of senescence-associated heterochromatin foci (SAHF), which are regions of highly condensed chromatin structures observed in vitro and in vivo (23–26). As SAHF sequesters genes controlling proliferation and cell cycle, SAHF formation is an important step leading to the deepening of the senescent phenotype (23,26,27). Along these lines, an increase in expression of High Mobility Group AT-Hook 2 (HMGA2) is associated with the formation of SAHF (28). Furthermore, an important chromatin remodeling process during the establishment of cellular senescence is the formation of cytoplasmic chromatin fragments (CCFs). CCFs are heterochromatin structures that are extruded from the nucleus and processed by lysosomes, leading to the general loss of histones in the senescent cells (17). This extrusion process is facilitated by the disintegration of the nuclear membrane upon the repression of Lamin B1 protein expression, a rapid and early event in the deepening of the cellular senescent state. Lamin B1 downregulation triggers both global and local modifications in chromatin, inducing an extensive chromatin remodeling and consequently enhancing senescent characteristics and deepening of the senescent phenotype (14,17,29). Furthermore, senescent cells secrete cytokines such as interleukin (IL)-6 as part of the senescence-associated secretory phenotype (SASP) (3,30).
In the present report, using increasing concentrations of exogenous H2O2, we demonstrate that replication stress-dependent formation of endogenous DNA damage is responsible for the initiation and establishment of the senescent phenotype induced by sub-lethal oxidative stress. Moreover, we show the critical role of p53 in the inhibition of Rad51 and Lamin B1 but not in the increase in senescence-associated β-galactosidase (SA-β-Gal) activity and HMGA2 expression upon the establishment of the senescent state.
MATERIALS AND METHODS
Cell lines and cultures
L6 rat myoblasts were obtained from Dr Larry Fliegel (Department of Biochemistry, University of Alberta, Canada). L6 myoblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 0.25 mg/ml Geneticin (G418 sulfate) and 1 mM Gentamicin Sulfate at 37°C, with 5% CO2 in a humidified atmosphere. Human retinal pigmental cells-1 (RPE1-hTERT) cells were grown in DMEM-F12 medium, supplemented with 10% FBS and 1 mM Gentamicin sulfate at 37°C, with 5% CO2 in a humidified atmosphere. After 24 h of seeding, the cells were treated with desired H2O2 concentration (Merck-107209) for 1 h and then replenished with fresh media and collected at the respective time intervals as per the experimental requirements. In the case of synchronized condition, after 24 h of seeding, the cells (L6 and RPE1) were treated with 2 mM thymidine for 24 h to synchronize the cells at late G1 phase, followed by H2O2 treatment. L6 cells were also synchronized in G1 using 1 μM lovastatin (Merck Millipore #438185) and late G1 phase using 10 μM aphidicolin (Merck Millipore #178273). In the case of gene silencing experiments, after 24 h of seeding, the cells were maintained in SiRNA mixture for 6 h and then followed by 24 h of synchronization and 1 h of H2O2 treatment. The cell morphology images were captured using Nikon Eclipse TS100 as per the experimental requirements.
Reagents and chemicals
The chemicals used in the study and the corresponding catalog numbers are: hydrogen peroxide (H2O2) (Merck #107209), 1,10-phenanthroline monohydrate (Phen) (Sigma #131377), 4-hydroxy TEMPO (Tempol) (Sigma #176141), MnTMPyP (Calbiochem #475872), thymidine (Sigma #T1895), etoposide (Sigma #E1383), KU55933 (Selleck #S1092) and AZ20 (#S7050).
Crystal violet assay: cell viability
Briefly, after washing with 1× phosphate buffered saline (PBS), cells were stained with 0.5 ml crystal violet solution (0.75% (w/v) crystal violet, 50% (v/v) ethanol, 1.75% (v/v) formaldehyde, 0.25% (w/v) NaCl) for 10 min and then lysed with 1% sodium dodecyl sulphate (SDS)/PBS to solubilize the dye retained in the adherent cells. The absorbance was measured using Spectrafluor Plus spectrofluorometer (TECAN, GmbH, Grödig, Austria) at 595 nm.
SDS-PAGE and western blot analysis
Cell lysates were prepared in radio-immunoprecipitation assay (RIPA) lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM egtazic acid (EGTA), 1% Triton X-100, supplemented with 1 mM Na3VO4 (Sigma, S6508), 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell lysates were resolved by SDS-PAGE and probed with protein of interest using SuperSignal chemiluminescent substrate (Thermo Scientific, 34080) with Kodak Biomax MR X-ray film or Biorad ChemiDoc™ MP System. For re-probing of the same membrane for the detection of different proteins, membrane was stripped with Restore western blot stripping buffer (Thermo Scientific, 21059). The primary antibodies used: rabbit-γH2AX (Ser139) (#2577), rabbit-p-p53 (Ser15) (#9284), rabbit-total p53 (#9282) rabbit- β-tubulin (#2146) and horseradish peroxidase (HRP)-conjugated goat anti-rat secondary antibody (#7077) were purchased from Cell Signaling. Rabbit-p16INK4a (#sc-1207) antibody was purchased from Santa Cruz. Mouse-Rad51 (#ab1837), rabbit-Lamin B1 (#ab16048) and rabbit-MPG (#ab155092) were purchased from Abcam. Rabbit-HMGA2 antibody (#GTX100519) was purchased from GeneTex. Mouse-PAR antibody (#ALX-804–220-R100) was purchased from Enzo lifesciences. Mouse-RB (#554136) and mouse-p21 (#ab7960) were purchased from BD Bioscience. Mouse-β-actin antibody (#A5441) was purchased from Sigma. HRP-conjugated goat anti-mouse secondary antibody (#31430) was purchased from Pierce. HRP-conjugated goat anti-rabbit secondary antibody (#P0448) was purchased from DakoCytomation.
Immunofluorescence assay using confocal microscopy
Cells were seeded on coverslips and were fixed with 4% paraformaldehyde for 30 min at room temperature. After permeabilization with 0.2% TX-100 (Sigma, X100) for 10 min at room temperature, cells were incubated with primary antibody overnight at 4°C. The primary antibodies used: rabbit-Lamin B1 (#ab16048) was purchased from Abcam; Rabbit-γH2AX (Ser139) (#2577), rat-RPA32 (#2208), mouse-Lamin A\C (#4777) and rabbit-histone H3 (#4499) were purchased from Cell Signaling; Rabbit-Rad51 (#sc8349), rabbit-PML (#sc-H238) and rabbit anti-53BP1(#sc-22760) were purchased from Santa Cruz; Mouse-γH2AX (Ser139) (#05–636) was purchased from Merck millipore. After washing with PBS, cells were incubated with either one of the following secondary tagged antibodies: Rhodamine RedTM-X goat anti-rabbit IgG (#R6394), fluorescein isothiocyanate (FITC) goat anti-rabbit IgG (#65611), FITC goat anti-mouse IgG (#F2761), FITC goat anti-rat IgG (#A11006) and Hoechst 34580 (Molecular Probes, H21486) for 1 h at room temperature (RT). The images were captured using Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena, Germany) and pictures were analyzed with Carl Zeiss ZEN 2010 software.
Cell cycle analysis and PI staining
The cells were fixed with 90% methanol for 30 min at room temperature. The cells were then stained with 500 μl of propidium iodide (PI)/RNaseA staining solution for 40 min at 37°C. The stained cells were then acquired using FACSCanto II-PE channel to measure the PI intensities. Flow cytometry data were analyzed using Flowing software V 2.5.1 (http://www.flowingsoftware.com) or FlowJo software.
Measurement of senescence-associated SA-β-Gal activity using C12FDG staining
Senescence-associated β-galactosidase (SA-β-Gal) activity was measured as directed by the nature-protocols, Chainiaux et al. (31).
RNA interference (RNAi) assay
Cellular transfection of SiRNA was performed using Lipofectamine RNAiMAX (Invitrogen, 13778150) in Opti-MEM reduced serum medium (Invitrogen, 31985070) according to the manufacturer’s protocol. SiRNA for p53 and MPG (ON-TARGETplus SMARTpool—Rat TP53 and Rat MPG) were purchased from Dharmacon (Thermo Scientific), and control SiRNA (QIAGEN, Valencia, CA, USA) that is non-homologous to any known gene sequence was used as a negative control.
Neutral comet assay
The neutral comet assay and comet analysis were performed using in-house developed OpenComet V 1.3 software as described previously (32).
Measurement of rat interleukin-6 (IL-6) using ELISA
The conditioned media in the wells were collected at 72 h post-H2O2 treatment. Rat IL-6 secretion in the media was measured using rat IL-6 enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen, CA, USA) according to manufacturer’s protocol.
RNA isolation, reverse transcription and real-time PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen, #74104) according to the manufacturer’s instructions. Reverse transcription was performed using the TaqMan Reverse Transcription Reagents kit (Life technologies, N8080234). Real-time quantitative polymerase chain reaction (PCR) reaction was carried out with SYBR-Green (Applied Biosystems, #4309155), detection using ABI PRISM 7300 (Applied Biosystems). The sequences of the primers used for PCR were as follows: rat p53 (FP:GCTCCCCTGAAGACTGGATAA, RP: ATTAGGTGACCCTGTCGCTG), rat Lamin B1 (FP:AAGGCTCTCTACGAGACCGA, RP: TCCTTCTTAGCATAATTGAGCAGC), rat RAD 51 (FP:CTGCGAAGTGTGTTTGAGCC, RP:AGCCATTACTGTCTCGCAGC), rat 18S (FP:CATTCGAACGTC TGCCCT, RP:GTTTCTCAGGCTCCCTCTCC). Relative gene expression was obtained after normalization with endogenous 18S and determination of the difference in threshold cycle (Ct) between treated and untreated cells using 2-ΔΔCt method (33).
Chromatin extraction
The chromatin extraction was done as described previously (34). At the end of required time point, the cells were harvested and the cytosolic protein fraction was extracted by incubation in hypotonic buffer (10 mM hydroxyethyl piperazineethanesulfonic acid (HEPES), pH 7, 50 mM NaCl, 0.3 M sucrose, 0.5% Triton X-100, supplemented with protease inhibitor; Roche) for 15 min on ice and centrifuged at 9000 rpm for 5 min. The obtained supernatant was the cytosolic fraction and was transferred into a new tube. The remaining pellet was re-suspended with nuclear buffer (10 mM HEPES, pH 7, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40 and protease inhibitor cocktail) and incubated on ice for 10 min and then centrifuged at 14 000 rpm for 5 min. The supernatant obtained was the nuclear fraction. The remaining pellet was re-suspended in the chromatin lysis buffer (10 mM HEPES, pH 7, 500 mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitor cocktail), sonicated at low amplitude for 30 s and then centrifuged for 5 min at 14000 rpm; the supernatant obtained was the chromatin extract. The total protein concentration in the chromatin extract was determined using Coomassie Plus protein assay reagent (Pierce, Thermo Fisher Scientific Inc, Rockford, IL, USA), according to manufacturer’s instructions and then proteins of interest were detected using western blot.
ImageJ analysis
Nuclear morphometric analysis (NMA): the nuclear analysis of H2O2-treated cells were performed using the NMA plugin as directed by the authors (35).
Foci counting in H2O2-treated cells: the H2AX, 53BP1, Rad51 and RPA32 foci in H2O2-treated cells were counted using Focipicker 3D ImageJ plugin (http://rsb.info.nih.gov/ij/plugins/foci-picker3d/index.html) or FoCo a matlab-based software (https://sourceforge.net/projects/focicount) (36). The parameters of the both the plugins were adjusted dynamically as per the images requirements. A total of at least 50 nuclei were counted per treatment and per time point. The data were represented as average number of foci per nucleus.
Co-localization finder: the co-localization between RPA32 and 53BP1 foci was found using co-localization finder plugin (http://rsb.info.nih.gov/ij/plugins/colocalization-finder.html). The respective channel images representing the RPA32 and 53BP1 foci were used to find the overlapping pixels between the two proteins localization at the DNA damage foci and the Pearson’s correlation ratio, the masked and overlapping pixels were generated.
Metaphase chromosome spreads preparation
After 12 h of H2O2 treatment, colcemid to a final concentration of 150 ng/ml was added for 4 to 6 h. The cells were then collected and 6 ml of 75 mM KCl (pre-warmed to 37°C) was added dropwise for first 1 ml, while gently vortexing and then incubated for 16 min at 37°C. After spinning at 1000 rpm for 10 min, the pellet was re-suspended and 5 ml of fixative (3:1 solution of methanol:glacial acetic acid (freshly prepared)) was added and incubated for 20 min at 4°C. The fixative step was repeated another two times and after spinning at 1000 rpm for 10 min, the cell pellet was re-suspended with small volume of fixative and then added dropwise onto the slides and allowed to dry overnight. The slides were then stained with Hoechst and the images were captured using Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena, Germany) and pictures were analyzed with Carl Zeiss ZEN 2010 and ImageJ software.
Statistical analysis
Student’s t-test was performed when appropriate, using the Microsoft Excel software. The P-value (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001 and ns—not significant) of <0.05 considered significant. The values represent the average of at least three independent experiments and error bars represent ± SD of the mean.
RESULTS
Sub-lethal oxidative stress induces two waves of DNA damage, growth arrest and senescence-like morphology
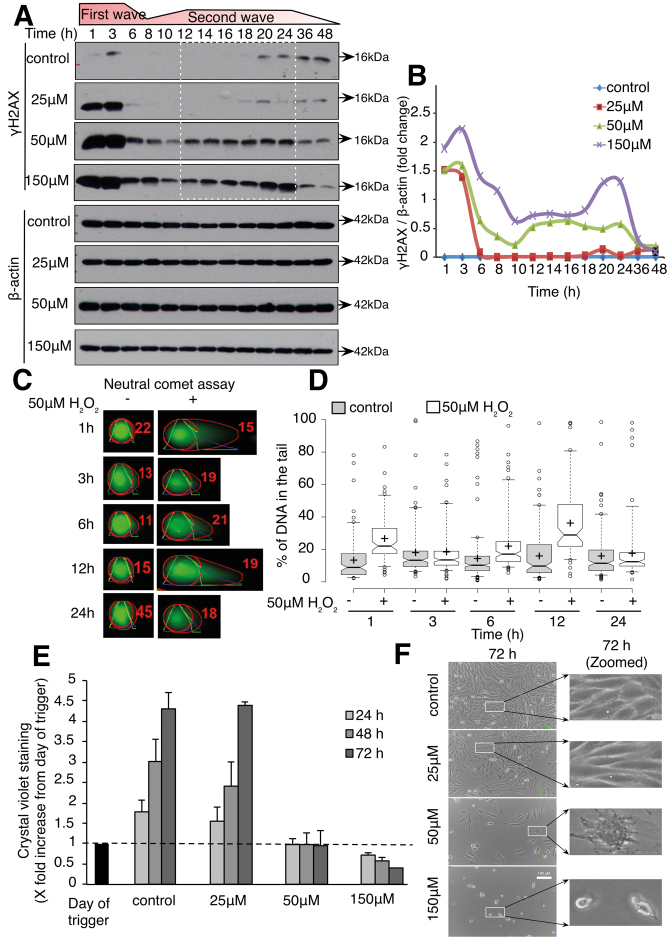
Exposure of L6 cells to 25, 50 and 150 μM of exogenous H2O2 resulted in a significant increase in γH2AX, 1 h after exposure to the oxidant (Figure 1A and B). Interestingly, while γH2AX receded to baseline level 6 h after the initial oxidative stress when 25 μM H2O2 was used, a second wave of γH2AX between 12 and 24 h was detected in cells exposed to 50 and 150 μM H2O2 (Figure 1A and B). In order to confirm that γH2AX detected using western blot reflected DNA damage, neutral comet assay was performed in cells exposed to 50 μM H2O2. Two peaks of DNA damage were detected at 1 and 12 h following cells’ exposure to the oxidant (Figure 1C and D). Interestingly, while cells exposed to 25 μM H2O2 showed a slight delay in the rate of proliferation following cells’ exposure to the oxidant, no significant change in the cells’ morphology could be detected up to 72 h after the redox stress (Figure 1E and F); however, increasing the concentration of H2O2 to 50 μM resulted in the inhibition of cells’ proliferation, as evidenced by absence of an increase in cell number as well as the acquisition of enlarged and flattened cells, a cell morphology reminiscent of a senescent-like phenotype. Interestingly, inhibition of the first wave of γH2AX using the iron chelator phenanthroline and two reactive oxygen species (ROS) scavengers Tempol and MnTMYp prevented the second increase in γH2AX in cells exposed to 50 μM H2O2 compared to control cells (Supplementary Figure S2A). Furthermore, pre-incubation of the cells with phenanthroline, Tempol or MnTMPyP rescued the cells’ growth arrest observed in control cells 72 h after exposure to the sub-lethal oxidative stress (Supplementary Figure S2E). Taken together, these results support that the first wave of γH2AX is required to drive the onset of the second wave of γH2AX and the cells’ growth arrest that follows. On the other hand, 150 μM H2O2 induced cell death with apoptotic characteristics such as cell size shrinkage (Figure 1E and F). Intrigued by the association between the appearance of a second wave of DNA damage and different cell fate, we set out to gain a deeper understanding of the origin of the second wave of DNA damage in cells exposed to 50 μM H2O2 and asked if the senescent-like morphology observed reflected a true cellular senescent state.
Figure 1.
Sub-lethal oxidative stress induces two waves of DNA damage, growth arrest and senescence-like morphology. L6 cells were treated with indicated concentration of H2O2 for 1 h and assessed for γH2AX and DNA damage by comet assay, cell growth and cell morphology. (A) Western blot of γH2AX kinetics in 25, 50 and 150 μM H2O2-treated cells. (B) Graph showing the densitometry plots of γH2AX from (A) in 25, 50 and 150 μM H2O2-treated cells. (C) Representative comets of Neutral comet assay measurement of DNA damage in control and H2O2-treated cells. (D) Percentage of DNA in the comet tails. Data are representative of three independent experiments, and at least 100 comets were scored for each time point. The box center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to 5th and 95th percentiles; outliers are represented by dots; crosses represent sample means. (E) Adherent cell density using crystal violet in 25, 50 and 150 μM H2O2-treated cells. Values are normalized to the day of trigger. (F) Cell morphology at 72 h observed using phase contrast microscope. Cell shape and morphology at 72 h were zoomed and highlighted in the second column.
DNA damage response pathways are activated upon the induction of DNA damage by a sub-lethal oxidative stress
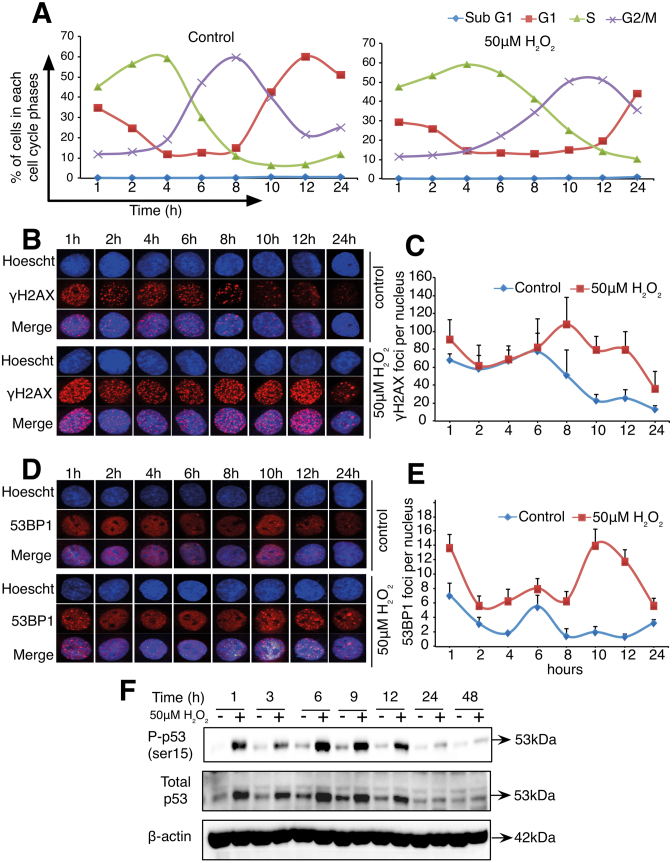
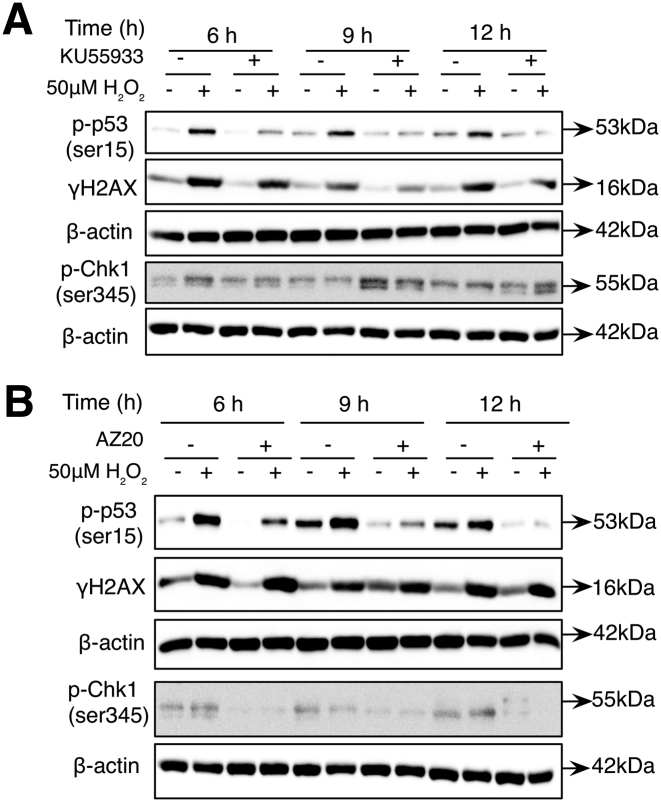
In order to gain a better understanding of the origin of the second wave of DNA damage in cells exposed to 50 μM H2O2, L6 cells were synchronized using lovastatin, aphidicolin or thymidine before the addition of the oxidant. Independent of the approach used to synchronize the cells, a consistent S and G2/M phase extension in response to H2O2 compared to control cells was observed (Figure 2A and Supplementary Figure S1A and C). Moreover, western blot analysis of γH2AX in lovastatin and aphidicolin synchronized cells or the detection of γH2AX foci in cells synchronized using thymidine block confirmed the induction of two waves of γH2AX in cells exposed to 50 μM H2O2 (Figure 2B and C; Supplementary Figure S1B and D). Interestingly, note that cell cycle analysis of non-synchronized cells exposed to 50 μM H2O2 also showed that the second increase in γH2AX (at 12 h) corresponded to an increase in cells at the late S to G2/M boundary (Supplementary Figure S9A and B). Moreover, in aphidicolin-synchronized cells exposed to a sub-lethal oxidative stress, detection of cyclin A1, a S-phase cyclin and the detection of cyclin B1, a G2\M cyclin show an overlap between cyclin A1 and cyclin B expression. Indeed, cyclin A1 is detected up to 14 h and cyclin B1 from 9 to 16 h following cells exposure to the sub-lethal oxidative stress (Supplementary Figure S1E and F). Taken together, these data confirmed that the second increase in γH2AX occurs at the S to G2/M boundary of the cell cycle (Supplementary Figure S1A and C). To further support DNA damage as the origin of the two waves of γH2AX, detection of the p53-binding protein 1 (53BP1), a well-known DDR factor recruited to nuclear structures at the site of DNA damage and phosphorylation of p53 at ser15 were then assessed in cells exposed to 50 μM H2O2. Similar to the results obtained with the detection of γH2AX, two waves of 53BP1 foci were detected in cells synchronized with the thymidine block technique. The first wave of 53BP1 foci occurred 1 h after the sub-lethal redox stress and the second wave of 53BP1 foci was detected at 10 h after the exposure of the cells to the oxidant (Figure 2D and E). In addition, two waves of p53 expression and p53 phosphorylation at ser15 were also detected in cells exposed to 50 μM H2O2 (Figure 2F). Finally, to assess if the ATM- or the ATR-dependent DDR pathways were activated at the time of the second wave of DNA damage, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 (ATR) kinases were inhibited before γH2AX and p53 phosphorylation were detected. Figure 3A shows that addition of the ATM inhibitor (KU55933) 3 h after the cells’ exposure to H2O2 prevented the increase in γH2AX and p53 phosphorylation at 6, 9 and 12 h after the initial exposure of the cells to the oxidant (Figure 3A). On the other hand, addition of the ATR inhibitor (AZ20) 2 h before each time point (6, 9 and 12 h) inhibited only the phosphorylation of p53 but not the increase in γH2AX (Figure 3B). As a control for the specificity of the inhibitors, it is shown that phosphorylation of CHK1 (S345) was only inhibited by AZ20 (Figure 3B) and not by KU55933 (Figure 3A).
Figure 2.
Sub-lethal oxidative stress induces two waves of γH2AX, 53BP1 foci and phosphorylated p53 (Ser15) in synchronized L6 cells. L6 cells were synchronized using thymidine before being treated with 50 μM H2O2 for 1 h and cell cycle analysis, γH2AX foci, 53BP1 foci, p53 expression and p53 phosphorylation were assessed. (A) Line plots capturing the percentage of cells in each cell cycle phase at the indicated time points. (B) Representative nucleus showing γH2AX foci in control and H2O2-treated cells. (C) Quantification of the average number of γH2AX foci per nuclei from the analysis of at least 30 to 40 nuclei. (D) Representative nucleus showing 53BP1 foci in control and H2O2-treated cells. (E) Quantification of the average number of 53BP1 foci per nuclei from the analysis of at least 30–40 nuclei. (F) Kinetics of phosphorylated p53 (ser15) and total p53 in control and 50 μM H2O2-treated cells. Plots shown in (C) and (E) are representative of at least two independent experiments and error bars represent ± SD of number of foci per nucleus counted within the experiment.
Figure 3.
ATM and ATR are activated after endogenous DNA damage induction. Cells were synchronized using thymidine before being treated with 50 μM H2O2 for 1 h. (A) Western blot analysis of the levels of γH2AX and phosphorylated p53 (ser15) at 6, 9 and 12 h following synchronized cells exposure to 50 μM H2O2 and the inhibition of ATM using KU55933 3 h after 50 μM H2O2 treatment. (B) Western blot analysis of γH2AX and phosphorylated p53 (ser15) after inhibition of ATR using AZ20 at the time of second wave of DNA damage (6–12 h). AZ20 was added 2 h before the collection of cells at the indicated time points.
The second wave of γH2AX in cells exposed to sub-lethal oxidative stress is due to an unresolved replication stress leading to replication forks collapse and the conversion of single-strand DNA breaks to double-strand DNA breaks
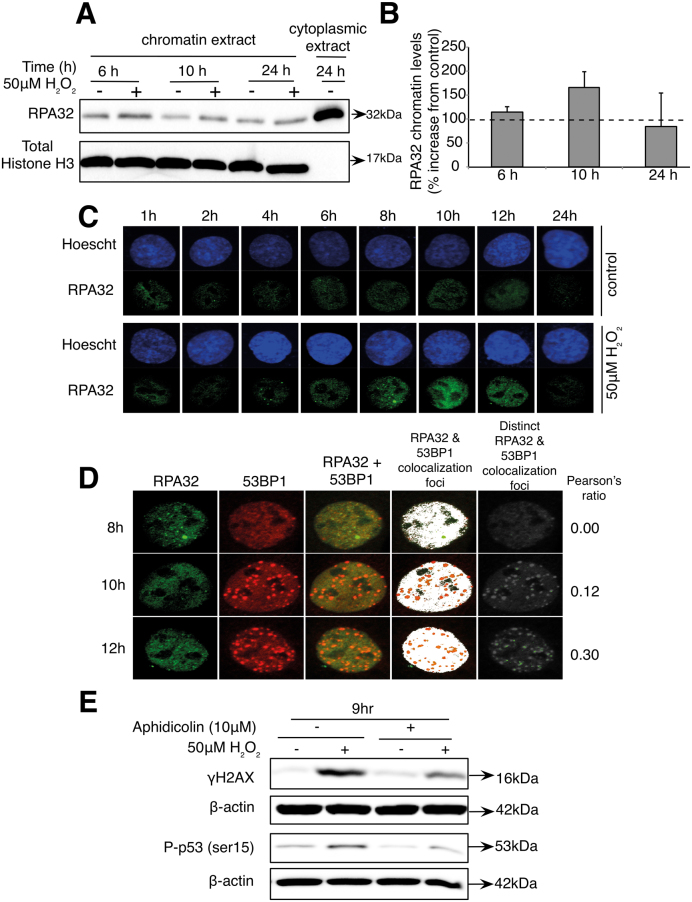
Interestingly, irrespective of the methods used to synchronize the cells, the second wave of γH2AX foci consistently corresponded with the cells’ transition between the S and the G2/M phases of the cell cycle. Moreover, the overlap of cyclin A1 and cyclin B1 detected in aphidicolin-synchronized cells further supported that the second increase in γH2AX occurred at the late S phase–G2/M boundary suggested that the second wave of DNA damage might have originated in S phase before the cells rolled into G2/M. We, hence, reasoned that the second wave of DNA damage detected in cells exposed to 50 μM H2O2 might result from the generation of endogenous DNA damage as a result of unresolved replication stress (9,19,37–41). The RPA complex is an essential regulator of eukaryotic DNA metabolism. RPA avidly binds to ssDNA through multiple oligonucleotide/oligosaccharide-binding folds and coordinates the recruitment and exchange of genome maintenance factors to regulate DNA replication, recombination and repair. At the time of the second wave of DNA damage, we show a transient recruitment of the replication protein A32 (RPA32) at the chromatin (Figure 4A and B). Furthermore, the detection of RPA32 nuclear foci from 4 to 12 h exposure to 50 μM H2O2 and the co-localization of RPA32 and 53BP1 (shown by the Pearson’s correlation ratio) support the presence of ssDNA and DSB at the time of the detection of the second wave of γH2AX (Figure 4C and D). Finally, incubation with aphidicolin, a DNA polα inhibitor, after 1 h of H2O2 treatment inhibited the second wave of γH2AX and the subsequent phosphorylation of p53 9 h after the initial oxidative stress. Together, these results provide evidence that the progression of the replication fork is required for the induction of the second wave of γH2AX (Figure 4E).
Figure 4.
Replicative stress induced by sub-lethal oxidative stress leads to the formation of DSB following replicative fork collapse. Cells were synchronized using thymidine before being treated with 50 μM H2O2 for 1 h. (A) Western blot showing the levels of RPA32 at the chromatin level at indicated time. (B) Densitometry plots of the chromatin level of RPA32 at 6, 10 and 24 h from (A). (C) Immunofluorescence images showing the levels of RPA32 foci in control and H2O2-treated cells in representative nucleus at indicated time points. (D) Co-localization analysis of 53BP1 and RPA32 foci. (E) Western blot of γH2AX and phosphorylated p53 (ser15) at 9 h, after addition of 10 μM aphidicolin (DNA polα inhibitor) 1 h after 50 μM H2O2 treatment.
Sub-lethal oxidative stress induces a state of cellular senescence
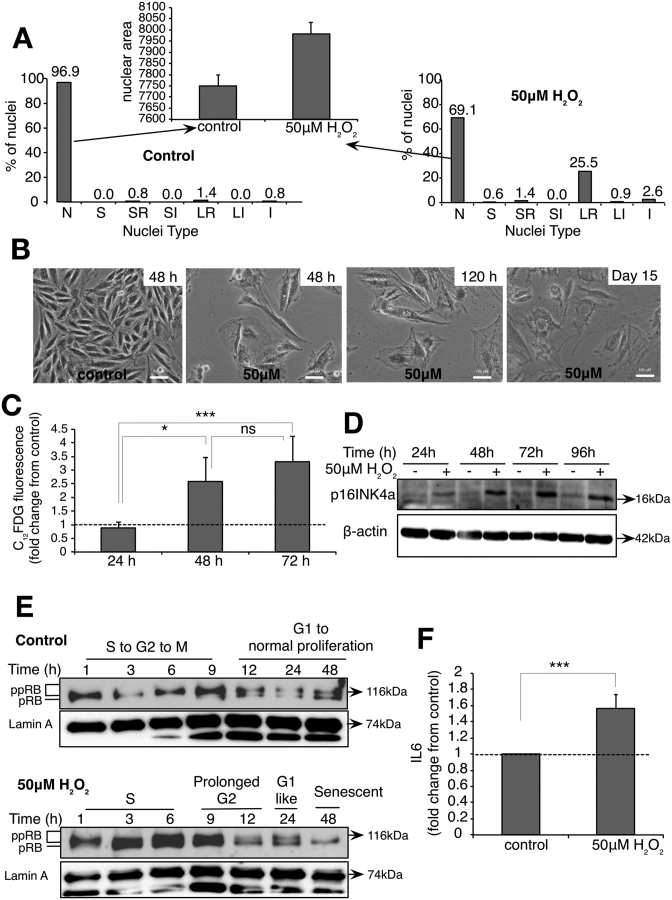
As shown in Figure 1, exposure of the cells to sub-lethal oxidative stress was associated with growth arrest and a senescent-like phenotype. Using an NMA that provides a direct and objective way of screening normal, senescent, apoptotic and nuclear irregularities that occur during failed mitosis or mitotic catastrophe, we show that 72 h following exposure to 50 μM H2O2, 25.5% of synchronized cells had a senescent nucleus, 1.4% of the nuclei were apoptotic and 2.6% showed a nucleus characteristic of mitotic catastrophe. Although this analytic tool predicted 69.1% of the cells to have normal nuclei following treatment with 50 μM H2O2, the average area of these nuclei was higher than the nuclei in control cells, indicating transition to the senescent phenotype (Figure 5A). Indeed, passage to a senescent phenotype is corroborated by the flat and enlarged morphology of cells at 48 h, 120 h and 15 days after exposure to 50 μM H2O2 (Figure 5B). Moreover, an increase in acidic SA-β-Gal activity (Figure 5C) and activation of p16–RB (Figure 5D and E), but not the p21 axis (Supplementary Figure S3A) and secretion of IL-6 (Figure 5F), the most prominent cytokine of the SASPs was detected 48 h after the initial oxidative stress.
Figure 5.
Exposure of L6 cells to 50 μM exogenous H2O2 induces senescence and senescence-associated phenotypic characteristics. Cells were synchronized using thymidine before being treated with 50 μM H2O2 for 1 h and senescence-associated characteristics were assessed. (A) NMA analysis of the nuclei of synchronized cells exposed to 50 μM H2O2 72 h post-H2O2 treatment. Nuclei types and its corresponding phases are described as below. N—normal (interphase), S—small (mitosis), SR—small regular (apoptosis), SI—small irregular (mitotic abnormality), LR—large regular (senescence), LI—large irregular (mitotic catastrophe) and I—irregular (mitotic catastrophe). (B) Fifty micromolars H2O2-treated cells acquired enlarged cell shape and size. (C) C12FDG fluorescence staining of SA-β-Gal activity in synchronized cells exposed to 50 μM H2O2 at the indicated time points. (D) Western blot showing the kinetics of p16INK4a expression at indicated time points. (E) Western blot showing the kinetics of hypo-phosphorylated RB (pRB) and hyper-phosphorylated levels of RB (ppRB) upon H2O2 treatment. Hypo-phosphorylated RB was observed in 50 μM H2O2 from 12 h and sustained up to 48 h (F). Secreted IL-6 levels in the media were measured at 72 h following the cells exposure to 50 μM H2O2 using ELISA. The data shown in (C) and (F) represent the mean of at least three independent experiments and the error represents ± SD. P-value (one-tailed, Student’s t-test, *P-value < 0.05, ***P-value < 0.001, ns—not significant) calculation is based on the comparison between the respective time points as indicated in the figures.
The cellular senescent state induced by sub-lethal oxidative stress is characterized by the formation of persistent DNA damage foci
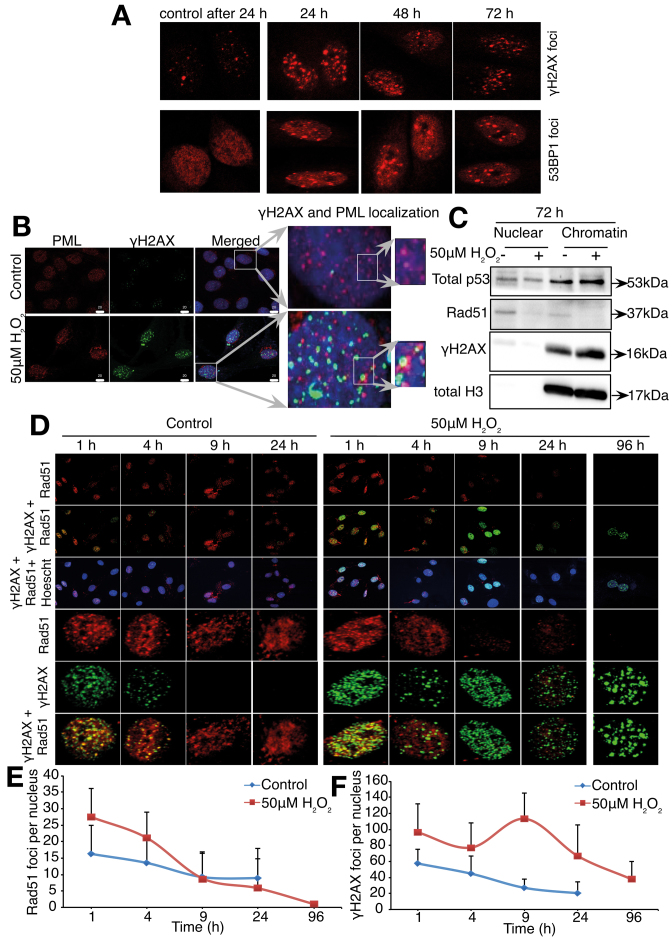
Severe or irreparable DNA damage causes cells to senesce with persistent DNA damage foci. These persistent changes precede the establishment of senescence-associated characteristics, including growth arrest (20,21,42) and SASP (3,43). In order to show that the senescent state induced by sub-lethal oxidative stress is also associated with these changes, the presence of persistent γH2AX and 53BP1 foci at 24, 48 and 72 h after the initial cells exposure to 50 μM H2O2 was assessed. Results show that persistent γH2AX and 53BP1 foci were detected in cells exposed to sub-lethal oxidative stress (Figure 6A). Persistent DNA damage foci/DNA-SCARS have been shown to exhibit promyelocytic leukemia (PML) nuclear bodies at their periphery and the absence of DNA repair protein Rad51 (21). Indeed, Figure 6B shows the co-localization of PML nuclear bodies at the periphery of γH2AX foci 72 h following exposure to the oxidant. In addition, persistent γH2AX foci concomitant with a decrease in Rad51 foci (Figure 6D–F) and repression of Rad51 expression at the whole chromatin level (Figure 6C) were detected 72 h after exposure of the cells to H2O2.
Figure 6.
Exposure of L6 cells to 50 μM exogenous H2O2 induced senescence-associated persistent foci/DNA-SCARS. Cells were synchronized in late G1 using thymidine before being treated with 50 μM H2O2 for 1 h and the appearance of senescence-associated persistent foci/DNA-SCARS assessed. (A) γH2AX and 53BP1 foci after 24, 48 and 72 h post-H2O2 treatment. (B) Immunofluorescence images showing the co-localization of PML and γH2AX 96 h after exposure to H2O2. (C) Total p53, Rad51 and γH2AX protein expression levels in a chromatin extract 72 h after cells exposure to 50 μM H2O2. Total histone H3 is used as a loading control. (D) Rad51 and γH2AX foci in control and 50 μM H2O2-treated cells at indicated time points. Representative immunofluorescence images of Rad51 and γH2AX foci in a nucleus are shown. Line plots showing the average of Rad51 foci (E) and γH2AX foci (F) from at least 30–40 nuclei at indicated time points are also shown. Error bars represent ± SD of number of foci per nucleus counted within the experiment.
The cellular senescent state induced by sub-lethal oxidative stress is characterized by an increase in HMGA2 expression and formation of CCF-like micronuclei
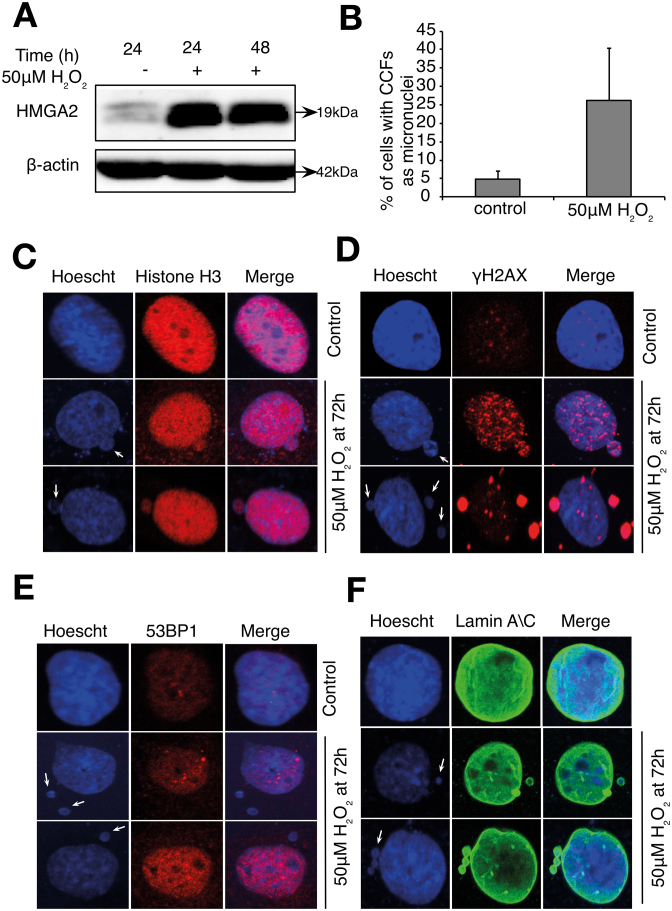
High-level expression of HMGA2 is sufficient to induce SAHF formation, an important step leading to the deepening of the senescent phenotype, as SAHF sequesters genes controlling proliferation and cell cycle, thus reinforcing permanent cell cycle arrest (26,28,44). As shown in Figure 7A, cells exposure to 50 μM H2O2 induces an increase in HMGA2 expression. Furthermore, extrusion of chromatin structures in the form of micronuclei staining positive for γH2AX and Histone H3 were also observed in cells treated with 50 μM H2O2 (Figure 7B–D). Interestingly, an increase in the chromatin structures observed in the cytoplasm did not stain for 53BP1 (Figure 7E), but were positive for Lamin A\C (Figure 7F), supporting these structures as micronuclei.
Figure 7.
Exposure of L6 cells to 50 μM exogenous H2O2 induces HMGA2 and extrusion of chromatin as micronuclei structures. Cells were synchronized in late G1 using thymidine before being treated with 50 μM H2O2 for 1 h and then observed for senescence-associated chromatin changes. (A) Western blot images showing the HMGA2 protein levels. (B) The number of nucleus with extruded chromatins are counted manually and percentage of cells with chromatin extrusions in control and H2O2-treated cells are shown in (A). The data shown represent the mean of at least three independent experiments and the error bars represent ± SD. (C and D) Immunofluorescence images showing the blebbing of chromatin from nucleus to cytoplasm at 72 h. Arrows indicate the blebbing and extruded chromatin. Presence of chromatin in CCFs is confirmed by occurrence of (C) histone H3 and (D) γH2AX in control and H2O2-treated cells at 72 h. (E) Absence of 53BP1 staining in extruded chromatin in 50 μM H2O2-treated cells at 72 h. (F) Presence of Lamin A\C staining in extruded chromatin in 50 μM H2O2-treated cells at 72 h.
Absence of p53 mitigates growth arrest and prevents the decrease in Rad51 and Lamin B1 expression
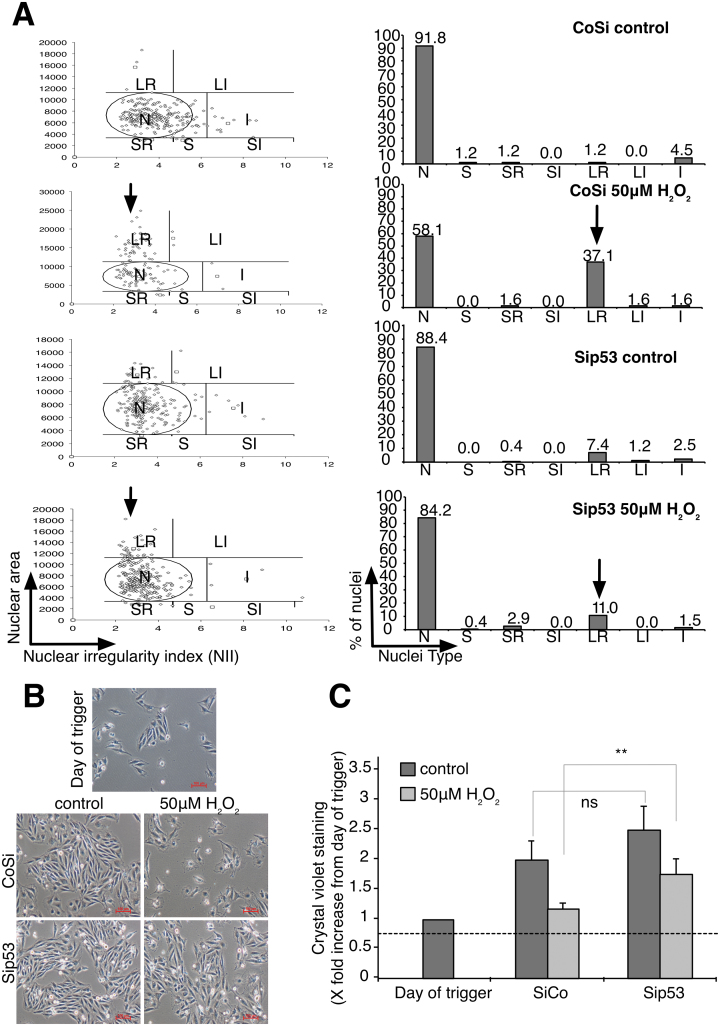
In the absence of p53, the percentage of cells showing signs of nuclear senescence and morphology, assessed by NMA, decreased significantly (Figure 8A) and cell growth arrest was mitigated (Figure 8B and C). Silencing p53 also prevented the repression of Rad51 protein (Figure 9A and B) as well as prevented the repression of Rad51 mRNA (Figure 9C). In addition, in agreement with the repression of Rad51 expression downstream of the second wave of DNA damage (Supplementary Figure S7A), inhibition of ATM 3 h after first wave of DNA damage prevented the phosphorylation of p53 and rescue of Rad51 expression at 9 and 12 h post-H2O2 treatment (Supplementary Figure S7B). Interestingly, absence of p53 did not affect the appearance of the second wave of γH2AX and the S phase delay despite the maintenance of Rad51 expression (Supplementary Figure S8A and B). Finally, silencing p53 expression prevented the repression of Lamin B1 protein and mRNA expression observed 6 h after cells exposure to 50 μM H2O2 (Figure 9D–F) and was accompanied by a decrease of Lamin B1 in the perinuclear region at 24 and 48 h (Figure 9G). On the other hand, a significant increase in genomic instability, as seen by an increase in micronuclei and aneuploid cells, was detected in p53 null cells exposed to 50 μM H2O2, compared to control cells (Supplementary Figure S4A–D). Interestingly, the absence of p53 did not have a notable effect on the SA-β-Gal activity (Supplementary Figure S4E) and HMGA2 expression (Supplementary Figure S4F).
Figure 8.
Endogenous DNA damage induced p53-dependent senescence growth arrest. Cells were synchronized with thymidine before being treated with 50 μM H2O2 for 1 h. (A) NMA analysis of the nuclei of untreated and 50 μM H2O2-treated synchronized cells at 72 h in CoSi and Sip53-transfected cells. Nuclei types and its corresponding phases are described as below. N—normal (interphase), S—small (mitosis), SR—small regular (apoptosis), SI—small irregular (mitotic abnormality), LR—large regular (senescence), LI—large irregular (mitotic catastrophe) and I—irregular (mitotic catastrophe). (B) Phase contrast images showing the cell morphology of untreated and 50 μM H2O2-treated cells in CoSi and Sip53-transfected cells 48 h following cells exposure to the oxidant. (C) Cells were stained with crystal violet to quantify adherent cell density of untreated and 50 μM H2O2-treated cells at 48 h upon silencing with CoSi and Sip53. Values are normalized to the day of trigger. The data shown in (C) represent the mean of at least three independent experiments and the error represents ± SD. P-value (one-tailed, Student’s t-test, **P-value < 0.01, ns—not significant) of the comparison groups are indicated in the figures.
Figure 9.
p53 transcriptionally repressed Rad51 and Lamin B1. Cells were synchronized using thymidine before being treated with 50 μM H2O2 for 1 h. (A) Western blot showing Rad51expression in CoSi and Sip53-transfected cells. (B) Rad51 protein expression at 24 and 48 h following cells exposure to 50 μM H2O2. (C) Rad51 mRNA expression in CoSi and Sip53-transfected cells 24 and 48 h after cells exposure to 50 μM H2O2. Data shown are the representative of four independent experiments and the error represents ± SD. (D) Western blot showing Lamin B1 expression in CoSi and Sip53-transfected cells 24 and 48 h following cells exposure to 50 μM H2O2. (E) Lamin B1 mRNA expression at 24 and 48 h upon silencing p53. Data shown are the representative of four independent experiments and the error represents ± SD. (F) Western blot showing Lamin B1 expression at indicated time points in 50 μM H2O2-treated cells. (G) Depletion of Lamin B1 in H2O2-treated cells at indicated time points, shown by immunofluorescence. White boxed nucleus are magnified to show the intensity of Lamin B1 at various time points indicated (right column). The Lamin B1 fluorescence intensity along the diameter of the nucleus is also quantified. Note the gradual decrease of Lamin B1 fluorescence intensity in 50 μM H2O2-treated cells (right bottom column).
Two waves of DNA damage followed by cellular senescence are not unique to L6 cells and sub-lethal oxidative stress
So far, our results support that endogenous DNA damage is responsible for the induction of cellular senescence in L6 cells exposed to sub-lethal oxidative stress. Supplementary Figure S5A and B shows that similar to what was observed in L6 cells, exposure of synchronized RPE1 cells to sub-lethal oxidative stress induced a prolonged S phase, two waves of DNA damage with the second wave coinciding with the transition between the S and the G2/M phase of the cell cycle (Supplementary Figure S5C and D), and establishment of cellular senescence detected by an increase in SA-β-Gal activity (Supplementary Figure S5E). Finally, supporting that other inducers of DNA damage that do not induce cell death might activate cellular senescence as seen with sub-lethal oxidative stress, we show that exposure of L6 cells to sub-lethal concentration of DNA topoisomerase II inhibitor, etoposide coincided with two waves of DNA damage and the induction of cell growth arrest (Supplementary Figure S6A–C).
DISCUSSION
The present report shows that exposure of cells to sub-lethal oxidative stress induces two waves of γH2AX and a cellular senescence state that is detected 48 h after the initial oxidative stress. While the formation of DSB is responsible for the two waves of γH2AX, the induction of the senescent state is supported by changes in cells morphology, an increase in SA-β-Gal activity, establishment of DNA-SCARS, formation of CCF-like micronuclei, activation of the p16–RB axis, an increase in HMGA2 expression as a marker of SAHF and secretion of IL-6 as one of the SASP cytokines.
Origin of the initial wave of γH2AX
It is well accepted that H2O2 can react with Fe2+ leading to base alterations that can lead to DNA damage. The two basic groups of complex DNA damage are DSBs and non-DSB oxidative clustered DNA lesion (OCDL). OCDLs are formed due to closely spaced lesions in the form of abasic sites, oxypyrimidines, oxypurines and single strand breaks (SSBs) (45,46). Interestingly, OCDLs can be converted to DSBs during the repair process when both strands are incised simultaneously in close proximity. In particular, activation of the base excision repair (BER) pathway has been shown to convert OCDLs to DSBs (45). Although in the present report we do not extensively address the nature of the first wave of DNA damage, we propose that similar to what has recently been shown by Sharma and colleagues (45) in DT40 cells exposed to 40 μM H2O2, the first wave of γH2AX detected after the exposure of L6 cells to 50 μM H2O2 can be explained by the conversion of OCDLs to DSBs. In support of this proposal, incubation of L6 cells with iron chelators and ROS scavengers prevented the rapid increase in γH2AX and the subsequent second wave of γH2AX after cells exposure to the oxidant (Supplementary Figure S2A and B). In addition, formation of SSBs is shown by the activation of Poly (ADP-ribose) polymerase (PARP) detected by an increase in Poly (ADP-ribose) (PAR) 1 h after the exposure of cells to 50 μM H2O2 (Supplementary Figure S2C). Finally, silencing N-methylpuridine DNA glycolsylase 1 (MPG1), one of the initiators of the BER pathway, partially reduced the initial H2AX phosphorylation after the cells exposure to 50 μM H2O2 (Supplementary Figure S2D). Partial reduction of the initial γH2AX upon silencing MPG1 also reduced the second wave of phosphorylated H2AX (Supplementary Figure S2D). Taken together, these results support that the first wave of γH2AX drives the onset of the second wave of γH2AX and the cells’ growth arrest that follows.
From an exogenous to an endogenous induction of DNA damage
Unlike other DNA damaging agents, H2O2 induces a variety of complex DNA ends including oxidized deoxyribose, DSBs, SSBs and OCDLs that need to be repaired to prevent potential replication stress when cells enter the replicative phase of the cell cycle. Contrary to the DNA damage induced by 25 μM H2O2 that was rapid and completely repaired only leading to a slight growth delay, the initial DNA damage induced by 50 μM H2O2 was only partially repaired. The partial repair of DNA damage following cells exposure to 50 μM H2O2 is first supported by the data showing that γH2AX does not go back to control level before the second wave is detected. Similarly, the number of 53BP1 foci detected in synchronized cells following exposure to 50 μM H2O2 stayed slightly higher than in control cells at the time preceding the second wave of γH2AX. Moreover, a delay in S phase is detected in synchronized cells exposed to 50 μM H2O2. Taken together, these data provide testimony that cells entered the S phase of the cell cycle with unrepaired DNA. Furthermore, despite the activation of the DDR pathways (including the ATR and ATM kinases), the inability to resolve the stress resulted in the stalling of the replication fork, replication fork collapse and the formation of DSB. Of note, the detection of RPA32 nuclear foci from 4 to 12 h in cells exposed to 50 μM H2O2 and presence of DSBs at the time of the detection of the second wave of γH2AX, as shown by the co-localization of RPA32 and 53BP1, demonstrate that replication fork collapse leading to DSB is the origin of the second wave of γH2AX upon sub-lethal oxidative stress. Furthermore, the second wave of γH2AX in cells exposed to sub-lethal oxidative stress corresponded with the junction between the extended S phase and entry into the G2/M phase of the cell cycle. The extended S phase is in agreement with replication stress and the extended G2/M suggests activation of checkpoints to prevent cells carrying DSB from entering mitosis. The latter is evidenced by the absence of histone H3 phosphorylation at ser10 in cells exposed to H2O2 (Supplementary Figure S3D). In addition, the number of cells in metaphase was significantly reduced upon exposure to 50 μM H2O2, compared to untreated cells (Supplementary Figure S3B and C). Hence, the clear demonstration of a replication stress and absence of mitotic entry as well as the necessity for replication fork progression for the formation of the second wave of γH2AX support that unresolved replication stress led to replication forks collapse.
Sub-lethal oxidative stress induces the formation of DNA-SCARS
Despite the recognition that acquisition of persistent DNA damage foci such as DNA-SCARS is a key feature of the decision to enter cellular senescence (5,16,21), no study has previously linked replication stress-induced DNA damage to DNA-SCARS formation. Formation of DNA-SCARS is accelerated, when the DNA repair proteins are inhibited, leading to an ineffective and incomplete DNA repair process. Rad51 is one such important protein that is absent from the DNA-SCARS structures. Interestingly, upon cells exposure to sub-lethal oxidative stress, we show that the expression of Rad51 is inhibited by p53. Moreover, the decrease in Rad51 resulted in the appearance of γH2AX foci without Rad51. DNA-SCARS also lack ssDNA-binding proteins like RPA, but harbor PML at the site of DNA damage foci. Both characteristics were detected in cells exposed to 50 μM H2O2. Taken together, the existence of 53BP1 and γH2AX foci without Rad51, repression of total RPA32 and chromatin level of RPA32, and localization of PML bodies at the periphery of DNA damage foci are in agreement with the formation of DNA-SCARS in response to 50 μM H2O2.
Sub-lethal oxidative stress and the extrusion of CCF-like micronuclei
Micronuclei structures induced by replication stress inducers contain non-repaired or mis-repaired DSBs (47,48). Increased micronuclei structures staining positive for γH2AX and Histone H3 were observed upon treatment with 50 μM H2O2. Interestingly, the increased chromatin structures observed in the cytoplasm did not stain for 53BP1, but were positive for Lamin A\C. These data suggest that the cytoplasmic chromatin structures detected in cells exposed to a sub-lethal redox stress resemble micronuclei, as reported previously (17,47,48).
Sub-lethal oxidative induces a p16–RB-dependent cellular senescence
The p53 target gene, p21, has often been considered critical for establishing senescence, whereas p16 may be more involved in the maintenance of the phenotype (7,49,50). In agreement with the role of p53 in the establishment of senescence by sub-lethal oxidative stress, two peaks of increase in p53 expression and p53 phosphorylation were detected in cells treated with 50 μM H2O2. Moreover, silencing p53 expression mitigated the cells growth arrest and the percentage of cells with senescent nuclei and morphology abnormality. However, the induction of senescence by exogenous H2O2 was not associated with a p53–p21 axis but with a p16–RB axis. It is interesting to note that the accumulation of p16 in tissues is implicated in cellular aging and clearance of p16-positive cells delays aging (51). Hence, the ability of a sub-lethal oxidative stress to induce a p16-dependent cellular senescence might explain the strong correlation between ROS and the development of an aging phenotype at both the cellular as well as at the organism level (52).
Senescence induced by sub-lethal oxidative stress requires the activation of p53–Rad51 and p53–Lamin B1 feedback loops
There is a plethora of genes activated by p53 that are associated with the establishment of the cellular senescent state (22). On the other hand, association of the transcriptional repression capacity of p53 and a cellular senescent phenotype is not well documented. In the present report, we show that following cells exposure to sub-lethal oxidative stress, p53 was responsible for the decrease in Rad51 and Lamin B1 expression. Rad51 is a key protein of the HR pathway involved in resolving replication stress (53). Silencing p53 rescued not only the growth arrest but also the expression of Rad51 in cells exposed to 50 μM H2O2. Knowing that absence of Rad51 is a characteristic of DNA-SCARS (21), we propose that upon a sub-lethal oxidative stress the repression of Rad51 by active p53 is critical for the formation of DNA-SCARS. In addition, reciprocally, DNA-SCARS formation is critical to maintain the low level of p53 activation necessary to maintain cell cycle arrest in senescent cells (21). On the other hand, Lamin B1 is a robust and easily detectable marker of senescence. Silencing Lamin B1 affects the proliferation rate of the cells (54) and contributes to chromatin extrusion (14,17). Activation of p53 or p16 has been shown to result in loss of Lamin B1 in senescent cells (14,54). However, in the present report repression of Lamin B1 expression was observed before the detection of an increase in p16 and the chromatin extrusion. Hence, we propose that in cells exposed to a sub-lethal oxidative stress, aggregation of DSB into micronuclei like structures or the formation of chromatin budding is initiated by the unresolved replication stress, while the p53-dependent repression of Lamin B1 might then facilitate the translocation of chromatin via micronuclei like structures to the cytoplasm. Furthermore, since Lamin B1 downregulation has been shown to activate p53 (54) and the expression of Lamin B1 and p53 levels were constitutively low in cells exposed to sub-lethal oxidative stress, similar to the p53–Rad51 feedback loop, we propose the establishment of a feedback mechanism between p53 and Lamin B1 to be critical to maintain growth arrest. Following the establishment of the p53–Rad51 and p53–Lamin B1 feedback loops, cells will then enter a deep senescent state, a key event to prevent the formation of tumorigenic cells. Indeed, in the absence of p53, cells exposed to 50 μM H2O2 showed severe genomic instability, aneuploidy and micronuclei formation (Supplementary Figure S4B–D). Interestingly, the absence of p53 did not have a notable effect on the SA-β-Gal activity and HMGA2 expression compared to control cells. These results suggest that p53-independent pathways might also be important to complete the redox stress-induced senescent phenotype. In particular, it might be interesting to better understand what is activating the formation of SAHF. Regarding the increase in SA-β-Gal, we have recently shown that sub-lethal oxidative stress increases lysosome biogenesis through a caspase 3/TFEB pathway (55). It could be interesting to decipher whether the increase in lysosome biogenesis could be linked to the increase in SA-β-Gal.
CONCLUSION
Our study contributes to the mechanistic understanding of the induction of cellular senescence following cells’ exposure to sub-lethal oxidative stress (Figure 10). Interestingly, the same pathways might be activated upon cells exposure to sub-lethal concentration of chemotherapeutic drug such as etoposide. In addition, evidence from the literature suggest that oncogenic stimuli that do not induce direct DNA damage also mediate senescence via replication stress-induced endogenous DNA damage (56,57). Hence, activation of cellular senescence due to the formation of endogenous DSB could be a pathway common to a variety of inducers of cellular senescence. Finally, establishing the role of replication stress induced endogenous DNA damage in the initiation and the establishment of cellular senescence following cells exposure to a sub-lethal oxidative stress is critical for a better understanding of the molecular mechanisms/dynamics involved in the induction of aging and cancer by ROS.
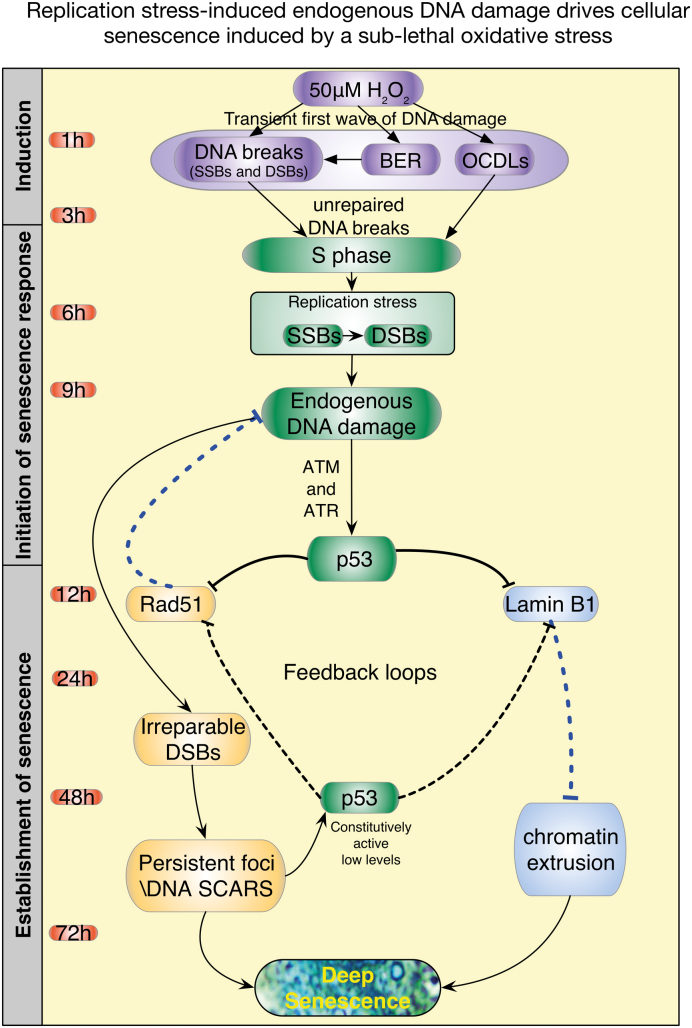
Figure 10.
Proposed model of the induction of cellular senescence by a sub-lethal oxidative stress. We propose that the acquisition of cellular senescence following cells’ exposure to a sub-lethal oxidative stress occurs in three phases. Phase 1 (1–3 h): upon exposure of cells to a sub-lethal oxidative stress, variety of complex DNA ends including oxidized deoxyriboses, DSBs, SSBs and OCDLs are generated. Phase 2 (4–12 h): despite the presence of unrepaired DNA, cells progress to S phase. Upon the progression to S phase, presence of unrepaired DNA damage induces replication stress. The inability to resolve the stress results in the stalling of the replication fork, replication fork collapse leading to the formation of DSB. At the same time activation of p53 inhibits the transcription of the HR repair protein Rad51 and the nuclear protein Lamin B1. Phase 3 (12–72 h): repression of Rad51 by active p53 is critical for the formation of DNA-SCARS. In addition, reciprocally, DNA-SCARS formation is critical to maintain the low level of p53 activation necessary to maintain cell cycle arrest. On the other hand, aggregation of DSB into micronuclei like structures or the formation of chromatin budding initiated by the unresolved replication stress associated with the p53-dependent repression of Lamin B1 facilitates the translocation of chromatin via micronuclei like structures to the cytoplasm. Furthermore, similar to the p53–Rad51 feedback loop, a feedback mechanism between p53 and Lamin B1 is critical to maintain growth arrest. Following the establishment of the p53–Rad51 and p53–Lamin B1 feedback loops, cells will then enter a deep senescent state, characterized by growth arrest, an increase in SA-β-Gal activity, activation of a p16-RB axis, establishment of DNA-SCARS, increase in HMGA2 expression as a marker of SAHF, formation of CCF-like micronuclei and secretion of IL-6.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Kathirvel Paramasivam and Ms Vishnupriya Manivannan for their help in performing experiments to complete the necessary revisions of the original manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education (MOE), Singapore (An AcR tier 1 FRC Grant) [T1–2013 to M.V.C.]; Biomedical Research Council of A*STAR (Agency of Science Technology and Research) (to U.S.); NGS Ph.D. Research Scholarship, National University of Singapore (to G.V.). Funding for open access charge: MOE, Singapore (AcR tier 1 FRC) (to M.V.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Mallette F.A., Gaumont-Leclerc M.-F., Ferbeyre G.. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007; 21:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pannu V., Rida P.C.G., Ogden A., Clewley R., Cheng A., Karna P., Lopus M., Mishra R.C., Zhou J., Aneja R.. Induction of robust de novo centrosome amplification, high-grade spindle multipolarity and metaphase catastrophe: a novel chemotherapeutic approach. Cell Death Dis. 2012; 3:e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodier F., Coppe J.-P., Patil C.K., Hoeijmakers W.A.M., Muñoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., Campisi J.. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009; 11:973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blazkova H., Krejcikova K., Moudry P., Frisan T., Hodny Z., Bartek J.. Bacterial intoxication evokes cellular senescence with persistent DNA damage and cytokine signalling. J. Cell. Mol. Med. 2009; 14:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodier F., Campisi J.. Four faces of cellular senescence. J. Cell Biol. 2011; 192:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knizhnik A.V., Roos W.P., Nikolova T., Quiros S., Tomaszowski K.-H., Christmann M., Kaina B.. Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS One. 2013; 8:e55665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G.. p53 dynamics control cell fate. Science. 2012; 336:1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Passos J.F., Nelson G., Wang C., Richter T., Simillion C., Proctor C.J., Miwa S., Olijslagers S., Hallinan J., Wipat A. et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 2010; 6:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fedor Y., Vignard J., NicolauI-Travers M.L., Bouter-Robinet E., Watrin C., Salles B., Mirey G.. From single-strand breaks to double-strand breaks during S-phase: a new mode of action of the Escherichia coli cytolethal distending toxin. Cell. Microbiol. 2013; 15:1–15. [DOI] [PubMed] [Google Scholar]
- 10. Luo Y., Zou P., Zou J., Wang J., Zhou D., Liu L.. Autophagy regulates ROS-induced cellular senescence via p21 in a p38 MAPKα dependent manner. Exp. Gerontol. 2011; 46:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vigneron A., Vousden K.H.. p53, ROS and senescence in the control of aging. Aging (Albany NY). 2010; 2:471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008; 9:619–631. [DOI] [PubMed] [Google Scholar]
- 13. Sarsour E.H., Kalen A.L., Goswami P.C.. Manganese superoxide dismutase regulates a redox cycle within the cell cycle. Antioxid. Redox Signal. 2013; 20:1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freund A., Laberge R.M., Demaria M., Campisi J.. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012; 23:2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blagosklonny M.V. Cell cycle arrest is not senescence. Aging (Albany NY). 2011; 3:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fumagalli M., Rossiello F., Clerici M., Barozzi S., Cittaro D., Kaplunov J.M., Bucci G., Dobreva M., Matti V., Beausejour C.M. et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012; 14:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivanov A., Pawlikowski J., Manoharan I., van Tuyn J., Nelson D.M., Rai T.S., Shah P.P., Hewitt G., Korolchuk V.I., Passos J.F. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013; 202:129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trachootham D., Lu W., Ogasawara M.A., Valle N.R.-D., Huang P.. Redox regulation of cell survival. Antioxid. Redox Signal. 2008; 10:1343–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marusyk A.A., Wheeler L.J.L., Mathews C.K.C., DeGregori J.J.. p53 mediates senescence-like arrest induced by chronic replicational stress. Mol. Cell. Biol. 2007; 27:5336–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbig U., Jobling W.A., Chen B.P.C., Chen D.J., Sedivy J.M.. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell. 2004; 14:501–513. [DOI] [PubMed] [Google Scholar]
- 21. Rodier F., Muñoz D.P., Teachenor R., Chu V., Le O., Bhaumik D., Coppé J.-P., Campeau E., Beauséjour C.M., Kim S.-H. et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011; 124:68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rufini A., Tucci P., Celardo I., Melino G.. Senescence and aging: the critical roles of p53. Oncogene. 2013; 32:5129–5143. [DOI] [PubMed] [Google Scholar]
- 23. van Deursen J.M. The role of senescent cells in ageing. Nature. 2014; 509:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeyapalan J.C., Ferreira M., Sedivy J.M., Herbig U.. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007; 128:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kreiling J.A., Tamamori-Adachi M., Sexton A.N., Jeyapalan J.C., Munoz-Najar U., Peterson A.L., Manivannan J., Rogers E.S., Pchelintsev N.A., Adams P.D. et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011; 10:292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W.. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003; 113:703–716. [DOI] [PubMed] [Google Scholar]
- 27. Baker D.J., Sedivy J.M.. Probing the depths of cellular senescence. J. Cell Biol. 2013; 202:11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narita M., Narita M., Krizhanovsky V., Nũnez S., Chicas A., Hearn S.A., Myers M.P., Lowe S.W.. A novel role for high-mobility group A proteins in cellular senescence and heterochromatin formation. Cell. 2006; 126:503–514. [DOI] [PubMed] [Google Scholar]
- 29. Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., McBryan T. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013; 27:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fumagalli M., Di Fagagna F.D.A.. SASPense and DDRama in cancer and ageing. Nat. Cell Biol. 2009; 11:921–923. [DOI] [PubMed] [Google Scholar]
- 31. Debacq-Chainiaux F., Erusalimsky J.D., Campisi J., Toussaint O.. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009; 4:1798–1806. [DOI] [PubMed] [Google Scholar]
- 32. Gyori B.M., Venkatachalam G., Thiagarajan P.S., Hsu D., Clement M.V.. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2014; 2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 34. Somyajit K., Basavaraju S., Scully R., Nagaraju G.. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol. Cell. Biol. 2013; 33:1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Filippi-Chiela E.C., Oliveira M.M., Jurkovski B., Callegari-Jacques S.M., da Silva V.D., Lenz G.. Nuclear morphometric analysis (NMA): screening of senescence, apoptosis and nuclear irregularities. PLoS One. 2012; 7:e42522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lapytsko A., Kollarovic G., Ivanova L., Studencka M., Schaber J.. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics. 2015; 16:3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karanam K., Kafri R., Loewer A., Lahav G.. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell. 2012; 47:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ha L. Generation of S phase-dependent DNA double-strand breaks by Cr(VI) exposure: involvement of ATM in Cr(VI) induction of -H2AX. Carcinogenesis. 2004; 25:2265–2274. [DOI] [PubMed] [Google Scholar]
- 39. Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marchetti M.A., Kumar S., Hartsuiker E., Maftahi M., Carr A.M., Freyer G.A., Burhans W.C., Huberman J.A.. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7472–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pascucci B., Russo M.T., Crescenzi M., Bignami M., Dogliotti E.. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005; 33:280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. D’adda Di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008; 8:512–522. [DOI] [PubMed] [Google Scholar]
- 43. Coppe J.-P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.-Y., Campisi J.. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008; 6:e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kosar M., Bartkova J., Hubackova S., Hodny Z., Lukas J., Bartek J.. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a). Cell Cycle. 2011; 10:457–468. [DOI] [PubMed] [Google Scholar]
- 45. Sharma V., Collins L.B., Chen T.-H., Herr N., Takeda S., Sun W., Swenberg J.A., Nakamura J.. Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations. Oncotarget. 2016; 10:25377–25390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kryston T.B., Georgiev A.B., Pissis P., Georgakilas A.G.. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res. 2011; 711:193–201. [DOI] [PubMed] [Google Scholar]
- 47. Terradas M., Martín M., Tusell L., Genescà A.. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair (Amst). 2009; 8:1225–1234. [DOI] [PubMed] [Google Scholar]
- 48. Xu B., Sun Z., Liu Z., Guo H., Liu Q., Jiang H., Zou Y., Gong Y., Tischfield J.A., Shao C.. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One. 2011; 6:e18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stein G.H., Drullinger L.F., Soulard A., Dulić V.. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 1999; 19:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Campisi J., D’adda Di Fagagna F.. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007; 8:729–740. [DOI] [PubMed] [Google Scholar]
- 51. Rayess H., Wang M.B., Srivatsan E.S.. Cellular senescence and tumor suppressor gene p16. Int. J. Cancer. 2012; 130:1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin N., Beach D., Gil J.. Ageing as developmental decay: insights from p16INK4a. Trends Mol. Med. 2014; 20:667–674. [DOI] [PubMed] [Google Scholar]
- 53. Thacker J. Homologous recombination repair. Encyclopedia of Cancer. 2011; 3:1725–1728. [Google Scholar]
- 54. Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D.. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011; 25:2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leow S.M., Chua S.X.S., Venkatachalam G., Shen L., Luo L., Clement M.V.. Sub-lethal oxidative stress induces lysosome biogenesis via a lysosomal membrane permeabilization-cathepsin-caspase 3-transcription factor EB-dependent pathway. Oncotarget. 2017; 8:16170–16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Di Micco R., Sulli G., Dobreva M., Liontos M., Botrugno O.A., Gargiulo G., dal Zuffo R., Matti V., d’Ario G., Montani E. et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 2011; 13:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre M., Nuciforo P.G., Bensimon A. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006; 444:638–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.