Abstract
New transcripts generated by RNA polymerase II (RNAPII) are generally processed in order to form mature mRNAs. Two key processing steps include a precise cleavage within the 3′ end of the pre-mRNA, and the subsequent polymerization of adenosines to produce the poly(A) tail. In yeast, these two functions are performed by a large multi-subunit complex that includes the Cleavage Factor IA (CF IA). The four proteins Pcf11, Clp1, Rna14 and Rna15 constitute the yeast CF IA, and of these, Pcf11 is structurally the least characterized. Here, we provide evidence for the binding of two Zn2+ atoms to Pcf11, bound to separate zinc-binding domains located on each side of the Clp1 recognition region. Additional structural characterization of the second zinc-binding domain shows that it forms an unusual zinc finger fold. We further demonstrate that the two domains are not mandatory for CF IA assembly nor RNA polymerase II transcription termination, but are rather involved to different extents in the pre-mRNA 3′-end processing mechanism. Our data thus contribute to a more complete understanding of the architecture and function of Pcf11 and its role within the yeast CF IA complex.
INTRODUCTION
Polyadenylation of mRNA 3′-ends is a co-transcriptional event that occurs for nearly all eukaryotic mRNA with the exception of histone mRNAs (1). This process relies on a two-step mechanism starting with cleavage of the pre-mRNA in the 3′-untranslated region (3′-UTR) (2–4). This first step is immediately followed by the polymerization of adenosines at the 3′-hydroxyl end of the upstream cleavage product, while the downstream fragment is degraded by Rat1 (5). In yeast, the polyadenylation machinery comprises two large multi-protein complexes: the Cleavage/polyadenylation Factor IA (CF IA) (6,7) and the Cleavage and Polyadenylation Factor (CPF). Additional factors are also required for the in vitro process such as Hrp1/Nab4 (6,8) and the poly(A)-binding proteins Pab1 and Nab2 (7,9–11).
The CF IA complex consists of the four proteins Rna14, Rna15, Pcf11 and Clp1. Each of these proteins consists of distinct domains that contribute to the function of CF IA, including assembly of the complex and interaction with other components of the processing machinery or target pre-mRNA. Investigation of these structured domains has directly led to insight into CF IA architecture and function. Solution studies and crystal structures of the Kluyveromyces lactis Rna14, as well as the orthologue CstF-77 from Encephalitozoon cuniculi and Mus musculus, have shown that Rna14 and CstF-77 self-associate into tight homodimers (12–15). The C-terminal region of Rna14 also forms a heteromeric complex with Rna15 as revealed by the solution structure of the Rna14–Rna15 minimal heterodimer (16) and the structure of the Rna14–Rna15 sub-complex from K. lactis (15). A three-helix bundle at the C-terminus of Rna15 has been shown to contact Pcf11 (17), and a short region in Pcf11 mediates association with Clp1 as observed in the structure of a Clp1–Pcf11 minimal complex (18,19). These contacts contribute to the correct assembly of CF IA, although they also dictate a more complicated stoichiometry versus a simple heterotetramer (20). Recent studies have proposed an overall model for CF IA (21), with a stoichiometry of 2:2:1:1 or 2:1:1:1 (21,22) respectively for Rna14:Rna15:Clp1:Pcf11, but the functional basis for such an asymmetry has not yet been described.
Protein domains within CF IA also enable interaction with the target pre-mRNA and components of the transcription and processing machinery. Molecular characterization of the RNA recognition motif (RRM) domain of Rna15, and the orthologue CstF-64, reveal aspects of pre-mRNA binding (23). Increased RNA sequence specificity has also been demonstrated with structural details of the auxiliary subunit Hrp1 (also known as CF IB or Nab4), and in particular within a Hrp1–Rna15–RNA ternary complex (24–26). A connection to the largest subunit of RNA polymerase II, Rpb1 is mediated by the C-terminal interacting domain (CID) of Pcf11 that binds to the Rpb1 C-terminal YSPTSPS heptamer repeats with phosphorylation on the serine in position 2 (27,28).
Despite the characterization of the CF IA domains described above, additional folded domains remain to be studied. This is most evident for Pcf11, for which only the N-terminal CID, the short Clp1-interacting region and recently, a zinc-binding domain (29) are described at the atomic level. In this study, we have analysed additional regions of Pcf11 and demonstrated that Pcf11 has two zinc-binding domains. Interestingly, mutations of these domains affect Pcf11 function to different extents. Mutation of the C-terminal zinc-binding motif has a dramatic impact on both yeast viability and pre-mRNA 3′-end processing, whereas mutations in the upstream zinc-binding domain has a small but noticeable effect on cell growth and 3′-end formation, suggesting different functions of these potentially similar motifs. The entire zinc finger region of Pcf11 is also capable of a non-specific interaction with RNA but only in the absence of Clp1. The C-terminal zinc-binding domain, which we find essential for pre-mRNA 3′-end processing, was also produced in isolation and was shown to contain flexible residues around an unusual zinc-binding fold. Altogether, this study provides a refined description of Pcf11 organization and gives novel insights into CF IA in mRNA 3′-end maturation.
MATERIALS AND METHODS
Cloning and rCF IA complex expression
Each full-length protein was obtained by PCR on yeast Saccharomyces cerevisiae genome and cloned into modified versions of pET-15b, pET28b, pCDF and pLysS bacterial expression plasmids (30,31). Integrity of each clone was verified by sequencing. For co-expression, BL21(DE3) (Novagen) were transformed with the relevant combination of plasmids and plated on Petri dishes with half of the recommended antibiotic concentration. Mini-cultures were grown in 10 ml of LB at 37°C with antibiotics and protein expression was induced by addition of a final concentration of 1 mM IPTG and overnight incubation at 15°C. Cells were harvested by centrifugation and lysed by sonication with buffer containing 1.5x PBS, 1 mM Mg(OAc)2, 0.1% NP-40, 20 mM imidazole, 10% (w/v) glycerol. After centrifugation, the supernatant was incubated with His-affinity resin (Sigma) for 30 min at 4°C. The resin was extensively washed and bound proteins were analysed by SDS-PAGE after addition of loading buffer.
Pull-down assays
After overnight induction of 10 ml culture with 0.1 mM IPTG, harvested cells resuspended in lysis buffer (1.5x PBS, 1 mM Mg(OAc)2, 0.1% NP-40, 20 mM Imidazole, 10% (w/v) glycerol) were lysed by 10 s sonication at 3 W. Crude extract was then centrifuged and soluble fraction incubated 30 min with 30 μl of His-Affinity Cobalt resin. After three washing steps with 1 ml of lysis buffer, beads were eluted with 3 volumes of lysis buffer supplemented with 250 mM imidazole and one-third of the sample was loaded on a SDS-polyacrylamide gel.
rCF IA complex purification
For large-scale protein purification, the plasmids expressing Pcf11 deleted from the residues 234 to 253 fused to a N-terminal His-tag, as well as a tri-cistronic construct encoding Rna15, Rna14 and Clp1 were transformed into Escherichia coli BL21 Rosetta 2 (DE3) cells. Two liters of Terrific Broth (TB) were inoculated with the relevant overnight pre-culture and grown to an OD of 1.5 and induced by addition of a final concentration of 1 mM IPTG and overnight incubation at 15°C. After cell harvesting, the cell pellet was resuspended in a buffer containing 25 mM Tris–HCl pH 7.5, 150 mM NaCl and lysed by three passes into an EmulsiFlex C3 (Avestin). The crude extract was centrifuged at 50 000 g for 1 h at 4°C. The supernatant was incubated with His-affinity resin (Sigma) and loaded onto an AK 16 column (GE Healthcare). Contaminants were removed by extensive wash with buffer A containing 25 mM Tris–HCl pH 7.5, 150 mM NaCl. Proteins were eluted from the column with a gradient of buffer A supplemented with 250 mM imidazole. The eluted proteins were then loaded onto a HP-Heparin 5 ml (GE Healthcare) and separated by a gradient of NaCl up to 500 mM in 25 mM Tris–HCl pH 7.5. The pool of fractions was concentrated and loaded onto a Superdex-200pg column (GE Healthcare) equilibrated in 25 mM Tris–HCl pH 7.5, 250 mM NaCl, 1 mM DTT.
Zinc-binding domains purification
The DNA sequence coding for residues 530–626 or 403–626 of Pcf11 were cloned into a modified pET-15b plasmid allowing the production of an N-terminally His6-GST-tagged proteins. The plasmids were transformed into E. coli BL21 (DE3). For large-scale protein purification, 1 L of TB was inoculated with the relevant overnight pre-culture and grown up to an OD of 1.5. After cell harvesting, the cell pellet was resuspended in a buffer containing 25 mM Tris–HCl pH 7.5, 150 mM and lysed by three passes into an EmulsiFlex C3 (Avestin). The crude extract was centrifuged at 50 000 g for 1 h at 4°C. The supernatant was incubated with His-affinity resin (Sigma) and loaded onto an AK 16 column (GE Healthcare). Contaminants were removed by extensive washes with a buffer A containing 25 mM Tris–HCl pH 7.5, 250 mM NaCl. Proteins were eluted from the column with a gradient of imidazole up to 250 mM supplemented in buffer A. The eluted proteins were cleaved with Tev protease to remove the His-GST tag over-night at 4°C. Then a Hi-prep 26/10 Desalting column was used to exchange the protein buffer into the Buffer A. The protein was then loaded on a His-affinity column to eliminate the His-GST tag and Tev protease. The zinc-finger domain was further purified using a High-resolution Superdex S-75pg gel-filtration column (GE Healthcare) equilibrated in 50 mM Tris (pH 7.5), 150 mM NaCl supplemented with 50 μM Zn(OAc)2. The purified protein samples were concentrated to 2.6 mg ml−1, frozen and stored at –80°C. The concentration was estimated from absorbance at 280 nm assuming absorption of 1.54 cm−1 for a 1 mg ml−1 solution as calculated from the construct sequence.
NMR spectroscopy
In addition to protein expressed in LB media at natural abundance, GST-tagged Pcf11 (530–626) was also expressed by using Rosetta2 (DE3) cells in M9 minimal media supplemented with 1 g/l [15N]-ammonium chloride, 2 g/l [13C]-glucose or with 2 g/l [10%-13C]-glucose and purified as with the natural abundance protein. Purified samples contained 100–360 μM untagged protein in 50 mM Tris (pH 7.5), 150 mM NaCl, 50 μM Zn(OAc)2 with either 10% or 99% (v/v) 2H2O. NMR spectra were acquired at 20°C on a 700 MHz or 800 MHz Bruker Avance III spectrometer, equipped with a room temperature or cryogenic triple resonance gradient probe, respectively. Data acquisition used TopSpin versions 2.1 and 3.2. Processing used NMRPipe/NMRDraw (32) and data analysis used Sparky (T.D. Goddard & D. G. Kneller, University of California, San Francisco, USA). Chemical shift assignment of the backbone 1H, 13C and 15N nuclei of Pcf11 for residues 548–603 relied on 2D 1H,15N-HSQC, 3D HNCO, 3D HNCACO, 3D HNCA, 3D CBCA(CO)NH, 3D HNCACB and 3D HNHA spectra. Aliphatic sidechain assignment used 2D 13C-HSQC, 3D (H)CCONH-TOCSY (14 ms mixing time), H(C)CH-TOCSY (14 ms mixing time) (H)CCH-TOCSY (14 ms mixing time). Stereospecific assignment of the γ1/γ2 methyl groups of valine and the δ1/δ2 methyl groups of leucine used the sample prepared with [10%-13C]-glucose (33). Aromatic sidechain assignment used 2D 1H,13C-HSQC and 1H,1H-NOESY (150 ms mixing time) spectra, with an additional long-range 1H,15N-HMBC spectrum to determine the protonation state of the δ1 and ε2 nitrogen atoms of the two histidine residues. The 1H, 13C and 15N chemical shift assignments have been deposited in the Biomolecular Magnetic Resonance Data Bank (http://bmrb.wisc.edu) under accession code 34061.
Structure calculation
The backbone 1HN, 1Hα, 13Cα, 13Cβ, 13C’ and 15NH chemical shift values of Pcf11 (530–626) provided Ψ and Φ angular restraints for 51 residues by using TALOS-N (34). In addition, 30 χ1 angular restraints were obtained. Two addition χ angle restraints for Val552 and Ile588 were based on analysis of 13C side chain resonances and the program Sider (35,36). Distance restraints were obtained from 2D 1H,1H-NOESY (150 ms mixing time), 3D 1H,1H-NOESY-13C-HMQC (150 ms mixing time) and 3D 1H,15N-HSQC–1H-NOESY (150 ms mixing time) with an initial set of 976, 701 and 338 crosspeaks, respectively. The final list of unambiguous restraints included 353 intraresidue, 160 sequential, 54 medium and 332 long range distances, with an additional list of 271 ambiguous distance restraints. The four distance and 13 dihedral zinc-binding geometry constraints were excluded from the initial structure calculation, and were only incorporated in the final annealing step performed in explicit water. The final ensemble contains the 15 lowest energy structures calculated by using CNS1.1 (37) implemented within ARIA1.2 (38). The structure ensemble has been deposited in the Protein Data Bank (http://www.ebi.ac.uk/pdbe) with PDB ID 5M9Z.
Atomic absorption spectroscopy
Protein samples were dialyzed against 20 mM Tris–HCl pH 8, 250 mM NaCl, 5 mM 2-mercaptoethanol, 0.1 mM EDTA at 4°C for 16 h. Measurement of metal content was performed on a Varian AA75 spectrophotometer at the appropriate wavelength and deduced from a standard calibration.
Mass spectrometry experiments
Pcf11 (530–626) constructs and Clp1-Pcf11 (403-626) subcomplexes were first buffer exchanged against a 250 mM ammonium acetate (NH4Ac) solution at pH 7.5 using gel filtration (Zeba 0.5 ml, Thermo Scientific, Rockford, IL, USA) and ultrafiltration spin columns (Vivaspin 500 MWCO 10 000, Sartorius, Göttingen, DE), respectively. Protein concentration was determined spectrophotometrically. NanoESI-MS characterization of Pcf11 variants was next generated on an electrospray TOF mass spectrometer (LCT, Waters, Manchester, UK) and on a Q-TOF (Synapt G2 HDMS, Waters, Manchester, UK), both equipped with an automated chip-based nanoESI source (Triversa Nanomate, Advion Biosciences, Ithaca, NY, USA) operating in the positive ion mode. External calibrations were generated from the multiple charged ions produced by a 2 μM horse heart myoglobin solution diluted in a 1:1 (v/v) water: acetonitrile mixture acidified with 1% (v/v) formic acid, and caesium iodide clusters diluted to 2 g/l in a 1:1 (v/v) water: isopropanol solution. Intact masses of individual Pcf11 constructs were first determined in denaturing conditions by diluting proteins to 2 μM in a 1:1 (v/v) water/acetonitrile mixture acidified with 1% (v/v) formic acid. Non-covalent complexes analysis in non-denaturing conditions was then performed by diluting samples to 5 μM in 250 mM NH4Ac buffer at pH 7.5. Fine-tuning of instrumental parameters was notably applied to preserve non-covalently bound species in the gas phase and ensure efficient ion transmission. Particularly, LCT and Synapt G2 source back pressures were respectively raised to 6.7 and 7.7 mbar using a throttling valve, while acceleration voltages applied on the sample cone were set to 120 and 80 V. Data processing was finally carried out on MassLynx 4.0 (Waters, Manchester, UK).
Purification and analysis of yeast 3′-end processing factors
Yeast extracts were prepared following the spheroplast procedure and tandem-affinity purification (TAP) of the factors was performed as described previously (10). The immunoblot analysis of 3′ processing complexes was done with equal amounts of purified factors run on 12% polyacrylamide-SDS gels and probed after transfer with rabbit polyclonal antibodies to Rna14 (1:3000), Rna15 (1:10000), Pcf11 (1:2000) and Clp1 (1:1000).
In vitro pre-mRNA 3′-end processing assays
Pre-mRNA 3′-end processing assays were performed with purified factors as described previously on the CYC1 or CYC1 pre-cleaved (CYC1pre) precursors radiolabeled in vitro with α-32P-UTP (10,39). Native CF IA (nCF IA) and nCPF were purified from yeast cells according to the TAP procedure (40) from strains expressing N-terminal TAP-tagged RNA15 and TAP-tagged FIP1, respectively (see Table 3 for the strains). The purification of nCPF was performed in a rna15-1 mutant background (inactive in this mutant form (39)) to ensure that no active nCF IA co-purifies and obscures the complementation assays with CF IA coming from native or recombinant origins (Figures 2 and 7). Hrp1/Nab4 (50 ng per assay) and Pab1 (140 ng per assay) were obtained from recombinant sources (10). Recombinant CF IA (rCF IA-ΔQ20) was obtained as described above. Reaction products of processing assays were analyzed on 6% polyacrylamide–8.3 M urea gels and visualized by phosphorimaging.
Table 3. Yeast trains used in this study.
| Strain | Genotype |
|---|---|
| YLM232 | MATa/α, pcf11Δ::HIS3/PCF11, ade2-1/ade2-1 leu2-3,112/leu2-3,112 ura3-1/ura3-1 trp1-1/trp1Δ his3-11,15/his3-11,15 can1-100 GAL+ |
| YLM233 | MATα, pcf11Δ::HIS3, pFL38-PCF11, ade2-1 leu2-3,112 ura3-1 trp1 his3-11,15 can1-100 GAL+ |
| YLM237 | MATα, pcf11Δ::HIS3, pFL36ΔSX-PCF11, ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YLM221 | MATa, [pNOP1]-TAP::RNA15-TRP1-Kl, ade2-1 leu2-3,112 ura3-1 trp1 his3-11,15 can1-100 GAL+ |
| YLM244 | MATa, pcf11Δ::HIS3, pFL36ΔSX-PCF11, [pNOP1]-TAP::RNA15-TRP1-Kl, ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YJG10 | MATα, pcf11Δ::HIS3, pFL36ΔSX-pcf11-18 (C421S,C424S), ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YJG23 | pcf11Δ::HIS3, pFL36ΔSX-pcf11-18 (C421S,C424S), [pNOP1]-TAP::RNA15-TRP1-Kl ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YJG12 | MATα, pcf11Δ::HIS3, pFL36-pcf11-19 (C564S,C567S), ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YJG24 | pcf11Δ::HIS3, pFL36-pcf11-19 (C564S,C567S), [pNOP1]-TAP::RNA15-TRP1-Kl, ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YLM239 | MATα, pcf11Δ::HIS3, pFL36ΔSX-pcf11ΔQ20, ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
| YJG35 | rna15-1, [pNOP1]-TAP::FIP1-TRP1-Kl, ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100 GAL+ |
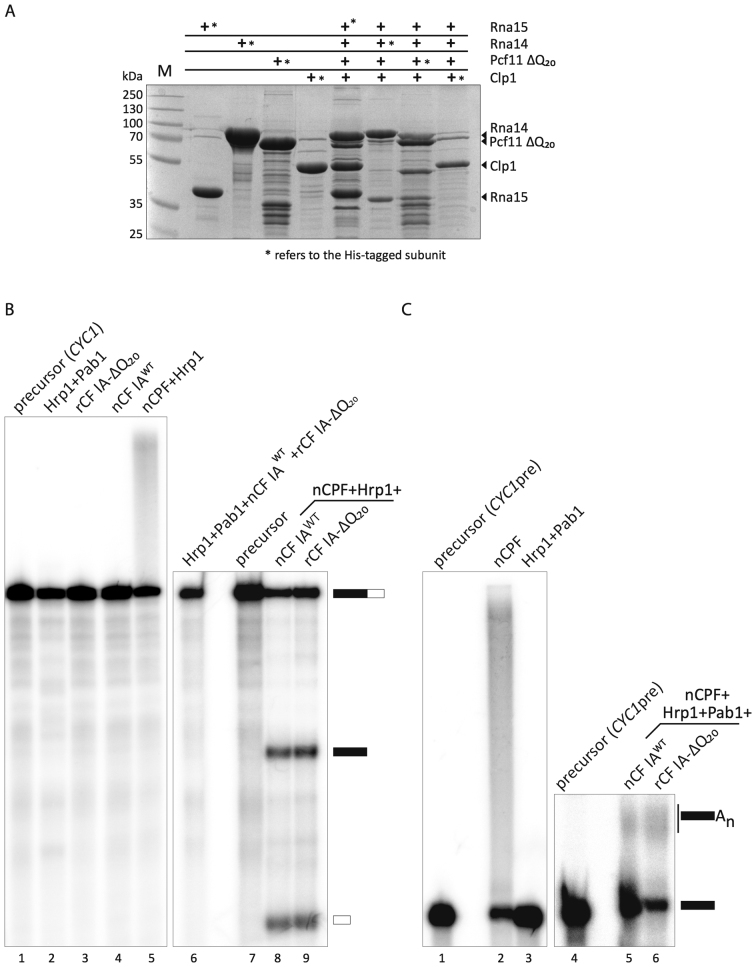
Figure 2.
Recombinant CF IA works efficiently in mRNA 3′-end formation. (A) CF IA was reconstituted by co-expression of the four CF IA subunits in E. coli. Each subunit was alternatively fused at its N-terminus with a hexahistidine tag allowing for protein purification. Pull-down experiments were performed from 10 ml cell culture after immobilization on affinity-resin. The bound proteins were eluted with imidazole and analyzed by a 10% SDS-PAGE. Proteins were revealed by Coomassie-blue staining. The asterisk (*) refers to the His-tagged subunit of CF IA. (B) In vitro cleavage assay with the CYC1 precursor was performed in the presence of native CPF (nCPF) and wild-type native CF IA (nCF IAWT) purified by TAP-tagging (lane 8) or recombinant CF IA (rCF IA-ΔQ20; lane 9). Hrp1 was included in the reaction to avoid cleavage at cryptic sites. Lanes 1 and 7: unreacted CYC1 precursor; lanes 1–6: negative controls showing the absence of cleavage activity of the different factors on their own. Lane 5 shows however the intrinsic and normal polyadenylation activity of Pap1-containing CPF. (C) In vitro polyadenylation assay of the CYC1 precleaved precursor (CYC1pre) with nCF IAWT (lane 5) or rCF IA-ΔQ20 (lane 6) and nCPF, Hrp1 and Pab1. The latter is required to control poly(A) tail elongation. Lanes 1 and 4: unreacted CYC1pre precursor; Lane 2: nCPF alone can polyadenylate unspecifically CYC1pre; lane 3: Hrp1 and Pab1 are inactive on their own.
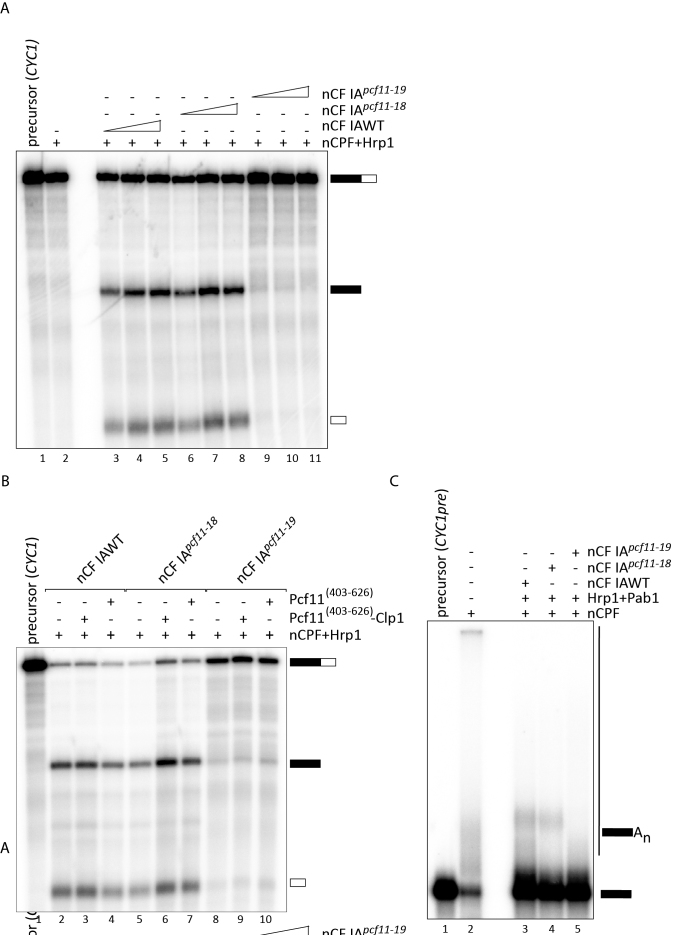
Figure 7.
Mutations in the zinc-binding motifs have distinct effects on 3′-end processing. (A) Pre-mRNA cleavage assay on the CYC1 precursor with TAP-purified factors from wild-type (nCF IAWT) and mutant (nCF IApcf11-18 and nCF IApcf11-19) cells. Lane 1: unreacted CYC1 precursor, lane 2: nCPF and Hrp1 were assayed as negative controls for the lack of cleavage activity in the absence of CF IA. Increasing amounts of nCF IAWT (0,5 μl, 1 μl and 2 μl; lanes 3–5) or nCF IApcf11-18 or nCF IApcf11-19 (2 μl, 4 μl and 8 μl lanes 6–8 and 9–11, respectively) were added to nCPF and Hrp1 to carry out the cleavage reaction. (B) Complementation of cleavage assays. Cleavage reactions were run as in (A) with 1 μl of nCF IAWT or 4 μl of nCF IApcf11-18 or nCF IApcf11-19. 100 ng of recombinant Pcf11[403-626]-Clp1 (lanes 3, 6, 9) or Pcf11[403-626] (lanes 4, 7, 10) were added to the reaction as indicated. Lane 1: unreacted CYC1 precursor. Lane 2: nCPF+Hrp1 were incubated with CYC1 as a negative control. (C) Polyadenylation assays performed on the CYC1 pre-cleaved precursor (CYC1pre) with the same amounts of nCF IAWT, nCF IApcf11-18 and nCF IApcf11-19 as in (B). Pab1 (140 ng) was added to the reaction to control de novo synthesis of poly(A) tails.
Yeast strains and plasmids
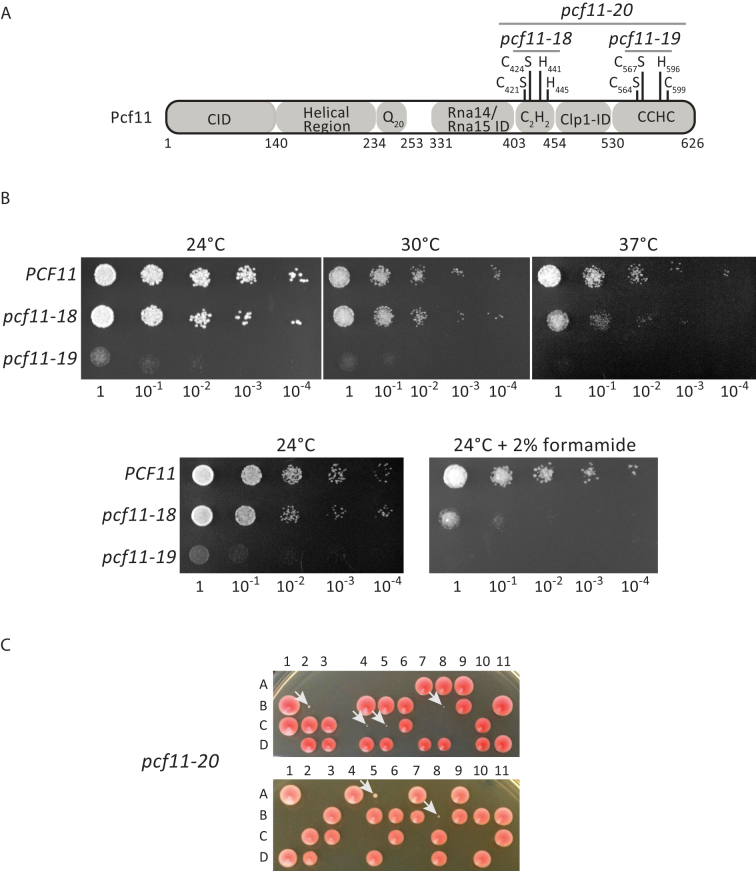
Strain YLM232 containing the pcf11Δ::HIS3 allele was constructed by replacing the TRP1 marker disrupting the PCF11 gene in strain NA50 (41) by the HIS3 marker. All of the yeast strains used is this study are listed in Table 3. pNA39 (URA3-ARS/CEN-AmpR (41)) was used to complement the pcf11Δ::HIS3 null allele in YLM233 strain. The PCF11 gene was also amplified by PCR with S. cerevisiae genomic DNA as template, from position –458 to +2566 (nucleotide +1 corresponding to the A of the start codon of PCF11 open reading frame) and cloned into the pFL36ΔSX plasmid (LEU2-ARS/CEN-AmpR); this plasmid was constructed as described in (41). Mutations of the two zinc-binding domains were obtained by site-directed mutagenesis and re-introduced in pFL36 or pFL36ΔSX by gap-repair in yeast (for pcf11-18) or by direct cloning (for pcf11-19), respectively. Plasmid pFL36ΔSX-pcf11-ΔQ20 was constructed by gap-repair. All of the constructs were verified by sequencing. Haploid strains expressing wild-type (YLM244) or mutant (YJG23, YJG24) plasmid-borne PCF11 alleles together with the TAP-tagged RNA15 gene were obtained by crossing YLM237, YJG10 and YJG12 with YLM221, respectively (see Table 3), and analysis of their progeny after meiosis. Mutant pcf11-20 expressing the Pcf11 protein mutated in the two zinc-binding domains was cloned in pFL36ΔSX and used to transform the diploid strain YLM232. Phenotypic analysis of the progeny after meiosis is described in Figure 1C.
Figure 1.
Effect of mutations of the Pcf11 zinc-binding residues on yeast viability. (A) Schematic diagram of Pcf11 domain organization. The zinc-binding residues are labelled as the point mutations and the relevant alleles. CID: RNAP II large subunit Carboxy-terminal domain Interacting Domain; ID: Interaction Domain. (B) Phenotypic analysis of the wild-type PCF11 and mutant pcf11-18 (C421S/C424S) and pcf11-19 (C564S/C567S) strains. Ten-fold serial dilutions were plated on rich medium (YPDA) and analysed at 24, 30 and 37°C, and at 24°C on YPDA containing 2% formamide. (C) Tetrad analysis after meiosis of two independent diploid strains of YLM232 (pcf11Δ::HIS3/PCF11) transformed with the plasmid-borne pcf11-20 allele (C421S/C424S/C564S/C567S). The four spores (A, B, C, D) coming from the same ascus (numbered horizontally) are arranged vertically. The slow growing spores corresponding to the pcf11-20 mutants are indicated with a white arrow (spores 2B, 4C, 5C, 8B from the first diploid, and spores 5A, 8B from the second diploid).
S1 nuclease protection assay
Plasmids used for the S1 nuclease protection assays were made as follows: a PCR fragment encompassing the sequence ranging from the Xho I site located at nucleotide +45 (according to the start of mature snR13 RNA) to nucleotide +93 in the TRS31 gene (+1 being the start of TRS31 ORF) was amplified from yeast genomic DNA and cloned into the pGEM4 vector between the EcoR I and BamH I restriction sites (Promega). This plasmid was digested with Xho I and Pvu II, the released fragment was purified on an agarose gel and 3′-end labeled with the Klenow enzyme and [α32P]-TTP (25 μCi/3000 Ci/mmol). Approximately 30 ng of the labeled DNA probe were hybridized with 30 μg of total RNAs extracted from each strain or 2 μg of control yeast tRNA and digested with 150 units of S1 nuclease (42). The protected fragments were resolved on 8% polyacrylamide–8.3 M urea gels and analyzed by phosphorimaging. The positive control RNA was obtained from a PCR fragment covering the whole snR13 mature transcription unit down to nucleotide +291 cloned between the EcoR I and HinD III restriction sites of pGEM4. The resulting plasmid was cut with EcoR I and served as template for the generation of a synthetic RNA by run-off transcription with T7 RNA polymerase.
UV-crosslinking assay
UV cross-linking reactions were performed in microtiter plates and contained (in 10 μl) 100 fmol of 32P-labeled CYC1 in 75 mM potassium acetate, 2 mM magnesium acetate, 0.01% NP-40, 1 mM DTT, 0.1 mg/ml purified BSA, 2 μM tRNA (0.5 μg), 5 μg total yeast RNA (when applicable) and 100 ng of recombinant proteins. The proteins were first incubated for 15 min at 30°C in the reaction mixture either alone or in the presence of 5 μg of total yeast RNA. The in vitro-labeled CYC1 transcript was then added and the incubation was further continued for 15 min at 30°C. The samples were irradiated twice at 500 mJ on ice in a UV Stratalinker, and then digested with RNase A (100 ng per reaction) for 30 min at 37°C. The proteins were mixed with 3 μl of SDS-loading buffer and separated on SDS–12% polyacrylamide gels. The gels were fixed, dried and visualized by phosphorimaging. To control that equal amounts of the different versions of the recombinant Pcf11[403–626] polypeptides were used for UV cross-linking, ∼600 ng of each recombinant proteins was loaded on a SDS–12% polyacrylamide gel and the proteins were compared after Coomassie staining (see Figure 9A).
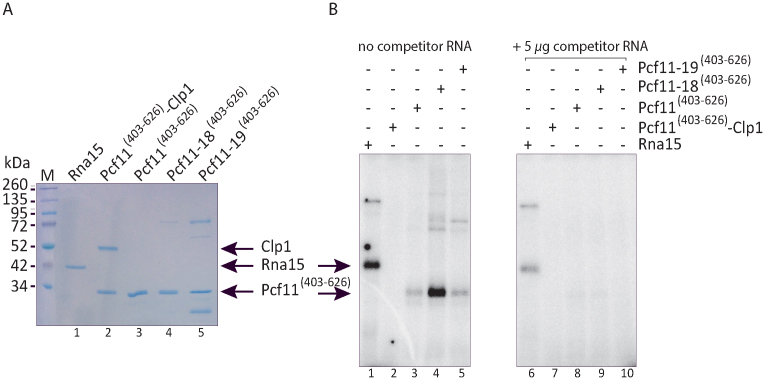
Figure 9.
The zinc-binding region can bind RNA in the absence of Clp1. (A) Equal amounts (600 ng) of recombinant proteins were loaded on a 12% SDS-polyacrylamide gel and stained with Coomassie. Lane 1: full-length Rna15; lane 2: Pcf11[403-626]-Clp1; lane 3: Pcf11[403-626]; lane 4: Pcf11-18[403–626] mutant (C421S/C424S); lane 5: Pcf11-19[403–626] mutant (C564S/C567S). (B) Equal amounts (100 ng) of recombinant proteins as described in (A) were incubated in the presence of 32P-labeled CYC1 transcript and subjected to UV cross-linking, in the absence (left panel) or presence of an excess of competitor yeast total RNA (5 μg; right panel). The position of the different polypeptides is indicated.
RESULTS
Putative zinc-binding domains in Pcf11 are required for cell viability
Sequence analyses of the yeast Pcf11 protein have previously revealed the presence of two zinc-finger motifs of the C2H2 and CCHC types neighboring the Clp1 interaction domain (Figure 1A; Supplementary Figure S1) (29,43,44). As a step towards experimental confirmation, we analysed whether these putative zinc-binding residues are important for cell viability. To do so, the PCF11 gene under its natural promoter was amplified by PCR and cloned into pFL36 and pFL38 centromeric plasmids. We next introduced point mutations in the PCF11 gene to target two potential zinc-binding residues within each region. Specifically, the conserved C421 and C424 residues within the first region, and independently the conserved C564 and C567 amino acids located in the second region, were mutated into serine and the resulting mutants, named pcf11-18 and pcf11-19, respectively, were assayed for growth at different temperatures (Figure 1B). Mutant pcf11-18 did not show any strong growth defect at physiological temperatures of 24°C or 30°C. In contrast, cell growth was significantly affected at 37°C and addition of 2% formamide in the medium exacerbated this phenotype at 24°C in comparison to the wild-type strain. Unlike the pcf11-18, pcf11-19 exhibited a strong growth defect at both 24°C and 30°C and turned out to be lethal at 37°C or in the presence of formamide at 24°C. We next constructed an allele that combined the mutations present in pcf11-18 and pcf11-19 such that both putative zinc-binding motifs were mutated. The resulting allele, pcf11-20, was cloned into a LEU2-marked plasmid and used to transform the heterozygous pcf11Δ::HIS3/PCF11 diploid strain YLM232 (see Materials and Methods and Table 3). After meiosis, only a few spores were observed showing a very strong slow-growth phenotype (Figure 1C; the spores are indicated with an arrow). These spores corresponded to the pcf11Δ::HIS3 null allele complemented with the pcf11-20 mutant since they grew on a synthetic medium lacking histidine and leucine. This mutant therefore has a much stronger growth defect in comparison to the pcf11-19 allele. Despite the centromeric nature of the plasmid carrying the pcf11-20 allele, only a few spores corresponding to the mutant were able to grow (spores 2B, 4C, 5C, 8B from the first diploid, and spores 5A, 8B from the second diploid, Figure 1C). After inspection under the microscope, many of the spores did not go beyond one or two cell divisions. Therefore, the pcf11-20 mutant conferred a strong germinative defect and a near-lethal phenotype for those spores which were able to escape germination.
In conclusion, the mutations of putative Pcf11 zinc-binding cysteines designed to abolish metal chelation in both the C2H2 and CCHC regions led to a clear growth defect, and thus supports a general need for Zn2+ binding in Pcf11 function.
Recombinant expression of CF IA
Prompted by the cell growth defects in the mutations described above and in order to circumvent native CF IA purification from these alleles, we first set-up a procedure for reconstituting S. cerevisiae CF IA from bacterially expressed proteins as previously described (22). This approach allows for the study of CF IA containing deletions of essential domains (e.g. Pcf11 CID) or point mutations that cannot be studied in vivo due to their lethal phenotype, and thus enable studies on CF IA architecture and pre-mRNA 3′-end processing. Briefly, each subunit was cloned into a plasmid carrying a resistance to a different antibiotic allowing for the introduction of up to four plasmids at a time into a single bacterial cell (30,31,45). Due to limited levels of protein expression of full-length Pcf11 (data not shown), we opted for a slightly altered version of Pcf11 in which a 20-aminoacid stretch of glutamine residues has been deleted (residues 234–253). This shorter version of Pcf11 is viable and has no detectable phenotype in yeast (Supplementary Figure S2), and will be referred to as Pcf11-ΔQ20.
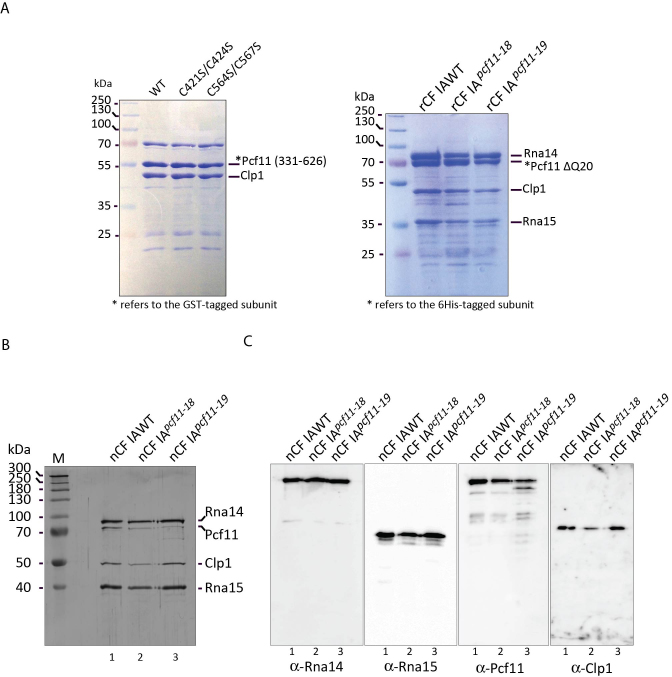
The expression of individual proteins was confirmed (Figure 2A). Subsequently, a four-subunit co-expression assay was attempted by swapping the His-tag alternatively to the N-terminus of each subunit. Each combination successfully retrieved the entire recombinant CF IA (rCF IA), however the best combination was found when Rna15 is His-tagged (Figure 2A). A robust purification procedure was then set-up in order to retrieve pure rCF IA-ΔQ20 complex for further experiments (see Materials and Methods).
rCF IA retains in vitro 3′-end processing activity
CF IA is essential in pre-mRNA 3′-end processing through its role in the positioning of the catalytically active CPF complex onto the pre-mRNA. We used an in vitro reconstituted cleavage and polyadenylation assay to test whether rCF IA is able to substitute for the yeast native complex (referred to here as nCF IA). As expected, nCF IA in combination with TAP-purified nCPF readily processed the CYC1 precursor in a cleavage-only reaction (where polyadenylation is inhibited with EDTA and Hrp1 added to control cleavage site selection (8); Figure 2B, lane 8), as well as in a polyadenylation reaction in which Hrp1 and Pab1 were also added (Figure 2C, lane 5). Note that none of the different factors used in the assays exhibit any processing activity on their own (Figure 2B, lanes 1–4, 6 and 2C, lane 3) except for CPF which is capable of non-specific polyadenylation activity on any RNA due to the presence of the poly(A) polymerase Pap1 as one of its intrinsic subunits as previously described in the literature (e.g. (40,46)) (Figure 2B, lane 5 and 2C, lane 2). With the recombinant complex, the cleavage and polyadenylation reactions appeared as efficient when rCF IA-ΔQ20 replaced native nCF IA in the assays (Figure 2B, lane 9, and 2C, lane 6, respectively). Therefore, Pcf11-ΔQ20 can efficiently work in recombinant rCF IA compared to native nCF IA, as far as in vitro 3′-end processing is concerned.
Pcf11 contains two Zn2+ atoms
rCF IA provides an ideal source to quantify the Zn2+ content in the complex by atomic absorption, as well as recombinant sub-complexes of Rna14–Rna15 and Pcf11–Clp1 heterodimers. Samples were analysed at 213.9 nm using the dialysis buffer as the baseline, and a titration curve ranging from 1.25 to 20 μM of ZnS04 solution was also prepared in the dialysis buffer. According to these measurements, only trace amounts of Zn2+ were detected in the Rna14–Rna15, whereas the rCF IA and Pcf11–Clp1 samples displayed significant Zn2+ content (Table 1). These data demonstrate the presence of natively bound Zn2+ atoms within rCF IA and most specifically within Pcf11, since the structure of Clp1 did not reveal any bound Zn2+ atoms (18,19).
Table 1. Zn2+ content analysis in recombinant CF IA.
| Protein | [Protein] (μM) | [Zn2+] (μM) | Ratio |
|---|---|---|---|
| Rna15–Rna14 | 3.14 | 0.5 | 0.16 |
| CF IA-ΔQ20 | 6.15 | 11 | 1.79 |
| Pcf11-ΔQ20–Clp1 | 17 | 23.5 | 1.38 |
To further identify an exact stoichiometry and to better define the zinc-binding regions in Pcf11, we used mass spectrometry analysis in non-denaturing conditions. We first designed a construct from residue 403 to the C-terminus (residue 626) that spans the two-potential zinc-binding regions. The protein was expressed in E. coli and purified to homogeneity and the accurate Zn2+ stoichiometry was determined by comparing the mass measured under non-denaturing or denaturing conditions (Figure 3). The mass spectrometry data obtained under non-denaturing conditions fit the theoretical value expected for Pcf11 (403–626) of 25 742.5 Da, which includes a G–H–M tripeptide at the N-terminus. Mass spectrometry measurement under native conditions shows an additional molecular mass of 130.5 Da corresponding to two bound Zn2+ atoms (Figure 3A). As a final test, the second C-terminal zinc-binding region of Pcf11 (residues 530–626) was expressed in isolation. The mass under denaturing conditions of 11 149.3 ± 0.1 Da again fits the theoretical value. Comparison to the denaturing conditions shows an additional molecular mass of 65.7 Da, attributed to a single bound Zn2+ atom (Figure 3B). Attempts to produce the other isolated zinc-binding region from residue 403–454 failed due to excessive instability. Nevertheless, the results demonstrate that Pcf11 harbors two zinc-binding regions: one Zn2+ binds at the C-terminus (from 530–626), and another is likely bound by the conserved amino acids within 403–454. Further insight into the architecture of these domains necessitated a structural approach.
Figure 3.
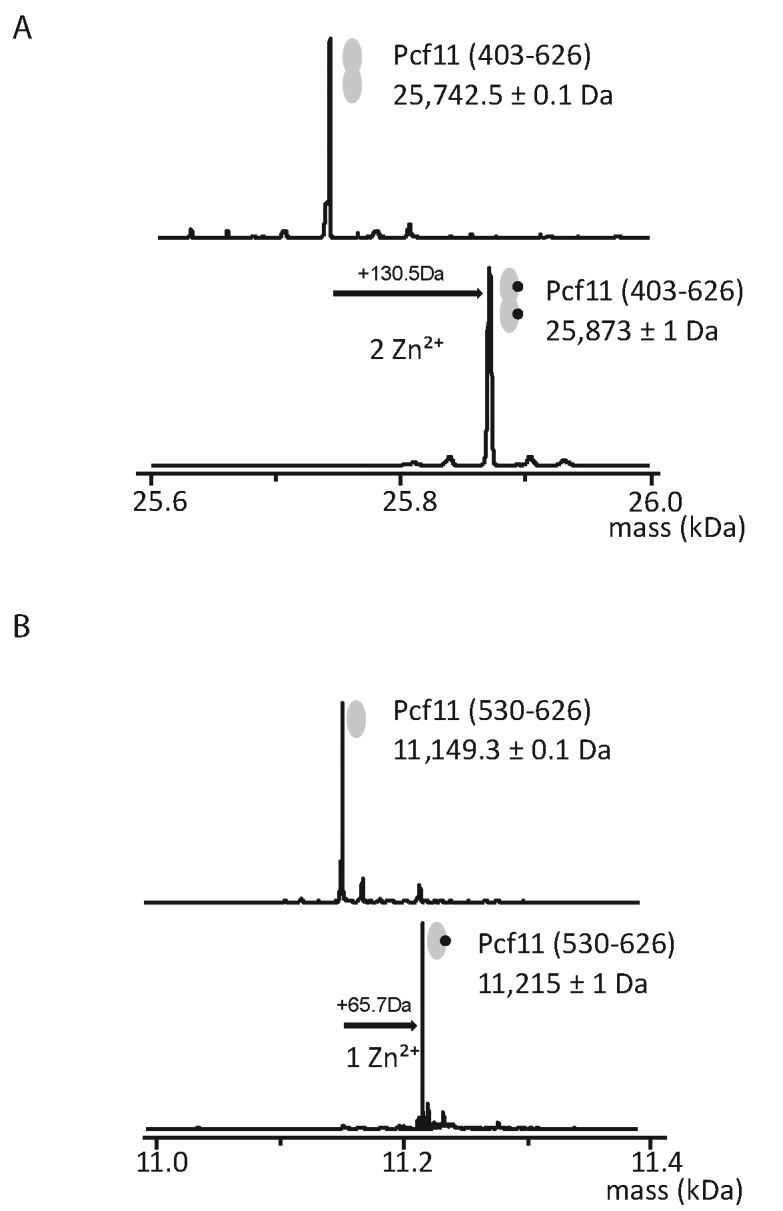
Pcf11 has two zinc-binding domains. Deconvoluted ESI mass spectra obtained from (A) Pcf11 (403–626) and (B) Pcf11 (530–626) constructs under denaturing (upper panels) and non-denaturing (lower panels) conditions. Mass differences between denaturing and non-denaturing spectra respectively correspond to the binding of two (+130.5Da) and one (+65.7Da) zinc atoms.
The Pcf11 C-terminal zinc-binding domain has a novel fold
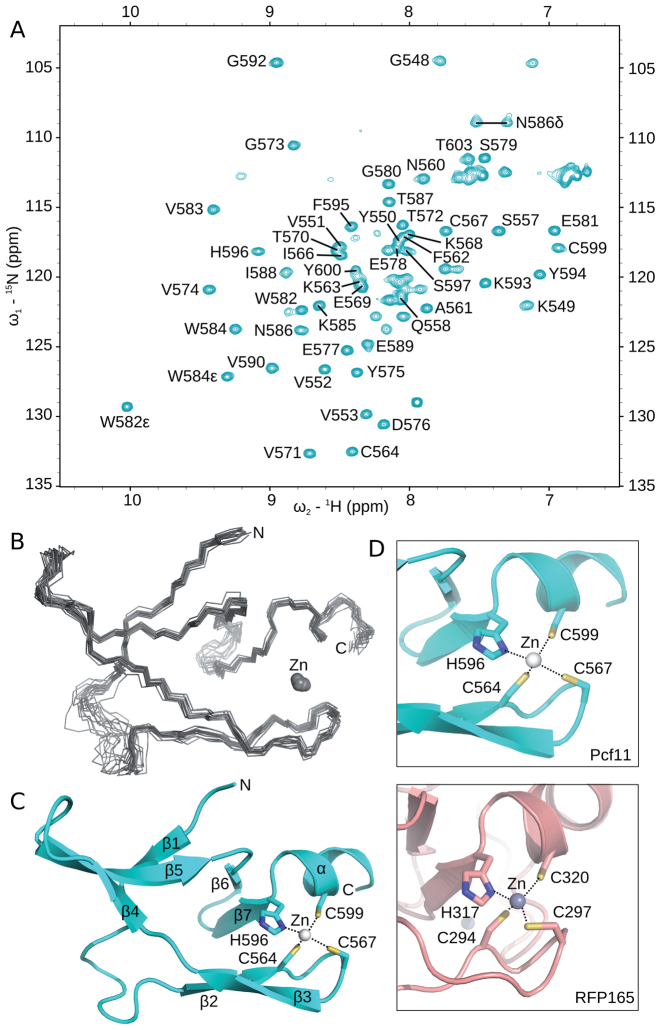
To obtain atomic information on both zinc-binding domains from Pcf11, we first attempted to determine a structure of the larger complex between Pcf11 (403–626) bound to full-length Clp1. Even though crystals diffracting to a limited resolution (<4 Å) were obtained, no electron density could be attributed to Pcf11 apart from the Clp1 interacting domain, as has been previously described (18,19). Due to the inability to produce the first zinc-binding domain in isolation, we focused on determining the structure of the second zinc-binding domain of Pcf11 (530–626). By using NMR spectroscopy, we found that the Pcf11 (530–626) construct indeed contains a well folded domain, as well as a significant number of residues that fall within the region typical of a conformationally disordered peptide (Figure 4A). It was also apparent that Zn2+ binding was required to maintain the overall fold, since the protein was unstable in the absence of added Zn2+ during purification. Although it was not possible to assign the chemical shift values of the N- and C-terminal residues, a central region from G548 to T603 was amenable to detailed structural analysis by NMR-derived dihedral and distance restraints (Figure 4B, Table 2 and Materials and Methods). To prevent bias of the Zn2+ binding residues during structure calculation, the protein fold was first determined without any Zn2+-based restraints. This calculated ensemble presented a consistent and favorable Zn2+ binding geometry formed by the side chains of C564, C567, H596 and C599. Therefore, a final refinement step in explicit water was performed, with the addition of a Zn2+ ion and a limited set of distance and dihedral restraints derived from an analysis of Zn2+ coordination in proteins (47). The overall structure is composed of seven β-strand elements and a single C-terminal α-helix (Figure 4C). The relatively large zinc-binding domain and high content of β-strands suggested an unusual zinc-binding fold. A recent structure of a similar region of Pcf11 from residues 538 to 608 (PDB ID 2NAX; (29)) displays the same overall fold, with a global RMSD between the two ensembles of 0.7 and 1.3 Å for the backbone and all heavy atoms, respectively, calculated by using SuperPose (48).
Figure 4.
Solution structure of the second zinc-binding domain. (A) 1H,15N HSQC of [15N]-labelled Pcf11 (530-626) in 50 mM Tris (pH 7.5) and 150 mM NaCl, collected at 298 K. Crosspeaks corresponding to residues G548 to T603 have been annotated. (B) Backbone traces for the ensemble of 15 structures calculated for residues 548–603. The Zn2+ ion is displayed as a sphere. (C) Cartoon representation of the structure closest to the mean, with secondary structure elements labelled and the Zn2+ ion shown as a sphere. The zinc-binding residues are shown in stick form and are labelled. (D) Comparison of the local region of structure similarity between the second zinc-binding domain of Pcf11 (top) to only one of the two zinc-binding sites of the RING finger proteins (bottom), in this case from RFP165 (PDB 5D0I; (50)).
Table 2. Statistics for the ensemble of 15 lowest energy structures of Pcf11 residues 548–603.
| NMR distance and dihedral restraints | |
|---|---|
| Distance restraints | |
| Total | 1170 |
| Intraresidue | 353 |
| Sequential (|i-j| = 1) | 160 |
| Medium range (1<|i-j|<5) | 54 |
| Long range (|i-j|>4) | 332 |
| Ambiguous | 271 |
| Dihedral angle restraints | |
| Φ | 51 |
| Ψ | 51 |
| χ | 32 |
| Zn2+ coordination restraints | |
| Distance | 4 |
| Dihedral | 13 |
| Structure statistics | |
| Violations (mean and SD) | |
| Distance restraints (Å) | 0.010 ± 0.001 |
| Dihedral angle restraints (°) | 0.76 ± 0.09 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.001 ± 0.000 |
| Bond angles (°) | 0.300 ± 0.005 |
| Impropers (°) | 0.22 ± 0.01 |
| Ramachandran plot (%)a,b | |
| Most favored regions | 91.0 (95.9)c |
| Additionally favored regions | 7.8 (4.1) |
| Generally allowed regions | 0.4 (0.0) |
| Disallowed regions | 0.8 (0.0) |
| Average pairwise rmsd (Å)a | |
| Backbone | 0.6 (0.4) |
| Heavy | 1.0 (0.9) |
A search of the available structures by using DALI (49) failed to find a close structural homologue, but instead identified local homology (Z-score > 2) to one of the two zinc-binding sites within the RING finger fold (Figure 4D). For example, comparison to the recent structure from RING finger protein 165 (RFP165; PDB ID 5D0I; (50)) reveals a common β-turn placement of the first two cysteine residues in a C-x-I-C motif (C564 and C567 in Pcf11; C294 and C297 in RFP165), as well as a helical location of the remaining H-x-x-C motif (H596 and C599 in Pcf11; H317 and C320 in RFP165). The rest of the Pcf11 fold has no structural similarity to the RING finger. As a result, it appears that the second zinc-binding region at the C-terminus of Pcf11 is a new structural class of the CCHC zinc-binding family (51). Other members of the CCHC family are mainly found in the nucleocapsid protein of retroviruses and are generally involved in packaging and early infection process. In eukaryotes, it is found in proteins binding RNA or single-stranded DNA.
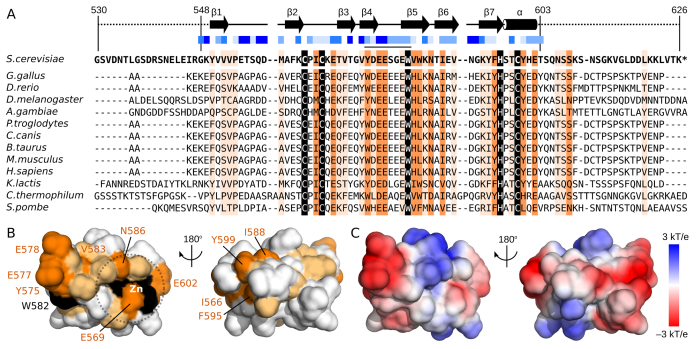
A sequence alignment was then used to identify putative functional residues within the second zinc-binding domain (Figure 5A). Outside of the CCHC residues, only W582 is identical in all 13 species. This residue is found to be >40% solvent exposed in the structural ensemble by using the NACCESS program (52) and is thus able to contribute to biomolecular interactions. By extending the alignment analysis to residues that share sidechain properties in all 13 species, several residues around the zinc-binding site are identified (Figure 5B) which may contribute directly to Zn2+ binding properties (e.g. E569, N585, E602). Opposite to the zinc-binding residues, the aromatic amino acid (Y575) and two acidic residues (E577 and E578) cluster with W582 (Figure 5B). This conserved surface therefore presents a region of four exposed sidechains anchored in the triple-stranded β-sheet, specifically within strands β4 and β5 as well as the turn that connects them. This area may provide a binding surface for interaction with CPF subunits. Comparison of this region to the surface charge properties (Figure 5C) emphasizes the acidic and exposed nature of this region.
Figure 5.
Surface residue conservation separate from the zinc-binding site. (A) Sequence alignment of the second zinc-binding domain of Pcf11 from 13 species. Boundaries of the construct used in NMR studies (residues 530–626) and of the calculated structure (residues 548–603) are indicated, as are the locations of β-strands (arrows) and the α-helix (cylinder). Solvent accessibility for sidechains are calculated by using NACCESS (52) and indicated with varying shades of blue squares from solid blue (>80% accessible) to white (<20% accessible). Residues with complete sequence identity or similarity is highlighted in black and orange, respectively. Residues with similarity to yeast Pcf11 in >75% of the sequences are highlighted in light orange. (B) Surface representation of Pcf11 (548–603) with residues coloured by conservation as in panel A. The conserved patch formed by residues Y575, E577, E578 and W582 as well as the conserved residues around the Zn2+ atom (E569, V583, N586, E602) are indicated. (C) Surface charge properties of Pcf11 (548-603) with the negatively charge acidic regions in red, and the positively charged basic regions in blue.
Zinc-binding mutations have little effect on CF IA assembly
Subunit–subunit interactions in CF IA have been reported through pairwise assays either with the two-hybrid analysis or by pairwise association/expression (13,16,17,20,41,43,53) giving rise to a refined interaction map in CF IA (26). However, interactions between two proteins that require a third stabilizing protein may not be detected, as for example with the PYM protein interaction with Mago and Y14 (54). We thus used our CF IA reconstitution assay to refine the subunit-subunit interaction map with respect to Pcf11, and to test whether the zinc-binding domains are necessary for CF IA assembly. We first tested the influence of double point mutations C421S/C424S (Pcf11-18) or C564S/C567S (Pcf11-19) in a pairwise coexpression assay between Pcf11 and Clp1 (Figure 6A, left panel). As observed, no significant decrease in binding between Pcf11 (331–626) and Clp1 was detected. We next addressed the effect of these double mutations on CF IA architecture (Figure 6A, right panel). The Pcf11 ΔQ20 protein carrying the double mutations C421S/C424S (Pcf11-18) or C564S/C567S (Pcf11-19) did not alter significantly the presence of any CF IA subunit in comparison to the wild-type in pull-down assays.
Figure 6.
The Pcf11 zinc-binding domains do not participate in CF IA assembly. (A) Pull-down assays of recombinant Pcf11–Clp1 and CF IA. The C421S/C424S point mutations and C564S/C567S point mutations were introduced into the ORF of Pcf11 (311–626) or Pcf11 ΔQ20. Pcf11:Clp1 or CF IA was pulled-down via a His-tag at the N-terminus of Pcf11. Proteins were eluted from the beads with imidazole and analysed by SDS-PAGE followed by Coomassie-Blue staining. Equivalent amounts of native wild-type and mutant TAP-purified CF IA were loaded on SDS–12%-polyacrylamide gels and subjected to silver staining (B) or Western blot (C) analyses. Lanes 1: 5 μl of purified nCF IAWT; lanes 2: 20 μl of nCF IApcf11-18, lanes 3: 20 μl of nCF IApcf11-19 were used. Polyclonal antibodies to each individual CF IA subunits were used to probe the Western blots.
We also tested the impact of the double cysteine mutants on the assembly of the native nCF IA complex (nCF IA). The mutant nCF IA factors were purified by the TAP-tagging method from strains YJG23 and YJG24, where Pcf11 was expressed from the pcf11-18 and pcf11-19 alleles, respectively (Figure 1 and Table 3). It should be noted that purifications of nCF IA from the mutant strains reproducibly retrieved lower amounts of factors compared to the wild-type strain for unknown reasons but likely related to the relative effects of the mutations on cell growth and viability. Nevertheless, all four subunits were present in nCF IA purified from the wild-type and mutant backgrounds (Figure 6B, lanes 1–3), but the bands corresponding to Pcf11 appeared slightly less abundant in the zinc-binding domain mutant alleles. Based on this silver-stained protein gels, equal amounts of factors were loaded on SDS-polyacrylamide gels and subjected to Western blot analyses with antibodies to all four subunits separately. The blots showed that, in comparison to equivalent signals detected with anti-Rna14 and anti-Rna15, the association of Pcf11 and also Clp1 was slightly impaired in the mutant factors (Figure 6C). With regard to nCF IA purified from the most affected mutant, the pcf11-19 allele, the Pcf11 mutant protein appeared to be more prone to degradation and was revealed as double bands yet both capable to associate with nCF IA (Figure 6B, lane 3 and Figure 6C, lane 3, panel α-Pcf11). Nevertheless, the consequence of cysteine mutations in the zinc-binding motifs on nCF IA assembly is not dramatic enough to explain their impact on pcf11-18 and particularly pcf11-19 growth phenotypes.
Mutations of the zinc-binding motifs impair mRNA 3′-end formation to different extents
We next examined the involvement of Pcf11 zinc-binding domains in mRNA 3′-end maturation in vitro. We assayed nCF IA factors purified from the wild-type and the mutant pcf11-18 and pcf11-19 strains in either cleavage or polyadenylation reactions at 30°C. As controls, no cleavage or polyadenylation activity was found with the purified factors on their own (Supplementary Figure S3). We took into account the lower amounts of nCF IA complexes retrieved from the mutant strains after TAP purification compared to wild-type as judged by the differences observed on protein gels (see Figure 6B and C) to carry out the assays. Moreover, to circumvent the variabilities observed in the Pcf11–Clp1 protein contents and their possible impact on 3′ processing activities, we performed the cleavage assays with increasing amounts of the different nCF IA factors. Efficient and very similar endonucleolytic cleavage of the CYC1 full-length precursor was obtained when nCF IAWT (Figure 7A, lanes 3–5) or nCF IApcf11-18 (Figure 7A, lanes 6–8) was used in the assay. Conversely, cleavage activity was undetectable with nCF IApcf11-19 no matter the amount of factor employed (Figure 7A, lanes 9–11). The possibility that the recombinant wild-type domain of Pcf11 could ameliorate the cleavage activity of mutant and wild-type nCF IA was tested. Starting from the same reaction conditions used in lanes 4, 7 and 10 of Figure 7A, we complemented the assays with 100 ng of either recombinant Pcf11[403-626]-Clp1 or Pcf11[403-626] alone. No significant improvement was observed with nCF IAWT or nCF IApcf11-18 with Pcf11[403-626]-Clp1 dimer but a slight inhibition could be noticed after addition of Pcf11[403-626] alone (Figure 7B, lanes 2–4 and 5–7). Cleavage assays with nCF IApcf11-19 was not dramatically changed with either proteins (Figure 7B, lanes 8–10). Therefore, reconstitution of an active factor or stimulation of the activity was not possible by simply complementing the assay with wild-type domains alone.
We then performed polyadenylation assays with the same wild-type and mutant factors. In these conditions, less efficient polyadenylation of the CYC1 pre-cleaved precursor was reproducibly obtained with nCF IApcf11-18 compared to the wild-type factor (Figure 7C, lanes 4 and 3). In contrast, the absence of characteristic polyadenylated product was observed with the nCF IApcf11-19 mutant factor (Figure 7C, lane 5). Therefore, the impact on pre-mRNA 3′-end processing in vitro may likely explain the growth defects seen when Pcf11 zinc-binding motifs are mutated. Noticeably, these deficiencies also parallel the amplitude of the growth defects observed with the two mutants (Figure 1B).
RNA polymerase II transcription termination in Pcf11 zinc-binding mutants
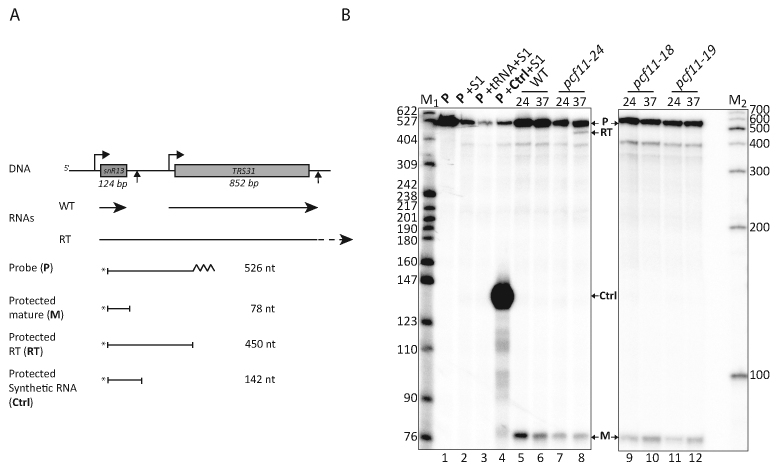
The C-terminal position of the two Pcf11 zinc-binding motifs would predict that they are not important for RNA polymerase II (RNAPII) transcription termination as it has been demonstrated that only mutations in the 266 N-terminal aminoacids of the protein affect termination (43). To test this assumption, an S1 nuclease protection assay was developed to map the 3′ ends of an RNAPII-transcribed gene coding for a stable snoRNA, snR13 (see Materials and Methods). Briefly, we designed a probe capable to hybridize with RNAs transcribed from the snr13-TRS31 region of the genome (Figure 8A). This 32P-5′-end-labeled probe (Figure 8B, lane 1) is sensitive to extensive degradation when subjected to S1 nuclease treatment when alone or in the presence of unspecific tRNA (Figure 8B, lanes 2–3). As a positive control, hybridization of the probe with a specific synthetic RNA of about 190 nucleotides transcribed from a region comprising part of the snR13 mature transcript downstream sequences, generated a predicted fragment of 142 nucleotides (Figure 8B, lane 4). Total RNAs extracted from the wild-type PCF11 strain cultivated at both 24°C or after a 1-hour shift to 37°C only produced a 78-nt product (M) in the assay representing the mature form of snr13 snoRNA (Figure 8B, lane 5–6). RNAs isolated from a temperature-sensitive allele, pcf11-24, containing mutations in the N-terminal domain of the Pcf11 protein (J. Guéguéniat et al. personal communication), produced the 78-nt product at both temperatures but also a 450-nt fragment (RT) at 37°C corresponding to read-through transcription downstream of the normal transcription termination site of snr13 (Figure 8B, lane 7–8). Mutations of the two Pcf11 zinc-binding motifs produced no detectable read-through transcript in the S1 nuclease protection assay but only the mature one (M), as anticipated from the localization of the domains affected by the mutations (Figure 8B, lanes 9–12). In conclusion, our data suggest that in contrast to the noticeable effect on mRNA 3′ end formation, the zinc-binding domains have likely no role in RNAPII transcription termination of snoRNA in vivo.
Figure 8.
The zinc-binding domains do not participate in RNA polymerase II transcription termination at snoRNA genes. (A) Schematic representation of the S1 nuclease protection assay to map transcription termination at snR13 transcription unit. (B) Detection of snR13 read-through products by S1 nuclease protection. The radiolabled probe (P, lane1) was incubated with S1 nuclease either alone (lane 2) or in the presence of non-specific RNAs (tRNA; lane 3), a specific RNA designed to protect a 142-nt fragment (lane 4), or total RNAs extracted from wild-type or mutant cells grown at 24°C or after a shift to 37°C during 60 min (lanes 5–12). The protected fragments were separated by electrophoresis on 8% polyacrylamide–8.3 M urea gels. The bands on the gels were indicated as diagrammed in (C). The labeled molecular weight makers (in number of nucleotides) are the Msp I-digested pBR322 (M1) and the 100-bp DNA ladder (Promega; M2).
Pcf11 zinc-finger domains can bind RNA in the absence of Clp1 with different avidities
The presence of zinc-binding motifs in polypeptides is sometimes associated with an ability to bind nucleic acids. This is often the case with proteins involved in RNA processing. Even though the structure of the Pcf11 C-terminal zinc-binding domain has no overall structural homology with classical zinc fingers, we tested whether the region of Pcf11 containing the two zinc-binding motifs can associate with RNA. After confirming that equal amounts of the Pcf11 zinc-binding domains were used with a Coomassie-stained gel (Figure 9A), we performed UV cross-linking experiments with uniformly 32P-labeled CYC1 substrate, using recombinant Rna15 as a positive control and testing recombinant Pcf11[403–626]-Clp1 (Figure 9B, lanes 1 and 2). As expected, a strong signal corresponding to Rna15 binding to the RNA was observed after SDS-PAGE and autoradiography of the gel (Figure 9B, lane 1). This interaction is due to the presence of a canonical RNP-type RNA-binding domain at the Rna15 N-terminus. Conversely, no signal was obtained in the same assay conditions with Pcf11[403–626]-Clp1 heterodimer (Figure 9B, lane 2). It has been previously shown that the region of Pcf11 encompassing the Clp1 binding site flanked by the two zinc finger domains binds not only Clp1 but also the RNA-binding protein and export factor Yra1 in a mutually exclusive mode (55). In competition experiments, the interaction between Yra1 and Pcf11 zinc-binding regions was lost after addition of Clp1 or RNA, despite the fact that the Yra1 RRM is not involved in the interaction between the proteins (56). To test whether the association of Pcf11[403–626] with Clp1 could mask an intrinsic capacity of the zinc finger domains to interact with RNA, we extended the UV cross-linking experiments to the Pcf11[403–626] region of the wild-type protein alone and of the corresponding mutant polypeptides Pcf11-18[403–626] and Pcf11-19[403–626] (Figure 9A, lanes 3–5). As hypothesized before, the absence of Clp1 revealed that the zinc-binding region of Pcf11 was able to effectively interact with CYC1 (Figure 9B, lane 3). Strikingly, the mutant Pcf11-18[403–626] polypeptide exhibited a strong signal comparable to that of Rna15 in the same assay conditions (Figure 9B, compare lane 4 and 1). Pcf11-19[403–626] was also prone to cross-link to the labeled RNA but in a manner similar to the wild-type protein (Figure 9B, lane 5 and 3). To evaluate the strength and/or specificity of Pcf11[403–626]–RNA interaction, we challenged CYC1 binding by incubating the proteins with an excess (5 μg) of total yeast RNA before adding the labeled transcript (see Materials and Methods). The right panel of Figure 9B showed that albeit diminished, Rna15 still exhibited a significant interaction with RNA (lane 6) but CYC1 interaction was lost for all of the Pcf11[403–626] versions (lanes 7–10). These experiments showed that Pcf11 zinc-binding domains have the capacity to bind non-specifically to RNA but only in the absence of another protein or RNA interactor.
DISCUSSION
In this article, we report the biochemical, biophysical and biological characterization of the two zinc-binding domains of Pcf11. The identified zinc-binding domains surround the Clp1 binding region (residues 454–530) and are located between residues 403–454 and 530–626, belonging to the C2H2 and CCHC class respectively (57). The zinc-binding domain spanning aminoacids 403–454 was the first to be identified (44) while both domains were more carefully depicted from the sequence of Pcf11 (43). Of note, the structural and functional analysis of the second zinc-binding domain was recently published (29). Mass spectrometry measurements demonstrate that each domain binds a single Zn2+ atom, and each zinc-binding domain displays a thermo-sensitive phenotype when two out of the four Zn2+ binding residues are mutated into serine. Simultaneous mutation of the two zinc-binding domains is close to lethality in yeast. We further demonstrate that these Pcf11 domains are not required for transcription termination in yeast and are involved to different degrees in pre-mRNA 3′-end processing in vitro.
CF IA bearing an altered version of Pcf11 in its first zinc-binding domain can efficiently cleave a pre-mRNA substrate and has only slightly impaired polyadenylation activity as compared with the wild-type factor. In contrast, the equivalent mutations in the second zinc-binding motif have a dramatic effect on both cell growth and 3′ processing. The overall assembly of the factor is mostly unchanged from the wildtype which is different than mutations in the Clp1 protein that abolished the association of Clp1 and Pcf11 with Rna14–Rna15 (53), or mutations in Rna14 or Rna15 which were described to impair CF IA subunit organization (16).
Along with this study, we also demonstrate that the poorly-conserved poly-glutamine extension of Pcf11 (residues 234–253), connecting the N-terminal CID to the Rna14–Rna15 binding domain, is not necessary for CF IA assembly. The poly-glutamine linker is also not required for mRNA 3′ end maturation, which likely explains the viability of pcf11 ΔQ20 mutant. As suggested by Amrani and colleagues (9), the poly-glutamine extension could confer a degree of freedom to the N-terminal and C-terminal domains, allowing them to fulfil independent functions. This possibility is reinforced by the observation that the region between residues 253 and 331 of Pcf11 is not otherwise necessary to assemble CF IA and may mainly function to extend the linker region by more than 75 residues. This type of organization is also observed for Nab2, implicated in the regulation of the poly(A) tail length (58,59). Nab2 also exhibits a repetition of 12 glutamine residues linking the N-terminal domain related to mRNA export function, and the RNA interacting region located at the central and C-terminal regions. Interestingly, deletion of the Q-rich region of Nab2 also has no impact on pre-mRNA 3′-end processing either (10).
We have also reported the solution structure of the second zinc-binding domain of Pcf11. Similar to the findings by the Varani and Moore labs (29) the overall fold does not exhibit similarity to other reported zinc-binding domains. So far only CCCH-type zinc knuckles have been characterized among the proteins involved in mRNA 3′-end maturation. This includes Mpe1, an essential protein required for both cleavage and polyadenylation, that contains a central zinc knuckle (60), Nab2 (58,59,61) and Yth1 that contains five zinc knuckle domains (62,63). It should be noted, however, that mutant analysis for zinc-binding residues in these domains, as performed for Pcf11, have not been described. Previously reported PCF11 alleles, such as pcf11-2 (E232G, D280G, C424R, S538G, F562S, S579P) or pcf11-9 (A66D, S190P, R198P, R227G, E354V, K435V) exhibit only a thermo-sensitive phenotype (9,43). Cysteine mutations within Mpe1 or Nab2 lead to a cold sensitive phenotype and no detectable effect on growth at 37°C (58,60). The phenotype difference reported in this article may also illustrate the functional independence of each zinc-binding domain. Indeed, for the five CCCH domains of Yth1 there is variation in function: ZF2 and ZF4 interact with RNA, ZF4 and ZF5 interact with Fip1, ZF1 and ZF4 interact with the endonuclease Ysh1/Brr5 and ZF1 and ZF2 are required for pre-mRNA cleavage (63).
Finally, Pcf11 is a key player in connecting RNAPII transcription termination, via its N-terminal CID, to pre-mRNA 3′-end cleavage, through its association to Clp1 and Rna14:Rna15 heterodimer. In this scheme, the Pcf11 zinc-binding domains appear to ensure the connection to pre-mRNA 3′-end processing of CF IA to CPF subunits. These zinc-binding domains may well play a regulatory role on CPF subunits. Indeed, the sole CPF is not able to cleave pre-mRNA in the absence of CF IA and Hrp1. In addition to the contacts within CF IA subunits, Pcf11 exhibits direct contacts to CPF subunits and other proteins. This is the case for Ysh1, Ydh1 and Yhh1 (64), Mpe1 (65), Ssu72 (66), Pti1 (67,68), Swd2 (69). Considering these interactions, the zinc-finger domains of Pcf11 could be seen as protein-protein interactors with specific CPF subunits. As Swd2 is not required for the cleavage and polyadenylation steps in vitro (69,70), yet can affect 3′-end formation of some mRNAs and snoRNAs in vivo, it is unlikely that a disruption of its interaction with Pcf11 could be responsible for the observed phenotypes.
Alternatively, but not in opposition, a putative role in RNA binding of the zinc-finger domain of Pcf11 has been suggested in our study. We showed that the region containing these motifs flanking the Clp1 interaction domain of Pcf11 can interact with RNA only in the absence of Clp1. This situation is reminiscent of Yra1 binding to the same region of Pcf11 as reported by David L. Bentley and coworkers (56). Tethering of Pcf11 to the pre-mRNA enhances binding of Yra1 to the C-terminal ZF1–Clp1-ID–ZF2 region of Pcf11 before transcription termination happens. Sub2 replaces Pcf11 and interacts with Yra1 which will cause the mRNA to be competent for export. In place of Yra1, Clp1 interacts with Pcf11 within the CF IA complex at the time of transcription termination (56). We propose that during these different exchange of interactions, a transient but efficient contact of Pcf11 with the elongating pre-mRNA occurs through the C-terminal ZF1–Clp1-ID–ZF2 domain and before Clp1 recruitment to maintain Pcf11 and associated proteins such as Rna14 and Rna15 on the pre-mRNA. It is also possible that Yra1 could act as a chaperone - as also proposed by Bentley and coworkers (56) - to induce a conformational change thus unmasking the Clp1-binding site of Pcf11.
As a result, the role of the C-terminal zinc-finger domain in pre-mRNA 3′-end processing is very likely direct and mediated through protein-protein contacts made with CPF subunits that are still to be uncovered. The RNA-binding capacity of the zinc-finger mutants is probably not responsible for the observed phenotypes on growth as the purified mutant factors both showed that assembly of the complexes is maintained. We cannot however rule out that a proportion of the mutant Pcf11-19 protein is not associated to the CF IA complex in vivo and thus may perturb RNA processing and export, leading to the strong growth phenotype associated with the pcf11-19 mutant.
In summary, we provide evidence that Pcf11 has two zinc-binding domains mandatory for Pcf11 function in vivo. Even though the exact role of each zinc-binding domain remains to be elucidated, both impact mRNA 3′-end formation to very different intensities when mutated. In comparison, transcription termination at snoRNA genes happens normally, and CF IA assembly is not affected. We propose a direct role in the interaction with specific CPF subunits or a possible function through RNA recognition during the process of coordinating 3′-end processing and mRNP export.
ACCESSION NUMBER
The structure ensemble has been deposited in the Protein Data Bank (http://www.ebi.ac.uk/pdbe) with PDB ID 5M9Z.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Yves Méchulam (Ecole Polytechnique, Palaiseau) for atomic absorption measurements. Access to NMR spectrometers, equipment and technical assistance were possible thanks to Axelle Grélard, Estelle Morvan and the structural biology platform at the Institut Européen de Chimie et Biologie (UMS 3033).
Footnotes
Present address: Julia Guéguéniat, Department of Molecular Genetics, The Ohio State University, Columbus, OH 43210, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Research Agency [ANR-12-BSV8-0016 to S.F., C.D.M., L.M.-S.]; Aquitaine regional government (to A.D. and S.F.); INSERM and the University of Bordeaux; Ministère de la Recherche Scientifique (to J.G.); Centre National de la Recherche Scientifique; Université de Strasbourg; Région Alsace (to J.S., S.C.); Centre National de la Recherche Scientifique [IR-RMN-THC Fr3050]. Funding for open access charge: INSERM, CNRS.
Conflict of interest statement. None declared.
REFERENCE
- 1. Marzluff W.F. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005; 17:274–280. [DOI] [PubMed] [Google Scholar]
- 2. Mandel C.R., Bai Y., Tong L.. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008; 65:1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Millevoi S., Vagner S.. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010; 38:2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y., Manley J.L.. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015; 29:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo W., Johnson A.W., Bentley D.L.. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006; 20:954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kessler M.M., Henry M.F., Shen E., Zhao J., Gross S., Silver P.A., Moore C.L.. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997; 11:2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minvielle-Sebastia L., Preker P.J., Wiederkehr T., Strahm Y., Keller W.. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Minvielle-Sebastia L., Beyer K., Krecic A.M., Hector R.E., Swanson M.S., Keller W.. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998; 17:7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amrani N., Minet M., Le Gouar M., Lacroute F., Wyers F.. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997; 17:3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viphakone N., Voisinet-Hakil F., Minvielle-Sebastia L.. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008; 36:2418–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn E.F., Hammell C.M., Hodge C.A., Cole C.N.. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005; 19:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bai Y., Auperin T.C., Chou C.Y., Chang G.G., Manley J.L., Tong L.. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell. 2007; 25:863–875. [DOI] [PubMed] [Google Scholar]
- 13. Legrand P., Pinaud N., Minvielle-Sebastia L., Fribourg S.. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007; 35:4515–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noble C.G., Walker P.A., Calder L.J., Taylor I.A.. Rna14–Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004; 32:3364–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paulson A.R., Tong L.. Crystal structure of the Rna14–Rna15 complex. RNA. 2012; 18:1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreno-Morcillo M., Minvielle-Sebastia L., Fribourg S., Mackereth C.D.. Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CF IA complex. Structure. 2011; 19:534–545. [DOI] [PubMed] [Google Scholar]
- 17. Qu X., Perez-Canadillas J.M., Agrawal S., De Baecke J., Cheng H., Varani G., Moore C.. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J. Biol. Chem. 2007; 282:2101–2115. [DOI] [PubMed] [Google Scholar]
- 18. Noble C.G., Beuth B., Taylor I.A.. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007; 35:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dupin A.F., Fribourg S.. Structural basis for ATP loss by Clp1p in a G135R mutant protein. Biochimie. 2014; 101:203–207. [DOI] [PubMed] [Google Scholar]
- 20. Gross S., Moore C.. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon J.M., Shikov S., Kuehner J.N., Liriano M., Lee E., Stafford W., Poulsen M.B., Harrison C., Moore C., Bohm A.. Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry. 2011; 50:10203–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stojko J., Dupin D., Chaignepain S., Beaurepaire L., Vallet-Courbin A., Van Dorsselaer A., Schmitter J.M., Minvielle-Sebastia L., Fribourg S., Cianférani S.. Structural characterization of the yeast CF IA complex through a combination of mass spectrometry appraoches. Int. J. Mass. Spectrom. 2016; doi:10.1016/j.ijms.2016.08.005. [Google Scholar]
- 23. Perez Canadillas J.M., Varani G.. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003; 22:2821–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leeper T.C., Qu X., Lu C., Moore C., Varani G.. Novel protein-protein contacts facilitate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J. Mol. Biol. 2010; 401:334–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez-Canadillas J.M. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006; 25:3167–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnwal R.P., Lee S.D., Moore C., Varani G.. Structural and biochemical analysis of the assembly and function of the yeast pre-mRNA 3′ end processing complex CF I. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:21342–21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meinhart A., Cramer P.. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004; 430:223–226. [DOI] [PubMed] [Google Scholar]
- 28. Noble C.G., Hollingworth D., Martin S.R., Ennis-Adeniran V., Smerdon S.J., Kelly G., Taylor I.A., Ramos A.. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 2005; 12:144–151. [DOI] [PubMed] [Google Scholar]
- 29. Yang F., Hsu P., Lee S.D., Yang W., Hoskinson D., Xu W., Moore C., Varani G.. The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3′-end processing. RNA. 2017; 23:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fribourg S., Romier C., Werten S., Gangloff Y.G., Poterszman A., Moras D.. Dissecting the interaction network of multiprotein complexes by pairwise coexpression of subunits in E. coli. J. Mol. Biol. 2001; 306:363–373. [DOI] [PubMed] [Google Scholar]
- 31. Diebold M.L., Fribourg S., Koch M., Metzger T., Romier C.. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011; 175:178–188. [DOI] [PubMed] [Google Scholar]
- 32. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 33. Senn H., Werner B., Messerle B.A., Weber C., TRaber R., Wüthrich K.. Stereospecific assignment of the methyl 1H NMR lines of valine and leucine in polypeptides by nonrandom 13C labelling. FEBS Lett. 1989; 249:113–118. [Google Scholar]
- 34. Shen Y., Bax A.. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013; 56:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen D.F., Neudecker P., Kay L.E.. Determination of isoleucine side-chain conformations in ground and excited states of proteins from chemical shifts. J. Am. Chem. Soc. 2010; 132:7589–7591. [DOI] [PubMed] [Google Scholar]
- 36. Hansen D.F., Kay L.E.. Determining valine side-chain rotamer conformations in proteins from methyl 13C chemical shifts: application to the 360 kDa half-proteasome. J. Am. Chem. Soc. 2011; 133:8272–8281. [DOI] [PubMed] [Google Scholar]
- 37. Brunger A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007; 2:2728–2733. [DOI] [PubMed] [Google Scholar]
- 38. Linge J.P., Habeck M., Rieping W., Nilges M.. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003; 19:315–316. [DOI] [PubMed] [Google Scholar]
- 39. Minvielle-Sebastia L., Preker P.J., Keller W.. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994; 266:1702–1705. [DOI] [PubMed] [Google Scholar]
- 40. Dheur S., Vo le T.A., Voisinet-Hakil F., Minet M., Schmitter J.M., Lacroute F., Wyers F., Minvielle-Sebastia L.. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 2003; 22:2831–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amrani N., Minet M., Wyers F., Dufour M.E., Aggerbeck L.P., Lacroute F.. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997; 17:1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sittman D.B., Graves R.A., Marzluff W.F.. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983; 11:6679–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sadowski M., Dichtl B., Hubner W., Keller W.. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003; 22:2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barilla D., Lee B.A., Proudfoot N.J.. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Romier C., Ben Jelloul M., Albeck S., Buchwald G., Busso D., Celie P.H., Christodoulou E., De Marco V., van Gerwen S., Knipscheer P. et al. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:1232–1242. [DOI] [PubMed] [Google Scholar]
- 46. Preker P.J., Ohnacker M., Minvielle-Sebastia L., Keller W.. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997; 16:4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang C., Vernon R., Lange O., Tyka M., Baker D.. Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Prot. Sci. 2010; 19:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maiti R., Van Domselaar G.H., Zhang H., Wishart D.S.. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004; 32:W590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright J.D., Mace P.D., Day C.L.. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat. Struct. Mol. Biol. 2016; 23:45–52. [DOI] [PubMed] [Google Scholar]
- 51. Krishna S.S., Majumdar I., Grishin N.V.. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003; 31:532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hubbard S.J., Thorton J.M.. NACCESS Computer Program, Department of Biochemistry and Molecular Biology, University College London. 1993. [Google Scholar]
- 53. Haddad R., Maurice F., Viphakone N., Voisinet-Hakil F., Fribourg S., Minvielle-Sebastia L.. An essential role for Clp1 in assembly of polyadenylation complex CF IA and Pol II transcription termination. Nucleic Acids Res. 2012; 40:1226–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bono F., Ebert J., Unterholzner L., Guttler T., Izaurralde E., Conti E.. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004; 5:304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson S.A., Cubberley G., Bentley D.L.. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 2009; 33:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnson S.A., Kim H., Erickson B., Bentley D.L.. The export factor Yra1 modulates mRNA 3′ end processing. Nat. Struct. Mol. Biol. 2011; 18:1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pabo C.O., Peisach E., Grant R.A.. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001; 70:313–340. [DOI] [PubMed] [Google Scholar]
- 58. Hector R.E., Nykamp K.R., Dheur S., Anderson J.T., Non P.J., Urbinati C.R., Wilson S.M., Minvielle-Sebastia L., Swanson M.S.. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002; 21:1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grant R.P., Marshall N.J., Yang J.C., Fasken M.B., Kelly S.M., Harreman M.T., Neuhaus D., Corbett A.H., Stewart M.. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J. Mol. Biol. 2008; 376:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee S.D., Moore C.L.. Efficient mRNA polyadenylation requires a ubiquitin-like domain, a zinc knuckle, and a RING finger domain, all contained in the Mpe1 protein. Mol. Cell. Biol. 2014; 34:3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aibara S., Gordon J.M., Riesterer A.S., McLaughlin S.H., Stewart M.. Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae. Nucleic Acids Res. 2017; 45:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barabino S.M., Hubner W., Jenny A., Minvielle-Sebastia L., Keller W.. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997; 11:1703–1716. [DOI] [PubMed] [Google Scholar]
- 63. Tacahashi Y., Helmling S., Moore C.L.. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 2003; 31:1744–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kyburz A., Sadowski M., Dichtl B., Keller W.. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 2003; 31:3936–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vo L.T., Minet M., Schmitter J.M., Lacroute F., Wyers F.. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol. Cell. Biol. 2001; 21:8346–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. He X., Khan A.U., Cheng H., Pappas D.L. Jr, Hampsey M., Moore C.L.. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003; 17:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Skaar D.A., Greenleaf A.L.. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell. 2002; 10:1429–1439. [DOI] [PubMed] [Google Scholar]
- 68. Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C.L., Greenblatt J.. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003; 278:33000–33010. [DOI] [PubMed] [Google Scholar]
- 69. Dichtl B., Aasland R., Keller W.. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004; 10:965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng H., He X., Moore C.. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 2004; 24:2932–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bhattacharya A., Tejero R., Montelione G.T.. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007; 66:778–795. [DOI] [PubMed] [Google Scholar]
- 72. Laskowski R.A., Moss D.S., Thornton J.M.. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993; 231:1049–1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.