Abstract
Changes in mature microRNA (miRNA) levels that occur downstream of signaling cascades play an important role during human development and disease. However, the regulation of primary microRNA (pri-miRNA) genes remains to be dissected in detail. To address this, we followed a data-driven approach and developed a transcript identification, validation and quantification pipeline for characterizing the regulatory domains of pri-miRNAs. Integration of 92 nascent transcriptomes and multilevel data from cells arising from ecto-, endo- and mesoderm lineages reveals cell type-specific expression patterns, allows fine-resolution mapping of transcription start sites (TSS) and identification of candidate regulatory regions. We show that inter- and intragenic pri-miRNA transcripts span vast genomic regions and active TSS locations differ across cell types, exemplified by the mir-29a∼29b-1, mir-100∼let-7a-2∼125b-1 and miR-221∼222 clusters. Considering the presence of multiple TSS as an important regulatory feature at miRNA loci, we developed a strategy to quantify differential TSS usage. We demonstrate that the TSS activities associate with cell type-specific super-enhancers, differential stimulus responsiveness and higher-order chromatin structure. These results pave the way for building detailed regulatory maps of miRNA loci.
INTRODUCTION
Cellular identity and functional state is reflected in the repertoire and concentrations of RNA species produced within each cell type. Many non-coding (ncRNA) genes encode for functional molecules that play a key role in transcriptional regulation, altering RNA synthesis, processing or degradation rates through regulation of chromatin dynamics and transcription factor (TF) binding, alternative splicing and transcript stability (1). Among the first characterized regulatory ncRNAs, miRNAs represent a cohort of functionally well-defined small RNAs that influence transcript translation and degradation (2,3). They have been shown to be transcribed by RNA polymerase II (RNA Pol II), often in loci containing multiple mature miRNA species that are termed miRNA clusters, capped, polyadenylated and co-transcriptionally spliced, similarly to their precursor messenger RNA counterparts (4,5). However, the mature ∼22 nt forms produced do not retain the transcription start sites (TSS) and the primary transcripts (pri-miRNA) have a short half-life, imposed through the transcription-coupled processing, making the characterization of miRNA genomic loci challenging using conventional RNA-seq methods. Therefore, our current understanding of miRNA expression patterns across cell types derives mainly from profiling the diversity of the mature miRNA forms (6,7, McCall et al. 2017, http://biorxiv.org/content/early/2017/03/24/120394). Recently, an elegant approach to capture pri-miRNAs was taken by inhibiting the effectors DROSHA and DGCR8 of the co-transcriptional Microprocessor complex, thereby allowing sequencing of uncleaved pri-miRNAs (8). Yet, this approach is difficult to apply for monitoring the activity of miRNA loci across cellular conditions. Identifying miRNA TSS based on histone modification data (9,10) would allow leveraging existing large data collections, such as made available by the ENCODE (11) and Roadmap Epigenomics (12) consortia. However, these data cannot define the TSS coordinates at high resolution.
Integrative analysis combining data types from different global assays is a powerful alternative for interrogating novel transcript types, including identification of ncRNA loci. Nucleotide resolution in defining the TSS could be achieved through integration with Capped Analysis of Gene Expression coupled with sequencing (CAGE-seq) data (9,13). Moreover, the genome-wide assay known as Global Run-On sequencing (GRO-seq) has emerged as a key technique to expose differential regulation of primary transcripts and regulatory ncRNAs through its specific design to measure the activity of RNA Pol II-driven transcription (14,15). Moreover, the GRO-seq signal is independent of the stability of the transcripts produced and captures the correlation between gene transcripts and enhancer activity (16,17).
The concomitant production of RNA at enhancers (eRNA) and gene regions opens the possibility to explore the regulatory architecture of miRNA loci across cell types. eRNAs arise at genomic regions associated with TFs and RNA Pol II and were discovered to promote TF binding, chromatin remodeling and enhancer looping, leading to enhanced target gene expression (18–21). Higher-order chromatin organization allows for enhancers to come into contact with promoters across wide distances. However, such looping is also confined by the chromatin architecture through insulator elements bound by the CCCTC-binding factor (CTCF), thereby organizing chromatin into topologically associated domains (TADs) (22). Furthermore, regulatory architecture of many cell identity genes is controlled by densely located regulatory elements, that occupy large genomic regions, called super enhancers (SEs) (23,24). Early studies of SEs performed in stem cells revealed that important pluripotency regulators were targeting these regions overseeing cell identity decisions (23).
Here, we present an adaptable data integration approach that detects pri-miRNA TSS at nucleotide resolution and use it to analyze the TSS-specific transcriptional output across commonly used human cell line models and primary cells in context of regulatory regions and chromatin architecture.
MATERIALS AND METHODS
GRO-seq assay
GRO-seq libraries were produced for A549, ARPE, HEK293T, HeLa, HepG2, hESC9, HUVEC, MRC5, NHA, T98G, SKOV3, THP-1 and U87 cells, to be integrated with our earlier data from HAEC, HUVEC, K562, LNCaP, Nalm6, REH, VCAP and other public GRO-seq samples (in AC16, H1-ESC, HCT116, IMR90, MCF7, Ramos, GM12004 and GM12750 cells). Cell lines were cultured following the ATCC guidelines and replicate samples were included to confirm reproducibility (see Supplementary Figure S1).
For the run-on assay, phosphate-buffered saline-washed cells were incubated in 10 ml of swelling buffer (10 mM Tris–HCl, 2 mM MgCl2, 3 mM CaCl2 and 2 U/ml SUPERase Inhibitor (Thermofisher, Carlsbad, CA, USA) RNAse inhibitor) for 5 min on ice. Cells were pelleted for 10 min at 400 × g and resuspended in 500 μl of swelling buffer supplemented with 10% glycerol. Subsequently, 500 μl of swelling buffer supplemented with 10% glycerol and 1% Igepal was added drop by drop to the cells under gentle vortexing. Nuclei were washed twice with lysis buffer (10 ml of swelling buffer supplemented with 0.5% Igepal and 10% glycerol), and once with 1 ml of freezing buffer (50 mM Tris–HCl pH 8.3, 40% glycerol, 5 mM MgCl2 and 0.1 mM ethylenediaminetetraacetic acid). Nuclei were counted, centrifuged at 900 × g for 6 min and suspended to a concentration of 5 million nuclei per 100 μl of freezing buffer, snap-frozen in LN2 and stored −80°C until run-on reactions. The nuclear run-on reaction buffer (NRO-RB; 496 mM KCl, 16.5 mM Tris–HCl, 8.25 mM MgCl2 and 1.65% Sarkosyl (Sigma-Aldrich, Steinheim, Germany) was pre-heated to 30°C. Then each ml of the NRO-RB was supplemented with 1.5 mM DTT, 750 mM adenosine triphosphate, 750 mM GTP, 4.5 mM CTP, 750 mM Br-UTP (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and 33 ml of SUPERase Inhibitor (Thermofisher, Carlsbad, CA, USA). A total of 50 μl of the supplemented NRO-RB was added to 100 μl of nuclei samples, thoroughly mixed and incubated for 5 min at 30°C. GRO-seq libraries were subsequently prepared as previously described (17). Briefly, the run-on products were treated with DNAse I according to the manufacturer's instructions (TURBO DNA-free Kit, Thermofisher, Carlsbad, CA, USA), base hydrolysed (RNA fragmentation reagent, Thermofisher, Carlsbad, CA, USA), end-repaired and then immuno-purified using anti-Br-UTP beads (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Subsequently, a poly-A tailing reaction (PolyA polymerase, New England Biolabs, Ipswich, MA, USA) was performed according to manufacturer’s instructions, followed by circularization and re-linearization. The cDNA templates were polymerase chain reaction (PCR) amplified (Illumina barcoding) for 11–14 cycles and size selected to 180–300 bp length. The ready libraries were quantified (Qubit dsDNA HS Assay Kit on a Qubit fluorometer, Thermofisher, Carlsbad, CA, USA) and pooled for 50 bp single-end sequencing with Illumina Hi-Seq2000 (GeneCore, EMBL Heidelberg, Germany).
Pre-processing GRO-seq data
A summary of all GRO-seq samples is presented in Supplementary Table S1. Raw reads for public GRO-seq data were acquired from the SRA database. GRO-Seq reads were trimmed using the HOMER v4.3 (http://homer.salk.edu/homer) (25) software (homerTools trim) to remove A-stretches originating from the library preparation. From the resulting sequences, those shorter than 25 bp were discarded. The quality of raw sequencing reads was controlled using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) (26) and bases with poor quality scores were trimmed (typically for read length of 50 requiring a minimum 97% of all bases in one read to have a minimum phred quality score of 10, otherwise adjusted based on read length) using the FastX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) available in the Galaxy platform (27). Aligning reads to the human hg19 reference genome version was preceded by removing reads mapping to rRNA regions (AbundantSequences as annotated by iGenomes) and blacklisted regions (unusual low or high mappability as defined by ENCODE, ribosomal and snoRNA loci from ENCODE and further manually curated for human genome (bed file available upon request)), all processed with the Bowtie version bowtie-0.12.7 (28). Up to two mismatches and up to three locations were accepted per read and the best alignment was reported. For visualization, reads were normalized to 107 reads to generate bedGraph and bigWig files. The bigWig files were further converted to track hubs and visualized as strand specific, MultiTracks on a custom Track Hub in the UCSC Genome browser. Previously unpublished data collected for the study has been deposited under the accession GSE92375.
Pre-processing ChIP-seq data
Active promoter and enhancer status can be distinguished based on the methylation status of histone 3 lysine 4 (H3K4) (11). Typically, trimethylation marks active promoters, while monomethylation can be used to distinguish enhancers. To compare these marker levels at candidate TSS locations, chromatin immunoprecipitation data (ChIP-seq) for H3K4me1 and H3K4me3 was downloaded from ENCODE (UwHistone and BroadHistone data available via ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/). The aligned .bam files were used for analysis. For cell lines not included into the ENCODE data collection, public data was acquired from the SRA database and processed as the GRO-seq data, with the exception that all reads that passed quality controlled were used for alignment. SE coordinates were downloaded from the dbSUPER (29) database.
Pre-processing CAGE-seq data
CAGE-seq peak scores were obtained from the FANTOM5 (30) consortium hg19 data release (hg19.cage_peak_phase1and2combined_counts.osc.txt). The scaling factors used for normalizing the scores across samples were obtained by upper quartile normalization (31) of the regions corresponding to annotated TSS. This method has been found robust for RNA-seq data normalization (32). Samples with <1 000 000 reads within annotated TSS regions were discarded. The normalization factors obtained were then used to normalize all the CAGE-seq peak scores in samples selected for analysis (refer to Supplementary Table S2).
Many of the peaks included in the peak matrix were found to locate to intragenic exon regions, representing a known problem in CAGE assays (13). To avoid false positives in downstream analysis, we compared the signal distribution between true TSS and exon-background and found that they could be separated on log-scale (data not shown). Gaussian mixture models were therefore selected as an approach to classify the normalized CAGE-seq scores into TSS or non-TSS signal. The Model-Based classification and density estimation was performed using the R package mclust 5.1 (MclustDA) (33). The models were fitted separately to log2 scores representing annotated TSS and annotated exons (that do not overlap any known TSS) from expressed gene loci (GRO-seq RPKM > 1) in three GM12878 replicate samples. The model was then used to classify all the normalized CAGE peaks across all samples. As an additional filtering step, any CAGE-seq peaks overlapping exons from gene body regions (i.e. not representing first exons) were required to pass the histone mark filtering step, as described below for de novo identified candidate TSS.
To identify TSS at 1-bp resolution, the FANTOM5 CAGE-seq datasets for each cell type were downloaded from the ftp resource (http://fantom.gsc.riken.jp/5/datafiles/latest/basic/) in .bam format and converted into bedGraph signal files visualizing the 5′ nt of each read (HOMER makeUCSCfile -tss option).
De novo identification of primary transcription units in each cell type
The miRNA maturation pathway is schematically illustrated in Figure 1A. The identification of pri-miRNA transcripts was performed in 26/27 cell types where matching CAGE-seq data were available. For the closely related cells GM12004 and GM12750, further referred to as GM pool, the GRO-seq samples were pooled for transcript detection. The workflow of the pipeline is depicted in Supplementary Figure S2A and tools and applied parameter settings are described below. Scripts and example data are available from https://bioinformatics.uef.fi/prima/.
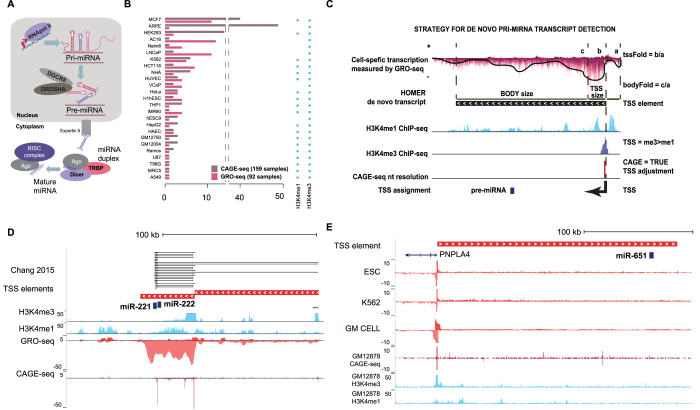
Figure 1.
Identification of active TSS from primary microRNA gene loci across human cell types. (A) Canonical miRNA maturation pathway. (B) Summary of the dataset assembled from GRO-seq, CAGE-seq (FANTOM5 Consortium) and histone ChIP-seq data (refer to Supplementary Table S1 for dataset identifiers). (C) Schematic depiction of the strand-specific TSS identification strategy by integration of GRO-seq (top), histone ChIP-seq (middle) and CAGE-seq (bottom track); the HOMER de novo transcript represents an exemplary transcription unit detected on the negative strand. (D) Two TSS detected by the integrated analysis from the hsa-miR-221∼222 locus (chrX:45, 550, 068–45 712 839) are shown, with the proximal (TSS1) and the distal (TSS2) sites located 23 140 and 104 431 nt, respectively, from the pre-miR-222; these TSS are also represented in the pri-miRNA transcript assembly by Chang et al. 2015. Signal combined from HUVEC samples is shown for each assay type. (E) A novel transcript overlapping the hsa-miR-651 pre-miRNA is detected on the plus strand, based on integrated analysis (visualized as in A) in ESC and multiple blood cell type (K562, GM cell), with a TSS that is located 199462 nt upstream from the precursor. The PNPLA4 gene encoded on the opposite strand shares the promoter (coordinates shown are chrX:7 862 384–8 132 720).
Transcript detection
HOMER v4.3 software suite was used for analyzing all uniquely mapped reads per cell type. The de novo transcript identification for each cell type was performed using the findPeaks.pl option -groseq, considering mappability gaps to avoid gaps (-uniqmap). The optimal choice of additional parameters that control the sensitivity were found to vary depending on sequencing depth and sample quality (refer to Supplementary Figure S1D and E). To improve the robustness of the transcript discovery in context of different sequencing depths per cell type, results from transcript detection were pooled from three separate runs where parameters that affect transcript detection sensitivity and specificity were adjusted (setting 1: minBodySize 1000 and minReadDepth 10, setting 2: minBodySize 1500 and minReadDepth 5, setting 3: minBodySize 900 and minReadDepth auto). Especially setting 2 helped increase the number of transcripts discovered in low depth libraries. Any transcripts matching blacklisted regions as specified in the GRO-seq pre-processing were discarded. Thereof, additional data (chromatin marks and CAGE-seq peaks) were used in filtering transcript candidates. First, ±500 bp around the TSS of novel transcript candidates were extracted and intersected with CAGE-seq peaks classified as TSS from all samples from the matching cell type. TSS with overlap in >1% of samples were kept and the start coordinate shifted to the CAGE-seq signal maximum, at nucleotide resolution.
Next, ChIP-seq signal for H3K4me1 and H3K4me3 signal level was quantified from ±500 bp around the putative TSS using annotatePeaks.pl with cpm normalization. A putative TSS passing the H3K4me3 > H3K4me1 filter was defined as a TSS where H3K4me3 levels were above 10 cpm and at least 5-fold higher than the H3K4me1 signal. For cell lines AC16, ARPE, hESC9, IMR90, MRC5, Nalm6 and VCAP the H3K4me1 data were not available. In this case, the H3K4me3 signal level was compared to the input or H3K27 acetylation ChIP-seq sample signal (see Supplementary Table S2). Transcript body was assembled from proximal transcript pieces (BEDTools cluster with parameters -d 500 -s). For typical distribution of gaps within annotated gene regions, refer to boxplots in Supplementary Figure S2A, step 2. Final transcript length above 5 kb was accepted. Finally, TSS-classified CAGE-seq peaks within de novo detected transcripts were included to resolve cases where the HOMER TSS identification did not accurately capture the transcript start (typically due to overlapping upstream eRNA signal). All transcript candidates passing the above specified TSS chromatin marker filter and CAGE-seq peak TSS filters were kept.
TSS evaluation across cell types
To minimize false positives, data from cell types with both H3K4me3 and H3K4me1 data were used to obtain quantification data for all found TSS. These data were used to filter away candidate TSS that had either weak support from CAGE-seq (not passing histone marker ratio) or consistent evidence of higher H3K4me1 signal (putative enhancers) in a matching region (90% overlap).
Defining genomic regions to evaluate the output of each TSS on primary transcription level
The identified primary transcription units were pooled across cell types and clustered (BEDTools cluster with parameters -s -d 0). Next, clusters were analyzed to distinguish the identified TSS and to define genomic regions that we refer to as TSS elements that allow quantification of the output of each TSS (refer to Supplementary Figure S2A, TSS element quantification). The TSS elements represent non-overlapping regions between the identified TSS in each cluster. Toward this end, TSS windows were specified at each identified TSS within ±1000 bp and then compared, to collapse nearby transcript starts. The most frequent start coordinate within each window was used to specify the start for the TSS element. Longest end for each transcript was also recorded across cell types. The element end coordinate was assigned the next TSS window start, until the cluster end. For the last element of the cluster, the end was assigned based on prominent drop in signal, as detected using change point analysis (R package changepoint (34): penalty BIC, method BinSeg, Q 1) from cell types in which the transcript had been discovered. The change point was analyzed from annotated transcript end until the cluster end. At novel loci, the end of the transcript corresponding to median tail length (based on annotation-matched transcripts) was analyzed. The furthest change point coordinate found was then chosen as the element end coordinate for expression level quantification. The resulting non-overlapping coordinates were then used for evaluating the contribution of each TSS to the primary transcript expression level.
Transcript annotation
For specifying precursor miRNA (pre-miRNA) locations, transcript annotations were retrieved from GENCODE (v19) and miRBase (v20) (35). Coding, long non-coding and miRNA annotations were distinguished. The .gtf or .gff files were converted to bed format using the R package GenomicFeatures (makeTxDbFromGFF, transcriptsBy or microRNAs). miRBase was integrated with GENCODE miRNA annotations to achieve a non-redundant catalog of pre-miRNA coordinates by substituting overlapping region annotation with the miRBase identifier. The overlap of TSS elements with pre-miRNA coordinates was used to detect candidate pri-miRNA transcripts (refer to Supplementary Table S3). The overlap to each TSS element was based on the coordinate range from the start until the longest end. The pri-miRNA TSS elements were further divided into intergenic and intragenic. Intragenic refers to miRNA species that share a TSS with coding genes. These were identified by overlapping annotated coding gene TSS with the pri-miRNA TSS element starts (±1000 bp). In gene-dense regions, the overlap based on longest end also resulted in likely false positive assignment of multiple coding genes as miRNA host genes. In these cases, only the TSS elements matching the closest coding gene were accepted. Candidate novel TSS for coding genes were also considered to represent miRNA host transcripts. They either represent TSS elements directly upstream or contained within annotated coding transcripts. It should be noted that due to annotation discrepancies, some miRNAs fall into either intra- or intergenic regions when consulting either RefSeq or GENCODE annotations (e.g. hsa-miR-196a-1 inside a putative HOXB7 transcription variant annotated in GENCODE). To compare the annotations to those reported in (8), we retrieved the .gtf for the RNA-seq based assembly of pri-miRNAs and overlapped them with pre-miRNA coordinates in a strand-specific manner using BEDTools (36) (limiting to miRBase annotations that were used in their work). Common pre-miRNA were defined based on overlapping transcript found using both approaches. Next, TSS regions (±1000 bp) were overlapped in a similar manner to detect additional common transcripts where the extension over the pre-miRNA was supported at least by one approach.
Quantification of pri-miRNA transcripts
Non-mappable coordinates, exons of coding genes and ribosomal RNA regions were removed from TSS elements prior to quantification using BEDTools (subtractBed) to exclude regions known to cause problems in quantification of GRO-seq data. Exon coordinates for coding genes were based on Refseq and UCSC knownGene annotations (November 2016). Exons were retrieved using the R/Bioconductor GenomicFeatures exonsBy command and overlapping coordinates were merged (BEDTools merge with parameters -s -c 6 -o distinct). The mappability file hg19_wgEncodeCrgMapabilityAlign50mer.bedGraph.gz was downloaded from UCSC table browser for 50mer alignments and processed to bed format to discard non-mappable regions. The non-overlapping pieces of each TSS element were quantified using HOMER (analyzeRepeats.pl with parameters -strand + -noadj -noCondensing -pc 3) and the read count summed. The lengths of the quantified region and total read counts in each library were used to report normalized signal levels (RPKM). The contribution of each TSS (TSSi) to the overall transcriptional activity in a given locus was determined by subtracting the signal level at the upstream element (TSSi+1), based on the RPKM values. The obtained values are referred to as differential TSS activity levels.
Quantification of mature miRNA transcripts
For RNA extraction, HUVEC were cultured as described in (37) and Nalm6 and A549 cells according to ATCC guidelines. RNA was extracted using phenol–chloroform extraction as in (38) with an overnight isopropanol precipitation step to capture short RNA species. Mature miRNA levels of hsa-miR-221–5p and hsa-miR-222–5p were quantified by RT-qPCR using the Exiqon miRCURY LNA Universal microRNA PCR assays (Exiqon A/S, Vedbaek, Denmark) according to manufacturer's instructions; normalization was performed to the UniSp6 synthetic RNA control template.
ENCODE CSHL short RNA-seq data from A549, GM12878, IMR90 and K652 total cell lysates was downloaded as mapped bam files (GSE24565). The BEDTools coverage tool was used for counting the reads on pre-miRNA loci with default settings (including strand specificity parameter -s). Reads were normalized to total library sizes and to quantified region length. Corresponding data from (McCall et al. 2017, http://biorxiv.org/content/early/2017/03/24/120394) (cpm data matrix) was included as a second dataset.
Sample set characterization
Spearman correlation of new GRO-seq samples was calculated. In accordance with ENCODE guidelines, the gene-level correlation of replicates within the new sample set was confirmed to be >0.9. In addition, the same criteria was met when correlating the new HUVEC samples generated with samples in our earlier study that represent the same condition (Supplementary Figure S1A–C).
To visualize sample similarities in two dimensions, the dimensionality reduction was performed using t-distributed Stochastic Neighbor Embedding (t-SNE) from R package Rtsne (with perplexity parameter set to 30), using either a geneset of pri-miRNA TSS element or similarly obtained TF TSS element quantification (log2 TPM normalized signal levels (39).
Hi-C data analysis
Hi-C data were processed as described previously (37). TADs were identified using HOMER v4.3, with ’findHiCDomains.pl’, using resolutions of 5 and 25 kb. This analysis is based on a statistic referred to as the ‘directionality index’, which describes the tendency of a given position to interact with either the chromatin upstream or downstream from its current position. The HOMER tool ‘analyzeHiC.pl’ was used to generate the heatmaps to visualize interaction frequencies.
Statistical test for pri-miRNA expression analysis
Non-normalized TSS element counts were used to perform differential expression analysis for LNCaP TNFα treatment (GSE83860). The BioConductor limma package was used to compute moderated t-statistics and false discovery rates (Benjamini-Hochberg) in order to identify significant changes between treatment and control.
Quantification of eRNA levels at candidate regulatory elements
Regulatory elements were quantified from GRO-seq libraries based on DNase I Hypersensitivity narrow peak coordinates from ENCODE (OpenChrom, Duke University, GSE32970). The peaks were extended ±250 bp from the peak center and merged using BEDTools merge to avoid redundancy. Peaks that were located in a 500-kbp window from both pri-miR-221∼222 TSS were quantified using HOMER v4.3 ‘analyzeRNA.pl’ across cell types. Peaks located within the TSS elements of pri-miR-221∼222 were quantified on the opposite strand of the pri-miRNA transcript. Peaks located outside of the pri-miRNA transcript were quantified on both strands (-strand both). All peaks were normalized by the length of the quantified region and the total number of mappable reads per GRO-seq library.
Additional data used in this study
Refer to Supplementary Table S2 listing additional data displayed in the figures. Transient transcriptome (TT)-seq (GSE75792) (40) and precision nuclear run-on (PRO)-seq (GSE96869) datasets were downloaded as mapped bam files and converted into bedGraph signal files for independent validation at selected pri-miRNA loci.
RESULTS
Integration of high-throughput sequencing data across cell types allow fine-resolution mapping of human pri-miRNA TSS
To allow profiling the functional state of miRNA primary transcription units across commonly used human cell lines, we have generated GRO-seq libraries for 19 cell types, and added publicly available datasets for 9 cells (in total 27 cell types, refer to Figure 1A for a schematic representation of the miRNA transcript synthesis and processing steps). We integrated these 92 nascent transcriptome samples with existing annotations and other gene regulatory data from ENCODE and FANTOM5 consortiums (Figure 1B, see also Supplementary Tables S1 and 2) to extend the genomic characterization of pri-miRNA loci, in a manner that is not limited by transcript stability (14).
In our analysis, pipeline candidate transcripts were first identified from cell-specific GRO-seq signal (see ‘Materials and Methods’ section). The exact TSS coordinate was then assigned based on CAGE-seq peaks (available in 26 cell types). The TSS status was further supported by the promoter histone methylation levels, through quantification of the H3K4me3 ratio to H3K4me1 (Figure 1C, see also Supplementary Figure S2A and ‘Materials and Methods’ section). In some regions, several closely localized TSS (within 1 kb) were obtained from different cell types. In this case, the most prevalent TSS was used in downstream analysis. In total, we identified 305 intergenic and 1242 intragenic active pri-miRNA TSS, corresponding to 415 and 1059 mature miRNAs from miRBase and GENCODE, respectively, that localize to 1177 genomic clusters. The evidence supporting the activity of each TSS based on the different genomic assays is summarized in Supplementary Table S3.
To benchmark the identification approach, we compared our results to the miRNA annotations (available for 1267/1871 miRNAs annotated in miRBase) obtained based on RNA-seq from DROSHA-DCGR8-depleted cell models (8). The 1142 common miRNAs include those with transcript covering the same pre-miRNA (1074) and 68 pre-miRNA sharing the same TSS (±1 kb) where overlap to pre-miRNA coordinates was confirmed based on the assembly in (8) (Supplementary Figure S2B). The mir-221∼222 locus represents a miRNA cluster that was captured with high confidence using both approaches (Figure 1D), whereas our more extensive cell collection allowed identification of novel pri-miRNA transcripts (149 in total), including a transcript spanning miR-651 that is specific to embryonic stem cells and blood cells (Figure 1E). The 125 pri-miRNA loci supported only by the assembly in (8) were found to correspond primarily to low expressed miRNA loci (Supplementary Figure S2C).
Long primary transcripts and existence of multiple TSS characterize complex miRNA loci
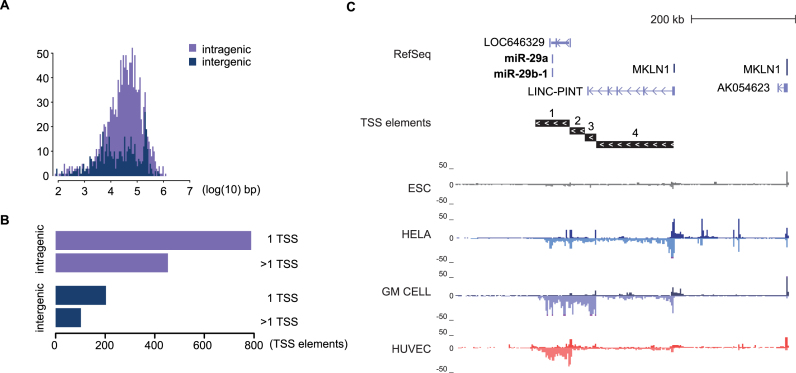
We next examined the locations of the TSS that were associated with each mature miRNA. Computational predictions of miRNA TSS have been based on closest promoter marker (H3K4me3) peaks. Here, the associated active transcriptional profiles provide evidence of new distal TSS that reside far away from the annotated pre-miRNA location, in line with the RNA-seq-based transcript assembly in (8). The log10 distance (in kb) from the annotated pre-miRNA location is shown in Figure 2A. Intergenic and intragenic miRNAs displayed a similar distance range, with majority of TSS at a 103–105 bp distance. However, the intergenic TSS distance distribution is notably more flattened (Figure 2A). More than one-third of pri-miRNA TSS elements are located in loci with multiple transcript variants (TV) at both inter- and intragenic loci (102/305 intergenic, 453/1242 intragenic TSS elements, Figure 2B, see also Supplementary Table S3). The miR-29a∼29b-1 cluster (Figure 2C and Supplementary Figure S3) exemplifies the complexity of intergenic miRNA loci: distinct TSS are active across cell types. Interestingly, the most distal TSS is shared with a lncRNA known as long intergenic non-protein coding RNA, p53 induced transcript, LINC-PINT and elongation from this TSS4 was confirmed from independent TT-seq and PRO-seq data available for K562 cells (Supplementary Figure S3C). The transcription from three proximal TSS on the other hand does not cover the lincRNA gene locus. These TSS for shorter TV have prominent activity across normal cells compared to ES or cancer cells that co-express both ncRNA species (Figure 2C and Supplementary Figure S3).
Figure 2.
pri-miRNA loci display complex patterns of distal TSS. (A) Distribution of pri-miRNA TSS log10 distance (in kb) relative to the miRNA precursor across all mature miRNA species is shown separately for intragenic (median distance 33.8 kb) and intergenic (median distance 20.4 kb) miRNA TSS, for miRNAs from miRBase and GENCODE. (B) Total numbers of intra- and intergenic pri-miRNA loci with one or multiple TSS elements found across all cell types (refer to Supplementary Table S3 listing miRNAs in each category), for miRNAs from miRBase and GENCODE. (C) GRO-seq signal across four cell types (ESC, HeLa, GM cells and HUVEC) at the complex pri-miRNA locus of hsa-mir-29a∼29b-1 cluster is shown (chr7:130 376 783–131 016 782). Four distinct TSS were identified, with the most distal (TSS4) being shared with the ncRNA LINC-PINT (FLJ43663).
Cell type-specific TSS usage at intergenic pri-miRNA loci
Based on the transcript identification results, approximately 20% of miRNA clusters have evidence of several TSS. Furthermore, active TSS locations within a miRNA locus were distinct when comparing between cell types (see Figures 1D and 2C; Supplementary Figure S3). Considering that this complex architecture may be an important regulatory feature at miRNA loci, we developed a strategy to quantify differential TSS usage (see ‘Materials and Methods’ section). The contribution of each TSS to the total transcriptional activity at the locus is exemplified at the mir-100∼let-7a-2∼mir-125b-1 locus in Figure 3A. In total six active TSS were found for this miRNA cluster by our approach. In order to reflect the contribution of each TSS to the observed signal levels in a given cell type, the locus was divided into six quantification elements that specify a unique region downstream of each TSS (the TSS numbering reflects increasing distance from the first annotated pre-miRNA start coordinate). The GRO-seq signal within each element was then quantified across the cell types (see ‘Materials and Methods’ section).
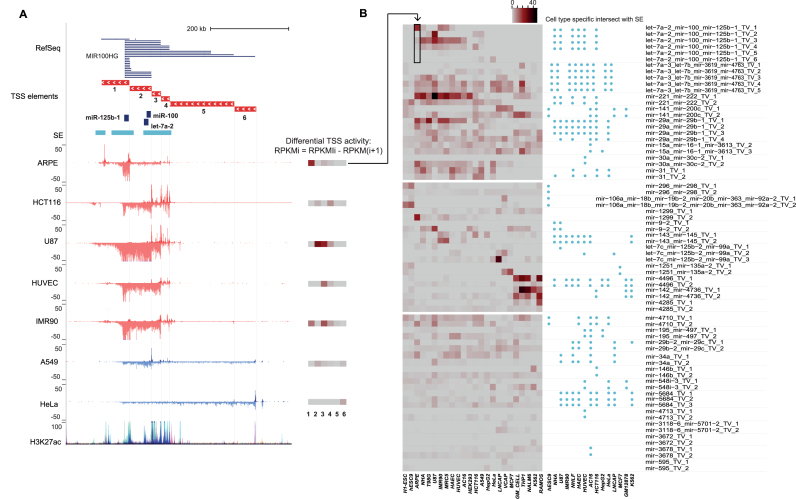
Figure 3.
Quantification of TSS elements that are used for cell type specifically. (A) The use of cell type-specific TSS can be quantified based on the TSS-elements (see ‘Materials and Methods’ section), as exemplified at the hsa-mir-100∼let-7a-2∼mir-125b-1 locus (chr11:121 723 037–122 387 271) that harbors six differentially active TSS, as supported by the Refseq annotation track. The differential TSS activity calculated based on the GRO-seq signal difference between adjacent elements (see equation in the legend and ‘Materials and Methods’ section) is summarized as a heatmap next to each track, where darker color tones correspond to higher TSS activity (in RPKM). The TSS numbering increases with the distance from the pre-miRNA. Super-enhancer locations indicated above the GRO-seq tracks can be compared to the ENCODE H3K27ac track (both correspond to overlaid activity from multiple cell types). (B) Cross-lineage quantification of differential TSS activity (left) and proximity to cell type-specific SE (right, ±100 kb) at intergenic pri-miRNA loci with multiple TSS is shown. The transposed heatmap panel corresponding to the ARPE data shown in (A) is indicated by an arrow. The heatmap is organized into three sections: the upper part of the heatmap corresponds to miRNA loci with highly dynamic TSS activity across multiple cell types; the middle part shows miRNA loci with cell-specific high activity; the lower part corresponds to miRNAs with moderate to low expression level across cell types. The ordering of the cell types reflects their origin: H1-ESC (stem cell); the neuronal lineage cell types ARPE, NHA (normal), T98G and U87 (cancer); mesoendodermal cell types IMR90, MRC5 (fibroblast), HAEC, HUVEC (endothelial), AC16 (cardiomyocyte) and HEK293T (kidney); cancer cell lines HCT116, A549, HEPG2, HeLa, LNCaP, VCAP, MCF7; blood lineage cell types (GM lymphoblastoid, normal), THP1, Nalm6, K562 and Ramos (cancer).
Similar to TFs, miRNAs are considered to be important regulators of cell fate. As a first confirmation, we tested whether the miRNA locus activity, as reflected by the quantified GRO-seq signal within each TSS element, could distinguish between the cell types analyzed. The dimensionality reduction (t-SNE) results generated using miRNA loci segregate the cell types in a comparable manner to the result obtained based on TF loci, as shown in Supplementary Figure S4.
Next, the contribution of each TSS (TSSi) to the overall transcriptional activity in a given locus was determined by subtracting the signal level at the upstream element (TSSi+1), based on the RPKM values. The TSS activity levels are summarized as heatmap bars next to the GRO-seq signal tracks (Figure 3A).
As can be seen from the signal tracks (Figure 3A), the most distal TSS (TSS6) is active only in A549 and HeLa cells. In HeLa, the signal remains constant across the locus, indicating that none of the other TSS are active. In comparison, at TSS1 location, a signal increase occurs in APRE and IMR90 cells, indicating its active state. Similarly, by comparing the signal before and after TSS2, HCT116, U87 and A549 utilize this TSS, while the constant signal level in ARPE and HUVEC indicates that the TSS is inactive. These patterns are readily distinguished from the differential TSS activity heatmaps shown next to the signal tracks.
Near TSS2, 3 and 4, the pri-miRNA signal on the minus strand is overlapped by multiple signal peaks on the opposite strand. Such peaks are indicative of strong enhancers, and co-localize with H3K27ac peaks. In confirmation, we retrieved annotated SE coordinates across multiple cell types from dbSUPER (Supplementary Table S2): TSS1–4 with highest quantified activity levels reside in close proximity to SE (Figure 3A).
Next, we utilized our quantification approach to compare miRNA TSS activities across the GRO-seq cell line collection. The result for miRBase intergenic miRNAs with multiple TSS is shown in Figure 3B. The 30 miRNA clusters are sorted into three groups. The mir-100∼let-7a-2∼mir-125b-1 locus is among the first group of miRNA loci that show cell type-specific differences in TSS utilization. In the middle group, the miRNA loci are highly expressed in only a small subset of cell types, including the ESC-restricted mir-106a cluster (41). The last group shows medium to low activity miRNA loci with less distinct cell- or TSS-specific activity profile. We further analyzed the presence of SEs in the vicinity (±100 kb) of the pri-miRNA TSS. The co-localization with these strong enhancers is visualized next to the heatmap, and shows a pattern that resembles the TSS activity profile with low activity miRNA loci lacking SE and cell-specific activity corresponding to cell-specific SE detection.
Distinct super-enhancers correlate with the activity of alternative TSS at the mir-221∼222 locus
To further elucidate the regulatory architecture at a miRNA locus, we selected the mir-221∼222 locus for more detailed examination, where SE locations across different cell types indicated six highly active regulatory regions in the vicinity (Figures 4A and 3B). The two alternative TSS found have differential activity across cell types (including normal and cancer cell types): the more distal TSS (TSS2) has lower and more constant level of expression across the cell types analyzed (Figure 3B), while TSS1 activity is highly dynamic and among the most active miRNA TSS overall. The mature miRNA levels, in agreement, were highest in IMR90 cells with high TSS1 activity (see Supplementary Figure S5 showing short RNA-seq data and RT-qPCR validation in multiple cell types for miR-221∼222, miR-100∼let-7a-2∼miR-125b-1 and miR-29a∼29b-1 clusters).
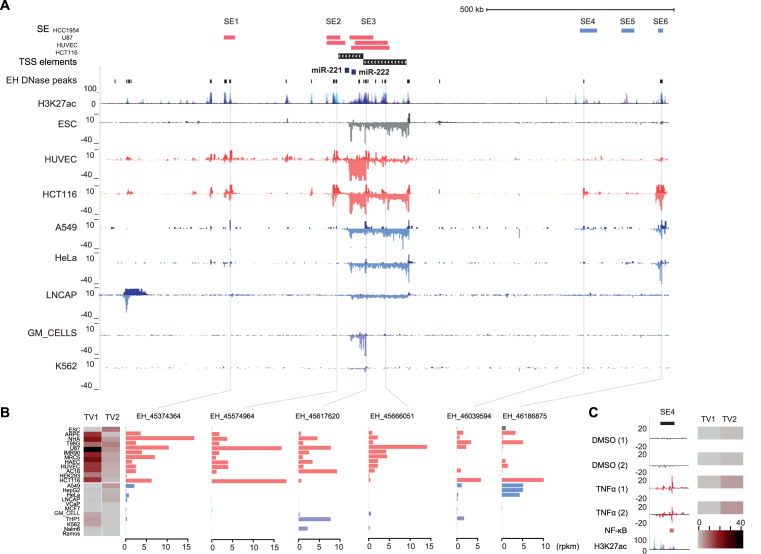
Figure 4.
hsa-mir-221∼222 expression is regulated by two TSS with distinct correlation to enhancer usage. (A) The super-enhancer track indicates the locations of six SEs found within ±500 kb from the two hsa-miR-221∼222 TSS in HCC1954, U87, HUVEC and HCT116. Sub-regions corresponding to individual active enhancers (see ‘Materials and Methods’ section) within the SEs and across the locus are indicated based on ENCODE DNAse-seq peaks. The ENCODE H3K27ac track provides additional confirmation for the locations of candidate regulatory regions (signal overlaid from multiple cell types). The GRO-seq signal from the hsa-miR-221∼222 cluster locus (chrX:45 169 213–46 205 597) is shown from human primary and cancer cell lines representing differential TSS activity: HUVEC, HCT116, GM cells and K562 represent cell types with active TSS1; ESC, A549, HeLa and LNCaP represent cell types with only TSS2 activity. (B) The differential TSS usage (shown as heatmap, as in Figure 3) can be compared to eRNA levels at representative enhancers from each SE region (see also Supplementary Figure S6). (C) GRO-seq signal from the TNFα-stimulated LNCaP cells is shown from the activated SE4 region (see also Supplementary Figure S6 showing the whole locus). NF-κB binding as measured in the same condition (GM12878 cells) and layered H3K27ac are shown below the GRO-seq tracks.
Even when the same mature miRNA level could be achieved by alternative TSS, their cell-specific activity and stimulus responsiveness may be completely different. Genome-wide mapping of nascent RNA opens a unique dimension where activity of regulatory regions can be inferred through investigating the eRNA patterns within enhancer regions. To characterize enhancer activity within the mir-221∼222 locus, we retrieved a list of DNase peaks (±500 kb from each TSS) from ENCODE across multiple cell types. eRNA expression centered at each peak (±500 bp) was then quantified based on the GRO-seq dataset. The results for representative enhancers within the six SE regions are shown in Figure 4B (all enhancers are shown in Supplementary Figure S6). The enhancer activity can be compared to the heatmap on the left showing the differential TSS activity. The enhancers in the immediate vicinity and downstream TSS1 (SE1–3) are highly active in cell types with predominant TSS1 activity. In contrast, SE4 and SE6 have a broader activity pattern, closely matching TSS2 activity (SE5, not shown, had modest eRNA levels across all cell types analyzed). A subset of enhancers within SE3 were active in the blood cell types that only showed weak TSS2 activity (Figure 4 and Supplementary Figure S6).
To further investigate whether the two TSS would be differentially responsive to stimuli, we performed differential expression analysis comparing the read counts at miRNA TSS elements between different treatment conditions in the GRO-seq dataset (refer to Supplementary Table S1). In LNCaP cells, the most significant change in pri-miRNA transcription upon TNFα treatment was found at the mir-221∼222 locus (adjusted P-value 2.40E-37), increasing by 2.40-fold the activity of TSS2 (TSS1 not active). Accordingly, the eRNA profile at SE4 (Figure 4C) showed a rapid activation, while SE1–3 remained unresponsive (Supplementary Figure S7). In further confirmation, the activated enhancer region within SE4 contains an NFkB binding site (GM12878 cell ChIP-seq data from ENCODE is shown).
Chromatin organization reveals TAD boundaries at pri-miRNA loci with differential TSS activity
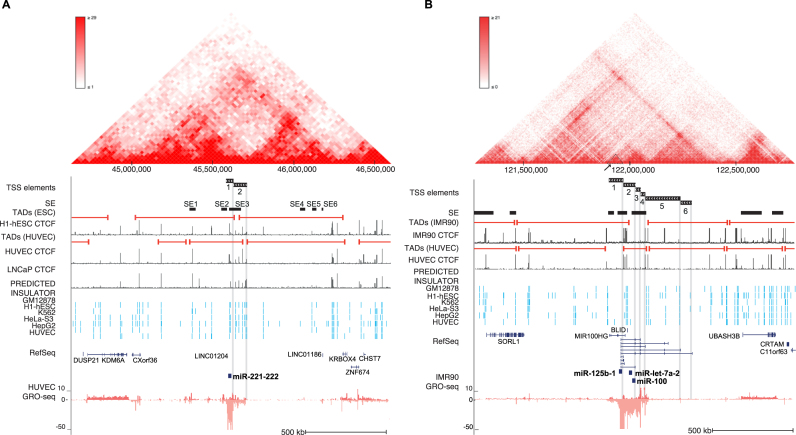
Based on the differential TSS activity pattern observed at multiple miRNA loci, we hypothesized that the higher-order chromatin structure might restrict contacts between the TSS and regulatory elements and thereby contribute to the cell- and stimulus-specific TSS activation patterns. We used insulator annotations and CTCF binding profiles from ENCODE chromatin segmentation, together with Hi-C data from three cell types (H1-hESC, HUVEC and GM12878) (42–44) to define TAD boundaries and analyzed these at miRNA clusters with multiple TSS.
The genomic region around the mir-221∼222 locus is shown in Figure 5A. A TAD boundary co-localizing with several CTCF ChIP-seq peaks and multiple nearby insulator segments was detected between the mir-221∼222 TSS (supported by data from H1-hESC, HUVEC and LNCaP cells). The downstream region from TSS1 containing SE1–3 has a distinct interaction pattern from the upstream region harboring SE4–6 (heatmap, data from HUVEC shown above the tracks). Similarly, the six TSS at mir-100∼let-7a-2∼mir-125b-1 locus are separated by TAD boundaries (Figure 5B): the low activity distal TSS (mir-100∼let-7a-2∼mir-125b-1 TSS5 and 6) are separated from the SE regions in vicinity of TSS1–4 (see also Figure 3A). Prominent CTCF peaks and insulator annotations from several cell types were also found between the mir-100∼let-7a-2∼mir-125b-1 TSS1 and mir-100∼let-7a-2∼mir-125b-1 TSS2–4. Closer examination of the Hi-C interaction pattern (a sub-triangle, highlighted in Figure 5B) indicated that this mir-125b-1-specific TSS may also segregate into its own regulatory domain, matching its more lineage-specific activity pattern (Figure 3A).
Figure 5.
Chromatin architecture contributes to alternative TSS usage at pri-miRNA loci. (A) Chromatin structure at the hsa-miR-221∼222 locus (chrX:44 625 000–46 600 000) in human endothelial cells. The Hi-C interaction frequency is shown as a triangle heatmap above (25 kb resolution). TAD domains found at 5 kb resolution from HUVEC and ESC are shown. TAD boundaries can be compared to the CTCF signal peaks (ENCODE) from corresponding cell types. The SE track indicates the locations of SE detected in all analyzed cell types. The location of TSS elements (matching different TADs) is highlighted. (B) Chromatin structure is shown at the hsa-mir-100∼let-7a-2∼mir-125b-1 locus (chr11:121 270 000–122 775 000) based on human fibroblasts (IMR90) data (5 kb resolution), as in (A). TAD domain boundaries (5 kb resolution) calculated from IMR90 and HUVEC, and the corresponding CTCF (ENCODE) signal are shown as in (A). The SE track indicates the locations of SEs detected in all analyzed cell types.
DISCUSSION
Using nascent transcriptomes and data integration we validated here that pri-miRNA display complex genomic organization with one or several TSS that can localize several hundred thousand bp upstream from the pre-miRNA coordinates. These results are consistent across 27 commonly used human cell line models and primary cells. We found that usage of multiple TSS is common within intergenic pri-miRNA genes and study in detail three such complex loci: mir-29a∼29b-1, mir-100∼let-7a-2∼125b-1 and mir-221∼222. The observed complexity of miRNA loci is in agreement with previous findings in multiple species (8,9). Focusing on the locus of mir-221∼222, where the identified TSS have also been previously validated using RACE (8), we demonstrated that the activity of the two TSS used by this cluster correlates with distinct enhancers. In prostate cancer cells (LNCaP), inflammatory activation results in higher eRNA transcription within a TAD harboring the distal TSS increasing its transcriptional output 2.4-fold, whilst the other TSS remained unresponsive. Our results suggest that multiple miRNA regulatory domains are similarly constrained by chromatin architecture, giving rise to complex cell- and response-specific regulation of miRNA expression.
Mapping total RNA sequenced from whole-cell or subcellular compartments back to the genome has shown that primary transcripts can derive from up to 75% of unique genomic locations (45). Currently the GENCODE database Version 25 (March 2016 freeze, GRCh38) annotates 7258 small non-coding RNA genes (46). However, only the sequence and location of precursor hairpins has been annotated for majority of miRNA loci. Rapid processing and maturation of ncRNAs into a heterogenous population of mature species presents a common difficulty for their detection using microarray hybridization or RNA-seq. Here, we used nascent transcriptomes (GRO-seq) that precede the processing mechanisms, revealing active transcription at miRNA loci. Our analysis confirmed the utilization of distal pri-miRNA promoters and alternative TSS, agreeing with results from eight cell lines depleted from DGCR8/DROSHA (8). The data types integrated here extend the characterization of miRNA loci across common human cell line models with further supporting evidence for TSS status based on CAGE-seq and the H3K4 methylation levels, and for transcript structures based on the GRO-seq signal profile. As each genome-wide assay has both strengths and weaknesses, it is important to jointly consider the evidence at each candidate TSS. In many miRNA loci, the previous RNA-seq-based assembly included more TSS than those confirmed by our CAGE- or GRO-seq profiles, possibly reflecting low transcript levels or false positives from the assembly algorithm. Our approach, on the other hand has less confidence in assigning transcript end coordinates, as the active Pol II transcriptional signal trails down at gene ends. Similarly, while CAGE-seq is a sensitive and high-resolution method to detect TSS (13), the signals at highly expressed gene exon regions can often be assigned as false positives. We tried to mitigate this issue using a mixture model for CAGE-peak scores and requiring that peaks at exons were additionally supported by the H3K4me histone mark status. The histone marks alone would not allow strand-specific analysis, and have low (kb) resolution to distinguish TSS locations. We further utilized the ratio between the H3K4me1 and H4K4me3 to distinguish between gene and enhancer transcripts. However, in some cases this distinction did not agree across the cell types. Instead, the typical bidirectional short (few kb) eRNA transcripts showed an extended TV in a subset of cells, some of which that spanned pre-miRNA locations. However, the mature miRNA derived from these loci represented low abundance or poorly confirmed miRNA species. Further experimental validation would be necessary to elucidate the role of extended eRNAs as pri-miRNA transcripts.
The presence of multiple TSS is a key feature defining the complex pri-miRNA loci. Moreover, it is possible that some of the transcripts may generate other ncRNA species with additional regulatory role in distinct cell types. This is the case at the mir-29a∼mir-29b-1 locus where the longest pri-miRNA TV shares a TSS with the lincRNA LINC-PINT that in mouse (Lncpint) has been shown to promote cellular proliferation; in contrast, human LINC-PINT was found to act as a negative regulator of proliferation and survival in HCT116 and A549 cells (47). According to our results, this discrepancy in its role merits further investigation. The GRO-seq data show that the shorter TV have prominent TSS activity across normal cells, with low transcriptional activity overlapping LINC-PINT. In contrast, ES or cancer cells co-express both ncRNA species at low to moderate levels (Figure 2C and Supplementary Figure S3), with relatively high expression in A549. In both human and mouse, LINC-PINT expression has been found to be regulated by p53 activation. Since miR-29 enhances p53 activity through repressing the negative loop between PI3K-AKT-MDM2 and p53 (48), it would be important to study the contribution of both ncRNA species and to distinguish between cell and cancer types that express the long or shorter TV at this locus.
The differential TSS activation could be a major feature governed by cell type-specific usage of regulatory elements and TF-mediated regulation. In order to dissect this regulatory complexity, we developed a quantification approach that allows analysis of the TSS-specific transcriptional output. Our analysis revealed a highly cell-specific pattern of TSS usage across majority of intergenic pri-miRNA loci and presented evidence that this could be linked with the lineage-specific transcriptional programs that establishes prominent enhancer activation in the vicinity of active miRNA TSS, as is evident from high degree of overlap with super-enhancers.
The small, 21–22 nt long miRNAs have been established as key regulators that themselves contribute to cell-fate decisions and fine-tune transcriptional responses, yet regulation of miRNA transcription has remained elusive. The analysis of eRNA transcription (18) in context of the miRNA locus can further pinpoint active regulatory regions. Focusing on the regulatory regions within the dynamically active mir-221∼222 locus, we analyzed the six super-enhancer regions and a larger set of candidate enhancers ±500 kb from each TSS to distinguish cell-specific eRNA transcription patterns that closely matched the differential activity of the two alternative TSS. We show that the strong transcriptional activity of TSS1 is reflected in the high eRNA levels at SE1-SE3. Using DNAse-seq peak centers to obtain higher resolution within SE enabled elucidating differential activation patterns of sub-regions upon stimuli. We found that the distal mir-221∼222 TSS2, correlating with SE4 and 6, was responsive to TNFα stimulation, with concomitant eRNA increase at the NF-κB bound region within SE4. This inflammatory mechanism modulating expression at the mir-221∼222 locus should be further investigated, as these miRNAs have been found to increase cellular proliferation potential by targeting CDKN1B (p27Kip1) in human prostate cancer cell lines (49).
Chromatin architecture is known to limit the contacts between TSS and regulatory elements. Hi-C data provide a comprehensive view of genome-wide chromatin interactions (50). We show here that the correlation of SE activity at the mir-221∼222 locus with a specific TSS could be physically orchestrated by TAD organization. TADs represent sub-regions with frequent contacts that are often bounded by CTCF binding sites and insulator elements. At the mir-100∼let-7a-2∼125b-1 cluster, where TSS1–4 have also previously been validated (8), the distal TSS were separated by a TAD boundary from more proximal TSS which co-localize with a SE region. This was reflected in dramatically different TSS activity levels. Furthermore, the unique transcription start site for mir-125b-1 (TSS1) that resides between the clustered miRNA species may enable cells to bypass the polycistronic transcription of each miRNA. In Drosophila, let-7 and miR-125 were found to function during two distinct stages, development and adulthood, rather than acting at the same time, based on differential modulation of their levels in a model of retinal neurodegeneration (51). Among the human cell types studied, here, the retinal cell line ARPE exclusively utilized the short mir-125b-1-specific TSS. Elevated mir-125b levels have also been associated with pathological states involving fibrosis and negative regulation of p53 (52,53). Accordingly, this TSS was also active in the fibroblast cells IMR90 and MRC5. The differential role of the clustered miRNA emphasizes the importance to distinguish between alternative TSS usage across cell types and their responsiveness to different stress stimuli. The approach and results presented here pave the way for building detailed regulatory maps of miRNA loci. Moreover, the ability of GRO-seq to detect different ncRNA species encourages application of this approach to elucidate the regulatory complexity at other ncRNA loci.
In summary, the findings presented link the multiple TV found at pri-miRNA loci with characterization of differential TSS usage across cell types and demonstrate that chromatin architecture is important in defining pri-miRNA regulatory domains.
ACCESSION NUMBER
Data have been deposited in GEO under the accession GSE92375.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the EMBL GeneCore sequencing team for sequencing service provided, the Bioinformatics Center at the University of Eastern Finland for data analysis infrastructure and Dr Björn Schwalb for kindly providing help with the TT-seq dataset. Authors acknowledge Fondation du Pélican de Marie et Pierre Hippert-Faber (Luxembourg) for graduate fellowship to M.B.L.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland [276634 to M.H., 295094 to M.B.L., 294073 to M.U.K]; Sigrid Juselius Foundation; Finnish Foundation for Cardiovascular Research; Finnish Cultural Foundation; University of Eastern Finland, School of Medicine; Fondation du Pélican de Marie et Pierre Hippert-Faber (Luxembourg) Graduate Fellowship (to M.B.L.). Funding for open access charge: Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cech T.R., Steitz J.A.. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014; 157:77–94. [DOI] [PubMed] [Google Scholar]
- 2. Ameres S.L., Zamore P.D.. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013; 14:475–488. [DOI] [PubMed] [Google Scholar]
- 3. Fang W., Bartel D.P.. The menu of features that define primary MicroRNAs and enable de novo design of microRNA genes. Mol. Cell. 2015; 60:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J.. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008; 15:902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin S., Yu Y., Reed R.. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Sci. Rep. 2015; 5:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vitsios D.M., Davis M.P., van Dongen S., Enright A.J.. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 2017; 45:1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang T.C., Pertea M., Lee S., Salzberg S.L., Mendell J.T.. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 2015; 25:1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nepal C., Coolen M., Hadzhiev Y., Cussigh D., Mydel P., Steen V.M., Carninci P., Andersen J.B., Bally-Cuif L., Müller F. et al. Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs. Nucleic Acids Res. 2016; 44:3070–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J. et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008; 134:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010; 28:1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanamori-Katayama M., Itoh M., Kawaji H., Lassmann T., Katayama S., Kojima M., Bertin N., Kaiho A., Ninomiya N., Daub K. et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011; 21:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Core L.J., Waterfall J.J., Lis J.T.. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008; 322:1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011; 474:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaikkonen M.U., Lam M.T., Glass C.K.. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011; 90:430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teppo S., Laukkanen S., Liuksiala T, Nordlund J., Oittinen M., Teittinen K, Grönroos T., St-Onge P., Sinnett D., Syvänen A.C. et al. Genome-wide repression of eRNA and target gene loci by the ETV6-RUNX1 fusion in acute leukemia. Genome Res. 2016; 26:1468–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013; 51:310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mousavi K., Zare H., Dell’orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V.. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013; 51:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R.. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013; 494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh C.L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.S., Chen S. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krueger F., Andrews S., Osborne C.S.. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One. 2011; 6:e16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenkom A Galaxy Team . Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010; 26:1783–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khan A., Zhang X.. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016; 44:D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S, Hori F., Ishikawa-Kato S. et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015; 16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bullard J.H., Purdom E., Hansen K.D., Dudoit S.. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics. 2010; 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dillies M.A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., Keime C., Marot G., Castel D., Estelle J. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brie. Bioinform. 2013; 14:671–683. [DOI] [PubMed] [Google Scholar]
- 33. Fraley F., Raftery A.E., Murphy T.B., Scrucca L.. mclust Version 4 for R: Normal Mixture Modeling for Model-Based Clustering, Classification, and Density Estimation. Technical report No. 597. 2012; Department of Statistics, University of Washington. [Google Scholar]
- 34. Killick R., Eckley I.A.. changepoint: an R Package for Changepoint Analysis. J. Stat. Softw. 2014; 58:1–19. [Google Scholar]
- 35. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaikkonen M.U., Niskanen H., Romanoski C.E., Kansanen E., Kivelä A.M., Laitalainen J., Heinz S., Benner C., Glass C.K., Ylä-Herttuala S.. Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization. Nucleic Acids Res. 2014; 42:12570–12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouvy-Liivrand M., Heinäniemi M., John E., Schneider J.G., Sauter T., Sinkkonen L.. Combinatorial regulation of lipoprotein lipase by microRNAs during mouse adipogenesis. RNA Biol. 2014; 11:76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van der Maaten L, Hinton G.E.. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008; 9:2579–2605. [Google Scholar]
- 40. Schwalb B., Michel M., Zacher B., Frühauf K., Demel C., Tresch A., Gagneur J., Cramer P.. TT-seq maps the human transient transcriptome. Science. 2017; 352:1225–1228. [DOI] [PubMed] [Google Scholar]
- 41. Li Z., Yang C.S., Nakashima K., Rana T.M.. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011; 30:823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Aiden E.L.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015; 518:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marín-Béjar O., Marchese F.P., Athie A., Sánchez Y., González J., Segura V., Huang L., Moreno I., Navarro A., Monzó M. et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013; 14:R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park S.Y., Lee J.H., Ha M., Nam J.W., Kim V.N.. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 2009; 16:23–29. [DOI] [PubMed] [Google Scholar]
- 49. Garofalo M., Quintavalle C., Romano G., Croce C.M., Condorelli G.. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr. Mol. Med. 2012; 12:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chawla G., Deosthale P., Childress S., Wu Y.C., Sokol N.S.. A let-7-to-miR-125 microRNA switch regulates neuronal integrity and lifespan in Drosophila. PLoS Genet. 2016; 12:e1006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nagpal V., Rai R., Place A.T., Murphy S.B., Verma S.K., Ghosh A.K., Vaughan D.E.. MiR-125b is critical for fibroblast-to-myofibroblast transition and cardiac fibrosis. Circulation. 2016; 133:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Le M.T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H.F., Lim B.. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009; 23:862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.