Abstract
MicroRNAs (miRNAs) are involved in many biological pathways, and detecting miRNAs accurately is critical for diagnosing a variety of diseases including cancer. However, most current methods for miRNA detection require lengthy sample preparation and amplification steps that can bias the results. In addition, lack of specificity and reproducibility give rise to various challenges in detection of circulating miRNAs in biological samples. In this work, we applied the Single Molecule Array (Simoa) technique to develop an ultra-sensitive sandwich assay for direct detection of multiple miRNAs without pre-amplification. We successfully detected miRNAs at femtomolar concentrations (with limits of detection [LODs] ranging from 1 to 30 fM) and high specificity (distinguishing miRNAs with a single nucleotide mismatch). This method was effective against a range of diverse target sequences, suggesting a general approach for miRNA detection. To demonstrate the practical application of this technique, we detected miRNAs in a variety of sample types including human serum and total RNA. The high sensitivity and simple workflow of the Simoa method represent excellent advantages for miRNA-based diagnostics of human diseases.
INTRODUCTION
MicroRNAs (miRNAs) constitute a class of short non-coding regulatory RNAs that play a major role in control of gene expression by repressing protein synthesis at the post-transcriptional level (1). As key components of gene expression regulation, miRNAs are involved in many biological pathways and thus represent a rich source of biological information (2,3). miRNAs can have immense regulatory power—one miRNA can regulate expression of hundreds of messenger RNAs (mRNAs) (4). miRNAs also regulate a broad range of cellular pathways—of particular interest are pathways associated with development and cancer (5–8). Aberrant expression of certain miRNAs, such as miR-21, miR-141 and the let-7 family, has been implicated in various types of cancer (9–11). Due to their associations with cancer, as well as other disease states, miRNAs have attracted wide interest as biomarkers (12,13). Circulating miRNAs with diagnostic utility have been detected in many biological samples including blood, saliva, and urine (13–16).
Detection of miRNAs is challenging for several reasons. miRNA biogenesis is a complex process that includes the presence of precursor (pre) and primary (pri) miRNAs (17) as well as sequence isoforms known as isomiRs, which give rise to miRNAs of varying lengths and sequences (18). Additionally, miRNAs that belong to the same family may have a high degree of homology, posing a challenge for discrimination of miRNAs. Furthermore, miRNAs are present at low levels, comprising ∼0.01% of the total RNA mass in a given sample (19). In many cases, additional extraction and purification steps are necessary to isolate the miRNA fraction, but each additional processing step risks further degradation and loss of the miRNA of interest. Thus, an optimal method for miRNA detection must have minimal sample handling and processing steps, high specificity for differentiating similar miRNAs, and high sensitivity for detecting miRNAs present at low concentrations.
Currently RT-qPCR is the gold-standard method for miRNA detection, largely due to its low limit of detection (LOD). A variety of PCR-based strategies have been developed for detecting the expression levels of miRNAs in clinical samples (20,21). However, the requirement for reverse transcription necessitates an additional step that introduces sample loss and substantial variation into the assay (22). In addition, RT-qPCR suffers from target-based amplification bias and other issues that complicate quantification. To avoid amplification biases, a number of amplification-free ‘direct detection’ techniques have been applied for miRNA detection, such as electrochemical sensors (23,24), and nanotechnology-based approaches (25). Although some of these recently developed direct detection techniques are highly sensitive, it remains challenging to quantitatively measure homologous miRNAs that are present in low concentrations.
Many miRNA detection methods rely on hybridization of the target miRNA to complementary probes. The northern blot has been widely used to detect miRNAs (26,27) but has major drawbacks including low sensitivity and low throughput. Microarrays have been used to overcome limitations in throughput and provide relative expression of thousands of miRNAs in a sample (28). Disadvantages of microarrays include the need for reverse transcription and enzymatic pre-amplification. More recently, next-generation sequencing (NGS) has been widely used to survey miRNAs (29,30). NGS also requires amplification to detect low abundance targets; however, in some cases, molecular barcoding can be implemented to overcome amplification biases (31). NGS methods facilitate miRNA discovery and allow variations in miRNA sequence to be identified. Despite their advantages, most methods described do not provide quantitative measurements with high specificity and sensitivity and minimal sample processing.
To overcome these limitations, we applied the Single Molecule Array (Simoa) platform, a technology capable of detecting single biomolecules with ultra-high sensitivity (32,33). This ultra-sensitive detection technique has been previously demonstrated for a variety of protein biomarkers (34,35) as well as for DNA (36), but has not yet been successfully applied to miRNA detection. In this study, we developed Simoa assays to measure concentrations of several miRNAs with clinical relevance. To validate the method, we detected synthetic miRNAs spiked into human serum samples and endogenous miRNAs in total RNA samples derived from cell lysates. We successfully measured single molecules of miRNA with high sensitivity (LODs in the low femtomolar range) and specificity (distinguishing miRNAs containing a single nucleotide difference). Our method involves a simple workflow and delivers quantitative and highly sensitive results without reverse transcription or target amplification.
MATERIALS AND METHODS
Probe design
Locked nucleic acid (LNA) capture and detection probes were designed to be partially complementary to their intended target miRNA. The length of each probe and the placement of bases within each LNA probe were then selected based on the following criteria: (i) consistent melting temperature for both capture and detection probe; (2) strong predicted binding between each probe and the target sequence; (3) low cross-reactivity between the capture and detection probes (in the absence of target). For each target miRNA, proposed designs for the candidate capture and detection probes were checked using the LNA Oligo Tm Prediction and LNA Oligo Optimizer tools on the Exiqon website (https://www.exiqon.com/oligo-tools). Probe designs were manually iterated to maximize the ratio of predicted target binding/predicted capture-detector binding and to ensure the secondary structure score for capture-detector hybridization remained low (below 20). Considerations for the complementarity and melting temperature of capture and detection probes as applied to the general population of human miRNA are described in Supplementary Figures S4–S6 and Supplementary Table S4.
Covalent coupling of capture probes to paramagnetic microbeads
Custom-made LNA capture probes were purchased from Exiqon. Carboxylated 2.7 μm paramagnetic beads, non-encoded for single-plex assays and dye-encoded (488, 647 and 700 nm) for multiplex assays, were purchased from Quanterix. 5 × 108 beads were washed three times with 0.01 M NaOH. 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (sulfo-NHS) were reconstituted in 2-(N-morpholino)ethanesulfonic acid (MES) buffer (50 mM, pH 6.0) to a final concentration of 50 mg/ml. 100 μl of each of the EDC and sulfo-NHS were added to the beads. The beads were activated on a shaker for 30 min. After activation, the beads were washed once with coupling buffer (1× phosphate buffered saline [PBS], 0.5 M NaCl, 0.1% Tween 20, pH 7.4). 20 nmol of the capture probe were diluted into 200 μl of coupling buffer and added to the beads. The beads were incubated at room temperature with shaking for three hours. The beads were then washed with wash buffer (1× PBS, 1% Tween 20) and incubated in 200 μl of quenching buffer (100 mM Tris–HCl, pH 7.4) with shaking for 45 min. The beads were washed twice with wash buffer and incubated in 200 μl of blocking buffer (1× PBS, 1% bovine serum albumin [BSA]) with shaking for 45 min. The beads were then washed three times with wash buffer and resuspended in a bead storage buffer (50 mM Tris–HCl, 150 mM NaCl, 10 mM ethylenediaminetetraacetic acid [EDTA], 0.1% Tween 20 and 1% BSA). The beads were counted using a Beckman–Coulter multisizer. Two three-plex Simoa assays were developed. The first three-plex assay simultaneously measured let-7a, let-7b and let7c, which were coupled to 488, 647 and 700 nm dye-encoded beads, respectively. The second three-plex assay simultaneously measured miR-21, miR-141 and miR-16 coupled to 488, 647 and 700 nm dye-encoded beads, respectively.
Setup of miRNA Simoa assays
Synthetic target miRNAs were purchased from IDT. Synthetic miRNAs were serially diluted in hybridization buffer (5× saline-sodium citrate [SSC] in diethyl pyrocarbonate [DEPC]-treated water) to desired concentrations. Capture beads (prepared as described above) were diluted to a concentration of 50 000 beads/μl for single-plex assays and 90 000 beads/μl for three-plex assays (with 30 000/μl beads per target) in hybridization buffer. Biotinylated LNA-modified detection probes were purchased from Exiqon and diluted to a concentration of 20 nM in hybridization buffer. 100 μl of sample, 10 μl capture beads and 10 μl of biotinylated detection probes were added to a low binding 96-well plate (Corning, CLS3651) and incubated at 50°C, with the exception that the let-7 multiplex assay was incubated at 55°C, with shaking for 2 h. The beads were then washed eight times with System Wash Buffer 1 (Quanterix), warmed to 50°C, using a microplate washer (BioTek). Streptavidin-β-galactosidase (SβG) Concentrate (Quanterix) was diluted to 200 pM in SβG Diluent (Quanterix). 100 μl of SβG was added to each well and the plate was incubated at room temperature with shaking for 20 min. The beads were then washed eight times with System Wash Buffer 1. The enzyme-labeled beads were then reconstituted in a dilution buffer (1× sodium chloride–sodium phosphate–EDTA [SSPE] and 1.6% dextran sulfate in DEPC-treated water), transferred onto a new 96-well plate (Quanterix) and loaded onto the HD-1 Analyzer (Quanterix) for analysis. All samples were measured in triplicate unless otherwise noted. Resorufin β-d-galactopyranoside (RGP), Wash Buffer 1, Wash Buffer 2 and Simoa Sealing Oil were purchased from Quanterix and loaded onto the Simoa HD-1 Analyzer based on the manufacturer's instructions. In the HD-1 Analyzer software, the assay was defined based on the ‘Acute care assay neat 2.0’ with an incubation time of one cadence.
Direct detection of miRNA in human serum using Simoa assays
Healthy human serum samples were purchased from BioReclamationIVT. Sodium dodecyl sulfate (SDS, final w/v 2%) and Proteinase K (final 0.16 U/ml) (New England Biolabs) were added to the serum samples. The serum samples were vortexed and incubated at room temperature for 15 min. The serum samples were then heated to 90°C for 2 min, diluted in hybridization buffer and spiked with synthetic miRNA to desired concentrations.
Detection of microRNAs in total RNA samples using Simoa assays
Human Lung Total RNA (ThermoFisher, AM7968) was serially diluted 10-fold in hybridization buffer and tested using the three-plex Simoa assay for miR-21, miR-141 and miR-16. To determine concentrations of miRNAs in these samples, a calibration curve for each target miRNA was fit to a 4PL nonlinear regression with a 1/y2 weighting factor, and unknown values were interpolated using the Simoa HD-1 Analyzer Software (Quanterix).
Detection of microRNAs in total RNA using qPCR
The following RT-qPCR reagents were purchased from ThermoFisher: qPCR Taqman assays for miR-21, miR-141 and miR-16 (4440886), TaqMan Universal Master Mix II (4440043), and TaqMan MicroRNA Reverse Transcription Kit (4366596). Reverse transcription of known standards and four dilutions of Human Lung Total RNA, corresponding to 100, 10, 1 and 0.1 ng of total RNA, was performed in triplicate. qPCR was performed based on the manufacturer's instructions using a CFX96 real-time PCR system and CFX Manager Software for data analysis (Biorad).
RESULTS
Simoa assay with a bead-based sandwich protocol
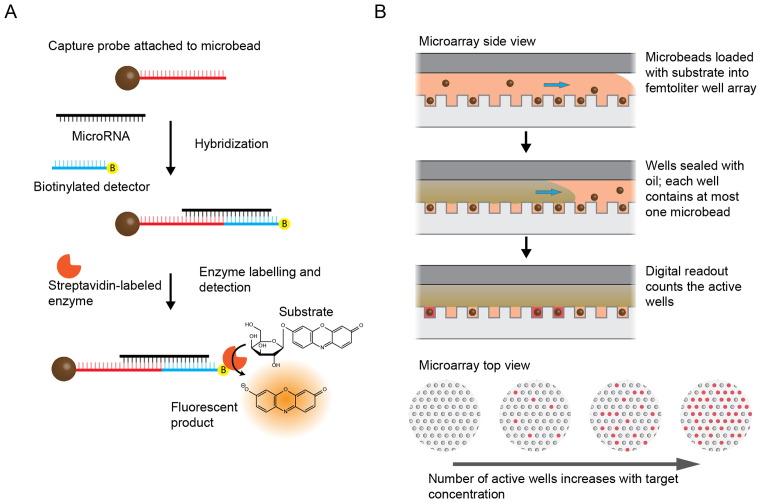
We applied the Simoa assay to miRNA detection by developing a bead-based sandwich protocol (Figure 1A). To increase hybridization specificity, we used LNA-modified capture and detection probes with sequences complementary to either 11 or 12 bases of the target miRNA (Supplementary Table S1). Before performing the Simoa assay, LNA-modified capture probes specific to a target miRNA were covalently coupled to paramagnetic microbeads. A sample containing the target miRNA was incubated with the capture probe-coupled microbeads and biotinylated detection probes, forming a sandwich complex. The beads were then washed to remove unbound miRNA. The beads labelled with both target miRNA and biotinylated detection probes were then labelled with an enzyme, SβG, via biotin–streptavidin interaction and detected by enzymatic readout in the Simoa platform (Figure 1B). The signal from the assay was measured in units of average enzyme per bead (AEB), as previously described (33).
Figure 1.
Schematic of miRNA detection using Simoa. (A) Overview of the sandwich protocol. Capture probes were covalently coupled to microbeads, and then incubated with target miRNA and biotinylated detection probes to form a sandwich complex. The beads were washed and incubated with SβG and RGP (enzyme and substrate) to produce a fluorescent product. (B) Detection was performed in the Simoa format as previously reported (32). As shown in the side view, following hybridization, microbeads suspended in fluorogenic substrate were loaded onto an array of femtoliter-size wells. After loading, the wells were sealed with oil, resulting in an array of isolated reaction chambers each of which contained either zero or one bead. If the enzyme-labeled complex was present on a bead, it generated a fluorescent product resulting in a detectable fluorescent signal. The array was imaged and analyzed to determine the total number of beads, and the number of ‘active’ wells was counted to calculate the average enzyme per bead (AEB). As shown in the top view, the number of active wells increased with increasing target concentration.
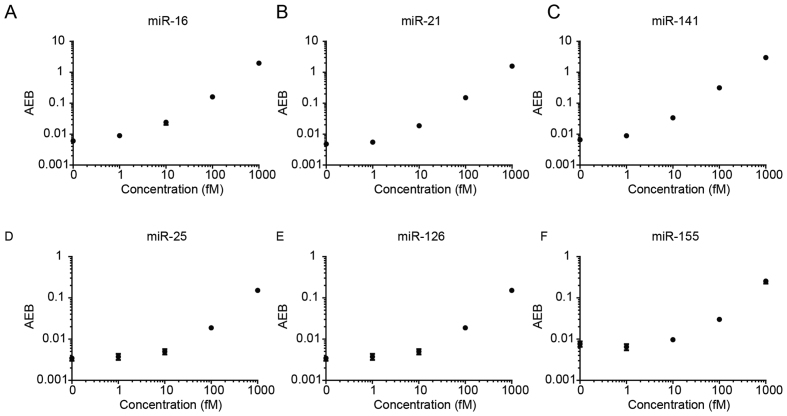
To test the performance of the assay, we selected several target miRNAs that have been previously associated with cancer. These targets include miR-16, miR-21, miR-141, miR-25, miR-126 and miR-155. The melting temperatures of these miRNAs are representative of the general population of miRNAs (Supplementary Table S4). Known concentrations of each target miRNA were then measured using the singleplex assays. As shown in Figure 2, we obtained similar performance for all assays, with limits of detection of 1–30 fM.
Figure 2.
Calibration curves for various miRNAs. LODs were calculated as three standard deviations above the blank for each assay: (A) miR-16 (LOD 0.76 fM), (B) miR-21 (LOD 1.60 fM), (C) miR-141 (LOD 0.58 fM), (D) miR-25 (LOD 27.34 fM), (E) miR-126 (LOD 8.94 fM) and (F) miR-155 (LOD 4.37 fM).
Multiplexed detection of miRNAs
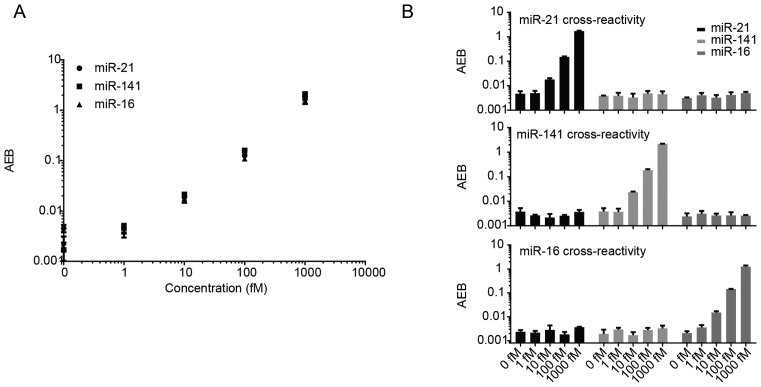
Simultaneous detection of multiple different target miRNAs in a single sample increases throughput and requires less sample volume compared to detection of each target individually. When making multiplexed measurements, the presence of multiple target miRNAs in a sample can potentially introduce cross-reactivity that limits the practical utility of an assay. We chose three widely-used miRNA biomarkers, miR-16, miR-21 and miR-141, to test our direct detection approach in a multiplex format. To enable multiplexing, we used paramagnetic beads labelled with different fluorescent dyes to produce distinct bead subpopulations. Each subpopulation of beads was then further modified with capture probes for a specific miRNA. After incubation with three specific biotinylated detection probes, we measured three miRNAs simultaneously with an average limit of detection of ∼10 fM (Figure 3A), demonstrating that multiplexing does not compromise sensitivity. To assess specificity, we tested the multiplex assay with increasing concentrations of each target miRNA individually (Figure 3B). Results showed this multiplex assay demonstrated high specificity for its intended targets, with minimal off-target signals even at high concentrations.
Figure 3.
Three-plex Simoa assay for miR-21, miR-141 and miR-16, and cross reactivity profiles. (A) Multiplex assay for the simultaneous direct detection of three different miRNAs. (B–D) Cross-reactivity of the multiplex assay in the presence of (B) only miR-21, (C) only miR-141 and (D) only miR-16.
Multiplexed detection of homologous miRNAs with a single nucleotide mismatch
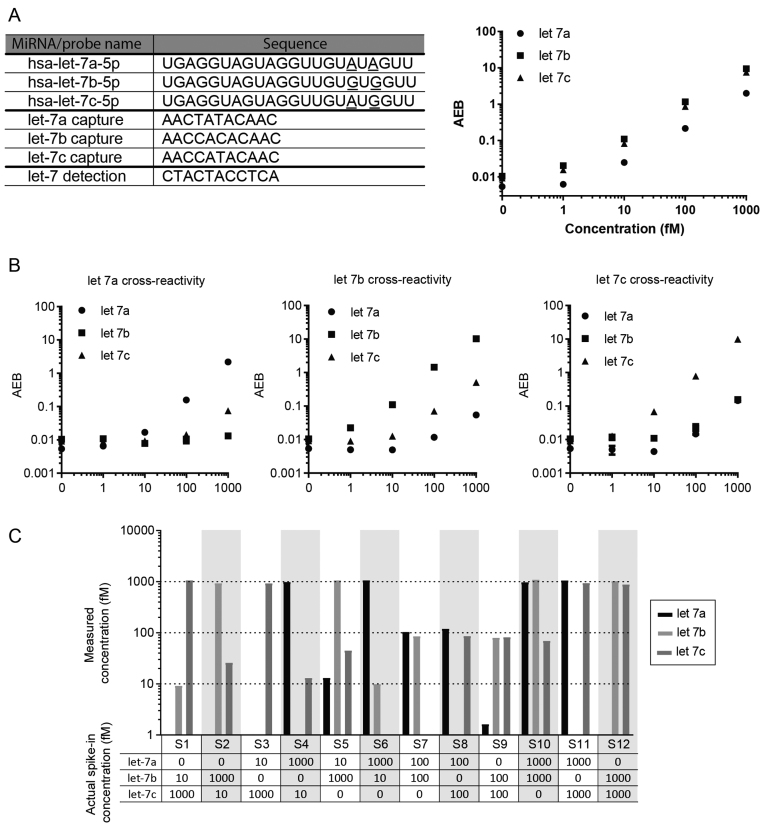
A major challenge in miRNA detection is distinguishing between miRNAs with very similar sequences. The challenge of cross-reactivity becomes especially pronounced when performing multiplexed measurements, as the number of probes and potential off-targets increases. To evaluate the specificity of the Simoa direct detection approach against highly homologous miRNAs, we tested miRNAs that differed by only one or two nucleotides. As representative target miRNAs with highly similar sequences, we chose three members from the human let-7 family: let-7a, let-7b and let-7c. We designed 20 different capture probes, specific to let-7c, with varying melting temperatures and number of LNA bases (Supplementary Table S2). We tested the selectivity of each probe against increasing concentrations (0, 1, 10 and 100 fM) of let-7c, and also tested the specificity of these capture probes against 100 fM of let-7b, which differs by a single nucleotide (Supplementary Figure S1). This screen was performed at room temperature and four hours of incubation. To identify probes with low cross-reactivity, we compared the on-target to off-target ratio (signal at 100 fM of let-7c over signal of 100 fM of let-7b, Supplementary Figure S2). Notably, probe 1, which does not contain any LNA bases, was the least cross-reactive. We then compared the signal to noise ratio (signal at 100 fM of let7c over signal of the blank) and selected probes 12, 17, 18 and 19 for further optimization. We tested the effects of incubation time and temperature on assay performance. For these four probes, we observed that higher specificity was obtained at 60°C, while the assay signal was substantially lower at 65°C (closer to the melting temperature of the probes). We chose to use probe 12 for further multiplexed assay development. We also note that we did not observe a clear correlation between the performance of the Simoa assay, the number of LNA residues, and the predicted melting temperature of the probes.
Based on these results, we developed a three-plex Simoa assay to measure let-7a, let-7b and let-7c simultaneously (Figure 4A). For the let-7a and let-7b probes, we chose a probe design in which the LNA bases were placed in the same positions as the let-7c probe. We spiked in varying concentrations of let-7a, let-7b and let-7c, in the absence of the other two targets, to determine the cross-reactivity of each target miRNA against the different capture probes (Figure 4B). We then tested mixed samples of varying concentrations of let-7a, let-7b and let-7c (Figure 4C). The initial measurements of these mixed samples were not accurate due to cross-reactivity (Supplementary Figure S3). To compensate for the cross-reactivity of off-target miRNAs, we applied a correction to the signal by fitting the corresponding cross-reactivity curve (Supplementary Figure S3 and Table S3). The resulting measurements of target miRNAs were in good agreement with the spiked-in concentrations. In samples S5 and S10, measurements of let-7c were substantially higher than the actual spike-in concentrations even after correcting for cross-reactivity. Nevertheless, we were able to accurately quantify the concentrations of let-7a, let-7b and let-7c in most of these samples.
Figure 4.
Three-plex Simoa assay for let-7a, let-7b and let-7c and cross-reactivity profiles. (A) Target, capture, and detection probe sequences used in this assay, and multiplex assay results for the direct detection of three different miRNAs simultaneously. (B–D) Cross-reactivity of the multiplex assay in the presence of (B) only let-7a, (C) only let-7b and (D) only let-7c. (E) Detection of let-7a, let-7b and let-7c in 12 samples using the three-plex Simoa assay. Actual spike-in concentrations for each of let-7a, let-7b and let-7c in the samples (S1–S12) are given in the table, and measured concentrations (after compensating for off-target signal as shown in Supplementary Figure S3 and Supplementary Table S3) are indicated in the plot.
Measuring miRNA concentrations spiked into human serum
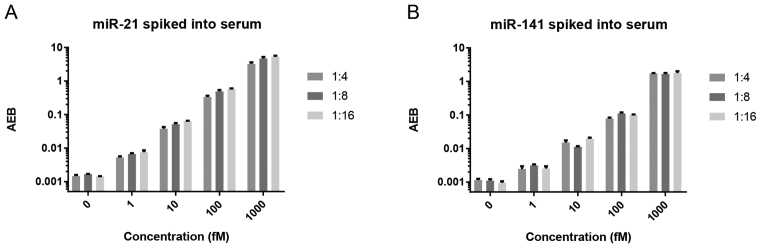
Detection of circulating miRNAs in serum is a promising strategy for minimally-invasive diagnostics. While measurements of miRNAs in serum often involve a preliminary RNA isolation step, we explored the potential for detecting miRNAs directly in serum without RNA isolation. One major challenge for direct detection of miRNAs in serum is the influence of matrix effects that may interfere with the assay. To evaluate the use of the Simoa direct detection assay with serum samples, we spiked known concentrations of miR-21 and miR-141 into healthy human serum. When miRNA was spiked directly into untreated serum, the miRNA was undetectable, presumably due to degradation (data not shown). However, when we pre-treated the serum with Proteinase K and SDS, followed by heating, the spiked-in miRNAs were detectable, as previously reported (37). As shown in Figure 5, further dilutions of serum in hybridization buffer had a relatively small effect on the measurements. These results suggest that, at least for dilution factors below 1:4, matrix effects from the serum do not interfere substantially with the detection process. Notably, 1 fM of spiked-in miRNA was detectable in the serum matrix.
Figure 5.
Direct detection of (A) miR-21 and (B) miR-141 spiked into human serum at varying dilutions.
miRNA detection in total RNA
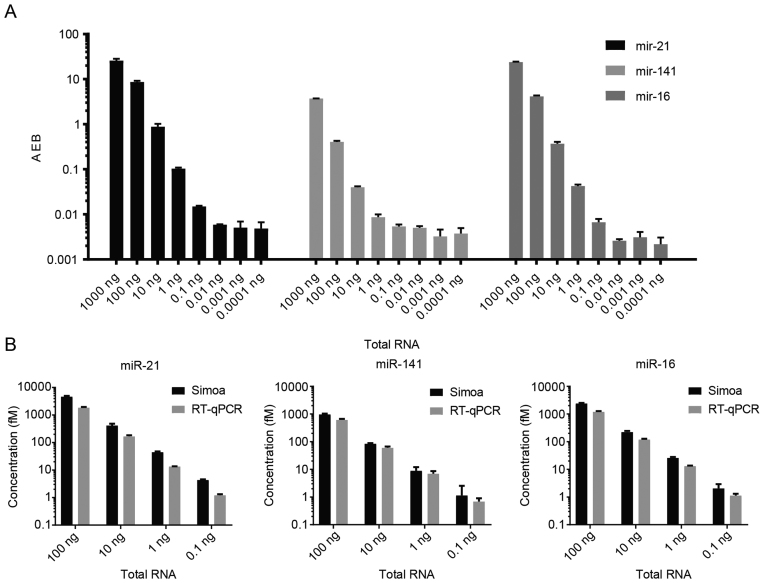
Accurate quantification of miRNAs in total RNA is useful for applications in both fundamental biology and clinical diagnostics. To explore the utility of the Simoa assay for detecting miRNAs in total RNA, we tested a commercially purchased sample of total RNA isolated from cell lysates. We serially diluted each of these samples ten-fold and measured miR-21, miR-141 and miR-16 using the three-plex Simoa assay. As shown in Figure 6A, endogenous miRNAs were detectable and readily quantifiable, exhibiting a linear trend that corresponded to the decreasing concentration of total RNA. The miRNAs were measured over a wide range, which spanned four orders of magnitude, and with a low LOD of ∼10 fM, corresponding to 0.1 ng of total RNA. As a control, we also tested a subset of samples against the beads from the let-7 multiplex assay to confirm that the signal was not due to non-specific binding (data not shown). To confirm the accuracy of the results, we used RT-qPCR against a set of known standards for use as a calibration curve and a subset of the samples that were previously tested using the Simoa assay. As shown in Figure 6B, the RT-qPCR results were in good agreement with the Simoa measurements for both relative and absolute quantification.
Figure 6.
Direct detection of miR-21, miR-141 and miR-16 in a total RNA sample. Samples were serially diluted and concentrations were measured using the three-plex Simoa assay. (A) AEB values of the samples at varying amounts of total RNA. (B) Concentrations of miR-21, miR-141 and miR-16 in varying amounts of total RNA measured using RT-qPCR and the three-plex Simoa assay.
DISCUSSION
The emergence of miRNAs as potential biomarkers for cancer and other diseases necessitates new approaches to detect miRNAs with high sensitivity and specificity. RT-qPCR remains the gold standard method for nucleic acid detection, but suffers from target amplification bias, sample loss due to reverse transcription, and lack of multiplexing capabilities. In addition, miRNAs represent particularly challenging targets for RT-qPCR due to their short sequence length, requiring nonstandard primers. Direct detection of miRNAs is a promising alternative to RT-qPCR but it is often difficult to detect low concentrations of miRNAs without target amplification. In this study, we developed a direct detection assay, based on the Simoa technology, which is capable of extremely high-sensitivity measurements of miRNAs. The Simoa platform has been used to detect protein biomarkers at subfemtomolar concentrations but the method reported here is the first implementation of Simoa for miRNA detection. There are several advantages to the Simoa format, including high sensitivity, specificity, minimal processing steps and multiplexing capabilities.
The Simoa direct detection approach can be used to measure a wide range of miRNAs. Several considerations must be taken into account when designing the LNA-modified capture and detection probes. Flexibility in probe design is limited due to the short length of miRNAs and thus the complementary sequence of the capture and detection probe pairs is pre-determined by the sequence of the target miRNA. LNA-modified probes can be used to increase specificity and have frequently been used for specific hybridization and multiplexed detection of miRNAs (26,27,38). A general LNA probe design strategy requires consideration of self-complementarity and cross-hybridization with other probes, which must be avoided to reduce the background signal. These considerations are particularly important in multiplexed assays, in which the number of probes and potential cross-reactive off targets increases. Additionally, it is important to ensure that the melting temperature for both capture and detection probes is similar. Due to the short length of miRNAs, it is often not possible to obtain similar melting temperatures for the capture and detection probes. We have demonstrated high sensitivity of our Simoa direct detection approach even when the melting temperatures of the capture and detection probes differed by over 30°C, as exemplified by the miR-155 assay (Supplemental Table S4). Additional information on sequence complementarity and melting temperatures for miRNA probes is provided in Supplementary Figures S4–S6.
Another challenge with miRNA detection is the ability to distinguish between homologous miRNAs. Sequence analysis of the general population of mature human miRNAs against the capture and detection probes used in this study revealed that over 96% of the probes contain three or more mismatches (Supplemental Figure S4). Additionally, in our Simoa direct detection approach, both capture and detection probes must bind to the target miRNA to produce a signal. Consequently, we do not expect cross-reactivity from off-target miRNAs to have substantial effects when using the Simoa approach to measure miRNAs. Some miRNA families, such as the let-7 family, have a high degree of homology; in these cases, the signal arising from cross-hybridization must be accounted for (Figure 4 and Supplemental Figure S3). It may also be challenging to detect IsomiRs, a class of miRNAs with minor sequence variations including substitutions, deletions, insertions, and 5′ or 3′ end cleavage, using the described method. Nevertheless, using the Simoa approach, we successfully measured highly homologous miRNAs that differed by only one or two nucleotides.
We sought to apply the Simoa-based direct detection approach to measure miRNAs in serum without a separate isolation step. We observed that spiked-in miRNA was undetectable in untreated serum; however, when the serum was pre-treated, 1 fM of spiked miRNAs was detectable (Figure 5). This result is consistent with previous findings that show exogenous miRNAs are undetectable upon addition to serum or blood, while endogenous miRNAs are stable (37). We also successfully demonstrated detection of three different miRNAs simultaneously in samples containing as little as 0.1 ng total RNA. When we compared the accuracy of our direct detection approach to the current gold standard tool, RT-qPCR, the results were in good agreement (Figure 6). Thus, our Simoa direct detection approach can provide highly sensitive, multiplexed, and accurate quantification of miRNAs.
The direct hybridization approach described here does not require pre-labelling, reverse transcription, or amplification steps. The total time required for the assay is ∼5 h, including about 1.5 h of ‘hands on’ time. Furthermore, multiplexing capabilities enable measurements of several miRNAs simultaneously and thus enhance efficiency. The simple and automated nature of this assay suggests that it can easily scale up to higher throughput for routine testing. Additionally, the high sensitivity and specificity of the Simoa direct detection approach make it a promising tool for miRNA detection.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Albert Tai of the Tufts University genomics core facility for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US Department of Defense BC100510 [W81XWH-11-1-0814]; Defense Advanced Research Projects Agency (DARPA) [HR0011-12-2-0001].
Conflict of interest statement. D.R.W. is the scientific founder and a board member of Quanterix Corp., a company that is currently commercializing a version of the Single Molecule Array (Simoa) technology.
REFERENCES
- 1. Guo H., Ingolia N.T., Weissman J.S., Bartel D.P.. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNA target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P.. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 6. Lin S., Gregory R.I.. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015; 15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes J., Peruzzi P.P., Lawler S.. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014; 20:460–469. [DOI] [PubMed] [Google Scholar]
- 8. Zhang B., Pan X., Cobb G.P., Anderson T.A.. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007; 302:1–12. [DOI] [PubMed] [Google Scholar]
- 9. Asangani I.A., Rasheed S.A.K., Nikolova D.A, Leupold J.H., Colburn N.H., Post S., Allgayer H.. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008; 27:2128–2136. [DOI] [PubMed] [Google Scholar]
- 10. Cheng H., Zhang L., Cogdell D.E., Zheng H., Schetter A.J., Nykter M., Harris C.C., Chen K., Hamilton S.R., Zhang W.. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011; 6:e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004; 64:3753–3756. [DOI] [PubMed] [Google Scholar]
- 12. Hayes J., Peruzzi P.P., Lawler S.. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014; 20:460–469. [DOI] [PubMed] [Google Scholar]
- 13. Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mar-Aguilar F., Mendoza-Ramírez J.A., Malagón-Santiago I., Espino-Silva P.K., Santuario-Facio S.K., Ruiz-Flores P., Rodríguez-Padilla C., Reséndez-Pérez D.. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers. 2013; 34:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18:997–1006. [DOI] [PubMed] [Google Scholar]
- 16. Kosaka N., Iguchi H., Ochiya T.. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010; 101:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winter J., Jung S., Keller S., Gregory R.I., Diederichs S.. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009; 11:228–234. [DOI] [PubMed] [Google Scholar]
- 18. Neilsen C.T., Goodall G.J., Bracken C.P.. IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012; 28:544–549. [DOI] [PubMed] [Google Scholar]
- 19. Kaufman E.J., Miska E.A.. The microRNAs of Caenorhabditis elegans. Semin. Cell Dev. Biol. 2010; 21:728–737. [DOI] [PubMed] [Google Scholar]
- 20. Benes V., Castoldi M.. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010; 50:244–249. [DOI] [PubMed] [Google Scholar]
- 21. Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 23. Yang C., Shi K., Dou B., Xiang Y., Chai Y., Yuan R.. In situ DNA-templated synthesis of silver nanoclusters for ultrasensitive and label-free electrochemical detection of MicroRNA. ACS Appl. Mater. Interfaces. 2015; 7:1188–1193. [DOI] [PubMed] [Google Scholar]
- 24. Lu N., Gao A., Dai P., Song S., Fan C., Wang Y., Li T.. CMOS-compatible silicon nanowire field-effect transistors for ultrasensitive and label-free microRNAs sensing. Small. 2014; 10:2022–2028. [DOI] [PubMed] [Google Scholar]
- 25. Husale S., Persson H.H.J., Sahin O.. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature. 2009; 462:1075–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Várallyay E., Burgyán J., Havelda Z.. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008; 3:190–196. [DOI] [PubMed] [Google Scholar]
- 27. Válóczi A., Hornyik C., Varga N., Burgyán J., Kauppinen S., Havelda Z.. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004; 32:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu C.-G., Calin G.A., Volinia S., Croce C.M.. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008; 3:563–578. [DOI] [PubMed] [Google Scholar]
- 29. Yang Q., Lin J., Liu M., Li R., Tian B., Zhang X., Xu B., Liu M., Zhang X., Li Y. et al. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016; 2:e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tam S., de Borja R., Tsao M.-S., McPherson J.D.. Robust global microRNA expression profiling using next-generation sequencing technologies. Lab. Investig. 2014; 94:350–358. [DOI] [PubMed] [Google Scholar]
- 31. Alon S., Vigneault F., Eminaga S., Christodoulou D.C., Seidman J.G., Church G.M., Eisenberg E.. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011; 21:1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010; 28:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rissin D.M., Fournier D.R., Piech T., Kan C.W., Campbell T.G., Song L., Chang L., Rivnak A.J., Patel P.P., Provuncher G.K. et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal. Chem. 2011; 83:2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu D., Milutinovic M.D., Walt D.R.. Single molecule array (Simoa) assay with optimal antibody pairs for cytokine detection in human serum samples. Analyst. 2015; 140:6277–6282. [DOI] [PubMed] [Google Scholar]
- 35. Song L., Hanlon D.W., Chang L., Provuncher G.K., Kan C.W., Campbell T.G., Fournier D.R., Ferrell E.P., Rivnak A.J., Pink B.A. et al. Single molecule measurements of tumor necrosis factor alpha and interleukin-6 in the plasma of patients with Crohn's disease. J. Immunol. Methods. 2011; 372:177–186. [DOI] [PubMed] [Google Scholar]
- 36. Song L., Shan D., Zhao M., Pink B.A., Minnehan K.A., York L., Gardel M., Sullivan S., Phillips A.F., Hayman R.B. et al. Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal. Chem. 2013; 85:1932–1939. [DOI] [PubMed] [Google Scholar]
- 37. Johnson-Buck A., Su X., Giraldez M.D., Zhao M., Tewari M., Walter N.G.. Kinetic fingerprinting to identify and count single nucleic acids. Nat. Biotechnol. 2015; 33:730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee J.M., Jung Y.. Two-temperature hybridization for microarray detection of label-free MicroRNAs with attomole detection and superior specificity. Angew. Chemie - Int. Ed. 2011; 50:12487–12490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.