Abstract
Scleractinian corals are the foundation species of the coral-reef ecosystem. Their calcium carbonate skeletons form extensive structures that are home to millions of species, making coral reefs one of the most diverse ecosystems of our planet. However, our understanding of how reef-building corals have evolved the ability to calcify and become the ecosystem builders they are today is hampered by uncertain relationships within their subclass Hexacorallia. Corallimorpharians have been proposed to originate from a complex scleractinian ancestor that lost the ability to calcify in response to increasing ocean acidification, suggesting the possibility for corals to lose and gain the ability to calcify in response to increasing ocean acidification. Here, we employed a phylogenomic approach using whole-genome data from six hexacorallian species to resolve the evolutionary relationship between reef-building corals and their noncalcifying relatives. Phylogenetic analysis based on 1,421 single-copy orthologs, as well as gene presence/absence and synteny information, converged on the same topologies, showing strong support for scleractinian monophyly and a corallimorpharian sister clade. Our broad phylogenomic approach using sequence-based and sequence-independent analyses provides unambiguous evidence for the monophyly of scleractinian corals and the rejection of corallimorpharians as descendants of a complex coral ancestor.
Keywords: naked corals, Scleractinia, Corallimorpharia, phylogenomics, evolution, calcification
Introduction
Scleractinian corals form the large bicarbonate structures that constitute the foundation of the coral reef ecosystem. Their evolutionary history traces back to the early Triassic around 245 Ma, a time of high diversification within this order when multiple coral clades appear in the fossil record for the first time (Simpson etal. 2011; Park etal. 2012). However, in contrast to fossil evidence, molecular analyses of the evolutionary history of Scleractinia remain inconclusive, with some extant deep-water families suggesting the evolutionary roots of this order to potentially date as far back as approximately 425 Ma (Stolarski etal. 2011). Phylogenetic analyses of extant corals using different genetic markers and methods clearly identify two distinct clades, termed the “Complex” and the “Robust” clades (Romano and Palumbi 1996; Romano and Cairns 2000; Chen etal. 2002; Le Goff-Vitry etal. 2004; Medina etal. 2006; Fukami etal. 2008; Stolarski etal. 2011; Kitahara etal. 2014; Lin etal. 2014; ). However, the precise phylogenetic relationships of scleractinian corals within the Hexacorallia has further been elusive due to contradicting phylogenies derived from phylogenetic analyses using different molecular markers and evolutionary models (Medina etal. 2006; Kitahara etal. 2014). Yet, understanding the evolutionary history of these organisms is imperative if we aim to understand their evolutionary history and resilience in light of climate change.
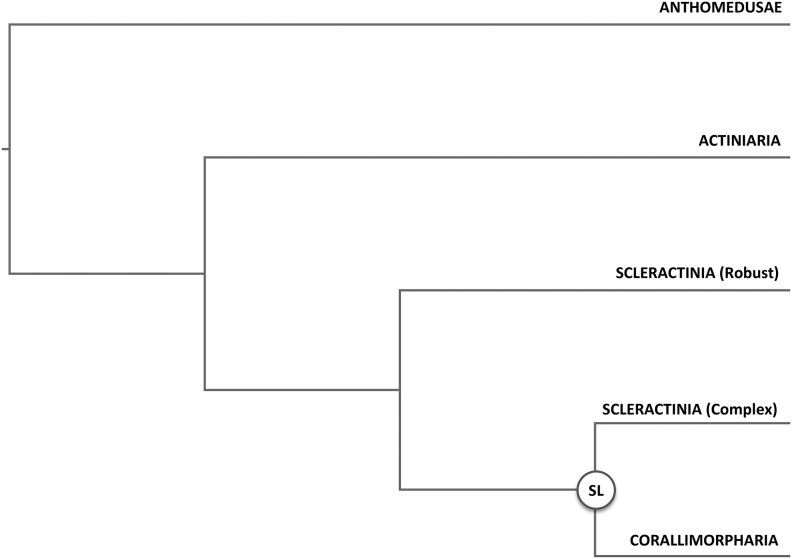
Of special interest is the phylogenetic relationship of scleractinian corals to the order Corallimorpharia (Kitahara etal. 2014). Corallimorpharia, colloquially termed “false corals”, are closely related to Scleractinia (Dunn 1982), but unlike reef-building corals, they do not possess a calcareous skeleton. Based on phylogenetic analyses of complete mitochondrial genomes it was proposed that Corallimorpharia evolved from a complex coral ancestor approximately 110–132 Ma (fig. 1), suggesting the loss of the calcium carbonate skeleton in response to increased oceanic CO2 prevalent during this time period (Medina etal. 2006). This finding provided strong support for the so-called “naked coral” hypothesis, which was first coined by Stanley and Fautin (Stanley and Fautin 2001) and proposes that corals have lost and reevolved skeletons repeatedly during the middle Triassic. The importance of this hypothesis lays in its implications as a potential mechanism for corals to escape extinction from aragonite skeletal dissolution during periods of increased CO2 levels such as those projected by future climate change scenarios.
Fig. 1.
—Phylogenetic relationships according to the “naked coral” hypothesis. (SL) marks the putative evolutionary origin of Corallimorpharia from a complex coral ancestor through “skeleton loss.”
Despite the strong support for this hypothesis provided by mitochondrial genome-based phylogenetic analyses (Chen etal. 1995; Medina etal. 2006), other studies using different markers and techniques reported contradicting results (Daly etal. 2003; Brugler and France 2007; Fukami etal. 2008; Aranda etal. 2012). Recently, various studies (Stolarski etal. 2011; Kayal etal. 2013; Kitahara etal. 2014; Lin etal. 2014, 2016) have addressed this discrepancy in detail by analyzing mitochondrial nucleotide and amino-acid-based alignments using different evolutionary models, showing that certain models allowed the recovery of Scleractinia as monophyletic group even when using mitochondrial markers. Although these newer studies kept challenging the idea of a potential complex coral origin of Corallimorpharia, the dearth of genome sequences did not allow rigorous testing of the hypothesis using nonsequence-based phylogenomic approaches. To overcome these limitations, we used a multipronged approach including phylogenetic analyses of nuclear-encoded genes as well as genome-wide presence/absence information and synteny conservation (supplementary fig. S1, Supplementary Material online) of genomes from six hexacorallian species including the Actiniaria Nematostella vectensis (Putnam etal. 2007) and Aiptasia pallida (Baumgarten etal. 2015), the Corallimorpharia Amplexidiscus fenestrafer and Discosoma sp. (Wang etal. 2017) as well as the complex scleractinian coral Acropora digitifera (Shinzato etal. 2011) and the robust coral Stylophora pistillata.
Results and Discussion
Nuclear-Encoded Genes Support Scleractinian Monophyly
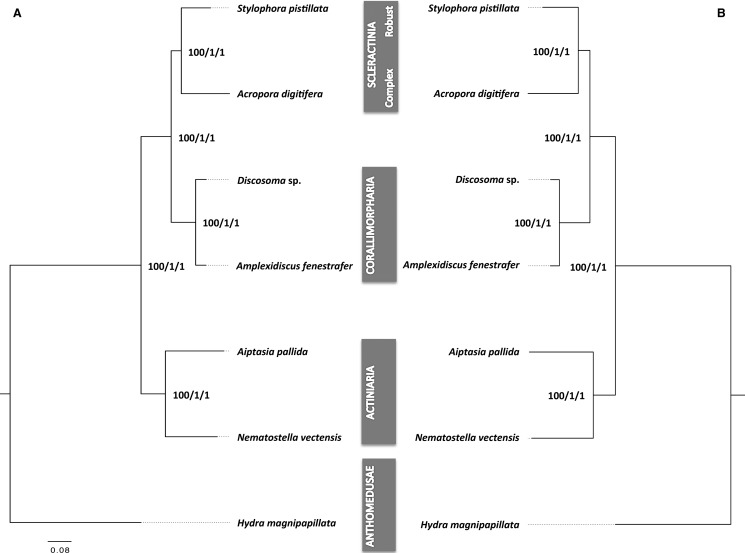
Phylogenetic analyses of nuclear encoded genes were performed on a set of single-copy orthologs on both the amino acid (aa) and nucleotide (nt) level. To this end, we first identified a suitable set of single-copy orthologs using OrthoMCL (Li etal. 2003) and the genome-encoded protein sets of N. vectensis, A. pallida, A. fenestrafer, Discosoma sp., A. digitifera, and S. pistillata as well as the hydrozoan Hydra magnipapillata which served as an outgroup in our analyses. Using this approach, we identified 1,421 single-copy orthologs that were used to select suitable subsets of orthologs for aa and nt-based phylogenetic analyses using Maximum Likelihood (ML) as well as Bayesian Inference (BI)-based methods in combination with different evolutionary models.
For the aa based analysis, we concatenated aligned sequences from 1,021 selected single copy orthologs that passed the filtering process (see material and methods), providing a supermatrix with 179,381 aa positions for phylogenetic reconstruction. Given the ongoing discussions with regard to the most appropriate substitution model for the inference of deep evolutionary splits (Pisani etal. 2015), we performed ML and BI-based phylogenetic analyses using different evolutionary substitution models (see also Materials and Methods). ML trees were constructed with RAxML using the LG + I+G + F (supplementary fig. S2A, Supplementary Material online) as determined by ProtTest (Darriba etal. 2011). BI analyses were performed with MrBayes using the LG + I+G +F (supplementary fig. S2B, Supplementary Material online) as well as Phylobayes using the CAT-LG (supplementary fig. S3A, Supplementary Material online), CAT-GTR (supplementary fig. S3B, Supplementary Material online), and CAT-Poisson models (supplementary fig. S3C, Supplementary Material online). The topologies of both the ML as well as the BI-based trees were identical and consistently showed maximum support for all nodes, independent of the substitution model used (fig. 2A). All trees recovered Scleractinia as monophyletic group with Corallimorpharia as its sister group. We also generated independent trees for each of the orthologous genes and obtained a consensus tree (majority rule) (supplementary fig. S4, Supplementary Material online). Interestingly, 96% of the single ortholog trees supported the monophyly of Corallimorpharia, but only 62% of the trees supported the monophyly of Scleractinia.
Fig. 2.
—(A) Phylogenetic analyses based on amino acid sequences of single-copy genes. (B) Phylogenetic analysis based on nucleotide sequences of coding single-copy genes. Values on the nodes correspond to branch support from RAxML, MrBayes, and PhyloBayes. Node support values depicted here were identical across all evolutionary models used.
Phylogenetic analyses of nucleotide sequence alignments followed the same general procedure. The nt super matrix was based on 1,255 selected single-copy orthologs that passed filtering, providing a total of 668,245 positions. Similar to the aa analysis, we used ML and BI-based methods and tested different evolutionary models in order to account for potential biases due to long-branch effects (see Material and Methods). Based on initial model tests using JModelTest2, we selected the GTR + I+G as the best model for further ML analyses with RAxML (supplementary fig. S5A, Supplementary Material online). BI-based phylogenetic reconstructions were performed with MrBayes using the GTR + I+G model (supplementary fig. S5B, Supplementary Material online) and Phylobayes to infer phylogenetic trees using the CAT-Poisson (supplementary fig. S6A, Supplementary Material online), and CAT-GTR model (supplementary fig. S6B, Supplementary Material online). All nucleotide-based analyses (fig. 2B) recovered the same topologies as the aa-based tree with maximum bootstrap and posterior probability support for all nodes independent of the method or model used.
Sequence-Independent Analyses Support Scleractinian Monophyly
The use of sequence data to resolve phylogenetic relationships can produce controversial results when analyzing deep evolutionary splits due to biases such as long-branch attraction or the choice of evolutionary models to determine evolutionary relationships, even in the presence of whole-genome information (Philippe etal. 2011). To address this problem, we also performed sequence-independent phylogenetic analyses using information on the presence/absence of ortholog groups as well as a synteny conservation.
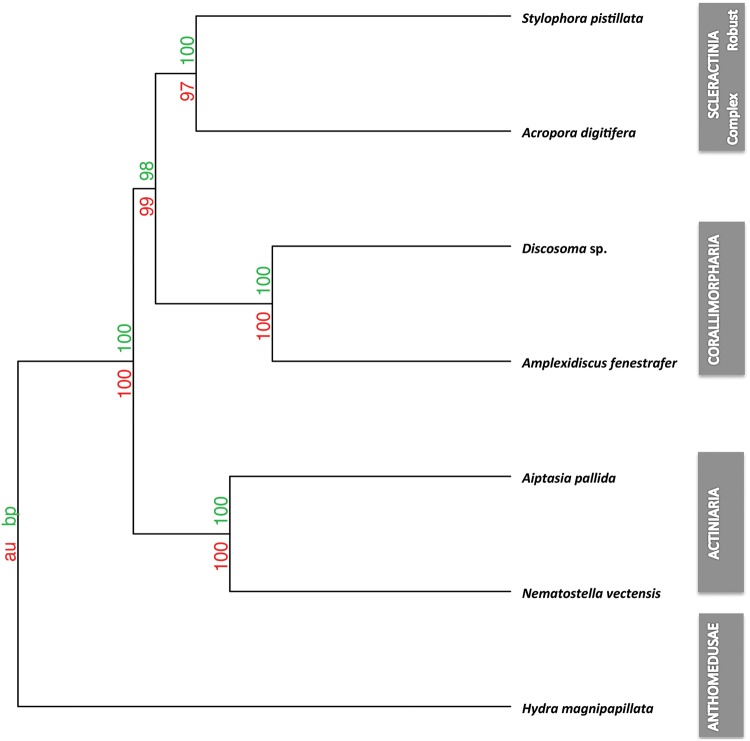
Based on our ortholog analysis, we identified 21,718 ortholog groups that were used to generate a distance matrix in R using the binary method (Borg and Groenen 2005). Based on this matrix we reconstructed putative phylogenetic relationships through hierarchical clustering of calculated distances across the six Hexacorallia genomes and our outgroup H. magnipapillata. To provide further statistical support for the nodes, we calculated the approximately unbiased probability and bootstrap probability using pvclust and 1,000 bootstraps. The recovered topology was identical to the phylogenetic analyses using nuclear-encoded single-copy orthologs and showed very high statistical support for all branches (fig. 3). Scleractinian monophyly was also recovered with strong statistical support when analyzing the presence/absence information using the binary F81-like model implemented in MrBayes, after applying an ascertainment bias correction by removing genes present in fewer than two species as suggested by Pisani (Pisani etal. 2015) (supplementary fig. S7, Supplementary Material online). Additionally, we also recovered the same tree topology using phylogenetic reconstruction based on the number of paralogs identified for the different ortholog/paralog groups (supplementary fig. S8, Supplementary Material online).
Fig. 3.
—Phylogenetic analyses based on gene presence/absence. The node supports are AU (Approximately Unbiased) P values and BP (Bootstrap Probability) values. The distance was calculated using binary methods and hierarchical cluster analysis with 1,000 bootstraps using the average Hcluster method.
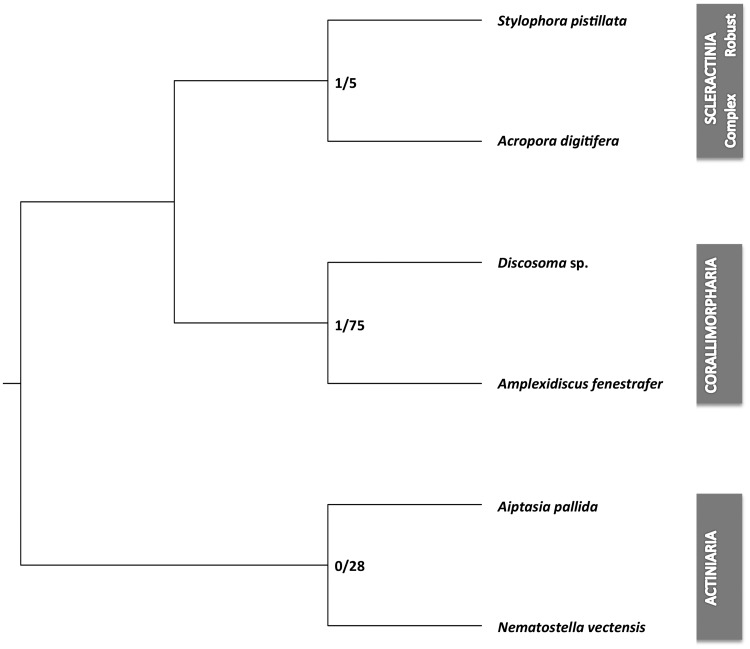
In order to derive further sequence-independent evidence we analyzed synteny conservation across the six genomes using the phylogenetic reconstruction tool PhyChro. Due to the phylogenetic distance and the dearth of conserved synteny groups in the H. magnipapillata genome, we omitted the outgroup from this analysis. Briefly, we first identified synteny blocks using SynChro (Drillon etal. 2014) with different block stringency parameters (Delta 1–6). In the following step, we analyzed the different outputs using PhyChro to identify “incompatible” block adjacencies between all genome pairs and from them deduce “pairs of incompatible groups of genomes” (PIGGs). If a block order (A, B) is shared by genome G1 and at least one other genome while block order (A, C) is shared by G2 and at least one other genome, we got a PIGG. From its set of PIGGs, PhyChro reconstructs the most parsimonious tree (where each PIGG can be “explained” by a unique rearrangement).
A common tree was found for Delta 2–5 (other deltas 1, 6 show only partial reconstructions), showing strong support for Scleractinia as a monophyletic group (fig. 4). The Scleractinia group (S. pistillata and A. digitifera) was, as expected, the weakest group in this analysis. It was also the last group identified in the recursive tree reconstruction, after the Actiniaria and Corallimorpharia. However, we identified only one PIGG that contradicted scleractinian monophyly, which is illustrated by the number of contradicting PIGGs (fig. 4) and the number of PIGGs supporting the branch of the Scleractinia group. It should be noted that this specific PIGG also contradicted corallimorpharian monophyly, which casts general doubt on its validity. Consequently, we conclude that scleractinian monophyly is the most parsimonious explanation for our results.
Fig. 4.
—Phylogenetic analyses based on synteny conservation. The unrooted tree (represented rooted as in figures 1 and 2 for more clarity) represents the topology obtained by PhyChro for Delta 2–5. The values on the branches correspond to the total number of PIGGs (pairs of incompatible groups of genomes) contradicting the branch/the total number of PIGGs supporting the branch obtained for Delta 2–5.
Corallimorpharia Are Not “Naked Corals”
Our phylogenetic analyses were based on a subset of stringently selected, high-confidence nuclear encoded single-copy orthologs that provided the longest alignment used to date to validate the phylogenetic relationship of scleractinian corals and corallimorpharians (Kitahara etal. 2014; Lin etal. 2016). The results obtained are consistent across all analyses independent of the sequence type, method or model used and recover Scleractinia as monophyletic group with Corallimorpharia as a close sister clade with maximum support for each node. Although the limited number of species used in our phylogenetic analyses is of potential concern, it should be noted that our results are in line with a recent study based on 15 Hexacorallia transcriptomes, including three Corallimorpharia species (Lin etal. 2016). More importantly, though, we show that our sequence-based results are strongly supported by two separate sequence-independent analyses that produced identical tree topologies with very strong support. Although our synteny-based analysis recovered Scleractinia as the weakest group, we did not find a single incompatibility allowing for a different grouping of these species and no alternative tree topologies were recovered.
The use of presence/absence and synteny information has previously been employed to provide additional support for sequence-based phylogenies in other organisms (Tian etal. 2012; Ryan etal. 2013; Pisani etal. 2015), but represents a novelty in the field of coral research due to the dearth of genomic data, although this is expected to change (Voolstra etal. 2015). Similar to sequence-based phylogenetic reconstruction methods these analyses are not entirely free of biases and potential errors (Pisani etal. 2015). However, the use of two sequence-independent approaches and two genomes for each of the orders analyzed provides additional confidence and support for our results.
Although all phylogenetic reconstruction methods are subject to biases and errors, we are confident that our whole-genome approach combining sequence and nonsequence-based approaches provides the most comprehensive study of the “naked coral” hypothesis to date. Although each analysis on its own might be associated with method-inherent biases, the consistent outcome in their sum provides unambiguous evidence for the monophyly of extent scleractinian corals and the rejection of Corallimorpharia as “naked corals.”
Our findings therefore show that corallimorpharians did not evolve from a complex coral ancestor but rather from a common ancestor of complex and robust corals. Consequently Corallimorpharia do not qualify as proof of the “naked coral” hypothesis, and it remains to be shown if scleractinian corals can indeed lose and regain the ability to calcify over geological timescales. Experimental approaches on selected coral species have shown that reef-building corals can indeed lose the ability to calcify under extreme conditions (Fine and Tchernov 2007) and recover; however, it remains to be shown if these responses observed in short-term experiments allow coral species to survive for evolutionary relevant periods. Although corallimorpharians do not appear to be the paragon of coral survival in light of increasing ocean acidification, they still represent their closest living, noncalcifying relatives to date, making them the best candidates for future studies aiming at understanding the evolution of corals and their traits, in particular calcification.
Materials and Methods
Data Collection and Preparation
The complete genome, and protein coding sequences of Nematostella vectensis, Aiptasia pallida, and Acropora digitifera, were obtained from NCBI (NCBI Resource Coordinators 2013). Genome data for Hydra magnipapillata was collected through Ensembl (www.ensembl.org) (Flicek etal. 2012) while the genomes of Stylophora pistillata, Amplexidiscus fenestrafer, and Discosoma sp. are available for download at http://corallimorpharia.reefgenomics.org (Liew etal. 2016).
In order to prepare the gene models for the subsequent comparative analyses, we discarded alternative isoforms and only selected the longest gene model for each locus. Further, we removed proteins with fewer than ten amino acids or low sequence quality (>20% stop codons, >20% nonstandard amino acids). This resulted in 26,908, 29,253, 21,327, 23,144, 23,232, 27,385, 32,338 genes for N. vectensis, A. pallida, A. fenestrafer, Discosoma sp., A. digitifera, S. pistillata, and H. magnipapillata, respectively.
After preparing the final gene and corresponding protein sets we ran OrthoMCL (Li etal. 2003) using an e-value cut-off of 10−5 to create groups of orthologs and paralogs across all seven genomes that were subsequently assigned to the latest OrthoMCL-DB 4 (http://orthomcl.org/orthomcl/.) (Chen etal. 2006) for further validation. Ortholog groups were annotated using the Pfam and BLAST database (Punta etal. 2012), and only groups with a single ortholog per species and a common Pfam domain, if available, were selected. This resulted in a set of 1,421 single-copy orthologs for subsequent phylogenetic analyses (supplementary table S1, Supplementary Material online).
Nuclear Single-Copy Ortholog Phylogenies
In order to analyze the phylogenetic relationships between the seven species, we used the previously selected single-copy genes derived from the ortholog analysis and performed separate phylogenetic analyses on both the amino acid (aa) and the nucleotide (nt) sequences.
For the aa-based analyses, we first aligned the sequences with MUSCLE 3.8.31(Edgar 2004). All alignments were optimized manually and trimmed to remove poor sequence alignments with Gblocks (Talavera and Castresana 2007) in order to exclude divergent regions and sequences (minimum length of block five, low conserved position 50%, and high flank position sequences 85%) (Talavera and Castresana 2007). After removal of low-scoring alignment sites and ambiguous aligned segments, we concatenated the remaining 1,021 alignments (supplementary table S2, Supplementary Material online), generating a supermatrix of 179,381 aa positions.
For the subsequent phylogenetic analyses, we first determined the most appropriate substitution model using ProtTestv2.4 (Darriba etal. 2011). In the following, we performed analyses using both Maximum Likelihood (ML) as well as Bayesian Inference (BI)-based methods. For ML inference, we ran the PTHREADS version of RAxML 8.1.22 (Stamatakis 2014) using the LG + I+G + F (LG substitution model under empirical amino acid frequencies [+F], estimation of the proportion of invariable sites [+I], and GAMMA model of rate heterogeneity [+G]) and 1,000 bootstrap replicates (−# 1000).
For the BI-based phylogenies, we used the parallel (MPI) version of MrBayes 3.2.6 (Ronquist etal. 2012) to construct a phylogenetic tree using the model LG + I+G + F (BIC = 6204664.29) determined by ProtTest. We ran at least 500 samples from the posterior probability distribution, and set four runs each with 5,000,000 (ngen = 5,000,000, samplefreq = 500, nruns = 4, printfreq = 500, nchains = 4). In addition, we also ran PhyloBayes-MPI (Lartillot etal. 2013) to infer phylogenetic trees using a range of different models, including the CAT, CAT-GTR, CAT-Poisson, and GTR model, which were shown to be less sensitive to saturation and long-branch attraction artifacts (Lartillot etal. 2007). All BI-based analyses were checked for convergence.
The phylogenetic analysis of the nucleotide sequences of single-copy orthologs followed the same overall approach as for the previously described protein sequences. We first constructed multiple alignments using MUSCLE, followed by removal of low-quality alignments with Gblocks using default parameters. Phylogenetic analyses were performed using ML and BI-based methods, which were carried out on the concatenated nucleotide alignments obtained from 1,255 single-copy orthologs that passed quality filtering (supplementary table S3, Supplementary Material online). To determine the most appropriate evolutionary model, we performed jModelTest 2.1.10 (Posada 2008). The ML analysis was performed using RAxML using the rapid hill-climbing algorithm with the GTRGAMAI model and 1,000 bootstrap replicates. Bayesian inference was performed using MrBayes 3.2.6 with 4 runs, 4 chains, and 5,000,000 generations, choosing GTR + I+G as best model. In addition, we also ran PhyloBayes with the CAT-Poisson, and CAT-GTR model. All BI-based analyses were checked for convergence.
The resulting trees were visualized and analyzed using Figtree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
Presence/Absence Analysis
Homologs, including orthologs and paralogs, were identified using OrthoMCL as described above. This analysis identified 21,718 orthology groups that were used to generate a binary presence/absence matrix (supplementary table S4, Supplementary Material online). Based on this matrix we calculated the distances between species using the binary (aka asymmetric binary) method in R and conducted a hierarchical cluster analysis using the average hclust method. Node supports were calculated as Approximately Unbiased P values (AU) and Bootstrap Probabilities (BP) using the R package Pvclust (Suzuki and Shimodaira 2006) and 1,000 bootstraps. To further provide a probabilistic analysis of the presence/absence information, we ran MrBayes using the binary restriction-site model and a discrete gamma distribution with four site rate categories (rates = gamma) after applying an ascertainment bias correction by removing genes present in fewer than two species (Pisani etal. 2015). In addition, we also reconstructed a phylogenetic tree using the number of paralogs identified for each species in the previously defined ortholog/paralog groups. Briefly, we calculated the distance based on the number of paralogs using the “canberra” method implemented in the R since the binary method used for the presence/absence data is not suitable for count data. The results were then clustered using the “average hclust” method and node supports were calculated using the R package Pvclust as described above.
Synteny-Based Phylogeny
Synteny conservation analyses were performed using the programs SynChro (Drillon etal. 2014) and PhyChro from the CHROnicle package (Drillon 2013). In a first step we identified synteny blocks with SynChro using multiple “Delta” (block stringency parameter) values ranging from 1 to 6. Subsequently, we analyzed the SynChro outputs using PhyChro in order to reconstruct phylogenetic relationships based on chromosomal rearrangement signals. Briefly, PhyChro looks for “incompatible” blocks adjacencies between two genomes (such as, for instance, the adjacency of the blocks A and B in a genome G1, and the adjacency of the blocks A and C in G2). Once these “incompatible” blocks adjacencies or “breakpoints” are identified between G1 and G2, PhyChro looks into the other genomes G3.Gn, to see if one or the other would not share the adjacency (A, B) or the adjacency (A, C). If (A, B) is shared by G1 and at least one other genome, (A, C) by G2 and at least one other genome also, we got what we called a “PIGG”: a pair of incompatible groups of genomes. Based on the parsimony principle, PhyChro tries to reconstruct a phylogenetic tree where each PIGG can be associated with a unique rearrangement that can be associated with the branch(es) that separates the two groups of genomes. PhyChro can be obtained upon request from guenoladrillon@gmail.com.
With extreme Delta values (Delta = 1 and Delta = 6), block synteny information was too scarce to resolve all groups. Other Delta values resulted in fully resolved trees, showing a common topology (which by the way included the partial trees reconstructed for the extreme Delta values). The resulting tree was parsimonious with only one PIGG out of the 263 PIGGs identified across the six analyses that could not be associated to a unique path of branches, that is, to a unique rearrangement. To incorporate an additional measure of branch support, we further provide the number of PIGGs rejecting a branch as well as the number of PIGGs supporting this branch. A PIGG is said to reject a branch when this branch implies at least two rearrangements to “explain” the PIGG and a PIGG is said to support a branch when the rearrangement to which it is associated must have occurred on this specific branch. Figure 3 shows the total number of PIGGs rejecting/supporting a branch obtained with the different Deltas 2–5.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgment
Research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). We thank Yi Jin Liew for help with the bioinformatic analyses and helpful discussions that greatly improved the manuscript.
Author Contributions
M.A. designed and conceived the study. M.A. and C.R.V. contributed reagents/materials/analysis tools. X.W., G.D., T.R., and M.A. analyzed the data. X.W. and M.A. wrote the manuscript with input from all authors. All authors agree to be held accountable for the content herein and gave approval for publication.
Literature Cited
- Aranda M, DeSalvo MK, Bayer T, Medina M, Voolstra CR.. 2012. Evolutionary insights into scleractinian corals using comparative genomic hybridizations. BMC Genomics 13(1): 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112(38): 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg I, Groenen PJ.. 2005. Modern multidimensional scaling: Theory and applications: New York: Springer Science & Business Media. [Google Scholar]
- Brugler MR, France SC.. 2007. The complete mitochondrial genome of the black coral Chrysopathes formosa (Cnidaria: Anthozoa: Antipatharia) supports classification of antipatharians within the subclass Hexacorallia. Mol Phylogenet Evol. 42(3): 776–788. [DOI] [PubMed] [Google Scholar]
- Chen CA, Odorico DM, Tenlohuis M, Veron JEN, Miller DJ.. 1995. Systematic relationships within the Anthozoa (Cnidaria, Anthozoa) using the 5'-end of the 28s Rdna. Mol Phylogenet Evol. 4(2): 175–183. [DOI] [PubMed] [Google Scholar]
- Chen CA, Wallace CC, Wolstenholme J.. 2002. Analysis of the mitochondrial 12S rRNA gene supports a two-clade hypothesis of the evolutionary history of scleractinian corals. Mol Phylogenet Evol. 23(2): 137–149. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS.. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34(90001): D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M, Fautin DG, Cappola VA.. 2003. Systematics of the Hexacorallia (Cnidaria: Anthozoa). Zool J Linn Soc. 139(3): 419–437. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8): 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillon G. 2013. Analyse combinatoire des réarrangements chromosomiques et reconstruction des génomes ancestraux chez les eucaryotes. PhD diss., Paris 6.
- Drillon G, Carbone A, Fischer G.. 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9(3): e92621.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DF. 1982. Sexual reproduction of two intertidal sea anemones (Coelenterata: Actiniaria) in Malaysia. Biotropica 14(4): 262–271. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine M, Tchernov D.. 2007. Scleractinian coral species survive and recover from decalcification. Science 315(5820): 1811.. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. 2012. Ensembl 2012. Nucleic Acids Res. 40(Database issue): D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami H, et al. 2008. Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PLoS One 3(9): e3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Roure B, Philippe H, Collins AG, Lavrov DV.. 2013. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol Biol. 13(1): 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, et al. 2014. The “naked coral” hypothesis revisited—evidence for and against scleractinian monophyly. PLoS One 9(4): e94774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H.. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 7(Suppl 1): S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4): 611–615. [DOI] [PubMed] [Google Scholar]
- Le Goff-Vitry M, Rogers A, Baglow D.. 2004. A deep-sea slant on the molecular phylogeny of the Scleractinia. Mol Phylogenet Evol. 30(1): 167–177. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9): 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew YJ, Aranda M, Voolstra CR.. 2016. Reefgenomics.Org - a repository for marine genomics data. Database (Oxford). 2016: doi: 10.1093/database/baw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, et al. 2016. Corallimorpharians are not “naked corals”: insights into relationships between Scleractinia and Corallimorpharia from phylogenomic analyses. PeerJ 4: e2463.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, et al. 2014. Mitochondrial genome rearrangements in the scleractinia/corallimorpharia complex: implications for coral phylogeny. Genome Biol Evol. 6(5): 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Collins AG, Takaoka TL, Kuehl JV, Boore JL.. 2006. Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci U S A. 103(24): 9096–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators 2013. Database resources of the national center for biotechnology information. Nucleic Acids Res. 41(D1): D8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, et al. 2012. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol. 62(1): 329–345. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9(3): e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, et al. 2015. Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci U S A. 112(50): 15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7): 1253–1256. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40(Database issue): D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834): 86–94. [DOI] [PubMed] [Google Scholar]
- Romano S, Cairns SD.. 2000. Molecular phylogenetic hypotheses for the evolution of scleractinian corals. Bull Mar Sci. 67: 1043–1068. [Google Scholar]
- Romano SL, Palumbi SR.. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271(5249): 640–642. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3): 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342(6164): 1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360): 320–U382. [DOI] [PubMed] [Google Scholar]
- Simpson C, Kiessling W, Mewis H, Baron-Szabo RC, Müller J.. 2011. Evolutionary diversification of reef corals: a comparison of the molecular and fossil records. Evolution 65(11): 3274–3284. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley GD Jr, Fautin DG.. 2001. Paleontology and evolution. The origins of modern corals. Science 291(5510): 1913–1914. [DOI] [PubMed] [Google Scholar]
- Stolarski J, et al. 2011. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol Biol. 11: 316.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H.. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22(12): 1540–1542. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4): 564–577. [DOI] [PubMed] [Google Scholar]
- Tian CF, et al. 2012. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A. 109(22): 8629–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra CR, et al. 2015. The ReFuGe 2020 Consortium—using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front Mar Sci. 2: 68. [Google Scholar]
- Wang X, et al. 2017. Draft genomes of the corallimorpharians Amplexidiscus fenestrafer and Discosoma sp. Mol Ecol Resour. doi: 10.1111/1755-0998.12680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.