Abstract
Our knowledge of the biological mechanisms underlying complex human disease is largely incomplete. While Semantic Web technologies, such as the Web Ontology Language (OWL), provide powerful techniques for representing existing knowledge, well-established OWL reasoners are unable to account for missing or uncertain knowledge. The application of inductive inference methods, like machine learning and network inference are vital for extending our current knowledge. Therefore, robust methods which facilitate inductive inference on rich OWL-encoded knowledge are needed. Here, we propose OWL-NETS (NEtwork Transformation for Statistical learning), a novel computational method that reversibly abstracts OWL-encoded biomedical knowledge into a network representation tailored for network inference. Using several examples built with the Open Biomedical Ontologies, we show that OWL-NETS can leverage existing ontology-based knowledge representations and network inference methods to generate novel, biologically-relevant hypotheses. Further, the lossless transformation of OWL-NETS allows for seamless integration of inferred edges back into the original knowledge base, extending its coverage and completeness.
Keywords: Biological Ontologies, Knowledge Bases, Semantic Web, Machine Learning
1. Introduction
Network representations facilitate the understanding of complex biological mechanisms, and have been used extensively in biomedical research to represent phenomena ranging from metabolism, to protein-protein interactions, and drug-drug interaction networks.1–3 Inference over the structure of a network can provide insight and generate hypotheses regarding the functional relationships between network elements.4
The Web Ontology Language (OWL) is a Semantic Web standard for a network-based knowledge representation and reasoning framework that is highly expressive and that has been used to model complex biological knowledge.5 Inference on the Semantic Web has heavily relied on deductive and probabilistic reasoning. Deductive OWL reasoners work by inferring logical consequences from a set of explicitly asserted facts.6 Constrained by first-order predicate logic, description logic reasoners (e.g., ELK,7 FaCT++8) are unable to account for uncertainty or incomplete knowledge.9 To account for this limitation, probabilistic methods (e.g., PROWL,10 P-CLASSIC11) have been developed.12 Unfortunately, these methods can only be applied in situations where the accuracy of propositions is ambiguous rather than unknown due to incomplete knowledge.13 While inductive methods, like machine learning, are powerful tools for producing predictions that are not explicitly asserted,12 the scalability of ontology properties can significantly limited the utility of these techniques.14
Link prediction, an inductive learning method, predicts unobserved connections between the nodes of a network. Most biological network representations can be assumed to be incomplete, making link prediction a potentially valuable tool for knowledge discovery. The application of such algorithms to biological networks has correctly predicted important relationships, including protein-protein interactions,15 drug-target pairs,16 and regulatory gene interactions.17 Although OWL is a highly expressive representation language,18 its use comes at the cost of a structurally complex network. We hypothesize that the expressivity of OWL reduces the power of link prediction algorithms to identify novel, biologically important insights. We further hypothesize that an abstraction of OWL networks will negate this power loss, providing a novel means to infer knowledge from available OWL resources.
In the field of biomedical informatics, abstraction networks have been used to obtain an alternative view of a terminology/ontology by reducing the complexity of its underlying structure.19,20 Primarily developed to assess the quality of clinical terminologies and ontologies, these methods leverage the underlying terminology/ontology structure to combine subsets of nodes with similar attributes (e.g., data or object properties, and relations).19,21–23 To the best of our knowledge, there are no existing abstraction methods designed to create network representations from complex OWL-encoded knowledge for the purpose of network inference.
We propose OWL-NETS (NEtwork Transformation for Statistical learning), a novel computational method that reversibly abstracts OWL-encoded biomedical knowledge into a network representation tailored for network inference. Using several examples built with the Open Biomedical Ontologies, we demonstrate that OWL-NETS results in networks with significantly different properties than their corresponding OWL representations. We also show that OWL-NETS can be used to leverage existing ontology-based knowledge representations and network inference methods to generate novel, biologically-relevant hypotheses. Further, the lossless transformation of OWL-NETS allows for seamless integration of inferred edges back into the knowledge base, extending its coverage and completeness.
2. Methods
OWL-NETS is implemented in Python (v2.7) and can be run from a simple GUI or from the command line. While primarily developed for use with OWL, the program can be easily extended for use with other Semantic Web technologies by modifying two primary assumptions:
A knowledge source contains identifiers that directly represent biologically meaningful concepts (e.g., GO:0001525 is the identifier for the biological process of angiogenesis). Within OWL-NETS, biologically meaningful concepts are referred to as ’NETS nodes’.
A knowledge source uses restrictions to specify the existence of biologically important relations between pairs of biological concepts (e.g., proteins are restricted to participate in angiogenesis). In OWL, restrictions provide a way to make the definition of a class more specific (e.g., proteins, specifically, protein kinases, participate in phosphorylation). Within OWL-NETS, biologically important relations are referred to as ’NETS edges’.
The OWL-NETS methodology is described using an example query that investigates disease-associated proteins that participate in angiogenesis (Figure 1; pseudocode for Steps 1–4 in Supplementary Material). The method takes a SPARQL query as input and outputs a directed OWL-NETS abstraction network, directed OWL representation, or both. To improve processing efficiency, the majority of the computational workload is performed on the input SPARQL query (Steps 1-3) rather than the resulting output.
Fig. 1.
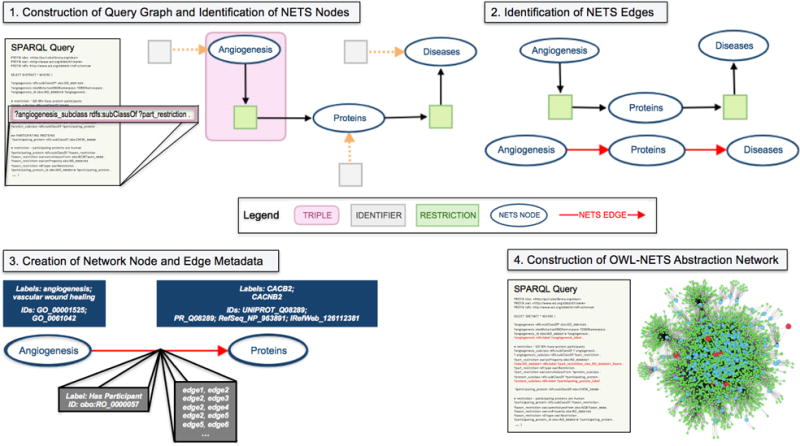
Overview of the OWL-NETS Method.
Construction of Query Graph and Identification of NETS Nodes: The input SPARQL query is used to create a directed query graph, where each triple (one of which is identified in Figure 1 in the query and in the graph with by a pink box) represents a directed edge in the query graph. The query graph is then searched for NETS nodes (Figure 1 shows identifier nodes pointing to NETS nodes via dashed orange arrows). NETS nodes in Figure 1 include: Angiogenesis, Proteins, and Diseases.
Identification of NETS Edges: The query graph is searched for restrictions (shown in Figure 1 as light green nodes). The NETS node that is reachable from the restriction node’s out-edges is the target of the NETS edge. In Figure 1, one of the green restriction nodes, that is pointed to by Proteins has an out-edge that points towards Diseases, thus an arrow is drawn from Proteins to Diseases. NETS edges are shown in the figure with a red arrow. When both NETS nodes can be reached from a restriction node, arrows pointing in both directions are drawn between the NETS nodes (i.e., nodeA → nodeB and nodeB → nodeA).
Creation of Network Node and Edge Metadata: Identifiers and labels for each NETS node and edge (shown in Figure 1 as dark blue and gray boxes) are stored as network metadata. This metadata is needed to transform the OWL-NETS abstraction network back into the OWL representation which facilitates the seamless integration of inferred edges back into the knowledge base, extending its coverage and completeness.
Construction of OWL-NETS Abstraction Network: Steps 1-3 gather information from the query graph that is needed to construct the OWL-NETS abstraction network. The final step augments the original query with this information. The red lines shown in the example SPARQL query, under Step 4, demonstrate the addition of NETS node and edge metadata to the query. The augmented SPARQL query is then run against an endpoint. The endpoint results are then used to construct the OWL-NETS abstraction network.
Supplemental material (including definitions and acronyms used throughout the paper), source code, and example data for exploring OWL-NETS can be found on GitHub (https://callahantiff.github.io/owl-nets/).
2.1. Biomedical Use Cases
To test the utility of OWL-NETS, we make use of the Knowledge Base Of Biology (KaBOB),24 an open-source ontology-based semantically integrated knowledge base of biomedical data. Currently, KaBOB contains 13 sources of biomedical data on humans as well as seven model organisms in a representation grounded in 17 Open Biomedical Ontologies (OBOs). The queries that led to the development of OWL-NETS provide the use cases for the current work. The queries, along with their data sources (all data downloaded on March 2016, except Reactome which was downloaded on November 2015) are described below:
Query 1: Human proteins localized to cellular and extracellular components and locations (Uniprot, Gene Ontology).
Query 2: Protein targets of drugs that interact with trametinib (Uniprot, Protein Ontology, RefSeq, IRefWeb, DrugBank).
Query 3: Protein targets of 100,000 drug-drug interactions and the pathways in which the proteins participate (Uniprot, DrugBank, Reactome).
Differences in the network properties of the OWL representation and OWL-NETS abstraction networks were explored using 100 protein localization networks (Query 1), each with 50 proteins, generated uniformly at random. Mann-Whitney U tests were then used to determine if the mean of each network property, measured across the 100 networks, significantly differed by network representation. In addition to generating basic network properties, the power-law fit of the complementary cumulative distribution function (CCDF) was calculated for the network representations generated from Queries 2 and 3. All network properties were calculated on undirected network representations.
2.2. Link Prediction Procedures
Given an undirected, unweighted network G(N, L), where N is the set of nodes and L is the set of observed edges between these nodes, the universal set of all possible edges U is . The set of nonexistent edges (i.e., the set of edges that don’t currently exist in the network) is U - L. From the observed network G, a uniformly random set of edges Ltesting was removed and the remaining edges Ltraining were used as the training network.25,26 Each link prediction algorithm was then run over the training network. The ability of each algorithm to recover the edges that were purposefully removed, Ltesting using only the information present in Ltraining, was then evaluated. The fraction of edges removed from the original network included 0.05, 0.10, 0.30, 0.50, 0.70, 0.90, and 0.95. For each removed fraction of edges, 100 iterations were run.
Ten similarity-based link prediction algorithms were run on networks resulting from Queries 2 and 3. In general, link prediction algorithms assign a similarity score to all non-observed edges in a network. The predicted edges with the highest scores are the most likely to exist.26 Both local (i.e., node-level) and global (i.e., path-level) similarity link prediction algorithms were evaluated. The details regarding these algorithms are provided in Section 2 of Supplementary Material.
2.2.1. Evaluation of Link Prediction Algorithm Performance
The area under the receiver operating characteristic curve (AUC)27 and top-L precision were used to evaluate link prediction algorithm performance.26
- AUC: The probability that a randomly chosen predicted edge that was purposely removed (true positive) has a higher score than a randomly chosen nonexistent predicted edge (true negative), where n′ is the number of comparisons for which the randomly chosen true positive was higher than the randomly chosen true negative, n″ is the number of comparisons for which the randomly chosen true positive and true negative had the same score, and n is the total number of comparisons:
(1) - Top-L Precision: Given a list of predicted edges sorted by score, the ratio of edges that were purposely removed LTP (true positives) among all predicted edges L:
(2)
The best performing algorithm was chosen using the highest average AUC (over 100 iterations) when removing a fraction of 0.5 edges from the original network. The time to run 100 iterations of each link prediction algorithm on each of the network representations from Query 2 was also evaluated. Algorithms were run in parallel on a machine running macOS Sierra with 16 GB of RAM and a 2.7 GHz processor with 8 cores.
2.2.2. Evaluation of Inferred Edges
The best-performing link prediction algorithm was re-run (this time exposing the algorithm to all existing edges in the network) and the highest-scoring edges for each network were evaluated via expert consultation and extensive literature review by a PhD-level biologist (author ALS). Additionally, an OWL reasoner (HermiT,28 via Protégé v5.1.1) was run on the OWL representation from Query 2 to demonstrate that deductive inference resulted in different predicted assertions than the link prediction algorithms.
3. Results
3.1. Comparison of Network Properties
Properties of the network representations from Query 1 are shown in Table S1 of Supplementary Material. On average, the OWL representation networks had significantly more nodes and edges, larger diameters, higher heterogeneity, a larger number of shortest paths, shorter average path lengths, more disassortative structures, and more cliques than the OWL-NETS abstraction networks. In contrast, the OWL-NETS abstraction networks had a larger average degree and a greater average clustering coefficient. Network properties are defined in Supplementary Material.
The OWL representation and OWL-NETS abstraction networks built from running Query 2 are visualized in Figure 2. The OWL representation network (left) contained 840 nodes and 1,426 directed edges. In comparison, the OWL-NETS abstraction network (middle) contained 59 nodes and 65 directed edges. The OWL-NETS abstraction network had a smaller average degree (2.00 vs. 3.42) than the OWL representation networks. As shown in the third plot (right), both network representations had good to moderate power-law fit. This is consistent with the fit reported in the literature for other heavy-tailed biological networks.29,30 Additional network properties can be found in Table S2 of Supplementary Material.
Fig. 2.
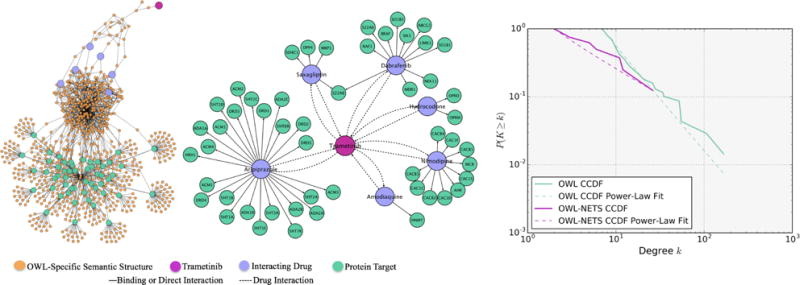
Query 2 OWL representation network (Left); OWL-NETS abstraction network (middle); and CCDF power-law fit (right). NETS nodes shown in magenta, purple, and teal. Orange ’OWL-Specific Semantic Structure’ nodes are needed to create valid OWL expressions and are only part of the OWL representation networks. NETS edge relations shown in solid and dashed lines.
The OWL representation and OWL-NETS abstraction networks built from Query 3 are visualized in Figure 3. The OWL representation network (left) contained 22,679 nodes and 33,848 directed edges. In comparison, the OWL-NETS abstraction network (middle) contained 1,783 nodes and 7,253 directed edges. The OWL-NETS abstraction network had nine connected components (as shown in the figure as one large network surrounded by eight smaller networks), with the largest connected component containing 1,702 nodes and 7,111 directed edges. The largest connected component of the OWL-NETS abstraction network had a larger average degree (4.42 vs. 2.98) and a longer average path length (6.54 vs. 4.13) than the OWL representation. As shown in the third plot (right), both networks had moderate to poor power-law fits. Similar to Query 2, the OWL representation network had a worse fit than the OWL-NETS abstraction network. See Table S2 (Supplemental Material) for additional network properties.
Fig. 3.
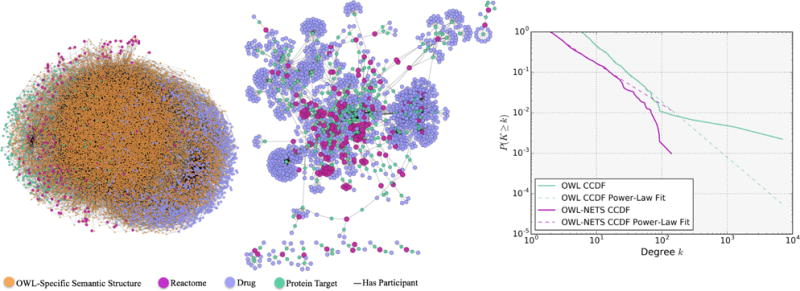
Query 3 OWL representation network (Left); OWL-NETS Abstraction Network (middle); and CCDF power-law fit (right). Node size is only for visualization. NETS nodes shown in magenta, purple, and teal. Orange ’OWL-Specific Semantic Structure’ nodes are needed to create valid OWL expressions and are only part of the OWL representation networks. NETS edge relations shown in solid and dashed lines.
3.2. Link Prediction Algorithm Performance
The results from performing link prediction on the network representations for Query 2 are shown in Figure S1 (Supplementary Material).
There were 350,944 nonexistent edges in the OWL representation network and 1,652 nonexistent edges in the OWL-NETS abstraction network. The average AUC scores for the OWL representation network, for all fractions of removed edges, ranged between 0.50 and 0.62. The highest average AUC was found for the Rooted PageRank algorithm when removing a fraction of 0.05 edges from the network (0.66). The average precision for algorithms across all fractions of removed edges was between 0.0001 and 0.001. For the OWL-NETS abstraction network, the average AUC scores for the best performing algorithms, across all fractions of removed edges, ranked between 0.50 and 0.92. The highest average AUC was found for the Katz algorithm when removing a fraction of 0.05 edges from the network (0.92). The average precision for algorithms across all fractions of removed edges was between 0.001 and 0.04.
The OWL-NETS abstraction network from Query 3 was also explored using the link prediction algorithms (Figure S2, Supplementary Material). There were 257,123,333 nonexistent edges in the OWL representation network and 1,584,713 nonexistent edges in the OWL-NETS abstraction network. The Katz (0.51-0.78), Shortest Path (0.51-0.75), Degree Product (0.58-0.73), and Rooted PageRank (0.64-0.67) algorithms consistently had a higher average AUC across all fractions of removed edges, compared to the other algorithms. Average precision values were similar to Query 2.
The total run time of each algorithm over the 100 iterations varied widely between the network representations from Query 2 (Table S3, Supplementary Material). On average, across all algorithms the OWL-NETS abstraction networks completed 7.5-1185.7 times faster than the OWL representation networks. This is expected given the drastic difference in the number of non-existent edges.
3.2.1. Inferred Edges
For Query 2, the Rooted PageRank algorithm performed best on the OWL representation; however, evidence could be found for only one of the predicted edges (HRH1-ADA1A occur in the same calcium signaling KEGG pathway) when run on all edges in the network. Additionally, running HermiT over the OWL representation network resulted in no inferred axioms.
Running the top-performing Katz algorithm on the full OWL-NETS abstraction network produced dramatically better results. From extensive literature review, direct or indirect evidence of a meaningful biological association was found in support of 50% of the top 20 predicted edges (Table 1). Examples of direct evidence found to validate predicted edges include amodiaquine and nimodipine, which have been shown to experimentally to regulate the expression of voltage-dependent calcium channel L-type alpha 1 C subunits31 and HNMT,32 respectively. Indirect biological evidence was found to support the edge between amodiaquine and AHR, which was substantiated by their shared relationship to the CYP1a1 enzyme33,34 and the amodiaquine-OPRM1 edge, which is supported by the reported relationship between pruritus (itching) induced by quinolones (a family of anti-malarial drugs including amodiaquine) and its pharmacologic treatment with naltrexone, an OPRM1 antagonist.35 Interestingly, many of the top 20 edges were related to each other by common biological themes such as histamines and the opioid receptor system. Additionally, as demonstrated by the results in Table 1, DPP4 appears to link seemingly unrelated groups of drugs including narcotics (e.g., hydrocodone), those used in the treatment of malaria (e.g., amodiaquine), and diabetes (e.e., saxagliptin). Therefore, it is possible that the examination of predicted edges can reveal biological mechanisms underlying the interactions of drugs with other drugs and targets.
Table 1.
Top scoring edges from Query 2 OWL-NETS abstraction network (n=10 edges)
| Node 1 | Node 2 | Description |
|---|---|---|
| amodiaquinea | DPP4a | Middle East Respiratory Syndrome-Coronavirus (MERS-CoV) gains entry into cells via DPP4 and amodiaquine has activity against MERS-CoV.36 |
| amodiaquinea | CACNA1Cb | Amodiaquine-treated mice have decreased expression of voltage-dependent calcium channel L-type alpha 1 C subunits in their livers.31 |
| amodiaquinea | CACNA1Db | Amodiaquine-treated mice have decreased expression of voltage-dependent calcium channel L-type alpha 1 C subunits in their livers.31 |
| nimodipinea | HNMTb | A small molecule screen demonstrated that nimodipine caused increased HNMT expression in cultured human cells.32 |
| amodiaquinea | OPRM1b | Quinolone-based antimalarials can induce generalized pruritus (itch), which can be treated with the mu-opioid receptor (OPRM1) antagonist naltrexone.35 |
| amodiaquinea | AHRb | Amodiaquine is metabolized by CYP1a1. CYP1a1 is induced by signaling through AhR.33,34 |
| hydrocodonea | HNMTb | HNMT regulates histamine release and opiates, like hydrocodone, induce histamine release.37 |
| hydrocodonea | DPP4b | DPP4 cleaves dietary gliadin into opioid peptides that can activate mu-opioid receptors. Hydrocodone activates mu-opioid receptors.38,39 |
| hydrocodonea | MRP1b | MRP1 is involved in maintaining the blood-brain barrier. Downregulation of MRP1 increases the analgesic effect of systemic morphine in mice and rats by decreasing the blood-brain barrier. Hydrocodone is a synthetic opioid.40 |
| saxagliptina | OPRM1b | Saxagliptin inhibits DPP4, which contributes to the cleavage of dietary gliadin into opioid peptides. The gliadin opioid peptide, gliadinomorphin-7, can activate the mu-opioid receptor (OPRM1).38,39 |
DrugBank entity (DrugBank ID used for experimental compounds);
Uniprot entity (gene symbol is shown to preserve space).
Evidence from the literature could be found to support 75% of the top 20 scoring edges predicted by the Katz algorithm on the OWL-NETS abstraction network generated from Query 3 (selected examples of which are presented in Table S4, Supplementary Material). The predicted edge between AG-1067, a derivative of probucol (an anti-hyperlipidemic drug), and MMP2 is supported by experimental evidence that probucol decreases the expression and activity of MMP2 in a mouse model.41 Similarly, the predicted edge between DB04513 and RAF1 is substantiated by evidence that calmodulin 1, which is the target of experimental drug N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (DB04513), modulates signaling through the Ras/Raf/MEK/ERK signaling pathway.42 Two additional edges between celiprolol and CYCS and between Reactome pathway 1454838 and transferrin were found to be biologically related through disease processes including hypertension,43,44 and multiple myeloma.45,46 The predicted edge between experimental drug 2-[formyl(hydroxy)amino]methyl-4-methylpentanoic acid (DB03683) and APAF1 was supported by their shared association with MMP9.47
4. Discussion
Networks representing biomedical mechanisms constantly evolve; the addition of new edges within a network may symbolize important interactions and provide valuable insight into its underlying biology.48 Investigating new edges within these networks provides a methodology for generating novel hypotheses. While OWL provides powerful techniques for representing existing knowledge, well-established OWL reasoners are unable to account for missing or uncertain knowledge. Further, the structural complexity of OWL reduces the effectiveness of certain types of network inference. To address these limitations we developed OWL-NETS, a novel computational method that reversibly abstracts OWL-encoded biomedical knowledge into a network representation tailored for network inference. To the best of our knowledge, there are no existing network abstraction methods designed to create network representations from OWL-encoded knowledge sources to facilitate network inference.
Existing network abstraction methods reduce the structural complexity of a terminology/ontology by aggregating nodes with similar attributes or properties.21–23,49 An abstraction network is considered useful if it is significantly smaller than the original terminology/ontology without losing structure and content.20 The goal of OWL-NETS is to collapse the nodes and edges that are necessary to logically represent relationships between biological entities in OWL, but are not themselves biologically meaningful and interfere with network inference. In contrast to existing methods, the reduced size of the OWL-NETS abstraction network, relative to its original OWL representation, is not predictive of its usefulness for inference. In fact, too much aggregation may result in a network whose properties are no better for inference than the original OWL representation. More importantly, existing network abstraction methods were not designed for network inference; combining nodes having the same attributes or properties could inadvertently mask important biological relations of the resulting abstraction networks.
This work is not without limitations. Relying on literature review, even if by a domain expert, does not provide the most robust evaluation of inferred edges. Future work will include a collaboration where results can be evaluated experimentally. Additionally, the current work evaluated relatively simple, unipartite networks. Future work will explore more complex types of biological network representations, such as bipartite and multiplex networks. We are also developing methods for adding edge weight to the OWL-NETS abstraction networks to indicate the amount and/or quality of the confidence/evidence of the connection between the biological entities. Finally, exploration of more complex link prediction algorithms that can accommodate directed networks as well as alternative methods for predicting missing edges (e.g., community detection methods50) is also a focus of future work.
5. Conclusions
OWL-NETS is a novel abstraction network methodology that generates semantically rich network representations that are easily consumed by network inference algorithms. OWL-NETS is easy to configure and can be modified for use with other knowledge sources leveraging Semantic Web technologies. When running link prediction algorithms over OWL-NETS we provided expert-verified evidence from the literature for 50-75% of inferred edges. By leveraging many knowledge sources in a representation tailored for network inference, OWL-NETS has a unique ability to recognize existing, natural patterns in the biological world that have not yet been identified, which would be readily testable in the laboratory environment.
Supplementary Material
Acknowledgments
We thank Marc Daya and Laura Stevens as well as Drs. Anis Karimpour-Fard, Daniel McShan, and Carsten Goerg for their feedback on the development of OWL-NETS. We also thank Ann Cirincione and Raja Cholan for their review of the manuscript.
Funding
This work was supported by the National Library of Medicine Training Grant T15 LM009451, as well as R01 LM009254 and R01LM008111 to LH.
Contributor Information
Tiffany J. Callahan, Computational Bioscience Program, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA.
William A. Baumgartner, Jr., Computational Bioscience Program, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA
Michael Bada, Computational Bioscience Program, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA.
Adrianne L. Stefanski, Department of Pulmonary Sciences and Critical Care, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA
Ignacio Tripodi, Interdisciplinary Quantitative Biology, University of Colorado Boulder, Boulder, CO 80309, USA.
Elizabeth K. White, Computational Bioscience Program, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA
Lawrence E. Hunter, Computational Bioscience Program, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA
References
- 1.Ravasz E, Somera A, Mongru DA, Oltvai ZN, Barabsi AL. Science. 2002;297:1551. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 2.Bultinck J, Lievens S, Tavernier J. Curr Pharm Des. 2012;18:4619. doi: 10.2174/138161212802651562. [DOI] [PubMed] [Google Scholar]
- 3.Lee M, Park K, Kim D. BMC Syst Biol. 2013;7(Suppl 3):S4. doi: 10.1186/1752-0509-7-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert R. Plant Cell. 2007;19:3327. doi: 10.1105/tpc.107.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O. W. Group. Web ontology language (owl) 2012 https://www.w3.org/OWL/
- 6.Dentler K, Cornet R, Teije A, de Keizer N. Sem Web. 2011;1:1. [Google Scholar]
- 7.Kazakov Y, Krotzsch M, Simancik F. J Autom Reasoning. 2014;53:1. [Google Scholar]
- 8.Tsarkov D, Horrocks I. Fact++ description logic reasoner: System description. Proceedings IJCAR-2006. 2006 [Google Scholar]
- 9.Carvalho R, Laskey K, Costa PD. Peer J Comp Sci. 2016;2:77. [Google Scholar]
- 10.Costa PCGD, Laskey KB, Laskey KJ. A Bayesian ontology language for the semantic web. Springer; Berlin, Heidlberg: [Google Scholar]
- 11.Koller D, Levy A, Pfeffer A. P-classic: a tractable probablistic description logic. Proceedings AAAI-97. 1997 [Google Scholar]
- 12.Rettinger A, Lsch U, Tresp V, d’Amato C, Fanizzi N. Data Min Knowl Disc. 2012;24:613. [Google Scholar]
- 13.Mohammadhassanzadeh H, Woensel WV, Abidi SR, Abidi SS. BioData Mining. 2017;10:7. doi: 10.1186/s13040-017-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baader F, Calvanese D, McGuiness D, Nardi D, Patel-Schneider P, editors. The Description Logic Handbook: Theory, Implementation and Applications. Cambridge University Press; 2003. [Google Scholar]
- 15.Wang H, Huang H, Ding C, Nie F. Soft Computing Ontologies Sem Web. 2013;20:344. [Google Scholar]
- 16.Hopkins AL. Nat Chem Biol. 2008;4:682. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 17.Watkinson J, Liang KC, Wang X, Zheng T, Anastassiou D. Ann N Y Acad Sci. 2009;1158:302. doi: 10.1111/j.1749-6632.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 18.Haarslev V, Moller R. An owl reasoning agent for the semantic web. Proceedings of the International Workshop on Applications, Products and Services of Web-based Support Systems, in conjunction with 2003 IEEE/WIC International Conference on Web Intelligence. 2003 [Google Scholar]
- 19.Halper M, Gu H, Perl Y, Ochs C. Artif Intell Med. 2015;64:1. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochs C, He Z, Zheng L, Geller J, Perl Y, Hripcsak G, Musen MA. J Med Bioinfom. 2016;61:63. doi: 10.1016/j.jbi.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. J Am Med Inform Assoc. 2000;7:66. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. J Biomed Inform. 2007;40:561. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Halper M, Wei D, Perl Y, Geller J. J Biomed Inform. 2012;45:15. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livingston KM, Bada M, Baumgartner WA, Hunter LE. BMC Bioinformatics. 2015;16:126. doi: 10.1186/s12859-015-0559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clauset A, Moore C, Newman MEJ. Nature. 2008;453:98. doi: 10.1038/nature06830. [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Zhou T. Physica A Journal. 2011;390:1150. [Google Scholar]
- 27.Hanley JA, McNeil BJ. Radiology. 1982;143:29. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 28.Glimm B, Horrocks I, Motik B, Stoilos G, Wang Z. J Autom Reasoning. 2014;53:245. [Google Scholar]
- 29.Towlson EK, Vertes PE, Ahnert SE, Schafer WR, Bullmore ET. J Neurosci. 2013;33:6380. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. PLoS Comput Biol. 2011;7:e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra SK, Singh P, Rath SK. Malar J. 2011;10:109. doi: 10.1186/1475-2875-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. Science. 2006;313:1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 33.Gil JP. Pharmacogenomics. 2008;9:1385. doi: 10.2217/14622416.9.10.1385. [DOI] [PubMed] [Google Scholar]
- 34.Johansson T, Jurva U, Grnberg G, Weidolf L, Masimirembwa C. Drug Metab Dispos. 2009;37:571. doi: 10.1124/dmd.108.025171. [DOI] [PubMed] [Google Scholar]
- 35.Ajayi AA, Kolawole BA, Udoh SJ. Int J Dermatol. 2004;43:972. doi: 10.1111/j.1365-4632.2004.02347.x. [DOI] [PubMed] [Google Scholar]
- 36.Pillaiyar T, Manickam M, Jung SH. Med Chem. 2015;5:361. [Google Scholar]
- 37.Baldo BA, Pham NH. Anaesth Intensive Care. 2012;40:216. doi: 10.1177/0310057X1204000204. [DOI] [PubMed] [Google Scholar]
- 38.Pruimboom L, de Punder K. J Health Popul Nutr. 2015;33:24. doi: 10.1186/s41043-015-0032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivedi MS, Shah JS, Al-Mughairy S, Hodgson NW, Simms B, Trooskens GA, Van Criekinge W, Deth RC. J Nutr Biochem. 2014;25:1011. doi: 10.1016/j.jnutbio.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su W, Pasternak GW. Synapse. 2013;67:609. doi: 10.1002/syn.21667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu BJ, Girolamo ND, Beck K, Hanratty CG, Choy K, Hou JY, Ward MR, Stocker R. J Pharmacol Exp Ther. 2007;321:477. doi: 10.1124/jpet.106.118612. [DOI] [PubMed] [Google Scholar]
- 42.Agell N, Bachs O, Rocamora N, Villalonga P. Cell Signal. 2002;14:649. doi: 10.1016/s0898-6568(02)00007-4. [DOI] [PubMed] [Google Scholar]
- 43.Venditti CP, Harris MC, Huff D, Peterside I, Munson D, Weber HS, Rome J, Kaye EM, Shanske S, Sacconi S. J Inherit Metab Dis. 2004;27:735. doi: 10.1023/B:BOLI.0000045711.89888.5e. [DOI] [PubMed] [Google Scholar]
- 44.Ying WZ, Sanders PW. Kidney Int. 2001;59:662. doi: 10.1046/j.1523-1755.2001.059002662.x. [DOI] [PubMed] [Google Scholar]
- 45.Arendt BK, Walters DK, Wu X, Tschumper RC, Huddleston PM, Henderson KJ, Dispenzieri A, Jelinek DF. Leukemia. 2012;26:2286. doi: 10.1038/leu.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.VanderWall K, Daniels-Wells TR, Penichet M, Lichtenstein A. J Inherit Metab Dis. 2013;18:449. doi: 10.1615/critrevoncog.2013007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gondi CS, Kandhukuri N, Dinh DH, Olivero WC, Gujrati M, Rao JS. Int J Oncol. 2008;33:783. [PMC free article] [PubMed] [Google Scholar]
- 48.Liben-Nowell D, Kleinberg J. J Am Soc Inf Sci. 2007;58:1019. [Google Scholar]
- 49.Gu H, Cimino JJ, Halper M, Geller J, Perl Y. Proc AMIA Annu Fall Symp. 1996;1996:275. [Google Scholar]
- 50.Hric D, Peixoto TP, Fortunato S. Phys Rev X. 2016;6:031038. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


