Abstract
In most eukaryotes, RNA silencing is an adaptive immune system regulating key biological processes including antiviral defense. To evade this response, viruses of plants, worms and insects have evolved viral suppressors of RNA silencing proteins (VSRs). Various VSRs, such as P1 from Sweet potato mild mottle virus (SPMMV), inhibit the activity of RNA-induced silencing complexes (RISCs) including an ARGONAUTE (AGO) protein loaded with a small RNA. However, the specific mechanisms explaining this class of inhibition are unknown. Here, we show that SPMMV P1 interacts with AGO1 and AGO2 from Arabidopsis thaliana, but solely interferes with AGO1 function. Moreover, a mutational analysis of a newly identified zinc finger domain in P1 revealed that this domain could represent an effector domain as it is required for P1 suppressor activity but not for AGO1 binding. Finally, a comparative analysis of the target RNA binding capacity of AGO1 in the presence of wild-type or suppressor-defective P1 forms revealed that P1 blocks target RNA binding to AGO1. Our results describe the negative regulation of RISC, the small RNA containing molecular machine.
INTRODUCTION
RNA silencing is a sequence specific mechanism that regulates gene expression in almost all eukaryotes, and controls key biological processes such as development, heterochromatin formation, stress and antiviral responses (1–4). It is triggered by double-stranded (ds) RNA (dsRNA) of different length and origin, which is processed into 21–26 nt small RNA (sRNA) duplexes by an RNase III-type exonuclease of the DICER family (5). The ARGONAUTE (AGO)-containing RISC (RNA-induced silencing complex) protein complex incorporates one of the strands of the sRNA duplex which serves as guide RNA for RISC to recognize and target single-stranded RNA (ssRNA) with high sequence complementarity (6–8).
Antiviral RNA silencing has been described in worms, insects, plants and humans as a robust cellular mechanism in charge of clearing infecting viruses (9–13). However, diverse proteins interfering with the host antiviral RNA silencing machinery have been identified in insect, plant and fungus-infecting viruses (14–16). These proteins, so-called viral suppressors of RNA silencing (VSRs), can block RNA silencing pathways at multiple levels (16). For instance, dsRNA-binding VSRs such as Turnip crinkle virus p38, Barley stripe mosaic virus γB and Drosophila C virus 1A inhibit sRNA biogenesis by targeting viral dsRNA replication intermediates and viral ssRNAs with ds regions (11,15). Other VSRs bind directly to sRNAs and inhibit RISC loading through a sRNA sequestration mechanism (15–17). In plants, RISC loading is also hampered by VSRs through the transcriptional or post-translational regulation of AGO1 protein levels. For example, several VSRs upregulate miR168 expression upon viral infection, causing AGO1 mRNA translational repression and subsequent decrease of AGO1 protein levels (18,19). Other examples of VSRs destabilizing AGO1 post-translationally include Polerovirus P0, Tomato ringspot virus (ToRSV) coat protein (CP) and Potexvirus P25 (20–24). In the case of P0, this protein contains a minimal F-box motif (similar to plant ubiquitin E3 ligases) which directs AGO1 ubiquitination unless AGO1 is loaded with sRNAs (22). Indeed P0 induces the autophagy pathway (25), while P25 and ToRSV CP direct the proteasomal degradation of plant AGO1 (21,23). Finally, VSRs can also inhibit pre-assembled RISCs (including an AGO protein and a sRNA) as shown for Cricket paralysis virus (CrPV) CrPV-1A (26) and Nora virus VP1 (27) both targeting Drosophila melanogaster AGO2, Lettuce necrotic yellows virus P phosphoprotein targeting AGO1 and AGO2 (28), and Sweet potato mild mottle virus (SPMMV) P1 inhibiting small interfering RNA (siRNA)- and microRNA (miRNA)-driven pre-assembled RISC activity by interacting with AGO1 through its WG/GW domain rich in glycine and tryptophan residues (29). However, the specific molecular mechanisms by which VSRs inhibit pre-assembled RISCs are still largely unknown.
AGO proteins are the core effector components of RISC complexes (8). In plants, ten AGOs have been identified in the model plant Arabidopsis thaliana (Arabidopsis) (30,31). Several of them are involved in antiviral defense in a cooperative or redundant manner, with AGO1 and AGO2 being the main antiviral AGOs in Arabidopsis and other plant species (32). In the present work, we used an agroinfiltration-based in vivo system in Nicotiana benthamiana to study the mechanism by which SPMMV P1 inhibit pre-assembled RISCs. We show that SPMMV P1 interacts with AGO1 and AGO2, but solely interferes with AGO1 function thus indicating that AGO binding is insufficient for P1-mediated inhibition. We also identified a putative zinc finger domain in P1 that was essential for P1 suppressor activity but not for AGO1 binding, as shown through a functional analysis of several P1 forms with mutations affecting residues of the zinc finger domain. Finally, a comparative analysis of the target RNA binding capacity of AGO1 in the presence of wild-type or suppressor-defective P1 forms showed that SPMMV P1 blocks target RNA binding to AGO1. These results describe a new mechanism of action for a VSR based on the inhibition of AGO binding to target RNA, which might help to better understand SPMMV pathogenicity.
MATERIALS AND METHODS
DNA constructs
The P11–395 DNA fragment was amplified by PCR using appropriate primers, then cloned into the pJET1.2/blunt vector. All mutants were obtained using the Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific) and the P11–395 in pJET1.2/blunt DNA as template. Primers are listed in Supplementary Figure S1.
P11-395 and mutant ORFs were cloned into the binary expression vector pBIN-Flag (33) to have a Flag-tag at the N-terminal of the fusion protein. 35S:HA-AGO1, 35S:HA-AGO1-DAH, 35S:HA-AGO2, 35S:HA-AGO2-DAD, 35S:TAS1c, 35S:TAS1c-A388T, 35S:amiR173-5΄A and 35S:amiR173 constructs were described before (34–37).
Plant materials
Nicotiana benthamiana plants were grown at 23°C in a plant growth chamber under a photoperiod of 16 h light/8 h dark. Leaves from plants of ∼21 days old were infiltrated.
Agroinfiltration
To test P1 mutants for silencing suppressor activity, transient expression assay in N. benthamiana leaves using Agrobacterium tumefaciens C58 strain was done as described (29). To test the inhibitory effect of P1 on AGO proteins, all cultures including A. tumefaciens strains bearing plasmid constructs were grown until they reached an OD600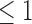 , resuspended in half volume of inducing solution (10 mM MES pH=5.8, 10 mM MgCl2, 0.15 mM acetosyringon), then diluted for infiltration at an OD600 = 0.3 each.
, resuspended in half volume of inducing solution (10 mM MES pH=5.8, 10 mM MgCl2, 0.15 mM acetosyringon), then diluted for infiltration at an OD600 = 0.3 each.
RNA analysis
RNA was isolated with Trizol reagent (Sigma-Aldrich) according to manufacturer's instructions. Four μg of total RNA was separated by 2.2 M formaldehyde and 1.2% agarose gels, and blotted to Amersham Hybond-N (GE Healthcare) membrane. Membranes were hybridized with P32-labeled GFP or TAS1c DNA probes.
Protein analysis
Protein input and IP extracts were separated on 8–10% PAGE gels before protein transfer to Immobilon-P membrane (Millipore). Anti-GFP (Invitrogen), anti-HA (Roche) and M2 anti-Flag (Sigma-Aldrich) antibodies were used to detect GFP, HA- and Flag-tagged proteins, respectively.
Native protein immunoprecipitation
Native protein immunoprecipitations were carried out as described (29) with minor modifications. For inputs 0.2–1% of the input and 10% of the eluates were loaded to the SDS PAGE gels.
RNA immunoprecipitation
Five hundred mg of N. benthamiana infiltrated leaves were used. Leaves were collected 44 h after infiltration, and were fixed in 1% formaldehyde for 30 min under vacuum. Fixation was stopped with 125 mM of glycine under vacuum for 25 min. Leaves were washed three times with ice-cold sterile water, dried, and submerged into frozen liquid nitrogen before proceeding to immunoprecipitation or stored at –80°C as described (38).
Frozen leaves were homogenized in 0.5 ml/g lysis buffer (15 mM Tris–HCl, pH 8, 150 mM NaCl, 1% Triton-X (w/v), 1 mM EDTA). Cell debris were pelleted by two centrifugation steps at 13 000 rpm for 5 min each at 4°C. Clarified lysates were incubated overnight at 4°C with 20 μl of anti-Flag M2 affinity gel (Sigma-Aldrich). Beads were washed three times for 5 min with 20 mM Tris–HCl, pH 8, 500 mM NaCl, 1% (w/v) Triton-X and 2 mM EDTA and twice with 20 mM Tris–HCl, pH 8 and 2 mM EDTA. Two elution steps of 10 min at 50°C were done with 120 μl of 50 mM Tris–HCl, pH 7, 2.5 mM EDTA, 10 mM DTT and 1% SDS buffer. Input fractions were collected from the cleared lysates after the second centrifugation. One hundred microliters were saved from each sample and stored at –80°C. They were supplemented to 10 mM DTT, 0.1% SDS and 2.5 mM EDTA. Both input and IP fractions were processed in parallel. After crosslinking reversal, samples were sequentially treated with DNaseI (Thermo Scientific) for 15 min at 37°C, digested with proteinase K (Thermo Scientific), extracted with saturated phenol: chloroform and chloroform, then precipitated with ethanol. Extracted RNA fractions were reverse-transcribed using the TaqMan MicroRNA Reverse Transcription Kit (AB) according to manufacturer's instructions, and 1/15th of the cDNA was analyzed by semi-quantitative PCR.
Quantitative reverse transcriptase PCR (qRT-PCR)
The N. benthamiana cDNA sequence (SubjectID Niben101Scf05245g01007.1) encoding for a putative AGO2 protein and the cDNA (SubjectID Niben101Scf04639g06007.1) encoding for the putative elongation factor 1 (EF-1) were used to design primers for qRT-PCR (Supplementary Figure S1). The particular SubjectIDs refer to predicted cDNAs of the N. benthamiana Genome v.1.0.1 at the Boyce Thompson Institute (bti.cornell.edu). RNA was isolated from three independent biological replicates for each construct with Trizol reagent (Sigma-Aldrich), then DNAseI treated (Thermo Scientific) and quantified with the NanoDrop™ 2000 Spectrophotometer (Thermo Scientific). 0.2 μg total RNA (OD260/280 ratio ≥ 1.8) was reverse-transcribed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) using AGO2-1rev and EF1rev primers at 10 μM final concentration in a 15 μl reaction volume according to the manufacturer's instructions. qRT-PCR was performed using the CFX 96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Samples were run in triplicates to ensure the integrity of amplification. Each reaction mixture consisted of 0.5 μl cDNA, 5 μl SYBR Select Master Mix (Applied Biosystems) in a 10 μl reaction volume. Both AGO2 and EF1 forward and reverse primers were used at 10 μM final concentration (Supplementary Figure S1). The thermal profile of the reaction was an initial denaturation at 95°C for 3 min, followed by 40 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Fluorescence acquisition was performed after each cycle. AGO2 and EF1 primers were expected to amplify a single product of 107 and 135 nt in size. AGO2 mRNA expression level was calculated with the 2−ΔΔCt method, and plotted as fold change of relative mean (±SD) expression normalized to the expression of N. benthamiana EF1. Groups were compared using one-way ANOVA followed by Bonferroni post hoc test (*P < 0.05).
5΄ RACE
RNA was isolated with Trizol reagent (Sigma-Aldrich). 5΄ RNA ligase-mediated RACE was performed with the GeneRacer™ Kit (Thermo Fisher) according to the manufacturer's instructions and TAS1 specific primers (described in (34)). 5΄RACE products were gel purified, cloned, screened for inserts, and sequenced (20 clones per sample). The number of independent 3΄ end product was at least two times more in samples where miR173 driven cleavage product was detected than in control samples.
Densitometry analysis
Densitometry analyses were done using autoradiography from Northern blots corresponding to three independent experiments. Specifically, the bands corresponding to full-length TAS1c/TAS1c-A388T/GFP mRNA were quantified with the GelAnalyzer2010a program, and for each sample, the mean and corresponding standard deviation were plotted in a bar graph. Loss of the full length TAS1 mRNA was measured, because the lower band contains mir173 dependent target cleavage products and results of non-specific TAS1 mRNA degradation.
RESULTS
SPMMV P1 inhibits AGO1 but not AGO2 cleavage activity in vivo
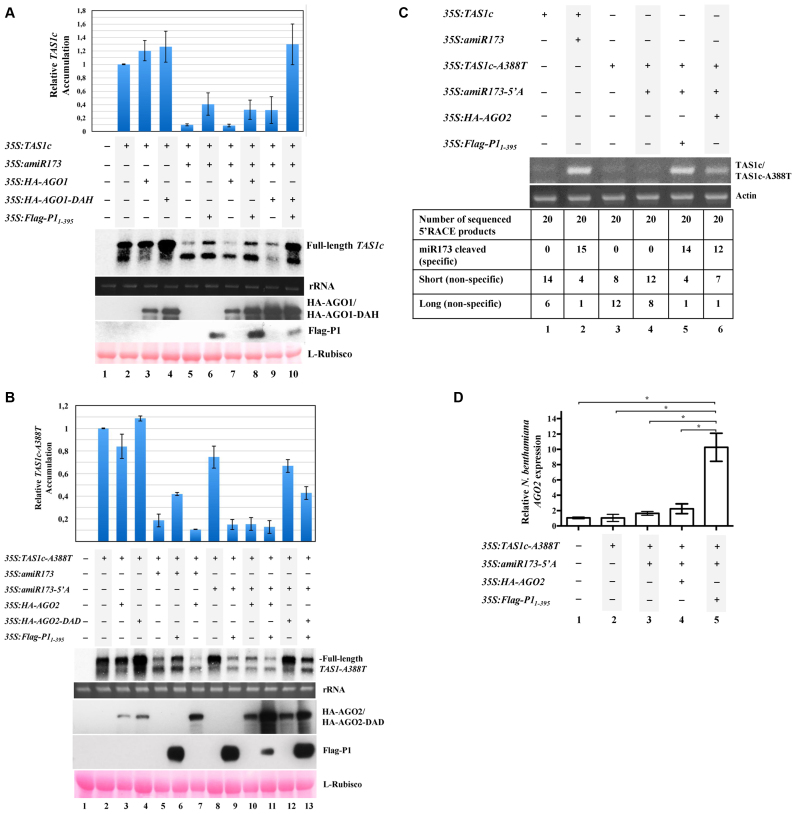
Because AGO1 and AGO2 are the two main plant antiviral AGOs, we decided to test the inhibitory activity of SPMMV P1 against these particular AGOs. In particular, the effect of P1 on target RNA cleavage was analyzed for each AGO. First, the effect of P1 on AGO1 cleavage activity was analyzed using the previously described TAS1c/miR173/AGO1 in vivo system (34). In this system, the coagroinfiltration in N. benthamiana leaves of constructs expressing Arabidopsis miR173 and TAS1c leads to cleavage of TAS1c transcripts by endogenous AGO1 through miR173 association (34). Importantly, TAS1c target cleavage is slightly increased or inhibited when coexpressing Arabidopsis wild-type or slicer-deficient AGO1 forms, respectively, with miR173- and TAS1c-expressing constructs (34). Here, several constructs to express TAS1c transcripts (35S:TAS1c), miR173 (35:amiR173), wild-type or slicer-deficient HA-tagged AGO1 forms (35S:HA-AGO1 and 35S:HA-AGO1-DAH, respectively) and Flag-tagged P1 (35S:Flag-P11-395, including only the first 395 residues of P1) were agroinfiltrated in different combinations in N. benthamiana leaves as shown in Figure 1A. Note that Flag-P11–395 has similar suppressor activity to full-length P1, but is able to escape from degradation (Supplementary Figure S2). As expected, the accumulation of TAS1c mRNA decreased when miR173 was coexpressed (Figure 1A, lane 5), independently of 35S:HA-AGO1 expression (Figure 1 lane 7). In contrast, when SPMMV P1 was present, TAS1c mRNA destabilization was compromised as observed in samples expressing 35S:TAS1c/35S:amiR173/35S:Flag-P11-395 or 35S:TAS1c/35S:amiR173/35S:HA-AGO1/35S:Flag-P11-395 samples (Figure 1A, compare lane 5 with lane 6, and lane 7 with lane 8). In any case, these results indicate that the inhibition of the cleavage activity of both endogenous and exogenous AGO1 is P1-dependent. As expected, TAS1c target RNA cleavage occurred in a lesser extent in samples co-agroinfiltrated with 35S:TAS1c/35S:amiR173/35S:HA-AGO1-DAH compared to samples co-agroinfiltrated with 35S:TAS1c/35S:amiR173 or 35S:TAS1c/35S:amiRNA/35S:HA-AGO1 (Figure 1A, compare lane 9 with lane 5 and lane 7). However, coexpression of 35S:TAS1c/35S:amiR173/35S:HA-AGO1-DAH/35S:Flag-P11-395 greatly inhibited TAS1c target cleavage most likely due to the combined effect of both P1 silencing suppressor activity and HA-AGO1-DAH-induced target RNA stabilization (Figure 1A, lane 10). Altogether, these results suggest that SPMMV P1 inhibits AGO1 cleavage activity.
Figure 1.
SPMMV P1 inhibits AGO1, but not AGO2. (A) N. benthamiana leaves were agroinfiltrated with 35S:TAS1c, 35S:amiR173, 35S:HA-AGO1, 35S:HA-AGO1-DAH and 35S:Flag-P11-395 as indicated. TAS1c mRNA cleavage was analyzed by Northern blotting. Protein extracts were analyzed by SDS-PAGE followed by anti-HA or anti-Flag Western blotting to detect HA-AGO1/HA-AGO1-DAH or Flag-P11–395, respectively. Ponceau staining shows equal loading of proteins. Top panel, mean (n = 3) relative TAS1c transcript level and ±SD (lane 2 = 1.0 for TAS1c transcript). One representative blot from three biological replicates is shown. (B) N. benthamiana leaves were agroinfiltrated with 35S:TAS1c-A388T, 35S:amiR173-5΄A, 35S:amiR173, 35S:HA-AGO2, 35S:HA-AGO2-DAD and 35S:Flag-P11-395 as indicated. TAS1c-A388T mRNA cleavage was analyzed by Northern blotting. Protein extracts were analyzed by SDS-PAGE followed by anti-HA or anti-Flag Western blotting to detect HA-AGO2/HA-AGO2-DAD or Flag-P11–395, respectively. Ponceau staining shows equal loading of proteins. Top panel, mean (n = 3) relative TAS1c-A388T transcript level and ±SD (lane 2 = 1.0 for TAS1c-A388T transcript). One representative blot from three biological replicates is shown. (C) 5΄ RACE product of TAS1c (lane 2) and and TAS1c-A388T (lanes 5 and 6) cleaved by amiR173/AGO1 and amiR173-5΄A/AGO2 complexes, respectively. In lanes 1, 3 and 4, amplification of fragments different from the miRNA cleaved products is observed. Amplification of N. benthamiana actin mRNA is used as control. (D) N. benthamiana leaves were agroinfiltrated with 35S:TAS1c-A388T, 35S:amiR173-5΄A, 35S:HA-AGO2 and 35S:Flag-P11-395, as indicated. N. benthamiana AGO2 mRNA expression was normalized to that of the endogenous elongation factor 1 (EF1), and represented as relative AGO2 mRNA expression. A mock-infiltrated control sample was used as the calibrator (relative AGO2 mRNA expression = 1.0). Three independent biological replicates of each treatment were carried out. For each biological replicate, two parallel samples were analyzed. The * indicates statistically significant differences between groups according to a one-way ANOVA followed by Bonferroni post hoc test (*P < 0.05).
Second, we explored if SPMMV P1 could also inhibit AGO2 activity by using the previously described TAS1c-A388T/miR173-5΄A/AGO2 in vivo system in N. benthamiana (34). This system is a modification of the above described TAS1c/miR173/AGO1 system to allow for the functional analysis of AGO2 cleavage activity. In this system, exogenous AGO2 associates with miR173-5΄A—a modified version of miR173 containing an AGO2-preferred 5΄A—to target and cleave TAS1c-A388T transcripts containing a substitution at the end of the miR173 target site to maintain full base-pairing with miR173-5΄A (34). Thus, several constructs to express TAS1c-A388T transcripts (35S:TAS1c-A388T), miR173-5΄A (35S:amiR173-5΄A), wild-type or slicer deficient HA-tagged AGO2 forms (35S:HA-AGO2 and 35S:HA-AGO2-DAD, respectively) and Flag-tagged P1 (35S:Flag-P11-395) were agroinfiltrated in different combinations in N. benthamiana leaves as shown in Figure 1B.
In agreement with previous results (34), TAS1c-A388T target RNA cleavage occurred when 35S:TAS1c-A388T was coexpressed with both amiR173-5΄A and HA-AGO2 (Figure 1B, compare lanes 8 and 10). However, no TAS1c-A388T target RNA cleavage was observed when 35S:TAS1c-A388T was coexpressed uniquely with 35S:amiR173-5΄A, that is, in the absence of exogenous AGO2 (Figure 1B, lane 8) indicating the lack of endogenous AGO2 activity in the infiltrated leaves under these conditions. Interestingly, TAS1c-A388T mRNA destabilization was not inhibited when 35S:TAS1c-A388T/35S:amiR173-5΄A/35S:HA-AGO2 were coexpressed with 35S:Flag-P11-395, but rather enhanced, when compared to samples coexpressing 35S:TAS1c-A388T/35S:amiR173-5΄A/35S:HA-AGO2 but not 35S:Flag-P11-395 (Figure 1B, lanes 10 and 11 respectively). This result suggests that SPMMV P1 does not inhibit the catalytic activity of AGO2. Strikingly, amiR173-5΄A-dependent TAS1c-A388T transcript destabilization occurred in samples expressing Flag-P11-395 but lacking HA-AGO2 (Figure 1B, lane 9), which suggests a P1-dependent induction of AGO2 activity (see below). Furthermore, the expression of HA-AGO2-DAD stabilized TAS1c-A388T transcript levels both in the presence and absence of miR173-5΄A (Figure 1B, lanes 4 and 12), except when Flag-P11-395 was also coexpressed (Figure 1B, lane 13). Finally, as described before (34), TAS1c-A388T target RNA cleavage was not observed when 35S:TAS1c-A388T, 35S:TAS1c-A388T/35S:HA-AGO2 or 35S:TAS1c-A388T/35S:HA-AGO2-DAD were co-agroinfiltrated (Figure 1B, lanes 2–4). However, TAS1c-A388T target RNA cleavage occurred when 35S:TAS1c-A388T was co-agroinfiltrated with 35S:amiR173, most likely through miR173 association with endogenous AGO1 (Figure 1B, lane 5). Moreover, amiR173-dependent TAS1c-A388T destabilization was inhibited in the presence of SPMMV Flag-P11-395 (Figure 1B, lane 6) but not in the presence of HA-AGO2 (which does not associate with miR173) (34) (Figure 1B, lane 7) supporting the idea that only endogenous AGO1 was hindered by SPMMV P1.
Intriguingly, Northern blot analyses revealed a shorter form of TAS1c/TAS1c-A388T transcript in all samples infiltrated with 35S:TAS1c or 35S:TAS1c-A388T (Figure 1 A and B, Northern panels). To address the nature of this shorter fragment, we performed 5΄ RNA ligase-mediated rapid amplification of cDNA ends (5΄ RACE) analysis on a subset of samples from Figure 1A and B. As reported before (34) miRNA-dependent 3΄ cleavage products were detected in samples expressing 35S:TAS1c/35S:amiR173 (Figure 1C, lane 2) or 35S:TAS1c-A388T/35S:amiR173-5΄A/35S:HA-AGO2 (Figure 1C, lane 6), and corresponded to TAS1c or TAS1c-A388T cleavage products resulting from the action of endogenous AGO1/miR173 and exogenous AGO2/amiR173-5΄A respectively. In agreement with our in vivo assay (Figure 1B lane 9), the 3΄ cleavage product was also detected in the sample expressing TAS1c-A388T/ amiR173-5΄A with Flag-P11-395, but lacking HA-AGO2 (Figure 1C lane 5). Moreover, TAS1c/TAS1c-A388T derived fragments, slightly different in size (both shorter and longer) compared to the 221 bp fragment corresponding to miR173/AGO1 or amiR173-5΄A/AGO2 dependent target cleavage were also detected in all samples analyzed (Figure 1C). This result indicates that some non-specific degradation of TAS1c/TAS1c-A388T transcripts occurred in the in vivo assays under our experimental conditions. The ratio of miR173 specific/non-specific products observed in samples where TAS1c/TAS1c-A388T cleavage was expected (Figure 1C, lanes 2, 5 and 6) revealed that miR173/amiR173-5΄A-dependent target RNA cleavage was predominant over non-specific degradation.
Finally, as stated above, TAS1c-A388T cleavage activity upon Flag-P11–395 infiltration but in the absence of exogenous AGO2 suggests the induction of endogenous N. benthamiana AGO2 in our in vivo assays (Figure 1B and C). To verify the induction of endogenous, quantitative reverse transcriptase PCR (qRT-PCR) detecting solely the N. benthamiana AGO2 was carried out (Figure 1D). Significant AGO2 mRNA accumulation was observed solely upon 35S:TAS1c-A388T/35S:amiR173-5΄A/35S:Flag-P11–395 infiltration (Figure 1D, lane 5), but not in the non-infiltrated control (Figure 1D, lane 1), nor in samples where 35S:TAS1c-A388T, 35S:TAS1c-A388T/35S:amiR173-5΄A, or 35S:TAS1c-A388T/35S:amiR173-5΄A/35S:HA-AGO2 were co-infiltrated (Figure 1D, lanes 2–4). Collectively, all these results suggest that SPMMV P1 inhibits AGO1 and promotes AGO2 activity in vivo.
AGO/P1 interaction is necessary but not sufficient for P1-dependent inhibition of AGO cleavage
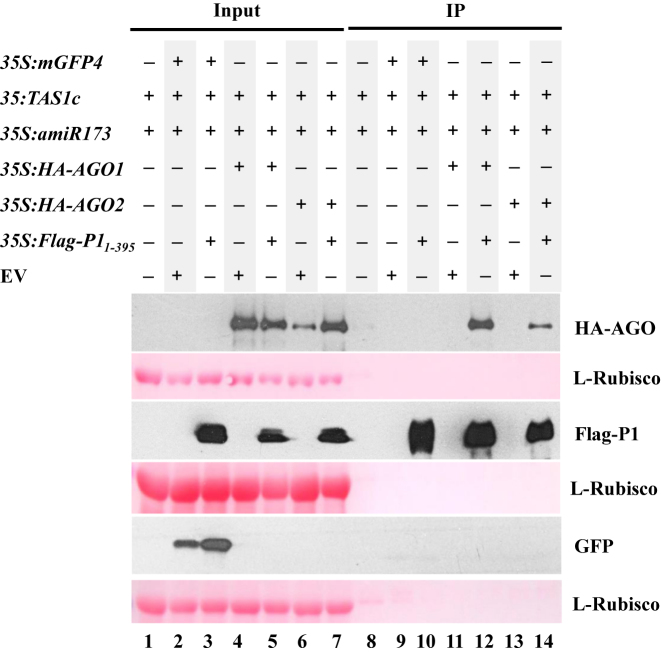
We recently reported that SPMMV P1 physically interacts with AGO1 through its WG/GW motif, and that this interaction is absolutely required for P1-dependent inhibition of AGO1 activity (29). Thus, because SPMMV P1 did not inhibit AGO2 activity (Figure 1B), we hypothesized that AGO2 and SPMMV P1 may not interact with each other. To test P1-AGO2 interaction, we set up an agroinfiltration assay in N. benthamiana in which SPMMV Flag-P11–395 was transiently co-expressed with HA-AGO2. 35S:Flag-P11-395 was also co-agroinfiltrated with 35S:HA-AGO1 or 35S:mGFP4 as interacting and non-interacting controls respectively. To further confirm the specificity of the interactions, 35S:HA-AGO1, 35S:HA-AGO2 and 35S:mGFP4 were also co-agroinfiltrated with an empty vector construct (EV) as shown in Figure 2. All samples were subjected to protein immunoprecipitation (IP) using an anti-Flag antibody to pull down Flag-P11–395 and analyze P1 binding to mGFP4, AGO1 and AGO2. Immunoblot analysis using appropriate antibodies to detect the different assayed proteins extracted from infiltrated leaves confirmed that all expressed proteins were detected in their corresponding input fractions (Figure 2, lanes 2–7). Importantly, GFP was not detected in the IP fractions of samples co-expressing 35S:mGFP4 and 35S:Flag-P11-395 suggesting that GFP does not interact with Flag-P11–395, and thus confirming the specificity of the assay (Figure 2, lane 10). In contrast, HA-AGO1 was detected in the IP fractions of samples co-expressing 35S:HA-AGO1 and 35S:Flag-P11–395 (Figure 2, lane 12) confirming that P1 interacts with AGO1, as reported before (29). Surprisingly, HA-AGO2 was detected in immunoprecipitates of samples co-expressing 35S:HA-AGO2 and 35S:Flag-P11–395 (Figure 2, lane 14). This results suggests that Flag-P11–395 interacts with HA-AGO2 as well, but in a lesser extent than HA-AGO1 did based on the lower amount of HA-AGO2 compared to HA-AGO1 detected in IP fractions (Figure 2, compare lanes 12 and 14). Taken together, these results show that SPMMV P1 binds to both AGO1 and AGO2 but only inhibits AGO1 function. Hence, it seems that AGO binding by P1 per se is not sufficient for inhibiting AGO activity.
Figure 2.
AGO2 interacts with SPMMV P1. 35S:mGPF4, 35S:HA-AGO1, 35S:HA-AGO2, 35S:amiR173, 35S:amiR173-5΄A were co-agroinfiltrated with 35S:Flag-P11–395, as indicated. Input and IP protein fractions were analyzed by SDS-PAGE followed by anti-HA, anti-Flag or anti-GFP Western blotting to detect HA-AGO1/HA-AGO2, Flag-P11–395 and GFP, respectively. Ponceau staining shows equal loading of proteins.
SPMMV P1 contains a zinc finger motif that contributes to its silencing suppressor activity
Because AGO binding by P1 seems necessary but not sufficient for inhibiting AGO activity, we hypothesized that other determinant(s) included in the P1 sequence might be responsible for the AGO1- (but not AGO2) specific inhibitory activity of SPMMV P1. The in silico analysis of the P1 sequence revealed the presence of a Cys4 zinc finger motif in SPMMV P1 composed by four Cys residues at amino acid positions 88, 91, 103, 106. This motif is apparently conserved in P1 proteins of potyviruses infecting sweet potato (39). Notably, SPMMV P1 contains an additional Cys residue at position 85.
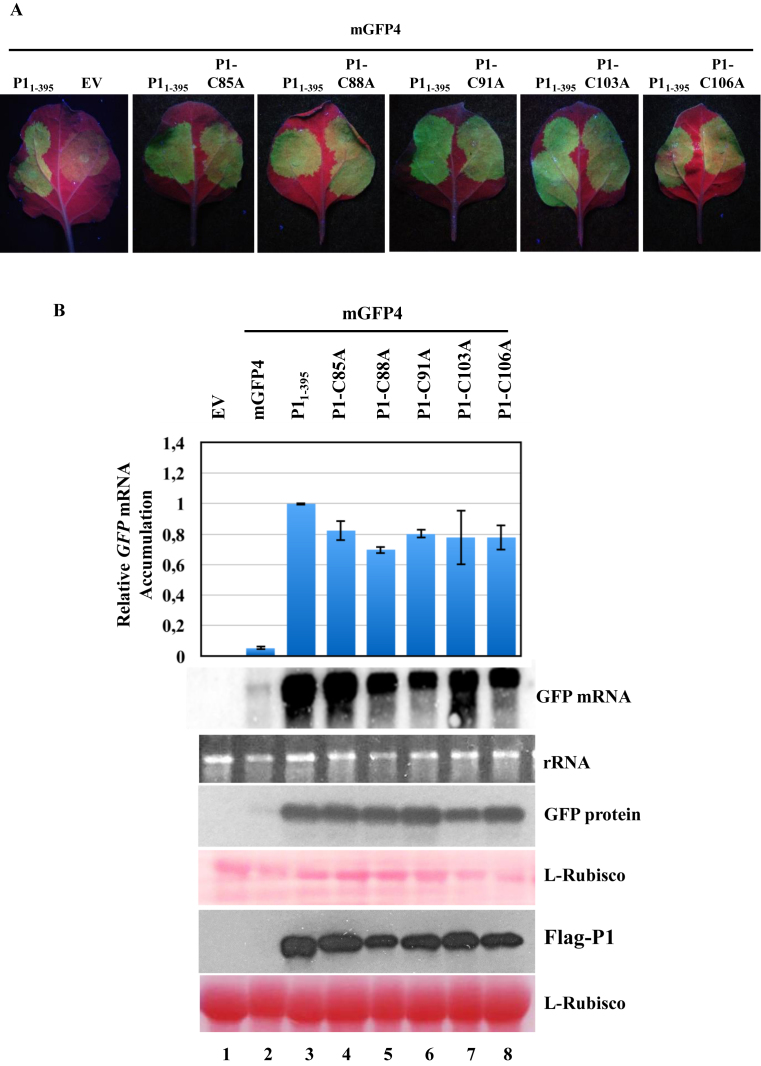
To study the significance of the putative zinc finger motif in the silencing suppressor activity of SPMMV P1, each of the five Cys residues was mutated independently to Ala by site-directed mutagenesis using 35S:Flag-P11–395 as template. The following P1 single mutant constructs were generated: 35S:P1-C85A, 35S:P1-C88A, 35S:P1-C91A, 35S:P1-C103A and 35S:P1-C106A. The silencing suppressor activity of wild-type and mutant P1 constructs was analyzed in N. benthamiana by co-agroinfiltrating the 35S:mGFP4 reporter independently with each P1 construct. GFP fluorescence was monitored for 72 h post-agroinfiltration (Figure 3A). As expected, a faint fluorescence was observed in negative control plants co-agroinfiltrated with 35S:mGFP4 and with EV indicating the silencing of the GFP mRNA. In contrast, a bright fluorescence was detected in leaf patches co-agroinfiltrated with 35S:mGFP4 and 35S:Flag-P11–395 indicating a lack of silencing of the GFP mRNA most likely due to a strong silencing suppressor activity by SPMMV P1. Notably, the bright fluorescence observed in patches co-agroinfiltrated with 35S:mGFP4 and each of the P1 mutants (Figure 3A, patches on the right side of the leaf) indicates that P1 silencing suppressor activity was not affected by any of the individual Cys to Ala mutations (Figure 3A). In agreement with visual examination, Northern and Western blot analyses revealed a slightly less GFP mRNA and protein accumulation in patches co-agroinfiltrated with 35S:mGFP4 and with any of the P1 mutant constructs, than that of 35S:Flag-P11–395 and 35S:mGFP4 infiltrated control sample (Figure 3B, lanes 3–8). Moreover, Western blot analysis revealed that P1 Cys to Ala mutants expressed at similar level than wild-type P11–395. Our results indicated that individual Cys to Ala mutants possess VSR activity comparable to wild-type P1 (Figure 3A; Supplementary Figure S2A).
Figure 3.
Mutational analysis of the zinc finger domain present in SPMMV P1. (A) 35S:P1-C85A, 35S:P1-C88A, 35S:P1-C91A, 35S:P1-C103A and 35S:P1-C106A respectively were coinfiltrated with 35S:mGFP4. Photos were taken under UV light at 3 days post agroinfiltration. (B) GFP mRNA was detected by Northern blotting. Extracts of infiltrated leaves were subjected to SDS-PAGE followed by immunoblotting with the anti-GFP antibody. Flag-tagged P1 proteins were detected by Western blotting using the M2-anti Flag antibody. Ponceau staining shows equal loading of proteins in both Western blots. Top panel, mean (n = 3) relative mGFP4 transcript level and ±SD (lane 3 = 1.0 for mGFP4 transcript). One representative blot from three biological replicates is shown.
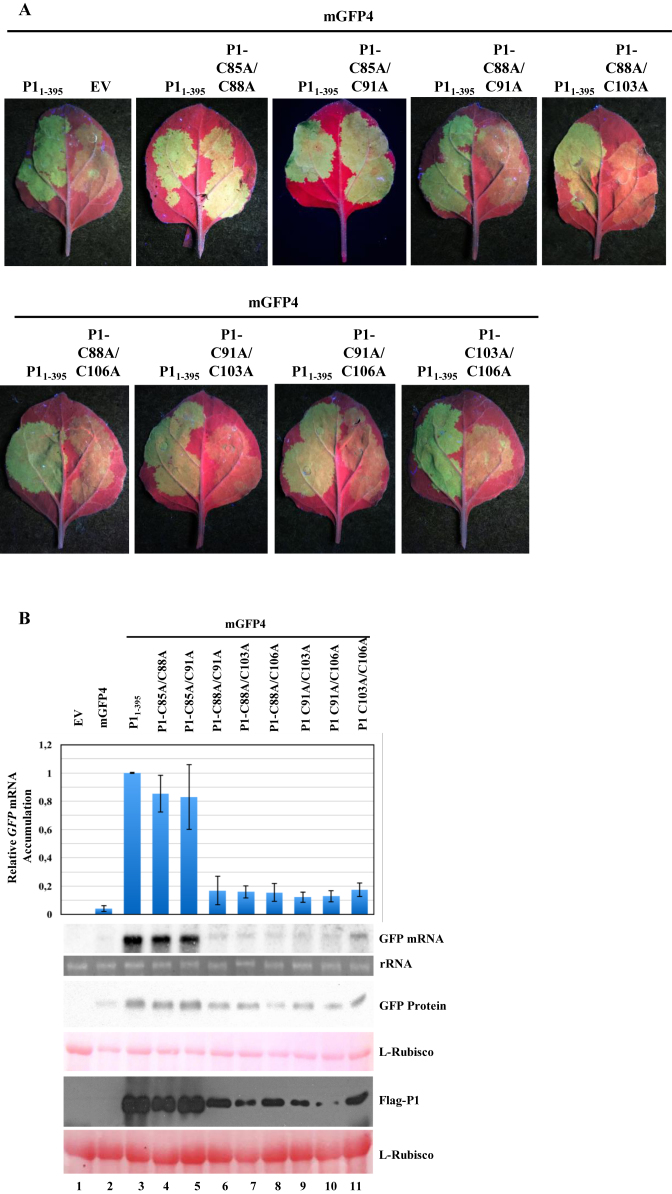
Similarly, we generated all possible combinations of Cys to Ala P1 double mutant constructs involving the C88, C91, C103 and C106 conserved residues (35S:P1-C88A/C91A, 35S:P1-C88A/C103A, 35S:P1-C88A/C106A, 35S:P1-C91A/C103A, 35S:P1C91A/C106A and 35S:P1-C103A/C106A), and two additional double mutant constructs including the C85A mutation (35S:P1-C85A/C88A and 35S:P1-C85A/C91A). The suppressor activity of each of the P1 double mutant constructs was tested as described above in the P1 single mutant analysis. Based on both visual observations of GFP fluorescence, molecular examination of GFP and P1 double mutant expression, our results revealed that converting any of two conserved Cys residues to Ala in the predicted zinc finger seriously compromised the silencing suppressor activity, which might account for reduced GFP and P1 double mutant expression (Figure 4B, lanes 6–11). P1-C85A/C88A and P1-C85A/C91A double mutants showed comparable suppressor activity and expression level (Figure 4B, lanes 4 and 5) to that of Flag-P11–395, and behaved similarly than P1-C88A and P1-C91A single mutants. Collectively, these results indicate that Cys residues at positions 88, 91, 103 and 106—but not at position 85—are likely involved in the formation of a putative zinc finger in SPMMV P1 which is required for the silencing suppression function of this particular VSR.
Figure 4.
The putative zinc finger is required for P1 activity. (A) 35S:Flag-P11-395, 35S:P1-C85A/C88A, 35S:P1-C85A/C91A, 35S:P1-C88A/C91A, 35S:P1-C88A/C103A, 35S:P1-C88A/C106A, 35S:P1-C91A/C103A, 35S:P1-C91A/C106A and 35S:P1-C103A/C106A were co-agroinfiltrated with 35S:mGFP4. Photos were taken under UV light at 3 days post agroinfiltration. (B) GFP mRNA was detected by Northern blotting. Extracts of infiltrated leaves were analyzed by SDS-PAGE followed by immunoblot with anti-GFP antibody. Flag-tagged P1 proteins were detected by Western blotting using anti-Flag antibody in both Western blots. Ponceau staining shows equal loading of proteins in both Western blots. Top panel, mean (n = 3) relative mGFP4 transcript level and ±SD (lane 3 = 1.0 for mGFP4 transcript). One representative blot from three biological replicates is shown.
P1 silencing suppressor and AGO1 binding functions can be uncoupled
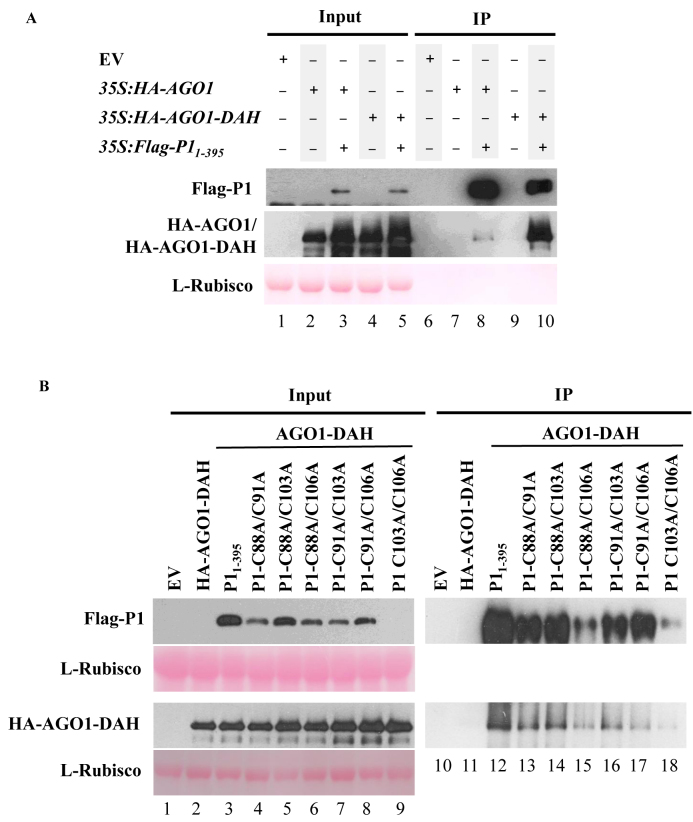
P1 WG/GW motifs are required for both P1 silencing suppression and AGO1 binding functions (29). Because the putative zinc finger motif in P1 is also required for P1 suppressor activity, we wondered if it was also involved in AGO1 binding. First, we set up a preliminary experiment to compare the strength of the interactions between Flag-P11–395 and HA-AGO1 or HA-AGO1-DAH. To that purpose, 35S:HA-AGO1 and 35S:HA-AGO1-DAH were agroinfiltrated alone or together with 35S:FlagP11-395 in N. benthamiana leaves (Figure 5A). Samples were analyzed as described before for results shown in Figure 2. Here, results suggest that Flag-P11–395 interacts stronger with HA-AGO1-DAH than with HA-AGO1, as HA-AGO1-DAH protein accumulation was significantly higher than HA-AGO1 accumulation in the corresponding IP fractions (Figure 5A, compare lane 8 with lane 10). Thus, HA-AGO1-DAH was selected for subsequent experiments.
Figure 5.
The zinc finger in SPMMV P1 is not required for AGO1 binding. (A) P1 binds to AGO1-DAH. 35S:amiR173, 35S:HA-AGO1, 35S:HA-AGO1-DAH, 35S:Flag-P11-395 were agroinfiltrated in N. benthamiana leaves as indicated. Flag IP was carried out, then input and IP fractions were analyzed by Western blot to detect P11–395, AGO1 and AGO1-DAH proteins. Ponceau staining shows equal loading of proteins. (B) 35S:Flag-P11-395, 35S:P1-C88A/9C1A, 35S:P1-C88A/C103A, 35S:P1-C88A/C106A, 35S:P1-C91A/C103A, 35S:P1-C91/C106A and 35S:P1-C103A/C106A were coinfiltrated with 35S:HA-AGO1-DAH. Input and IP protein fractions were analyzed by SDS-PAGE followed by anti-HA or anti-Flag immunoblot to detect HA-AGO1-DAH or Flag-P11–395 proteins, respectively. Ponceau staining shows equal loading of proteins. One representative blot from three biological replicates is shown.
Next, 35S:HA-AGO1-DAH was co-agroinfiltrated with wild-type or double mutant P1 constructs in N. benthamiana leaves, then P1 proteins were immunoprecipitated and analyzed by Western blot. All HA-AGO1-DAH, wild-type and mutant P1 proteins could be detected, but the P1-C103A/C106A mutants expressed at much lower level in the input (Figure 5B, lanes 1–9). Analysis of the IP fractions indicated that P1 double mutants were pulled down in varying amounts, which did not always correlate with the expression level in the input fractions. As expected, HA-AGO1-DAH accumulated to high levels in the IP fraction of samples including 35S:HA-AGO1-DAH and 35S:Flag-P11–395 thus suggesting that SPMMV interacts with HA-AGO1-DAH (Figure 5B, lane 12). Strikingly, AGO1-DAH was also detected in IP fractions of samples including each of the P1 double mutant constructs suggesting that indeed all P1 double mutants interact with HA-AGO1-DAH (Figure 5B, lanes 13–18). Interestingly, in each case, the proportion of HA-AGO1-DAH versus P1 accumulating in the IP fraction was similar for wild-type and for each of the P1 double mutants (except for P1-C91A/C106A) suggesting that the majority of P1 forms bind to HA-AGO1-DAH with similar strength. Collectively, these results suggest that the putative zinc finger in SPMMV P1 is required for suppressor activity but not for AGO1 binding. Therefore, these two P1 functions can be uncoupled.
SPMMV P1 inhibits target RNA association with AGO1
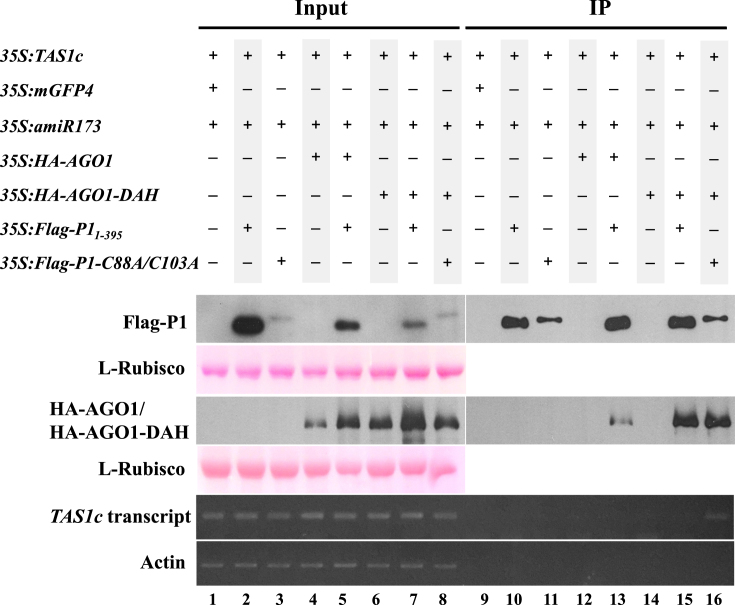
SPMMV P1 was previously shown to inhibit the activity of AGO1 complexes loaded with miRNA or viral siRNA (vsiRNA) (29), but the specific molecular mechanism explaining the functional inhibition of AGO1 was not described. We hypothesized that P1 could be inhibiting target RNA binding by sRNA-loaded AGO1. To test this hypothesis, we used the miR173/TAS1c/AGO1 system described in Figure 1. For that purpose 35S:TAS1c, 35S:miR173, 35S:mGFP4, 35S:HA-AGO1, 35S:HA-AGO1-DAH, 35S:Flag-P11–395 and 35S:P1-C88A/C103A constructs were agroinfiltrated in different combinations in N. benthamiana leaves as shown in Figure 6. Extracts from agroinfiltrated leaves were subjected to anti-flag immunoprecipitation to pull down both Flag-P11-395 and P1-C91A/C103A forms. Input (Figure 6, lanes 1–8) and IP fractions (Figure 6, lanes 9–16) were analyzed by Western blot to detect the corresponding expressed proteins, and by RT-PCR to detect TAS1c target RNA.
Figure 6.
Target RNA binding is inhibited by SPMMV P1. 35S:TAS1c1, 35S:mGFP4, 35S:amiR173, 35S:HA-AGO1, 35S:HA-AGO1-DAH, 35S:Flag-P11–395 and 35S:P1-C88A/C103A respectively were infiltrated into N. benthamiana leaves as indicated. To detect protein–protein interactions native extracts were analyzed, while for RNA immunoprecipitations infiltrated leaves were crosslinked with formaldehyde prior to extract preparation. Input and IP protein fractions were analyzed by SDS-PAGE followed by anti-HA or anti-Flag Western blot to detect HA-AGO1/HA-AGO1-DAH or Flag-P11–395 proteins, respectively. Ponceau staining shows equal loading of proteins. Input and IP RNA fractions where analyzed by semi-quantitative RT-PCR to detect TAS1c or actin mRNAs. One representative blot from three biological replicates is shown.
All expressed and control proteins and RNAs were detected in the input fractions (Figure 6, lanes 1–8). TAS1c transcript was not detected in the IP fraction of samples including 35S:TAS1c/35S:amiR173/35S:mGFP4 indicating that no unspecific TAS1c binding to anti-Flag beads occurred (Figure 6, lane 9). P11–395 and P1-C88A/C103A were detected in the IP fraction of samples including 35S:amiR173/35S:TAS1c/35S:Flag-P11–395 and 35S:TAS1c/35S:miR173/35S:P1-C88A/C103A, respectively, but no TAS1c target could be detected in any of these fractions suggesting that P11–395 and P1-C88A/C 103A do not bind TAS1c transcripts (Figure 6, lanes 10 and 11). As expected, in the absence of Flag-P11–395, neither HA-AGO1 nor HA-AGO1-DAH were detected in samples including 35S:amiR173/35S:TAS1c/35S:HA-AGO1 or 35S:amiR173/35S:TAS1c/35S:HA-AGO1-DAH IP fractions, respectively (Figure 6, lanes 12 and 14), and no TAS1c transcript was detected in any of the two IP fractions. As expected, in the presence of Flag-P11–395, TAS1c mRNA was not detected in 35S:amiR173/35S:TAS1c/35S:HA-AGO1/35S:Flag-P11–395 immunoprecipitates (Figure 6, lane 13). Importantly, the HA-AGO1-DAH slicer-deficient mutant stably binds to TAS1c target RNA when coexpressed with miR173 in N. benthamiana (34). However, in 35S:amiR173/35S:TAS1c/35S:HA-AGO1-DAH/35S:Flag-P11–395 samples, where HA-AGO1-DAH is complexed with P1, no target RNA could be detected in the immunoprecipitates (Figure 6, lane 15) suggesting that TAS1c mRNA targeting is inhibited by P1. In contrast, TAS1c mRNA was detected in 35S:amiR173/35S:TAS1c/35S:HA-AGO1-DAH/35S:P1-C88A/C103A immunoprecipitates. Indeed, it seems that HA-AGO1-DAH interacted with Flag-P11–395 and P1-C88A/C103A with similar strength, but only HA-AGO1-DAH complexes including P1-C88A/C103A complex associated with TAS1c mRNA (Figure 6, compare lane 15 and 16). Finally, we could not amplify the endogenous actin mRNA from any of the IP fractions indicating that no unspecific RNA binding occurred in this experiment. Collectively, these results indicate that the AGO1-binding but suppressor-deficient P1-C88A/C103A zinc finger mutant does not interfere with target RNA binding by HA-AGO1-DAH (Figure 6, lane 16), thus indicating that the zinc finger motif in the SPMMV P1 protein is absolutely required for the inhibition of target RNA binding by HA-AGO1-DAH. In conclusion, the data presented here supports the idea that SPMMV P1 inhibits AGO1 function by precluding target RNA association with AGO1.
DISCUSSION
Differential effect of P1 on AGO1 and AGO2
An in vivo assay in N. benthamiana was used to analyze the effect of SPMMV P1 on the cleavage and target RNA binding activities of Arabidopsis AGO1 and AGO2, the two main plant antiviral AGOs. We observed that SPMMV P1 inhibits both endogenous and overexpressed AGO1 but not AGO2 cleavage function. Indeed, we noticed that P1 enhanced endogenous AGO2 activity at the transcriptional level, however, AGO2 activity could be induced by P1 derepressing the AGO1/miR403-mediated silencing of AGO2 mRNA (40). Interestingly, P1 could not inhibit either endogenous or overexpressed AGO2-mediated cleavage despite that both proteins seem to interact with each other according to our coimmunoprecipitation experiments. Although the amino acid similarity between Arabidopsis AGO1 and AGO2 is rather low (33%), a common protein domain involved in P1 binding might be included in the sequence of these two AGOs. Pertinent to this context, it is known that AGO proteins contain a conserved pocket for binding sRNAs and Trp residues of the WG/GW proteins (41) (42–44). Therefore, it is possible that the conserved WG/GW domains of P1 (29) are also involved in P1 interaction with AGO2.
The zinc finger motif in SPMMV P1 uncouples the silencing suppressor activity and AGO1 binding functions of P1
Cys4-type zinc finger motifs are typically present in transcription factors and RNA binding proteins (45). The mutation of residues in this type of zinc finger motifs can lead to suppression of the protein function, as observed with the individual conversion of Cys residues to Ser residues in the adenoviral Cys4 zinc finger-containing E1A transcription factor that abolished mutant transactivation function (46). Here, a series of SPMMV P1 forms with Cys to Ala mutations in the four conserved residues of a putative zinc finger motif were analyzed. Only SPMMV P1 zinc finger double mutants showed reduced suppressor activity compared to wild-type P1. Zinc finger motifs have been found in other VSRs. For instance, the AC2 protein from Mungbean yellow mosaic virus-Vigna (MYMV) lost transactivator, DNA binding and VSR activity when its zinc finger motif was mutated (47). Also, the zinc finger motif in the p14 protein of Beet necrotic yellow vein virus (BNYVV) and of Beet soil-borne mosaic virus (BSBMV) was required for VSR activity and long-distance movement (48).
The WG/GW domains of SPMMV P1 were shown to be involved in both AGO1 binding and VSR activity (29). In contrast, results presented here show that suppressor-deficient P1 double mutants are still able to interact with AGO1 indicating that the VSR and AGO1 binding functions of P1 can be uncoupled. These results also suggest that the zinc finger motif of P1 is indeed an effector domain. Similarly, in proteins including WG/GW domains such as GW182, KTF1, Tas3 and RNA Pol IV, the AGO binding and the effector functions were mapped to different domains (49–54). Thus, the modular architecture of proteins including WG/GW domains might explain how these proteins could play positive or negative role in RNA silencing.
Interestingly, a subset of WG/GW proteins also contain zinc finger domains. For example, in the Arabidopsis NERD protein, the reiterated WG/WG domains are separated from the putative zinc finger motif at the C-terminal end of the protein. In another example, the CnjBp protein of the ciliate Tetrahymena thermophila also contains a zinc finger motif in this case surrounded by two reiterated WG/GW domains (55). Unfortunately, the biological role of the NERD or CnjBp zinc finger-containing proteins has not been elucidated yet (55,56).
Molecular mechanism of P1-mediated inhibition of AGO1 activity
We previously reported that the SPMMV P1 silencing suppressor inhibits pre-assembled RISC activity by binding to AGO1 via its conserved WG/GW motifs (29). Here, we used an in vivo agroinfiltration system in N. benthamiana, followed by protein/RNA immunoprecipitation and Northern, Western and RT-PCR analyses to show that indeed SPMMV P1 blocks target RNA binding to AGO1. This particular mode of action of P1 represents a novel silencing suppression mechanism for a VSR.
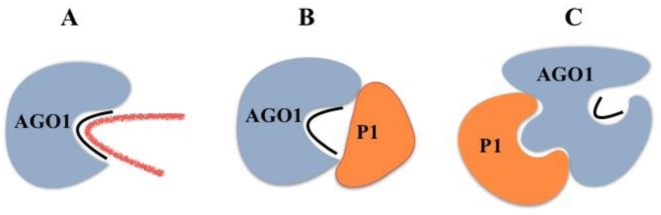
We propose a structural model that could explain how SPMMV P1 suppresses AGO1 but not AGO2-mediated silencing (Figure 7). In this model, the zinc finger domain of P1 might compete with target RNA (substrate) for binding to AGO1-sRNA binary complexes. The seed region (nucleotides 2–7) of the sRNA plays a key role in target RNA recognition of human AGO2 (57). Structural studies have also reported that in human AGO2–miRNA binary complexes, the helix-7 of the AGO2 L-2 domain is inserted between nucleotides 6 and 7 of the sRNA that brakes the A-form of the guide RNA leading to the inhibition of target RNA-guide RNA interaction. However, in the trimeric complex representing the target bound state, the helix-7 (which is conserved between animal and plant AGO proteins) is shifted upon seed pairing resulting in the relaxation of the kink (57). It is possible that the effector domain of P1 does not allow helix-7 displacement by freezing AGO1 in a target unbound state. Alternatively, the P1 zinc finger might cover the first half of the central cleft of the AGO1-miRNA binary complex, which nucleates the seed region of the guide RNA and by making RNA-protein and/or protein–protein interactions in the central cleft, SPMMV P1 could inhibit target RNA-sRNA interaction (Figure 7B). Finally, P1 could also act in a non-competitive way by altering the conformation of the AGO1-sRNA binary complex which, in turn, could distort the central cleft impeding target RNA binding (Figure 7C).
Figure 7.
Model for P1 silencing suppression mechanism. (A) The AGO1-sRNA binary complex binds target RNA leading to RNA cleavage or translational inhibition. (B) P1 interferes with target RNA association in a competitive way. (C) P1 interferes with target RNA association in a non-competitive way.
Considering P1 zinc finger as the effector domain, the competitive and the non-competitive way of inhibition might be regulated by protein–protein and/or protein–nucleic acid interactions. For example, diverse examples show that zinc finger motifs can promote specific protein–protein interactions to regulate transcription, proteolysis or cellular hypoxic stress (58). Regulation of nucleoprotein complexes could also be controlled by protein–nucleic acid interactions either in a nucleotide sequence specific or non-specific ways. For example, the zinc finger protein TFIIIA controls translation in a non-specific way by binding to the sugar-phosphate backbone of the 5S RNA included in the large ribosomal subunit (45). Since AGOs bind to the sugar-phosphate backbone of sRNAs (42), it is therefore unlikely that P1 interferes with target RNA association in a non-specific way of RNA binding. In contrast, the zinc finger domain of tristetraprolin protein binds mRNAs by recognizing the AU rich elements at the 3΄ UTR to facilitate mRNA degradation, thus inhibiting translation of some certain cytokine and chemokine mRNAs (59). The sequence specific inhibition of target RNA association with pre-assembled AGO1 complexes is not likely to completely block AGO1-directed RNA silencing because of the sequence heterogeneity of AGO1-bound sRNAs. In agreement with this idea, we did not observe interaction between P1 and target RNA in our coimmunoprecipitation experiment described in Figure 6. Therefore, we find more likely that the SPMMV P1 inhibitory mechanism might be based on either competitive or non-competitive way via specific protein–protein interactions (Figure 7B and C).
The functional output of the P1 dependent inhibition of AGO1 in plants is reminiscent to that of the regulation of miRNA-driven RNA silencing by poly(ADP-ribose) polymerases (PARP) (60). Acute stress increases the poly(ADP-ribosylation) level of AGO1–4 proteins by PARPs. ADP-ribosylation creates a strong negatively charged environment, which might antagonize with target RNA bound to the AGO–miRNA complex leading to the miRNA–target RNA dissociation, or could interfere with target RNA binding by steric hindrance leading to reduced RNA silencing activity (60). The mechanism of action of SPMMV P1 seems to differ from that of the poly(ADP-ribosylation) because of the inhibition of AGO function. Hence, the results reported here represent a novel molecular mechanism explaining the inhibition of pre-assembled RISCs.
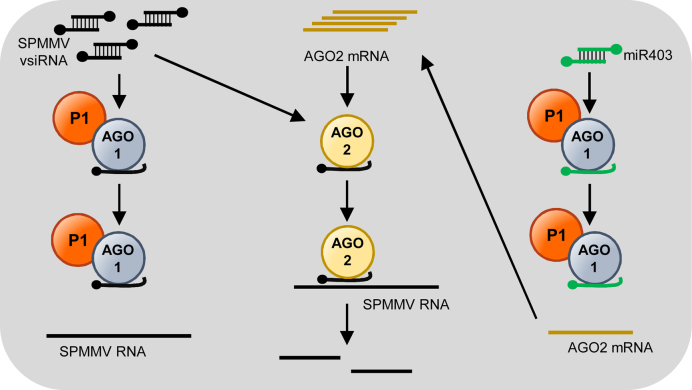
A model for SPMMV pathogenicity
Single SPMMV infection of sweet potato leaves causes moderate symptoms manifested in mild vein chlorosis and mottling. Symptoms last 2–4 weeks, and SPMMV cannot be detected in newly developed leaves as a consequence of plant recovery from virus infection (61). According to our previous model, we hypothesized that at early stages of SPMMV infections the existing AGO1–miRNA complexes sequester P1 from the de novo vsRNA-containing RISC complexes, leading to mild symptoms and recovery (29). In the light of the results presented here, we can postulate a new model that might better explain SPMMV pathogenicity (Figure 8). In this model, SPMMV replication results in vsRNA and P1 protein production. SPMMV P1 association with AGO1–vsRNA complexes might inhibit viral RNA targeting by pre-assembled AGO1 complexes. Also, P1 seems to induce AGO2 mRNA accumulation, which could lead to higher AGO2 protein levels in virus infected cells. Although translation of the AGO2 mRNA is repressed by miR403/AGO1 complexes (40), SPMMV P1-dependent inhibition of AGO1 function might lead to derepression of miR403/AGO1-mediated cleavage of AGO2 mRNA. In this scenario, where AGO1-mediated antiviral silencing is suppressed and AGO2 activity against SPMMV RNA is induced, AGO2 may be active against SPMMV RNA and thus could be acting as a second defense layer as proposed before for Cucumber mosaic virus and Turnip crinkle virus (40). More recent reports have confirmed that AGO2 is indeed the primary antiviral AGO in certain plant viruses not being targeted by AGO1 such as Tobacco rattle virus (62) and Turnip mosaic virus (34,63). Finally, it seems that SPMMV P1 can bind to AGO2 but somehow it is not able to inhibit AGO2 activity. The biological significance of the P1-AGO2 interaction is still to be determined.
Figure 8.
Model for SPMMV pathogenicity based on P1 inhibition of target RNA binding by pre-assembled AGO1 complexes. P1 binding to AGO1 complexes loaded with SPMMV-derived vsiRNAs could inhibit SPMMV RNA targeting by preventing its association with pre-assembled AGO1 complxes. In addition, P1 binding to AGO1/miR403 complexes could prevent their association with AGO2 mRNA binding, leading to derepression of AGO2 mRNA. AGO2 would overaccumulate and load SPMMV vsiRNAs to target complementary SPMMV RNAs. For simplicity, P1 interaction with AGO2 is omitted in this model.
Supplementary Material
ACKNOWLEDGEMENTS
Authors thank James C. Carrington for providing the 35S:HA-AGO1, 35S:HA-AGO1-DAH, 35S:HA-AGO2, 35S:HA-AGO2-DAD, 35S:TAS1c, 35S:TAS1c-A388T, 35S:amiR173-5΄A and 35S:amiR173 constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hungarian Scientific Research Fund (OTKA) [K91042, NN107787, NN11024 to L.L.]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska Curie [655841 to A.C.]. Funding for open access charge: OTKA [NN11024].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sayed D., Abdellatif M.. Micrornas in development and disease. Physiol. Rev. 2011; 91:827–887. [DOI] [PubMed] [Google Scholar]
- 2. Martin R.C., Liu P.P., Goloviznina N.A., Nonogaki H.. microRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J. Exp. Bot. 2010; 61:2229–2234. [DOI] [PubMed] [Google Scholar]
- 3. Baulcombe D.C., Molnar A.. Crystal structure of p19–a universal suppressor of RNA silencing. Trends Biochem. Sci. 2004; 29:279–281. [DOI] [PubMed] [Google Scholar]
- 4. Matzke M.A., Mosher R.A.. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014; 15:394. [DOI] [PubMed] [Google Scholar]
- 5. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 6. Ender C., Meister G.. Argonaute proteins at a glance. J. Cell Sci. 2010; 123:1819–1823. [DOI] [PubMed] [Google Scholar]
- 7. Hutvagner G., Simard M.J., Mello C.C., Zamore P.D.. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004; 2:E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 9. Wilkins C., Dishongh R., Moore S.C., Whitt M.A., Chow M., Machaca K.. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005; 436:1044–1047. [DOI] [PubMed] [Google Scholar]
- 10. Li S., Liu L., Zhuang X., Yu Y., Liu X., Cui X., Ji L., Pan Z., Cao X., Mo B. et al. . MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013; 153:562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Rij R.P., Saleh M.C., Berry B., Foo C., Houk A., Antoniewski C., Andino R.. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006; 20:2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baulcombe D. Viral suppression of systemic silencing. Trends Microbiol. 2002; 10:306–308. [DOI] [PubMed] [Google Scholar]
- 13. Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W., Voinnet O.. Antiviral RNA interference in mammalian cells. Science. 2013; 342:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bronkhorst A.W., van Rij R.P.. The long and short of antiviral defense: small RNA-based immunity in insects. Curr. Opin. Virol. 2014; 7:19–28. [DOI] [PubMed] [Google Scholar]
- 15. Burgyan J., Havelda Z.. Viral suppressors of RNA silencing. Trends Plant Sci. 2011; 16:265–272. [DOI] [PubMed] [Google Scholar]
- 16. Csorba T., Kontra L., Burgyan J.. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015; 479-480:85–103. [DOI] [PubMed] [Google Scholar]
- 17. Pumplin N., Voinnet O.. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013; 11:745–760. [DOI] [PubMed] [Google Scholar]
- 18. Varallyay E., Havelda Z.. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 2013; 14:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varallyay E., Valoczi A., Agyi A., Burgyan J., Havelda Z.. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010; 29:3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baumberger N., Tsai C.H., Lie M., Havecker E., Baulcombe D.C.. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 2007; 17:1609–1614. [DOI] [PubMed] [Google Scholar]
- 21. Chiu M.H., Chen I.H., Baulcombe D.C., Tsai C.H.. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010; 11:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Csorba T., Lozsa R., Hutvagner G., Burgyan J.. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010; 62:463–472. [DOI] [PubMed] [Google Scholar]
- 23. Karran R.A., Sanfacon H.. Tomato ringspot virus coat protein binds to ARGONAUTE 1 and suppresses the translation repression of a reporter gene. Mol. Plant Microbe Interact. 2014; 27:933–943. [DOI] [PubMed] [Google Scholar]
- 24. Pazhouhandeh M., Dieterle M., Marrocco K., Lechner E., Berry B., Brault V., Hemmer O., Kretsch T., Richards K.E., Genschik P. et al. . F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P.. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nayak A., Berry B., Tassetto M., Kunitomi M., Acevedo A., Deng C., Krutchinsky A., Gross J., Antoniewski C., Andino R.. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010; 17:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Mierlo J.T., Bronkhorst A.W., Overheul G.J., Sadanandan S.A., Ekstrom J.O., Heestermans M., Hultmark D., Antoniewski C., van Rij R.P.. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012; 8:e1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mann K.S., Johnson K.N., Carroll B.J., Dietzgen R.G.. Cytorhabdovirus P protein suppresses RISC-mediated cleavage and RNA silencing amplification in planta. Virology. 2016; 490:27–40. [DOI] [PubMed] [Google Scholar]
- 29. Giner A., Lakatos L., Garcia-Chapa M., Lopez-Moya J.J., Burgyan J.. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010; 6:e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008; 13:350–358. [DOI] [PubMed] [Google Scholar]
- 31. Mallory A., Vaucheret H.. Form, Function, and Regulation of ARGONAUTE Proteins. Plant Cell. 2010; 22:3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carbonell A., Carrington J.C.. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015; 27:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kertesz S., Kerenyi Z., Merai Z., Bartos I., Palfy T., Barta E., Silhavy D.. Both introns and long 3΄-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006; 34:6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C.. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012; 24:3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C.. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010; 17:U997–U111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C.. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008; 133:128–141. [DOI] [PubMed] [Google Scholar]
- 37. Carbonell A., Takeda A., Fahlgren N., Johnson S.C., Cuperus J.T., Carrington J.C.. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014; 165:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Terzi L.C., Simpson G.G.. Arabidopsis RNA immunoprecipitation. Plant J. 2009; 59:163–168. [DOI] [PubMed] [Google Scholar]
- 39. Li F., Xu D., Abad J., Li R.. Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2. Virus Genes. 2012; 45:118–125. [DOI] [PubMed] [Google Scholar]
- 40. Harvey J.J., Lewsey M.G., Patel K., Westwood J., Heimstadt S., Carr J.P., Baulcombe D.C.. An antiviral defense role of AGO2 in plants. PLoS One. 2011; 6:e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G.. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007; 14:897–903. [DOI] [PubMed] [Google Scholar]
- 42. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell. Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 45. Brown K.M., Chu C.Y., Rana T.M.. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005; 12:469–470. [DOI] [PubMed] [Google Scholar]
- 46. Webster L.C., Zhang K., Chance B., Ayene I., Culp J.S., Huang W.J., Wu F.Y., Ricciardi R.P.. Conversion of the E1A Cys4 zinc finger to a nonfunctional His2,Cys2 zinc finger by a single point mutation. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:9989–9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trinks D., Rajeswaran R., Shivaprasad P.V., Akbergenov R., Oakeley E.J., Veluthambi K., Hohn T., Pooggin M.M.. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005; 79:2517–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chiba S., Hleibieh K., Delbianco A., Klein E., Ratti C., Ziegler-Graff V., Bouzoubaa S., Gilmer D.. The benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing-suppression activity. Mol. Plant-Microbe Interact. 2013; 26:168–181. [DOI] [PubMed] [Google Scholar]
- 49. Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T.. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009; 10:649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chekulaeva M., Filipowicz W., Parker R.. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009; 15:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T.. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007; 21:2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He X.J., Hsu Y.F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.S., Jin H., Zhu J.K.. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009; 137:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Till S., Ladurner A.G.. RNA Pol IV plays catch with Argonaute 4. Cell. 2007; 131:643–645. [DOI] [PubMed] [Google Scholar]
- 54. Zipprich J.T., Bhattacharyya S., Mathys H., Filipowicz W.. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bednenko J., Noto T., DeSouza L.V., Siu K.W., Pearlman R.E., Mochizuki K., Gorovsky M.A.. Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement. Mol. Cell. Biol. 2009; 29:5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pontier D., Picart C., Roudier F., Garcia D., Lahmy S., Azevedo J., Alart E., Laudie M., Karlowski W.M., Cooke R. et al. . NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell. 2012; 48:121–132. [DOI] [PubMed] [Google Scholar]
- 57. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gamsjaeger R., Liew C.K., Loughlin F.E., Crossley M., Mackay J.P.. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007; 32:63–70. [DOI] [PubMed] [Google Scholar]
- 59. Hall T.M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005; 15:367–373. [DOI] [PubMed] [Google Scholar]
- 60. Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P.. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011; 42:489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mukasa S.B., Rubaihayo P.R., Valkonen J.P.T.. Interactions between a crinivirus, an ipomovirus and a potyvirus in coinfected sweetpotato plants. Plant Pathol. 2006; 55:458–467. [Google Scholar]
- 62. Ma X., Nicole M.C., Meteignier L.V., Hong N., Wang G., Moffett P.. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015; 66:919–932. [DOI] [PubMed] [Google Scholar]
- 63. Garcia-Ruiz H., Carbonell A., Hoyer J.S., Fahlgren N., Gilbert K.B., Takeda A., Giampetruzzi A., Garcia Ruiz M.T., McGinn M.G., Lowery N. et al. . Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 2015; 11:e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.