Abstract
Fanconi anemia (FA) is a recessive genetic disorder characterized by congenital abnormalities, progressive bone-marrow failure, and cancer susceptibility. The FA pathway consists of at least 21 FANC genes (FANCA-FANCV), and the encoded protein products interact in a common cellular pathway to gain resistance against DNA interstrand crosslinks. After DNA damage, FANCD2 is monoubiquitinated and accumulates on chromatin. FANCD2 plays a central role in the FA pathway, using yet unidentified DNA binding regions. By using synthetic peptide mapping and DNA binding screen by electromobility shift assays, we found that FANCD2 bears two major DNA binding domains predominantly consisting of evolutionary conserved lysine residues. Furthermore, one domain at the N-terminus of FANCD2 bears also nuclear localization sequences for the protein. Mutations in the bifunctional DNA binding/NLS domain lead to a reduction in FANCD2 monoubiquitination and increase in mitomycin C sensitivity. Such phenotypes are not fully rescued by fusion with an heterologous NLS, which enable separation of DNA binding and nuclear import functions within this domain that are necessary for FANCD2 functions. Collectively, our results enlighten the importance of DNA binding and NLS residues in FANCD2 to activate an efficient FA pathway.
INTRODUCTION
Fanconi anemia (FA) is a rare cancer-predisposing and developmental-associated genetic syndrome characterized by bone marrow failure and cellular hypersensitivity to DNA crosslinking agents (1,2). The FA pathway consists of 21 distinct and mostly autosomal genes (FANCA, -B (X-lined), -C, -D1, -D2, -E, -F, -G, -I, -J, -L, -M, -N, -O, -P, -Q, -R, -S, -T, -U and -V), whose protein products participate in the common cellular pathway of DNA interstrand cross-links (ICLs) repair in conjunction with FA-associated proteins (FAAP24, FAAP100, MHF1 and MHF2)(2–5).
Detection of ICLs begins with the recruitment of FANCM and FAAP proteins to stalled replication forks. This triggers the assembly and anchorage to chromatin of an eight-subunit FA core complex (6), containing the E3 ubiquitin ligase FANCL to monoubiquitinate FANCD2 and FANCI on Lysine-561 (K-561) and K-523 respectively (7–10). Monoubiquitination of FANCD2 and FANCI is a pivotal step in the activation of the FA pathway and is essential for localization of these proteins to ICL damage sites within chromatin, where they function together as a protein complex (the ID2 complex) to direct downstream repair steps. The ID2 complex is also phosphorylated by ATR and/or ATM kinase, which not only facilitates its monoubiquitination by the FA core complex, but also serves as a converging point between the FA and BRCA pathways (8,11–14). The ID2 complex serves as the molecular platform to recruit redundant, structure-specific nucleases and TLS polymerases to unhook and bypass the ICLs respectively (1,15). Phosphorylated and monoubiquitinated FANCI and FANCD2 also co-localize with BRCA1 at DNA repair site and other downstream FA proteins associated with DSB repair, such as FANCN (PALB2, partner and localizer of BRCA2), FANCD1 (BRCA2, homologous recombination mediator), FANCJ (BRIP1, a helicase) and RAD51C (a member of RAD51 gene family implicated in HR) (16). Deubiquitination of FANCD2 (8) and FANCI proteins by the multisubunit protein complex USP1-UAF1 is required for the completion of the FA pathway (17).
The molecular entities surrounding the pivotal modification step of the ID2 complex (18) is central to the FA pathway activation and its regulation. The ID2 complex per se is not a good substrate for monoubiquitination in vitro by FANCL, which is in accordance with the published crystal structure of the ID2 complex depicting solvent inaccessibility of the lysine targeted for monoubiquitination (8). Interestingly, in Xenopus egg extracts, FANCD2 monoubiquitination is stimulated by the presence of linear and branched double-stranded DNA (19). Studies with purified chicken FANCD2 also showed that its monoubiquitination is stimulated by the presence of various DNA substrates such as linear single-stranded DNA (ssDNA) and branched double-stranded DNA, with maximum stimulation being achieved with 5′ flapped DNA, which mimics the arrested replication fork (20). Recent studies with purified human FANCD2 showed the failure of chromatinized and unstructured ssDNA to stimulate monoubiquitination as compared to duplex-branched DNA (21). The DNA-mediated stimulation required the presence of FANCI, showing that FANCD2 monoubiquitination may occur within the ID2 complex (20). Enlightening studies with purified native FA core complex from chicken DT40 cells clearly showed DNA-mediated stimulation of FANCD2 monoubiquitination but not of FANCI (22). Cumulatively, there is a strong association between DNA binding and monoubiquitination of FANCD2. DNA binding was the first biochemical activity described for FANCD2 (23). The DNA binding activity was then attributed to the full-length protein as no specific DNA-binding regions were identifiable using truncation mutations restricted to the C-terminus of the protein.
In the present study, we used a thorough truncation strategy, combined with protein purification and DNA-binding assays as a biochemical approach to demonstrate the presence of six evolutionary semi-conserved DNA-binding residues in an important FA protein FANCD2. Point mutations of the DNA-binding residues in the full-length protein resulted in a 10-fold reduction in DNA binding activity of FANCD2. Interestingly, investigation of the mutations in a cellular context revealed an impairment in the nuclear localization of FANCD2. Previously Boisvert et al. reported the presence of a NLS in the amino-terminal 58 amino acids of FANCD2, without pinpointing the specific residues involved in FANCD2 nuclear localization (24). Here, we demonstrate the presence of bifunctional NLS and DNA binding residues in FANCD2. The identified NLS can promote the nuclear localization of green fluorescent protein (GFP). The NLS-defective mutants failed to rescue mitomycin C (MMC) sensitivity of patient derived FA-D2 cells. Moreover, the NLS- defective mutant was unable to complement the monoubiquitination and chromatin association defects of FANCI and FANCD2 in FA-D2 cells. In addition, complementation of NLS-defective mutant by a heterologous NLS resulted in defective FANCD2 monoubiquitination, and showed significantly reduced levels of MMC-induced CtIP foci. Complementation of FA-D2 cells with the DNA binding mutant fused to the heterologous NLS failed to efficiently rescue MMC sensitivity. Our work provides further insights in the regulation of FANCD2, a crucial FA protein, through newly identified domains.
MATERIALS AND METHODS
Cell lines
U2OS, HEK293T and HeLa cells were maintained in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin/streptomycin. PD20 FA-D2 cells were cultured in MEM-alpha supplemented with 20% (v/v) FBS and 1% penicillin/streptomycin. These cells are known to harbour hypomorphic mutation in the FANCD2 gene leading to production of severely truncated protein product (25). To generate stable U2OS or PD20 FA-D2 cell lines expressing WT or mutant FANCD2, FA-D2 cells were infected with pLentiPGKBLASTDEST lentivirus, followed by selection with 2.5 μg/ml of blasticidin in MEM-alpha supplemented with 20% (v/v) FBS and 1% penicillin/streptomycin.
DNA constructs
FANCD2 domains were amplified by PCR (oligonucleotide containing a His-tag in C-terminal) and sub-cloned in the vector pGEX6P.1 containing a GST-tag in N-terminal for the fusion protein (GE HealthCare Life Sciences). FANCD2 WT full-length constructs were cloned by PCR amplification in a modified pFASTBAC1 plasmid (Invitrogen) encoding His/Strep-tags. FANCD2 full-length mutants were generated by site-directed mutagenesis on the WT FANCD2 construct using the Q5® Site-Directed Mutagenesis Kit (NEB). Primers sequences corresponding to the F1 and FANCI NLS were annealed and cloned in pEGFP-C1 (Clontech laboratories). The pEGFP-C1-SYT1 plasmid was a kind gift from the laboratory of Dr Thomas Moss (CHU de Québec). The full-length FANCD2 WT and NLS defective mutant constructs were subcloned from pFASTBAC1 to pLentiPGKBlastDEST (Addgene No.19095). A citrine (GFP derivative) tag was cloned into the C-terminal of FAND2WT and mutant constructs.
Antibodies and immunoblotting
For immunoblotting, cells were washed once with PBS and then lysed in a buffer containing 10 mM HEPES pH 7.4, 10 mM KCl, 1% (v/v) TritonX-100 and 1 mM EDTA. Proteins were resolved by migrating in NuPage 3–8% (w/v) Tris-acetate gel and transferred to nitrocellulose membrane. Commercial antibodies used were rabbit polyclonal FANCD2 antibody (NB100–182; Novus Biologicals), rabbit polyclonal GFP antibody (ab290; Abcam), rabbit polyclonal Histone3 antibody (ab1791; Abcam), mouse monoclonal FANCI antibody (A7; Sigma-Aldrich), mouse monoclonal FANCD2 antibody (GTX30142; GeneTex), and mouse monoclonal GAPDH antibody (MGC127711; Fitzgerald).
RNA interference
The siRNAs were synthesized by Qiagen and directed against the 3′UTR of FANCD2 with the following target sequence 5′ CTGGCTCAGGATTCTAATGTA 3′. Transfection of siRNA was performed using Dharmafect transfection reagent (Thermo Scientific), according to the manufacturer's protocol with minor modifications. In brief, cells were seeded in six-well plates at 2 × 105 cells, 16 h before transfection. For each transfection, 10 μl of Dharmafect was diluted with serum and antibiotic-free media DMEM (Gibco), kept at room temperature (RT) for 5 min and mixed with 200 μl of serum and antibiotic-free DMEM containing 6 μl of 20 uM siRNAs. The mixture was then incubated at RT for 30 min and added dropwise to the cells containing 1.6 ml of antibiotic free DMEM media. The cells were harvested 48 h post-transfection.
Protein purification
Recombinant GST-FANCD2 fragments-His fusions were purified from Escherichia coli BL21 (DE3) RP strain (Stratagene) and grown at 37°C in the LB medium supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. At A600 = 0.4, 80 μg/ml ampicillin was added to the medium, and at A600 = 0.8, 0.1 mM IPTG was added to the culture and incubated at 16°C overnight (16 h). The cells were harvested by centrifugation and the cell pellet was resuspended in 40 ml GST buffer (PBS 1×, 150 mM KCl, 1% Triton X-100, 0,5 mM DTT, 1 mM PMSF, 0.019 UIT/ml Aprotinin, 1 μg/ml Leupeptin). The suspension was sonicated three times for 30 s and insoluble material was removed by centrifugation (18 000 rpm for 30 min in a Sorvall SS34 rotor). An amount of 2 ml of glutathione sepharose beads (GE Healthcare) was added to the supernatant and incubated 2 h at 4°C. The beads were washed four times with GST buffer and two times with PreScission washing buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.05% (v/v) Tween 20). The FANCD2 fragments were eluted by cleavage with PreScission protease (80 U/ml, GE Healthcare) overnight at 4°C. The supernatant was dialyzed against Talon buffer (50 mM NaHPO4 pH 7, 500 mM NaCl, 10% (v/v) glycerol, 0.05% (v/v) Triton X-100, 5 mM imidazole). Then 2 ml of Talon resin (Clontech) was added and incubated for 30 min at 4°C. The resin was washed three times with 10 ml of Talon washing buffer (50 mM NaHPO4 pH 7, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100, 40 mM imidazole). FANCD2 fragments proteins were eluted in Talon buffer containing 500 mM imidazole and dialyzed in three successive steps in storage buffer (20 mM Tris–HCl pH 8.0, 10% glycerol, 0.05% Tween 20, 1 mM DTT) containing 500, 250 and 100 mM NaCl. The full-length FANCD2 proteins (FANCD2 WT and the mutant forms) were purified from baculovirus-infected SF9 cells. Recombinant baculoviruses were produced by Bac-to-Bac expression system as user manual (Invitrogen), Sf9 insect cells (500 ml culture) were infected with the different baculoviruses for 2 days at 27°C. The cell pellet was resuspended in 50 ml of Talon buffer containing PMSF (1 mM), Aprotinin (0.019 TIU/ml) and Leupeptin (1 μg/ml). The suspension was lysed using a Dounce homogenizer (10 strokes), sonicated three times for 30 s at 10% amplitude, and then homogenized a second time. Insoluble material was removed by centrifugation (18 000 rpm for 30 min in a Sorvall SS34 rotor). The supernatant was loaded on a 5 ml Talon column (Clonetech) and washed stepwise with P-buffer containing 30 mM and 50 mM imidazole. The proteins were then eluted with a linear gradient of 0.05–0.5 M imidazole in Talon buffer. The fractions containing the protein were identified by SDS-PAGE and dialysed in the storage buffer. The proteins were then concentrated using an Amicon ultra-15 column (Millipore) and stored at –80°C.
DNA substrates and DNA-binding assays
The oligonucleotides sequences used to synthesize the indicated DNA substrates are mentioned in Supplementary Table S1A and combination used to generate specific DNA probes are listed in Supplementary Table S1B. DNA substrates used were generated with purified oligonucleotides as described (39). Briefly, all the substrates were prepared by annealing reaction carried out by slowly cooling from 95 to 12°C. The DNA binding reactions (10 μl) contained 32P-labeled DNA oligonucleotides (20 nM nucleotides of each substrate) and the indicated concentrations of FANCD2 full-length or fragments in MOPS buffer (25 mM MOPS at pH 7.0, 60mM KCl, 0.2% Tween 20, 2 mM DTT and 1 mM MnCl2). Reaction mixtures were incubated at 37°C for 10 min and then protein–DNA complexes were fixed with 0.2% (v/v) glutaraldehyde for 15 min. The reactions were subjected to electrophoresis on an 8% TBE1X-acrylamide gel and 32P-labeled DNA was visualized by autoradiography.
Dot-blot analysis of the DNA binding domains
Peptides were synthesized by JPT Peptide Technologies (Berlin, Germany) and spotted on a cellulose-β-alanine-membrane. The membrane was washed three times with TBS + 0.05% Tween 20 at room temperature with gentle agitation. It was then incubated for one supplemental hour in TBS-T followed by one hour in TBS-T containing 100 nM of ssDNA probe. It was then washed extensively in TBS-T, air-dried and subjected to autoradiography.
Sub-cellular fractionation
Cellular fractionation of PD20 cells was performed according to Boisvert et al. (24) with minor modifications. Briefly, cells were lysed in the Buffer A (10 mM PIPES pH 6.8, 300 mM sucrose, 125 mM NaCI, 5mM MgCl2, 1 mM EGTA and 0.5% (v/v) Triton-X-100) for 10 min on ice. Pellets were washed thrice with buffer A, resuspended in buffer containing 2% (w/v) SDS, 50 mM Tris–HCI pH7.4, 10 mM EDTA, boiled for 10 min and sonicated for 20 s at 10% amplitude using diagenode BIORUPTOR. Protein concentration of the samples was quantified by the Lowry protein assay and equivalent quantity of each samples was processed for the western blots.
Immunofluorescence and microscopy
For GFP localisation studies with NLS constructs, HeLa cells were transfected with Effectene (Qiagen) according to the manufacturer protocol and fixed with 2% (w/v) paraformaldehyde in PBS for 10 min. The coverslips were then mounted on glass slides with prolong gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Life technologies).
To perform CtIP foci formation, PD20 cells, complemented with empty vector or WT and mutant FANCD2 constructs, were grown on coverslips, washed once with 1× PBS and fixed with 4% (w/v) paraformaldehyde dissolved in 1× PBS. Cells were permeabilized with 0.5% (w/v) Triton X-100 in 1× PBS, washed once with 1X PBS, and blocked with blocking buffer (5% v/v goat serum in 1× PBS) for 45 min. Cells were then incubated with monoclonal anti-CtIP (clone 14-1, Active Motif) diluted (1:300) in the blocking buffer for 1 h 30 min. Cells were washed thrice for 10 min with 1× PBS and incubated in suitable secondary fluorescent antibody (Alexa) diluted in 1× PBS. Finally, the cells were washed thrice with 1× PBS. The coverslips were mounted as described earlier. The slides were visualized with the fluorescence microscope (Leica DMI6000B).
Fluorescence microscopy and laser-induced DNA DSBs
For nuclear localisation studies, PD20 cells were plated on poly-l-lysine treated glass coverslips and transfected with the Effectene transfection reagent. Post-transfection, cells were washed with PBS and fixed with 2% (w/v) paraformaldehyde in PBS for 10 min. Coverslips were washed three times for 10 min with PBS and were mounted onto glass slides with ProLong Gold antifade reagent containing DAPI (Life technologies) and processed for fluorescence microscopy. PD20 cells stably expressing FANCD2 WT and MYCD2-F1+F3 mutant were grown on glass-bottom dishes (MatTek Cop) and laser-induced DNA breaks were created as described earlier (26). The real-time recruitment was measured for each of the constructs. Cells were fixed and mounted as mentioned above.
DNA damage and cell survival assay
For MTT survival assays, PD20 cells complemented with WT and mutant FANCD2 constructs were plated in the Thermo Scientific Nunc 96-well optical bottom dish at 5000 cells per well. The cells were incubated for 6 h at 37°C with 5% CO2, treated with increasing concentration of freshly prepared MMC (0.5 mg/ml Stock, Sigma-Aldrich). The cells were incubated post treatment for 96 h at 37°C with 5% CO2. Cell survival was measured using manufacturer protocol by the MTT Cell Proliferation Assay kit of ATCC bio products.
For CellTiter-Glo Luminescent Cell Viability Assay (Promega), PD20 cells complemented with empty vector, WT, or mutant FANCD2 constructs were seeded in opaque-walled multiwall plates. Twenty-four hours later, cells were treated with indicated concentration of MMC for 24 h. Post-treatment, growth media containing MMC was replaced. Cell survival was measured 72 h later using manufacturer protocol.
RESULTS
FANCD2 bears two DNA binding regions
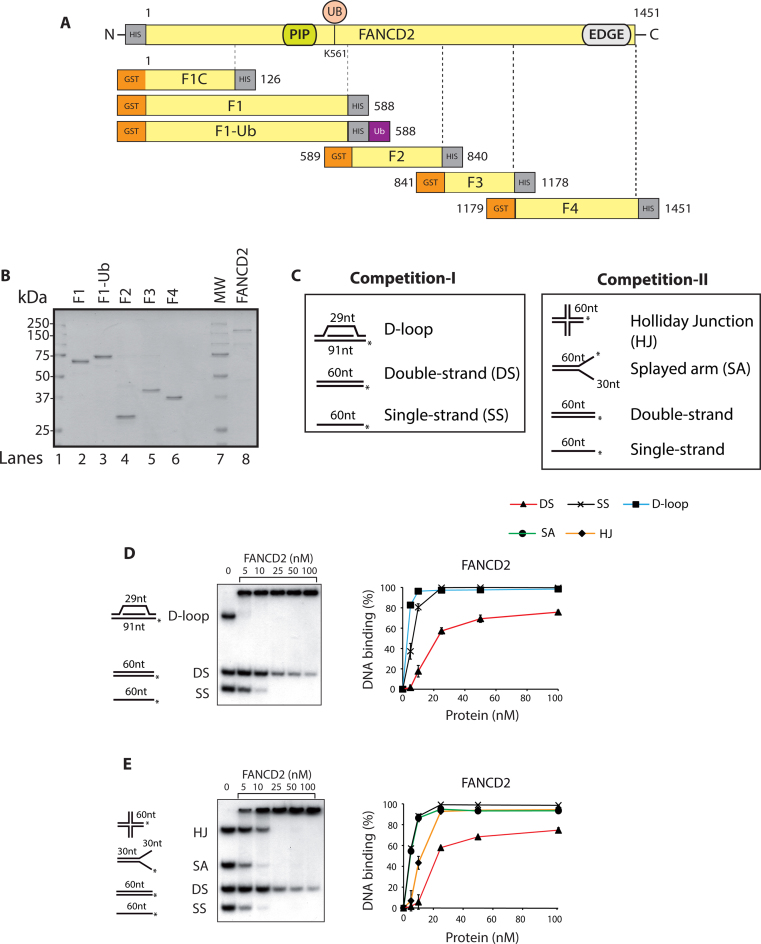
As DNA binding is important for FANCD2 monoubiquitination (19), we initially sought to discover the DNA binding residues of FANCD2. The structure-function analysis and position for each truncation with reference to full-length FANCD2 and associated tags are depicted in Figure 1A. Full-length human FANCD2 with an N-terminal His10 epitope was expressed in Spodoptera frugiperda (Sf9) insect cells and purified (Figure 1B, lane 8). FANCD2 truncations (named hereafter F1–F4) tagged with an N-terminal glutathione S-transferase (GST) tag and C-terminal His10 epitope were expressed in BL21 E. coli cells and purified with double-affinity GST-His purification as described before (Figure 1B, lanes 2, 4–6) (27). Since the bacterial cells are not competent for monoubiquitination, we tagged an ubiquitin moiety to the C-terminal of F1 to derive F1-Ub (Figure 1B, lane 3). For all the GST- tagged proteins used, the GST tag was cleaved off the protein using PreScission enzyme.
Figure 1.
DNA binding analysis of full length FANCD2. (A) Schematic representation of the FANCD2 protein indicating the PCNA interaction motif (PIP), and the carboxyl terminal with EDGE motif. Lysine 561 (K561) is a site of monoubiquitination. Below is the schematic diagram of FANCD2 fragments (F1C, F1-F4) with respect to the full-length FANCD2 to define DNA binding domains. F1-Ub is the ubiquitin coupled F1 fragment. (B) SDS-PAGE gel of purified full length FANCD2 (160 KDa) and fragments F1 (66 kDa), F1-Ub (74 kDa), F2 (28 kDa), F3 (38 kDa) and F4 (31 kDa). MW, molecular mass marker. (C) Competitive DNA binding assay-I and II with two different sets of DNA substrates. Displacement-loop (D-loop); double-strand DNA (DS); single-strand DNA (SS); Holliday Junction (HJ); splayed arms (SA); (*) indicates the 5′ radiolabeling. The DNA binding activity of the full-length FANCD2 was tested in both the competitions I (D) and II (E). Phosphorimager quantification of the percentage of the DNA probes shifted by FANCD2 is shown at the right panels. In both the competitions 20 nm of each DNA substrate was used. Error bars indicate SE from three independently performed experiments.
Previous reports including one from our lab have shown that FANCD2 is a DNA-binding protein with affinity towards branched substrates (23,28). Consistent with the literature, we found that FANCD2 clearly bound to diverse recombinant DNA substrates using competitive gel shift assays with two different sets of probes. In competition assay I, the proteins were incubated with displacement-loop (D-loop), single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA). In competitive assay II, the proteins were incubated with ssDNA, dsDNA, splayed arm (SA) and Holliday junction (HJ) (Figure 1C). In competitive DNA binding assay I, FANCD2 preferred binding to the D-loop followed by ssDNA and dsDNA (Figure 1D). In competition assay II, FANCD2 bound ssDNA and SA with very high affinity and to a lesser extent to HJ followed by dsDNA (Figure 1E). However, to our surprise, purified active FANCD2 could bind efficiently to the D-loop substrate, which might mimic the form of DNA template expected to occur in vivo after the fork reversal step (29). Consistent with this, FANCD2 has been shown to play a direct role in the replication fork dynamics (30–32). Starting from 25 nM, FANCD2 reaches saturation and binds all the recombinant substrates provided except dsDNA. These finding indicates that FANCD2 preferred branched DNA substrates over linear DNA substrates.
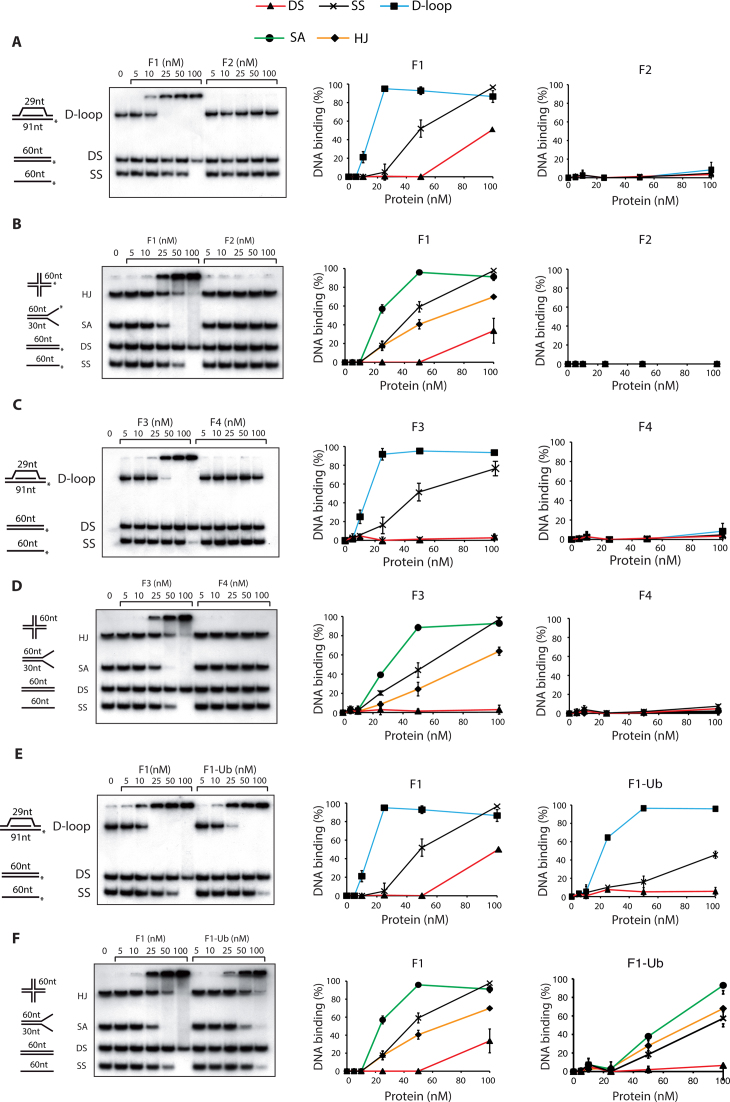
To determine the DNA-binding domains in FANCD2, we performed DNA binding assays with FANCD2 truncations F1–4 (Figure 1A) under same conditions as with full-length FANCD2. Interestingly, we found that the F1 and F3 domains were able to bind DNA in both competition assay I and II (Figure 2A–D). Remarkably, the F1 and F3 domains bound with high affinity to splayed arm, a structure resembling replication forks, compared to ssDNA (Figure 2B and D) while the full-length protein bound to both DNA with almost equal affinity (Figure 1E). In contrast, fragments F2 and F4 failed to show any DNA binding in both competition assays (Figure 2A–D).
Figure 2.
The F1 and F3 fragments are responsible for FANCD2 DNA binding to various substrates. DNA binding activity of F1 and F2 fragments in competition-I (A) and competition-II (B); DNA binding activity of F3 and F4 fragments in competition-I (C) and competition-II (D); DNA binding activity of F1 and F1-Ubiquitin to various DNA substrates in competition-I (E) and competition-II (F). The percentage of the DNA probes shifted in each of the competitions were quantified by phosphorimaging and shown in the right panels. Phosphorimager quantification of the percentage of the DNA probes shifted by FANCD2 is shown at the right panels. In both the competitions 20 nm of each DNA substrate was used. Error bars indicate S.E from three independently performed experiments.
To assess the effect of monoubiquitination on DNA binding activity, we compared the DNA binding activity of F1 with that of F1-Ub. In competitive assay I, F1 and F1-Ub showed comparative DNA binding, though F1-Ub bound dsDNA and ssDNA to a lesser extent. However, F1-Ub binding to D-loop was as efficient as that seen with the F1 fragment (Figure 2E). In competitive assay II, F1-Ub bound various substrates with lesser affinity as compared to non-ubiquitinated F1 (Figure 2F). Importantly, the ubiquitin moiety did not hinder the DNA binding activity of F1 fragment.
These observations strongly suggest that FANCD2 bears two DNA binding regions. The first domain, named F1, comprises amino acids 1–588 and the second, named F3, spans FANCD2 amino acids 841–1178. Further analysis of the F1 domain led to the identification of a smallest DNA binding region in F1 from 1 to 126 aa (referred as F1-C, Supplementary Table S2, line 1 and indicated in Figure 1A).
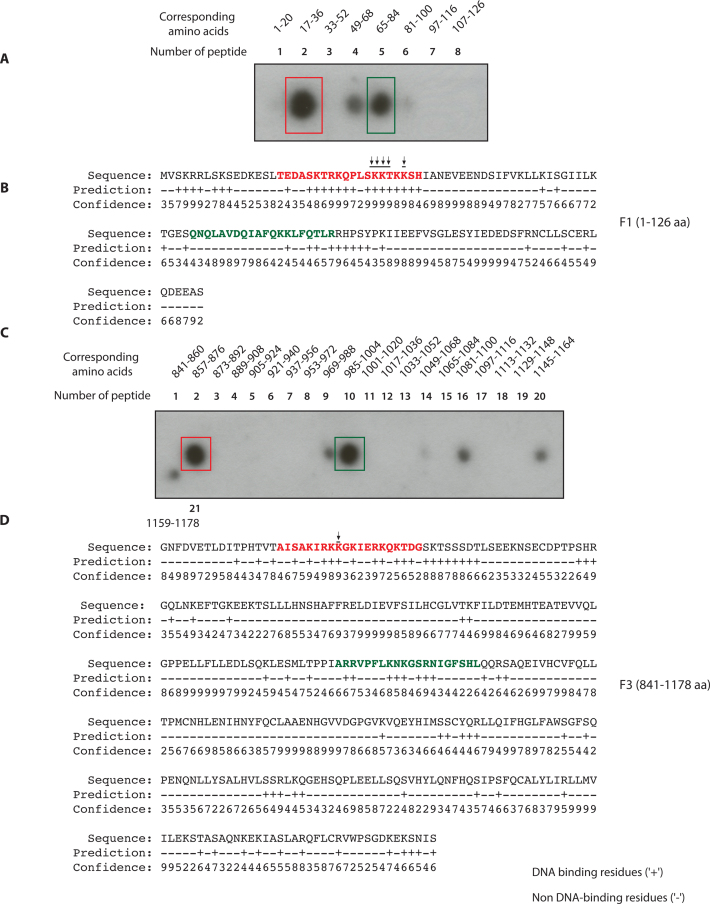
Identification of DNA binding residues through synthetic peptide binding approach
To identify the amino acids required for DNA binding in F1C and F3, we employed a synthetic peptide DNA binding assay. Briefly, synthetic peptides of twenty amino acids of length and with an overlap of four amino acids were generated, covering the entire length of F1C (1–126aa) and F3 (841–1178aa) and immobilized on a membrane. These peptides were then scored for their ability to bind to ssDNA, a substrate that is more stable under this assay conditions compared to the branch substrates used before. Two peptides in F1 (peptide 2, amino acids 17–36 indicated with a red box; and peptide 5, amino acids 65–84 indicated with a green box) lead to intense DNA binding (Figure 3A). A minor signal was detected in peptide 4 (amino acids 49–68). The bioinformatic analysis by DNA binding prediction algorithm BindN (http://bioinfo.ggc.org/bindn/) predicted a few DNA binding residues in F1C with high scores (Figure 3B). The sequences corresponding to the peptide 2 (red letters Figure 3B), and 5 (green letters Figure 3B), were considered for further mutagenesis studies. Sequences in peptide 4 were not showing high BindN probability as DNA binding residues and were therefore ruled out. Similarly, the synthetic peptide DNA binding approach was performed for F3 (Figure 3C). We found the presence of two intense regions for DNA binding corresponding to peptide 2 (amino acids 857–876 indicated with red box) and peptide 10 (amino acids 985–1004 indicated with green box). The high scoring DNA binding residues in the F3 fragment were predicted by BindN (Figure 3D). The sequences corresponding to the peptide 2 (red letters Figure 3D) and 10 (green letters Figure 3D) were considered for further mutagenesis.
Figure 3.
Identification of FANCD2 DNA binding residues by synthetic peptide spot assays and bioinformatic analyses (A) The synthetic peptides of 20 aa length with overlap of four amino acids (aa) were generated corresponding to F1 fragment aa 1–126. These peptides were spotted on a nitrocellulose membrane and tested for binding with radiolabeled single-strand DNA. (B) BindN prediction of highly probable DNA binding residues in F1 (high score hits are in red and lower score in green). (C) The aforementioned synthetic peptide approach was adapted for F3. The number of peptides and the corresponding amino acid number are indicated. It was seen that F3 had two intense regions corresponding to second peptide (861–876 aa) and tenth peptide (985–1004 aa). (D) Hit map of the highly probable and predicted DNA binding residues in F3 domain by BindN analysis. Peptide 2 (marked by red color) and peptide 10 (marked by green color) are shown.
Identification of residues involved in DNA binding
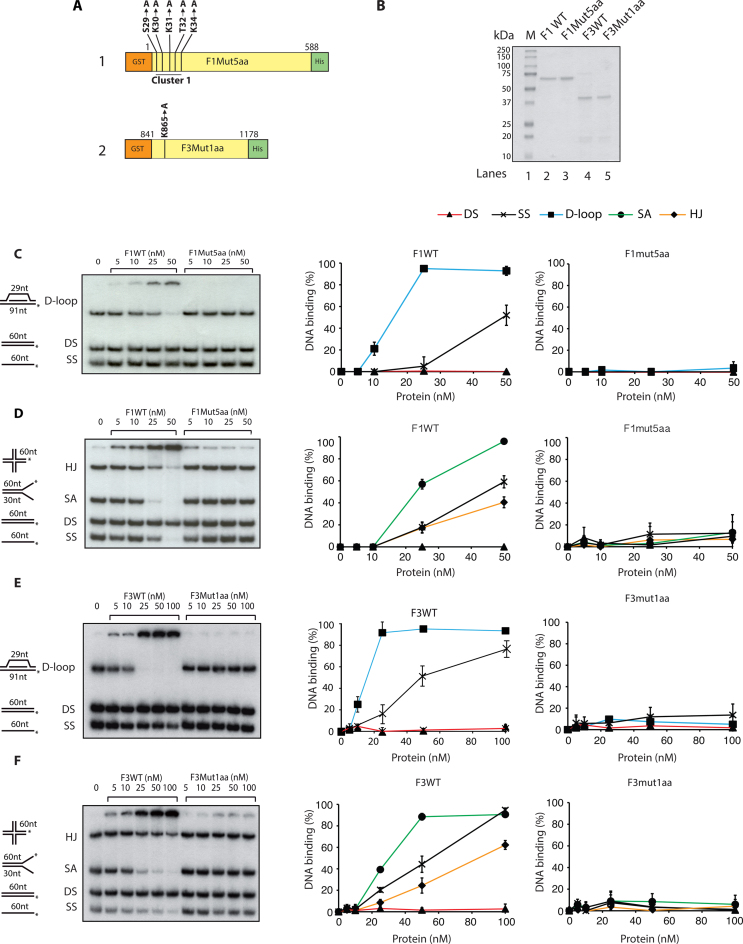
For further mutagenesis and DNA binding assays we utilized the sequences in F1C and F3 that bound ssDNA and those predicted by BindN approach. Mutation to alanine of the eight amino acids corresponding to peptide 2 (Supplementary Table S2, line 5) in F1C resulted in loss of the DNA binding. However, mutations in the sequences corresponding to peptide 5 (Supplementary Table S2, line 6 and referred as cluster2 in Supplementary Figure S1A) in F1C led to DNA binding and thus this region was precluded from further analysis. We then set out to screen the minimum number of mutations required to lose the DNA binding activity of F1 domain. The output of the screen is shown (Figure 4A). The F1 domain having substitute mutations in S29, K30, K31, T32 and K34 (shown by arrowheads in Figure 3B), referred to as F1Mut5aa (Figure 4A), was purified from E. coli BL21 cells with the corresponding wild type F1 domain (Figure 4B). Interestingly, mutations in the five residues (referred as Cluster1) listed above completely abrogated DNA binding activity of the F1 domain in both the competition assay I and II (Figure 4C and D, right panels). To dissect out more specifically, we engineered F1 mutant with four amino acids substitutions (S29A, K30A, K31A and T32A; Supplementary Table S2, line 8), which resulted in reduced but not abrogation of F1 DNA binding. From these data, we concluded that the five residues S29, K30, K31, T32 and K34 of cluster 1 are the important DNA binding residues in F1 domain.
Figure 4.
Loss of DNA binding upon mutations of amino acid residues in FANCD2 F1 and F3 fragments. (A) Diagram representation of F1 (1) and F3 (2) domains indicating point mutations generated by site-specific mutagenesis. Cluster1 in F1 indicates the substitution of five amino acids (aa) (S29A, K30A, K31A, T32A, K34A) and here by would be referred as F1Mut5aa. (B) Coomassie blue staining of purified F1WT (66 kDa), F1Mut5aa (66 kDa), F3WT (38 kDa), F3Mut1aa (38 kDa) proteins. M, molecular mass marker. DNA binding assays of F1WT and F1Mut5aa in competition-I (C) and competition-II (D). DNA binding quantifications are shown at the right. DNA binding activity of F3WT and F3Mut1aa in competition-I (E) and competition-II (F). DNA binding quantifications are shown at the right. In both the competitions 20 nm of each DNA substrate was used. Error bars indicate S.E from three independent experiments.
We adopted similar approach with the second DNA binding region, F3. The five amino acids in peptide 2, i.e. K861, R863, K864, K865 and K867, were mutated to alanine in F3, which resulted in abrogation of DNA binding activity of F3 fragment (Supplementary Table S2, line 12). Subsequently, we generated a six amino acid substituent mutant of F3 in the region of peptide 10 (Supplementary Table S2, line 15), which still bound to the DNA, leading to the exclusion of this region from further analysis. Moreover, substituent mutants of F3 fragment in peptide 2 (Supplementary Table S2, line 13) that bear mutations in the three amino acids i.e. R863, K864 and K865 lacked DNA binding activity. This led us to the conclusion that K861 and K867 were dispensable for the DNA binding of the F3 domain. The single amino acid substitution at K865 (referred as F3Mut1aa Figure 4A and shown by arrowheads in Figure 3D) resulted in the abrogation of DNA binding activity in both the competitive assay I and II (Figure 4E and F). These finding helped us to conclude that K865 is the important DNA binding residue in the F3 domain.
Charge dependency of FANCD2 DNA binding
On one hand, it is a widely established fact that non-sequence specific DNA-protein interaction could be the result of the electrostatic attraction, since the negative charge attributed by the phosphate group is evenly distributed along the length of the DNA molecule (33,34). On the other hand, side chains of polar amino acids such as arginine, lysine, serine and threonine provide a wide window for hydrogen bonding (35). Since the important DNA binding residues identified in FANCD2 mainly consisted of polar lysine, threonine and serine, it was very important to prove that the charge cluster formed by these residues were truly responsible for DNA binding. To address the question of charge involvement, we generated F1CPep2 (K-R), a charge-preserving lysine to arginine mutant in the cluster1 of F1C (Supplementary Figure S1A, 2). To abolish the positive charge from cluster1, we generated the F1Cpep2 (K-A) mutant (Supplementary Figure S1A, 1). The mutations done in F1Cpep5 (K-A) are referred as cluster2 in order to differentiate from cluster1; indeed these residues were the high scoring residues in the BindN analysis (Figure 3B). All the mutants listed above were expressed and purified from E. coli BL21 cells (Supplementary Figure S1B). In this experiment, DNA probes from both the competitions I and II were combined in the single reaction to provide the better competitions and for technical simplicity. As expected, charge-disrupting mutations in F1Cpep5 (K-A) had no effect on the DNA binding of the F1C fragment, moreover charge-preserving mutant F1CPep2 (K-R) regained its DNA binding ability as compared to the charge-disrupting mutant F1Cpep2 (K-A) (Supplementary Figure S1C and D). To correlate this finding from F1C to the complete length of F1, we decided to generate the charge-preserving F1Mut5aa (K-R) mutant (Supplementary Figure S1E and F). As anticipated, F1Mut5aa (K-R) mutant regained its DNA binding ability compared to charge-disrupting F1Mut5aa (K-A) mutant (Supplementary Figure S1G and H). Thus, the positive charges in Cluster1 are responsible for F1 DNA binding activity.
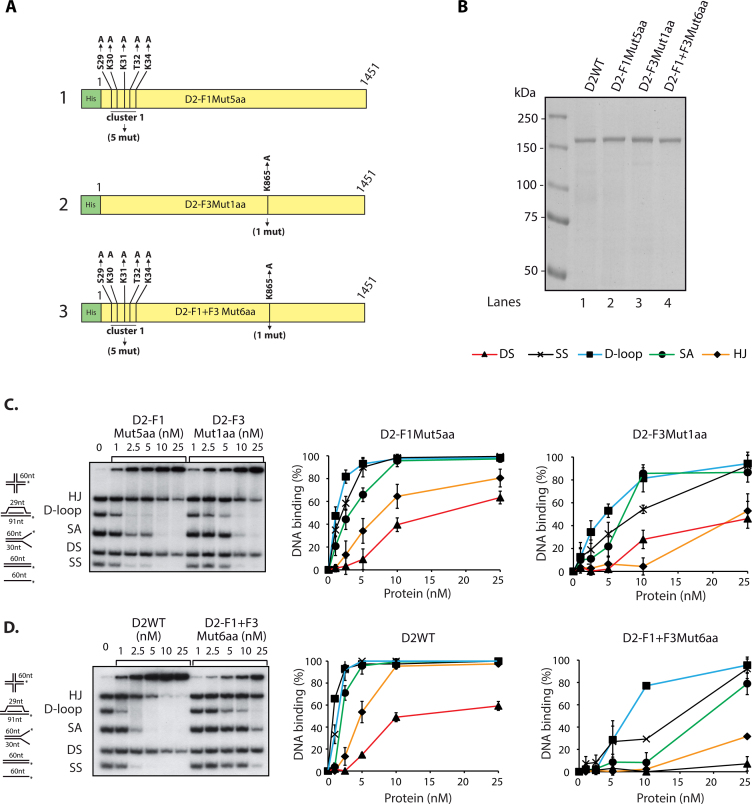
DNA binding activity of the full-length FANCD2 protein
The way domains fold individually as compared to the full-length protein may differ such that mutations performed on domains might have different effects on the whole protein. To address this question, we generated three different substitute mutants in the full-length FANCD2 protein by site-directed mutagenesis. We first mutated Cluster1 to generate D2-F1Mut5aa (Figure 5A, 1), K865 to generate D2-F3Mut1aa (Figure 5A, 2) and combination of the both referred as D2-F1+F3Mut6aa (Figure 5A, 3). The corresponding proteins were purified from SF9 insect cells using Talon affinity combined FPLC chromatography (Figure 5B). To preclude the possibility that mutations could induce a drastic conformational change at the protein level, we verified that purified FANCI still interacted with the purified mutants. No appreciable difference in the ability of FANCD2 WT or mutants to interact with FANCI was observed (Supplementary Figure S2A and B). The DNA binding capacity of individual mutant D2-F1Mut5aa, D2- F3Mut1aa and D2-F1+F3Mut6aa was tested as mentioned before. Interestingly, D2-F1Mut5aa and D2-F3Mut1aa showed a 2-fold and 5-fold reduction, respectively, in DNA binding compared to FANCD2-WT, as judged from gel shift assay and quantification (Figure 5C and D). More strikingly, the double mutant D2-F1+F3Mut6aa had an ∼10-fold reduction in overall DNA binding compared to FANCD2-WT (Figure 5D). Altogether, these results clearly suggest that the identified residues in F1 and F3 affect full-length FANCD2 DNA binding.
Figure 5.
Mutation of Cluster1 and F3Mut1aa results in reduction of FANCD2 DNA binding. (A) Schematic representation of full length FANCD2 mutant constructs D2-F1Mut5aa (1), D2-F3Mut1aa (2) and D2-F1+F3Mut6aa (3). (B) Coomassie blue staining of purified full length FANCD2 (D2WT) (160 kDa), D2-F1Mut5aa, D2-F3Mut1aa and D2-F1+F3Mut6aa. M, molecular mass marker. DNA binding activity to various recombinant DNA substrates of D2-F1Mut5aa and D2-F3Mut1aa (C) or D2WT and D2-F1+F3Mut6aa (D). The percentage of DNA probes shifted were quantified by phosphorimager, right panels. In both the competitions 20 nm of each DNA substrate was used. Error bars indicate S.E from three independent experiments.
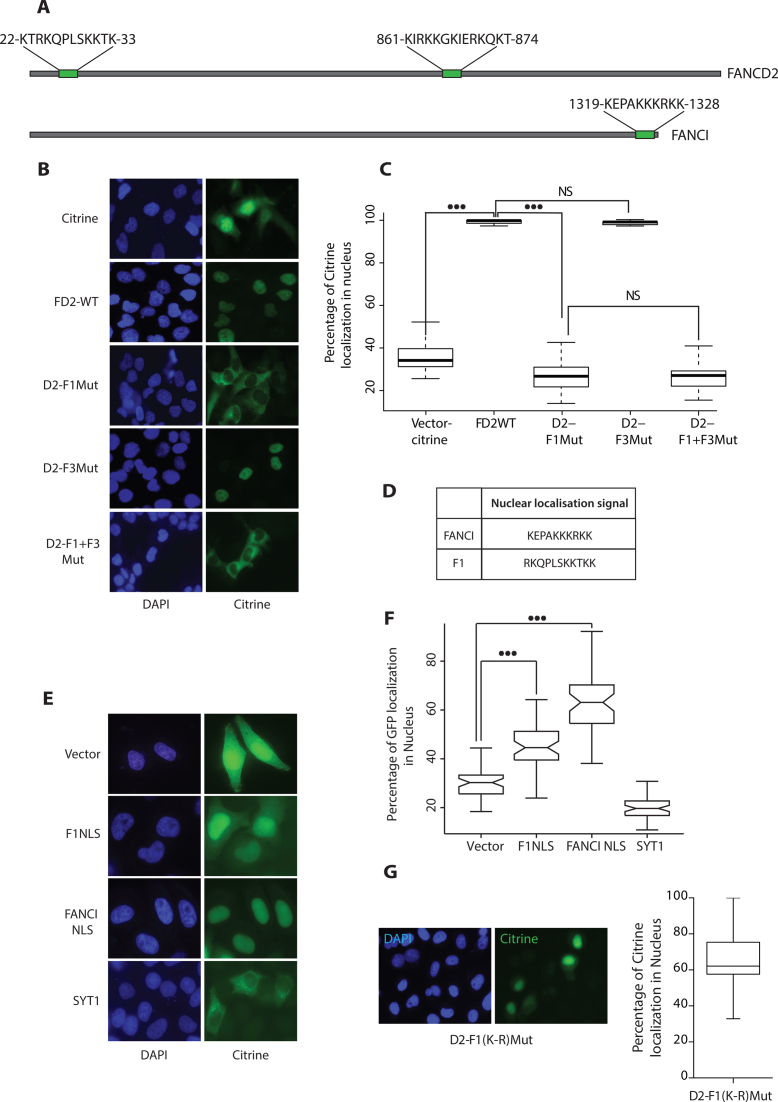
Mutation of the five F1 DNA binding residues abrogated FANCD2 nuclear localization
In an initial attempt to investigate the functional significance of FANCD2 DNA binding domains in human cells, we observed that deletion of the F1 domain (1–125aa) from the full-length FANCD2 led to an aberrant localization of the protein marked by diffuse staining in cytoplasm (data not shown). This observation was consistent with the demonstration of the presence of a NLS within first 58 N-terminal amino acids of FANCD2 (24). In silico analysis of the nuclear localization sequences in FANCD2 by NLStradamus speculated two distant NLS in the protein (Figure 6A). Since there are many NLS prediction protocols available and various predicted positions, we used FANCI as a positive control for this analysis. As already described in the literature, FANCI has one NLS at its extreme C-terminus (36), which was accurately predicted by NLStradamus (Figure 6A). Astonishingly, the predicted FANCD2 NLSs were located in the identified DNA binding residues (Figure 6A). To determine whether mutations of these DNA binding residues had an effect on nuclear localization, we generated citrine (modified GFP)-tagged FANCD2 WT and missense mutants in plentiPGKBLASTDEST (W524–1) plasmid (mutations were introduced as shown in Figure 5A). Upon transfection in FA-D2 cell line, we noticed that D2-F1mut and D2-F1+F3mut displayed defective nuclear localization (Figure 6B). Contrarily, D2-F3Mut and FANCD2WT were exclusively nuclear (Figure 6B). This observation ruled out the involvement of a F3 domain region in nuclear localization. Quantification by Volocity 6.3 High performance 3D imaging software of the percentage of citrine expressed in the individual cell nucleus clearly showed a defect in the nuclear localization of D2-F1Mut (Figure 6C). We noticed a basal level of citrine localization in the nucleus due to passive transport of the 26.9 kDa molecule via the nuclear pore complex (37) (Figure 6C). We then assessed whether the DNA binding sequence, now referred as F1 NLS, could drive the nuclear localization of the GFP protein. We therefore generated GFP-tagged NLS constructs based on the sequences predicted by NLStradamus (Figure 6D). We used the 47.5 kDa calcium-sensing protein Synaptotagmin (Syt1), involved in vesicular transport, and FANCI-NLS tagged to eGFP as cytoplasmic and nuclear expression controls, respectively (Figure 6E) (38). The quantification of eGFP expressed in the nucleus of an individual cell versus the total expression clearly showed that F1-NLS could drive the expression of eGFP in the nucleus (Figure 6F). These results strongly suggest that the F1Cluster functions not only in DNA binding but also as a nuclear localization sequence. To assess a role of charge for nuclear localization within the F1 cluster, we generated full-length D2-F1(K-R)Mut harboring charge preserving K to R mutations in cluster 1 as depicted in (Supplementary Figure S1A). As expected from previous studies on other proteins, by preserving charge in NLS restored FANCD2 into the nucleus (Figure 6G) (39).
Figure 6.
DNA binding residues in region F1 acts as a Nuclear Localization Signal for FANCD2. (A) Represents the hit map of the putative NLS sequences using the NLS prediction tool (www.moseslab.csb.utoronto.ca/NLStradamus) over full length FANCD2 and FANCI. (B) FANCD2-deficient cells (FA-D2) were transfected with the indicated citrine tagged FANCD2 expression constructs. Twenty-four hours post transfection, cells were fixed and stained with 4,6-diaminidino-2-phenyllindole (DAPI) to mark the nucleus. (C) A total of 60 cells transfected with the citrine vector, FANCD2 WT, F1Mut, F3Mut, F1+F3Mut and were scored and box plotted for percentage of citrine expressed in the nucleus. (D) NLS sequences used for F1 and FANCI. (E) NLS sequences predicted in (A) were fused to eGFP and transfected in HeLa cells. peGFP-C1 and SYT1 were used as vector and a cytoplasmic control. Twenty-four hours post transfection, cells were fixed and stained with 4,6-diaminidino-2-phenyllindole (DAPI) to mark the nucleus. Representative images of each expression constructs are shown. (F) A total of 60 cells were counted for F1NLS, FANCI NLS, pEGFP-C1, and SYT1 were scored and box plotted for percentage of eGFP expressed in the nucleus. Wilcoxon's test was employed, P-value<0.01 represents significant difference indicated by (***). (G) FA-D2 cells were transfected with the D2-F1(K-R)Mut as described in B and a total of 60 cells were scored and box plotted for percentage of citrine expressed in the nucleus.
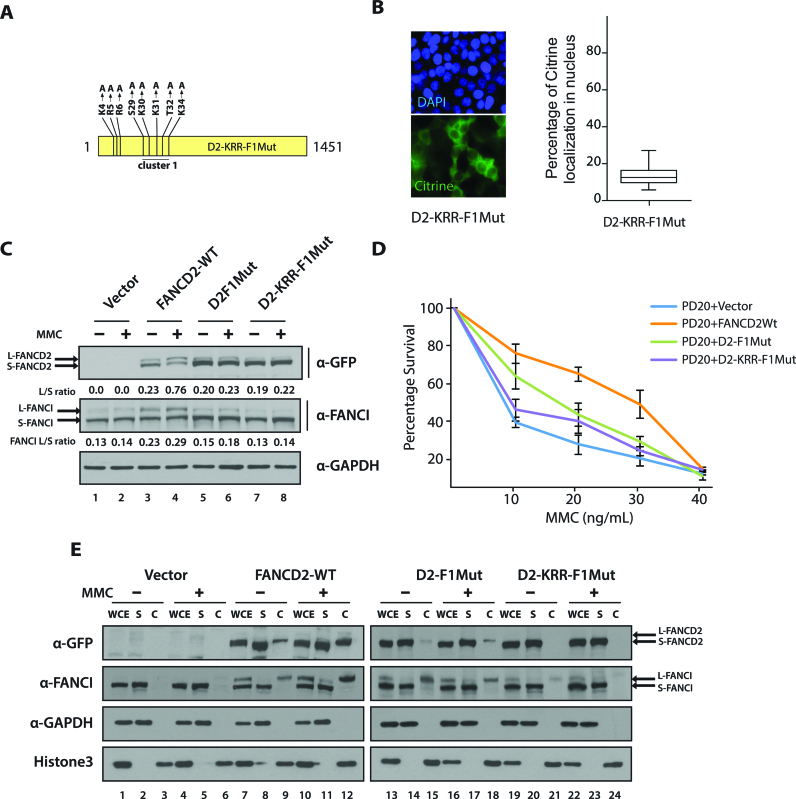
The NLS-defective mutants fail to rescue the FA phenotype of patient-derived FA-D2 cells
A pivotal step in the FA-BRCA pathway is the monoubiquitination of FANCD2 and FANCI. Therefore, we sought to examine these important modifications in FA-D2 cells expressing the NLS-defective mutant. Boisvert et al. have studied the role of a block of positively charged amino acid K-3, R-4 and R-5 (KRR) on FANCD2 nuclear localization (24). We speculated that F1NLS might work in a concerted manner with the KRR sequence. We generated a citrine-tagged FANCD2 full-length mutant by site-specific mutagenesis in the region of Cluster1 and KRR (referred as D2-KRR-F1Mut Figure 7A). As expected, D2-KRR-F1Mut expressed predominantly in the cytoplasm even more than D2F1Mut (compare Figure 7B with D2-F1Mut in Figure 6C). FA-D2 cells stably expressing the empty vector, FANCD2WT, D2F1Mut or D2-KRR-F1Mut were generated. We observed that FA-D2 cells complemented with the empty vector were defective both in spontaneous and damage-induced FANCI monoubiquitination (Figure 7C, lanes 1, 2). This observation shows that even if FA-D2 cells contain residual FANCD2, this is not sufficient to support efficient FANCI monoubiquitination. Indeed, the monoubiquitination of FANCI and FANCD2 is interdependent (7,10). Thus, FA-D2 cells expressing FANCD2 WT showed efficient damage-induced FANCD2 monoubiquitination and rescued the defective FANCI monoubiquitination phenotype of FA-D2 cells (Figure 7C, lanes 3, 4). Strikingly, both NLS-defective mutants, i.e. D2-F1Mut and D2-KRR-F1Mut, had significant impact on FANCD2 and FANCI monoubiquitination as compared to FA-D2 cells expressing FANCD2 WT (Figure 7C, lanes 6,8). Complementation with NLS-defective D2-F1Mut resulted in a slight increment of FANCI monoubiquitination (Figure 7C, lanes 5,6), however the increase was not as marked as that seen for D2WT (Figure 7C compare lanes 3, 4 to 5, 6). D2-KRR-F1Mut failed to rescue FANCI monoubiquitination as seen with FA-D2 cells expressing the vector alone (Figure 7C compare lanes 7, 8 to 1, 2).
Figure 7.
NLS defective mutants fail to rescue FA phenotype of patient derived FA-D2 cells. (A) Diagram of D2-KRR-F1Mut construct. (B) A total of seventy-five FA-D2 cells expressing D2-KRR-F1Mut were scored and box plotted for percentage of citrine expressed in the nucleus. (C) FA-D2 cells stably expressing vector alone, FANCD2WT, D2-F1Mut or D2-KRR-F1Mut were untreated or treated with 80 ng/ml of MMC for 24h. FANCD2 or FANCI monoubiquitination was assessed by western blot. The L/S ratios between the monoubiquitinated form (L) and unmodified from of FANCD2 or FANCI (S) are shown. (D) FA-D2 cells complemented with above mentioned constructs were treated with indicated concentration of MMC for 5 days. Post incubation, cell survival was estimated by MTT assay and plotted as shown. Error bar represents the S.E from three independent experiments. (E) FA-D2 cells stably expressing FANCD2WT, D2-F1Mut or D2-KRR-F1Mut were treated as aforementioned. The cell pellets were fractionated into soluble (S) and chromatin-associated (C) fractions. Fractions were immunoblotted with antibodies against eGFP, FANCI, GAPDH and Histone H3. WCE, represents unfractionated whole cell extract.
FA-D2 cells, like any other FA patient-derived cells are sensitive to ICL-induced genotoxicity and to other DNA damaging agents (40). Therefore, we sought to examine the ability of the NLS-defective constructs to rescue the MMC hypersensitivity phenotype of FA-D2 cells (Figure 7D). In the clastogenic assay, FA-D2 cells expressing FANCD2 WT rescued MMC hypersensitivity, whereas D2-F1Mut partially rescued MMC sensitivity. Notably, FA-D2 cells expressing the D2-KRR-F1 mutant showed even greater decrease in survival than those expressing D2-F1Mut. Additionally, chromatin fractionation experiments revealed that FA-D2 cells are completely defective in both spontaneous and damage-induced FANCI chromatin association (Figure 7E, lanes 3, 6). FA-D2 cells complemented with FANCD2WT restored FANCI bound to the chromatin (Figure 7D, lanes 9,12). FA-D2 cells complemented with D2F1Mut had much reduced FANCD2 and FANCI binding to the chromatin (Figure 7E, compare lanes 9, 12 with 15, 18). The level of FANCD2 and FANCI chromatin association was further reduced in D2-KRR-F1Mut (Figure 7E, lanes 21, 24). Our results thus suggest that the F1 NLS and KRR NLS act in a concerted manner for FANCD2 binding to chromatin and sensitivity to MMC.
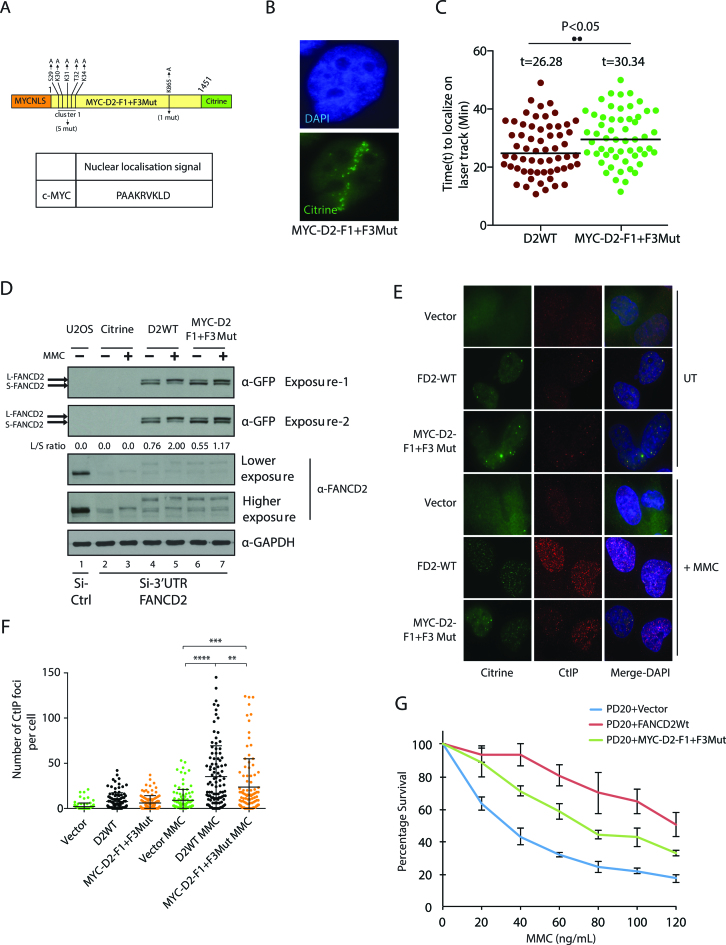
Addition of an exogenous NLS on FANCD2 DNA binding domain mutant does not efficiently restore FANCD2 monoubiquitination, foci formation and mitomycin C resistance
Our results suggested that FANCD2 contains bifunctional NLS and DNA binding sequences. Since our DNA binding mutant was defective in nuclear localization, we aimed to verify whether addition of heterologous NLS such as c-MYC would rescue the phenotype of the D2-F1+F3 mutant. The DNA binding domains of c-MYC has been reported and does not overlap with its NLS (41). Thus, we generated a D2-F1+F3 DNA binding mutant fused with an heterologous NLS of c-MYC (Figure 8A). The resulting construct, MYC-D2-F1+F3Mut, localized to the nucleus and laser-induced breaks (Figure 8B, Supplementary Figure S3A), with slower kinetics than wild-type FANCD2 (Figure 8C). The endogenous FANCD2 was depleted with siRNA against 3′UTR and complemented with wild-type FANCD2 or MYC-F1+F3. Strikingly, U2OS cells expressing MYC-F1+F3 had defective monoubiquitination in both unperturbed and DNA cross-linked cells as compared to wild-type FANCD2 (appreciate the appearance of the slower migrating S-FANCD2 in Figure 8D, lanes 6, 7).
Figure 8.
FANCD2 DNA binding mutant MYC-D2-F1+F3Mut exhibit impaired function in the FA pathway although it is relocalized to the nucleus by an heterologous NLS. (A) Diagram of MYC-D2-F1+F3Mut construct. The c-MYC NLS sequence was tagged to the N-terminal FANCD2-F1+F3-Mut-citrine construct to produce MYC-D2 F1+F3Mut. (B) MYC-D2-F1+F3Mut localizes to laser-induced DSBs. (C) Kinetics of recruitment of MYC-D2-F1+F3Mut compared to wild-type FANCD2. (D) U2OS cells stably expressing vector, MYC-D2-F1+F3Mut (also fused to citrine), and D2WT-citrine were transfected with control or 3′-UTR FANCD2 siRNA. Eight hours post second transfection; cells were untreated or treated with 150 ng/ml of MMC for 24 h. Whole cell extracts were immunoblotted with anti-GFP, anti-FANCD2 and anti-GAPDH antibodies. FANCD2 monoubiquitination was assessed from whole cell extracts by western blot with anti-GFP antibody. The L/S ratio between the monoubiquitinated form (L) and unmodified from of FANCD2 (S) is shown. (E) PD20 cells transfected with empty vector, MYC-D2-F1+F3Mut-citrine and D2-WT-citrine were used to score CtIP foci formation post-treatment with 80 ng/mL of MMC for 24 hours. Immunofluorescence against CtIP was performed and representative images are shown. (F) The number of CtIP foci shown in (E) was scored. The number of CtIP foci from three independent experiments were quantified and plotted as shown. Mann-–Whitney was employed, (**) represents significant difference with a P-value = 0.0021, (***) represents significant difference with a P-value = 0.0003 and (****) represents significant difference with a P-value <0.0001. (G) PD20 cells stably expressing mentioned constructs above were treated with the indicated concentration of MMC for 24 h. Seventy two hours post-incubation, cell survival was estimated with CellTiter-Glo Luminescent Cell Viability Assay. Error bar represents the S.E. from three independent experiments.
It was previously shown that relocalization of CtIP to damaged chromatin is dependent on FANCD2 (42,43). Therefore, we analyzed the formation of MMC-induced CtIP foci formation of the DNA binding mutant MYC-D2-F1+F3Mut compared to wild-type FANCD2. We confirmed that both proteins were expressed at similar levels (Supplementary Figure S3B). The DNA binding mutant was defective as compared to wild-type FANCD2 (Figure 8E and F). Moreover, PD20 cells expressing MYC-D2-F1+F3Mut were more sensitive to MMC than PD20 complemented with wild-type FANCD2 (Figure 8G). Altogether, the introduction of an exogenous NLS on D2-F1+F3Mut did not fully rescue the phenotypes of FANCD2 deficient cells. We conclude that both DNA binding and nuclear localization activities associated with the F1 and F3 regions are important for efficient FANCD2 functions and therefore proper execution of the FA pathway.
DISCUSSION
FANCD2 is a central FA protein, however, the biochemical entities involved in the critical events leading to its monoubiquitination are still poorly understood. Through a biochemical screen, we identified six polar amino acid residues in the full-length FANCD2 protein required for efficient DNA binding. In attempt to study the role of these DNA binding residues in the cellular response to DNA cross-links we have discovered the presence of a nuclear localization sequence. This discovery led to an unexpected finding, FANCD2 NLS is embedded in a DNA binding domain, hence highlighting important regulatory mechanisms. To our knowledge, this is the first FA protein to share bifunctional DNA binding and NLS amino acid sequences.
A previous study from Boisvert et al. (24) has demonstrated the presence of a functional NLS within the first 58 amino acids at the amino terminus of FANCD2. However, it remained to be identified which specific residues were involved in the nuclear localization of the protein. Here, we determined that mutation of residues S-29, K-30, K-31, T-32 and K-34, a block of conserved amino acids at the N-terminal region that is evolutionary conserved (Supplementary Figure S4), resulted not only in the reduction of DNA binding but also in the impairment of nuclear localization of the FANCD2 protein. The mutations in this cluster of amino acid also resulted in the defective monoubiquitination of FANCD2. At cellular level, FA-D2 cells expressing NLS-defective FANCD2 were sensitive to MMC. The NLS defective-mutant not only failed to localize to the chromatin but also had defective FANCI chromatin localization. Moreover, these mutations resulted in the failure to rescue FANCI monoubiquitination and chromatin association as well as MMC sensitivity of FA-D2 cells.
Our data suggest the existence of two NLS in FANCD2, encompassed by the KRR region and F1 DNA binding domain. Both blocks are important for effective FANCD2 nuclear localization as the rescue of the typical FA phenotypes of FA-D2 cells was much less effective in the double NLS mutant. Indeed, fusion of amino acids 24–34 of FANCD2 to the N-terminal of GFP could drive its nuclear expression. FANCD2 is a large protein of 1451 amino acids necessitating an active nuclear transport, which has been demonstrated to be mediated via an importin α/β-dependent mechanism. While the classical bipartite NLS separated by mutations tolerant 10–12 amino acids as exemplified by the nucleoplasmin NLS, there are reports of unconventional bipartite and tripartite NLSs. Indeed, few proteins possess both unconventional monopartite and classical bipartite NLS as exemplified by the Influenza virus A nucleoprotein (44). The FANCD2 protein represents an example of the nonconventional yet classical type of NLS characterized by the involvement of stretch of K/R amino acids. The NLStradamus, a software shown to demonstrate higher precision over the existing NLS prediction tools, generated hits in the demonstrated NLS.
It has been demonstrated that FANCD2 has DNA binding capacity that is independent of its monoubiquitination status. In this study, we present the specific residues involved in the DNA binding of FANCD2, indeed the DNA binding remains unaffected by its monoubiquitination status. At the domain level, charge-disrupting mutations of the S-29, K-30, K-31, T-32, K-34 and K-865 residues completely abrogated DNA binding of the FANCD2 domains, while at the full-length protein level, it greatly reduced the DNA binding capacity. The presence or generation of other minor DNA binding residues upon protein folding at the three-dimensional structure level could explain the residual DNA binding in full-length DNA-binding mutants of FANCD2. These residues remain to be identified, but are most likely important only for residual DNA binding in the context of the mutated F1 and F3 domains. FANCD2 was shown to activate the transcription of tumor suppressor Tap63 by binding to its promoter (45). The DNA binding domain we identified binds to DNA without sequence specificity which is important as DNA damage from crosslinking agents could occur at any location in the genome. Thus, the presence of charge-dependent DNA binding is consistent for a DNA repair protein rather than canonical DNA binding domains found in transcription factors.
The crystal structure of the murine FANCD2-FANCI complex (8), and more recently the cryo-EM structure has been reported (18). In the crystal structure, the absence of the extreme N-terminal comprising the demonstrated NLS and the region surrounding K-865 (F3 region) precluded the speculation about the secondary structure in the region. Moreover, detailed FANCD2 domain architecture has been simulated from the structure of various FANCI-DNA complexes. Interestingly, the secondary structure prediction tool (http://www.sbg.bio.ic.ac.uk/phyre2) Phyre2 predicted that the F1 DNA binding sequences folds into α-helix with high confidence (Supplementary Figure S5).
A key molecular event in the Fanconi anemia pathway is the organization of a molecular platform by the monoubiquitinated FANCD2-FANCI complex. The molecular processes regulating the key event of monoubiquitination are of great interest to understand the details of the FA-BRCA pathway. The monoubiquitination of FANCD2 and FANCI is interdependent in vivo as well as in vitro. It is well-established that DNA is a biochemical entity required for the stimulation of FANCD2 monoubiquitination. The DNA-mediated stimulation requires the DNA binding activity of FANCI. Here, we demonstrated the presence of specific DNA binding residues in the FANCD2 protein. Indeed, FANCD2 has intrinsic DNA binding activity and this is independent of FANCI DNA binding activity. Our studies clearly show that FANCD2 DNA binding residues are important for efficient execution of the FA pathway but also coordination of downstream factors such as CtIP (42,43).
To our knowledge, FANCD2 protein is the first FA-BRCA protein showing bifunctional NLS-DNA binding residues. This suggests that evolution used some part of the DNA binding mechanism when compartmentalizing DNA binding proteins into the nucleus. In silico analysis published >15 years ago by Cokol et al. (46) predicted that there are many proteins with bifunctional DNA binding and NLS residues. Our results crystallize this prediction for a major player of the FA pathway.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Guy Poirier and Stéphane Richard for advice and mentorship, Patrick Sung for reagents and Tom Moss for the use of a SP5 microscope obtained with CFI funding (project 23598). We thank Ugo Déry, Céline Roques, Julien Vignard, Josiane Tremblay-Rochette, Marie-Michelle Genois, Isabelle Brodeur for help and discussions in the initial stages of this study. We thank K.J. Patel and members of the Fanconi anemia field for discussions and Isabelle Brodeur and Amélie Rodrigue for critically reviewing the manuscript. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Present address: Joshi Niraj, Department of Radiation Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02215, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Laval University Cancer Research Center HOG scholarship (to N.J.); FRQS MSc scholarship (to K.D.); NSERC (to S.B.); A.M.C is a Luc Bélanger scholar; JYM was a FRQS Chercheur National and is a FRQS Chair in DNA repair and genome stability; CIHR [363317]. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kottemann M.C., Smogorzewska A.. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013; 493:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moldovan G.L., D’Andrea A.D.. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009; 43:223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sawyer S.L., Tian L., Kahkonen M., Schwartzentruber J., Kircher M., Majewski J., Dyment D.A., Innes A.M., Boycott K.M., Moreau L.A. et al. . Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015; 5:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J.P., Minguillon J., Ramirez M.J., Pujol R. et al. . Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 2013; 92:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kashiyama K., Nakazawa Y., Pilz D.T., Guo C., Shimada M., Sasaki K., Fawcett H., Wing J.F., Lewin S.O., Carr L. et al. . Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 2013; 92:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang M., Kim J.M., Shiotani B., Yang K., Zou L., D’Andrea A.D.. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell. 2010; 39:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R. 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D. et al. . Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007; 129:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joo W., Xu G., Persky N.S., Smogorzewska A., Rudge D.G., Buzovetsky O., Elledge S.J., Pavletich N.P.. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011; 333:312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boisvert R.A., Howlett N.G.. The Fanconi anemia ID2 complex: dueling saxes at the crossroads. Cell Cycle. 2014; 13:2999–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sims A.E., Spiteri E., Sims R.J. 3rd, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H. et al. . FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007; 14:564–567. [DOI] [PubMed] [Google Scholar]
- 11. Andreassen P.R., D’Andrea A.D., Taniguchi T.. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004; 18:1958–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tomida J., Itaya A., Shigechi T., Unno J., Uchida E., Ikura M., Masuda Y., Matsuda S., Adachi J., Kobayashi M. et al. . A novel interplay between the Fanconi anemia core complex and ATR-ATRIP kinase during DNA cross-link repair. Nucleic Acids Res. 2013; 41:6930–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T. et al. . FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008; 15:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhi G., Wilson J.B., Chen X., Krause D.S., Xiao Y., Jones N.J., Kupfer G.M.. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009; 69:8775–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim H., D’Andrea A.D.. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012; 26:1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Andreassen P.R., D’Andrea A.D.. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 2004; 24:5850–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D.. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007; 28:786–797. [DOI] [PubMed] [Google Scholar]
- 18. Liang C.-C., Li Z., Lopez-Martinez D., Nicholson W.V., Vénien-Bryan C., Cohn M.A.. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat. Commun. 2016; 7:12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sobeck A., Stone S., Hoatlin M.E.. DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2. Mol. Cell. Biol. 2007; 27:4283–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato K., Toda K., Ishiai M., Takata M., Kurumizaka H.. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012; 40:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Longerich S., Kwon Y., Tsai M.S., Hlaing A.S., Kupfer G.M., Sung P.. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014; 42:5657–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajendra E., Oestergaard V.H., Langevin F., Wang M., Dornan G.L., Patel K.J., Passmore L.A.. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol. Cell. 2014; 54:858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park W.H., Margossian S., Horwitz A.A., Simons A.M., D’Andrea A.D., Parvin J.D.. Direct DNA binding activity of the Fanconi anemia D2 protein. J. Biol. Chem. 2005; 280:23593–23598. [DOI] [PubMed] [Google Scholar]
- 24. Boisvert R.A., Rego M.A., Azzinaro P.A., Mauro M., Howlett N.G.. Coordinate nuclear targeting of the FANCD2 and FANCI proteins via a FANCD2 nuclear localization signal. PLoS One. 2013; 8:e81387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D’Andrea A.D. et al. . Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell. 2001; 7:241–248. [DOI] [PubMed] [Google Scholar]
- 26. Dery U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.Y.. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell. Biol. 2008; 28:3058–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maity R., Pauty J., Krietsch J., Buisson R., Genois M.M., Masson J.Y.. GST-His purification: a two-step affinity purification protocol yielding full-length purified proteins. J. Vis. Exp. 2013; e50320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roques C., Coulombe Y., Delannoy M., Vignard J., Grossi S., Brodeur I., Rodrigue A., Gautier J., Stasiak A.Z., Stasiak A. et al. . MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J. 2009; 28:2400–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burkovics P., Sebesta M., Balogh D., Haracska L., Krejci L.. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 2014; 42:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaudhury I., Sareen A., Raghunandan M., Sobeck A.. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013; 41:6444–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lossaint G., Larroque M., Ribeyre C., Bec N., Larroque C., Decaillet C., Gari K., Constantinou A.. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell. 2013; 51:678–690. [DOI] [PubMed] [Google Scholar]
- 32. Yeo J.E., Lee E.H., Hendrickson E.A., Sobeck A.. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 2014; 23:3695–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nadassy K., Wodak S.J., Janin J.. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999; 38:1999–2017. [DOI] [PubMed] [Google Scholar]
- 34. Xiong Y., Sundaralingam M.. eLS. 2001; John Wiley & Sons, Ltd. [Google Scholar]
- 35. Luscombe N.M., Laskowski R.A., Thornton J.M.. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001; 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colnaghi L., Jones M.J., Cotto-Rios X.M., Schindler D., Hanenberg H., Huang T.T.. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood. 2011; 117:2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei X., Henke V.G., Strubing C., Brown E.B., Clapham D.E.. Real-time imaging of nuclear permeation by EGFP in single intact cells. Biophys J. 2003; 84:1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sudhof T.C. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat. Med. 2013; 19:1227–1231. [DOI] [PubMed] [Google Scholar]
- 39. Lee B.J., Cansizoglu A.E., Suel K.E., Louis T.H., Zhang Z., Chook Y.M.. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006; 126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Auerbach A.D., Wolman S.R.. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976; 261:494–496. [DOI] [PubMed] [Google Scholar]
- 41. Dang C.V., van Dam H., Buckmire M., Lee W.M.. DNA-binding domain of human c-Myc produced in Escherichia coli. Mol. Cell. Biol. 1989; 9:2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murina O., von Aesch C., Karakus U., Ferretti L.P., Bolck H.A., Hanggi K., Sartori A.A.. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014; 7:1030–1038. [DOI] [PubMed] [Google Scholar]
- 43. Unno J., Itaya A., Taoka M., Sato K., Tomida J., Sakai W., Sugasawa K., Ishiai M., Ikura T., Isobe T. et al. . FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014; 7:1039–1047. [DOI] [PubMed] [Google Scholar]
- 44. Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H.. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007; 282:5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park E., Kim H., Kim J.M., Primack B., Vidal-Cardenas S., Xu Y., Price B.D., Mills A.A., D’Andrea A.D.. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol. Cell. 2013; 50:908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cokol M., Nair R., Rost B.. Finding nuclear localization signals. EMBO Rep. 2000; 1:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.