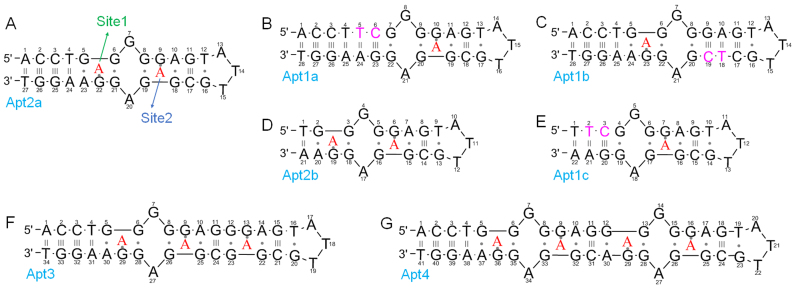Abstract
The DNA aptamer for adenosine (also for AMP and ATP) is a highly conserved sequence that has recurred in a few selections. It it a widely used model aptamer for biosensor development, and its nuclear magnetic resonance structure shows that each aptamer binds two AMP molecules. In this work, each binding site was individually removed by rational sequence design, while the remaining site still retained a similar binding affinity and specificity as confirmed by isothermal titration calorimetry. The thermodynamic parameters of binding are presented, and its biochemical implications are discussed. The number of binding sites can also be increased, and up to four sites are introduced in a single DNA sequence. Finally, the different sequences are made into fluorescent biosensors based on the structure-switching signaling aptamer design. The one-site aptamer has 3.8-fold higher sensitivity at lower adenosine concentration with a limit of detection of 9.1 μM adenosine, but weaker fluorescence signal at higher adenosine concentrations, consistent with a moderate cooperativity in the original aptamer. This work has offered insights into a classic aptamer for the relationship between the number of binding sites and sensitivity, and a shorter aptamer for improved biosensor design.
INTRODUCTION
Aptamers are single-stranded nucleic acids that can specifically bind to target molecules (1,2). Aptamers were isolated in test tubes and also found in mRNA known as riboswitches in many bacterial cells regulating gene expression (1–3). While aptamers can bind essentially any type of molecule or surface, those binding small molecules are particularly interesting, since antibodies are usually less effective in binding or detecting small molecules (4). With excellent ligand binding properties and programmability, aptamers have been extensively used in biosensor development (5–12).
The 27-nt DNA aptamer for adenosine is one of the most studied since its initial report in 1995 by Huizenga and Szostak (13). It has a similar affinity to a few adenosine derivatives including adenosine monophosphate (AMP), cyclic adenosine monophosphate (cAMP) and adenosine triphosphate (ATP), but it cannot bind other nucleosides such as guanosine (14). This aptamer has been used as a model system for extensive biophysical studies and for biosensor development (15–26). Nuclear magnetic resonance (NMR) shows two identical binding pockets in this aptamer, each binding one AMP molecule (27,28). Various spectroscopy studies were performed to understand its binding dynamics and cooperativity (29–31). This aptamer was later re-discovered using a completely different selection method by the Li lab (32), and thus it is a highly recurrent sequence.
It is quite intriguing that this aptamer binds two target molecules, while most other small molecule binding aptamers only bind one (33). For example, an RNA aptamer was selected to bind just one AMP (34), even though NMR showed that the DNA and RNA aptamers have the same mode of binding despite very different overall architectures (27). Herein, we are interested in understanding whether these two adenosine binding pockets are inter-dependent or not. In other words, we wished to test whether we can delete one of the binding pockets while still retaining binding. Tuning the number of binding sites has important analytical implications to affect biosensor sensitivity (35,36). To study this, we resorted to isothermal titration calorimetry (ITC) to measure binding thermodynamics and fluorescence spectroscopy to follow biosensor performance.
MATERIALS AND METHODS
Chemicals
All the DNA samples were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). The names and sequences of the DNAs are in the Supplementary Figure S1. Sodium chloride, magnesium chloride, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), adenosine and other nucleosides were from Mandel Scientific (Guelph, ON, Canada). Milli-Q water was used to prepare all the buffers and solutions.
ITC
ITC was performed using a VP-ITC microcalorimeter instrument (MicroCal). All the ITC tests used the following protocols unless otherwise specified. Prior to each measurement, each solution was degassed to remove air bubbles. An aptamer sample (10 μM) in buffer A (50 mM HEPES, pH 7.6, NaCl 100 mM and 5 mM MgCl2) was loaded in a 1.45 ml ITC cell at 25°C. Adenosine or other nucleosides (280 μl, 0.5 mM) in the same buffer was titrated into the cell through a syringe (10 μl each time, except for the first injection of 2 μl). The enthalpy (ΔH) and binding constant (Ka) were obtained through fitting the titration curves to a one-site binding model using the Origin software. The dissociation constant (Kd) values were calculated from 1/Ka and ΔG = −RT ln(Ka), where R is the gas constant. ΔS was calculated from ΔG = ΔH − TΔS. The Wiseman coefficient (c-value) was calculated from c = n⋅[aptamer]/Kd, where n is the number of binding site (37,38). To ensure data accuracy, all the ITC results with c-value below 1 were re-titrated with higher aptamer concentration (20 μM) or at lower temperatures to obtain higher c-values.
Binding cooperativity
The binding cooperativity was evaluated by extracting the Hill coefficient (h). The sum of enthalpy changes (ΔH) of different aptamers produced by the total concentration of titrated adenosine was fitted using Equation (1):
 |
(1) |
in which Bmax is the maximum binding enthalpy changes, [Ade] is the concentration of titrated adenosine, and h is the Hill coefficient representative of positive cooperativity (h > 1), or non-cooperative (h = 1) between different adenosine binding sites.
Biosensor detection
A total of 40 nM F-DNA, 60 nM aptamers containing DNA and 80 nM Q-DNA were mixed in the buffer A. This 1: 1.5: 2 ratio was chosen to ensure a low background signal. These DNAs were dissolved in buffer A and heated to 85°C, then cooling down slowly to 25°C at a rate of ∼1°C/min to form the sensor complex. Different concentrations of adenosine or other nucleosides were added and the fluorescence intensity was recorded on a Varian Eclipse fluorescence spectrometer (Agilent Technologies, Santa Clara, CA, USA) with excitation at 490 nm and emission at 520 nm at 25°C. Multiple samples of the free sensor without adenosine were measured to calculate the standard deviation of the background variation (σ). Then the initial slope of the calibration curve was calculated for determining the detection limit (3σ/slope).
RESULTS AND DISCUSSION
Aptamer binding is retained after removing one site
The secondary structure of the originally selected adenosine aptamer is shown in Figure 1A. This wild-type aptamer is named Apt2a, where the ‘2’ describes its binding two adenosine molecules. To facilitate discussion, each nucleotide in the aptamer is numbered. The two binding sites that are quite symmetrically arranged (the red colored ‘A’ denotes for the target adenosine). We call the site near G22 site 1 and the one near G9 site 2. According to its NMR structure, each adenosine interacts with two nearby nucleotides via hydrogen bonding and it also stacks on a guanine by a reversed Hoogsteen mismatch (27). The two sites have identical binding interactions and they are separated by non-canonical base pairs.
Figure 1.
The secondary structures of (A) the wild-type adenosine aptamers (Apt2a), and the mutants (B–G). The wild-type aptamer has two binding sites (1 and 2). (B) Site 1 is removed; (C) site 2 removed; (D) three base pairs deleted from the wild-type; (E) three base pairs deleted from Apt1a. Extended aptamers with (F) 3 and (G) 4 adenosine binding sites. The red color ‘A’ represents the bound adenosine. The nucleotides in pink color are the mutated to remove adenosine binding sites. The number in the name of each sequence indicates the number of adenosine binding sites.
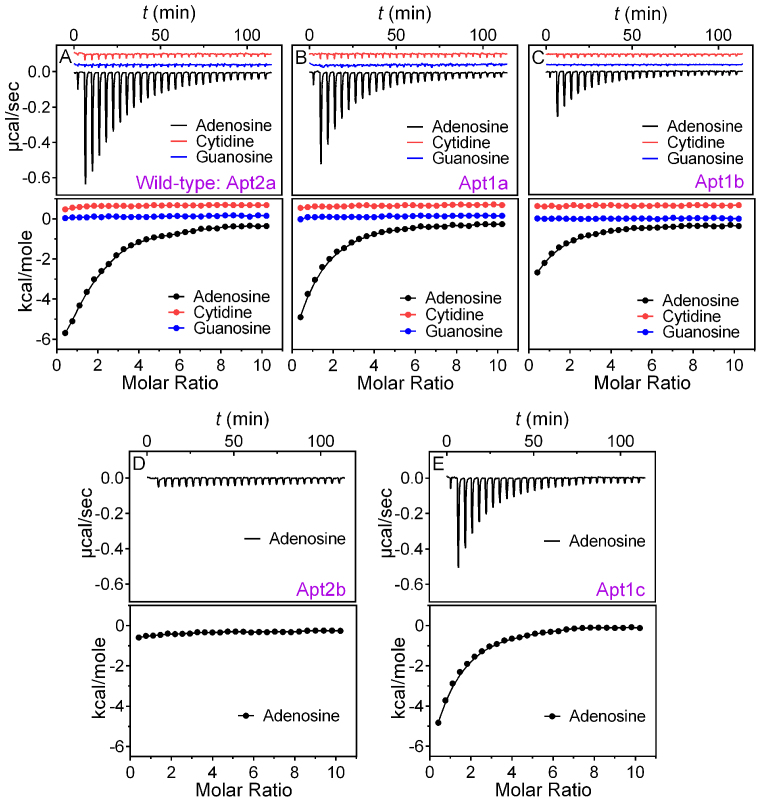
To measure binding, we used ITC, since it is a label-free technique providing rich thermodynamic information for aptamer binding (39,40). Adenosine was first titrated into the wild-type aptamer and the heat of each injection was followed over time (Figure 2A, black trace in top panel). The reaction was exothermic and we integrated the heat profile (Figure 2A, black dots in lower panel) to directly obtain the reaction enthalpy (ΔH), binding stoichiometry (n) and dissociation constant (Kd), which allowed further calculation of ΔG and ΔS. All the thermodynamic parameters are presented in Table 1 (the first row). The Kd of the wild-type aptamer is 16.4 μM. While this is higher than that in the original paper (6 μM adenosine) reported using the centrifugation filter method (13), it agrees well with that from isocratic elution (13 μM ATP) (41) and is tighter than that from microscale thermophoresis (28.9 μM adenosine diphosphate, ADP) (42). Indeed, each aptamer binds 2.1 ± 0.2 adenosine molecules, consistent with the structural biology literature (27).
Figure 2.
ITC traces and integrated heat of (A) the wild-type aptamer Atp2a, and (B) the Apt1a and (C) Apt1b mutants binding adenosine, cytidine and guanosine. ITC titrations of the shortened aptamers (D) Apt2b and (E) Apt1c binding adenosine.
Table 1. Binding thermodynamic values of the wild-type aptamer and the mutants.
| Aptamer name | n | K a (× 104 M−1) | K d (μM) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | ΔS (cal K−1mol−1) |
|---|---|---|---|---|---|---|
| Apt2a (WT) | 2.1 ± 0.2 | 6.1 ± 0.5 | 16.4 ±1.4 | −6.5 ±0.1 | −14.1 ± 0.5 | −25.7 ±1.6 |
| Apt1a | 1.1 ± 0.1 | 8.3 ± 0.2 | 12.0 ±0.3 | −6.7 ±0.04 | −10.2 ± 0.8 | −12.0 ±2.6 |
| Apt1b | 0.8 ± 0.2 | 7.1 ± 0.4 | 14.1 ±0.8 | −6.5 ±0.03 | −6.3 ± 0.4 | 0.7 ±1.2 |
| Apt2b | -a | −0.5 ± 0.03 | - | |||
| Apt1c | 1.0 ± 0.2 | 8.3 ± 0.4 | 12.0 ±0.6 | −6.7 ±0.1 | −12.5 ± 0.2 | −18.6 ±0.6 |
| Apt3 | 3.1 ± 0.2 | 9.1 ± 0.5 | 11.0 ±0.6 | −6.7 ±0.03 | −8.6 ± 0.4 | −6.4 ±1.2 |
| Apt4 | 3.8 ± 0.4 | 10.2 ± 0.5 | 9.8 ±0.5 | −6.8 ±0.2 | −8.4 ± 0.4 | −5.5 ±1.2 |
Notes: Data obtained using a one-site binding model.
aVery weak binding (Ka < 1000 M−1) not detectable by ITC. The errors listed here were from the standard deviation of at least two independent measurements. The fitting error for each ITC trace is in general below 8%.
After confirming binding of the wild-type, the first question we wanted to answer was whether we could remove one of the binding sites while still retaining the binding affinity and specificity of the other. To test this, we introduced a G5T mutant to the wild-type and also inserted a C base to pair with G22. As a result, two more base pairs were added and this mutant was called Apt1a (Figure 1B). We reason these additional base pairs should eliminate binding site 1.
For the Apt1a mutant (Figure 2B), the amount of heat released was smaller and the entropy loss was also smaller, resulting in a similar Kd (12.0 μM, Table 1). The number of binding sites on each aptamer indeed reduced to 1.1 ± 0.1, consistent with our design of removing one binding site. For both the wild-type and Apt1a, binding was specific, since titrating cytosine or guanosine gave no heat response (Figure 2A and B, red and blue traces). Therefore, we successfully eliminated binding site 1, while retained binding of the remaining site 2. This is important since for the first time we experimentally demonstrated that a one-site aptamer can be engineered for this very important and classic aptamer.
Thermodynamic equivalency of the two sites
While the two sites are symmetrical and should have the same binding property, we still tested it experimentally to understand their thermodynamic equivalency. Using the same method, we sealed site 2 while still retained site 1 by designing Apt1b (Figure 1C). From ITC (Figure 2C), an even smaller heat release and entropy loss was measured. But interestingly, a similar Kd (14.1 μM, Table 1) was still retained. The Apt1b also has the expected one-site binding stoichiometry of 0.8 ± 0.2. This study confirms that the two binding sites have identical binding affinity, which is not surprising from their structure biology perspective.
What's interesting is the enthalpy-entropy compensation. For a DNA hybridization reaction, one expects heat release and entropy loss. Although aptamer binding is not a classic DNA hybridization reaction, it usually involves base pair formation or similar interactions. It appears adenosine's binding to Apt1b has fewer base pair formation compared to its binding to Apt1a. This could be explained by the fact that Apt1a has only six base pairs, while the rest of the sequences do not form a stable structure in the absence of adenosine. After binding adenosine, a large structural change to a more rigid one is observed. For Apt1b, however, two stable base paired regions exist and a smaller structural change is needed for binding. For the wild-type aptamer, the number of base pairs formed is the largest in these three sequences, and it indeed has the highest heat release. We illustrated this structural change upon adenosine binding in Supplementary Figure S2. Taken together, while these two sites are equivalent in Kd, they differ in entropy and enthalpy of the binding reaction. Similar to Apt1a, Apt1b is also specific for adenosine since cytosine and guanosine yielded no heat (Figure 2C, red and blue traces).
Relationship of the two binding sites
In the above experiments, we removed each binding pocket by introducing new base pairs and replacing non-canonical base pairs with Watson–Crick base pairs. This way, the structural stabilization role of the removed binding site was still retained. Next, we tried to remove binding sites by cutting base pairs, thus disrupting the stable structures. For example, we deleted the first three base pairs from the wild-type Apt2a aptamer. The resulting Apt2b mutant (Figure 1D) completely lost binding (Figure 2D), which was attributable to that the entire DNA molecule was unfolded with only two Watson-Crick pairs. These two base pairs cannot stabilize binding site 1, and site 2 cannot form as a result. This experiment shows that disrupting the first site led to a full loss of binding for the second site, even though the second site was formally intact.
Taken together, for the wild-type aptamer, either site can bind adenosine first and binding either one can stabilize the other site. There is unlikely to be a preference in the order of binding based on their identical Kd. Since ITC measures only binding thermodynamics, a kinetic preference cannot be excluded. In Apt1a and Apt1b, the stabilization role of the removed site was replaced by DNA base pairing and this is perfectly fine for the remaining site to bind normally.
ITC data quality
The quality of ITC data is often described by the Wiseman coefficient c = n⋅[aptamer]/Kd, where n is the number of binding site. In the original paper in 1989 (38), it was concluded that c needs to be >1 for reliable ITC experiments. In our above titrations, the c-value ranged from 0.6 to 0.8 for the engineered one-site aptamers. However, the Wiseman paper was amended in 2003 by Turnbull and Daranas articulating that the c-value can be smaller than one as long as a high analyte concentration (adenosine in our case) is used to ensure complete binding is achieved (37). In our case, for the one-site binding, we used an adenosine-to-aptamer ratio of 10:1 and all titrations went completion and thus our data are of sufficient quality.
Based on its definition, one way to increase the c-value is to increase the aptamer concentration. When we increased the aptamer concentration from 10 to 20 μM, the c-value for the wild-type aptamer increased from 1.2 to 2.1 (not doubled) due to the increased Kd value (the number of binding sites remained 2, Supplementary Figure S3 and Table S1). This increase Kd might be attributed to inter-aptamer interaction at higher aptamer concentrations. Therefore, 10 μM aptamer appears to be an optimal concentration for this experiment.
Effect of temperature
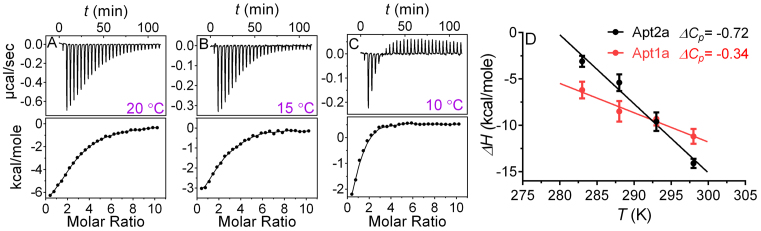
To gain further insights, we also tried lowering temperature to 20, 15 and 10°C. At 10°C, the binding affinity increased significantly (Kd = 4.6 ± 0.4 μM, Figure 3A–C and Supplementary Table S1), leading to a much better c-value of 4.1 ± 0.8. It is interesting to note that the final spikes run upward indicating heat absorption after saturated aptamer binding. This is attributable to the dilution of adenosine that released some of the base stacking.
Figure 3.
ITC trace and integrated heat of 10 μM of wild-type aptamer (Apt2a) binding to adenosine at different temperatures: (A) 20°C, (B) 15°C and (C) 10°C. Related thermodynamic parameters are shown in Supplementary Table S1. (D) The enthalpy changes (ΔH) of 10 μM of Apt2a and Apt1a binding adenosine (0.5 mM) as a function of temperature. ΔCp (unit: kcal mol−1 K−1) was obtained from the linearly fitted slope (ΔH/ΔT).
With the data at different temperatures, we calculated the heat capacity change (ΔCp) of the wild-type aptamer Apt2a and the one-site aptamer Apt1a (Figure 3D). The ΔCp of these two aptamers were both negative indicating induced fitting, which is typical of aptamer binding (33). The one-site Apt1a has a smaller ΔCp (−0.34 kcal mol−1 K−1) than that of the two-site Apt2a (−0.72 kcal mol−1 K−1). This result is also consistent with the entropy changes from ITC (Table 1 and Supplementary Figure S2). This can be easily rationalized by the smaller conformational change of Apt1a described in Supplementary Figure S2. A few other literature reported aptamer also showed a similar trend (43,44). For example, a long-stem cocaine aptamer has a smaller structural change (ΔCp= −0.5 kcal mol−1 K−1), while a shorter one has a larger change (ΔCp= −0.9 kcal mol−1 K−1) (45).
Analytical implications
This aptamer has been extensively used as a model for biosensor development (15–24). The specific recognition of adenosine, for example, was used to screen inhibitors for adenosine deaminase (46). Measuring ATP inside cells (47–49), or in serum and was also reported (21,50). We believe our discovery here has interesting analytical applications. By eliminating one binding site, it is possible to use even shorter aptamers. For example, while the wild-type aptamer cannot be truncated, we can truncate three base pairs from the Apt1a mutant to make Apt1c (Figure 1E), which still retained its binding as determined by ITC (Figure 2E, Table 1). It has a similar Kd value and binds just one adenosine. Compared to the 27-nt wild-type, this mutant has only 21 nt.
This adenosine aptamer has been a subject of intense mutation and splitting studies to design new and better biosensors. For example, some early work involved splitting the aptamer near the T14 position. The two aptamer halves can assemble into the full binding complex in the presence of adenosine (51–53). The length of the stem was systematically varied using an aptamer beacon design (18). All these modifications still have kept both binding sites. In another paper, only half of the split aptamer was reported to have some affinity toward ATP (54). However, our ITC measurement of other half aptamers showed quite weak binding, although it can be rescued by molecular imprinting (55). Our work here is the first attempt to test the feasibility of just having one-site binding for this aptamer. Due to the very short requirement of DNA sequence, we believe this is an excellent system for understanding aptamer binding and biosensor design (56). Reducing the number of binding sites can also make more sensitive biosensors (vide infra).
Implication for SELEX
Systematic evolution of ligands by exponential enrichment (SELEX) is a very powerful technique to isolate aptamers. It is also called in vitro selection by some researchers. This adenosine aptamer motif has been reported a few times in different labs (13,32). We carefully examined all the published sequences. In both cases, the same conserved sequence was obtained, binding two adenosine molecules. Out of over 100 sequences, none of them matched with the one-site mutants we tested here, even though the one-site mutants bind just as well with a similar Kd. From statistic calculation, the chance of evolving a single-site aptamer is much higher since they require fewer conserved nucleotides. In the original selection by Szostak, the target molecule was immobilized. As such, it is even more difficult to bind two targets (e.g. the immobilization density has to be extremely high so that two molecules can come within sub-nm distance to bind together).
This is the only example of small molecule binding aptamers obtained via in vitro selection that can bind two target molecules. The RNA aptamer for AMP was also reported in multiple in vitro selection experiments, which binds only one target molecule (34). Structural biology studies have indicated that the RNA and DNA aptamers use the same mechanism for AMP binding, yet they have different number of binding sites. Therefore, an interesting and intriguing question is then why no one-site aptamers were found so far.
While it is difficult to know for sure the reason for selecting such a two-site aptamer, we have the following speculations. (i) One possibility is that sequences such as that shown in Figure 1B with a long and very stable stem might be less competitive in polymerase chain reaction amplification. However, this problem can be easily solved by having fewer base pairs as shown in Figure 1E or introducing some mismatches. (ii) This study suggests that the minimal binding sequence is quite small. Those SELEX experiments used a much longer random region. Therefore, the library needs to find an effective way to hide the redundant sequences. Introducing an additional site might use a few more nucleotides. (iii) For the immobilized target, having two binding sites will enhance the avidity of binding via polyvalent effect. However, the other selection by the Li lab was performed using the structure-switching method. We showed here that one site binding works just fine for this purpose. Therefore, there might be some more fundamental reason that we are not yet understood.
More binding sites can be introduced
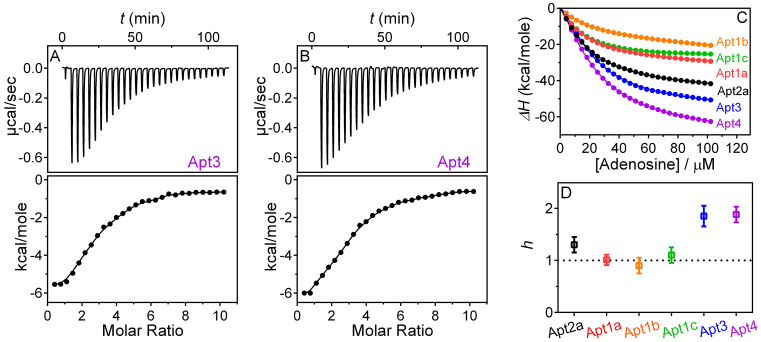
After demonstrating reducing the number of site from two to one, and understanding the requirement for binding, we next tested whether we can increase the binding site. For this purpose, two additional aptamers were designed (Apt3 and Apt4, see Figure 1F and G for sequence) and they contained three and four adenosine binding sites, respectively. Indeed as shown in ITC (Figure 4A and B), the binding stoichiometry is as designed in both cases. The Kd value still remained similar to be around 10 μM.
Figure 4.
ITC traces and integrated heat of the engineered aptamers with (A) 3 and (B) 4 adenosine binding sites. The thermodynamic constants are listed in Table 1. (C) The total enthalpy change (ΔH) of different aptamers as a function of added adenosine. (D) The Hill coefficients, h, fitted from (C) using Equation (1); h > 1 represents positively cooperative and h = 1 represents non-cooperative behavior between binding sites.
Binding sites cooperativity
Given the relationship between the two sites, an intriguing observation is that the two sites did not show an obvious cooperativity of binding. Our ITC trace for the wild-type aptamer was fitted well with two identical binding sites. Most previous binding studies on this aptamer also showed a similar binding curve instead of a sigmoidal curve expected for cooperative binding.
By fitting the above ITC data directly, we obtained thermodynamic parameters of binding (57). Next, we want to quantitatively compare binding cooperativity of these aptamers. We constructed the binding curves by calculating the cumulative heat, and used Equation (1) in the ‘Materials and Methods’ section to fit the data (Figure 4C). The Hill-coefficients are plotted in Figure 4D. It is interesting to note that all the three one-site mutants have a Hill-coefficient of around 1, while the wild-type is around 1.2. Therefore, these two site have a very moderate cooperativity. The Apt3 and Apt4 aptamers have a Hill-coefficient of nearly 2, and this is likely due to the proximity of the two sets of binding sites.
Only a few examples are known for aptamers binding multiple small molecule targets. Another one is the glycine riboswitch binding two ligands simultaneously (58). However, this riboswitch is much more complex in structure, making it difficult for a systematic biding study via mutation. This riboswitch has a Hill-coefficient of 1.64 and thus its binding of glycine is more cooperative than the adenosine binding here.
Improved biosensor sensitivity
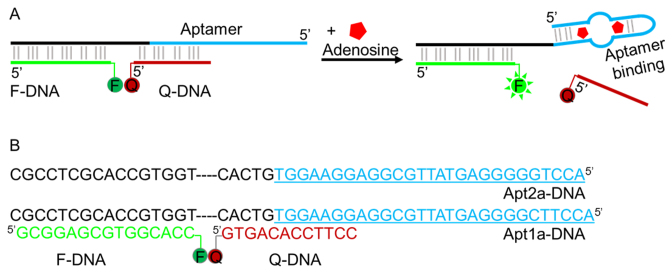
For an aptamer cooperatively binding multiple molecules, its sensitivity is lower at low analyte concentration. Even though the wild-type adenosine aptamer is only weakly cooperative, the one-site aptamers might still have better sensitivity. To test this idea, the structure-switching aptamer method is used here as shown in Figure 5A (17,59). The aptamer sequence was extended to hybridize with a fluorophore-labeled fragment (F-DNA). In addition, part of the extension and part of the aptamer together (Apt-DNA) hybridized to a quencher-labeled fragment (Q-DNA). In the presence of adenosine, the aptamer folds to release the quencher strand resulting in fluorescence enhancement. We are interested in comparing Apt2a and Apt1a, since they have very similar binding affinity for adenosine but a different number of binding sites. The exact sequences of the sensors are shown in Figure 5B.
Figure 5.
(A) A scheme of structure-switching signal aptamer. Binding of adenosine releases the quencher-labeled fragment and produces enhanced fluorescence. The scheme describes the wild-type aptamer binding two adenosine molecules. For the Apt1a mutant, only one adenosine is bound. (B) The DNA sequences of the two sensors tested.
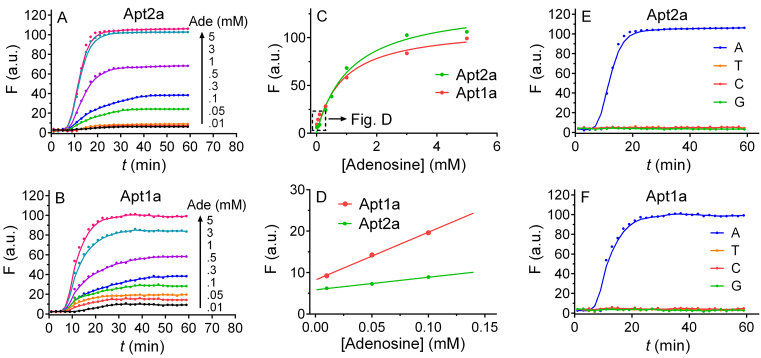
When adenosine was titrated into the sensor containing the wild-type aptamer (Figure 6A), a time-dependent fluorescence was observed and the amount of enhancement was related to the concentration of adenosine, allowing quantitative analysis. After 30 min, each sample reached a plateau and we plotted the maximal fluorescence as a function of adenosine concentration (Figure 6C). From this, we determined the limit of detection to be 28.9 μM adenosine based on the 3σ/slope calculation, where σ is the standard deviation of the background variation measured from multiple sensor samples without adding adenosine (Figure 6D).
Figure 6.
Biosensor performance. The sensor fluorescence kinetics with (A) the wild-type Apt2a aptamer and (B) the one site mutant Apt1a as a function of adenosine concentration (0.01–5 mM). The fluorescence intensity of the two sensors after 30 min as a function of (C) high (0.3–5 mM) and (D) low (10–100 μM) adenosine concentration. The fluorescence kinetics of (E) the Apt2a aptamer and (F) the Apt1a aptamer contained sensors after adding other nucleosides: guanosine (G), cytidine (C) and thymidine (T) (5 mM) with the background fluorescence subtracted.
When the Apt1a mutant was used, a similar response was observed (Figure 6B). It is interesting to note though, the initial fluorescence increase at low adenosine concentrations (e.g. <0.2 mM) was higher for the mutant, although its final fluorescence was lower (Figure 6D). The slope of the curve, which measures the sensitivity of the sensor, is 3.8-fold higher for the Apt1a mutant than the wild-type Apt2a. This might be another evidence of cooperative binding required for the wild-type. In the mutant, the limit of detection was 9.1 μM adenosine since the initial increase was higher. In both cases, binding was specific and none of other nucleosides gave much signal (Figure 6E and F). With this simple sensor design, we have demonstrated an important analytical application of this model aptamer.
CONCLUSION
In summary, we systematically measured the binding of a classical and important DNA aptamer. This aptamer has been studied for over 20 years and it has significantly fueled the development of aptamer-based biosensors. In all the previous studies, the wild-type aptamer with two target binding sites was used. While in vitro selection has always resulted in two adjacent binding pockets, this work shows that single pocket aptamers can also work with a similar binding affinity and specificity. Since the binding of the one-site aptamer is non-cooperative, a biosensor made with it has a better sensitivity at low analyte concentrations. This study has led to new insights into this important aptamer and has resulted in better biosensor.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) [386326]. Funding for open access charge: NSERC [386326].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 2. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 3. Winkler W., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. [DOI] [PubMed] [Google Scholar]
- 4. Winkler W.C., Breaker R.R.. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005; 59:487–517. [DOI] [PubMed] [Google Scholar]
- 5. Liu J., Cao Z., Lu Y.. Functional nucleic acid sensors. Chem. Rev. 2009; 109:1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li D., Song S.P., Fan C.H.. Target-responsive structural switching for nucleic acid-based sensors. Acc. Chem. Res. 2010; 43:631–641. [DOI] [PubMed] [Google Scholar]
- 7. Tan W.H., Donovan M.J., Jiang J.H.. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013; 113:2842–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao W., Brook M.A., Li Y.. Design of gold nanoparticle-based colorimetric biosensing assays. Chembiochem. 2008; 9:2363–2371. [DOI] [PubMed] [Google Scholar]
- 9. Zhang H., Li F., Dever B., Li X.-F., Le X.C.. DNA-mediated homogeneous binding assays for nucleic acids and proteins. Chem. Rev. 2013; 113:2812–2841. [DOI] [PubMed] [Google Scholar]
- 10. Wilner O.I., Willner I.. Functionalized DNA nanostructures. Chem. Rev. 2012; 112:2528–2556. [DOI] [PubMed] [Google Scholar]
- 11. Cho E.J., Lee J.-W., Ellington A.D.. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009; 2:241–264. [DOI] [PubMed] [Google Scholar]
- 12. Xiang Y., Lu Y.. DNA as sensors and imaging agents for metal ions. Inorg. Chem. 2014; 53:1925–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huizenga D.E., Szostak J.W.. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995; 34:656–665. [DOI] [PubMed] [Google Scholar]
- 14. Deng Q., German I., Buchanan D., Kennedy R.T.. Retention and separation of adenosine and analogues by affinity chromatography with an aptamer stationary phase. Anal. Chem. 2001; 73:5415–5421. [DOI] [PubMed] [Google Scholar]
- 15. Zuo X., Xiao Y., Plaxco K.W.. High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J. Am. Chem. Soc. 2009; 131:6944–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zuo X., Song S., Zhang J., Pan D., Wang L., Fan C.. A target-responsive electrochemical aptamer switch (TREAS) for reagentless detection of nanomolar ATP. J. Am. Chem. Soc. 2007; 129:1042–1043. [DOI] [PubMed] [Google Scholar]
- 17. Nutiu R., Li Y.. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003; 125:4771–4778. [DOI] [PubMed] [Google Scholar]
- 18. Urata H., Nomura K., Wada S.-I., Akagi M.. Fluorescent-labeled single-strand ATP aptamer DNA: Chemo- and enantio-selectivity in sensing adenosine. Biochem. Biophys. Res. Comm. 2007; 360:459–463. [DOI] [PubMed] [Google Scholar]
- 19. Liu J., Lu Y.. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem., Int. Ed. 2006; 45:90–94. [DOI] [PubMed] [Google Scholar]
- 20. Xu W., Lu Y.. A smart magnetic resonance imaging contrast agent responsive to adenosine based on a DNA aptamer-conjugated gadolinium complex. Chem. Commun. 2011; 47:4998–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L.-L., Ge P., Selvin P.R., Lu Y.. Direct detection of adenosine in undiluted serum using a luminescent aptamer sensor attached to a terbium complex. Anal. Chem. 2012; 84:7852–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stojanovic M.N., Kolpashchikov D.M.. Modular aptameric sensors. J. Am. Chem. Soc. 2004; 126:9266–9270. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Tang L.H., Li Z.H., Lin Y.H., Li J.H.. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells. Nat. Protoc. 2014; 9:1944–1955. [DOI] [PubMed] [Google Scholar]
- 24. Meng H.-M., Zhang X., Lv Y., Zhao Z., Wang N.-N., Fu T., Fan H., Liang H., Qiu L., Zhu G. et al. DNA dendrimer: An efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014; 8:6171–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang S.B., Hu R., Hu P., Wu Z.S., Shen G.L., Yu R.Q.. Blank peak current-suppressed electrochemical aptameric sensing platform for highly sensitive signal-on detection of small molecule. Nucleic Acids Res. 2010; 38:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boese B.J., Breaker R.R.. In vitro selection and characterization of cellulose-binding DNA aptamers. Nucleic Acids Res. 2007; 35:6378–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin C.H., Patel D.J.. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997; 4:817–832. [DOI] [PubMed] [Google Scholar]
- 28. Nonin-Lecomte S., Lin C.H., Patel D.J.. Additional hydrogen bonds and base-pair kinetics in the symmetrical amp-DNA aptamer complex. Biophys. J. 2001; 81:3422–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia T., Yuan J., Fang X.. Conformational dynamics of an ATP-binding DNA aptamer: a single-molecule study. J. Phys. Chem. B. 2013; 117:14994–15003. [DOI] [PubMed] [Google Scholar]
- 30. Armstrong R.E., Strouse G.F.. Rationally manipulating aptamer binding affinities in a stem-loop molecular beacon. Bioconjug. Chem. 2014; 25:1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakamura I., Shi A.-C., Nutiu R., Yu J.M.Y., Li Y.. Kinetics of signaling-DNA-aptamer ATP binding. Phys. Rev. E. 2009; 79:031906. [DOI] [PubMed] [Google Scholar]
- 32. Nutiu R., Li Y.. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. 2005; 44:1061–1065. [DOI] [PubMed] [Google Scholar]
- 33. Hermann T., Patel D.J.. Adaptive recognition by nucleic acid aptamers. Science. 2000; 287:820–825. [DOI] [PubMed] [Google Scholar]
- 34. Sassanfar M., Szostak J.W.. An RNA motif that binds ATP. Nature. 1993; 364:550–553. [DOI] [PubMed] [Google Scholar]
- 35. Ricci F., Vallee-Belisle A., Simon A.J., Porchetta A., Plaxco K.W.. Using nature's “tricks” to rationally tune the binding properties of biomolecular receptors. Acc. Chem. Res. 2016; 49:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simon A.J., Vallee-Belisle A., Ricci F., Watkins H.M., Plaxco K.W.. Using the population-shift mechanism to rationally introduce “Hill-type” cooperativity into a normally non-cooperative receptor. Angew. Chem. Int. Ed. 2014; 53:9471–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turnbull W.B., Daranas A.H.. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry?. J. Am. Chem. Soc. 2003; 125:14859–14866. [DOI] [PubMed] [Google Scholar]
- 38. Wiseman T., Williston S., Brandts J.F., Lin L.-N.. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989; 179:131–137. [DOI] [PubMed] [Google Scholar]
- 39. Neves M.A.D., Reinstein O., Saad M., Johnson P.E.. Defining the secondary structural requirements of a cocaine-binding aptamer by a thermodynamic and mutation study. Biophys. Chem. 2010; 153:9–16. [DOI] [PubMed] [Google Scholar]
- 40. Bernard Da Costa J., Dieckmann T.. Entropy and Mg2+ control ligand affinity and specificity in the malachite green binding RNA aptamer. Mol. Biosyst. 2011; 7:2156–2163. [DOI] [PubMed] [Google Scholar]
- 41. Jhaveri S.D., Kirby R., Conrad R., Maglott E.J., Bowser M., Kennedy R.T., Glick G., Ellington A.D.. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc. 2000; 122:2469–2473. [Google Scholar]
- 42. Entzian C., Schubert T.. Studying small molecule–aptamer interactions using microscale thermophoresis (MST). Methods. 2016; 97:27–34. [DOI] [PubMed] [Google Scholar]
- 43. Sokoloski J.E., Dombrowski S.E., Bevilacqua P.C.. Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer. Biochemistry. 2012; 51:565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reinstein O., Yoo M., Han C., Palmo T., Beckham S.A., Wilce M.C.J., Johnson P.E.. Quinine binding by the cocaine-binding aptamer. Thermodynamic and hydrodynamic analysis of high-affinity binding of an off-target ligand. Biochemistry. 2013; 52:8652–8662. [DOI] [PubMed] [Google Scholar]
- 45. Neves M.A.D., Reinstein O., Johnson P.E.. Defining a stem length-dependent binding mechanism for the cocaine-binding aptamer. A combined NMR and calorimetry study. Biochemistry. 2010; 49:8478–8487. [DOI] [PubMed] [Google Scholar]
- 46. Elowe N.H., Nutiu R., Allali-Hassani A., Cechetto J.D., Hughes D.W., Li Y., Brown E.D.. Small-molecule screening made simple for a difficult target with a signaling nucleic acid aptamer that reports on deaminase activity. Angew. Chem., Int. Ed. 2006; 45:5648–5652. [DOI] [PubMed] [Google Scholar]
- 47. Zheng D., Seferos D.S., Giljohann D.A., Patel P.C., Mirkin C.A.. Aptamer nano-flares for molecular detection in living cells. Nano Lett. 2009; 9:3258–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y., Li Z.H., Hu D.H., Lin C.T., Li J.H., Lin Y.H.. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J. Am. Chem. Soc. 2010; 132:9274–9276. [DOI] [PubMed] [Google Scholar]
- 49. Tan X., Chen T., Xiong X., Mao Y., Zhu G., Yasun E., Li C., Zhu Z., Tan W.. Semiquantification of ATP in live cells using nonspecific desorption of DNA from graphene oxide as the internal reference. Anal. Chem. 2012; 84:8622–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang P.J.J., Liu J.W.. Flow cytometry-assisted detection of adenosine in serum with an immobilized aptamer sensor. Anal. Chem. 2010; 82:4020–4026. [DOI] [PubMed] [Google Scholar]
- 51. Stojanovic M.N., de Prada P., Landry D.W.. Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc. 2000; 122:11547–11548. [DOI] [PubMed] [Google Scholar]
- 52. Li F., Zhang J., Cao X.N., Wang L.H., Li D., Song S.P., Ye B.C., Fan C.H.. Adenosine detection by using gold nanoparticles and designed aptamer sequences. Analyst. 2009; 134:1355–1360. [DOI] [PubMed] [Google Scholar]
- 53. Dave N., Liu J.. Biomimetic sensing based on chemically induced assembly of a signaling DNA aptamer on a fluid bilayer membrane. Chem. Commun. 2012; 48:3718–3720. [DOI] [PubMed] [Google Scholar]
- 54. Li S., Chen D., Zhou Q., Wang W., Gao L., Jiang J., Liang H., Liu Y., Liang G., Cui H.. A general chemiluminescence strategy for measuring aptamer–target binding and target concentration. Anal. Chem. 2014; 86:5559–5566. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Z., Liu J.. Molecularly imprinted polymers with DNA aptamer fragments as macromonomers. ACS Appl. Mater. Inter. 2016; 8:6371–6378. [DOI] [PubMed] [Google Scholar]
- 56. Vallee-Belisle A., Ricci F., Plaxco K.W.. Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range. J. Am. Chem. Soc. 2012; 134:2876–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Velazquez-Campoy A., Goñi G., Peregrina J.R., Medina M.. Exact analysis of heterotropic interactions in proteins: characterization of cooperative ligand binding by isothermal titration calorimetry. Biophys. J. 2006; 91:1887–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 59. Nutiu R., Li Y.. Structure-switching signaling aptamers: Transducing molecular recognition into fluorescence signaling. Chem. Eur. J. 2004; 10:1868–1876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.