Abstract
Geosmithia morbida is an emerging fungal pathogen which serves as a model for examining the evolutionary processes behind pathogenicity because it is one of two known pathogens within a genus of mostly saprophytic, beetle-associated, fungi. This pathogen causes thousand cankers disease in black walnut trees and is vectored into the host via the walnut twig beetle. Geosmithia morbida was first detected in western United States and currently threatens the timber industry concentrated in eastern United States. We sequenced the genomes of G. morbida in a previous study and two nonpathogenic Geosmithia species in this work and compared these species to other fungal pathogens and nonpathogens to identify genes under positive selection in G. morbida that may be associated with pathogenicity. Geosmithia morbida possesses one of the smallest genomes among the fungal species observed in this study, and one of the smallest fungal pathogen genomes to date. The enzymatic profile in this pathogen is very similar to its nonpathogenic relatives. Our findings indicate that genome reduction or retention of a smaller genome may be an important adaptative force during the evolution of a specialized lifestyle in fungal species that occupy a specificniche, such as beetle vectored tree pathogens. We also present potential genes under selection in G. morbida that could be important for adaptation to a pathogenic lifestyle.
Keywords: Geosmithia morbida, pathogenicity, tree pathogen, thousand cankers disease
Introduction
Uncovering the specific genetic and molecular events behind the evolution of novel traits such as pathogenicity in fungal species has been a long-standing objective of pathologists. Geosmithia (Ascomycota: Hypocreales), a genus first proposed in 1979 for fungi that were formerly placed in genus Penicillium (Pitt 1979), serves a paradigm for examining the processes contributing to the evolution of pathogenicity. Geosmithia species are filamentous fungi that most commonly associate with phloeophagous bark beetles (Kolařík et al. 2005, 2011), although some Geosmithia fungi, such as G. eupagioceri and G. microcorthyli, are known to affiliate with ambrosia beetles (Kolařík and Jankowiak 2013). Geosmithia species and their beetle associates occupy a variety of hosts, including pines, oaks, junipers, and walnut trees (Kolařík et al. 2007; Kolařík and Kirkendall 2010; Kolařík and Jankowiak 2013). The ecology and diversity of symbiosis between these fungi and their beetle associates is poorly understood, but investigators are beginning to explore such relationships (Kolařík et al. 2007; Kolařík and Jankowiak 2013). Although most species in Geosmithia are saprotrophic, two species were recently determined to be pathogenic—Geosmithia pallida (Lynch et al. 2014) and Geosmithia morbida (Tisserat et al. 2009), on coast live oak (Quercus agrifolia) and black walnut (Juglans nigra), respectively. However, both of these species live saprophytically in association with bark beetles and other tree hosts. It is still unclear what mechanisms allow these species of Geosmithia to be pathogenic to a new host whereas other members of the genus remain saprobes.
Geosmithia morbida causes thousand cankers disease (TCD) in J. nigra (eastern black walnut). Although no evidence of TCD has been detected in other Juglans to date, several species, such as J. californica, J. cinerea, J. hindsii, J. regia, are also susceptible to the pathogen (Utley et al. 2013). The fungus is most often vectored into its hosts by Pityophthorus juglandis, commonly known as the walnut twig beetle (WTB) (Kolařík et al. 2011). Unusual mortality of J. nigra was first noted in Colorado, USA in 2001. Since then, nine western states (CO, WA, OR, ID, NV, UT, CA, NM, and AZ) and seven eastern states (PA, OH, IN, MD, VA, TN, and NC) have reported TCD in one or more locations (Zerillo et al. 2014). This increase in TCD is likely a consequence of the expansion of WTB’s geographic range. WTB was present in only four counties of California, Arizona, and New Mexico in the 1960s, however, as of 2014, the beetle has been detected in over 115 counties in the western and eastern United States (Rugman-Jones et al. 2015).
The origin of this pathogen is not clear. However, it has been hypothesized that G. morbida may have undergone a host shift from J. major (Arizona black walnut) to a more naïve host, J. nigra, because the fungus does not cause disease in the Arizona black walnut, and neither WTB nor G. morbida were observed in the native range of J. nigra until 2010 (Zerillo et al. 2014). Juglans nigra is not indigenous to western United States but was planted throughout the region as an ornamental species. An alternative prediction based on G. morbida population genetic data is that the origin of G. morbida and WTB are the walnut populations of southern California, where the pathogen has been isolated from both healthy and diseased J. californica trees (Zerillo et al. 2014).
Early symptoms of infection by G. morbida include yellowing, wilting, and thinning of the foliage followed by branch dieback and tree death within 2–3 years after the initial infestation (Tisserat et al. 2009; Kolařík et al. 2011). Little is known about the specific means G. morbida employs for initiating and maintaining the infection, or what benefits, if any, the fungus imparts to the WTB vector. However, previous studies have demonstrated that fungal pathogens that occupy ecological niches similar to G. morbida must be capable of enduring and combating toxic host environments used by plants to resist infection. For instance, Grosmannia clavigera, a fungal symbiont of the mountain pine beetle (Dendroctonus ponderosae), can detoxify metabolites such as terpenoids and phenolics produced by the host as defense mechanisms (DiGuistini et al. 2011).
We recently developed a reference genome of G. morbida that consisted of 73 scaffolds totaling 26.5 Mb in length (Schuelke et al. 2016). This genome represents one of the smallest fungal tree pathogen genomes reported to date. Rapid changes in genome size have accompanied dramatic biological changes in newly emerged fungal and oomycete species (Raffaele and Kamoun 2012; Adhikari et al. 2013). In fungi, a link has been observed between genome expansion and evolution of pathogenicity (Raffaele and Kamoun 2012). Genome expansions were associated with parasitism in general and increased pathogenicity and virulence in several fungal lineages (Spanu et al. 2010). Previous genome sequencing of G. morbida (Schuelke et al. 2016) showed that this newly emerged fungal pathogen has a smaller genome than several of its closely related nonpathogenic relatives in the Hypocreales. Hence, it is possible that G. morbida may have taken an evolutionary path to pathogenicity that has not been characterized previously in plant-associated fungi.
The arrival of new pathogens, frequently referred to as Black Swan events due to their perceived unpredictability, represent a significant threat to native and agriculturally important tree species (Ploetz et al. 2013). Thus, beetle-associated symbionts that have switched to pathogenic lifestyles represent excellent models for investigating the evolution of pathogenicity and its relationship to genome size. Although the genus Geosmithia is distributed worldwide, G. morbida, and more recently, G. pallida, are the first members of the genus to be described as plant pathogens among the 60 known nonpathogenic species (Kolařík and Kirkendall 2010; Kolařík et al. 2011; Lynch et al. 2014).
In this work, we compare the reference genome of the pathogenic and host-specific species G. morbida with two closely related nonpathogenic generalist species, Geosmithia flava and Geosmithia putterillii. Based on this comparison, we identify genes under positive selection that may be involved in the specialization of a pathogenic life strategy that depends on a single beetle vector and a narrow, but potentially expanding, host range. We also present a species phylogeny estimated using single-copy orthologs that confirms the placement of Geosmithia species in the order Hypocreales, and that their closest fungal relative is Acremonium chrysogenum. The primary goal of this study was to gain insight into the evolution of pathogenicity within G. morbida. We also investigated the presence of convergent evolution in G. morbida and G. clavigera, two tree pathogens vectored into their hosts via beetle symbionts.
Materials and Methods
DNA Extraction and Sequencing
The CTAB method delineated by the Joint Genome Institute was used to extract DNA for genome sequencing from lyophilized mycelium of Geosmithia flava and Geosmithia putterillii (Kohler et al. 2011). Table 1 lists genetic, geographic, and host information for each Geosmithia species used in this study. Total DNA concentration was measured with Nanodrop, and DNA sequencing was conducted at Purdue University Genomics Core Facility in West Lafayette, Indiana. DNA libraries were prepared using the paired-end Illumina Truseq protocol and sequenced on an Illumina HiSeq 2500 using a single lane. Mean insert sizes for G. flava and G. putterillii were 477 and 513 bp, correspondingly. The remaining sequencing statistics are listed in table 2.
Table 1.
Species, Geographic Origins, and Host Information for Geosmithia morbida, Geosmithia flava, and Geosmithia putterillii
| Species | Pathogen | Isolate | Geographic Origins | Host |
|---|---|---|---|---|
| G. morbidaa | Yes | 1262 | California | J. californica |
| G. flava | No | CCF3333 | Czech Republic | Castenea sativa |
| G. putterillii | No | CCF4204 | California | J. californica |
This isolate is the reference genome. The details of assembly for this genome are discussed in Schuelke et al. (2016).
Table 2.
Statistics for Sequence Data from Isolates of Geosmithia morbida, Geosmithia flava, and Geosmithia putterillii
| Species | Total Read Pairs | Est. Coverage | ||
|---|---|---|---|---|
| G. morbida | 14,013,863a | 20,674,289b | 109×a | 160×b |
| G. flava | 16,183,281 | 102× | ||
| G. putterillii | 19,711,745 | 131× | ||
These values are for paired-end read data for G. morbida from Schuelke et al. (2016).
These values are for mate-pair read data for G. morbida from Schuelke et al. (2016).
Preprocessing Sequence Data
The raw paired-end reads for G. flava and G. putterillii were corrected using BFC (version r181) (Li 2015). BFC utilizes a combination of hash table and bloom-filter to count k-mers for a given read and correct errors in that read based on the k-mer support. Because BFC requires interleaved reads as input, khmer 1.1 was leveraged to interleave as well as split the paired-end reads before and after the error correction stage, respectively (Crusoe et al. 2015). Next, low quality bases and adapters in error corrected reads were trimmed with Trimmomatic, version 0.32, using a Phred threshold of 4 (Bolger et al. 2014).
Assembly Construction
Genome assemblies were constructed with ABySS 1.9.0 using four k-mer sizes (61, 71, 81, and 91) (Simpson et al. 2009). The resulting assemblies were evaluated using BUSCO (v1.1b1) (Simao et al. 2015), a tool which assess completeness based on the presence of universal single-copy orthologs within fungi. Length-based statistics were generated with QUAST v2.3 (Gurevich et al. 2013). Final assemblies were manually chosen based on length-based and genome completeness statistics. Furthermore, the raw reads of G. flava and G. putterillii were mapped back to their corresponding genomes using BWA version 0.7.9a-r786 (Li and Durbin 2009) to assess the quality of the chosen assemblies.
Structural and Functional Annotation
We utilized the automated annotation software Maker version 2.31.8 (Cantarel et al. 2008) to functionally annotate the genomes of G. flava and G. putterillii. We used two of the three gene prediction tools available within the pipeline SNAP (released 2013, Korf 2004) and Augustus 2.5.5 (Stanke et al. 2006). SNAP was trained using gff files generated by CEGMA v2.5 (a program similar to BUSCO) (Parra et al. 2007). Augustus was trained with Fusarium solani protein models (v2.0.26) downloaded from Ensembl Fungi (Kersey et al. 2016). The protein sequences generated by the structural annotation were blasted against the Swiss-Prot database (Boutet et al. 2016) to functionally annotate the genomes of G. flava and G. putterillii. We assessed the completeness of the final transcript sets using BUSCO (v1.1b1) and fungal data set 9 provided with this software.
Assessing Repetitive Elements Profile
To evaluate the repetitive elements profile of G. flava and G. putterillii, we masked the interspersed repeats within the assembled genomes with RepeatMasker 4.0.5 (Smit et al. 1996) using the sensitive mode and default values as arguments.
In most fungi, repeat-induced point mutations are a defense mechanism against the propogation of repetitive genetic elements and tend to favor the conversion of cytosines to thymines resulting in AT-rich regions (Selker 2002). Therefore, we also examined whether Geosmithia species have undergone repeat-induced point mutations using OcculterCut v1.1 (Testa et al. 2016).
Identifying Putative Genes Involved in Host–Pathogen Interactions
To search for putative genes contributing to pathogenicity, we conducted a BLASTp (v2.2.28+) (Altschul et al. 1990) search with an e-value threshold of 1e–6 against the PHI-base 4.0 database (Winnenburg et al. 2006) that includes known genes implicated in pathogenicity. We also assessed how many of the peptides with PHI-base hits were cysteine-rich proteins based on methods described in Kim et al. (2016). A protein was considered cysteine-rich if the length was 300 or less amino acids and the cysteine percentage was at least 3% of the total length. Further, we identified proteins that contain signal peptides and lack transmembrane domains in each Geosmithia species as well as their close relative A. chrysogenum with SignalP 4.1 and TMHMM 2.0 using default parameters (Krogh et al. 2001; Peterson et al. 2011).
Identifying Species-Specific Genes
To identify unique genes present in G. morbida, we performed an all-versus-all BLASTp search among the three Geosmithia species and A. chrysogenum with Orthofinder version 0.3.0 (Emms and Kelly 2015). Using a custom Python script, we analyzed homology among the four fungal species.
Identifying Carbohydrate-Active Proteins and Peptidases
To identify enzymes capable of degrading carbohydrate molecules in species belonging to Hypocreales and G. clavigera, we performed a HMMER 3.1b1 (Eddy 1998) search against the CAZy database (Lombard et al. 2014) released July 2015 and filtered the results following the developer’s recommendations. Finally, we profiled the proteolytic enzymes present in species using the MEROPS database 10.0 (Rawlings et al. 2016).
Identifying Secondary Metabolite Biosynthesis Gene Clusters
To identify gene clusters involved in secondary metabolite biosynthesis, we used antiSMASH 3.0 (Weber et al. 2015) to locate such gene clusters in the genomes. The analysis was conducted with default settings with the “DNA of eukaryotic origin” option.
Phylogenetic Analysis
Taxon Sampling
In order to determine phylogenetic position of Geosmithia, we combined the predicted peptide sequences from three Geosmithia species described here with the predicted peptide sequences of an additional 17 fungal genomes that represent the breadth of pathogens and nonpathogens within Ascomycota. Our data set contained eleven pathogens and nine nonpathogens (table 3).
Table 3.
Fungal Species Used for Phylogenetic Analysis in This Study
| Species | Class | Order | Ecological Role | Download Source | References |
|---|---|---|---|---|---|
| G. morbida | Sordariomycetes | Hypocreales | Pathogen | — | Schuelke et al. (2016) |
| G. flava | Sordariomycetes | Hypocreales | Nonpathogen | — | – |
| G. putterillii | Sordariomycetes | Hypocreales | Nonpathogen | — | – |
| A. chrysogenum | Sordariomycetes | Hypocreales | Beneficial | FungalEnsembl | Terfehr et al. (2014) |
| S. grisellum | Sordariomycetes | Hypocreales | Saprotrophic | JGI | Used with permission |
| Trichoderma virens | Sordariomycetes | Hypocreales | Mycoparasite | JGI | Kubicek et al. (2011) |
| Trichoderma reesei | Sordariomycetes | Hypocreales | Saprotrophic | FungalEnsembl | Martinez et al. (2008) |
| Escovopsis weberi | Sordariomycetes | Hypocreales | Mycoparasite | EnsemblGenomes | de Man et al. (2016) |
| Ustilaginoidea virens | Sordariomycetes | Hypocreales | Biotrophic pathogen | FungalEnsembl | Zhang et al. (2014) |
| C. militaris | Sordariomycetes | Hypocreales | Insect pathogen | FungalEnsembl | Zheng et al. (2011) |
| M. inundatum | Sordariomycetes | Hypocreales | Saprotrophic | JGI | Used with permission |
| F. solani | Sordariomycetes | Hypocreales | Necrotrophic pathogen | FungalEnsembl | Coleman et al. (2009) |
| Fusarium graminearum | Sordariomycetes | Hypocreales | Necrotrophic pathogen | FungalEnsembl | Trail et al. (2003), Cuomo et al. (2007) and Ma et al. (2010) |
| C. platani | Sordariomycetes | Microascales | Pathogen | FungalEnsembl | Belbahri (2015) |
| Neurospora crassa | Sordariomycetes | Sordariales | Saprotrophic | FungalEnsembl | Galagan et al. (2003) |
| Chaetomium globosum | Sordariomycetes | Sordariales | Saprotrophic | JGI | Berka et al. (2011) |
| Grosmannia clavigera | Sordariomycetes | Ophiostomatales | Pathogen | FungalEnsembl | DiGuistini et al. (2011) |
| Eutypa lata | Sordariomycetes | Xylariales | Pathogen | JGI | Blanco-Ulate et al. (2013) |
| Botrytis cinerea | Leotiomycetes | Helotiales | Necrotrophic pathogen | FungalEmsebl | Amselem et al. (2011) and Staats and van Kan (2012) |
| Oidiodendron maius | Leotiomycetes | Incertae sedis | Mycorrhizal | JGI | Kohler et al. (2015) |
Note.—The species in bold were utilized for positive selection analysis.
Inferring Orthology
Orthologous peptide sequences among the 20 fungal genomes were determined using OrthoFinder version 0.3.0 (Emms and Kelly 2015). All-versus-all BLASTp (2.2.28+, Altschul et al. 1990) searches were performed among a set of protein coding genes to infer orthogroups and aligned using MAFFT (v7.123b, Katoh and Standley 2013). These orthogroups may contain paralogs as well as orthologs, and because data sets rich in paralogs can confound phylogenomic analysis, the orthogroup alignment files produced by OrthoFinder were parsed to recover only those that contained single-copy orthologs from each of the 20 species. This resulted in 1,916 total orthogroups with 100% taxon occupancy.
Trimming Alignments
For each alignment, regions that contained gap rich sites were removed using –gappyout option in trimAl v1.4.rev15 (Capella-Gutiérrez et al. 2009). Next, all files containing orthogroups were renamed so the respective headers among these files were identical and individual alignments were concatenated. Concatenation resulted in a fasta file containing all 1,916 partitions with 1,054,662 amino acid sites at 100% taxon occupancy. This initial alignment was further filtered using MARE (v.0.1.2) (Misof et al. 2013), which reduced the data matrix to 633 partitions and 247,627 sites. Next, the best-fit substitution models for each partition and a global partitioning scheme were determined with PartitionFinder (v1.1.1) using hcluster clustering algorithm and default parameters (Lanfear et al. 2014).
Constructing Phylogeny
Maximum likelihood (ML) analysis was conducted in RaxML v 8.1.20 (Stamatakis 2014) leveraging the partitioning scheme determined by PartitionFinder. The ML tree and 200 bootstrap replicates were performed in a single analysis using the –f a option. In addition, we estimated the phylogenies of each of the 633 individual partitions under the LG model and used these topologies to estimate internode certainty (IC) and tree certainty (TC) measures (Salichos et al. 2014). Bayesian Markov Chain Monte Carlo (BMCMC) analysis was performed in MrBayes 3.2.6 (Ronquist et al. 2012) and PhyloBayes 4.1 (Lartillot et al. 2009). For MrBayes analysis, we truncated the alignment to contain the maximum number of positions allowed in MrBayes (89,999 sites), specified the mixed amino acid model prior and, based on preliminary analyses of larger data sets (not shown), ran the tree search for 215,000 generations. After testing for convergence, a consensus tree was generated after discarding 25% of the run as burnin using the sumt command. The nexus file, including MrBayes block, provides other details of the MrBayes analysis (supplementary Methods S1, Supplementary Material online). PhyloBayes analyses were performed under the CAT-GTR model and the posterior distributions of two independent runs were assessed with a burn-in of 1,000 and capturing every 2 trees using the bpcomp program. The complete data matrix and scripts used in these analyses can be found in our online repository.
Detecting Genes under Positive Selection
To identify genes under positive selection in G. morbida, we compared G. morbida with all nonpathogens from the aforementioned 20 fungi used to estimate the species tree. Among this batch of 10 fungal species, we detected 22,908 protein orthogroups using OrthoFinder that contained paralogs as well as orthologs. Of these, only 9,560 orthogroups were alignable with MAFFT because many groups consisted of only one sequence from a single species (Katoh and Standley 2013). A total of 3,327 orthogroups, composed of single-copy orthologs, were filtered and corresponding coding DNA sequences for each peptide in these partitions were extracted using custom scripts that can be found online.
The coding DNA sequences were then aligned with MACSE v1.01.b (Ranwez et al. 2011). This Java-based utility accounts for frameshifts and premature stop codons in coding sequences during the alignment process and outputs aligned protein and nucleotide sequences. In order to filter out alignments with frameshifts and internal stop codons, we utilized a program called PAL2NAL v14 (Suyama et al. 2006). This software searches for complementary regions between multiple protein alignments and the corresponding coding DNA sequences, and omits any problematic codons from the output file. This cleaning step reduced the number of 3,327 orthogroups to 2,798 that were used for detecting genes under selective pressures.
The branch-site model (BSM) in the CodeML program of package PAML v4.8 was used for selection analysis (Yang 2007). BSM permits ω (dN/dS) to vary among sites and branches permitting the identification of specific branches and sites subjected to selection. We computed two models in order to calculate and compare the likelihood values: a null model with a fixed ω value of 1 and an alternative model that estimates ω in the foreground branch, which is G. morbida in our case. In the effort to reduce false positives, we implemented the Benjamini–Hochberg correction method when comparing likelihood ratios for null and alternative models using a P-value threshold of 0.05. We performed similar BLAST searches as mentioned previously to characterize the functions of these proteins and identify proteins with signal peptides and transmembrane domains.
The above procedures were repeated for detecting genes under selection in G. clavigera because this fungal pathogen plays an ecological role similar to G. morbida. By performing these analyses, we sought to uncover genes under adaptive evolution in both beetle-vectored tree pathogens.
Results
Assembly Features
We recently assembled a reference genome for a G. morbida strain isolated from J. californica in Southern California (Schuelke et al. 2016). The reference contained 73 scaffolds with an estimated size of 26.5 Mb. By using the MAKER annotation pipeline, we predicted 6,273 protein models in this reference in-silico (Cantarel et al. 2008). In this work, we sequenced strains of G. flava and G. putterillii at approximately 102× and 131× coverage, respectively. The G. flava assembly was composed of 1,819 scaffolds totaling 29.47 Mb in length, and the G. putterillii genome contained 320 scaffolds extending 29.99 Mb. Geosmithia flava and G. putterillii totaled 6,976 and 7,086 protein models, respectively. Both genomes contained 98% of the single-copy orthologs present in more than 90% of the fungal species. Nearly all of the raw reads (97% and 98%) mapped back to G. flava and G. putterillii genome assemblies, respectively (table 4). These statistics indicated that our genome assemblies are high quality and complete.
Table 4.
Length-Based Statistics for Geosmithia morbida, Geosmithia flava, and Geosmithia putterillii Generated with QUAST v2.3
| Species | Est. Genome Size (Mb) | k-mer for ABySS Assembly | Scaffold Count | Largest Scaffold | NG50 | LG50 | Genome Completeness | Predicted Proteins | Transcript Completenessb |
|---|---|---|---|---|---|---|---|---|---|
| G. morbida | 26.5 | NAa | 73 | 2,597,956 | 1,305,468 | 7 | 98 | 6,273 | 93 |
| G. flava | 29.6 | 91 | 1,819 | 1,534,325 | 460,430 | 22 | 98 | 6,976 | 94 |
| G. putterillii | 30.0 | 91 | 320 | 2,758,267 | 1,379,352 | 9 | 98 | 7,086 | 94 |
Note.—The average GC content for G. morbida, G. flava, and G. putterillii equals 54%, 52%, and 55.5%, respectively. All genome completeness values were produced with BUSCO v1.1b1. These percentages represent genes that are complete and not duplicated or fragmented. NG50 is the scaffold length such that considering scaffolds of equal or longer length produce 50% of the bases of the reference genome. LG50 is the number of scaffolds with length NG50.
Genome assembly for G. morbida was constructed using AllPaths-LG (v49414). See Schuelke et al. (2016) for further details.
These percentages were computed using the fungal data set 9 provided with BUSCO.
An estimated 0.80% of G. morbida reference genome sequence represented repeats, whereas 0.63% and 0.64% of the sequences in G. flava and G. putterillii consisted of repetitive elements. There were 60, 42, and 15 DNA transposons in G. morbida, G. flava, and G. putterillii, respectively. Furthermore, G. morbida possessed only 152 retroelements, whereas G. flava and G. putterillii had 401 and 214 of such elements, correspondingly (table 5).
Table 5.
Repetitive Elements Profile of Geosmithia Species Generated with RepeatMasker v4.0.5
| Genome Size (Mb) | GC (%) | Bases Masked (%) | No. of Retroelements | No. of DNA Transposons | |
|---|---|---|---|---|---|
| G. morbida | 26.5 | 54.30 | 0.81 | 152 | 60 |
| G. flava | 29.6 | 51.87 | 0.63 | 401 | 42 |
| G. putterillii | 30.0 | 55.47 | 0.64 | 214 | 15 |
We also looked for the presence of repeat-induced point mutations within each Geosmithia species by searching for AT-rich regions. In many fungi, AT-rich regions are associated with such mutations that favor the transition of cytosines to thymines resulting in a lower GC content. We did not find AT-rich regions in any of the three Geosmithia species (supplementary figs. S2–S4, Supplementary Material online).
Identifying Putative Genes Involved in Pathogenicity
Approximately 32%, 34%, and 35% of the total proteins in G. morbida, G. flava, and G. putterillii respectively shared significant homology with protein sequences in the PHI-base 4.0 database. The number of unknown proteins with hits in the PHI-base database was similar for G. morbida (26), G. flava (28), and G. putterillii (36). The full BLASTp search results against the PHI-base database for G. morbida, G. flava, and G. putterillii are available in the supporting material (supplementary table S1, Supplementary Material online). Only 20 proteins with PHI-base hits were cysteine-rich peptides in all three species.
Identifying Species-Specific Genes
An analysis of orthologous groups (OGs) between the three Geosmithia species and A. chrysogenum was completed to determine whether a significant loss in OGs in any of the Geosmithia species has taken place. The three Geosmithia species and A. chrysogenum contained a total of 9,065 OGs. Among the set of homologous genes, there were 4,655 single copy orthologs. A. chrysogenum contained 2,338 species-specific genes, of which seven genes were paralogous. Geosmithia morbida possessed 73 unique genes whereas, G. putterilli and G. flava had 161 and 146 species-specific genes. The two nonpathogenic Geosmithia species did not contain any paralogs, however G. morbida had three unique genes present in five copies. Based on a functional search against NCBI’s nonredundant database, the three genes encode hydantoinase B/oxoprolinase, aldehyde dehydrogenase, and ABC-2 type transporter.
A total of 205 OGs were present in A. chrysogenum, G. putterillii, and G. flava, but not in G. morbida. This is likely due to a species-specific gene loss events in G. morbida, as it was the most parsimonious scenario. At the same time, only 90 and 88 OGs are specifically missing in G. putterillii and G. flava, respectively. This suggests that G. morbida has experienced a higher OG loss rate than the other two Geosmithia species.
Identifying Putative Secreted Proteins
A total of 349, 403, and 395 proteins in G. morbida, G. flava, and G. putterillii contained signal peptides, respectively. Of these putative signal peptide-containing proteins in G. morbida, 27 (7.7%) encoded proteins with unknown function, whereas G. flava and G. putterillii contained 29 (7.2%) and 30 (7.6%) unknown proteins, respectively. The difference in percentage of unknown proteins with signal peptides was minimal among the three genomes. For each species, proteins containing signal peptides were subjected to a membrane protein topology search using TMHMM v2.0. There were 237, 281, and 283 proteins in G. morbida, G. flava, and G. putterillii that lacked any transmembrane protein domains.
Profiling Carbohydrate-Active Enzymes and Peptidases
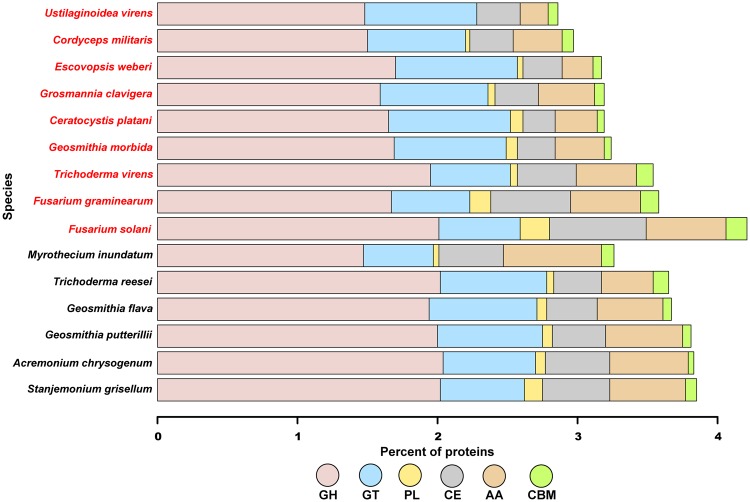
Carbohydrate-active enzymes (CAZymes) break down plant structural components, enabling initiation and establishment of infection. We assessed the CAZymatic profile of all species in the order Hypocreales, Geosmithia species, and G. clavigera (fig. 1). The glycoside hydrolase (GH) family members dominated all protein models, followed by glycosyltransferase (GT) family. The two most prominent families among all fungal species were GH3 and GH16 (supplementary table S2, Supplementary Material online). GH3 hydrolases are involved in cell wall degradation and overcoming the host immune system, and GH16 enzymes fulfill a wide range of cellular functions including transporting amino acids. The third most representative family was GH18; however G. morbida only contained four of these enzymes. In contrast, this number for other species ranges from 9 to 31 enzymes. Along with acetylglucosaminidases, family GH18 harbors chitinases that assist in the production of carbon and nitrogen. In terms of other CAZyme families, all fungi except F. solani have a similar overall distribution. Fusarium solani contains more CAZymes than any other pathogen or nonpathogen.
Fig. 1.
—CAZymes distribution for Geosmithia species, other Hypocreales, and C. platani. The species in red are pathogens, whereas the names in black are nonpathogens. CAZymes were identified with HMMer searches of dbCAN peptide models. GH, glycoside hydrolases; GT, glycosylTransferases; PL, polysaccharide lyases; CE, carbohydrate esterases; AA, auxiliary activities enzymes; CBM, carbohydrate-binding molecules.
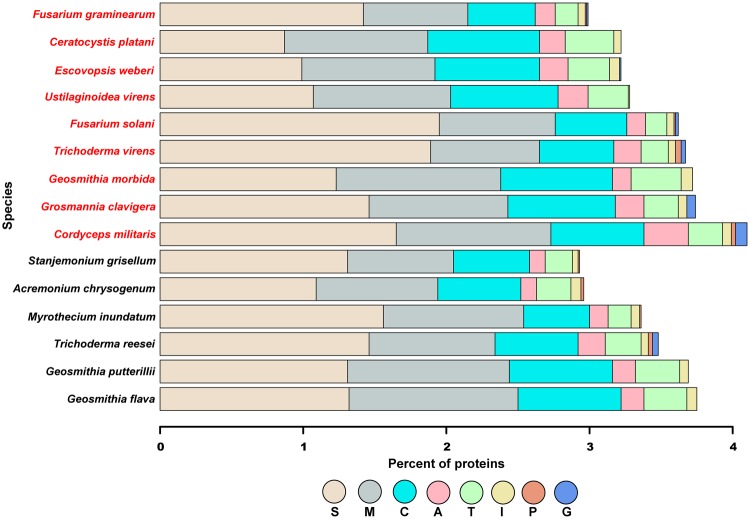
In addition to profiling CAZymes, we performed a BLAST search against the peptidase database—Merops v10.0 (Rawlings et al. 2016)— for each Hypocreales, Ceratocystis platani, and G. clavigera. Among the pathogens, G. morbida has the third highest percent of predicted proteases after Cordyceps militaris (insect pathogen) and G. clavigera (fig. 2 and supplementary table S3, Supplementary Material online). Moreover, G. flava and G. putterillii have the largest percent of peptidases among the nonpathogenic fungi. All three Geosmithia species illustrate similar proteolytic profiles and contain no glutamic and mixed peptidases.
Fig. 2.
—Proteolytic enzymes distribution for Geosmithia species, other Hypocreales, and C. platani. The species in red are pathogens, whereas the names in black are nonpathogens. Proteases were identified using BLASTp searches against the MEROPs database v10. S, serine; M, metallo; C, cysteine; A, aspartic; T, threonine; I, inhibitors; P, mixed; G, glutamic.
Smaller Repertoire of Secondary Metabolite Biosynthesis Gene Clusters in Geosmithia Species
Contrary to primary metabolites, secondary metabolites are not necessary for normal fungal growth and development (Fox and Howlett 2008). Although their role is not completely known, it is most likely they help the organism survive its ecological niche (Fox and Howlett 2008). To examine the capacity for secondary metabolite biosynthesis in Geosmithia and two most closely related species, we identified gene clusters responsible for such biosynthesis using antiSMASH (Weber et al. 2015). Although the two closely related species, Acremonium chrysogenum and Stanjemonium grisellum, have large repertoires of such gene clusters (42 and 60, respectively), the Geosmithia species have much smaller repetoires, ranging from 14 to 19 gene clusters (supplementary table S4, Supplementary Material online). Within the Geosmithia genus, G. morbida has an even smaller number of such gene clusters than G. putterilli and G. flava (14 vs 16 and 19, supplementary table S4, Supplementary Material online).
Furthermore, to assess the impact of a fragmented genome on clusters detection, we plotted N50 values of these genomes against the total cluster numbers. We found no evidence of a positive correlation between the two, indicating that a smaller N50 does not lead to under-detection of gene clusters (supplementary fig. S1, Supplementary Material online).
Inferring Phylogeny
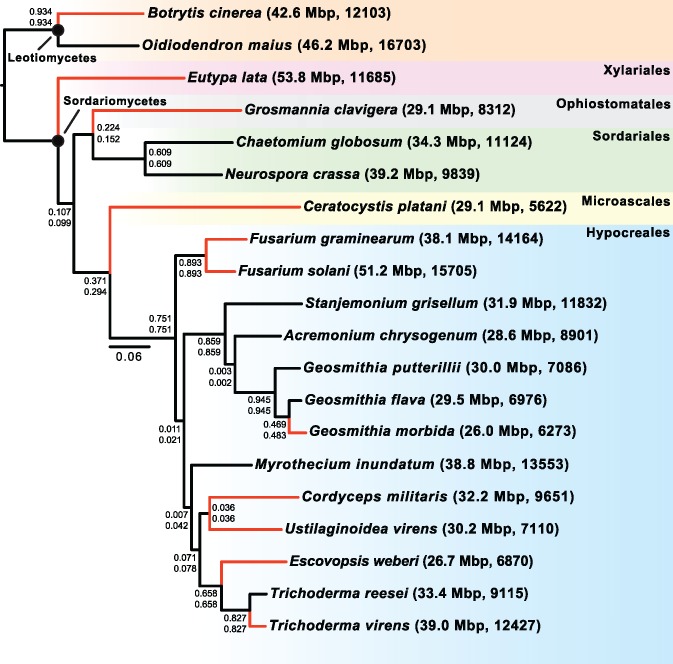
Even though Geosmithia was first established as a genus in 1979, it has only recently been described in depth. One of the objectives in this study was to uncover the phylogenetic relationship between Geosmithia species and other fungal pathogens using coding DNA sequence data. In order to determine the broader evolutionary history of Geosmithia species, we constructed ML and BMCMC phylogenies using genome-scale data from G. morbida, G. putterillii, G. flava, and 17 additional fungal taxa (table 3). Our data set consisted of 11 pathogens and 9 nonpathogens. After trimming and filtering, our data matrix comprised of 633 partitions and 247,627 amino acid sites. The topologies generated under ML and BMCMC were identical, and all nodes in both analyses received bootstrap support of 100% (ML) and posterior probabilities of 1.0 (BMCMC). Our MrBayes analysis reached convergence after 215,000 generations with the average standard deviation of split frequencies = 0.000000. Phylobayes analyses had a max diff score of 0.178571 after 2,061 cycles. However, analyses using the standard IC/TC algorithm (Salichos et al. 2014) in RAxML 8.1.2 (Stamatakis 2014) produced a TC value of only 0.457, indicating a significant degree of discordance among topologies for individual partitions. In addition, IC and IC All scores reveal a number of poorly recovered internodes among topologies individual partitions (fig. 3). Taken together, our phylogenomic analyses provide strong support for the monophyly of the Hypocreales and the position of Geosmithia within it and is consistent with previous reports (Fitzpatrick et al. 2006, Wang et al. 2009). The Geosmithia species form a well-supported monophyletic clade with two nonpathogenic fungi, A. chrysogenum and S. grisellum, suggesting that the common ancestor shared among these species was likely not a pathogen.
Fig. 3.
—ML was estimated with RAxML (Stamatakis 2014) using a scheme determined by PartitionFinder (Lanfear et al. 2014). The IC (top) and IC All (bottom) scores are also presented for each node. This topology is identical to the BMCMC phylogeny constructed in MrBayes (Ronquist et al. 2012). All nodes in ML and BMCMC analyses receive maximum support of 1. The black circles symbolize classes. The color-shaded boxes at the right of the figure denote the orders within each class. The first and second numbers in parentheses represent the genome sizes in Mb and the number of predict protein models, respectively. Black and red branches correspond to nonpathogens and pathogens, respectively, which span multiple orders.
Genes under Positive Selection
In order to understand the molecular basis of pathogenicity in G. morbida, we sought to detect genes under positive selection by leveraging the BSM in PAML’s codeml program (4.8). Geosmithia morbida was selected as the foreground branch. Our results showed 38 genes to be under positive selection using an adjusted P-value < 0.05. Next, we performed a functional search for each protein by blasting the peptide sequences against the NCBI nonredundant and pfam databases. We determined that several were involved in catabolic activity, gene regulation, and cellular transport (table 6 and supplementary table S4, Supplementary Material online).
Table 6.
Functional Analyses of Genes under Positive Selection in Geosmithia morbida Detected by the Branch-Site Model in PAML 4.8
| Gene Number | Function | dN/dS | Transmembrane Domain (N) |
|---|---|---|---|
| 3078 | Takes part in intracellular signaling, protein recruitment to various membranes | 2.04 | 0 |
| 2666 | Involved in receptor-mediated endocytosis and vesicle trafficking | 2.01 | 0 |
| 563 | Unclear function | 1.94 | 1 |
| 2194 | Unknown function | 1.94 | 0 |
| 801 | Catalyzes the transfer of electrons from ferrocytochrome c to oxygen converting the cytochrome c into water | 1.93 | 1 |
| 3944 | Involved in methylation and have a wide range of substrate specificity | 1.90 | 5 |
| 5058 | Involved in ubiquitination of proteins target for degradation | 1.90 | 0 |
| 1843 | Involved in heat-shock response | 1.86 | 0 |
| 521 | Involved in damage DNA binding and repair | 1.85 | 0 |
| 5111 | Involved in receptor-mediated endocytosis and vesicle trafficking | 1.84 | 0 |
| 4128 | Catalyzes the hydrolysis of esters | 1.84 | 0 |
| 923 | Hydrolases the peptide bond at the C-terminus of ubiquitin | 1.83 | 1 |
| 4405 | Involved in transport and metabolism of lipids | 1.83 | 1 |
| 3137 | Part of proteins with diverse functions such as cell-cycle regulators, signal transducers, transcriptional initiators | 1.78 | 0 |
| 4359 | Unknown function | 1.73 | 2 |
| 5639 | Involved in rRNA synthesis | 1.67 | 0 |
| 5 | Involved in vesicular transport | 1.63 | 0 |
| 624 | Involved in transfer of glucose molecules that are part of a larger glycosylation machinery | 1.62 | 9 |
| 3929 | Unknown function but associates with GRAM domain found in glucosyltransferases and other membrane affiliated proteins | 1.61 | 0 |
| 1456 | Involved in DNA repair and replication | 1.59 | 0 |
| 4829 | Form cAMP | 1.59 | 0 |
| 254 | Major ATP transporters | 1.59 | 2 |
| 4888 | Unknown function | 1.54 | 0 |
| 5426 | Hydrolyzes nonubiquitinated peptides | 1.54 | 0 |
| 5709 | Transcription factors | 1.50 | 0 |
| 859 | May be involved in the timing of nuclear migration | 1.50 | 0 |
| 5703 | Cleave peptide bonds in other proteins | 1.47 | 6 |
| 5255 | Heat shock protein involved in induced stress response to ethanol | 1.46 | 3 |
| 5704 | Regulates gene expression during oxidative stress caused by the host plant | 1.46 | 0 |
| 2485 | Transfer phosphates | 1.39 | 0 |
| 6116 | Hydratase and/or isomerase | 1.38 | 0 |
| 5266 | Breaks down actin, cell membrane deformations | 1.34 | 0 |
| 5000 | Catalyzes the first step in histidine biosynthesis | 1.34 | 0 |
| 3326 | Involved in de novo synthesis of nucleotide purine | 1.32 | 0 |
| 2142 | E2 enzymes that catalyze the binding of activated ubiquitin to the substrate protein. The substrate proteins are targeted for degradation by the proteasome | 1.24 | 0 |
| 581 | Ribosomal protein | 1.17 | 0 |
| 5948 | Involved in initiation of transcription | 1.14 | 1 |
| 3700 | Part of the TOM complex that recognizes and regulates the transport of mitochondrial precursor molecules from the cytosol to the intracellular space of the mitochondrion | 1.03 | 0 |
Note.—The gene number corresponds to the sequence ID in the G. morbida protein file available at DRYAD. The P-values for each dN/dS ratio is < 0.05. dN/dS is the ratio of nonsynonymous substitutions to synonymous changes.
Genes under Adaptive Evolution in Beetle-Vectored Fungal Pathogens
In addition to detecting genes under selective pressures in G. morbida, we performed the same selection analysis for Grosmannia clavigera to identify overlapping proteins that may help explain adaptations leading to the ecological role these two beetle-vectored fungi play. We found that G. clavigera possessed 42 positively selected genes that shared protein domains with only two of the 38 genes predicted to be under selection in G. morbida. The two overlapping motifs are methyltransferase and protein kinase domains. Our KEGG analysis exhibited no common pathways between G. morbida and G. clavigera. The complete table displaying BLASTp results for genes under positive selection in G. clavigera is available in the supporting material (supplementary table S6, Supplementary Material online).
Discussion
This study aims to provide insight into the evolution of pathogenicity within G. morbida, a beetle vectored pathogen that is the causal agent of TCD in Julgans species. Our results indicate that the genome size, gene number, transposon number, and secondary metabolite profile of both pathogenic and nonpathogenic species of Geosmithia are very similar. This is not specifically unexpected as all three species occupy similar niches as beetle-vectored fungi associated with trees. The only difference in life history strategy is that G. flava and G. putterilli have a much large host range and are primarily saprotroph that exist on dead or dying trees including coniferous and deciduous species, whereas G. morbida has a pathogenic life strategy and primarily infects members of the genus Juglans. The main distinction between G. morbida and the nonpathogenic species amounts to 38 genes found to be under positive selection in the former, of which several have known function in pathogenicity in other plant pathogenic fungi. Although small in numbers, it may be these genes that separate the pathogenic G. morbida from its saprotrophic congeners.
The Smaller Genome Size of Geosmithia Species
In contrast to other species in the phylogeny (fig. 3), fungi associated with trees either as pathogens or saprophytes (Geosmithia species, G. clavigera, and C. platani) had reduced genomes and gene content. We predict this smaller genome and gene content could potentially be a result of evolving specialized lifestyles to occupy a specialized niche. For instance, all three Geosmithia species and G. clavigera are vectored into their respective hosts via bark beetles, which may result in strong selection on the genetic variability of the fungi because they must adapt to their vectors and hosts simultaneously. A recent study characterizing the genome of mycoparasite Escovopsis weberi showed that specialized pathogens tend to have smaller genomes and predicted protein sets because they lack genes that are not required beyond their restricted niche when compared to closely related generalists (de Man et al. 2016). Our results agree with this finding because G. morbida has a more specialized beetle vector (P. juglandis) and plant host range (Juglans species) in comparison to G. putterilli and G. flava which can be found on a variety of trees species including both gymnosperms and angiosperms (Kolařík and Jankowiak 2013), and can be vectored by multiple beetle species. Both G. morbida and E. weberi represent a significant contrast to previous reports that have documented the importance of genome expansion with the evolution of pathogenicity (Adhikari et al. 2013; Raffaele and Kamoun 2012). Furthermore, our results are supported by prior findings which showed that gene loss and gain can lead to a more specialized lifestyle in bacterial and eukaryotic lineages (Ochman and Moran 2001; Lawrence 2005). Another example is the obligate ectomycorrhizal symbiont Amanita bisporigera, which was found to lack many plant cell-wall-degrading enzymes suggesting that these genes may no longer be required for A. bisporigera’s specialized lifestyle (Nagendran et al. 2009).
Genome reduction or retention of a smaller genome is an important evolutionary mechanism that propels divergence of species and more often than not enables adaptation to specific environments. Although smaller genomes are more frequent in prokaryotes, it is not uncommon among eukaryotes including fungal species (Ochman and Moran 2001; Nagendran et al. 2009; Spanu et al. 2010).
Mobile Genetic Elements
Although G. morbida has a smaller genome size, the pathogen possesses a slightly higher percentage of repeats than G. flava and G. putterillii (table 5). The extent to which mobile genetic elements affect genome evolution in Geosmithia is unknown, but mobile genetic elements may be influential drivers of adaptive evolution in G. morbida. They are known to be responsible for genomic rearrangements and expansion, horizontal gene transfer and generation of new genes (Casacuberta and González 2013; Stukenbrock and Croll 2014). For example, Fusarium oxysporum has a genome nearly 60 Mb in length and contains 16.83 Mb of repetitive sequences (Ma et al. 2010). Although G. morbida harbors fewer mobile genetic elements than fungal species such as F. oxysporum, it is possible that such elements have contributed to the evolution of pathogenicity in Geosmithia via horizontal gene transfer and/or changes in gene numbers. Understanding the role of mobile genetic elements within genus Geosmithia may be key in discovering the genetic basis behind the evolution of pathogenicity.
Repeat-Induced Point Mutations
None of the Geosmithia species contain AT-rich regions which are an indicator of repeat-induced point mutations. The paucity of substrate (repetitive genetic elements) could explain the lack of such mutations: if the genome contains low repeats, then repeat-induced point mutations will also occur at a smaller rate.
Species-Specific Genes
We identified three genes unique to G. morbida which are all involved in stress responses that can be induced by the host immune system during the infection process. For example, aldehyde dehydrogenases are part of a large protein family that detoxify aldehydes and alcohols in all organisms including fungal species (Asiimwe et al. 2012). Hydantoinase B/oxoprolinase is involved in the synthesis of glutathione, a compound essential for basic cellular functions but also important in cellular defense against oxidative stress (Pócsi et al. 2004). Glutathione has been shown to chelate damaging metal ions by inhibiting their spread in the cell (Pócsi et al. 2004), and to prevent the accumulation of H2O2 in Paxillus involutus (Ott et al. 2002). Finally, ATP-binding cassette (ABC) proteins belong to an especially large family of proteins that regulates transport of substances across the cellular membrane. In pathogenic fungi, they are involved in drug resistance and in the production of defense molecules (Krattinger et al. 2009; Wang et al. 2013; Karlsson et al. 2015).
Carbohydrate-Active Enzymes and Peptidases
Although one might expect the pathogen G. morbida to possess more carbohydrate binding enzymes than its nonpathogenic relatives, our results indicated that all fungi except F. solani have a similar overall distribution (fig. 1). Fusarium solani contains more CAZymes than any other pathogen or nonpathogen. This Fusarium species is a generalist necrotrophic pathogen that is believed to possess more CAZymes than biotrophic and hemibiotrophic fungi. This discrepancy may be due to the fact that necrotrophic pathogens require an extensive toolkit to promote host cell death as quickly as possible; whereas biotrophs need to keep the host alive, and dispensing large number of degradative enzymes can be detrimental to that aim (Zhao et al. 2013).
In addition to CAZymes, we searched for peptidases. Geosmithia morbida has the third highest percent of predicted proteases after C. militaris (insect pathogen) and G. clavigera (fig. 2 and supplementary table S3, Supplementary Material online). Moreover, G. flava and G. putterillii have the largest percent of peptidases among the nonpathogenic fungi. All three Geosmithia species illustrate similar proteolytic profiles and contain no glutamic and mixed peptidases. These results were expected because all three Geosmithia species are closely related. Furthermore, given that these species are plant affiliates (except C. militaris), the ability to degrade lignin and cellulose is an important life history trait that is conserved throughout fungal pathogens, but perhaps did not give rise to pathogenicity in G. morbida.
Transmembrane Protein and Effector Genes
Transmembrane proteins are important mediators between a host and its pathogens during microbial invasion. Fungal pathogens either penetrate a surface or enter the host through a wound or opening such as stomata in order to gain access to the nutrients in the plant (Chisholm et al. 2006). Once the infiltration process is completed, pathogens are exposed to host plasma membrane receptors that detect pathogen-associated molecular patterns (PAMP) and induce PAMP-triggered immunity (PTI) to prevent further proliferation of the microbe. Transmembrane proteins expressed by a fungal pathogen are crucial during PTI because they are responsible for suppressing PTI directly or by secreting effector molecules, which contain signal peptides necessary for proper targeting and transport (Chisholm et al. 2006; Boller and He 2009). Our analysis of the 38 proteins under positive selection showed that 11 of these possess at least one or more transmembrane domains. Although nearly 30% of the positively selected genes identified were membrane bound, a similar proportion of nonselected genes in G. morbida were membrane associated, indicating this result is not strong evidence that interactions with the host surface are drivers of evolution within G. morbida. Among proteins under selection, we found no protein that contained a signal peptide, indicating none of these proteins are secretory.
Secondary Metabolite Biosynthesis Gene Clusters in Geosmithia Species
In comparison to the two closely related species, A. chrysogenum and S. grisellum, the Geosmithia species have much smaller repetoires of secondary metabolite biosynthesis gene clusters, with G. morbida having even fewer such gene clusters than G. putterilli and G. flava (supplementary table S5, Supplementary Material online). Geosmithia morbida also has a more specialized ecological niche, associating with only one beetle vector (P. juglandis) and a narrow plant host range (Juglans species), whereas G. putterilli and G. flava are associated with a variety of tree species and multiple beetle species (Kolařík and Jankowiak 2013). Having a narrow host range and a smaller arsenal of secondary metabolite biosynthesis gene clusters may be advantageous to G. morbida so that fewer genes trigger PTI within the plant and its system does not build immunity rapidly.
Genes under Positive Selection
Our analyses identified several genes under positive selection in G. morbida that encode for proteins that have been implicated in pathogenicity in other fungal pathogens (table 6). For instance, a Cullin3-like protein was predicted to be under positive selection. These proteins belong to a group of structurally similar molecules involved in protein degradation, such as the Skp-Cullin-F-box (SCF) ubiquitin ligase complex (Cardozo and Pagano 2004; Pintard et al. 2004). Furthermore, a ubiquitin-conjugating enzyme (E2) that interacts with cullin3 to prepare substrate for degradation, also had a dN/dS > 1, indicating that both genes are under positive selection within G. morbida. Although little is known regarding the precise functional abilities of these complexes, it is possible these proteins are involved in pathogenicity of G. morbida. Previous studies have also implicated ubiquitin ligase complexes in infection and disease development (Duyvesteijn et al. 2005; Han et al. 2007).
Our analysis also revealed a regulatory protein homologous to basic leucine zipper (bZIP) transcription factors was under selection. The bZIP proteins are similar to AP-1 transcription factors and monitor several developmental and physiological processes including oxidative stress responses in eukaryotes (Corrêa et al. 2008). Fungal pathogens such as the rice blast fungus Magnaporthe oryzae express AP1-like transcription factor called MoAP1 that contains bZIP domain. MoAP1 is highly active during infection and is translocated from the cytoplasm to the nucleus in response to oxidative stress induced by H2O2 (Guo et al. 2011). MoAP1 regulates enzymes such as laccase and glutamate decarboxylase that are involved in lignin breakdown and metabolism of γ-aminobutyric acid, respectively (Solomon and Oliver 2002; Baldrian 2005; Janusz et al. 2013). Some of the other positively selected genes include ABC transporter, proteases, proteins involved in apoptosis, and proteins related to DNA replication and repair. As previously mentioned, ABC transporters are important mediators that aid in protection against plant defenses as well as natural toxic compounds (Krattinger et al. 2009; Wang et al. 2013; Karlsson et al. 2015; Lo Presti et al. 2015). Apoptosis or programmed cell death helps establish resistance during host–microbe interactions, helps organisms cope with oxidative environments, and may even be essential for infection (Veneault-Fourrey et al. 2006; Kabbage et al. 2013). In fungal species, proteins involved in DNA replication and repair are essential for the formation and penetration of appressorial structures into the host cell (Son et al. 2016). Only five of the 38 genes with evidence of selection encoded proteins with unknown functions. These positively selected genes may also be involved in the evolution and adaptation of G. morbida.
A similar analysis comparaing genes under positive selection in G. clavigera and G. morbida found only two genes to be under positive selection in both organisms. These findings emphasize that evolutionary forces act differently on divergent populations. All fungal pathogens face dissimilar environmental challenges and associate with different hosts both spatially and temporally. Even closely related organisms can be highly distinct molecularly. For instance, the fungi responsible for the Dutch elm disease—Ophiostoma ulmi and Ophiostoma novo-ulmi—differ in their genetic composition and virulence despite their strong evolutionary relationship (Brasier 2001; Khoshraftar et al. 2013; Comeau et al. 2015).
In conclusion, our study provides insight into the potential mechanisms that may be behind the evolution of pathogenicity within the genus Geosmithia. We also identified a small set of genes that may be contributing to pathogenicity of G. morbida. Functional experiments and analyses of the expression levels of these genes during infection as compared to gene expression of a nonpathogen would shed light on the mechanisms influencing pathogen evolution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The use of trade names is for the information and convenience of the reader and does not imply official endorsement or approval by the United States Department of Agriculture or the Forest Service of any product to the exclusion of others that may be suitable. This project was funded by the USDA Forest Service, Forest Health Monitoring, and partial funding was provided by the New Hampshire Agricultural Experiment Station. Special thanks to Dr. Miroslav Kolarik for providing isolates of Geosmithia flava and Geosmithia putterilli. We are also grateful to Dr. Joseph Spatafora and his team for giving us permission to utilize sequence data for Stanjemonium grisellum and Myrothecium inundatum. Finally, we thank the 1000 Fungal Genomes Project for being a valuable source of genetic data.
Author Contributions
T.A.S. conceived, designed, and performed the experiments and wrote the manuscript. A.W. assembled and annotated the genomes. G.W. performed key analysis and interpreted the results. He also designed and implemented secondary metabolite analyses, wrote part of the manuscript and reviewed the paper. K.B. conceived and designed the study, wrote and reviewed the manuscript. He also conceived funding. D.C.P. designed and implemented the phylogenetic methods in this study and reviewed the manuscript. K.W. conveived funding and designed the experiments. He also wrote and reviewed the manuscript. M.D.M. conceived and designed the study, developed analyses pipelines and edited the manuscript.
Literature Cited
- Adhikari BN, et al. 2013. Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS One 8(10):e75072.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamouti SM, et al. 2014. Comparative genomics of the pine pathogens and beetle symbionts in the genus Grosmannia. Mol Biol Evol. 31(6):1454–1474.http://dx.doi.org/10.1093/molbev/msu102 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amselem J, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7(8):e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiimwe T, Krause K, Schlunk I, Kothe E.. 2012. Modulation of ethanol stress tolerance by aldehyde dehydrogenase in the mycorrhizal fungus Tricholoma vaccinum. Mycorrhiza 22(6):471–484.http://dx.doi.org/10.1007/s00572-011-0424-9 [DOI] [PubMed] [Google Scholar]
- Baldrian P. 2006. Fungal laccases-occurrence and properties. FEMS Microbiol Rev. 30(2):215–242.http://dx.doi.org/10.1111/j.1574-4976.2005.00010.x [DOI] [PubMed] [Google Scholar]
- Belbahri L. 2015. Genome sequence of Ceratocystis platani, a major pathogen of plane trees. [WWW document] Available from: http://www.ncbi.nlm.nih.gov/nuccore/814603118 [accessed 2015 Dec 15].
- Berka RM, et al. 2011. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 29(10):922–927.http://dx.doi.org/10.1038/nbt.1976 [DOI] [PubMed] [Google Scholar]
- Blanco-Ulate B, Rolshausen PE, Cantu D.. 2013. Draft genome sequence of the grapevine dieback fungus Eutypa lata UCR-EL1. Genome Announc. 1(3):e00228–e00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, He SY.. 2009. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324(5928):742–744.http://dx.doi.org/10.1126/science.1171647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E, et al. 2016. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374:23–54. Available from http:www.uniprot.org/ [accessed 2015 May 6]. [DOI] [PubMed] [Google Scholar]
- Brasier CM. 2001. Rapid evolution of introduced plant pathogens via interspecific hybridization hybridization is leading to rapid evolution of Dutch elm disease and other fungal plant pathogens. Bioscience 51:123–133.http://dx.doi.org/10.1641/0006-3568(2001)051[0123:REOIPP]2.0.CO;2 [Google Scholar]
- Cantarel BL, et al. 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196.http://dx.doi.org/10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M.. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Cell Biol. 5(9):739–751.http://dx.doi.org/10.1038/nrm1471 [DOI] [PubMed] [Google Scholar]
- Casacuberta E, González J.. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22(6):1503–1517. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ.. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4):803–814.http://dx.doi.org/10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Coleman JJ, et al. 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5(8):e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, et al. 2015. Functional annotation of the Ophiostoma novo-ulmi genome: insights into the phytopathogenicity of the fungal agent of Dutch elm disease. Genome Biol Evol. 7(2):410–430.http://dx.doi.org/10.1093/gbe/evu281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa LG, et al. 2008. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3(8):e2944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusoe MR, et al. 2015. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Research 4:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, et al. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317(5843):1400–1402.http://dx.doi.org/10.1126/science.1143708 [DOI] [PubMed] [Google Scholar]
- de Man TJ, et al. 2016. Small genome of the fungus Escovopsis weberi, a specialized disease agent of ant agriculture. Proc Natl Acad Sci USA. 113(13):3567–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGuistini S, et al. 2011. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc Natl Acad Sci USA. 108(6):2504–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyvesteijn RG, et al. 2005. Frp1 is a Fusarium oxysporum F‐box protein required for pathogenicity on tomato. Mol Microbiol. 57(4):1051–1063. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14(9):755–763.http://dx.doi.org/10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G.. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 6:99.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EM, Howlett BJ.. 2008. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 11(6):481–487.http://dx.doi.org/10.1016/j.mib.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868.http://dx.doi.org/10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- Guo M, et al. 2011. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7(2):e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075.http://dx.doi.org/10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YK, Kim MD, Lee SH, Yun SH, Lee YW.. 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol Microbiol. 63(3):768–779. [DOI] [PubMed] [Google Scholar]
- Janusz G, Kucharzyk KH, Pawlik A, Staszczak M, Paszczynski AJ.. 2013. Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzyme Microb Technol. 52(1):1–12. [DOI] [PubMed] [Google Scholar]
- Kabbage M, Williams B, Dickman MB.. 2013. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9(4):e1003287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, et al. 2015. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol Evol. 7(2):465–480.http://dx.doi.org/10.1093/gbe/evu292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, et al. 2016. Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44(D1):D574–D580. [WWW document] Available from: http://fungi.ensembl.org/index.html [accessed 2015 Nov 14]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshraftar S, et al. 2013. Sequencing and annotation of the Ophiostoma ulmi genome. BMC Genomics 14:162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KT, et al. 2016. Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front Plant Sci. 7:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Francis M, Costa M.. 2011. High quality genomic DNA extraction using CTAB and Qiagen genomic-tip (version 2). [WWW document] Available from: http://1000.fungalgenomes.org/home/wp-content/uploads/2013/02/genomicDNAProtocol-AK0511.pdf [accessed 2015 Dec 12].
- Kohler A, et al. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 47(4):410–415.http://dx.doi.org/10.1038/ng.3223 [DOI] [PubMed] [Google Scholar]
- Kolařík M, Freeland E, Utley C, Tisserat N.. 2011. Geosmithia morbida sp. nov., a new phytopathogenic species living in symbiosis with the walnut twig beetle (Pityophthorus juglandis) on Juglans in USA. Mycologia 103:325–332. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Jankowiak R.. 2013. Vector affinity and diversity of geosmithia fungi living on subcortical insects inhabiting Pinaceae species in Central and Northeastern Europe. Microb Ecol. 66(3):682–700. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Kirkendall LR.. 2010. Evidence for a new lineage of primary ambrosia fungi in Geosmithia Pitt (Ascomycota: Hypocreales). Fungal Biol. 114(8):676–689. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Kostovcik M, Pazoutova S.. 2007. Host range and diversity of the genus Geosmithia (Ascomycota: Hypocreales) living in association with bark beetles in the Mediterranean area. Mycol Res. 111:1298–1310. [DOI] [PubMed] [Google Scholar]
- Kolařík M, Kubatova A, van Cepicka I, Pazoutova S, Srutka P.. 2005. A complex of three new white-spored, sympatric, and host range limited Geosmithia species. Mycol Res. 109(12):1323–1336. [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):1..http://dx.doi.org/10.1186/1471-2105-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, et al. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323(5919):1360–1363.http://dx.doi.org/10.1126/science.1166453 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kubicek CP, et al. 2011. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12(4):R40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A.. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288.http://dx.doi.org/10.1093/bioinformatics/btp368 [DOI] [PubMed] [Google Scholar]
- Lawrence JG. 2005. Common themes in the genome strategies of pathogens. Curr Opin Genet Dev. 15(6):584–588.http://dx.doi.org/10.1016/j.gde.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760.http://dx.doi.org/10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2015. BFC: correcting Illumina sequencing errors. Bioinformatics 31(17):2885–2887.http://dx.doi.org/10.1093/bioinformatics/btv290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, et al. 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 66:513–545.http://dx.doi.org/10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B.. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42(Database issue):D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SC, et al. 2014. First report of Geosmithia pallida causing foamy bark canker, a new disease on coast live oak (Quercus agrifolia), in association with Pseudopityophthorus pubipennis in California. Plant Dis. 98(9):1276..http://dx.doi.org/10.1094/PDIS-03-14-0273-PDN [DOI] [PubMed] [Google Scholar]
- Ma LJ, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464(7287):367–373.http://dx.doi.org/10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, et al. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol. 26:553–560.http://dx.doi.org/10.1038/nbt1403 [DOI] [PubMed] [Google Scholar]
- Misof B, et al. 2013. Selecting informative subsets of sparse supermatrices increases the chance to find correct trees. BMC Bioinformatics 14:348.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendran S, Hallen-Adams HE, Paper JM, Aslam N, Walton JD.. 2009. Reduced genomic potential for secreted plant cell-wall-degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Genet Biol. 46(5):427–435. [DOI] [PubMed] [Google Scholar]
- Ochman H, Moran NA.. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292(5519):1096–1099.http://dx.doi.org/10.1126/science.1058543 [DOI] [PubMed] [Google Scholar]
- Ott T, Fritz E, Polle A, Schützendübel A.. 2002. Characterisation of antioxidative systems in the ectomycorrhiza-building basidiomycete Paxillus involutus (Bartsch) Fr. and its reaction to cadmium. FEMS Microbiol Ecol. 42(3):359–366. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067.http://dx.doi.org/10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8(10):785–786.http://dx.doi.org/10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M.. 2004. Cullin‐based ubiquitin ligases: Cul3–BTB complexes join the family. EMBO J. 23(8):1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI. 1979. Geosmithia, gen. nov. for Penicillium lavendulum and related species. Can J Bot. 57(19):2021–2030.http://dx.doi.org/10.1139/b79-252 [Google Scholar]
- Ploetz RC, Hulcr J, Wingfield MJ, De Beer ZW.. 2013. Destructive tree diseases associated with ambrosia and bark beetles: black swan events in tree pathology? Plant Dis. 97(7):856–872. [DOI] [PubMed] [Google Scholar]
- Pócsi I, Prade RA, Penninckx MJ.. 2004. Glutathione, altruistic metabolite in fungi. Adv Microb Physiol. 49:1–76. [DOI] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S.. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 10(6):417–430. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP, Murphy WJ.. 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One 6(9):e22594.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn RD.. 2016. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44:D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.http://dx.doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugman-Jones PF, Seybold SJ, Graves AD, Stouthamer R, Etges WJ.. 2015. Phylogeography of the walnut twig beetle, Pityophthorus juglandis, the vector of thousand cankers disease in North American walnut trees. PLoS One 10(2):e118264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Stamatakis A, Rokas A.. 2014. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol Biol Evol. 31(5):1261–1271.http://dx.doi.org/10.1093/molbev/msu061 [DOI] [PubMed] [Google Scholar]
- Schuelke TA, Westbrook A, Broders K, Woeste K, MacManes MD.. 2016. De novo genome assembly of Geosmithia morbida, the causal agent of thousand cankers disease. PeerJ 4:e1952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. 2002. Repeat-induced gene silencing in fungi. Adv Genet. 46:439–450. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):1–3. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.http://dx.doi.org/10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 1996. RepeatMasker. [WWW document] Available from: http://www.repeatmasker.org [accessed 2015 Jun 12].
- Solomon PS, Oliver RP.. 2002. Evidence that γ-aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta 214(3):414–420. [DOI] [PubMed] [Google Scholar]
- Son H, et al. 2016. A novel transcription factor gene FHS1 is involved in the DNA damage response in Fusarium graminearum. Sci Rep. 6:21572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, et al. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330(6010):1543–1546.http://dx.doi.org/10.1126/science.1194573 [DOI] [PubMed] [Google Scholar]
- Staats M, van Kan JA.. 2012. Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot Cell 11(11):1413–1414.http://dx.doi.org/10.1128/EC.00164-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313.http://dx.doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Croll D.. 2014. The evolving fungal genome. Fungal Biol Rev. 28(1):1–2.http://dx.doi.org/10.1016/j.fbr.2014.02.001 [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terfehr D, et al. 2014. Genome sequence and annotation of Acremonium chrysogenum, producer of the β-lactam antibiotic cephalosporin C. Genome Announc. 2(5):e00948-14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa AC, Oliver RP, Hane JK.. 2016. OcculterCut: a comprehensive survey of AT-rich regions in fungal genomes. Genome Biol Evol. 8(6):2044–2064.http://dx.doi.org/10.1093/gbe/evw121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserat N, Cranshaw W, Leatherman D, Utley C, Alexander K.. 2009. Black walnut mortality in colorado caused by the walnut twig beetle and thousand cankers disease. Plant Health Prog. 1–10. doi:10.1094/PHP-2009-0811-01-RS. [Google Scholar]
- Trail F, Xu JR, San Miguel P, Halgren RG, Kistler HC.. 2003. Analysis of expressed sequence tags from Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet Biol. 38(2):187–197.http://dx.doi.org/10.1016/S1087-1845(02)00529-7 [DOI] [PubMed] [Google Scholar]
- Utley C, et al. 2013. Susceptibility of walnut and hickory species to Geosmithia morbida. Plant Dis. 97(5):601–607.http://dx.doi.org/10.1094/PDIS-07-12-0636-RE [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ.. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312(5773):580–583. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu Z, Gao L, Hao B.. 2009. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol Biol. 9:195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2013. A specialized ABC efflux transporter GcABC‐G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle‐associated fungal pathogen of pine trees. New Phytol. 197(3):886–898. [DOI] [PubMed] [Google Scholar]
- Weber T, et al. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43(W1):W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnenburg R, et al. 2006. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34:D459–D464. [WWW document] Available from: http://www.phi-base.org/ [accessed 2015 Nov 22]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591.http://dx.doi.org/10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zerillo MM, et al. 2014. Population structure of Geosmithia morbida, the causal agent of thousand cankers disease of walnut trees in the United States. PLoS One 9(11):e112847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. 2014. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun. 5:3849. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu J.. 2013. Erratum to: comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 15:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, et al. 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 12(11):R116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.